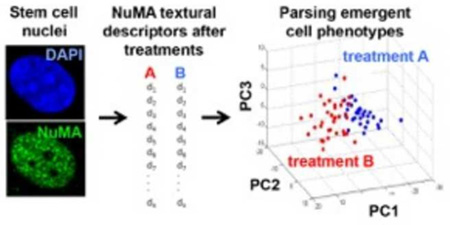
Abstract
Stem and progenitor cells that exhibit significant regenerative potential and critical roles in cancer initiation and progression remain difficult to characterize. Cell fates are determined by reciprocal signaling between the cell microenvironment and the nucleus; hence parameters derived from nuclear remodeling are ideal candidates for stem/progenitor cell characterization. Here we applied high-content, single cell analysis of nuclear shape and organization to examine stem and progenitor cells destined to distinct differentiation endpoints, yet undistinguishable by conventional methods. Nuclear descriptors defined through image informatics classified mesenchymal stem cells poised to either adipogenic or osteogenic differentiation, and oligodendrocyte precursors isolated from different regions of the brain and destined to distinct astrocyte subtypes. Nuclear descriptors also revealed early changes in stem cells after chemical oncogenesis, allowing the identification of a class of cancer-mitigating biomaterials. To capture the metrology of nuclear changes, we developed a simple and quantitative “imaging-derived” parsing index, which reflects the dynamic evolution of the high-dimensional space of nuclear organizational features. A comparative analysis of parsing outcomes via either nuclear shape or textural metrics of the nuclear structural protein NuMA indicates the nuclear shape alone is a weak phenotypic predictor. In contrast, variations in the NuMA organization parsed emergent cell phenotypes and discerned emergent stages of stem cell transformation, supporting a prognosticating role for this protein in the outcomes of nuclear functions.
Keywords: stem cells, differentiation, oncogenic transformation, nuclear, organization, imaging, image informatics, mesenchymal stem cells, biomaterials
Graphical Abstract
Introduction
Breakthroughs in stem cell biology have the potential to advance tissue repair and regeneration after injury or disease [1]. They also hold promise for the treatment of disorders caused by imbalanced stem cell differentiation such as cancer and osteoporosis [2, 3]. Stem cell signaling programs are complex and vary across tissues and developmental processes. The ability to accurately assess stem cell responses to their environments and identify their fate early is therefore extremely challenging [4–6]. Identifying phenotypes among heterogeneous populations in vivo and predicting single cell responses to biomaterials in vitro may provide new insights into differentiation mechanisms and facilitate purification and testing in cell cultures. However, most current approaches to characterize stem cells are limited to population-based analyses and require differentiation to occur, rather than pluripotent stages.
Our group recently reported that subtle quantifiable differences in cellular organization capture early variations in human mesenchymal stem cells (MSCs), as the cells commit to osteogenic or adipogenic lineages [7, 8]. These changes occur prior to detectable phenotypic marker expression. Since the cell nucleus is the terminal gateway of outside-in signaling, including mechanotransduction of microenvironmental cues, the premise for this work is that accurate descriptors of emerging stem cell phenotypes may be derived from nuclear shape and organization.
Nuclear shape is defined by the conformation of the lamina and by the physical properties of the nucleoplasm. The nuclear envelope is connected to cytoskeletal actin, microtubules, and intermediate filaments that interact with the lamina via the linker of nucleoskeleton and cytoskeleton (LINC) complexes [9]. Changes in actin organization during stem cell differentiation [8] may therefore alter nuclear shape. Differentiation is further characterized by extensive remodeling of chromatin - one of the major determinants of nuclear rheological properties [10]. Nuclei isolated from human embryonic stem cells are, for instance, more deformable and stiffer compared to nuclei from differentiated cells [10]. However, to our knowledge it remains unknown whether nuclear shape per se would be a sensitive marker to distinguish stem cells during MSC lineage commitment, as well as in other contexts.
In addition to the overall nuclear shape, changes in nuclear organization may mirror different stages of stem cell differentiation. The cell nucleus is devoid of an internal membrane system and does not contain classic skeletal fibers. Yet, highly dynamic compartments found in the nucleoplasm regulate key nuclear processes such as the spatiotemporal control of genome expression and maintenance. For instance, splicing speckles labeled with the SRm160 splicing coactivator were shown to increase in size and become less numerous as mammary epithelial cells differentiate in response to extracellular matrix (ECM) signals [11]. In addition, we showed recently that high-content imaging of nuclear speckles labeled by SC-35 identifies changes in nuclear organization in response to microenvironmental cues, including growth factors, surface topologies, and microscale topographies [12].
The nuclear mitotic apparatus (NuMA) protein is a nucleoskeletal element expressed throughout the nucleoplasm. NuMA is diffusely distributed in the nucleus of undifferentiated mammary epithelial cells and in patient-derived tumors, whereas it forms distinct focal patterns in the differentiated mammary gland, the intestinal epithelium, as well as in differentiated lens epithelial cells [11, 13, 14, unpublished data]. We previously analyzed combinations of morphometric readouts derived from actin, nuclear shape, and NuMA to classify MSC phenotypes [7]. The present study was designed to test whether the higher-order nuclear organization of NuMA per se parses and predicts emergent stem and progenitor cell fates across multiple contexts.
Materials and methods
Human mesenchymal stem cell culture and transformation
MSCs were obtained from Lonza and cultured according to the supplier’s recommendations and reagents. Cells were received at passage 1 and used between passages 13 and 19. Oncogenic transformation was induced with periodic treatments with nickel(II) sulfate (NiSO4) [15]. Briefly, 24h after plating, cells were treated for 48h with a NiSO4 solution (36–72 µM) in MSC cell culture medium (Lonza). This incubation medium was then replaced with regular medium for 48h; cells were treated a second time with NiSO4 for 72h, incubated 72h in regular cell culture medium, and used for analysis. Genetically transformed MSCs (termed tMSC in this manuscript) were obtained from Dr. Richard Gorlick at the Children’s Hospital at Montefiore, Bronx, New York. These cells were immortalized with hTERT expression and transformed following p53 inactivation and activation of K-Ras and C-Myc [16]. The osteogenic and adipogenic induction as well as phenotypic characterization of MSCs were performed as described previously [8]. Briefly, cells were cultured for two weeks either in adipogenic medium or in osteogenic medium, then stained for alkaline phosphatase activity and lipid contents with Fast Blue RR/napthol (Sigma) and Oil Red O (Sigma), respectively. To image nuclei in live cells, MSCs were incubated with CellLight Nucleus-GFP (Invitrogen; 30 particles per cell for 12h) to express SV40NLS-GFP.
Assessment of MSC transformation
Fluorescence in situ hybridization (FISH) was used to detect transcripts from the human telomerase holoenzyme (hTR), in order to assess the degree of transformation in MSC populations. A cadmium selenium (CdSe)-conjugated quantum dot probe complementary to hTR mRNA (5'-NH2(CH2)12-T*C*T*C*AGTTAGGG*T*T*A*G; peak emission = 594 nm) was obtained from Roche Applied Science and used for FISH assays according to the manufacturer’s protocol. FISH signals were imaged using a Leica TCS SP2 confocal microscope and mean fluorescence intensities were calculated for each cell after background subtraction. A Transformation Index, defined as the hTR expression relative to tMSCs, was calculated for each treatment. This index ranges between 0 and 1, with 0 corresponding to untransformed (untreated) MSCs and 1 corresponding to fully transformed tMSCs. BrdU assays were performed according to the manufacturer’s instructions (Invitrogen). Expression of cell surface markers was quantified by flow cytometry, using CD44-FITC, Stro-1-PE, and CD133-APC antibodies (eBioscience). 100,000 cells were incubated with the antibodies for (15 minutes, room temperature) and analyzed with a BD Facscalibur™ flow cytometer.
Polymeric substrates for MSC cultures
Tyrosine-derived polycarbonates [17], polymethacrylates [18], and poly(L-lactic acid) (Resomer L-206; Boehringer Ingelheim; Ridgefield, CT) were spin-coated onto cover glass using established protocols [19]. MSCs were seeded on polymer-coated substrates at a density of 20,000 cells per cm2, prior to NiSO4 treatment, for either 3 or 11 days. At day 3, cells were immunostained with NuMA antibodies and imaged to calculate the Parsing Index (described below). Telomerase expression was measured at day 11.
Isolation and culture of oligodendrocyte precursor cells
Oligodendrocyte precursor cells (OPCs) were isolated from P6–P7 rat neonate brains and cultured as previously described [20]. After removing the cerebellum and olfactory bulb, the corpus collosum (CC) was carefully separated from the hippocampus, striatum, and cerebral cortices. To obtain OPCs from the cerebral cortex (CX), the dorsal cortical tissue anterior to the underlying hippocampus was taken after removal of the meninges and separation of the hippocampus, CC, and other ventral and anterior brain tissues. Lastly, to obtain OPCs from the optic nerve (ON) sections were performed between the region ~1 mm from the eye and ~2 mm before the convergence of the optic chiasm. The meninges were removed and tissues were sliced with a scalpel prior to enzymatic and mechanical tissue separation. After extraction, OPCs were maintained in base culture medium consisting of Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with Sato components (0.0286% v/v bovine serum albumin pathocyte, 0.2 µM progesterone, 100 µM putrecine, 0.2 µM sodium selenite), 50 µg/ml human apo-transferrin, and 1 µg/ml bovine pancreas insulin. Following dissection and purification, OPCs were cultured in base medium supplemented with 10 ng/ml recombinant human platele-tderived growth factor-AA (rhPDGF-AA; Peprotech) at 37 °C in a humidified incubator with 10% CO2. Cell culture substrates used for OPC culture were coated with poly-L-lysine. For NuMAbased image analysis, freshly isolated OPCs were plated onto LabTek chamber slides (Thermo Scientific) at densities of 50,000 cells/cm2. Cells were allowed to adhere for 4h. Then, samples were processed for immunofluorescence analysis.
Analysis of OPCs after long-term clonal selection
The day following isolation and purification, OPCs were plated at a low density (4–10 cells/cm2) in medium with rhPDGF-AA. Cell adhesion, viability, and density were verified after 24h of culture. Thereafter, cells were either maintained in base medium with 10 ng/ml PDGF-AA supporting proliferation, or cultured in base medium supplemented with 1 ng/ml PDGF-AA and 0.49 nM thyroid hormones (T3/T4) for differentiation. The media were changed on days 3 and 5. Cells were fixed on day 7 and immunostained for A2B5 and GalC. Nuclei were counterstained with Hoescht and the corresponding signals were used to identify and count cells in each colony. The proportion of OPCs (A2B5 positive and GalC negative) and of oligodendrocytes (A2B5 negative and GalC positive) was calculated for each individual colony based on the immunofluorescence signals. Colony probability was calculated as the number of colonies exhibiting a certain trait (i.e., number of oligodendrocytes and number of OPCs per colony) divided by the total number of colonies for a given treatment condition.
Immunofluorescence staining
MSCs and OPCs were fixed with 4% paraformaldehyde for 10 minutes and incubated 1h in blocking and permeabilization solution [0.1% Triton X-100 (Sigma) and 5% normal goat serum (MP Biomedicals) in phosphate-buffered saline (PBS)]. Primary antibodies, diluted 1:500 in blocking/permeabilization solution, were incubated overnight at 4 °C and detected using antirabbit and -mouse secondary antibodies conjugated to Alexa Fluor 488 (Invitrogen; 1:250 dilutions; 2h incubations at room temperature). To identify cell nuclei, samples were counterstained with 1 µg/ml 4',6-diamidino-2-phenylindole (DAPI) in PBS. The primary antibodies used for immunofluorescence were against the A2B5 antigen (Millipore, MAB312), galactocerebroside (GalC ; Millipore, Ab142), NuMA (Abcam, ab36999), and topoisomerase II-α (Abcam, ab75765).
High-content imaging and morphotextural informatics
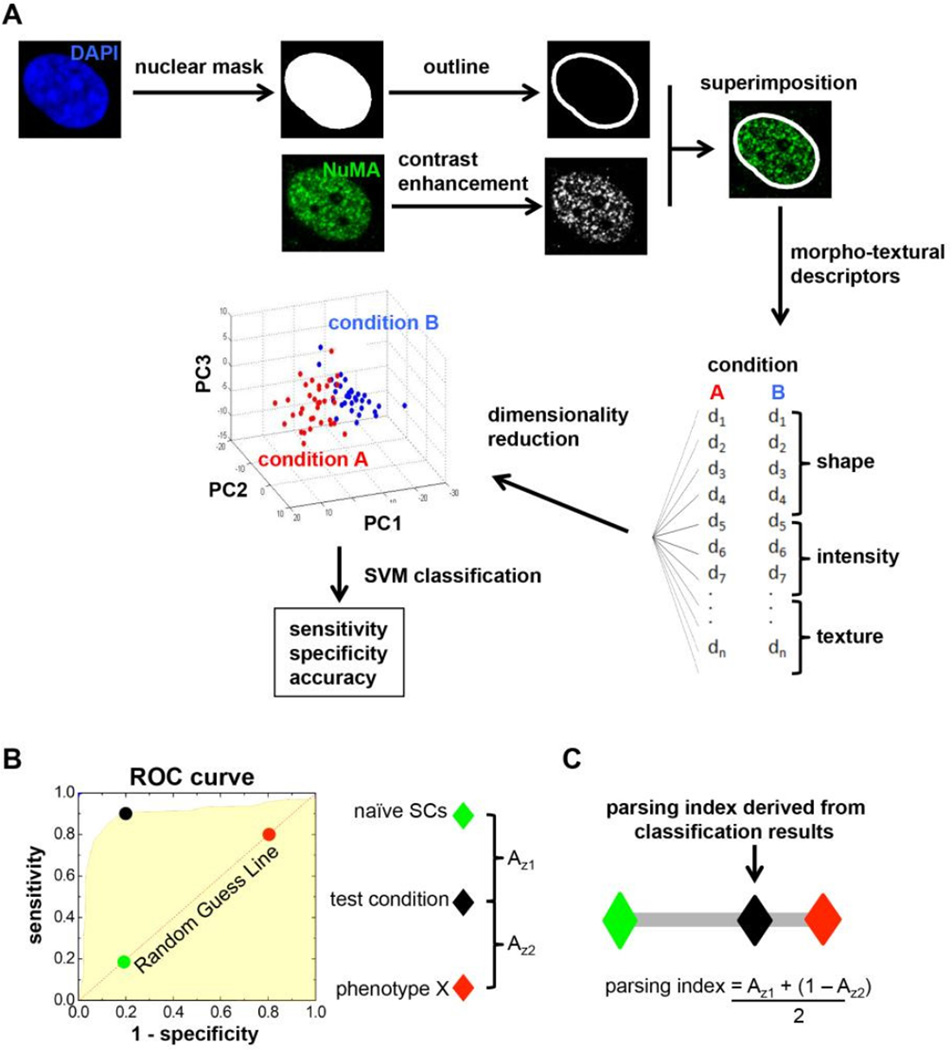
Samples stained with NuMA antibodies and with a nuclear dye (DAPI) were imaged with a 63× glycerin immersion objective (NA = 1.3) mounted on a multiphoton TCS SP2 confocal microscope. To analyze NuMA signal, binary masks were generated from DAPI images and were superimposed with the corresponding NuMA images (Fig. 1A). The ImagePro Plus (Media Cybernetics) software was then used to obtain first order nuclear shape and higher-order NuMA descriptors for each cell. Descriptors included in the analyses were related to nuclear shape, signal intensity (expression level of NuMA), and organization/texture of the protein (describing the spatial distribution and location of NuMA within the nucleus). A complete list of shape and texture descriptors used in this study can be found in Supplementary Table 1. After computation of the 43 shape, intensity, and organization/texture descriptors for each cell, principal component analysis was used to reduce the high dimensionality of the descriptor space into three principal components (PCs), visualized in 3D plots. In these plots, each axis (PC) is derived from a linear combination of a group of descriptors (Fig 1A). To identify differences across conditions, the PCs underwent a classification scheme which utilized a support vector machine (SVM) with a randomized two-fold cross validation on individual data sets. Three parameters (sensitivity, specificity, and accuracy) were calculated using SVM and used to evaluate the performance of the classification. Sensitivity and specificity represent the proportion of actual positives and actual negatives that are correctly identified as such, respectively, whereas accuracy is defined as the sum of the actual positives and actual negatives divided by the total number of cells analyzed in both groups. High classification and prediction are demarcated by high values of sensitivity and specificity, while low values of sensitivity and specificity indicate poor classification and discrimination between two datasets.
Figure 1. Analysis workflow for the quantification of morphotextural nuclear descriptors.
(A) Images of cell nuclei labeled with DAPI (blue) are used to generate masks that are superimposed with contrast-enhanced confocal images of NuMA immunostaining (green) to extract 43 different descriptor values. The descriptors characterize nuclear shape, NuMA intensity, and NuMA texture for each individual nucleus. Dimensionality reduction by principal component analysis (PCA) is applied to generate linear combinations of descriptors from any of the three categories. PCA results are visualized in 3D plots of principal components (PC) with a color-coded system for each treatment condition. Support vector machine (SVM) classification is used to calculate sensitivity, specificity, and accuracy to evaluate the performance of the classification. High values of sensitivity, specificity, and accuracy indicate high classification performance. (B) Shifting a hyperplane that best separates two experimental conditions using PCA (e.g., naïve stem cells and differentiated phenotype ‘X’) generates a receiver operator curve (ROC). The ROC represents an estimate of how different the test set (black) is from the two defined conditions (green and red). (C) A Parsing Index is derived by combining the area under the ROC curves between test and reference conditions (i.e., Az1 and Az2). This index evaluates how similar the test set is relative to the trained classifier conditions. The Parsing Index value for the test set ranges between 0 and 1, where 0 (green diamond) and 1 (red diamond) correspond to the two reference conditions.
A Parsing Index was developed in this study to better visualize and quantify the robustness of classifications based on morphotextural analyses (Fig 1B–C). For this approach, treatments are compared with ‘positive’ and ‘negative’ controls in the SVM classifier. For example, to assess the extent of differentiated cell differentiation in a test condition, the negative and positive datasets correspond to naïve, undifferentiated cells and fully differentiated cells, respectively. To generate Parsing Indexes, sensitivity and specificity values are calculated for the negative, positive, and test datasets. Next, sensitivity and specificity values are used to generate a “Receiver Operating Curve” (ROC), such that the area under the ROC (Azi) is an estimate of how different the test condition is from the negative and positive controls (black, green, and red circles in Fig. 1B). To calculate the Parsing Index for a test condition, Az1 (ROC between test condition and negative control) and Az2 (ROC between test condition and positive control) are used to estimate the location of the test set (black diamond) with respect to the negative (green diamond) and positive (red diamond) controls (Fig 1C). Such indexing allows the emergent cell lineage state to be assessed in relation to the degree of lineage restriction.
Results
Early identification of mesenchymal stem cell differentiation with high-content imaging of nuclear descriptors
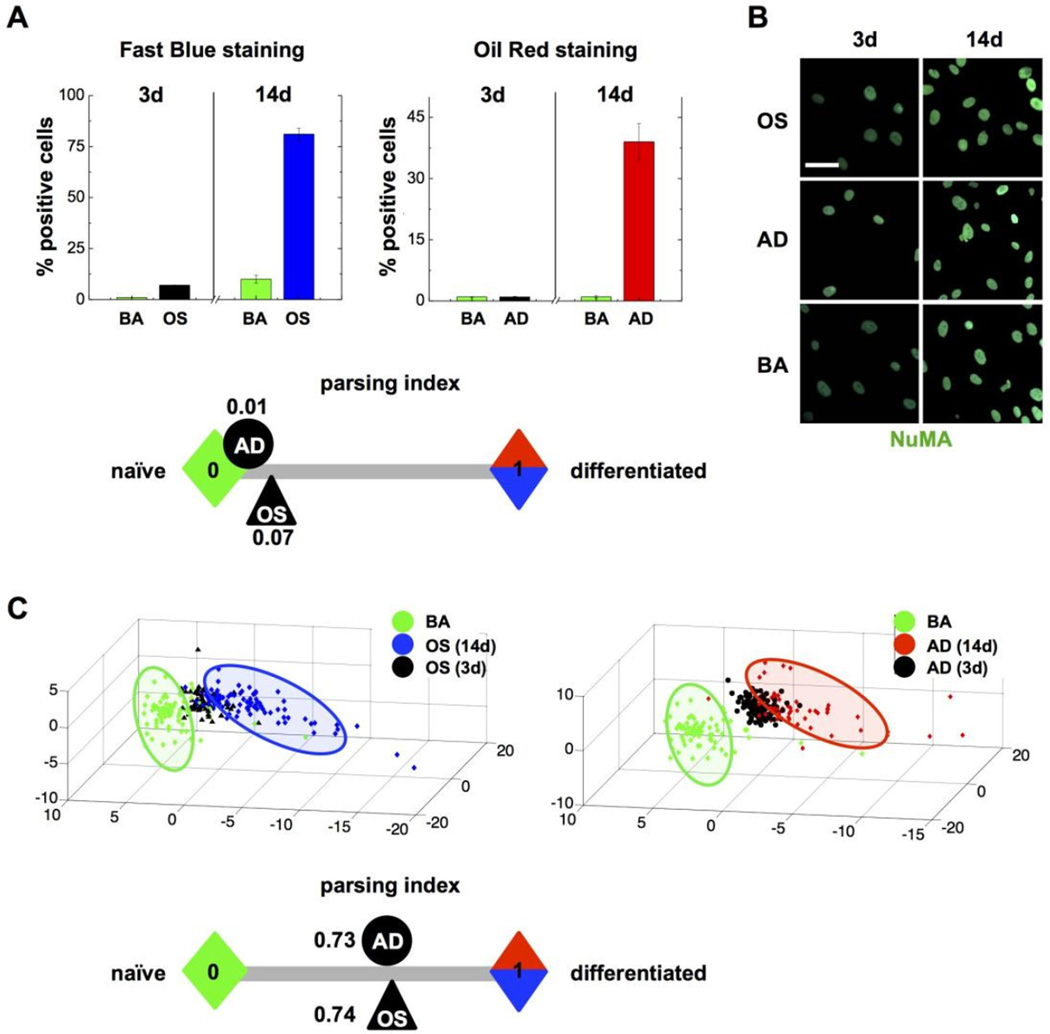
Human mesenchymal stem cells (MSCs) derived from the bone marrow differentiate in vitro into osetogenic (OS) and adipogenic (AD) cell lineages depending on cell density and growth factor exposure. Cells committed to these linages are phenotypically distinct, notably with high alkaline phosphatase activity after OS differentiation, and increased lipid content in the AD lineage. These divergent phenotypes can easily be parsed after 14 days in culture using conventional cytological stains (Fig. 2A). However, cells poised to the OS or AD linages are indistinguishable with cytological dyes at the early stage of culture. We showed previously that high-content image analysis of the actin cytoskeleton [8] or of composite staining of actin, DNA, and the nuclear structural protein NuMA [7] classifies cells early (72h) during OS and AD differentiation - well before gross morphological and biochemical changes take place. High-content analysis of NuMA signals distinguishes MSCs with high vs. low expression of the transcription factors Oct-4 and Sox2 that maintain pluripotent MSCs [21]. We therefore analyzed MSC differentiating towards the OS and AD lineages using nuclear shape descriptors, as well as texture descriptors derived from NuMA staining (see the analysis pipeline in Fig. 1). For both OS and AD treatments, nuclear shape and NuMA descriptors, reduced for visualization to three principal components, clearly parsed differentiating cells from controls in basal medium (Fig. 2B–C).
Figure 2. High-content analysis of nuclear descriptors identifies MSC lineage commitment.
(A) Proportion of MSCs stained positively with cytological dyes for osteogenesis (Fast Blue) and for adipogenesis (Oil Red) after 3 or 14 days in either osteogenic (OS), adipogenic (AD) or basal (BA) medium. (B) Representative confocal images of NuMA immunostaining in MSCs treated as in (A); scale bar, 25 µm. (C) High-content analysis of nuclear shape (from DAPI signals) and nuclear organization (NuMA texture descriptors) in MSCs treated with OS, AD, and BA media for 3 or 14 days. Parsing Indexes are calculated in (A) and (C) and schematically displayed to visualize the efficacy of the phenotypical assays at 3 days.
To better assess similarity/dissimilarity between treatment conditions, we developed a Parsing Index whose values range between 0 and 1 - each extreme corresponding to fully distinct phenotypes (see Material and methods and Fig. 1B–C). In this case, zero represents undifferentiated, naïve MSCs, whereas one corresponds to MSCs fully differentiated to the OS or AD lineages after 14 days of culture. Parsing Indexes were computed after 72h in culture. At this time point, while the indexes derived from conventional cytological stains were close to zero for both OS and AD, the indexes derived from the high-content nuclear descriptors were clearly distinct from the undifferentiated stage (0.74 and 0.73 for cells in osteogenic medium and in adipogenic medium, respectively). Hence, nuclear descriptors are sufficient for the early distinction of MSCs committed to distinct differentiation paths, and MSC commitment can precisely be quantified using Parsing Indexes derived from the high-content imaging data.
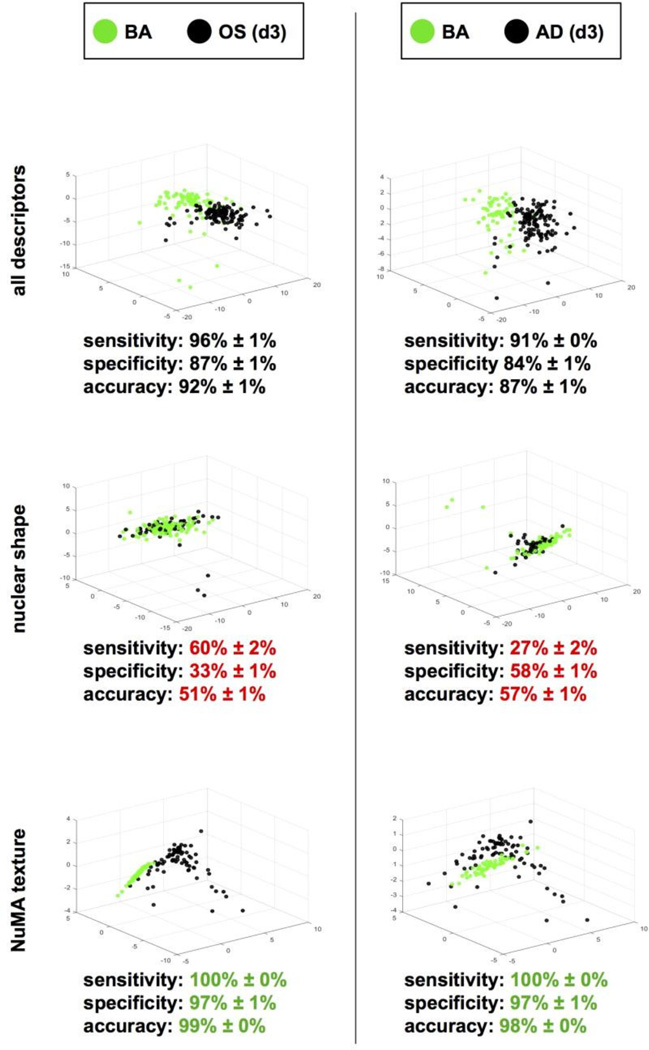
The classification of the OS and AD phenotypes in Fig. 2C regroups very different morphotextural aspects of the cell nucleus: Its shape and internal organization. To determine which of these aspects contributes to our ability to distinguish emergent and established MSC phenotypes, the datasets collected after 3 or 14 days of osteogenic and adipogenic treatments were reanalyzed using descriptors corresponding either to nuclear shape (DAPI signals) or to nuclear organization (NuMA staining) (Fig. 3). As expected, both classification methods were highly accurate to parse fully differentiated MSCs (day 14; data not shown). For MSCs at an early differentiation stage (day 3), nuclear shape descriptors poorly separated the treatment conditions, whereas NuMA descriptors alone were different, suggesting that changes in nuclear organization take place early during MSC differentiation.
Figure 3. NuMA texture distinguishes MSC phenotypes.
High-content morphotextural analysis of nuclear descriptors in MSCs treated for 3 days with osteogenic (OS), adipogenic (AD) or basal (BA) medium. All descriptors (top panels) or subsets of descriptors corresponding to nuclear shape (DAPI; middle panels) and NuMA texture (bottom panels) were used for classification. Classifying metrics are shown. Red and green highlights identify worse and better classifications relative to the complete descriptor pool, respectively.
NuMA descriptors distinguish oligodendrocyte precursors located in different brain regions
The trilineage differentiation of MSCs (adipocytes, osteoblasts and chondroblasts) yields widely different phenotypes, easily distinguishable using morphotextural analysis of the actin cytoskeleton [8] and the shape and internal organization of the cell nucleus (Fig. 2), as well as classical cytological assays detecting changes in cellular composition and enzymatic activities – albeit at later time points. To test whether the morphotextural descriptors derived from NuMA have the capacity to classify more subtle differences in phenotype, we analyzed oligodendrocyte precursor cells (OPCs) derived from distinct brain regions as a model system of antigenically identical stem/progenitor cells. In developing and adult brains, OPCs differentiate into glial cells producing myelin, the major component of white matter [22]. But OPCs from diverse regions of the brain have intrinsic differences such as distinct timing of myelination. Currently, only laborious assays of proliferation and differentiation profiles can tell apart different OPC populations in vitro [20, 23].
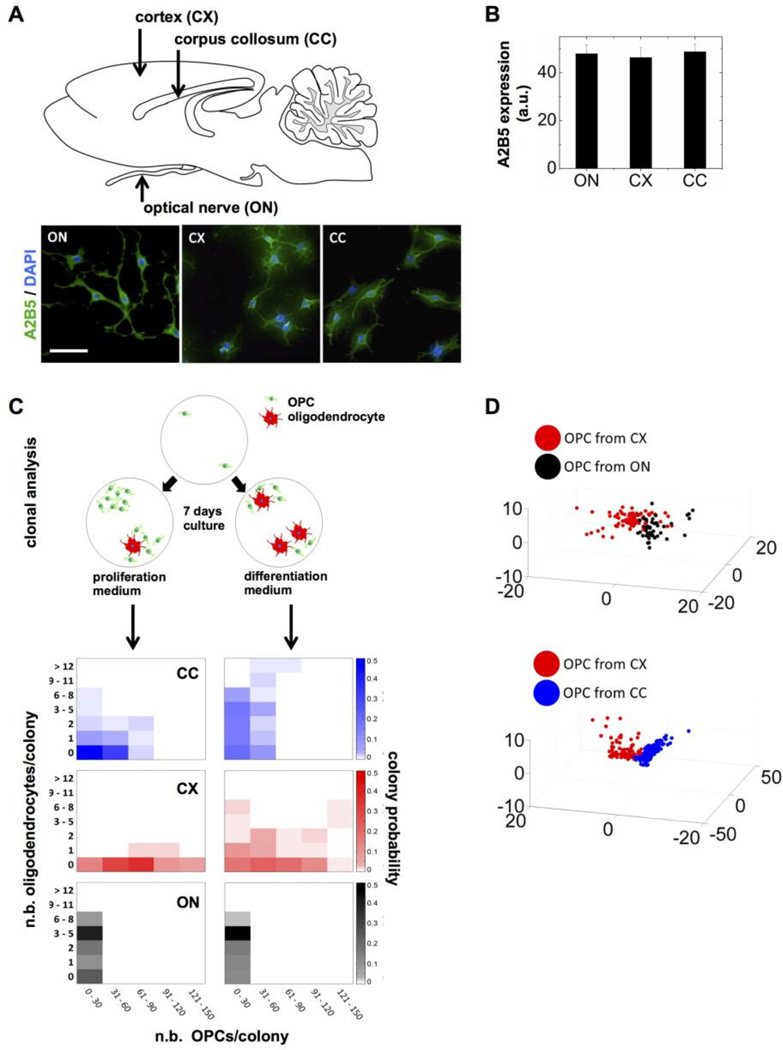
OPCs were isolated from the cerebral cortex (CX), the corpus collosum (CC), and the optic nerve (ON) from neonatal rats, then attached onto glass coverslips and analyzed immediately. We first confirmed purity based on high expression of the A2B5 antigen that was observed in more than 99% of CX-, CC-, and ON-derived OPCs (Fig. 4A). A2B5 staining was indistinguishable among OPCs from the different brain locations and the marker was expressed at similar levels (Fig. 4B). We also verified absence of galactocerebroside expression (GalC, a marker for differentiated oligodendrocytes) in purified OPCs. As expected [20], differences in the relative proportion of OPCs and oligodendrocytes were observed after seven days in culture when comparing primary cells isolated from different brain regions (Fig. 4C). Notably, CC precursors were less proliferative and more prone to differentiate than CX precursors.
Figure 4. High-content analysis of nuclear descriptors distinguishes OPCs from different brain regions.
(A) Diagram of a sagittal section of the neonatal rat brain highlighting the different regions from which OPCs were isolated. Representative images of freshly isolated OPCs stained with A2B5 antibodies and DAPI are shown; scale bar, 100 μm. (B) Quantification of A2B5 expression based on staining intensity in OPCs isolated from the optic nerve (ON), the cortex (CX), and the corpus collosum (CC). (C) Clonal analysis of OPCs isolated from the ON (black), CX (red) and CC (blue). The OPCs were plated at clonal density and cultured in proliferation and differentiation media for seven days to assess the number and proportion of OPCs (A2B5 positive staining) and of differentiated oligodendrocytes (GalC positive). Each densitogram plot represents 100 clones, where the x, y1, and y2 axes indicate the number of OPCs, oligodendrocytes, and fractional number of clones with a particular composition (indicated by brightness of the corresponding color), respectively. (D) High-content analysis of nuclear descriptors. OPCs were fixed and stained with DAPI and NuMA antibodies 4 hours after isolation. The 3D plots (bottom) represent principal component analyses of nuclear shape combined with NuMA descriptors.
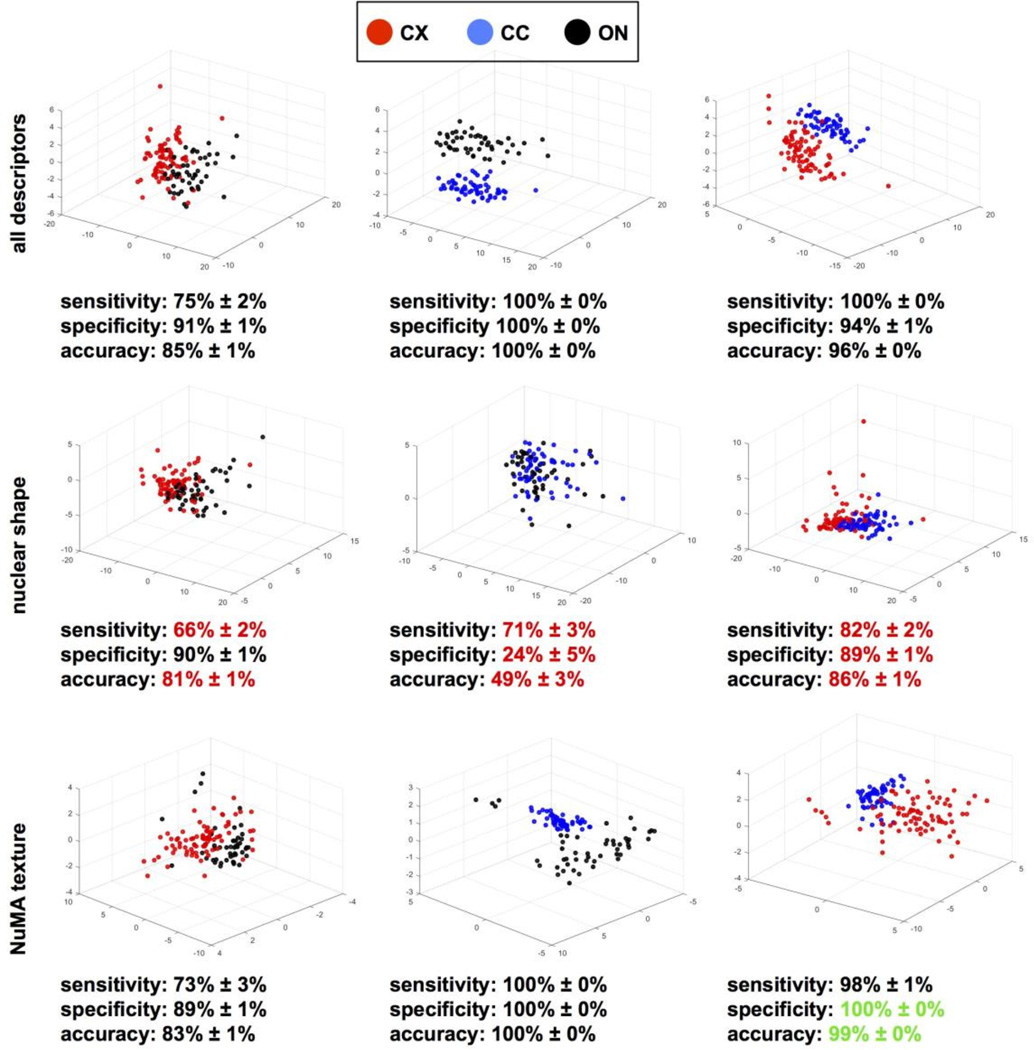
Morphotextural analysis with descriptors derived from nuclear shape and from NuMA staining distinguished OPCs directly upon isolation from the different brain locations (Fig. 4D). Effective separation was visualized in three-dimensional feature spaces after principal component analyses and evidenced by high classification performance (86.8% sensitivity and 80.7% specificity for CX vs. ON; 94.7% sensitivity and 91.7% specificity for CC vs. CX). To tease apart the relative contribution of nuclear shape and nuclear organization probed with NuMA texture descriptors, we repeated the analysis of CX, CC and ON OPCs, this time using only either shape descriptors (30) or texture descriptors (13) (Fig. 5). While classification was consistently less accurate using shape descriptors compared to the analyses with all descriptors, classification based on NuMA texture proved as accurate as classification with the entire descriptor set, indicating that the subnuclear distribution of NuMA is a major discriminating feature of OPCs poised to subtly different phenotypic outcomes.
Figure 5. NuMA texture parses OPCs from different origins.
NuMA staining and DAPI nuclear conterstain of OPCs isolated from the optic nerve (ON), the cortex (CX) and the corpus collosum (CC) were used for high-content morphotextural analysis using all (top panels) or subsets of descriptors corresponding to nuclear shape (middle panels) and NuMA texture (bottom panels). Classifying metrics are shown. Red and green highlights identify worse and better classifications relative to the complete descriptor pool, respectively.
Morphotextural descriptors derived from NuMA predict oncogenic transformation of MSCs
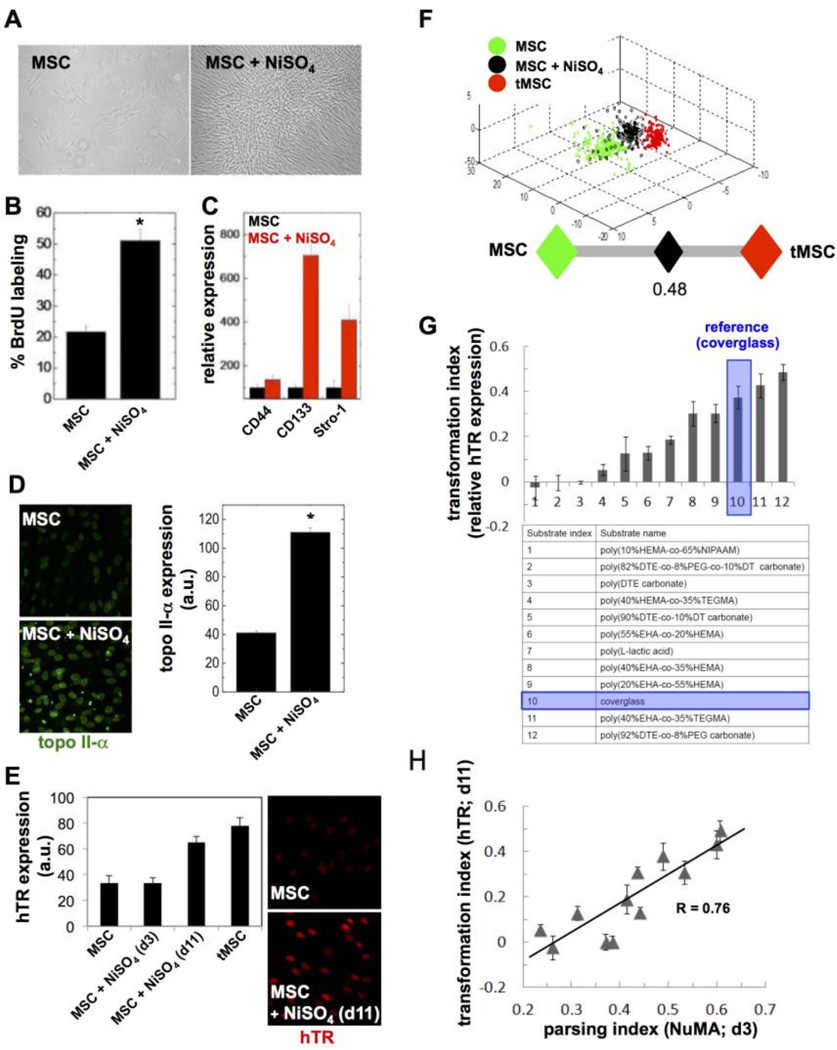
Having established that nuclear shape and organization derived from NuMA staining predicts MSC and OPC differentiation, we examined whether similar descriptors can differentiate stem cells undergoing transformation following nickel sulfate (NiSO4) treatment, which is used as a model of oncogenesis [15, 24]. We chose a chemical oncogenesis method since this approach enabled us to assess phenotypes at earlier time points compared to genetic transformation. Treatment with NiSO4 is reported to induce genomic instability [25] and to increase the expression of alpha4, a key regulator of the protein phosphatase 2A highly expressed in human cancers [26]. In addition, we measured increased proliferation, higher expression of topoisomerase II-α, and altered cell surface marker profiles, consistent with oncogenesis by NiSO4 in MSCs (Fig. 6A–D). We also assessed telomerase (hTR) expression as an indicator of MSC transformation. While hTR transcript abundance was similar in control and NiSO4-treated cells after 3 days of culture, hTR expression was increased in NiSO4-treated cells after an extended (11 days) culture period (Fig. 6E). As expected, hTR was also found to be upregulated in fully transformed MSCs (tMSCs; [16]) used as a benchmark for comparisons. Nuclear morphotextural descriptors extracted from MSCs, NiSO4-treated MSCs, and tMSCs could discriminate between the three conditions with high specificity, sensitivity, and accuracy (Fig. 6F and 7A, top panels). In particular, the Parsing Index approach developed and validated in the previous section clearly showed that nuclear shape and/or organization in cells exposed to the carcinogen is mid-way between that of unchallenged and fully transformed cells (Fig. 6F).
Figure 6. Oncogenic transformation of MSCs detected by high-content imaging of the nucleus.
(A) Phase contrast images of MSCs, before (left) or after (right) the oncogenic NiSO4 treatment. (B) Proliferation of NiSO4-treated MSCs and controls measured using BrdU labeling after 3 days in culture. (C) Expression of the cell surface markers CD44, CD133, and Stro-1 quantified by flow cytometry in untreated and NiSO4-treated MSCs. (D) Expression of topoisomerase II-α, measured by immunostaining in MSCs and NiSO4-treated MSCs after 3 days in culture. (E) Telomerase (hTR) expression detected by FISH either 3 or 11 days after ending the NiSO4 treatment, as well as in untreated MSCs and in transformed MSCs (tMSCs). Fluorescence signal intensities are quantified in the bar graph and representative images are shown. (F) High-content analysis of nuclear descriptors derived from MSCs, NiSO4-treated MSCs (3 days after treatment), and tMSCs. Classification is visualized in 3D space after principal component analysis as well as with the parsing index. (G) Transformation indexes for MSC cells cultured on different biomaterial substrates (listed in the table) after NiSO4 exposure. The indexes represent hTR expression relative to untransformed MSCs (value 0) and tMSCs (value 1). hTR analysis was performed 11 days after ending the oncogenic treatment. (H) Highcontent imaging parsing indexes plotted against Transformation Indexes for MSCs cultured on different substrates as in (G). R, Pearson correlation coefficient. *, P < 0.01.
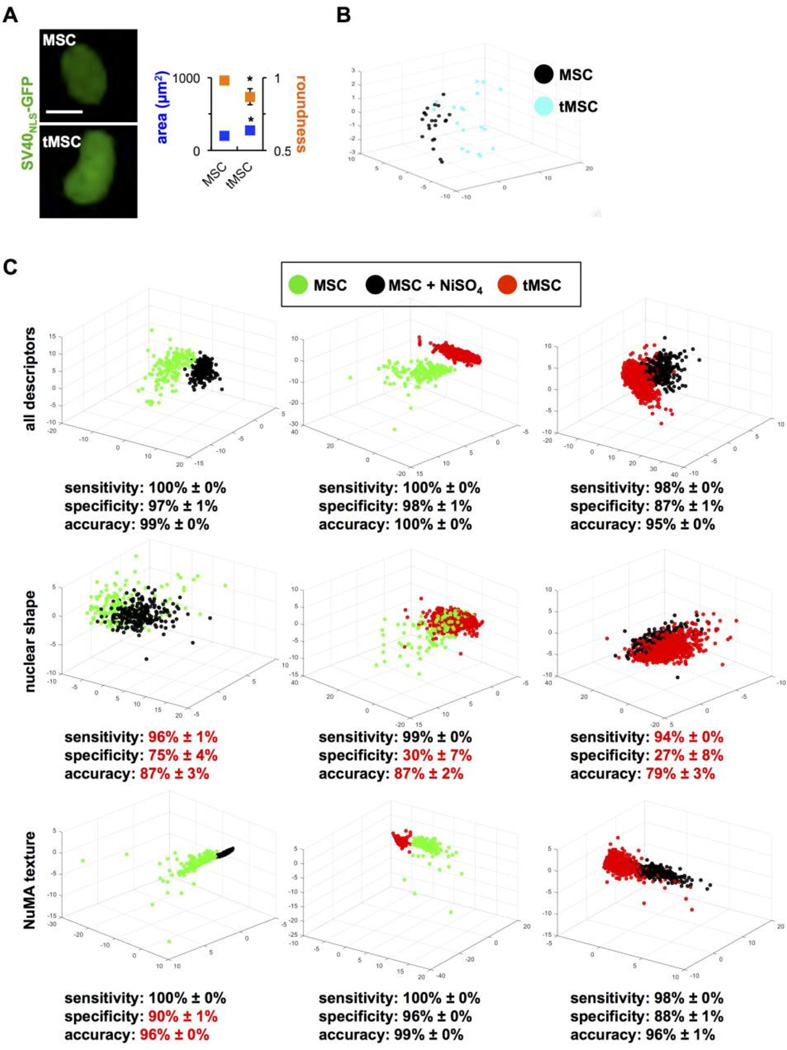
Figure 7. NuMA textural descriptors parse transformed and transforming MSCs.
(A) Classic shape parameters (nuclear size and roundness) computed from GFP signals in normal and transformed MSCs transduced with SV40NLS-GFP to label cell nuclei. *, P < 0.001. (B) Highcontent morphotextural analysis based on NuMA and DNA staining of MSCs, NiSO4-treated MSCs (three days after treatment), and tMSCs. All descriptors were used for the analyses in the top panels, whereas a subset of nuclear shape or NuMA texture descriptors were used for the analyses in the middle and lower panels, respectively. Classifying metrics are shown, with red highlights identifying worse classification compared to the full pool of descriptors.
In physiologically normal and pathological contexts, divergent cell phenotypes are never found in isolation, but rather combined, leading to intrinsic tissue heterogeneity, which is particularly relevant to cancer progression [27]. To determine if morphotextural nuclear descriptors distinguish phenotypes within a heterogeneous population, we analyzed nuclear descriptors derived from cocultures of normal and transformed MSCs (Supplementary Fig. S1). We took advantage of the distinct levels of actin filaments in MSCs and tMSC (lower in the latter) to verify the classification. Morphotextural nuclear descriptors could parse the two cell types, albeit less efficiently than in monocultures, suggesting that reciprocal influences between normal and transformed cells may lead to phenotypical uniformization.
Interactions between cells and their ECM define the structured integration of microenvironmental cues [28]. As a corollary, responses to oncogenic stimuli are strongly influenced by the composition and physical properties of the ECM and the characteristics of the substratum. Therefore, we tested the stem cell transformative effects of different engineered biomaterials designed as candidate substrates for the replacement of resected, cancerous bone or for the implantation of stem cells with osteogenic potential. We sought to use the nuclear descriptors of stem cells to demarcate the capacity of these biomaterials to mitigate or promote NiSO4-induced transformation of MSCs. For this purpose, we used a combinatorial library of biocompatible polymers (Supplementary Fig. S2) and assessed transformation of MSC on these substrates based on hTR expression and nuclear descriptors. Compared to cells on coverglass used as the reference, nine polymers reduced or abolished hTR upregulation by the carcinogen, whereas two polymers increased hTR expression after NiSO4-treatment, (Fig. 6G). Importantly, hTR expression tightly correlated (Pearson correlation coefficient = 0.76) with the Parsing Index values computed with nuclear descriptors for cell populations on each biomaterial substrate (Fig. 6H). These results demonstrate that descriptors of nuclear shape and/or NuMA texture predict MSC susceptibility to oncogenic transformation.
Pathologists routinely use nuclear shape and size (nuclear grade) to stage tumors. As expected, transformed MSCs had larger, more irregular nuclei compared to MSCs, visualized by imaging nuclei in live cells expressing GFP fused to the SV40 nuclear localization signal (Fig. 7A). This result was confirmed by high-content analysis, using the shape-based pool of descriptors (Fig. 7B). Next, to define the relative contribution of nuclear shape vs. organization to phenotypic parsing in the context of early MSC transformation, we took the same approach as for OPCs and MSCs, namely calculated classification metrics (sensitivity, specificity, accuracy) based on either class of descriptors (Fig. 7C). Similar to the OPC and MSC classifications, shape descriptors poorly differentiated non-transformed MSCs from partially transformed MSCs. In particular, specificity values significantly decreased for all comparisons when only nuclear shape descriptors were analyzed, indicating a high number of false positives. In contrast, NuMA textural descriptors efficaciously classified MSCs, tMSCs, and MSCs shortly after carcinogen exposure.
Discussion
The high-content profiling methodology used for the present study detects minute variations in nuclear shape and organization in stem cells poised to different phenotypes although these cells are essentially indistinguishable with conventional assays. The remarkable power of this single-cell approach is its ability to glean and predict emergent cellular behaviors in heterogeneous cell populations. The analyses reported herein extend our previous observation that the overall morphological attributes of actin cytoskeleton predicts mesenchymal stem cell lineage commitment [8]. Tensegrity models supported by a large body of experimental evidence predict mechanical coupling of intranuclear elements to the nuclear envelope, the cytoskeleton, and the ECM [29]. This dynamic structural continuum enables reciprocity between nuclear functions and the cell’s microenvironment [28, 30]. It was therefore expected that emergent mesenchymal stem cell phenotypes distinguishable based on cytoskeletal organization [8] or based on a composite pool of cytoskeletal and nuclear descriptors [7] would also be differentiated solely based on nuclear shape and organization (Fig. 2). Our analysis of oligodendrocyte precursor cells from different brain regions (Fig. 4) further indicates that high-content analysis of nuclear shape and organization is likely to be generally applicable to predict and sort phenotypes in (stem) cells. In these different contexts, nuclear features parsed cell behaviors earlier and more reliably than conventional assays.
The morphotextural profiling approach is distinct from classic machine learning methods that distinguish between image features appearing clearly different to the human eye, such as prominent heterochromatin domains in mouse vs. human nuclei [31] or grossly different nuclear morphologies (e.g., hepatocytes vs. fibroblasts; [32]). In contrast, high-content image profiling yields an unbiased assessment that does not rely on any preconception or biased supervision. The approach parses subtle differences, typically not apparent to visual observation. A drawback of the approach is the challenge to conceptualize differences detected in highdimensional descriptor spaces. A comparative analysis of subsets of descriptors (as done here for nuclear shape vs. texture) is one approach to address this challenge.
From our experiments, the nuclear organization stands out as a powerful readout of stem cell fates. It will be interesting to compare in future studies the various phenotypical classifications achieved with different nuclear proteins, individually or in combination. Here, we focused on the structural protein NuMA, an abundant component of the nucleus in eukaryotic cells. Beyond its well characterized role in mitosis [33], mounting evidence indicates that NuMA plays important roles in cell differentiation [11, 13, 14], apoptosis [34, 35], DNA repair [36, 37], and cancer initiation and progression [14, unpublished observations]. While differentiated cells exhibit clear differences in nuclear size and shape, nuclear shape descriptors alone were inadequate to robustly classify emergent phenotypes (Fig. 3 and 5). In contrast, a relatively small number (13) of texture descriptors derived from NuMA staining could parse distinct MSC and OPC fates. The results mirror our previous finding that textural analysis of nuclear splicing speckles (SC-35), a feature of nuclear organization intimately related to gene expression control, efficaciously classified MSCs poised to different lineages, whereas Lamin A staining, which essentially delineates the area of the nucleus, did not [12]. Our observations highlight the importance of nuclear organization in general and of the NuMA protein in particular in the regulation of key cellular processes that are precursors to changes in cell phenotypes and behaviors. Specifically, we propose that expression and subnuclear distribution of NuMA regulates transcription programs, including those of stemness-related genes. We showed previously that NuMA is present in the chromatin fraction and influences chromatin organization [14].
Specifically, NuMA-derived descriptors classified cells with high and low expression of the stem cell markers Oct-4 and Sox2 in human embryonic stem cells and in induced pluripotent stem cells, respectively [21]. Finally, transcriptome profiling of differentiating MSCs showed that NuMA is upregulated after osteogenic induction and downregulated after adipogenic induction in comparison to naïve MSCs [7]. The different NuMA textures measured in MSCs committed to become osteoblasts and adipocytes, as well as in oligodendrocyte progenitor cells with distinct self-renewal capabilities and differentiation patterns, further supports this hypothesis. Pathologists use the histological assessment of the nuclear grade, typically, the evaluation of the size and shape of nuclei, to broadly classify normal and diseased tissues. As expected, we measured slightly divergent nuclear shape descriptors in normal and transformed MSCs that matched measurements with traditional shape parameters (nuclear size and roundness) (Fig. 7A). Yet, similarly to differentiating stem/progenitor cells, the NuMA organizational texture proved to be a more robust classifier for cell transformation. This observation is in agreement with the differential radial localization of NuMA measured in non-neoplastic vs. malignant breast epithelial cells [38]. Cellular transformation is driven by genomic instability, and we recently showed that NuMA localization responds to DNA damage induction [36, 37]. However, since nickel sulfate is a weak inducer of DNA damage, it is likely that its effect on NuMA also reflects oncogenic alterations beyond DNA breaks, including altered transcriptional profiles under stress conditions.
Stem cells hold tremendous promise for regenerative medicine, yet two major bottlenecks restrict their clinical application. The purification and amplification of stem cells from heterogeneous sources requires rigorous characterization, yet most conventional assays rely on cell populations and/or fail to discern divergent phenotypes early. There is also a concern that stem cell amplification may lead to transformation in specific contexts, leading to cancer initiation or recurrence. High levels of regenerative activity of stem cells can indeed result in stem cell transformation [6, 14, 39, 40]. Therefore, biocompatible growth substrates promoting stem cell proliferation while inhibiting their transformation are highly desirable. Using a small library of polymeric biomaterials, we identified divergent effects of different chemistries on chemical oncogenesis (Fig. 6). For example, tyrosine-derived polycarbonates containing negatively charged desaminotyrosyl-tyrosine (DT) reduced transformation by nickel sulfate, whereas polycarbonates with polyethylene glycol (PEG) enhanced NiSO4-induced oncogenesis. While elucidating the underlying mechanisms is beyond the scope of this study, we hypothesize that the biomaterials modulate exogenous and intracellular reactive oxygen species (ROS) levels, and thus influence oncogenesis [41–43]. In an earlier study, we showed that PEGcontaining substrates induce ROS production, whereas DT-containing polymers have the opposite effect [19]. Alternatively, electrostatic binding of nickel ions to the DT polymer surface may reduce nickel bioavailability and toxicity. Irrespective of the underlying mechanisms controlling cytotoxicity, we predict further applications of morphotextural analyses in the characterization of biomaterials for implantation and for in vitro expansion of stem cell populations for applications in regenerative medicine. We also anticipate applications to study and diagnose cancer stem cell emergence and amplification during cancer initiation and progression. Finally, with induced pluripotent stem cells (iPSCs) revolutionizing research and treatment options for common and rare diseases, we envision applying morphotextural analyses of nuclear organization for characterizing differentiation protocols, predicting genomic instability [44], and studying tumor heterogeneity with cancer-derived iPSCs [45].
Supplementary Material
Highlights.
High-content analysis of nuclear shape and organization classify stem and progenitor cells poised to distinct lineage differentiation.
Early oncogenic changes in mesenchymal stem cells (MSCs) are also detected with nuclear descriptors, and a new class of cancer-mitigating biomaterials was identified.
Textural metrics of the nuclear structural protein NuMA are sufficient to parse emergent cell phenotypes.
Acknowledgments
We would like to thank Dr. Richard Gorlick from the Children’s Hospital at Montefiore in Bronx, New York for providing us with genetically transformed MSCs used in the oncogenic transformation studies. This work was funded by the National Institutes of Health (P41EB001046 to P.V.M, J.K., and M.L.B.; R00CA163957 to P.-A.V; and R01CA112017 to S.A.L.), the National Science Foundation (DGE 0801620), and the NJ Stem Cell Technology CORE grants (both to P.V.M.). We gratefully acknowledge the contributions of Matthew D. Treiser, MD, PhD., of Rutgers, whose original work established the foundations for this study.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Klimanskaya I, Rosenthal N, Lanza R. Derive and conquer: sourcing and differentiating stem cells for therapeutic applications. Nat Rev Drug Discov. 2008;7(2):131–142. doi: 10.1038/nrd2403. [DOI] [PubMed] [Google Scholar]
- 2.Rangel MC, Bertolette D, Castro NP, Klauzinska M, Cuttitta F, Salomon DS. Developmental signaling pathways regulating mammary stem cells and contributing to the etiology of triple-negative breast cancer. Breast Cancer Res Treat. 2016;156(2):211–226. doi: 10.1007/s10549-016-3746-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antebi B, Pelled G, Gazit D. Stem cell therapy for osteoporosis. Curr Osteoporos Rep. 2014;12(1):41–47. doi: 10.1007/s11914-013-0184-x. [DOI] [PubMed] [Google Scholar]
- 4.Chamberlain G, Fox J, Ashton B, Middleton J. Concise review: mesenchymal stem cells: their phenotype, differentiation capacity, immunological features, and potential for homing. Stem Cells. 2007;25(11):2739–2749. doi: 10.1634/stemcells.2007-0197. [DOI] [PubMed] [Google Scholar]
- 5.Tysnes BB. Tumor-initiating and -propagating cells: cells that we would like to identify and control. Neoplasia. 2010;12(7):506–515. doi: 10.1593/neo.10290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tolar J, et al. Sarcoma derived from cultured mesenchymal stem cells. Stem Cells. 2007;25(2):371–379. doi: 10.1634/stemcells.2005-0620. [DOI] [PubMed] [Google Scholar]
- 7.Liu E, Gordonov S, Treiser MD, Moghe PV. Parsing the early cytoskeletal and nuclear organizational cues that demarcate stem cell lineages. Cell cycle. 2010;9(11):2108–2117. doi: 10.4161/cc.9.11.11864. [DOI] [PubMed] [Google Scholar]
- 8.Treiser MD, Yang EH, Gordonov S, Cohen DM, Androulakis IP, Kohn J, Chen CS, Moghe PV. Cytoskeleton-based forecasting of stem cell lineage fates. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(2):610–615. doi: 10.1073/pnas.0909597107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simon DN, Wilson KL. The nucleoskeleton as a genome-associated dynamic 'network of networks'. Nat Rev Mol Cell Biol. 2011;12(11):695–708. doi: 10.1038/nrm3207. [DOI] [PubMed] [Google Scholar]
- 10.Pajerowski JD, Dahl KN, Zhong FL, Sammak PJ, Discher DE. Physical plasticity of the nucleus in stem cell differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(40):15619–15624. doi: 10.1073/pnas.0702576104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lelievre SA, Weaver VM, Nickerson JA, Larabell CA, Bhaumik A, Petersen OW, Bissell MJ. Tissue phenotype depends on reciprocal interactions between the extracellular matrix and the structural organization of the nucleus. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(25):14711–14716. doi: 10.1073/pnas.95.25.14711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vega SL, Dhaliwal A, Arvind V, Patel PJ, Beijer NR, de Boer J, Murthy NS, Kohn J, Moghe PV. Organizational metrics of interchromatin speckle factor domains: integrative classifier for stem cell adhesion & lineage signaling. Integrative biology. 2015;7(4):435–446. doi: 10.1039/c4ib00281d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gribbon C, Dahm R, Prescott AR, Quinlan RA. Association of the nuclear matrix component NuMA with the Cajal body and nuclear speckle compartments during transitions in transcriptional activity in lens cell differentiation. European journal of cell biology. 2002;81(10):557–566. doi: 10.1078/0171-9335-00275. [DOI] [PubMed] [Google Scholar]
- 14.Abad PC, Lewis J, Mian IS, Knowles DW, Sturgis J, Badve S, Xie J, Lelievre SA. NuMA influences higher order chromatin organization in human mammary epithelium. Molecular biology of the cell. 2007;18(2):348–361. doi: 10.1091/mbc.E06-06-0551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DiPaolo JA, Casto BC. Quantitative studies of in vitro morphological transformation of Syrian hamster cells by inorganic metal salts. Cancer research. 1979;39(3):1008–1013. [PubMed] [Google Scholar]
- 16.Li N, Yang R, Zhang W, Dorfman H, Rao P, Gorlick R. Genetically transforming human mesenchymal stem cells to sarcomas: changes in cellular phenotype and multilineage differentiation potential. Cancer. 2009;115(20):4795–4806. doi: 10.1002/cncr.24519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bourke SL, Kohn J. Polymers derived from the amino acid L-tyrosine: polycarbonates, polyarylates and copolymers with poly(ethylene glycol) Adv Drug Deliv Rev. 2003;55(4):447–466. doi: 10.1016/s0169-409x(03)00038-3. [DOI] [PubMed] [Google Scholar]
- 18.Kholodovych V, Gubskaya AV, Bohrer M, Harris N, Knight D, Kohn J, Welsh WJ. Prediction of biological response for large combinatorial libraries of biodegradable polymers: Polymethacrylates as a test case. Polymer. 2008;49(10):2435–2439. [Google Scholar]
- 19.Sung HJ, Chandra P, Treiser MD, Liu E, Iovine CP, Moghe PV, Kohn J. Synthetic polymeric substrates as potent pro-oxidant versus anti-oxidant regulators of cytoskeletal remodeling and cell apoptosis. J Cell Physiol. 2009;218(3):549–557. doi: 10.1002/jcp.21629. [DOI] [PubMed] [Google Scholar]
- 20.Power J, Mayer-Proschel M, Smith J, Noble M. Oligodendrocyte precursor cells from different brain regions express divergent properties consistent with the differing time courses of myelination in these regions. Dev Biol. 2002;245(2):362–375. doi: 10.1006/dbio.2002.0610. [DOI] [PubMed] [Google Scholar]
- 21.Vega SL, Liu E, Patel PJ, Kulesa AB, Carlson AL, Ma Y, Becker ML, Moghe PV. High-content imaging-based screening of microenvironment-induced changes to stem cells. J Biomol Screen. 2012;17(9):1151–1162. doi: 10.1177/1087057112453853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sher F, Balasubramaniyan V, Boddeke E, Copray S. Oligodendrocyte differentiation and implantation: new insights for remyelinating cell therapy. Curr Opin Neurol. 2008;21(5):607–614. doi: 10.1097/WCO.0b013e32830f1e50. [DOI] [PubMed] [Google Scholar]
- 23.Raff M, Apperly J, Kondo T, Tokumoto Y, Tang D. Timing cell-cycle exit and differentiation in oligodendrocyte development. Novartis Found Symp. 2001;237:100–107. doi: 10.1002/0470846666.ch9. discussion 107–12, 158–63. [DOI] [PubMed] [Google Scholar]
- 24.Rani AS, Qu DQ, Sidhu MK, Panagakos F, Shah V, Klein KM, Brown N, Pathak S, Kumar S. Transformation of immortal, non-tumorigenic osteoblast-like human osteosarcoma cells to the tumorigenic phenotype by nickel sulfate. Carcinogenesis. 1993;14(5):947–953. doi: 10.1093/carcin/14.5.947. [DOI] [PubMed] [Google Scholar]
- 25.Ohshima S. Induction of genetic instability and chromosomal instability by nickel sulfate in V79 Chinese hamster cells. Mutagenesis. 2003;18(2):133–137. doi: 10.1093/mutage/18.2.133. [DOI] [PubMed] [Google Scholar]
- 26.Chen LP, et al. alpha4 is highly expressed in carcinogen-transformed human cells and primary human cancers. Oncogene. 2011;30(26):2943–2953. doi: 10.1038/onc.2011.20. [DOI] [PubMed] [Google Scholar]
- 27.Lelievre S, Hodges KB, Vidi PA. Application of Theranostics to Measure and Treat Cell Heterogeneity in Cancer. In: Chen X, Wong S, editors. Cancer Theranostics. Elsevier Inc.: Academic Press; 2014. pp. 493–516. [Google Scholar]
- 28.Nelson CM, Bissell MJ. Of extracellular matrix, scaffolds, and signaling: tissue architecture regulates development, homeostasis, and cancer. Annu Rev Cell Dev Biol. 2006;22:287–309. doi: 10.1146/annurev.cellbio.22.010305.104315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ingber DE, Tensegrity I. Cell structure and hierarchical systems biology. J Cell Sci. 2003;116(Pt 7):1157–1173. doi: 10.1242/jcs.00359. [DOI] [PubMed] [Google Scholar]
- 30.Lelievre SA. Contributions of extracellular matrix signaling and tissue architecture to nuclear mechanisms and spatial organization of gene expression control. Biochim Biophys Acta. 2009;1790(9):925–935. doi: 10.1016/j.bbagen.2009.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cunha GR, Vanderslice KD. Identification in histological sections of species origin of cells from mouse, rat and human. Stain Technol. 1984;59(1):7–12. doi: 10.3109/10520298409113823. [DOI] [PubMed] [Google Scholar]
- 32.Logan DJ, Shan J, Bhatia SN, Carpenter AE. Quantifying co-cultured cell phenotypes in high-throughput using pixel-based classification. Methods. 2016;96:6–11. doi: 10.1016/j.ymeth.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Radulescu AE, Cleveland DW. NuMA after 30 years: the matrix revisited. Trends Cell Biol. 2010;20(4):214–222. doi: 10.1016/j.tcb.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weaver VM, Carson CE, Walker PR, Chaly N, Lach B, Raymond Y, Brown DL, Sikorska M. Degradation of nuclear matrix and DNA cleavage in apoptotic thymocytes. Journal of cell science. 1996;109(Pt 1):45–56. doi: 10.1242/jcs.109.1.45. [DOI] [PubMed] [Google Scholar]
- 35.Taimen P, Kallajoki M. NuMA and nuclear lamins behave differently in Fas-mediated apoptosis. J Cell Sci. 2003;116(Pt 3):571–583. doi: 10.1242/jcs.00227. [DOI] [PubMed] [Google Scholar]
- 36.Vidi PA, Chandramouly G, Gray M, Wang L, Liu E, Kim JJ, Roukos V, Bissell MJ, Moghe PV, Lelievre SA. Interconnected contribution of tissue morphogenesis and the nuclear protein NuMA to the DNA damage response. Journal of cell science. 2012;125(Pt 2):350–361. doi: 10.1242/jcs.089177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vidi PA, Liu J, Salles D, Jayaraman S, Dorfman G, Gray M, Abad P, Moghe PV, Irudayaraj JM, Wiesmuller L, Lelievre SA. NuMA promotes homologous recombination repair by regulating the accumulation of the ISWI ATPase SNF2h at DNA breaks. Nucleic acids research. 2014;42(10):6365–6379. doi: 10.1093/nar/gku296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Knowles DW, Sudar D, Bator-Kelly C, Bissell MJ, Lelievre SA. Automated local bright feature image analysis of nuclear protein distribution identifies changes in tissue phenotype. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(12):4445–4450. doi: 10.1073/pnas.0509944102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burness ML, Sipkins DA. The stem cell niche in health and malignancy. Seminars in cancer biology. 2010;20(2):107–115. doi: 10.1016/j.semcancer.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 40.Rosland GV, Svendsen A, Torsvik A, Sobala E, McCormack E, Immervoll H, Mysliwietz J, Tonn JC, Goldbrunner R, Lonning PE, Bjerkvig R, Schichor C. Long-term cultures of bone marrow-derived human mesenchymal stem cells frequently undergo spontaneous malignant transformation. Cancer research. 2009;69(13):5331–5339. doi: 10.1158/0008-5472.CAN-08-4630. [DOI] [PubMed] [Google Scholar]
- 41.Valko M, Rhodes CJ, Moncol J, Izakovic M, Mazur M. Free radicals, metals and antioxidants in oxidative stress-induced cancer. Chemico-biological interactions. 2006;160(1):1–40. doi: 10.1016/j.cbi.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 42.Behrend L, Henderson G, Zwacka RM. Reactive oxygen species in oncogenic transformation. Biochemical Society transactions. 2003;31(Pt 6):1441–1444. doi: 10.1042/bst0311441. [DOI] [PubMed] [Google Scholar]
- 43.Fruehauf JP, Meyskens FL., Jr Reactive oxygen species: a breath of life or death? Clinical cancer research: an official journal of the American Association for Cancer Research. 2007;13(3):789–794. doi: 10.1158/1078-0432.CCR-06-2082. [DOI] [PubMed] [Google Scholar]
- 44.Yoshihara M, Hayashizaki Y, Murakawa Y. Genomic Instability of iPSCs: Challenges Towards Their Clinical Applications. Stem Cell Rev. 2016 doi: 10.1007/s12015-016-9680-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Izgi K, Canatan H, Iskender B. Current status in cancer cell reprogramming and its clinical implications. J Cancer Res Clin Oncol. 2016 doi: 10.1007/s00432-016-2258-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.