Summary
CRSsNP is a heterogenous disease but type 2 inflammation in CRSsNP was more common than type 1 inflammation among patients in Chicago, Illinois. Distinct therapeutic strategies may be needed depending on the type of inflammation found in CRSsNP.
Keywords: Chronic rhinosinusitis without nasal polyps; Eosinophils; Interferon-γ; IL-13, IL-17A, Type 2 inflammation
To the Editor
Although it is accepted that chronic rhinosinusitis with nasal polyps (CRSwNP) is characterized by type 2 inflammation with pronounced eosinophilia and the presence of high levels of IL-5 and IL-13 in Western countries, the mechanism of inflammation in non-polypoid CRS (CRSsNP) is poorly understood.1, 2 Initial studies by Van Zele et al. in Belgium, suggested that CRSsNP is characterized by type 1 inflammation on the basis of elevation of IFN-γ.3 While several papers from the same group have confirmed these findings,4 other groups, including our own, have been unable to find elevation of IFN-γ in CRSsNP.5, 6 We therefore examined potential differences in experimental design between these studies. Studies that found type 1 inflammation in CRSsNP compared inferior turbinate (IT) tissue from controls, ethmoid tissue (ET) from CRSsNP and nasal polyp (NP) tissue from CRSwNP.3, 4, 7 In contrast, those that did not find type 1 inflammation compared uncinate tissue (UT) from controls, CRSsNP and CRSwNP, and NPs.5, 6 It was thus unclear whether reported IFN-γ elevations were due to differences in sampled anatomy or countries.
To clarify patterns of inflammatory cytokines in CRSsNP in our study population, we collected IT, UT, and ET from control patients and patients with CRSsNP and CRSwNP (see Fig. E1 and Table E1 and E2), and determined the presence of IFN-γ by real-time RT-PCR. Detailed methods are given in the Online Repository. We found that IFN-γ was not significantly elevated in CRSsNP when compared to controls or CRSwNP within the same tissue type (Fig. 1A). We also compared CRSsNP ET to control IT or CRSwNP NP as previously reported.3, 4 However, IFN-γ was not elevated in ET from CRSsNP (Fig. 1B). We also analyzed a marker of eosinophilia, Charcot-Leyden crystal galectin (CLC, also known as eosinophil lysophospholipase), type 2 cytokines, IL-5 and IL-13, and the type 3 cytokine IL-17A. Since we found significant differences in the levels of CLC and IL-17A between control and CRSsNP in ET and not in IT or UT (Fig. 1 and E2), we focused our further analysis on ET. We represent further analysis using dot plots to better illustrate the inflammatory patterns in individual specimens.
Fig. 1. Messenger RNAs for markers of type 2 and 3 inflammation were elevated in CRSsNP.
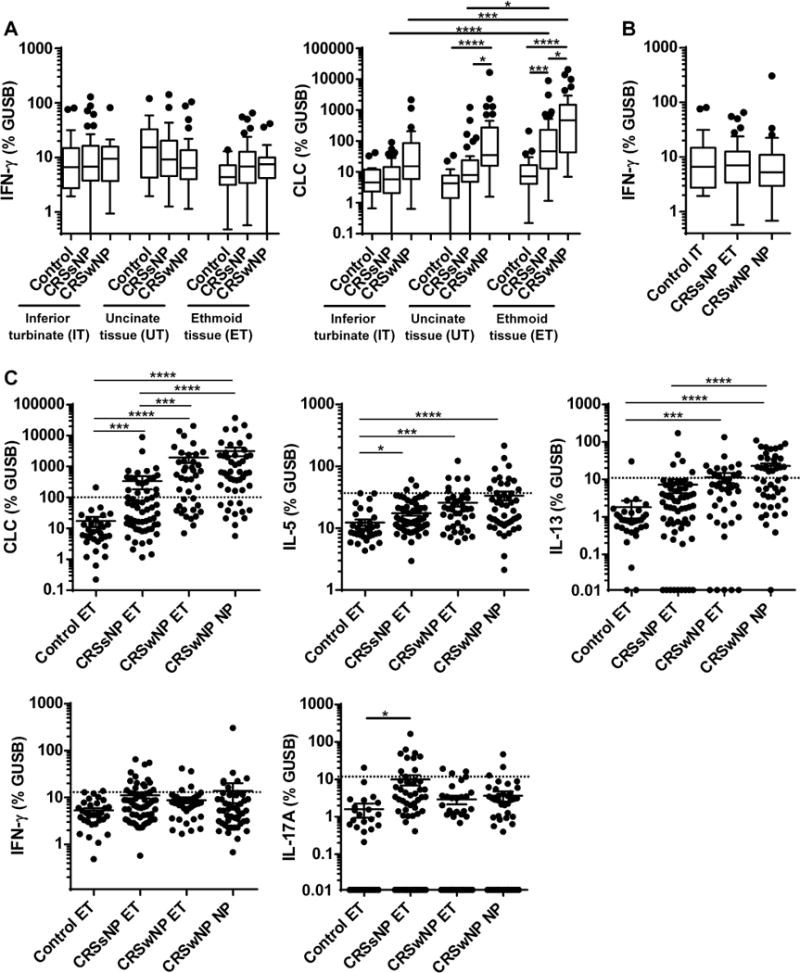
Total RNA was extracted from whole tissue of control IT (n=19), control UT (n=21), control ET (n=33), CRSsNP IT (n=53), CRSsNP UT (n=44), CRSsNP ET (n=61), CRSwNP IT (n=28), CRSwNP UT (n=29), CRSwNP ET (n=40) and CRSwNP NP (n=48). Expression of mRNAs for IFN-γ, CLC, IL-5, IL-13 and IL-17A was analyzed using real-time RT-PCR. Gene expression was normalized to a housekeeping gene, β-glucuronidase (GUSB), and expression levels were shown as % expression of GUSB. Results are shown as medians (25% to 75% interquartile ranges) (A, B) or mean ± SEM (C). Dotted line indicates the threshold based on the 95th percentile expression in control ET (CLC: 96.4, IL-5: 36.5, IL-13: 11.2, IFN-γ: 13.2, IL-17A: 12.1) (C). In order to display undetectable data, we plotted 0 as 0.01 in the log scaled figures (C). * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001, by one-way ANOVA.
On further analysis, CLC, IL-5 and IL-13 were expectedly significantly higher in CRSwNP ET compared to control ET (Fig. 1C). However, the levels of CLC, IL-5 and IL-17A were also significantly elevated in CRSsNP ET compared to control ET (Fig. 1C). CLC expression positively correlated with IL-5 (r=0.3652, P=0.0037) and IL-13 (r=0.7262, P<0.0001), but not with IL-17A or IFN-γ in the CRSsNP ET (n=61, not shown).
We next established thresholds for defining each inflammatory subtype using the 95th percentile of expression in control ETs.8 Using these thresholds, 23%, 36% and 15% of CRSsNP ET showed type 1, 2 and 3 inflammation, based on the expression of IFN-γ, CLC and IL-17A, respectively (Fig. 1C). In contrast, CRSwNP ET and NP had higher frequencies of type 2 inflammation (65% and 77%) but lower frequencies of type 1 (10% and 15%) or 3 (8% and 6%) inflammation (Fig. 1C). Interestingly, minor subsets of CRSsNP donors had mixed inflammation: 8% showed type 1 and 2 mixed inflammation, 7% showed type 1 and 3 mixed inflammation, and a single donor showed all three types in ETs (Fig. E3). Also noteworthy was that 43% of CRSsNP donors did not have elevated type 1, 2 or 3 inflammation in ETs (Fig. E3). We initially hypothesized that the type of inflammation in CRSsNP might be correlated with clinical comorbidities like atopy or asthma. However, we found no significant differences in the levels of inflammation markers between atopic and non-atopic patients or between asthmatic and non-asthmatic patients in CRSsNP (not shown).
We further confirmed our findings at the protein level. We generated protein tissue extracts from ETs and NPs and measured eosinophil cationic protein (ECP) and cytokine levels by ELISA and Luminex respectively (Table E2). Similar to our RT-PCR results, IFN-γ protein was not elevated but markers of type 2 (ECP, IL-5 and IL-13) and 3 (IL-17A) inflammation were significantly elevated in CRSsNP ET compared to control ET (Fig. 2). Using the 95th percentile of protein expression in control ETs to define inflammatory subtypes, we found that 5%, 46% and 16% of CRSsNP ET showed type 1, 2 and 3 inflammation, based on the expression of IFN-γ, ECP and IL-17A, respectively (Fig. 2). Although the frequency of type 2 and 3 inflammation was similar to that established using mRNA expression, the frequency of type 1 inflammation was lower when using protein measures (Fig. 2). One possible explanation is the decreased sensitivity of the IFN-γ protein detection system compared to real-time RT-PCR. We therefore further analyzed the data classifying only by type 2 and 3 inflammation in CRSsNP ET. We found that 37% showed type 2, 7% showed type 3, 8% showed mixed type 2 and 3 inflammation, and 47% of CRSsNP donors showed neither type 2 nor 3 inflammation in ETs (Fig. E4).
Fig. 2. Type 2 and 3 cytokines were elevated in CRSsNP tissue protein extracts.
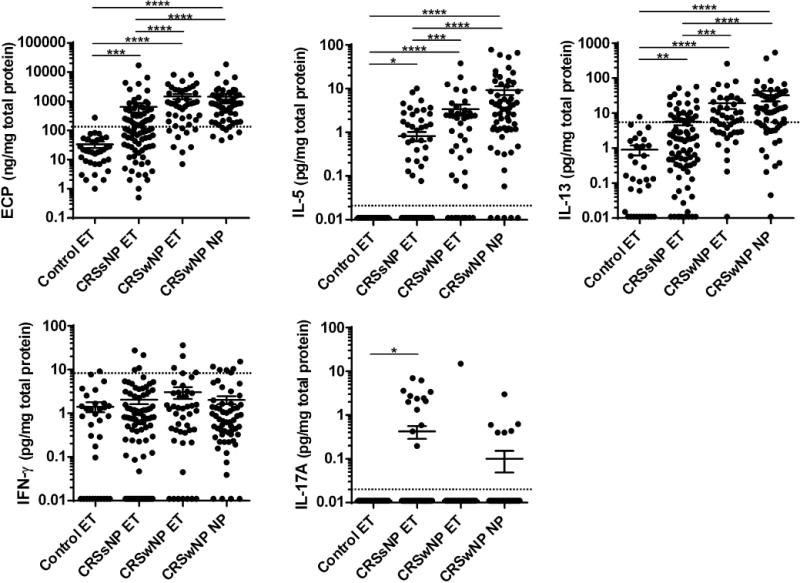
Protein extracts were generated from ET of control (n=34), CRSsNP (n=83) and CRSwNP (n=45) and from NPs (n=60). Expression of ECP, IL-5, IL-13, IFN-γ and IL-17A proteins in tissue homogenates was measured using ELISA and Luminex. The protein concentrations were normalized to the concentration of total protein. Dotted line indicates the threshold based on the 95th percentile expression in control ET (ECP: 131.5 ng/mg, IL-5: 0.02 pg/mg (detectable), IL-13: 5.5 pg/mg, IFN-γ: 8.1 pg/mg, IL-17A: 0.02 pg/mg (detectable)). In order to display undetectable data, we plotted 0 as 0.01 in the log scaled figures. * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001, by one-way ANOVA.
Our study has some limitations. Patient matched IT, UT and ET were not available from all patients due to variations in the extent of surgery, the size of surgically resected tissue and quality of RNA extracted. Subsequently, the numbers of specimens available from each anatomic location and for analysis by protein or RT-PCR were variable. Our study also recruited patients undergoing surgery in a tertiary care practice in Chicago. Thus, whether our results are applicable to the general population, or only tertiary care populations in the United States, would require further multi-institutional studies. Nonetheless, we report data from a larger number of samples than most other published studies and had sufficient control ET to define a 95th percentile threshold for inflammatory subtyping. Our results also suggest that CRSsNP cannot be generalized as a type 1 inflammatory condition since approximately 40% and 15% of CRSsNP patients demonstrated type 2 and 3 inflammation, respectively. We also further find that over 40 % of CRSsNP patients did not show the signature for type 1, 2 or 3 inflammation in ET, IT or UT (Fig. 1, E3 and E4 and not shown). Further studies will be required to identify the nature of the inflammation in these patients.
Using flow cytometry, Derycke et al. have reported heterogenous T-helper populations in CRSsNP, with Th1 being the most common population in control, CRSsNP and CRSwNP populations.9 Th2 cells were extremely rare in the CRSsNP tissue studied. In contrast, while we also report heterogeneous inflammatory patterns in CRSsNP, type 2 inflammation was most common. While Derycke et al. used stimulated cells to identify the presence of T-helper subsets in tissue,9 our study evaluated relative ongoing expression of inflammatory cytokines and eosinophil granule proteins. Since these studies utilize different biomarkers for each inflammatory subtype and have disparate methods for defining an inflammatory subtype, they are not directly comparable. The methodologic differences may indeed lead to the different results.
Very recently, two groups published on patterns of inflammation in CRS. In support of our findings, Konig et al. found that IFN-γ was not elevated in nasal secretion of German patients with CRSsNP.10 Tomassen et al. proposed 10 endotypes of CRS based on inflammatory patterns found in a multicenter study in Europe.11 This study also discovered heterogenous inflammation in CRSsNP and the overall frequency of IFN-γ, IL-5 and IL-17A high populations in CRSsNP was 20, 30 and 11%, respectively, which similar to our current study. Together, these results may indicate our findings may be applicable to CRSsNP patients in both the United States and Europe.
In conclusion, we report here that CRSsNP is a heterogenous disease and the overall frequency of type 2 inflammation is higher than type 1 inflammation in our United States based population. In light of emerging therapies targeting type 2 inflammatory mechanisms, our findings indicate that further studies will be needed to better identify type 2 CRSsNP patients for tailored therapeutic strategies.
Supplementary Material
Acknowledgments
This research was supported in part by NIH grants, R01 AI104733, U19 AI106683, K23 DC012067 and R37 HL068546 and by a grant from the Ernest S. Bazley Foundation.
Abbreviations
- CLC
Charcot-Leyden crystal galectin
- CRS
Chronic rhinosinusitis
- CRSsNP
CRS without nasal polyps
- CRSwNP
CRS with nasal polyps
- ECP
Eosinophil cationic protein
- ET
Ethmoid tissue
- IT
Inferior turbinate
- NP
Nasal polyp
- UT
Uncinate tissue
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests: The authors declare no conflict of interest as to the interpretation and presentation of this manuscript.
References
- 1.Fokkens WJ, Lund VJ, Mullol J, Bachert C, Alobid I, Baroody F, et al. European Position Paper on Rhinosinusitis and Nasal Polyps 2012. Rhinol Suppl. 2012;3:1–298. [PubMed] [Google Scholar]
- 2.Kato A. Immunopathology of chronic rhinosinusitis. Allergol Int. 2015;64:121–30. doi: 10.1016/j.alit.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Zele T, Claeys S, Gevaert P, Van Maele G, Holtappels G, Van Cauwenberge P, et al. Differentiation of chronic sinus diseases by measurement of inflammatory mediators. Allergy. 2006;61:1280–9. doi: 10.1111/j.1398-9995.2006.01225.x. [DOI] [PubMed] [Google Scholar]
- 4.Van Bruaene N, Perez-Novo CA, Basinski TM, Van Zele T, Holtappels G, De Ruyck N, et al. T-cell regulation in chronic paranasal sinus disease. J Allergy Clin Immunol. 2008;121:1435–41, 41 e1–3. doi: 10.1016/j.jaci.2008.02.018. [DOI] [PubMed] [Google Scholar]
- 5.Nagarkar DR, Poposki JA, Tan BK, Comeau MR, Peters AT, Hulse KE, et al. Thymic stromal lymphopoietin activity is increased in nasal polyps of patients with chronic rhinosinusitis. J Allergy Clin Immunol. 2013;132:593–600 e12. doi: 10.1016/j.jaci.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stevens WW, Ocampo CJ, Berdnikovs S, Sakashita M, Mahdavinia M, Suh L, et al. Cytokines in Chronic Rhinosinusitis. Role in Eosinophilia and Aspirin-exacerbated Respiratory Disease. Am J Respir Crit Care Med. 2015;192:682–94. doi: 10.1164/rccm.201412-2278OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao PP, Li HB, Wang BF, Wang SB, You XJ, Cui YH, et al. Distinct immunopathologic characteristics of various types of chronic rhinosinusitis in adult Chinese. J Allergy Clin Immunol. 2009;124:478–84, 84 e1–2. doi: 10.1016/j.jaci.2009.05.017. [DOI] [PubMed] [Google Scholar]
- 8.Thompson CF, Price CP, Huang JH, Min JY, Suh LA, Shintani-Smith S, et al. A pilot study of symptom profiles from a polyp vs an eosinophilic-based classification of chronic rhinosinusitis. Int Forum Allergy Rhinol. 2016;6:500–7. doi: 10.1002/alr.21687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Derycke L, Eyerich S, Van Crombruggen K, Perez-Novo C, Holtappels G, Deruyck N, et al. Mixed T helper cell signatures in chronic rhinosinusitis with and without polyps. PLoS One. 2014;9:e97581. doi: 10.1371/journal.pone.0097581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Konig K, Klemens C, Haack M, Nicolo MS, Becker S, Kramer MF, et al. Cytokine patterns in nasal secretion of non-atopic patients distinguish between chronic rhinosinusitis with or without nasal polys. Allergy Asthma Clin Immunol. 2016;12:19. doi: 10.1186/s13223-016-0123-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tomassen P, Vandeplas G, Van Zele T, Cardell LO, Arebro J, Olze H, et al. Inflammatory endotypes of chronic rhinosinusitis based on cluster analysis of biomarkers. J Allergy Clin Immunol. 2016;137:1449–56 e4. doi: 10.1016/j.jaci.2015.12.1324. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


