Abstract
The seeds of the Ricinus communis (Castor bean) plant are the source of the economically important commodity castor oil. Castor seeds also contain the proteins ricin and R. communis agglutinin (RCA), two toxic lectins that are hazardous to human health. Radial immunodiffusion (RID) and the enzyme linked immunosorbent assay (ELISA) are two antibody-based methods commonly used to quantify ricin and RCA; however, antibodies currently used in these methods cannot distinguish between ricin and RCA due to the high sequence homology of the respective proteins. In this study, a technique combining antibody-based affinity capture with liquid chromatography and multiple reaction monitoring (MRM) mass spectrometry (MS) was used to quantify the amounts of ricin and RCA independently in extracts prepared from the seeds of eighteen representative cultivars of R. communis which were propagated under identical conditions. Additionally, liquid chromatography and MRM-MS was used to determine rRNA N-glycosidase activity for each cultivar and the overall activity in these cultivars was compared to a purified ricin standard. Of the cultivars studied, the average ricin content was 9.3 mg/g seed, the average RCA content was 9.9 mg/g seed, and the enzymatic activity agreed with the activity of a purified ricin reference within 35% relative activity.
Keywords: Ricin, RCA, Ricinus communis, Cultivar, Quantification, Mass spectrometry
1. Introduction
The seed of the castor plant (Ricininus communis L.) is the only source of the economically important castor oil. Castor oil is used in numerous manufacturing processes to produce paints, varnishes, lubricants, plastics, cosmetics and an array of other products. Importantly, castor oil is the only commercially available source of ricinoleic acid, which has several unique properties that are exploited as a starting material for many chemical processes (Dyer et al., 2008; Mutlu and Meier, 2010; Ogunniyi, 2006). Recently, castor oil has been considered as a biofuel source, because of its solubility in short chain alcohols, which makes the transesterification step more facile (Conceição et al., 2007; Severino et al., 2012). However, castor oil production is complicated by the high abundance of the toxic proteins ricin and RCA.
Ricin is a highly toxic protein with a mass of ~64 kDa consisting of two subunits linked by a single disulfide bond (Lappi et al., 1978; Olsnes and Pihl, 1973). Ricin is a Type II ribosome-inactivating protein (RIP) belonging to the A-B family of toxins (dimeric) consisting of two functionally different subunits (Stirpe and Barbieri, 1986). The A-chain has rRNA N-glycosidase activity while the B-chain is a lectin (Olsnes and Pihl, 1973). RCA is very similar to ricin, containing an A- and B-chain coupled together by a single disulfide bridge to make a ~64 kDa monomer unit, and exists naturally as a non-covalently linked dimer, resulting in a ~120 kDa complex (Cawley et al., 1978; Hegde and Podder, 1998). RCA shares high sequence homology with ricin, with over 93% similarity between A-chains and 84% similarly between B-chains, but is much less toxic (Roberts et al., 1985). Ricin gains entry to the cell following the binding of the B-chain to galactose-containing surface glycoproteins and glycolipids. Once the entire toxin is inside the cell, the chains dissociate. Then, the A-chain binds to a specific site on the 28S RNA of the 60S ribosome irreversibly removing an adenine (depurination) from a position known as the sarcin/ricin tetraloop (Endo et al., 1991; Endo and Tsurugi, 1987; Olsnes et al., 1974). This effectively prevents the binding of elongation factor 2 to the ribosome and shuts down all protein synthesis leading to cell death (Nilsson and Nygard, 1986). Ricin is a potent toxin, where it has been reported that the mode of action is so efficient that one ricin molecule is sufficient to disable an entire cell (Halling et al., 1985), though the median LD50 for ricin varies by route of administration. For humans it is estimated between 5 and 10 µg/kg by inhalation of particles less than 5 µm, and 1–20 mg/kg by ingestion (Audi et al., 2005; Bradberry et al., 2003). The toxicity of RCA has been measured to be less than that of ricin by a factor of ~50–2000, where the range in the toxicity is due to the type of assay used (Cawley et al., 1978; Lin and Liu, 1986; Saltvedt, 1976; Zhan and Zhou, 2003). Currently there is no approved treatment available for ricin intoxication (Franz and Jaax, 1997).
Due to the toxicity of ricin, availability, and ease in which a crude preparation of ricin can be generated from R. communis seeds, there is a concern that this toxin could be used in a terrorist event (Burrows and Renner, 1999; Rotz et al., 2002; Sobel et al., 2002). In the United States it is legal to possess castor beans but the procurement of ≥100 mg of purified ricin is governed by the Select Agent Regulation (National Select Agent Registry, 2014). An aspect that has gained the attention of security agencies is the amount of crude ricin present in large scale castor oil extraction facilities. When the oil from castor beans has been extracted, a large amount of byproduct called a “mash” or “cake” remains. If inactivation of the mash is not complete or is neglected, large amounts of crude ricin (50,000 tons/year worldwide) could be available for potentially malevolent uses (Fredriksson et al., 2005; Smallshaw and Vitetta, 2012).
There is considerable interest in knowing the amount of ricin and other proteins that are typically found in castor beans. Antibody-based methods have been used to measure amounts of ricin in different cultivars, but the antibodies used in these techniques cannot differentiate ricin from RCA and therefore report the combined quantities of both proteins (Auld et al., 2003, 2001; Baldoni et al., 2011; Garber, 2008; Pinkerton, 1997; Pinkerton et al., 1999) leading to over-estimation of ricin. A method that does not suffer from the adverse effects of the high homology between ricin and RCA uses the reverse transcriptase polymerase chain reaction. This was used to individually monitor the gene expression of ricin and RCA during R. communis seed development (Chen et al., 2005). Methods involving mass spectrometry (MS) do not suffer from the selectivity issues that affect RID and ELISA and monitor the actual toxin itself rather than the DNA or RNA. MS can be used to detect and discriminate between ricin and RCA by exploiting the small number of amino acid sequence differences between the two proteins (Brinkworth, 2010; Fredriksson et al., 2005; Kalb and Barr, 2009; Kull et al., 2010; Ostin et al., 2007). We have employed a method that can be used to quantify ricin and RCA individually by combining antibody-based affinity capture, tryptic digestion, and peptide analysis by liquid chromatography isotope dilution mass spectrometry (IDMS) (McGrath et al., 2011). Using antibody-based affinity capture with tandem mass spectrometry and judiciously choosing peptides that are unique to ricin and RCA, we achieved specificity that is much higher than antibody-based methods such as RID and ELISA. In addition, MS has been used to estimate the toxicity of ricin by measuring the activity of the A-chain with a synthetic DNA mimic of the natural RNA substrate (Becher et al., 2007; Hines et al., 2004; Kalb and Barr, 2009). For the present study, these two MS approaches were combined to better understand the range of ricin and RCA concentrations naturally present in seed extracts of 18 different R. communis cultivars, which were chosen according to seed size and geographic location. First, the quantities of ricin and RCA were measured individually in extracts from the castor bean seeds. We subsequently correlated ricin and RCA concentration data with total N-glycosidase activity, measured by LC-MS/MS detection of adenine liberated from a DNA substrate. The information captured in these analyses adds to the growing pool of R. communis biology (Chan et al., 2010; Loss-Morais et al., 2013; Nogueira et al., 2012), the facilitation of finding and breeding castor cultivars with low or absent levels of ricin (Auld et al., 2003, 2001; Barnes et al., 2009; Khvostova, 1986; Pinkerton, 1997), and determination of refinement/purity of “white powder” preparations that were found in recent events such as the letters delivered to a United States Senator, a Judge, the Mayor of New York City, and the President of the United States (Geranios, 2013; Nelson, 2013; Weiner, 2013; Weisman, 2013).
2. Materials and methods
2.1. Safety precautions
Exposure to ricin is hazardous to human health. Experiments using ricin were all performed in a biosafety level-2 cabinet equipped with a HEPA filter; individuals handling ricin were equipped with personal protective gear and practiced universal safety protocols.
2.2. Reagents
Ricin, RCA and biotinylated and non-biotinylated polyclonal goat anti-ricinus communis antibody were obtained from Vector Laboratories (Burlingame, CA). Bovine Serum Albumin (BSA) standard reference material (SRM 927d) was obtained from the National Institute for Standards and Technology (NIST) (Gaithersburg, MD). Trypsin Gold, Mass Spectrometry Grade was purchased from Promega Corporation (Madison, WI). RapiGest SF Surfactant was purchased from Waters Corporation (Milford, MA). Dynabeads MyOne Streptavidin T1 magnetic beads were obtained from Life Technologies (Carlsbad, CA). Native heavy-labeled peptide standards were synthesized by Midwest Biotech, Inc. (Fishers, IN). The DNA substrate (5′-GCGCGAGAGCGC-3′) for measurement of N-glycosidase activity was synthesized by Integrated DNA Technologies (Coralville, IA). Adenine was purchased from Sigma–Aldrich (St. Louis, MO) and 13C-labeled adenine was purchased from Moravek Biochemicals and Radiochemicals (Brea, CA). Tris (2-carboxyethyl) phosphine (TCEP) was obtained from Thermo Fisher Scientific (Rockford, IL). HPLC grade water, HPLC grade water with 0.1% formic acid, acetonitrile, acetonitrile with 0.1% formic acid and Methanol (Honeywell, Burdick and Jackson) were obtained from VWR (West Chester, PA). All other chemicals were purchased from Sigma–Aldrich (St. Louis, MO) and were used without further purification.
2.3. Ricin and RCA native and isotopically-labeled peptides
Peptide sequences shown in Table 1 were selected from the A-and B-chains of ricin (P02879) and RCA (XP_002534220) to provide specificity and to discriminate between the respective chains. Native and 13C, 15N-labeled peptides received from Midwest Biotech, Inc. (Fishers, IN) were resuspended in 0.1% formic acid, aliquotted to approximately 45 pmol µL into ~800 vials, lyophilized, and stored at −80 °C until use. For accurate quantitative measurements, four randomly selected tubes from each set of peptide aliquots was subjected to amino acid analysis (AAA) using an in-house developed AAA method which was calibrated to amino acid standards obtained from NIST (Woolfitt et al., 2009).
Table 1.
Peptide sequences used for ricin and RCA quantification and detection. Peptide sequences are listed with corresponding isotopically heavy 13C,15N-labeled peptide standards with the labeled residues denoted with an (*). The monoisotopic precursor m/z are listed with the product ion transitions. Product ion transitions in bold were used for quantification while the others are used for confirmation.
| Quantification | Sequence | Precursor ion | Transition ions monitored | |||
|---|---|---|---|---|---|---|
| TA7, Ricin | VGLPINQR | 448.77 (+2) | 417.22 (+1) y3 | 530.35 (+1) y4 | 627.45 (+1) y5 | |
| VGLPINQ*R | 452.27 (+2) | 424.22 (+1) y3 | 537.35 (+1) y4 | 634.31 (+1) y5 | ||
| TA5, Ricin | LTTGADVR | 416.73 (+2) | 274.19 (+1) y2 | 517.27 (+1) y5 | 618.32 (+1) y6 | |
| LTTGADV*R | 419.73 (+2) | 280.19 (+1) y2 | 523.27 (+1) y5 | 624.32 (+1) y6 | ||
| TB19, Ricin | ASDPSLK | 717.38 (+1) | 260.20 (+1) y2 | 347.23 (+1) y3 | 444.28 (+1) y4 | |
| ASDPSL*K | 724.38 (+1) | 267.20 (+1) y2 | 354.23 (+1) y3 | 451.28 (+1) y4 | ||
| TA6, Ricin, RCA | HEIPVLPNR | 537.81 (+2) | 267.07 (+1) b2 | 695.25 (+1) y6 | 808.47 (+1) y7 | |
| HEIPVLP*NR | 540.81 (+2) | 267.09 (+1) b2 | 701.37 (+1) y6 | 814.49 (+1) y7 | ||
| TA5, RCA | SHLTTGGDVR | 521.77 (+2) | 225.10 (+1) b2 | 705.35 (+1) y7 | 818.44 (+1) y8 | |
| SHLTTGGDV*R | 524.77 (+2) | 225.10 (+1) b2 | 711.35 (+1) y7 | 824.44 (+1) y8 | ||
| TA7, RCA | VGLPISQR | 435.26 (+2) | 390.21 (+1) y3 | 503.30 (+1) y4 | 600.35 (+1) y5 | |
| VGLPISQ*R | 438.76 (+2) | 397.21 (+1) y3 | 510.30 (+1) y4 | 607.35 (+1) y5 | ||
| Detection | ||||||
| TB18, Ricin | NDGTILNLYSGLVLDVR | 931.50 (+2) | 274.23 (+1) y2 | 858.52 (+1) y8 | 1021.63 (+1) y9 | |
| TB18, RCA | NDGTILNLYNGLVLDVR | 945.01 (+2) | 274.23 (+1) y2 | 885.52 (+1) y8 | 1048.58 (+1) y9 | |
| (TA24–SS–TB1), Ricin, RCA | CAPPPSSQF ADVCMDPEPIVR |
759.01 (+3) | 484.32 Tb1 (+1) y4 | 662.26 TA24 (+1) y6 | 710.42 Tb1 (+1) y6 | 1055.8 TA24 (+2) b8 |
2.4. Measurement of protein mass
The molecular weight of ricin and RCA was determined by Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF MS). MALDI matrix was prepared by generating a saturated solution of Sinapinic Acid (SA) in a 70:30 acetonitrile/ddH2O with 0.01% TFA. The NIST BSA stock solution was diluted 1:1 with ddH2O. Sample preparation for the ricin consisted of mixing 3µL stock purified ricin (5 mg/mL) with 3 µL of 1:1 diluted NIST-BSA plus 6 µL saturated SA matrix solution. Sample preparation for the RCA consisted of mixing 6 µL of stock purified RCA (5 mg/mL) with 2 µL of 1:1 diluted NIST-BSA plus 8µL saturated SA matrix solution. A portion of the saturated SA was diluted 1:1 with ddH2O and 0.5 µL was applied to the MALDI sample plate and allowed to air dry at room temperature. The sample-SA mixture was then deposited onto the dried layer of diluted SA matrix and allowed to air dry at room temperature. Each spot was washed using 2.5 µL of 0.1 % TFA applied directly on top of the dried spot for 5 –10 s, which then was removed using a pipette. Once sample spots were dry, analysis was then performed on a 4800 + MALDI TOF/TOF Analyzer (Applied Biosystems, Carlsbad, CA) in linear mode. The NIST value for the measured molecular mass of the SRM 927d BSA reference material was 66,432 ± 7 Da and the signals from the BSA (M+1)1+ and (M+2)2+ charged ions were used to internally calibrate the time of flight spectra for the (M+1)1 ion of ricin and (M+2)2+ ion of RCA. 10 individual replicates were generated for ricin and for RCA analysis.
2.5. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and western blot analysis
Ricin 2 µg, 1 µg, and 0.5 µg protein (loaded per individual well) were separated using one-dimensional SDS-PAGE in precast Novex NuPAGE 4–12% bis-tris gels (Life Technologies, Carlsbad, CA). Broad molecular weight protein standards (10–250 kDa) were used as a size reference (Bio-Rad Laboratories, Hercules, CA). Denatured proteins were visually examined using a silver stain for mass spectrometry kit (Pierce, Rockford, IL). Furthermore to assess immunoreactivity, an unstained duplicate gel consisting of separated ricin was electroblotted using the iBlot dry blotting system (Life Technologies). The protein-immobilized PVDF blot was subsequently immunodetected with an affinity purified primary goat-anti ricinus communis antibody using the anti-goat Westernbreeze chromogenic kit (Life Technologies). Of note, to ensure maximal immunoreactivity, the primary antibody (5 mg/mL; diluted in 1% nonfat dry milk) was incubated over night at room temperature on a rocking platform and the next day, the remaining steps in the Westernbreeze chromogenic kit protocol were completed.
2.6. Generation of a peptide-based standard calibration curve
A peptide-based standard calibration curve was created to quantify the purified ricin and RCA in the commercial stock solutions. Individual native and labeled synthetic peptides used for quantification of ricin and RCA were solubilized in 0.1% formic acid with 1 pmol/ µL human [Glu1]-Fibrinopeptide B to generate a m 60 pmol/µL peptide solution. Equal volumes of the individual 60 pmol/µL solutions of the native peptides were mixed to create a 10 pmol/µL combined solution of all native standards. This procedure was repeated with the 13C,15N-labeled peptides to create a 10 pmol/µL combined solution of all labeled standards. The native peptide solution was serially diluted in 0.1% formic acid with 1 pmol/ µL human [Glu1]-Fibrinopeptide B to create an eleven point standard curve covering a concentration range from1 to 5000 fmol/ µL and the mixture of labeled peptides was diluted to a concentration of 50 fmol/µL to create the working stock of peptide internal standard. The internal standard solution was added to each of the native peptide levels at a ratio of 2:1 to generate the standard peptide calibration curve. Quantification of ricin was carried out using the peptide ions VGLPINQR and LTTGADVR for the A-chain and ASDPSLK for the B-chain. For quantification of RCA, only the A-chain peptide ions VGLPISQR and SHLTTGGDVR were used.
2.7. Identity and sourcing of R. communis cultivars
Seeds from 18 R. communis cultivars were obtained from J. Bradley Morris at the Plant Genetic Resources Conservation Unit, Agricultural Research Service (ARS), U.S. Department of Agriculture (USDA), in Griffin, Georgia. The choice of cultivar was based on geographic distribution and seed size (small, medium and large) (Hodge et al., 2013) from a total 1043 accessions in the castor germplasm collection. Propagation, harvesting and conservation of the cultivars were carried out according to Morris et al. (2011). Briefly, propagation was carried out such that 50 plants per cultivar occupied a 6 m2 area. To prevent cross pollination the inflorescences were covered preceding fertilization. When the seeds reached maturity, castor bean plants were harvested by hand then dried at 21 °C, and 25% relative humidity, for approximately one week and then threshed. Seeds were counted, weighed, and stored at 4 °C for distribution, and for longer term storage seeds were stored at −18 °C.
The 18 R. communis accessions used were: US Florida cv.: PI265508, Iran cv.: PI222265, Peru cv.: PI215770, Mexico cv.: PI165446, Morocco cv.: PI253621, Brazil cv.: PI202711, Cuba cv.: PI208839, US Virgin Islands cv.: PI209326, Afghanistan cv.: PI212115, Pakistan cv.: PI217539, Turkey cv.: PI167342, China cv.: PI436592, Former Soviet Union cv.: PI257654, Argentina cv.: PI219767, India cv.: PI173947, Puerto Rico cv.: PI209132, Hale cv.: PI624000, Syria cv.: PI181916. For use as negative controls in the quantification of ricin and RCA, seed extracts of two near neighbors to R. communis from the Euphorbiaceae family (67117) Adriana quadripartia (B & T World Seeds, Aigues-Vives, France) and (3403) Macaranga grandifolia (Top Tropicals, Ft. Meyers, Florida), and one species from the Phytolaccaceae family (39161) Phytolacca americana (B & T World Seeds, Aigues-Vives, France), were also analyzed. R. communis seeds var. gibsonii (Seed Saver Exchange, Decorah, IA) and var. zanzibarensis (Tropilab, Inc., St. Petersburg, FL) were used for evaluation of reproducibility of seed extraction.
2.8. Extraction of seeds
2.8.1. Initial extraction of seeds
Crude seed extracts were prepared by modifying a method provided by E.A.E. Garber (E.A.E. Garber, CFSAN, FDA, personal communication). Briefly, 100 seeds for each cultivar were weighed, placed in a small coffee grinder, and the edges taped to minimize aerosols. After grinding to a fine particle size, the ground seed material was carefully transferred to a 50 mL conical centrifuge tube containing phosphate-buffered saline with 0.1% Tween-20 (v/ v) (PBST 0.1%). The ratio of PBST 0.1% to whole seed weight was kept constant at 2.25 mL/g of seeds. Each tube was closed, sealed with parafilm, mixed on a vortex mixer, and placed on a rocker platform for 16–19 h in the dark at 4 °C, and finally centrifuged at 3000 rpm (GSA rotor) for 4 min. After centrifugation, the cloudy brownish middle between the lower debris and upper lipid layer was transferred into cryovials at 0.25 mL aliquots and stored at −80 °C until use.
2.8.2. Reproducibility of extraction from seeds
To determine if the amounts of ricin and RCA were affected by the extraction procedure, replicate extracts were each generated from the seeds using two varieties of R. communis. The extraction procedure described in 2.8.1. was used to create five individual replicates from the gibsonii seeds and three individual replicates from the zanzibarensis seeds. Six aliquots from each replicate were processed and analyzed using IDMS to determine the variation in amounts of ricin and RCA from the extraction procedure.
2.9. Coupling of magnetic beads with antibody
Biotinylated polyclonal ricin antibody was received as a 0.5 mg lyophilized powder and was dissolved in 0.5 mL of ddH2O (1 mg/ mL) prior to coupling to the magnetic beads. The streptavidin-coated magnetic beads were supplied at 10 mg/mL in a solution of phosphate buffered saline (PBS), pH 7.4, with 0.01% Tween-20 (v/ v) and 0.09% (w/v) NaN3 as preservative. The coupling of antibody to beads was prepared according to the manufacturer’s directions. A150 µL aliquot of antibody stock solution was combined with 9 mL of PBS and vortex mixed, then combined with 1 mL of beads in PBS and incubated at room temperature with gentle rotation for 1 h. After incubation the beads were washed twice with PBS and the supernatant removed. The beads were re-suspended in 1 mL of PBS containing 0.05% Tween-20 (v/v) (PBST 0.05%) and stored at 4 °C until use.
2.10. Preparation of protein standard calibration curve, quality control, and seed extract samples
Ricin and RCA protein standard calibration curve, quality control, and seed extract samples were generated using the Biomek NXP Laboratory Automation Workstation (Beckman Coulter, Brea, CA) with a 96 deep-well polypropylene plate. All dilutions were carried out using PBST 0.05%. For the purpose of direct comparison, all control and unknown samples were prepared as 500 µL aliquots. The stock solutions of 4.5 mg/mL purified Vector ricin and 4.9 mg/ mL RCA were manually diluted, respectively, to obtain solutions of 11.4 ng/µL for ricin and 24.8 ng/µL for RCA. Using the diluted stock, a seven point standard calibration series was constructed for each protein using the Biomek workstation: 0.003–0.286 ng/µL for ricin and 0.006–0.624 ng/µL for RCA. Using 500 µL from each concentration point, the calibration curve had a protein quantity of 1.4–143 ng of ricin/500 µL PBST 0.05% and 3.1–312 ng of RCA/ 500 µL PBST 0.05% which was subjected to antibody capture and analysis. The quality control samples (QCs) for ricin and RCA were generated by diluting the stock ricin and RCA solutions to produce a 114 ng/500 µL PBST 0.05% high QC and a 11.4 ng/500 µL PBST 0.05% low QC for ricin and a 249 ng/500 µL PBST 0.05% high QC and a 24.9 ng/500 µL PBST 0.05% low QC for RCA. Seed extracts were manually diluted 4000 fold in PBST 0.05%, then 500 µL aliquots from each diluted sample was subjected to antibody capture and analysis.
2.11. Antibody capture of ricin and RCA
The 96 deep-well plate containing 500 µL sample aliquots was loaded into a KingFisher 96 automated magnetic particle processor (Thermo Fisher Scientific Inc., Waltham, MA). The particle processor deposited 20 µL of antibody-coated magnetic beads into each sample containing well. The samples were continuously mixed for 1 h to allow the antigens to bind to the antibodies coupled to magnetic beads. After 1 h, the magnetic beads were retrieved from each of the samples and sequentially washed with 1 mL of PBST 0.05%, 1 mL of PBST 0.01%, 0.5 mL of PBS and 200 µL of ddH2O. The beads were then deposited into 100 µL of ddH2O in preparation for subsequent processing.
2.12. Tryptic digestion
2.12.1. Digestion of purified ricin and RCA
The concentration of the stock purified ricin and RCA was determined using IDMS analysis of tryptic peptides from the two proteins. Three sample preparation conditions were evaluated: (a) digestion with 4 µg of trypsin, (b) digestion with 40 µg of trypsin, and (c) digestion with 40 µg of trypsin with reduction and alkylation. The concentration of reducing and alkylating agents used in condition (c) were equimolar to the molar amount of disulfide bonds present in the ricin and RCA, respectively. This was done in an effort to limit the spread of non-specific alkylation to other residues which could shift the peptide ion m/z values being monitored. The acid labile surfactant RapiGest SF was dissolved in 500 mM ammonium bicarbonate containing 1 mM CaCl2 to generate a 0.3% solution. For each reaction, 10 µL of 0.3% RapiGest SF solution and 63 ng(1000 fmol)of ricinor 128 ng(1000 fmol)of RCA was added to several wells of two 96-well semi-skirted PCR trays (ThermoFisher Scientific, Waltham, MA). One tray was used for reduction/alkylation and the other without. The trays were sealed and mixed with an Eppendorf MixMate (Fisher Scientific, Inc. Pittsburgh, PA) vortex mixer, then placed into a Veriti 96 well thermal cycler (Applied Biosystems, Carlsbad, CA) and incubated for 15 min at 99 °C. After heating, the PCR plates were removed and allowed to cool for 2 min. For samples undergoing reduction and alkylation, 6 or 12 pmol of tris (2-carboxyethyl) phosphine (TCEP) were added to the ricin or RCA wells, respectively. The plate used for reduction/alkylation was mixed, capped and returned to the thermocycler for a 1 h incubation at 60 °C. Upon completion of incubation and cooling, 12 pmol and 24 pmol of iodoacetamide were added to the ricin or RCA samples, respectively, and the samples were allowed to incubate for 30 min at room temperature in the dark. Two microliters of 2 µg/µL or 20 µg/µL trypsin was added to each of the wells in appropriate plate. The plates were mixed on a vortex mixer and incubated for 1 h at 37 °C then cooled for 2 min, centrifuged, and 3 µL of 2 M HCl was added to degrade the Rapigest SF. The plates were again mixed and returned to the thermocycler where the samples were incubated for 1 h at 37 °C. After this incubation period, and before LC-MS/MS analysis, 4 µL of a solution of 50 fmol/µL heavy labeled internal standard was added to each sample.
2.12.2. Digestion of antibody-captured ricin and RCA
Following incubation of antibody-coupled magnetic beads with sample solutions, the Kingfisher 96-well magnetic PCR head (ThermoFisher Scientific, Waltham, MA), was used to manually transfer the beads to a 96-well semi-skirted PCR tray containing 10 µL of 0.3% RapiGest SF solution per well. The plate was sealed and vortex mixed, then placed into the thermocycler and incubated for 15 min at 99 °C. Following this step, all subsequent steps were identical to the tryptic digest procedure in Section 2.12.1 for digestion of the purified ricin plus the additional step where the magnetic beads were removed prior to sample analysis.
2.13. Liquid chromatography/mass spectrometry
Liquid chromatography and mass spectrometry (LC-MS) data were acquired using an Acquity UPLC system (Waters Corporation, Milford, MA) coupled to a Vantage triple quadrupole mass spectrometer (Thermo Fisher Scientific, Waltham, MA) using a 1 mm × 50 mm HSS T3 reverse phase column (Waters Corporation, Milford, MA). The reversed phase gradient solvents consisted of solvent A (0.1% formic acid in water) and solvent B (0.1% formic acid in acetonitrile). The gradient program initially started at an organic composition of 1% solution B. Between time zero and 3 min the gradient was ramped to 25% solution B, at 3.1 min ramped to 90% solution B and held at 90% solution B to 4.1 min. Between 4.1 min and 4.75 min the gradient was dropped back to 1% solution B and held until 6 min had elapsed. The flow rate used was variable: from 0 to 3.5 min, the flow rate was 200 µL/min, then it was increased to 300 µL/min from 3.5 to 4.75 min, and reduced to 200 µL/min for the remainder of the run. The UPLC system was equipped with a 20 µL loop and 9.5 µL of each sample was injected per analysis. The instrument was programed to acquire data in MRM mode for positive ions, where the transitions for each peptide are shown in Table 1. Instrument parameters consisted of a spray voltage of 3400 V, vaporizer temperature of 350 °C, a sheath gas pressure of 193 kPa (28 psi), an ion sweep gas pressure of 68.9 kPa (10 psi), an auxiliary gas pressure of 138 kPa (20 psi), and a capillary temperature of 320 °C. Collision energies and S-lens values were optimized for each native peptide by infusion of diluted peptide stock solutions. Xcalibur 2.2 software (Thermo Fisher Scientific, Waltham, MA) was used for instrument control and to process the data files.
2.14. Preparation of samples for quantification of enzyme activity
Once the concentration of ricin in the stock solutions had been determined for each cultivar extract, an aliquot from each was diluted to 5.7 ng/µL of ricin, and 40 ng of material from each diluted cultivar extract was subjected to the adenine release assay. Normalization of the quantity of ricin ensured that any differences measured in N-glycosidase activity would not be due to variations in quantity of ricin protein. Ricin protein calibration curves and QC samples were generated identically to the procedure described in Section 2.10 “Preparation of protein standard calibration curve, quality control, and seed extract samples.” However, for quantification of enzyme activity, the top point of the standard curve extended to 286 ng rather than 143 ng in 500 µL.
2.15. Adenine release activity assay
Ricin and RCA were captured on magnetic beads in the same manner as described above. The bound proteins were incubated with a DNA oligonucleotide substrate (5′-GCGCGAGAGCGC-3′) that mimics the natural ribosomal RNA target and contains the -GAGA-tetraloop structure. The N-glycosidase activity of ricin releases an adenine molecule, which can be monitored by LC-MS/MS. The magnetic beads were manually transferred from the ddH2O using a Kingfisher 96 well magnetic PCR head to a semi-skirted PCR tray where each of the wells contained 25 µL of activity buffer. The activity buffer consisted of 10 mM ammonium citrate and 1 mM ethylenediaminetetraacetic acid, pH 4.0, and contained 100 mM of DNA substrate. The plate was vortex mixed, placed into the thermocycler, and incubated for 4 h at 37 °C. After incubation, 5 µL of 7.5 pmol/ µL 13C-labeled adenine and 20 µL of 0.08% formic acid/0.1% acetic acid/0.08% ammonium hydroxide in ddH2O was added to each well. The plate was vortex mixed, the antibody-coated magnetic beads were removed using the Kingfisher 96 PCR magnet head, and the tray containing the sample supernatants was placed into the autosampler of the Acquity UPLC system. An in-line filter was used to help protect the column from any stray magnetic beads that may not have been removed from the sample wells.
2.16. Quantification of N-glycosidase activity by LC-MS/MS
The method used for measurement of N-glycosidase activity follows that of the method developed by Becher et al., (2007) with additional modifications. An Acquity UPLC with a 2.1 × 10 cm Ascentis Express C18 HPLC column (Sigma–Aldrich, St. Louis, MO) was coupled to an ABI 4000 Q-Trap mass spectrometer (Applied Biosystems, Carlsbad, CA). Solvent A consisted of 0.08% formic acid/ 0.1% acetic acid/0.08% ammonium hydroxide in ddH2O and solvent B consisted of 0.08% formic acid/0.1% acetic acid/0.08% ammonium hydroxide in methanol. The gradient duration was 7 min at a flow rate of 100 µL/min. Solvent B started at 5% and increased to 15% at 4 min, then increased to 75% at 4.5 min, and was held at 75% for 30 s before returning to 5% for the remaining time. The sample injection volume was set at 10 µL. Instrumental parameters for the mass spectrometer were as follows: the nitrogen curtain gas was set at 35 units at high collision gas flow; heated nebulizer flow was 45 units; and the nebulizing gas temperature was set at 500 °C. The ionization voltage was set at −4.5 kV and unit resolution was specified for quadrupole one and quadrupole three. The ABI 4000 Q-trap was programed to run in MRM mode to monitor the precursor/product transitions for adenine and the 13C-labeled adenine. The precursor/product transitions for adenine monitored were m/z 136 to 119, m/z 136 to 92 and m/z 136 to 65. The 13C-labeled adenine transitions monitored were m/z 141 to 124, m/z 141 to 96 and m/z 141 to 68. For quantification, the most abundant fragment ions of 119 (adenine) and 124 (13C-labeled adenine) were selected, which was the loss of ammonia (Nelson and McCloskey, 1992). Data acquisition and processing were performed with the ABI Analyst 1.5.2 software.
3. Results and discussion
3.1. Characterization of purified standards
3.1.1. Measurement of molecular weight of purified material
Purified ricin and RCA were used to generate protein standard calibration curves and quality controls (QCs). The use of a protein standard curve improves accuracy because all of the samples undergo the same processing which normalizes the effect of buffer, antibody capture conditions and digestion. A natural source of purified ricin and RCA are required to create these standard calibration curves. It is known that ricin can exist as different isoforms depending on glycosylation (Despeyroux et al., 2000; Duriez et al., 2008; Griffiths et al., 1995; Hegde and Podder, 1992; Helmy and Pieroni, 2000; Mise et al., 1977; Sehgal et al., 2011) and perhaps on the cultivar from which the ricin and RCA were obtained; therefore, it is necessary to characterize the purified material to determine the measured molecular weights and the actual concentrations of the ricin and RCA. The molecular weight of the protein as determined from the database sequence of ricin and RCA would not take into account any glycosylation of the two proteins. Further, the cultivar used to extract the purified ricin and RCA for this study was not known by the manufacturer, Vector Laboratories. To determine the measured molecular weights of the two proteins, the purified ricin and RCA were analyzed by linear mode MALDI-TOF MS. To improve measurement accuracy, NIST BSA was added to the samples so that internal calibration could be performed. MALDI matrix spot application followed the procedure of Onnerfjord et al. (1999) to remove salts and improve resolution. However, the resolution was not high enough to determine if any isoforms existed. The measured molecular weight of the purified ricin was determined to be 62,912 ± 29 Da and the measured molecular weight of the purified RCA was determined to be 127,553 ± 46 Da. (data not shown).
3.1.2. Determination of concentration of purified ricin and RCA
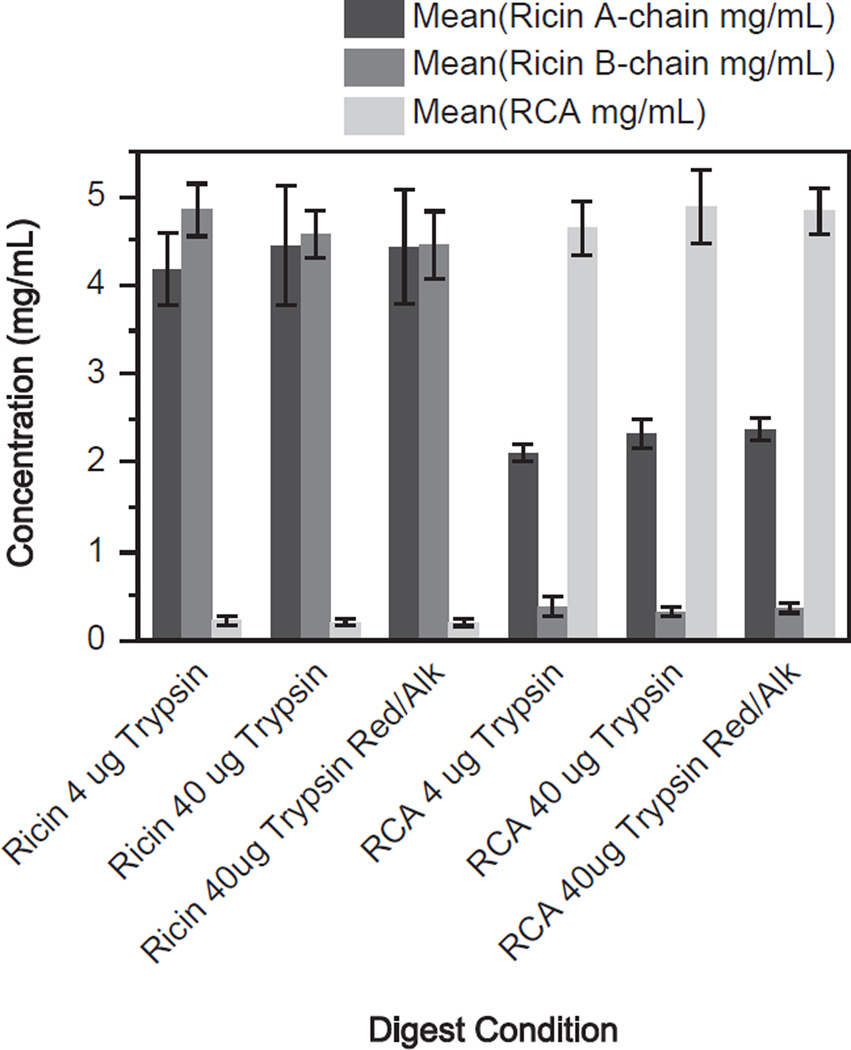
Determination of the concentration of the purified ricin and RCA was accomplished by digesting the two proteins with trypsin without antibody capture using the three digest conditions. According to the manufacturer’s information, the expected concentration of both the purified stock ricin and RCA solutions were each 5 mg/mL. Three peptides were chosen to uniquely quantify ricin: two peptides from the ricin A-chain, VGLPINQR and LTTGADVR, and one from the ricin B-chain, ASDPSLK. Two peptides were monitored for RCA, both from the A-chain, VGLPISQR and SHLTTGGDVR. Fig. 1 shows the measured concentration of the purified ricin and RCA under the tryptic digestion conditions of 4 µg, 40 µg and 40 µg with reduction/alkylation. These values were calculated from the peptide standard curve using the mean values for the two quantification peptides of the ricin A-chain, the single quantification peptide for the ricin B-chain and the two quantification peptides for RCA. Based on this information the overall measured concentration was determined to be 4.5 ± 0.6 mg/mL for ricin and 4.9 ± 0.4 mg/mL for RCA in the purchased protein stock solutions. The concentration values for ricin and RCA were used to generate the protein based standard calibration curves and the QCs in subsequent experiments. The results shown in Fig. 1 indicate that the use of 10x increase in trypsin quantity and reduction/alkylation did not provide any improvement in the digestion of the proteins. All three conditions produced almost identical amounts of the two proteins, indicating that maximum digestion had been reached with 4 mg trypsin and without reduction/alkylation. Because ricin and RCA were quantified individually in each sample using the protein-specific peptides, we could observe the presence of RCA in the ricin-purified sample and ricin in the RCA-purified sample. The ricin standard had a contamination level of 0.2 mg/mL of RCA, representing 4% of the total amount of material; the RCA standard had more significant contamination mainly due to ricin A-chain at a level of 2.3 mg/mL comprising 32% of the overall material but the ricin B-chain was only present at a level of 0.4 mg/mL comprising 7% of the total material.
Fig. 1.
Measurement of purified ricin and RCA standard concentrations by isotope dilution mass spectrometry. Three different tryptic digestion conditions using 4 µg, 40 µg and 40 µg with reduction and alkylation were used to digest ricin and RCA. Quantity of protein was determined using a peptide based standard calibration curve. The ricin A-chain peptides are denoted in dark gray, ricin B-chain peptide is medium gray and the RCA A-chain peptides are denoted in light gray. Error bars show one standard deviation from the mean for three replicates.
3.2. Quantification of ricin and RCA in R. communis cultivar extracts
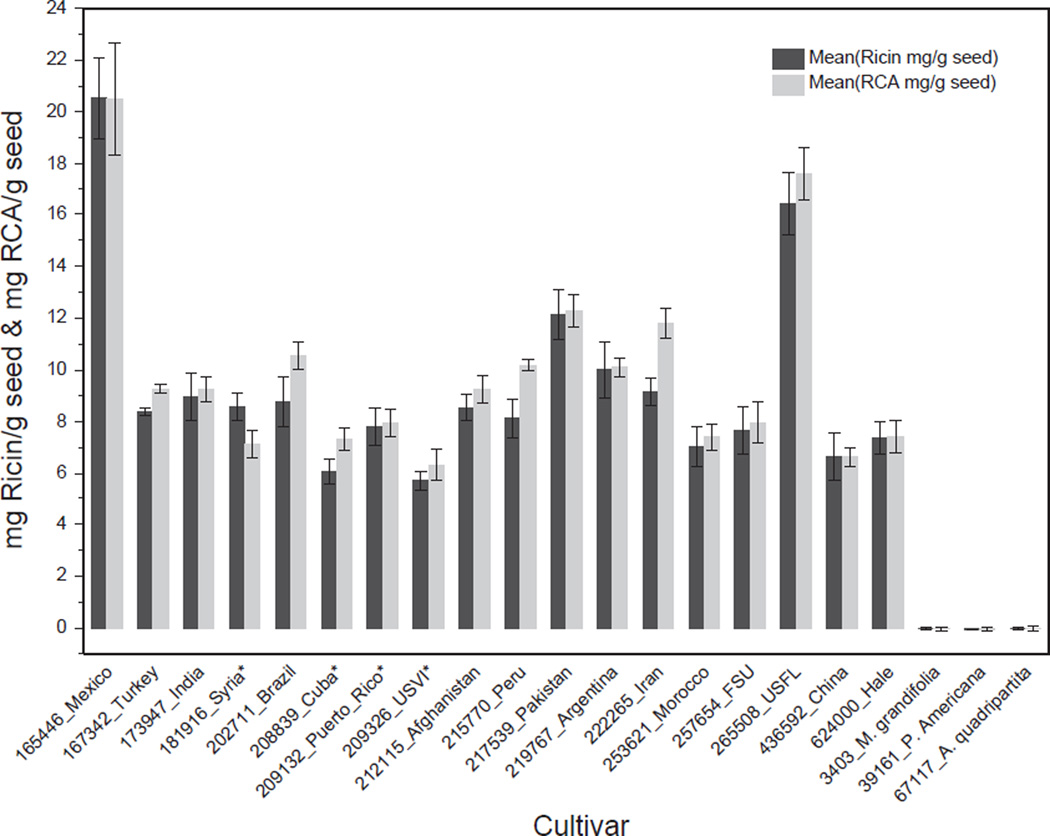
Once the concentration of the purified ricin and RCA standards were determined, the extracts from the 18 cultivars and the three negative controls were analyzed for ricin and RCA content. Fig. 2 shows the levels of ricin and RCA in mg/g of seed for each of the R. communis cultivars studied, measured using the mean of the two A-chain peptides for ricin and RCA. The mean amount of ricin in mg/g seed weight was 9.3 with a minimum of 5.7 mg/g and maximum of 20.5 mg/g. The mean amount of RCA in mg/g seed weight was 9.9 with a minimum of 6.3 mg/g and maximum of 20.5 mg/g. Signals for ricin and RCA were below the limit of detection of 1.0 ng/mL in the three control materials (near neighbors and non-family). The cultivars with highest levels of ricin and RCA were Mexico cv.: PI165446 and US Florida cv.: PI265508; the lowest levels of ricin and RCA were found in US Virgin Islands cv.: PI209326. Based on weight, the average ratio of ricin to RCA was 0.9 where the largest difference in protein quantity was a ricin-to-RCA ratio of 0.8 for Iran cv.: PI222265 and 1.2 for Syria cv.: PI181916. This data correlates well with observations of Harley et al., where the authors reported a 1:1 correlation between gram ricin/gram RCA throughout all stages of seed development (Harley and Beevers, 1986). These values are based on intact seed weights where the shell and oil have not been removed.
Fig. 2.
Measurement of the quantity of ricin and RCA in individual cultivars in mg/g of seed as determined by isotope dilution mass spectrometry. Dark gray denotes quantity of ricin calculated from the mean of the A-chain peptides VGLPINQR and LTTGADVR, light gray denotes quantity of RCA calculated from the mean of the A-chain peptides VGLPISQR and SHLTTGGDVR. The y-axis is the quantity of ricin or RCA in mg/g seed. Asterisk denotes small seed cultivars. Each value represents the mean of three replicates; error bars show one standard deviation from the mean.
3.3. Differences in ricin amount as calculated from the A-chain and the B-chain
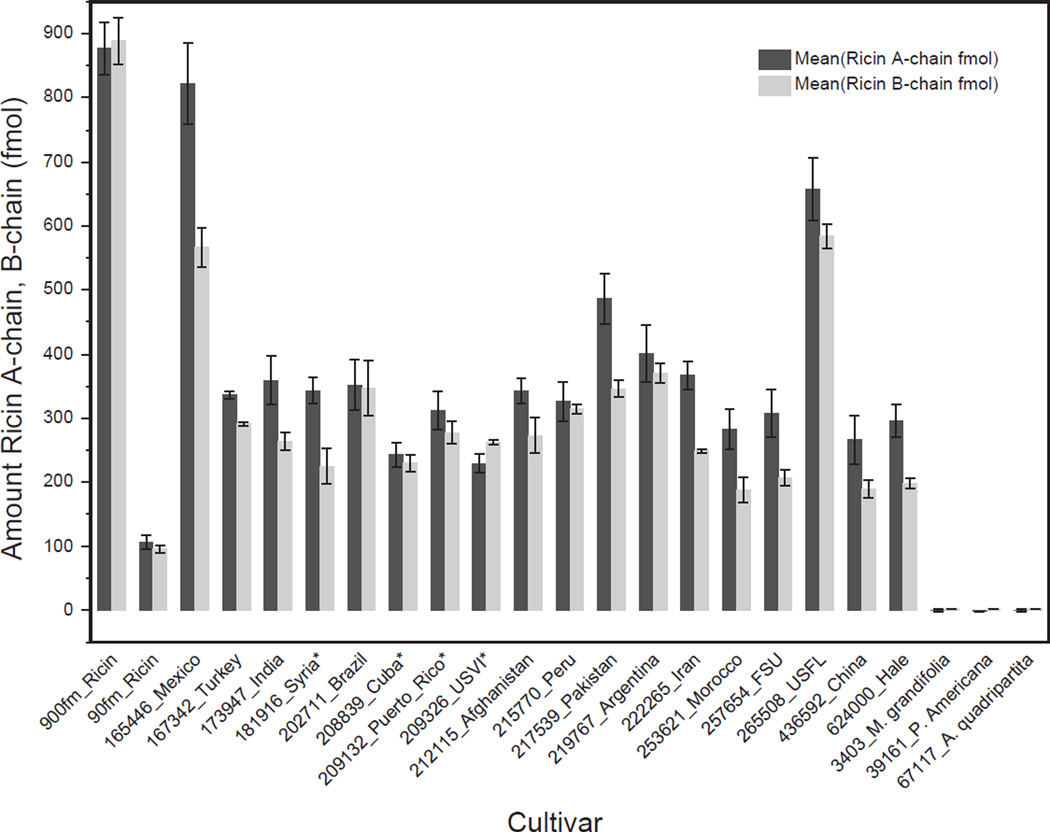
The B-chain peptide from ricin was not included in the calculation of ricin content because in most of the cultivars the mean B-chain value did not agree with the measured mean of the A-chain. Because ricin is composed of one A-chain and one B-chain connected by a disulfide bond, there should be a 1:1 correlation between the amounts of ricin calculated using the A-chain and the B-chain peptides. However, we observed that the quantity of ricin determined using the B-chain peptide was lower than that calculated from the A-chain peptides with the exception of cultivars Brazil cv.: PI202711 and Cuba cv.: PI208839 which had the same levels and US Virgin Islands cv.: PI209326 which had a greater level of B-chain relative to A-chain (Fig. 3). This difference between A-chain and B-chain was not observed with the purified ricin control (Fig. 3 (900 fmol_Ricin, 90 fmol_Ricin)). It is possible that cultivars with a ratio of B-chain/A-chain < 1 have a different B-chain sequence than the Vector ricin reference sequence, specifically, the part of the B-chain sequence containing the quantitation peptide ASDPSLK. It is known that there is genetic recombination between RCA and ricin genes (Ladin et al., 1987). This may result in the replacement of the ASDPSLK peptide from the B-chain of ricin with another sequence derived from RCA, which has been designated as ricin E (Araki and Funatsu, 1987; Mise et al., 1977). This form of ricin has been reported in some but not all “small seed” varieties where small seed cultivars are defined based on the weight of 100 seeds less than 20 g (B. Morris, ARS, USDA, personal communication). Within the set of cultivars we tested, there were four (Cuba cv.: PI208839, Puerto Rico cv.: PI209132, Syria cv.: PI181916, US Virgin Islands cv.: PI209326) whose seeds would be considered “small seeds.” However, there was no correlation between reduced levels of B-chain and seed size of the cultivars we examined in this study. In order to determine the contribution of ricin E, the two additional peptides of ricin DNTIR (TB31) and SNTDANQLWTLK (TB29), which are closer to the N-terminus of the B-chain, could be monitored and would be detected in a digest of either ricin E or ricin. If both ricin and ricin E were present, using the (TB31) or (TB29) peptides should provide the same calculated amount of ricin as the A-chain peptides. One caveat to this approach would be that a purified source of ricin E would be required to generate a protein standard calibration curve.
Fig. 3.
Quantity of ricin in fmol in individual cultivars as calculated from individual A- and B-chain peptides. The mean quantity of A-chain peptides LTTGADVR and VGLPINQR is denoted in dark gray. The mean quantity of B-chain peptide ASDPSLK is denoted in light gray. Control samples of stock purified ricin are shown at 90 and 900 fmol. Error bars represent one standard deviation from the mean for three individual replicates.
Alternatively, other expressed genes from the ricin/RCA family may contribute to the unequal ratio of A-to B-chain levels. Ricin and RCA originate from a multi-gene family (Cawley et al., 1978; Chan et al., 2010; Halling et al., 1985; Leshin et al., 2010; Tregear and Roberts, 1992), where 18 RIP-encoding genes have been identified in the R. communis genome. Of these genes, two designated as Rco_II3 (XP_002532191.1) and Rco_II4 (XP_002532193.1) have truncated B-chain domains where the B-chain peptide ASDPSLK is not present. Rco_II3 only contains the LTTGADVR A-chain peptide and Rco_II4 only contains the VGLPISQR A-chain peptide (Loss-Morais et al., 2013). Expression of Rco_II3 and Rco_II4 in mature seeds may affect the ratio of levels between the A- and B-chains as determined by the peptides used in this mass spectrometric approach. Experiments are underway to further determine why the quantities of the two ricin chains are different and what could be the source of the disparity.
3.4. Reproducibility of extraction from R. communis seeds
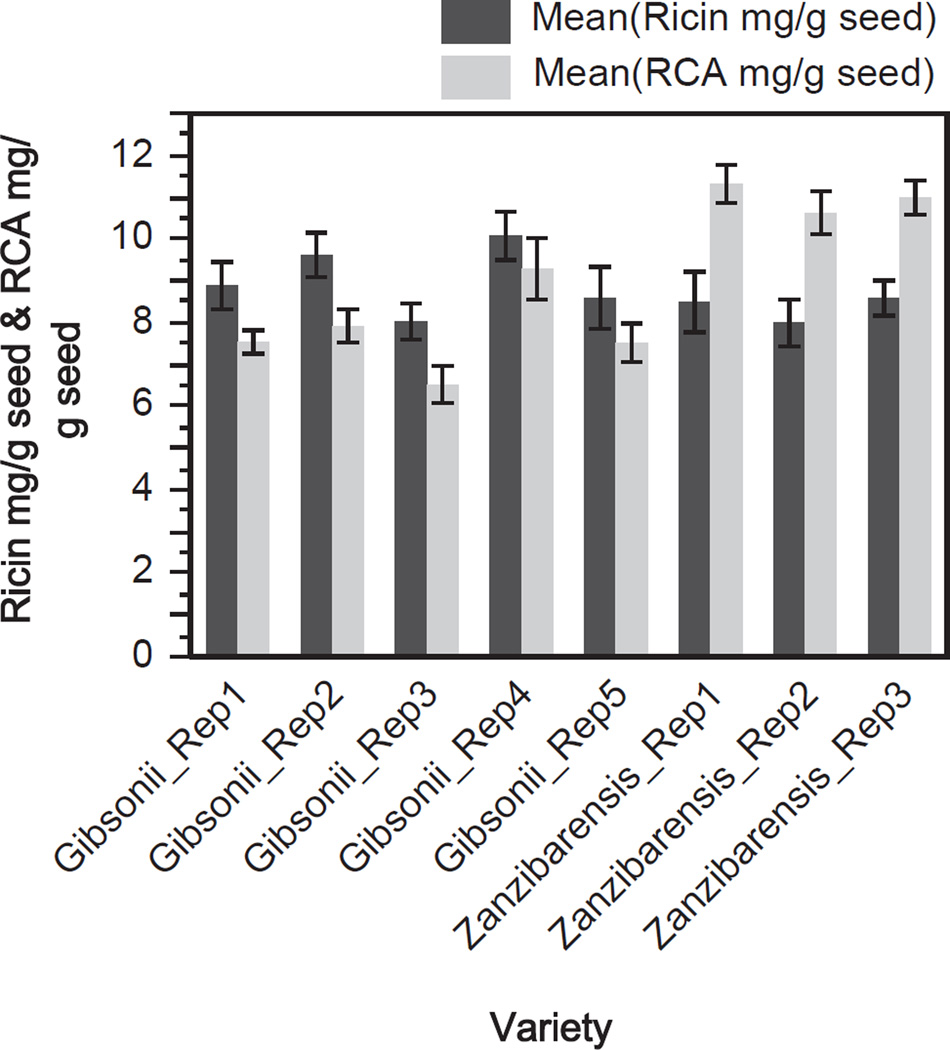
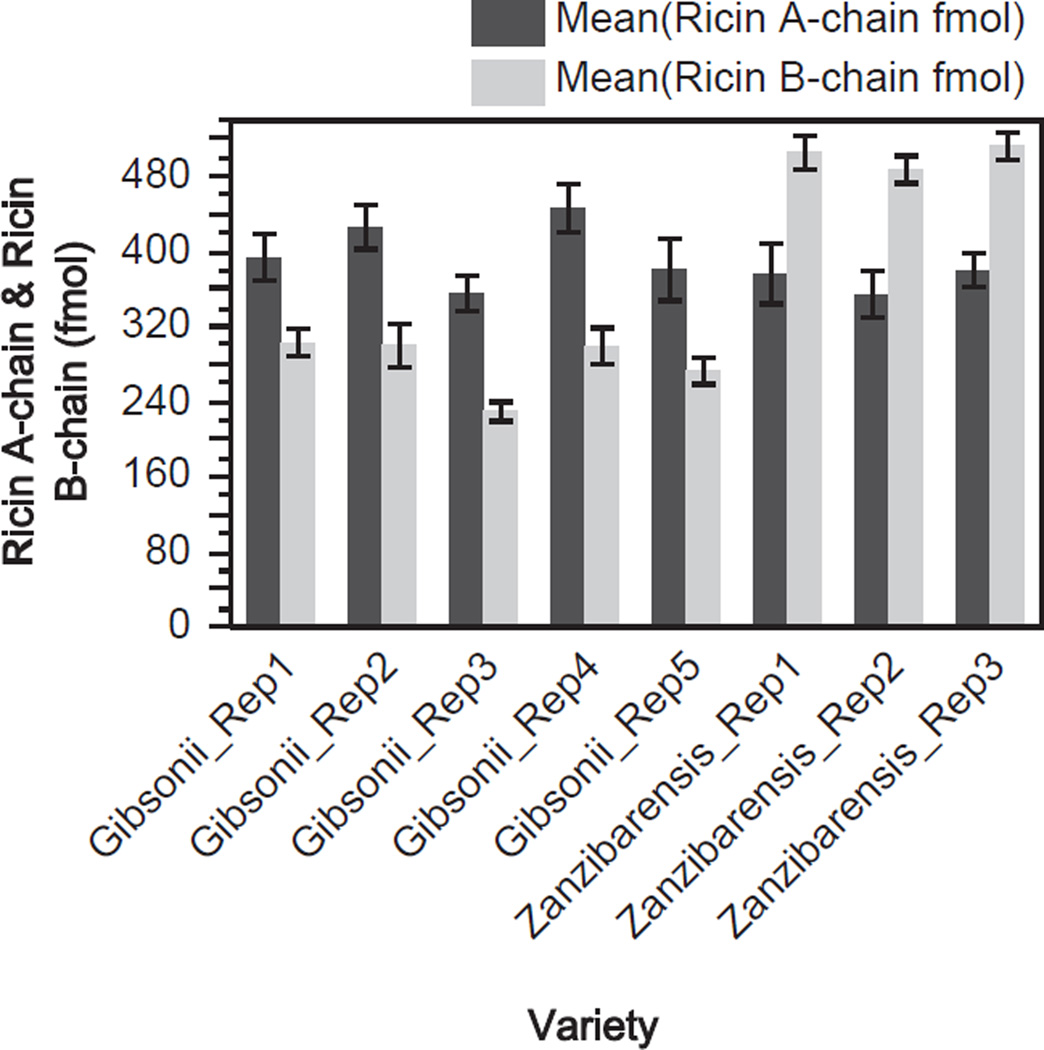
It is possible that R. communis grown in different conditions such as humid or dry climates may have an effect on the amount of ricin and RCA present in the seed and may contribute to variations inter- and intra-cultivar. To minimize the effects of environmental induced variation among the cultivars, R. communis seeds used in this study were obtained from the Plant Genetics Resources Conservation Unit at the ARS USDA. The seeds from each cultivar used in this study were propagated such that they would be exposed to identical growth, harvesting and storage conditions along with measures to prevent cross pollination. Therefore, the contributions from environment, harvesting and storage should have minimal impact on the levels of ricin and RCA across the cultivars. However, a source of variability that may have a measurable impact on the levels of ricin and RCA amounts could be due to the extraction procedure. To assess extraction reproducibility R. communis seeds were subjected to replicate extractions. Fig. 4 shows the amounts of ricin and RCA determined by IDMS per replicate extractions from the gibsonii and zanzibarensis seeds. For the gibsonii seeds the mean amount of ricin for the replicates was 9.0 mg/g seed with a relative standard deviation (RSD) of 9.0% and the mean amount of RCA was 7.8 mg/g seed with an RSD of 12.9%. For the zanzibarensis seeds the mean amount of ricin was 8.4 mg/g seed with an RSD of 3.8% and the mean amount of RCA was 11.0 mg/g seed with an RSD of 3.1%. Fig. 5 shows the amounts of ricin A- and B-chain per replicate for the two varieties of R. communis seeds. The mean for the gibsonii extracts for the A-chain of ricin was 400 fmol with an RSD of 9.0% and the mean B-chain amount was 281 fmol with an RSD of 11.1%. For the zanzibarensis replicate extracts the mean for the A-chain of ricin was 370 fmol with an RSD of 3.8% and the mean for the B-chain of ricin was 502 fmol with an RSD of 2.6%. The extracts from the gibsonii seeds had more variability in the amounts of ricin, RCA, ricin A- and B-chain than that of the zanzibarensis seed extracts. It is not known why there was increased variability for the gibsonii preparations versus the zanzibarensis extracts since each replicate of seeds underwent identical processing. It is interesting to note that from the entire set of seed extracts used in this study the zanzibarensis seeds had a much larger amount of RCA than ricin and a much larger amount of the B-chain of ricin versus the A-chain.
Fig. 4.
Quantification of ricin and RCA in replicate extractions from R. communis gibsonii and zanzibarensis seeds. Measured amount of ricin (dark gray) and RCA (light gray) in mg/g of seed in five individual replicate extractions from the gibsonii variety of seed and three individual extractions from the zanzibarensis seeds. Each value represents the mean of six individual IDMS replicates; error bars show one standard deviation from the mean.
Fig. 5.
Quantification of ricin A-chain and B-chain in replicate extractions from R. communis gibsonii and zanzibarensis seeds. Measured amount of ricin A-chain (dark gray) and B-chain (light gray) are in fmol from five individual replicate extractions from the gibsonii variety of seed and three individual extractions from the zanzibarensis seeds. Each value represents the mean of six individual IDMS replicates; error bars show one standard deviation from the mean.
3.5. Comparison of enzymatic activity in R. communis cultivars
Recent studies have suggested that there is low genetic diversity in the overall genome among R. communis cultivars (Allan et al., 2008; Foster et al., 2010; Rivarola et al., 2011). However, there is diversity among the genes in the ricin/RCA family, but it is not known if this diversity is present across cultivars and whether it would result in varying amount of N-glycosidase function (Halling et al., 1985). Several methods have been developed to detect and quantify the products of the ricin/RCA rRNA N-glycosidase reaction, ranging from colorimetric assays to mass spectrometry (Barbieri et al., 1997; Heisler et al., 2002; Hines et al., 2004; Kalb and Barr, 2009; Keener et al., 2008; Sturm and Schramm, 2009; Zamboni et al., 1989). Our activity assay is based on that of Becher et al., which measures adenine release from a DNA substrate with LC-MS/ MS detection, and incorporates the quantitation of adenine released by the action of the N-glycosidase (Becher et al., 2007). Ricin and RCA have N-glycosidase activity; however, the level of activity for RCA has been determined to be less than that of ricin (Cawley et al., 1978; Lin and Liu, 1986; Saltvedt, 1976; Zhan and Zhou, 2003).
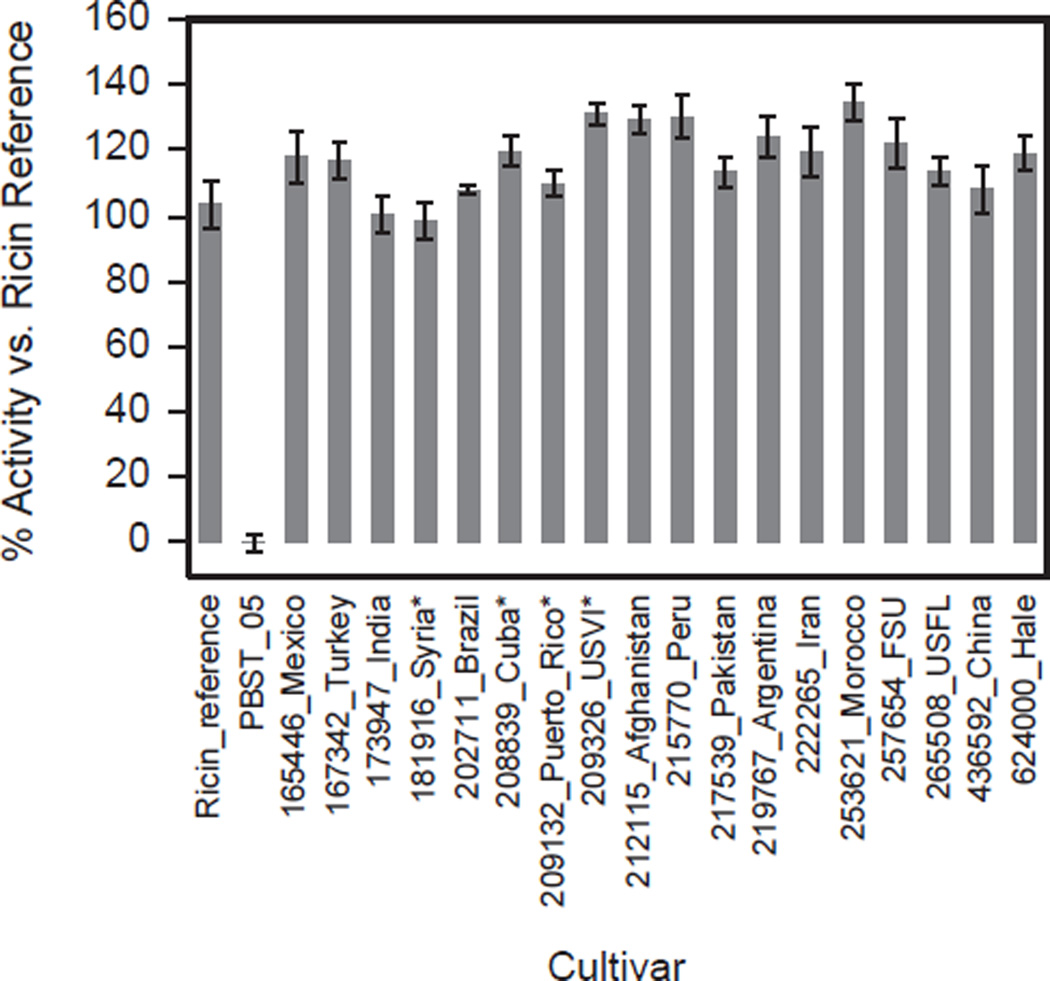
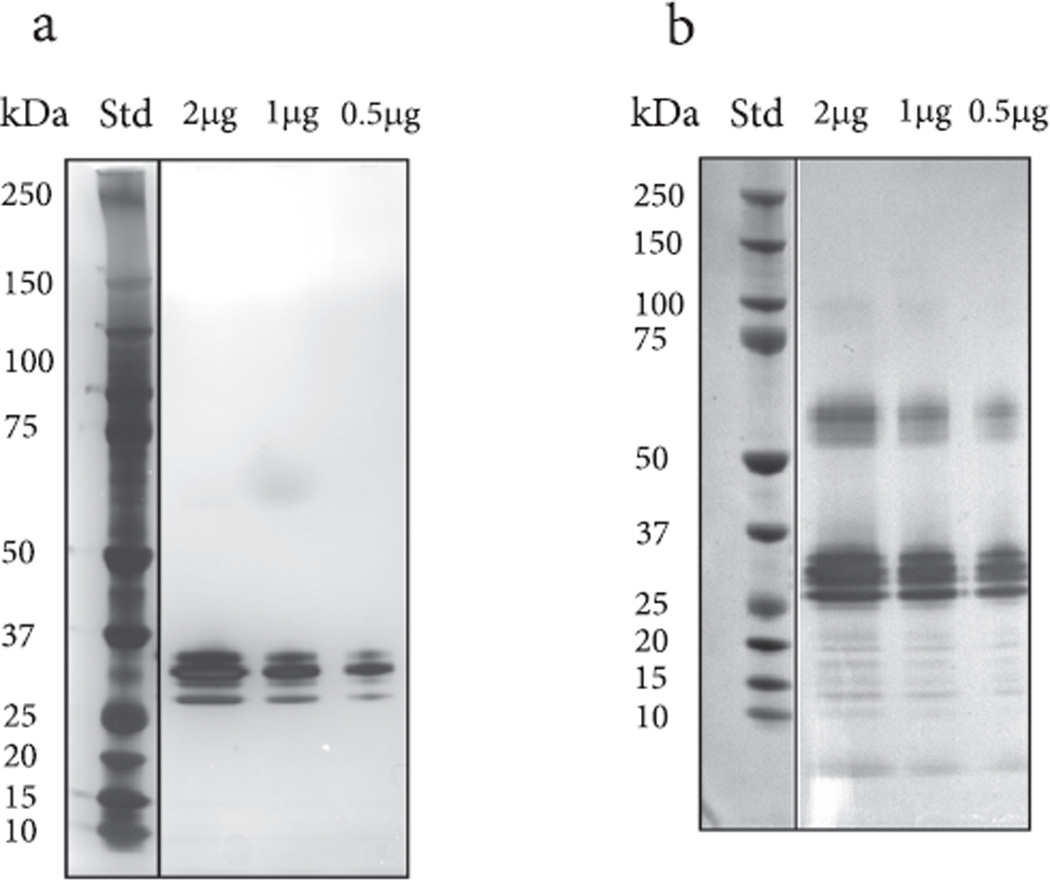
While the ricin and RCA protein quantitation can be independently traced back to peptide calibration standards, the activity-based quantitation is calibrated relative to the activity of the purified ricin protein standard, which itself contains a measurable amount of RCA. By measuring moles of ricin and RCA content of each cultivar independently by IDMS and measuring its N-glycosidase activity/moles of ricin relative to the ricin calibration standard, we were able to examine if there were any differences in overall activity/mole ricin between cultivars. In analysis of the data, we normalized the activity of each cultivar by dividing the measured activity of the cultivar by the measured activity of the ricin reference standard wherein this result was expressed as a percent. Fig. 6 shows a plot of the normalized overall N-glycosidase activity of each cultivar. The range of activity was 98–134% (average 118%) of the activity of the reference ricin material, with most seed extracts exhibiting a higher degree of enzymatic activity than the reference. The range of activity is wider than the error for the activity method, which is 1.2–6.7%. A number of explanations could be responsible for the bias towards higher levels of activity relative to the ricin reference. The bias could be due to the contribution of RCA and/or may also reflect the diminished activity of the purified ricin standard, which could be attributed to the multiple steps needed to obtain highly purified ricin from a castor bean extract. It is also possible that the ricin reference may have had impurities present which could suppress activity or have been degraded which in-turn could lead to less active material. With respect to the contribution of RCA to overall N-glycosidase activity, the quantitation of activity among the cultivars is based solely on the enzymatic activity/mole of the ricin reference material. This does not take into account any contribution of enzymatic activity from RCA which is estimated to be approximately 10% of ricin (Cawley et al., 1978). It was not possible to effectively assess the contribution of RCA because the RCA reference standard contained a substantial amount of ricin. If a pure standard of RCA was available the activity contributions of ricin and RCA could be estimated in the extracts of the cultivars. As such, the study was limited to use of ricin only to measure the overall N-glycosidase activity in the cultivars. To determine if enzymatic activity was impacted by the presence of impurities or if the ricin reference standard had degraded, a one dimensional SDS-PAGE gel was loaded with 0.5, 1 and 2 µg of the Vector ricin material and was silver stained to visualize the contents separated material. Fig. 7a shows the silver stained gel where several bands are visible between 25 and 37 kDa. These bands are expected to be the individual chains of ricin and possibly the individual chains of an isoform of ricin. No other bands were visible in each of the lanes which would preclude the presence of impurities unless impurities were co-migrating with the ricin bands. Subsequently, a Western blot was performed (Fig. 7b) to determine if any ricin degradation products were present. The results show a series of bands which were immuno-reactive to the antibody between 25 and 37 kDa which would correspond to the individual chains of ricin. Another series of bands between 50 and 75 kDa would likely represent un-denatured ricin while a third set of bands were observed between 10 and 20 kDa which suggest the products of degraded ricin. The presence of these lower bands which reacted to the antibody may indicate that the ricin reference material could have somewhat reduced enzymatic activity. Quantification of amount by IDMS would measure the total amount of ricin present in the standard material but it cannot differentiate between active and degraded ricin. This may be an additional reason why there is a bias towards higher N-glycosidase activity of the cultivar extracts versus the ricin reference. Among the cultivars, there are some differences in activity levels which could be due to differing glycosylation (Sehgal et al., 2011), sequence differences in ricin and RCA, or other components in the seed extracts that enhance the N-glycosidase activity. In general the overall N-glycosidase activity is relatively uniform and there were no large differences recorded. If this set of cultivars is representative of average N-glycosidase activity, it may be expected that all R. communis cultivars would show this degree of activity; however, a larger pool of cultivars should be evaluated for verification of this trend. This information could be important to law enforcement and industries that process castor beans in that they would not encounter a ricin preparation or a cultivar that would have an extraordinarily high level of activity.
Fig. 6.
Quantification of overall N-glycosidase activity for each cultivar seed extract using the adenine release assay. The N-glycosidase activity for each cultivar is reported as a percent relative to the purified ricin reference standard (Ricin_reference =100%). PBST_05 is the negative, ricin-free control. Each value represents the mean of three replicate analyses and error bars show one standard deviation from the mean.
Fig. 7.
a. Separation and visualization of the ricin reference standard by one dimensional SDS-PAGE and silver staining. Molecular weight markers are in the left lane and 2, 1 and 0.5µg of the reference ricin are in the subsequent lanes. Individual ricin chains are likely located between 25 and 37 kDa b. is a duplicate gel use in the Western blot analysis. A series of immuno-reactive bands to the goat anti-ricinus communis antibody are seen between 50 and 75 kDa, 25–37 kDa and 10–20 kDa.
4. Conclusion
We have applied a method for quantification of ricin and RCA in seed extracts of 18 R. communis cultivars and compared their overall N-glycosidase activity. This method builds upon previous work where the approach consists of two levels of selectivity: antibody capture of ricin and RCA to extract both proteins, and MRM-MS to quantify peptides derived from ricin and RCA or estimate the proteins’ N-glycosidase activity. In the described method, both A- and B-chains of ricin and RCA can be easily distinguished by performing a tryptic digestion and choosing unique peptides to monitor by MRM-MS. Quantification is enabled by the use of heavy isotope-labeled versions of the peptides as internal standards, and accuracy of quantification is improved by using a protein calibration standard curve. The empirically-determined concentration values for ricin and RCA were further used in protein calibration curves to more accurately determine the quantities of both proteins in cultivar samples.
Analysis of the cultivars confirmed that ricin and RCA were present in nearly equal amounts, approximately 10 mg/g seed, for all seeds studied. The quantity of ricin and RCA differed among the cultivars examined; the Mexican and US Florida cultivars contained almost twice as much ricin and RCA (20.5 and 16.4 mg ricin/g seed, 20.5 and 17.6 mg RCA/g seed respectively) as other cultivars while the US Virgin Island and the Cuba cultivars had the least amount of both proteins (5.7 and 6.1 mg ricin/g seed, 6.3 and 7.3 mg RCA/g seed respectively). Because this method monitors peptides from both the A- and B-chains of ricin, both chains could be quantified independently and compared to each other. The overall trend among the cultivars showed that there was a 21% lower amount of B-chain relative to the A-chain. Possible explanations could be the presence of a hybrid form of ricin known as ricin E and/or the contribution of additional gene products from the ricin/RCA gene family, which could affect the ricin A-/B-chain ratio. Further exploration among the cultivars may be able to determine why the variation exists.
Upon quantification of the amount of ricin and RCA in the cultivars, differences in overall N-glycosidase activity were assessed. This was facilitated by antibody capture of ricin/RCA and exposure to a synthetic DNA substrate that mimicked the natural target of ricin. The released adenine was monitored by MRM MS and included a heavy-isotope-labeled version of adenine as an internal standard. As in the quantification of ricin and RCA, a protein calibration curve using ricin was used to improve the accuracy of the activity assay by correcting for the matrix and the efficiency of capture by the antibody. Quantification of N-glycosidase activity revealed that, on average, the cultivars had 18% higher specific activity than that of the ricin standard. The explanation for the increased N-glycosidase activity of the cultivars could be as simple as the ricin standard used to generate the protein calibration curve may have reduced activity due to purification and storage conditions or there may be other factors involved. We are currently investigating the reasons for the differences in N-glycosidase activity.
An area of research that could benefit from this method would be the search for R. communis cultivars with low or absent levels of ricin. Currently, work in this area has involved the use of ELISA or RID to quantify ricin in different cultivars. Rather than use antibody-based quantification, which cannot distinguish ricin from RCA, the IDMS method would provide accurate values for the quantification of ricin and RCA separately and not as a combined estimate. Accurate information would enable researchers to better screen cultivars for low levels of ricin and to evaluate the success of cross breeding in hopes of further reducing ricin content.
One of the most important applications from a security standpoint is the accurate quantification of ricin in “white powder” preparations. Recent events have demonstrated that ricin has been, and continues to be, used as a threat agent; juxtaposed with the reality that castor seeds are an industrially important source of castor oil and castor plants are common ornamentals. Powdered preparations are of the most concern as these can range from simply crushed castor beans, a crude preparation based on recipes found from the Internet, or a highly purified powder with concentrated amounts of ricin. The more information that can be provided with respect to accurate analysis of these powders will help public health and law enforcement to make informed decisions.
Footnotes
Disclaimer
The ricin measurements from this study were from a naturally occurring substance not a manufactured substance with an enhanced toxicity. The study authors do not anticipate the results of this study would be used in an in appropriate manner. References in this article to any specific commercial products, process, service, manufacturer, or company do not constitute an endorsement or a recommendation by the U.S. Government or the Centers for Disease Control and Prevention. The findings and conclusions in this report are those of the authors and do not necessarily represent the views of CDC.
Ethical statement
No animal or human subjects were used in the study encompassed by the manuscript titled “Quantification of ricin, RCA and comparison of enzymatic activity in 18 Ricinus communis cultivars by Isotope Dilution Mass Spectrometry”.
Conflict of interest
The authors declare that there are no conflicts of interest.
Transparency document
Transparency document related to this article can be found online at http://dx.doi.org/10.1016/j.toxicon.2015.01.003.
References
- Allan G, Williams A, Rabinowicz PD, Chan AP, Ravel J, Keim P. Worldwide genotyping of castor bean germplasm (Ricinus communis L.) using AFLPs and SSRs. Genet. Resour. Crop Evol. 2008;55:365–378. [Google Scholar]
- Araki T, Funatsu G. The complete amino acid sequence of the B-chain of ricin E isolated from small-grain castor bean seeds. Ricin E is a gene recombination product of ricin D and Ricinus communis agglutinin. Biochim. Biophys. Acta. 1987;911:191–200. doi: 10.1016/0167-4838(87)90008-2. [DOI] [PubMed] [Google Scholar]
- Audi J, Belson M, Patel M, Schier J, Osterloh J. Ricin poisoning a comprehensive review. JAMA. 2005;294:2342–2351. doi: 10.1001/jama.294.18.2342. [DOI] [PubMed] [Google Scholar]
- Auld DL, Rolfe RD, McKeon TA. Development of castor with reduced toxicity. J. New Seeds. 2001;3:61–69. [Google Scholar]
- Auld DL, Pinkerton SD, Boroda E, Lombard KA, Murphy CK, Kenworthy KE, Becker WD, Rolfe RD, Ghetie V. Registration of TTU-LRC Castor germplasm with reduced levels of ricin and RCA120. Crop Sci. 2003;43:746–747. [Google Scholar]
- Baldoni AB, de Carvalho MH, Sousa NL, Nobrega MBD, Milani M, Aragao FJL. Variability of ricin content in mature seeds of castor bean. Pesq. Agropec. Bras. 2011;46:776–779. [Google Scholar]
- Barbieri L, Valbonesi P, Bonora E, Gorini P, Bolognesi A, Stirpe F. Poly-nucleotide:adenosine glycosidase activity of ribosome-inactivating proteins: effect on DNA, RNA and poly(A) Nucleic Acids Res. 1997;25:518–522. doi: 10.1093/nar/25.3.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes DJ, Baldwin BS, Braasch DA. Ricin accumulation and degradation during castor seed development and late germination. Ind. Crop Prod. 2009;30:254–258. [Google Scholar]
- Becher F, Duriez E, Volland H, Tabet J-C, Ezan E. Detection of functional ricin by immunoaffinity and liquid chromatography-tandem mass spectrometry. Anal. Chem. 2007;79:659–665. doi: 10.1021/ac061498b. [DOI] [PubMed] [Google Scholar]
- Bradberry SM, Dickers KJ, Rice P, Griffiths GD, Vale JA. Ricin poisoning. Toxicol. Rev. 2003;22:65–70. doi: 10.2165/00139709-200322010-00007. [DOI] [PubMed] [Google Scholar]
- Brinkworth CS. Identification of ricin in crude and purified extracts from castor beans using on-target tryptic digestion and MALDI mass spectrometry. Anal. Chem. 2010;82:5246–5252. doi: 10.1021/ac100650g. [DOI] [PubMed] [Google Scholar]
- Burrows WD, Renner SE. Biological warfare agents as threats to potable water. Environ. Health Perspect. 1999;107:975–984. doi: 10.1289/ehp.99107975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cawley DB, Hedblom ML, Houston LL. Homology between ricin and Ricinus communis agglutinin: amino terminal sequence analysis and protein synthesis inhibition studies. Arch. Biochem. Biophys. 1978;190:744–755. doi: 10.1016/0003-9861(78)90335-1. [DOI] [PubMed] [Google Scholar]
- Chan AP, Crabtree J, Zhao Q, Lorenzi H, Orvis J, Puiu D, Melake-Berhan A, Jones KM, Redman J, Chen G, Cahoon EB, Gedil M, Stanke M, Haas BJ, Wortman JR, Fraser-Liggett CM, Ravel J, Rabinowicz PD. Draft genome sequence of the oilseed species Ricinus communis . Nat. Biotechnol. 2010;28:951–956. doi: 10.1038/nbt.1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GQ, He X, McKeon TA. A simple and sensitive assay for distinguishing the expression of ricin and Ricinus communis agglutinin genes in developing castor seed (R. communis L.) J. Agric. Food Chem. 2005;53:2358–2361. doi: 10.1021/jf040405t. [DOI] [PubMed] [Google Scholar]
- Conceição MM, Candeia RA, Silva FC, Bezerra AF, Fernandes NJ, Jr, Souza AG. Thermoanalytical characterization of castor oil biodiesel. Renew. Sustain. Energy Rev. 2007;11:964–975. [Google Scholar]
- Despeyroux D, Walker N, Pearce M, Fisher M, McDonnell M, Bailey SC, Griffiths GD, Watts P. Characterization of ricin heterogeneity by electrospray mass spectrometry, capillary electrophoresis, and resonant mirror. Anal. Biochem. 2000;279:23–36. doi: 10.1006/abio.1999.4423. [DOI] [PubMed] [Google Scholar]
- Duriez E, Fenaille F, Tabet JC, Lamourette P, Hilaire D, Becher F, Ezan E. Detection of ricin in complex samples by immunocapture and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J. Proteome Res. 2008;7:4154–4163. doi: 10.1021/pr8003437. [DOI] [PubMed] [Google Scholar]
- Dyer JM, Stymne S, Green AG, Carlsson AS. High-value oils from plants. Plant J. Cell Mol. Biol. 2008;54:640–655. doi: 10.1111/j.1365-313X.2008.03430.x. [DOI] [PubMed] [Google Scholar]
- Endo Y, Tsurugi K. RNA N-glycosidase activity of ricin a-chain mechanism of action of the toxic lectin ricin on eukaryotic ribosomes. JBC. 1987;262:8128–8130. [PubMed] [Google Scholar]
- Endo Y, Glück A, Wool IG. Ribosomal RNA identity elements for ricin A-chain recognition and catalysis. JMB. 1991;221:193–207. doi: 10.1016/0022-2836(91)80214-f. [DOI] [PubMed] [Google Scholar]
- Foster JT, Allan GJ, Chan AP, Rabinowicz PD, Ravel J, Jackson PJ, Keim P. Single nucleotide polymorphisms for assessing genetic diversity in castor bean (Ricinus communis) BMC Plant Biol. 2010;10:13. doi: 10.1186/1471-2229-10-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz D, Jaax N. Ricin toxin (Chapter 32) In: Zajtchuk Russ, Army M., editors. Medical Aspects of Chemical and Biological Warfare. Washington, D.C: Office of the Surgeon General. Department of the Army, United States of America; 1997. pp. 631–642. Brigadier General. [Google Scholar]
- Fredriksson S-Ã, Hulst AG, Artursson E, deJong AL, Nilsson C, van Baar BLM. Forensic identification of neat ricin and of ricin from crude castor bean extracts by mass spectrometry. Anal. Chem. 2005;77:1545–1555. doi: 10.1021/ac048756u. [DOI] [PubMed] [Google Scholar]
- Garber EA. Toxicity and detection of ricin and abrin in beverages. J. Food Prot. 2008;71:1875–1883. doi: 10.4315/0362-028x-71.9.1875. [DOI] [PubMed] [Google Scholar]
- Geranios NK. FBI Searches Spokane Apartment in Ricin Letter Case. The Washington Post. 2013 [Google Scholar]
- Griffiths GD, Rice P, Allenby AC, Bailey SC, Upshall DG. Inhalation toxicology and histopathology of ricin and abrin toxins. Inhal. Toxicol. 1995;7:269–288. doi: 10.1177/096032719501400201. [DOI] [PubMed] [Google Scholar]
- Halling KC, Halling AC, Murray EE, Ladin BF, Houston LL, Weaver RF. Genomic cloning and characterization of a ricin gene from Ricinus communis . Nucleic Acids Res. 1985;13:8019–8033. doi: 10.1093/nar/13.22.8019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley SM, Beevers H. Lectins in castor bean seedlings. Plant Physiol. 1986;80:1–6. doi: 10.1104/pp.80.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde R, Podder SK. Studies on the variants of the protein toxins ricin and abrin. Eur. J. Biochem. 1992;204:155–164. doi: 10.1111/j.1432-1033.1992.tb16618.x. [DOI] [PubMed] [Google Scholar]
- Hegde R, Podder SK. Evolution of tetrameric lectin Ricinus communis agglutinin from two variant groups of ricin toxin dimers. Eur. J. Biochem. 1998;254:596–601. doi: 10.1046/j.1432-1327.1998.2540596.x. [DOI] [PubMed] [Google Scholar]
- Heisler I, Keller J, Tauber R, Sutherland M, Fuchs H. A colorimetric assay for the quantitation of free adenine applied to determine the enzymatic activity of ribosome-inactivating proteins. Anal. Biochem. 2002;302:114–122. doi: 10.1006/abio.2001.5527. [DOI] [PubMed] [Google Scholar]
- Helmy M, Pieroni G. RCA(60): purification and characterization of ricin D isoforms from Ricinus sanguineus. J. Plant Physiol. 2000;156:477–482. [Google Scholar]
- Hines H, Brueggemann E, Hale M. High-performance liquid hromatographye-mass selective detection assay for adenine released from a synthetic RNA substrate by ricin A chain. Anal. Biochem. 2004;330:119–122. doi: 10.1016/j.ab.2004.03.046. [DOI] [PubMed] [Google Scholar]
- Hodge DR, Prentice KW, Ramage JG, Prezioso S, Gauthier C, Swanson T, Hastings R, Basavanna U, Datta S, Sharma SK, Garber EA, Staab A, Pettit D, Drumgoole R, Swaney E, Estacio PL, Elder IA, Kovacs G, Morse BS, Kellogg RB, Stanker L, Morse SA, Pillai SP. Comprehensive laboratory evaluation of a highly specific lateral flow assay for the presumptive identification of ricin in suspicious white powders and environmental samples. Biosecurity Bioterror. Biodef. Strategy Pract. Sci. 2013;11:237–250. doi: 10.1089/bsp.2013.0053. [DOI] [PubMed] [Google Scholar]
- Kalb SR, Barr JR. Mass spectrometric detection of ricin and its activity in D.M. Schieltz et al. / T food and clinical samples. Anal. Chem. 2009;81:2037–2042. doi: 10.1021/ac802769s. [DOI] [PubMed] [Google Scholar]
- Keener WK, Rivera VR, Cho CY, Hale ML, Garber EA, Poli MA. Identification of the RNA N-glycosidase activity of ricin in castor bean extracts by an electrochemiluminescence-based assay. Anal. Biochem. 2008;378:87–89. doi: 10.1016/j.ab.2008.03.019. [DOI] [PubMed] [Google Scholar]
- Khvostova IV. In: Ricin: the Toxic Protein of Seed. Moshkin VA, editor. New Delhi, India: Castor. Amerind Publ; 1986. pp. 85–92. [Google Scholar]
- Kull S, Pauly D, Stormann B, Kirchner S, Stammler M, Dorner MB, Lasch P, Naumann D, Dorner BG. Multiplex detection of microbial and plant toxins by immunoaffinity enrichment and matrix-assisted laser desorption/ ionization mass spectrometry. Anal. Chem. 2010;82:2916–2924. doi: 10.1021/ac902909r. [DOI] [PubMed] [Google Scholar]
- Ladin BF, Murray EE, Halling AC, Halling KC, Tilakaratne N, Long GL, Houston LL, Weaver RF. Characterization of a cDNA encoding ricin E, a hybrid ricin-Ricinus communis agglutinin gene from the castor plant Ricinus communis . Plant Mol. Biol. 1987;9:287–295. doi: 10.1007/BF00166464. [DOI] [PubMed] [Google Scholar]
- Lappi DA, Kapmeyer W, Beglau JM, Kaplan NO. The disulfide bond connecting the chains of ricin. Proc. Natl. Acad. Sci. U. S. A. 1978;75:1096–1100. doi: 10.1073/pnas.75.3.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leshin J, Danielsen M, Credle JJ, Weeks A, O’Connell KP, Dretchen K. Characterization of ricin toxin family members from Ricinus communis . Toxicon. 2010;55:658661. doi: 10.1016/j.toxicon.2009.09.002. [DOI] [PubMed] [Google Scholar]
- Lin J-Y, Liu S-Y. Studies on the antitumor lectins isolated from the seeds of Ricinus communis (castor bean) Toxicon. 1986;24:757–765. doi: 10.1016/0041-0101(86)90100-5. [DOI] [PubMed] [Google Scholar]
- Loss-Morais G, Turchetto-Zolet AC, Etges M, Cagliari A, Körbes AP, Maraschin Fd.S, Margis-Pinheiro M, Margis R. Analysis of castor bean ribosome-inactivating proteins and their gene expression during seed development. Genet. Mol. Biol. 2013;36:74–86. doi: 10.1590/S1415-47572013005000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath SC, Schieltz DM, McWilliams LG, Pirkle JL, Barr JR. Detection and quantification of ricin in beverages using isotope dilution tandem mass spectrometry. Anal. Chem. 2011;83:2897–2905. doi: 10.1021/ac102571f. [DOI] [PubMed] [Google Scholar]
- Mise T, Funatsu G, Ishiguro M. Isolation and characterization of ricin E from castor beans. Agric. Biol. Chem. Tokyo. 1977;41:2041–2046. [Google Scholar]
- Morris JB, Wang ML, Morse SA. Ricinus. In: Kole C, editor. Wild Crop Relatives: Genomic and Breeding Resources: Oilseeds. first. Springer-Verlag; 2011. pp. 251–260. [Google Scholar]
- Mutlu H, Meier MAR. Castor oil as a renewable resource for the chemical industry. Eur. J. Lipid Sci. Tech. 2010;112:10–30. [Google Scholar]
- National Select Agent Registry. US Department of Health and Human Services. Atlanta, GA: US Department of Agriculture; 2014. 7 C.F.R. Part 331, 9 C.F.R. Part 121, 42 C.F.R. Part 73; p. 30333. [Google Scholar]
- Nelson S. Another Suspected Ricin Letter Mailed to President Obama. U.S. News and World Reports. 2013 [Google Scholar]
- Nelson C, McCloskey J. Collision-induced dissociation of adenine. J. Am. Chem. Soc. 1992;114:3661–3668. [Google Scholar]
- Nilsson L, Nygard O. The mechanism of the protein-synthesis elongation cycle in eukaryotes. Effect of ricin on the ribosomal interaction with elongation factors. Eur. J. Biochem. 1986;161:111–117. doi: 10.1111/j.1432-1033.1986.tb10130.x. [DOI] [PubMed] [Google Scholar]
- Nogueira FC, Palmisano G, Soares EL, Shah M, Soares AA, Roepstorff P, Campos FA, Domont GB. Proteomic profile of the nucellus of castor bean (Ricinus communis L.) seeds during development. J. Proteomics. 2012;75:1933–1939. doi: 10.1016/j.jprot.2012.01.002. [DOI] [PubMed] [Google Scholar]
- Ogunniyi DS. Castor oil: a vital industrial raw material. Bioresour. Technol. 2006;97:1086–1091. doi: 10.1016/j.biortech.2005.03.028. [DOI] [PubMed] [Google Scholar]
- Olsnes S, Pihl A. Different biological properties of the two constituent peptide chains of ricin, a toxic protein inhibiting protein synthesis. Biochemistry. 1973;12:3121–3126. doi: 10.1021/bi00740a028. [DOI] [PubMed] [Google Scholar]
- Olsnes S, Refsnes K, Pihl A. Mechanism of action of the toxic lectins abrin and ricin. Nature. 1974;249:627–631. doi: 10.1038/249627a0. [DOI] [PubMed] [Google Scholar]
- Onnerfjord P, Ekstrom S, Bergquist J, Nilsson J, Laurell T, Marko-Varga G. Homogeneous sample preparation for automated high throughput analysis with matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 1999;13:315–322. doi: 10.1002/(SICI)1097-0231(19990315)13:5<315::AID-RCM483>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Ostin A, Bergström T, Fredriksson S-A, Nilsson C. Solvent-assisted trypsin digestion of ricin for forensic identification by LC-ESI MS/MS. Anal. Chem. 2007;79:6271–6278. doi: 10.1021/ac0701740. [DOI] [PubMed] [Google Scholar]
- Pinkerton SD. Masters thesis. Lubbock, TX: Texas Tech University; 1997. Selection of Castor with Divergent Concentrations of Ricin and RCA Using Radial Immunodiffusion. [Google Scholar]
- Pinkerton SD, Rolfe R, Auld DL, Ghetie V, Lauterbach BF. Selection of castor for divergent concentrations of ricin and Ricinus communis agglutinin. Crop Sci. 1999;39:353–357. [Google Scholar]
- Rivarola M, Foster JT, Chan AP, Williams AL, Rice DW, Liu X, Melake-Berhan A, Huot Creasy H, Puiu D, Rosovitz MJ, Khouri HM, Beckstrom-Sternberg SM, Allan GJ, Keim P, Ravel J, Rabinowicz PD. Castor bean organelle genome sequencing and worldwide genetic diversity analysis. PLoS One. 2011;6:e21743. doi: 10.1371/journal.pone.0021743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts LM, Lamb FI, Pappin DJ, Lord JM. The primary sequence of Ricinus communis agglutinin. Comparison with ricin. JBC. 1985;260:15682–15686. [PubMed] [Google Scholar]
- Rotz LD, Khan AS, Lillibridge SR, Ostroff SM, Hughes JM. Public health assessment of potential biological terrorism agents. Emerg. Infect. Dis. 2002;8:225–230. doi: 10.3201/eid0802.010164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltvedt E. Structure and toxicity of pure Ricinus agglutinin. Biochim. Biophys. Acta (BBA) -Gen. Subj. 1976;451:536–548. doi: 10.1016/0304-4165(76)90149-5. [DOI] [PubMed] [Google Scholar]
- Sehgal P, Kumar O, Kameswararao M, Ravindran J, Khan M, Sharma S, Vijayaraghavan R, Prasad GB. Differential toxicity profile of ricin isoforms correlates with their glycosylation levels. Toxicology. 2011;282:56–67. doi: 10.1016/j.tox.2011.01.012. [DOI] [PubMed] [Google Scholar]
- Severino LS, Auld DL, Baldanzi M, Candido MJD, Chen G, Crosby W, Tan D, He XH, Lakshmamma P, Lavanya C, Machado OLT, Mielke T, Milani M, Miller TD, Morris JB, Morse SA, Navas AA, Soares DJ, Sofiatti V, Wang ML, Zanotto MD, Zieler H. A review on the challenges for increased production of castor. Agron. J. 2012;104:853–880. [Google Scholar]
- Smallshaw JE, Vitetta ES. Ricin vaccine development. Curr. Top. Microbiol. Immunol. 2012;357:259–272. doi: 10.1007/82_2011_156. [DOI] [PubMed] [Google Scholar]
- Sobel J, Khan AS, Swerdlow DL. Threat of a biological terrorist attack on the US food supply: the CDC perspective. Lancet. 2002;359:874–880. doi: 10.1016/S0140-6736(02)07947-3. [DOI] [PubMed] [Google Scholar]
- Stirpe F, Barbieri L. Ribosome-inactivating proteins up to date. FEBS Lett. 1986;195:1–8. doi: 10.1016/0014-5793(86)80118-1. [DOI] [PubMed] [Google Scholar]
- Sturm MB, Schramm VL. Detecting ricin: sensitive luminescent assay for ricin a-chain ribosome depurination kinetics. Anal. Chem. 2009;81:2847–2853. doi: 10.1021/ac8026433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tregear JW, Roberts LM. The lectin gene family of Ricinus communis: cloning of a functional ricin gene and three lectin pseudogenes. Plant Mol. Biol. 1992;18:515–525. doi: 10.1007/BF00040667. [DOI] [PubMed] [Google Scholar]
- Weiner R. Ricin-laced Letter Sent to Michael Bloomberg, Police Say. The Washington Post. 2013 [Google Scholar]
- Weisman J. Letter Mailed to Senator Tests Positive for Ricin. The New York Times. 2013 [Google Scholar]
- Woolfitt AR, Solano MI, Williams TL, Pirkle JL, Barr JR. Amino acid analysis of peptides using isobaric-tagged isotope dilution LC-MS/MS. Anal. Chem. 2009;81:3979–3985. doi: 10.1021/ac900367q. [DOI] [PubMed] [Google Scholar]
- Zamboni M, Brigotti M, Rambelli F, Montanaro L, Sperti S. High-pressure-liquid-chromatographic and fluorimetric methods for the determination of adenine released from ribosomes by ricin and gelonin. Biochem. J. 1989;259:639–643. doi: 10.1042/bj2590639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan J, Zhou P. A simplified method to evaluate the acute toxicity of ricin and ricinus agglutinin. Toxicology. 2003;186:119–123. doi: 10.1016/s0300-483x(02)00726-6. [DOI] [PubMed] [Google Scholar]