Abstract
RUNX2, an essential transcription factor for osteoblast differentiation and bone formation, is activated by ERK/MAP kinase-dependent phosphorylation. However, relationship between these early events and specific epigenetic modifications of chromatin during osteoblast differentiation have not been previously examined. Here, we explore these relationships using chromatin immunoprecipitation (ChIP) to detect chromatin modifications in RUNX2-binding regions of Bglap2 and Ibsp. Growth of MC3T3-E1c4 preosteoblast cells in differentiation conditions rapidly induced Bglap2 and lbsp mRNAs. For both genes, osteogenic stimulation increased chromatin-bound P-ERK, P-RUNX2, p300 and RNA polymerase II as well as histone H3K9 and H4K5 acetylation. The level of H3K4 di-methylation, another gene activation-associated histone mark, also increased. In contrast, levels of the gene repressive marks, H3K9 mono-, di-, and tri-methylation, in the same regions were reduced. Inhibition of MAP kinase signaling blocked differentiation-dependent chromatin modifications and Bglap2 and Ibsp expression. To evaluate the role of RUNX2 phosphorylation in these responses, RUNX2-deficient C3H10T1/2 cells were transduced with adenovirus encoding wild type or phosphorylation site mutant RUNX2 (RUNX2 S301A/S319A). Wild type RUNX2, but not the non-phosphorylated mutant, increased H3K9 and H4K5 acetylation as well as chromatin-associated P-ERK, p300 and polymerase II. Thus, RUNX2 phosphorylation is necessary for subsequent epigenetic changes required for osteoblast gene expression. Taken together, this study reveals a molecular mechanism through which osteogenic genes are controlled by a MAPK and P-RUNX2-dependent process involving epigenetic modifications of specific promoter regions.
Keywords: RUNX2, Histone Acetylation and Methylation, Gene Expression, Osteoblast Differentiation
Introduction
Epigenetic modifications of chromatin including histone acetylation and methylation have major effects on chromatin structure and gene expression that are critical for the differentiation of all cell types including osteoblasts. Histone acetylation, which is considered a hallmark of gene activation, is catalyzed by histone acetyltransferases (HATs) such as GCN5, PACAF, P300 and MOF (Grewal and Moazed, 2003). Conversely, histone deacetylases (HDACs) remove acetyl groups leading to chromatin condensation and gene repression(Verdone et al., 2005). Histone methylation has also been traditionally linked to gene repression although methylation at certain sites such as histone H3 at lysine 4, lysine 36 and lysine 79 are associated with gene activation(Wang et al., 2008). Like histone acetylation, methylation can be reversed by histone demethylases such as LSD1, LSD2, JMJC1, JMJD1(A,B,C), JMJD2 (A,B,C), JARID1 (A,B,C,D), FBXL 2(Cloos et al., 2008; Katz et al., 2014).
The MAP kinase pathway is an important mediator of the response of osteoprogenitor cells to extracellular matrix composition and stiffness, mechanical loading, hormone, growth factor and morphogen stimulation(Greenblatt et al., 2013; Kapur et al., 2005; Khatiwala et al., 2009; Li et al., 2012; Xiao et al., 2002a; Xiao et al., 2002b; Xiao et al., 2000; You et al., 2001). Osteoprogenitor-specific knock out of ERK1/2 (Mapk3/Mapk1) or overexpression of a dominant-negative MEK1 leads to reduced bone mass and delayed bone development while overexpression of constitutive-active MEK1 increases bone formation(Ge et al., 2007; Matsushita et al., 2009). Upon activation in osteoprogenitor cells, P-ERK translocates to the nucleus where it binds the RUNX2 transcription factor on the chromatin of target genes. P-ERK phosphorylates RUNX2 on specific serine residues that are necessary for induction of osteoblast-specific transcription(Ge et al., 2009; Li et al., 2010). The differentiation of osteoblast from mesenchymal progenitors has been associated with specific epigenetic modifications including histone acetylation, methylation and phosphorylation(Eslaminejad et al., 2013; Yasui et al., 2011). Recent analysis of osteoblast differentiation using ChIP-Seq identified gene clusters associated with transcription factors such as RUNX2 and C/EBPβ and specific histone modifications such as methylation of H3 lysine 4, acetylation of H3 lysine 9 and H4 lysine 5 and methylation of H3 lysine 27(Meyer et al., 2014a; Meyer et al., 2014b; Wu et al., 2014). However, the relationship between these epigenetic changes and the MAPK/RUNX2 pathway described above has not been previously described.
Here we explore this relationship by examining differentiation-related epigenetic changes associated with RUNX2-binding regions of two important osteoblast-related genes, Bglap2 and Ibsp. As will be shown, after initiation of differentiation, P-ERK phosphorylation of RUNX2 initiates a series of chromatin changes associated with RNA polymerase II recruitment and transcription.
Materials and Methods
Cell culture
Subclone 4 MC3T3-E1 (MC3T3-E1c4) preosteoblast cells were maintained in growth medium (GM) containing α-Minimal Essential Medium (α-MEM; Life Technologies, Inc., Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS; Gibco, Life technologies, USA) and 1% penicillin/streptomycin, as described previously(11). C3H10T1/2 cells were maintained in Dulbecco Modified Eagle Medium (DMEM, Gibco, Life technologies, USA) supplemented with 10% fetal bovine serum (FBS; Gibco, Life technologies, USA) and 1% penicillin/streptomycin. For differentiation studies, cells were plated at a density of 50,000 cells/cm2 and cultured in differentiation medium (DM) composed of GM containing 50 µg/ml ascorbic acid.
Antibodies
P-ERK, total ERK, histone H3 dimethyl lysine 4 (H3K4m), histone H3 mono- di- and trimethyl lysine 9 (H3K9m), histone H3 acetylated at lysine 9 (AcH3K9), histone H4 acetylated at lysine 5 (AcH4K5) and GAPDH antibodies were obtained from Cell Signaling. Pol II and p300/CBP antibodies were obtained from Santa Cruz Biotechnology. Total RUNX2 antibody was obtained from MBL. Anti-RUNX2-S319-P antibody was generated in the project laboratory(Ge et al., 2012).
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) assays were carried out as described previously(Li et al., 2010). Briefly, cells were cross-linked at room temperature with 1% formaldehyde for 10 minutes. Crude nuclei were then isolated and sheared by sonication. Chromatin (average size, 500 bp) was pre-cleared by incubation with protein A/G agarose beads (Santa-Cruz Biotechnology) followed by incubation with primary antibodies against P-ERK, RUNX2, RUNX2-S319-P, Pol II, P300, H3K4m, H3K9m, AcH3K9 and AcH4K5 respectively, and precipitated with A/G agarose beads. Input DNA and DNA from ChIP samples were measured by PCR (35 cycles) or by quantitative real-time PCR as previously described(Li et al., 2010). These latter measurements used previously described Taqman probes flanking OSE2a and OSE2b RUNX2 binding sites in Bglap2 or the transcribed region of Bglap2, a RUNX2 -binding site in the proximal Ibsp promoter or distal site in Ibsp that is devoid of Runx2 binding activity.
Quantitation of mRNA
Total RNA was extracted from cells using TRIzol reagent (Invitrogen) and cleaned with an RNeasy Mini Kit (Qiagen). Single strand cDNA was synthesized using Taqman reverse transcriptase (Applied Biosystems). Quantitative real-time polymerase chain reaction (PCR) measurement of Bglap2, Ibsp, and Gapdh mRNA was carried out as described previously(22) using predeveloped Taqman probes and an ABI PRISM 7700 sequence detector (Applied Biosystems). mRNA expression was normalized to Gapdh mRNA.
Western blot
Cells were harvested in lysis RIPA buffer (10 Mm Tris, Ph7.5, 5 mM EDTA, 1% Triton X-100, 150 mM NaCI, 0.1% SDS, PMSF, 1 × proteinase inhibitor cocktail, Sigma). Protein samples were separated by SDS-PAGE using 4% to 20% Novex Tris-Glycine gels (Life technologies, USA). After transfer to nitrocellulose membranes, samples were incubated with total RUNX2 (1:500 dilution), phospho-ERK (1:1000 dillution), total ERK (1:1000 dilution) or GAPDH (1:2000 dilution) antibodies, respectively, overnight at 4°C. A secondary antibody of sheep anti-mouse conjugated with horseradish perioxidse (GE Healthcare, UK) was used at 1:10000 dilution. Protein expression was detected by ECL (Amersham).
Statistical analysis
The results are presented as mean ± SE, with n = 3 per group for all comparisons. Statistical analysis was determined using a one-way ANOVA followed by Tukey's multiple-comparison test.
Results
Chromatin changes accompanying induction of osteoblast gene expression
Induction of osteoblast differentiation is accompanied by gradual (within 1–3 days) activation of ERK/MAP kinase activity. P-ERK translocates to the nucleus where it binds and phosphorylates RUNX2 at several sites including S319. These changes are followed by induction of osteoblast marker genes such as Bglap2 and Ibsp(Ge et al., 2012; Li et al., 2010). An example of this mRNA induction is shown in Figure 1 where MC3T3-E1c4 cells were maintained in growth (GM) or differentiation medium (DM) for 3, 6 or 9 days before RNA isolation. Induction of both mRNAs was detected by 3 days with levels continuing to increase through the 9 day time point.
Figure 1. Time course of Bglap2 and Ibsp mRNA expression during osteoblast differentiation.
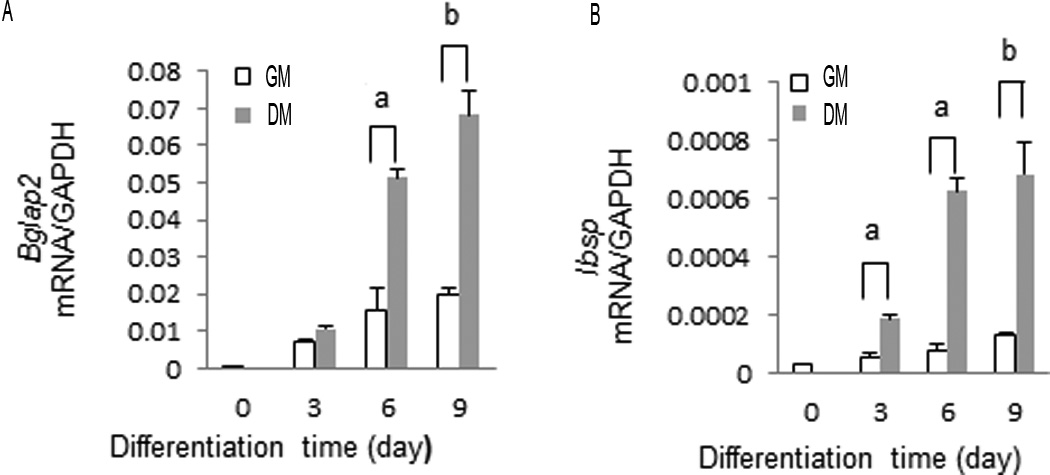
Preosteoblast MC3T3-E1 subclone 4 cells were cultured in growth medium (GM) or differentiation medium (DM) for the times indicated. Total RNA was isolated and Bglap2 and Ibsp mRNA levels measured by qRT-PCR.
To examine chromatin changes associated with Bglap2 and Ibsp mRNA induction, ChIP analysis was conducted using previously characterized RUNX2-binding regions of these two genes (shown in Fig. 2A) (Ducy and Karsenty, 1995; Roca et al., 2005). Bglap2 contains 2 RUNX2-binding enhancers in the proximal promoter at −130 bp (OSE2a) and at −605 bp (OSE2b), respectively. An additional primer pair detected binding to the transcribed region of Bglap2 at +400 bp (TSR). Primers were also designed to detect interactions with two regions of Ibsp, a region in the first 200 bp of the proximal promoter (Prox) containing two functional RUNX2-specific enhancers and a second non-functional cryptic RUNX2 consensus binding site at −1300 bp (Dist). This later site does not bind RUNX2 in ChIP assays, lacks enhancer activity and, therefore, serves as a negative control(Roca et al., 2005). Each site is sufficiently separated from adjacent sites such that it is not immunoprecipitated in the same chromatin fragment during ChIP analysis, thereby allowing each site to be analyzed separately(Li et al., 2010). ChIP analysis was conducted using cells grown for up to 6 days in GM or DM. Although time course studies revealed that early P-ERK binding, RUNX2 phosphorylation and epigenetic changes in chromatin could be detected after 2–3 days in DM (Suppl. Fig. 1), the 6 day time point was selected for detailed analysis because more robust epigenetic changes and strong induction of Bglap2 and Ibsp mRNAs were seen by this time. Figure 2B shows semi-quantitative PCR analysis of ChIP samples while Figure 2C provides more quantitative data using real-time PCR detection of ChIP DNA. Consistent with our previous report(Li et al., 2010), growth in DM stimulated the association of P-ERK with RUNX2-binding regions of Bglap2 (OSE2a, OSE2b) and Ibsp (BSPprox). In contrast, P-ERK did not bind to the transcribed region of Bglap2 (TSR) or distal region of Ibsp since these regions do not bind RUNX2, which is necessary for chromatin docking of P-ERK(Li et al., 2010). Although the differentiation condition did not significantly affect total bound RUNX2, chromatin-associated P-RUNX2, measured with an antibody that preferentially detects RUNX2-S319-P, was dramatically increased at all 3 sites (OSE2a, OSE2b and Prox) as would be expected if RUNX2 were being phosphorylated by the associated P-ERK. Coincident with these changes was the association of the HAT, p300, with chromatin, changes in histone acetylation and methylation, and increased binding of RNA polymerase II (Pol II). The histone marks selected for analysis were acetylation at histone H3 lysine 9 and H4 lysine 5 (AcH3K9 and AcH4K5), which are both associated with gene activation, mono-, di- or tri-methylation at H3 lysine 9 (detected with pan-H3K9m antibody) associated with repression and di-methylation at H3 lysine 4 (H4K5m) associated with gene activation(Grewal and Moazed, 2003)). Levels of AcH3K9 and AcH4K5 were elevated in OSE2a and OSE2b regions of Bglap2 and the proximal region of Ibsp (BSPprox). In addition, the activation-associated methylation mark, H4K5m, was increased while a consistent decline in the repressive mark, H3K9m, was observed at all 3 sites. Because levels of all marks were at low to undetectable levels in the TSR of Bglap2 and the distal region of Ibsp (Fig. 2B), these chromatin regions were not used in subsequent analysis.
Figure 2. Differentiation-dependent epigenetic changes associated with Runx2-binding regions in the proximal promoter regions of Bglap2 and Ibsp.
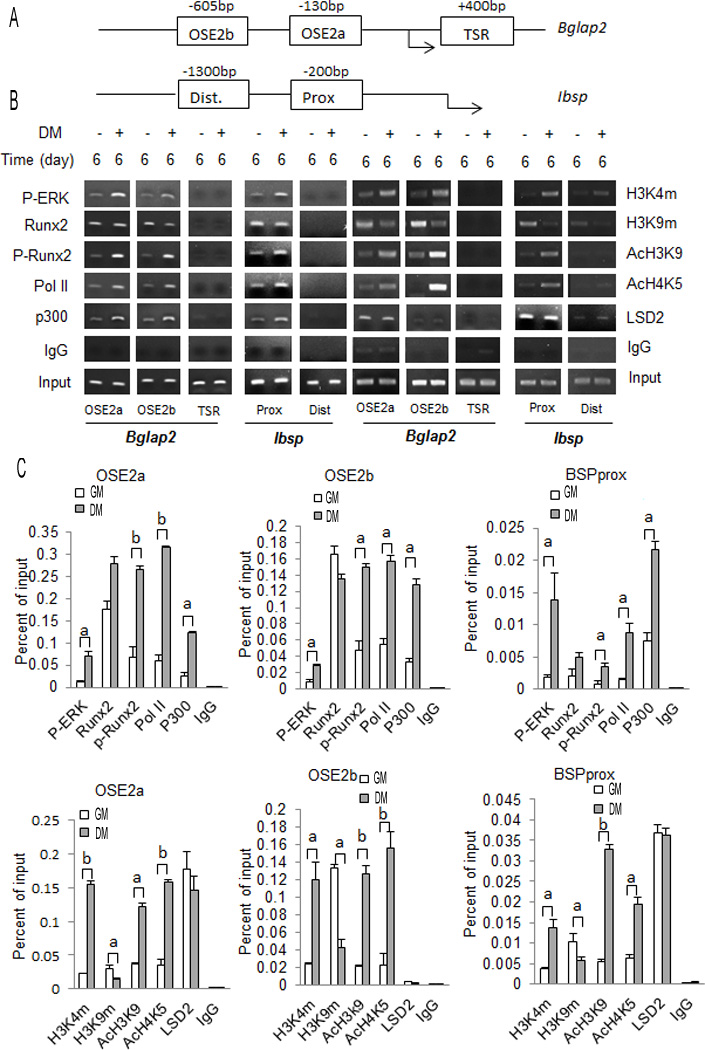
(A) Schematic of proximal promoters of Bglap2 and Ibsp showing regions amplified during ChIP analysis. OSE2a and OSE2b are functional Runx2 binding sites in Bglap2. A control region (Ct) in the transcribed region of Bglap2 is also shown. Ibsp contains distal (Dist) and proximal (Prox) consensus Runx2 binding sites, but only the proximal site is functional. (B, C) ChIP analysis using different amplified regions of Bglap2 and Ibsp shown in (A). MC3T3-E1c4 cells were cultured in growth (GM) or differentiation medium (DM) for 6 days. Isolated chromatin was immunoprecipitated with the indicated antibodies to determine the chromatin association of P-ERK, RUNX2, P-RUNX2, RNA polymerase II (Pol II), histone acetylase p300 (P300), histone H3 methylation at lysine 4 (H3K4m) and lysine 9 (H3K9m), histone H3 acetylation at lysine 9 (AcH3K9) and histone H4 acetylation at lysine 5 (AcH4K5). (B) Gel image of semi-quantitative PCR results. (C) Quantitative PCR analysis of ChIP results using TaqMan probes for OSE2a and OSE2b regions of Bglap2 and the Prox region of Ibsp. Significant differences between DM and GM samples: p ≤ 0.05 (a), p ≤ 0.01 (b), n=3.
As noted above, the differentiation condition increased association of p300 with RUNX2-binding regions of Bglap2 and Ibsp, which may explain the observed increases in histone acetylation. In an attempt to explain the observed increase in H3K4 dimethylation, we also measured chromatin levels of the histone demethylase, LSD2, which preferentially removes mono and dimethly groups from H3K4m(Katz et al., 2014). However, we did not observe any LSD2 binding changes between DM and GM suggesting that the observed methylation changes in H3K4m could not be explained by regulation of this demethylase.
MAP kinase activity is necessary for differentiation-dependent chromatin modifications
Previous studies established that ERK must be in its activated, phosphorylated form to bind RUNX2 on chromatin(Li et al., 2010; Li et al., 2012). However, the role of MAPK signaling in the differentiation-dependent chromatin changes described above has not been examined. To address this issue, cells were grown in DM for 6 days and then treated with the MAPK inhibitor, U0126, for 12 h. Previous work established this treatment time as being sufficient to block MAPK signaling and osteoblast gene expression while avoiding toxicity associated with chronic inhibitor exposure(Xiao et al., 2002a). This is confirmed in the results shown in Figure 3 A–C where U0126 treatment blocked MAPK activity (ERK phosphorylation) as well as Ibsp and Bglap2 mRNA induction. Inhibitor treatment also totally or partially reverse all the differentiation-related chromatin changes described in Figure 1. Thus, increases in chromatin-associated P-ERK, P-RUNX2, Pol II, p300 and the gene activation-associated histone marks H3K4m, AcH3K9 and AcH4K4 all returned to control levels in U0126-treated samples regardless of the chromatin site examined (Fig. 3, D–F). The differentiation-dependent reduction in the inhibitory chromatin marks detected with H3K9m antibody was also partially reversed by inhibitor treatment although this reversal was not complete and results varied between chromatin sites (compare H3K9m result for OSE2a-Panel D, with Panels E and F). Taken together, these results indicate that the MAPK activity is required for subsequent differentiation-related chromatin changes and gene expression.
Figure 3. MAP kinase activity is required for induction of osteoblast gene expression and chromatin-associated epigenetic changes.
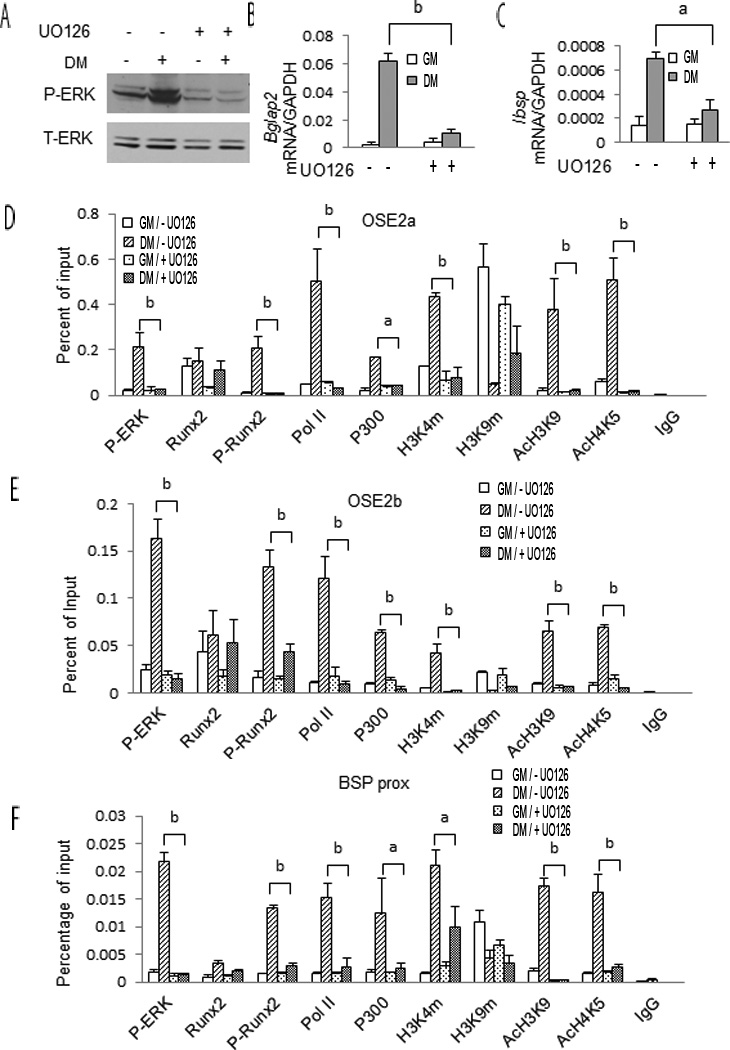
MC3T3-E1c4 cells were cultured in GM and DM for 6 days and then treated with 20 µg/ml U0126 for 12 hours. Cells were subsequently harvested for measurement of P-ERK and total ERK (A), Bglap2 and Ibsp mRNA (B, C) and ChIP analysis using the indicated antibodies (D-F). Statistics: Comparisons were conducted between cells grown in DM without UO126 and in DM with UO126. (a) p≤0.05, (b) p≤0.01, n=3.
RUNX2 phosphorylation is necessary for differentiation-related changes in chromatin occupancy and epigenetic marks
A final series of experiments evaluated the role of RUNX2 phosphorylation in differentiation-related chromatin changes. C3H10T1/2 cells, which have low endogenous RUNX2 levels, were transduced with adenovirus vectors expressing LacZ, wild type RUNX2 (RUNX2 WT) or a phosphorylation-deficient mutant containing serine to alanine mutations in at S301 and S319 (RUNX2 SA). As previously shown, phosphorylation at these sites is necessary for MAPK-dependent RUNX2 transcriptional activity(Ge et al., 2016; Ge et al., 2009). This is confirmed by the result shown in Fig. 4A where wild type RUNX2 strongly stimulated Bglap2 and Ibsp mRNAs when cells were grown in DM while the phosphorylation site mutant expressed at equivalent levels was much less active (Fig. 4AB).
Figure 4. Runx2 and Runx2 S301/S319 phosphorylation are required for induction of osteoblast gene expression and chromatin-associated epigenetic changes.
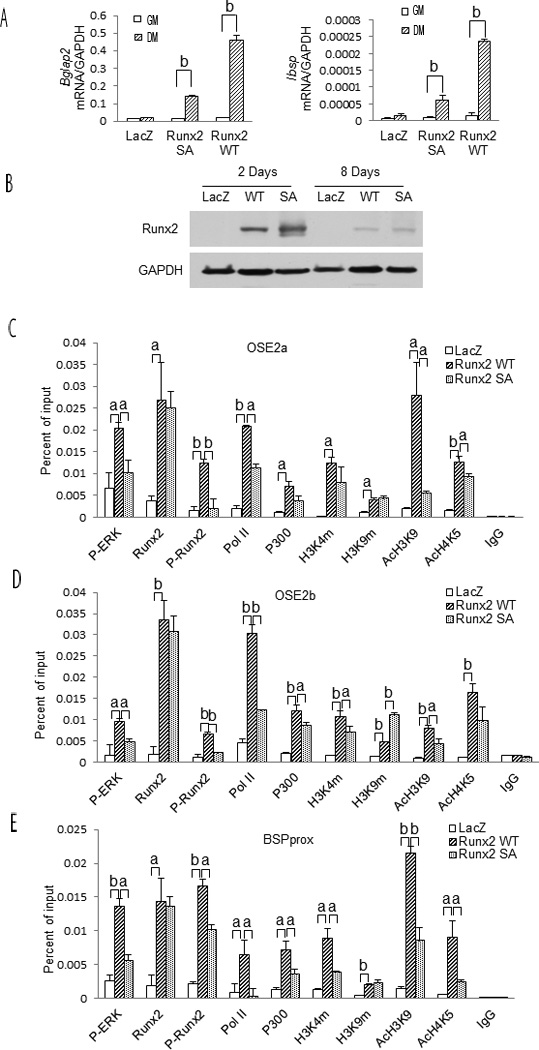
C3H10T1/2 cells were transduced with control adenovirus (LacZ), virus expressing wild type RUNX2 (Runx2 WT) or RUNX2 containing S301A, S319A mutations (Runx2 SA). After 2 days, cells were switched to GM or DM and cultured for an additional 6 days before analysis of gene expression (A). RUNX2 protein levels were measured in cells grown in DM for 2 or 8 days (B) while ChIP analysis used cells grown in DM for 6 days (C–E).
Statistics: Comparisons were conducted between groups indicated by horizontal bars. (a) p ≤ 0.05; (b), p ≤ 0.01, n = 3.
ChIP assays were next used to assess effects of RUNX2 and RUNX2 phosphorylation on recruitment of factors to chromatin and epigenetic marks (Fig. 4C–E). Equivalent binding of wild type or mutant RUNX2 was detected at OSE2a, OSE2b and BSPprox chromatin sites. However, dramatic differences in levels of other chromatin-associated factors were observed. In agreement with our previous report that RUNX2 is necessary for docking of P-ERK on chromatin, cells expressing wild type RUNX2 showed a dramatic increase in chromatin-associated P-ERK when compared with LacZ controls.(Li et al., 2010). However, P-ERK binding was reduced in cells expressing RUNX2 SA. As expected, the ChIP signal with the anti-RUNX2-S319-P antibody (P-Runx2) was greatly reduced in cells transduced with the RUNX2 SA mutant. Consistent with its enhanced transcriptional activity, wild type RUNX2 increased chromatin-associated p300, the gene activation-associated histone marks, H3K4m, AcH3K9 and AcH4K5, as well as Pol II while the RUNX2 SA mutant was significantly less active. Paradoxically, the inhibitory marks detected by the pan H3K9m antibody were actually increased in both wild type and RUNX SA groups. Nevertheless, the general conclusion from these studies is that RUNX2 must be phosphorylated at S301 and S319 to recruit accessory factors to chromatin leading to epigenetic changes necessary for transcriptional activation.
Discussion
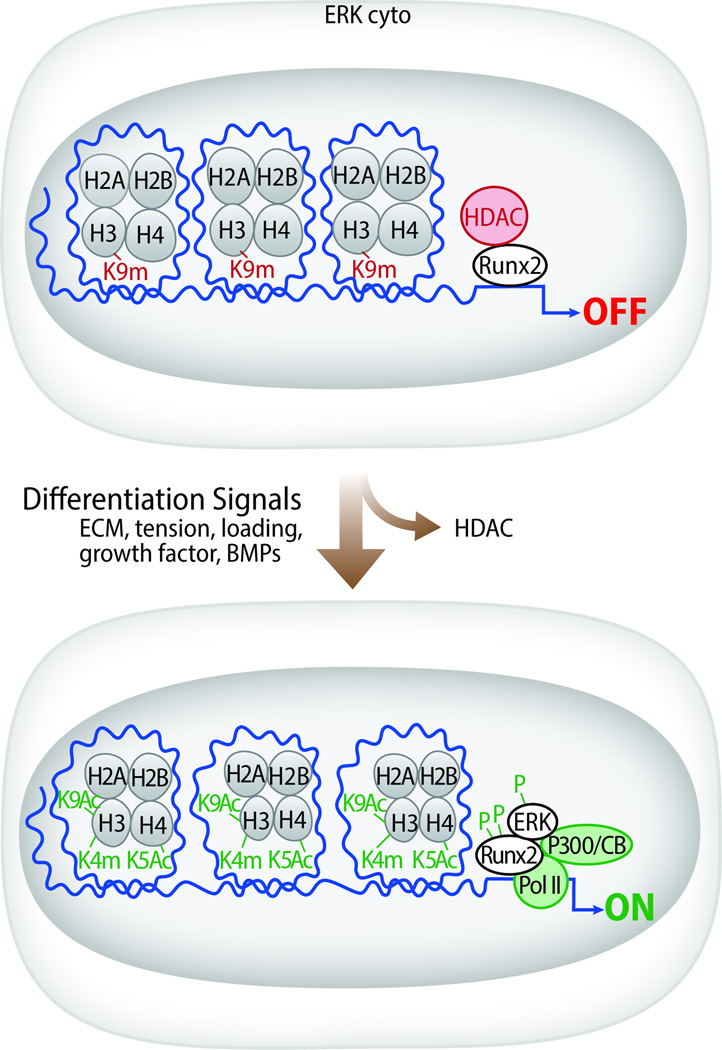
This study examined the relationship between RUNX2 phosphorylation and differentiation-dependent chromatin modifications of the proximal RUNX2-binding regions of Bglap2 and Ibsp. Initiation of differentiation was associated with translocation of P-ERK to the nucleus where it associated with and phosphorylated RUNX2 previously bound to specific enhancer-containing chromatin regions. These events were required for subsequent recruitment of p300/CBP to chromatin, increases in gene activation-associated epigenetic histone marks (H3K4m, AcH3K9 and AcH4K5), reduction in the repressive histone mark, H3K9m, RNA polymerase II recruitment and transcription. These findings support the model shown in Figure 5 where RUNX2 phosphorylation plays a pivotal role in recruiting chromatin-modifying factors to gene enhancers thereby allowing the epigenetic changes necessary for transcription and osteoblast differentiation.
Fig 5. Model showing differentiation-dependent changes in chromatin structure induced by ERK-dependent RUNX2 phosphorylation.
In undifferentiated preosteoblast cells, RUNX2 is already bound to relevant enhancer sites on chromatin together with histone H3 methylated at lysine 9 (H3K9m). Levels of histone H3 acetylation at lysine 9 and H4 acetylation at lysine 5 remain low consistent with the condensed state of chromatin. After induction of differentiation, P-ERK translocates to the nucleus where it binds and phosphorylates RUNX2 at S301 and S319. Phosphorylated RUNX2 facilitates binding of the histone acetyltransferase, p300, to chromatin leading to acetylation of H3 at lysine 9 and H4 at lysine 5. In addition, an as yet unidentified methyltransferase is also recruited resulting in methylation of histone H3 at lysine 4. Together, these epigenetic changes stimulate chromatin decondensation, increased RNA polymerase II recruitment and transcription of osteoblast genes.
Results of the present study are reminiscent of previous work from this laboratory that examined the role of RUNX2 phosphorylation in the response of osteoblasts and bone to mechanical loading(Li et al., 2012). Both exposure of preosteoblasts to fluid flow shear stress (FFSS) and ulnar loading of mice were shown to increase MAPK signaling and RUNX2 S319 phosphorylation. Similar to the present study, the ability of FFSS to stimulate osteoblast gene expression required MAPK activation and RUNX2 S301,S319 phosphorylation. Furthermore, these phosphorylation events were shown to be required for overall increases in histone acetylation although the specific histone acetylation marks involved were not identified nor was histone methylation examined.
As suggested in the present study, phosphorylation of RUNX2 may affect its ability to bind P-ERK and p300/CBP in that chromatin binding of these factors was dramatically reduced in cells treated with a MAPK inhibitor or transfected with a phosphorylation-deficient RUNX2 mutant (RUNX2 SA). Although a direct association of these factors with RUNX2 cannot be inferred from ChIP analysis, we previously obtained evidence using immunoprecipitation that ERK directly binds RUNX2 and identified a consensus MAPK-binding D site between amino acids 201 and 215 in the runt domain that was necessary for this binding(Ge et al., 2009). Interestingly, in that study mutation of S301 and S319 phosphorylation sites in RUNX2 (RUNX2 SA) did not affect its ability to bind ERK while in the present study, chromatin binding of P-ERK was reduced in cells transfected with RUNX2 SA versus wild type RUNX2 (Fig. 4). Since we previously establish that ERK does not bind the chromatin regions studied in the absence of RUNX2(Li et al., 2010), these results may reflect unique aspects of the in vivo chromatin environment that require intact RUNX2 phosphorylation sites for optimal P-ERK binding. There is also evidence from other laboratories that RUNX2 phosphorylation mediated by either p38 or ERK MAP kinases is important for complex formation with p300/CBP and Smads(Afzal et al., 2005; Greenblatt et al., 2010). In the former study, p38 MAPK-mediated phosphorylation of RUNX2 at several serine residues including the S319 ERK/MAPK site analyzed in the present study was shown to be necessary for binding to p300/CBP in immunoprecipitation assays. In the latter study, treatment with a MAP kinase inhibitor blocked the co-immunoprecipitation of RUNX2 with Smad1.
We do not currently know the full range of factors that are recruited to chromatin by P-RUNX2. From our analysis, it is clear that RUNX2 phosphorylation stimulates histone acetylation (AcH3K9, AcH4K5) and the activation-associated methylation mark, dimethyl H3K4, while suppressing mono, di and trimethyl H3K9. However, it is not known whether the observed changes in acetylation are explained by the recruitment of p300/CBP to chromatin. In fact, the ability of p300/CBP to stimulate RUNX2-dependent transcription of the osteocalcin gene was previously reported to not require HAT activity(Sierra et al., 2003), although HAT activity of p300/CBP was required for PTH induction of Mmp13(Boumah et al., 2009). Alternately, results could be explained by suppression of HDACs, some of which are known to associate with RUNX2(Jensen et al., 2008; McGee-Lawrence and Westendorf, 2011). Similarly, the nuclear factors responsible for the observed differentiation-related increases in H3K4 dimethylation and inhibition of H3K9 mono, di and trimethylation remain to be described. Because of its specificity for removing methyl groups from histone H3K4, chromatin levels of the histone demethylase, LSD2, were examined, but no differentiation-related changes were observed (Fig. 2). Although not examined in the present study, the demethylases KDM4B and KDM6B were previously shown to be required for osteogenic differentiation of human MSC(Ye et al., 2012). KDM4B selectively demethylates trimethyl H3K9 so it could be involved in the differentiation-related reduction of this histone mark. KDM6B demethylates trimethyl H3K27, which was not examined in our study. Clearly, further work will be required to define the breadth of nuclear factors recruited to chromatin by phosphorylated RUNX2 and relate them to the epigenetic changes observed.
Because the ChIP analysis in the present study was restricted to proximal regions of Bglap2 and Ibsp known to contain functional RUNX2-binding enhancers, it might be questioned whether our conclusions are relevant to understanding the role of RUNX2 phosphorylation at a genomic level. While a definitive answer to this question would require genomic analysis of P-RUNX2 distribution using ChIP-Seq, some insight into the broader relevance of our work can be gleaned by consideration of recent ChIP-Seq studies on total RUNX2 distribution that were conducted using differentiated osteoblasts(Meyer et al., 2014a; Wu et al., 2014). In both studies, chromatin-associated RUNX2 was broadly distributed throughout the genome with about 30% of RUNX2 occupancy in the promoter regions of putative target genes and 70% of RUNX2 located in non-promoter regions including intron, exon and intergenic regions. Most interestingly, RUNX2 binding sites were frequently in chromatin regions containing methylated histone H3K4 as well as acetylated histones H3K9 and H4K5, the same activating histone marks that were associated with and shown to require RUNX2 phosphorylation in the present study(Meyer et al., 2014a). Because ChIP-Seq analysis was conducted using differentiated osteoblasts, it is likely that much of the chromatin-associated RUNX2 detected was in the phosphorylated form. It can, therefore, be inferred that RUNX2 phosphorylation at S301 and S319 likely has broad effects through out the genome leading to local increases in activating histone marks, chromatin decondensation and gene expression.
Supplementary Material
Acknowledgments
Contract Grant Sponsor: NIH
Contract Grant Numbers: DE11723, K12 DE023574 (Junior Faculty Training Award, CG) and NIDDK Grant P30 DK092926 (Michigan Center for Diabetes Translational Research)
Footnotes
All authors state that they have no conflicts of interest
References
- Afzal F, Pratap J, Ito K, Ito Y, Stein JL, van Wijnen AJ, Stein GS, Lian JB, Javed A. Smad function and intranuclear targeting share a Runx2 motif required for osteogenic lineage induction and BMP2 responsive transcription. J Cell Physiol. 2005;204(1):63–72. doi: 10.1002/jcp.20258. [DOI] [PubMed] [Google Scholar]
- Boumah CE, Lee M, Selvamurugan N, Shimizu E, Partridge NC. Runx2 recruits p300 to mediate parathyroid hormone's effects on histone acetylation and transcriptional activation of the matrix metalloproteinase-13 gene. Mol Endocrinol. 2009;23(8):1255–1263. doi: 10.1210/me.2008-0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloos PA, Christensen J, Agger K, Helin K. Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes Dev. 2008;22(9):1115–1140. doi: 10.1101/gad.1652908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducy P, Karsenty G. Two distinct osteoblast-specific cis-acting elements control expression of a mouse osteocalcin gene. Mol Cell Biol. 1995;15(4):1858–1869. doi: 10.1128/mcb.15.4.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eslaminejad MB, Fani N, Shahhoseini M. Epigenetic regulation of osteogenic and chondrogenic differentiation of mesenchymal stem cells in culture. Cell J. 2013;15(1):1–10. [PMC free article] [PubMed] [Google Scholar]
- Ge C, Cawthorn WP, Li Y, Zhao G, MacDougald OA, Franceschi RT. Reciprocal Control of Osteogenic and Adipogenic Differentiation by ERK/MAP Kinase Phosphorylation of Runx2 and PPARgamma Transcription Factors. J Cell Physiol. 2016;231(3):587–596. doi: 10.1002/jcp.25102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge C, Xiao G, Jiang D, Franceschi RT. Critical role of the extracellular signal-regulated kinase-MAPK pathway in osteoblast differentiation and skeletal development. J Cell Biol. 2007;176(5):709–718. doi: 10.1083/jcb.200610046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge C, Xiao G, Jiang D, Yang Q, Hatch NE, Roca H, Franceschi RT. Identification and functional characterization of ERK/MAPK phosphorylation sites in the Runx2 transcription factor. J Biol Chem. 2009;284(47):32533–32543. doi: 10.1074/jbc.M109.040980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge C, Yang Q, Zhao G, Yu H, Kirkwood KL, Franceschi RT. Interactions between extracellular signal-regulated kinase 1/2 and p38 MAP kinase pathways in the control of RUNX2 phosphorylation and transcriptional activity. J Bone Miner Res. 2012;27(3):538–551. doi: 10.1002/jbmr.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblatt MB, Shim JH, Glimcher LH. Mitogen-Activated Protein Kinase Pathways in Osteoblasts. Annu Rev Cell Dev Biol. 2013 doi: 10.1146/annurev-cellbio-101512-122347. [DOI] [PubMed] [Google Scholar]
- Greenblatt MB, Shim JH, Zou W, Sitara D, Schweitzer M, Hu D, Lotinun S, Sano Y, Baron R, Park JM, Arthur S, Xie M, Schneider MD, Zhai B, Gygi S, Davis R, Glimcher LH. The p38 MAPK pathway is essential for skeletogenesis and bone homeostasis in mice. J Clin Invest. 2010;120(7):2457–2473. doi: 10.1172/JCI42285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI, Moazed D. Heterochromatin and epigenetic control of gene expression. Science. 2003;301(5634):798–802. doi: 10.1126/science.1086887. [DOI] [PubMed] [Google Scholar]
- Jensen ED, Schroeder TM, Bailey J, Gopalakrishnan R, Westendorf JJ. Histone deacetylase 7 associates with Runx2 and represses its activity during osteoblast maturation in a deacetylation-independent manner. J Bone Miner Res. 2008;23(3):361–372. doi: 10.1359/JBMR.071104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapur S, Mohan S, Baylink DJ, Lau KH. Fluid shear stress synergizes with insulin-like growth factor-I (IGF-I) on osteoblast proliferation through integrin-dependent activation of IGF-I mitogenic signaling pathway. J Biol Chem. 2005;280(20):20163–20170. doi: 10.1074/jbc.M501460200. [DOI] [PubMed] [Google Scholar]
- Katz TA, Vasilatos SN, Harrington E, Oesterreich S, Davidson NE, Huang Y. Inhibition of histone demethylase, LSD2 (KDM1B), attenuates DNA methylation and increases sensitivity to DNMT inhibitor-induced apoptosis in breast cancer cells. Breast Cancer Res Treat. 2014;146(1):99–108. doi: 10.1007/s10549-014-3012-9. [DOI] [PubMed] [Google Scholar]
- Khatiwala CB, Kim PD, Peyton SR, Putnam AJ. ECM compliance regulates osteogenesis by influencing MAPK signaling downstream of RhoA and ROCK. J Bone Miner Res. 2009;24(5):886–898. doi: 10.1359/JBMR.081240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Ge C, Franceschi RT. Differentiation-dependent association of phosphorylated extracellular signal-regulated kinase with the chromatin of osteoblast-related genes. J Bone Miner Res. 2010;25(1):154–163. doi: 10.1359/jbmr.090705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Ge C, Long JP, Begun DL, Rodriguez JA, Goldstein SA, Franceschi RT. Biomechanical stimulation of osteoblast gene expression requires phosphorylation of the RUNX2 transcription factor. J Bone Miner Res. 2012;27:1263–1274. doi: 10.1002/jbmr.1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita T, Chan YY, Kawanami A, Balmes G, Landreth GE, Murakami S. Extracellular signal-regulated kinase 1 (ERK1) and ERK2 play essential roles in osteoblast differentiation and in supporting osteoclastogenesis. Mol Cell Biol. 2009;29(21):5843–5857. doi: 10.1128/MCB.01549-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee-Lawrence ME, Westendorf JJ. Histone deacetylases in skeletal development and bone mass maintenance. Gene. 2011;474(1–2):1–11. doi: 10.1016/j.gene.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer MB, Benkusky NA, Lee CH, Pike JW. Genomic determinants of gene regulation by 1,25-dihydroxyvitamin D3 during osteoblast-lineage cell differentiation. J Biol Chem. 2014a;289(28):19539–19554. doi: 10.1074/jbc.M114.578104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer MB, Benkusky NA, Pike JW. The RUNX2 cistrome in osteoblasts: characterization, down-regulation following differentiation, and relationship to gene expression. J Biol Chem. 2014b;289(23):16016–16031. doi: 10.1074/jbc.M114.552216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca H, Phimphilai M, Gopalakrishnan R, Xiao G, Franceschi RT. Cooperative interactions between RUNX2 and homeodomain protein-binding sites are critical for the osteoblast-specific expression of the bone sialoprotein gene. J Biol Chem. 2005;280(35):30845–30855. doi: 10.1074/jbc.M503942200. [DOI] [PubMed] [Google Scholar]
- Sierra J, Villagra A, Paredes R, Cruzat F, Gutierrez S, Javed A, Arriagada G, Olate J, Imschenetzky M, Van Wijnen AJ, Lian JB, Stein GS, Stein JL, Montecino M. Regulation of the bone-specific osteocalcin gene by p300 requires Runx2/Cbfa1 and the vitamin D3 receptor but not p300 intrinsic histone acetyltransferase activity. Mol Cell Biol. 2003;23(9):3339–3351. doi: 10.1128/MCB.23.9.3339-3351.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdone L, Caserta M, Di Mauro E. Role of histone acetylation in the control of gene expression. Biochem Cell Biol. 2005;83(3):344–353. doi: 10.1139/o05-041. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, Zhao K. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40(7):897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Whitfield TW, Gordon JA, Dobson JR, Tai PW, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Genomic occupancy of Runx2 with global expression profiling identifies a novel dimension to control of osteoblastogenesis. Genome Biol. 2014;15(3):R52. doi: 10.1186/gb-2014-15-3-r52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao G, Gopalakrishnan R, Jiang D, Reith E, Benson MD, Franceschi RT. Bone morphogenetic proteins, extracellular matrix, and mitogen-activated protein kinase signaling pathways are required for osteoblast-specific gene expression and differentiation in MC3T3-E1 cells. J Bone Miner Res. 2002a;17(1):101–110. doi: 10.1359/jbmr.2002.17.1.101. [DOI] [PubMed] [Google Scholar]
- Xiao G, Jiang D, Gopalakrishnan R, Franceschi RT. Fibroblast growth factor 2 induction of the osteocalcin gene requires MAPK activity and phosphorylation of the osteoblast transcription factor, Cbfa1/Runx2. J Biol Chem. 2002b;277(39):36181–36187. doi: 10.1074/jbc.M206057200. [DOI] [PubMed] [Google Scholar]
- Xiao G, Jiang D, Thomas P, Benson MD, Guan K, Karsenty G, Franceschi RT. MAPK pathways activate and phosphorylate the osteoblast-specific transcription factor, Cbfa1. J Biol Chem. 2000;275(6):4453–4459. doi: 10.1074/jbc.275.6.4453. [DOI] [PubMed] [Google Scholar]
- Yasui T, Hirose J, Tsutsumi S, Nakamura K, Aburatani H, Tanaka S. Epigenetic regulation of osteoclast differentiation: possible involvement of Jmjd3 in the histone demethylation of Nfatc1. J Bone Miner Res. 2011;26(11):2665–2671. doi: 10.1002/jbmr.464. [DOI] [PubMed] [Google Scholar]
- Ye L, Fan Z, Yu B, Chang J, Al Hezaimi K, Zhou X, Park NH, Wang CY. Histone demethylases KDM4B and KDM6B promotes osteogenic differentiation of human MSCs. Cell Stem Cell. 2012;11(1):50–61. doi: 10.1016/j.stem.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You J, Reilly GC, Zhen X, Yellowley CE, Chen Q, Donahue HJ, Jacobs CR. Osteopontin gene regulation by oscillatory fluid flow via intracellular calcium mobilization and activation of mitogen-activated protein kinase in MC3T3-E1 osteoblasts. J Biol Chem. 2001;276(16):13365–13371. doi: 10.1074/jbc.M009846200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.