Abstract
DNA double-strand breaks are normal consequences of cell division and differentiation and must be repaired faithfully to maintain genome stability. Two mechanistically distinct pathways are known to efficiently repair double-strand breaks: homologous recombination and Ku-dependent non-homologous end joining. Recently, a third, less characterized repair mechanism, named microhomology-mediated end joining (MMEJ), has received increasing attention. MMEJ repairs DNA breaks via the use of substantial microhomology and always results in deletions. Furthermore, it probably contributes to oncogenic chromosome rearrangements and genetic variation in humans. Here, we summarize the genetic attributes of MMEJ from several model systems and discuss the relationship between MMEJ and ‘alternative end joining’. We propose a mechanistic model for MMEJ and highlight important questions for future research.
MMEJ – erroneous repair of DNA double-strand breaks
Of the many types of DNA damage, DNA double-strand breaks (DSBs) present a unique challenge to cells. On the one hand, DSBs are necessary for such vital processes as meiotic recombination [1] and vertebrate immune system development [2]. On the other hand, their misrepair can create mutations and promote genome instability, and an inability to repair DSBs results in cell death. Two mechanistically distinct sets of pathways have evolved to repair DSBs: (i) homologous recombination (HR; see Glossary); and (ii) non-homologous end joining (NHEJ). HR employs a homologous template as a source from which to copy lost DNA sequences, whereas NHEJ seals the break without relying on a template. HR and NHEJ operate in both competitive and collaborative manners, depending on the repair context and specific attributes of the broken DNA (for a review, see Ref. [3]).
Recent findings implicate a distinct, error-prone mechanism of end joining, dubbed ‘microhomology-mediated end joining’ (MMEJ), in the efficient repair of DSBs created during murine B lymphocyte development (for a review, see Ref. [4]). The foremost distinguishing property of MMEJ is the use of 5–25 base pair (bp) microhomologous sequences during the alignment of broken ends before joining, thereby resulting in deletions flanking the original break. MMEJ is also frequently associated with chromosome abnormalities such as deletions, translocations, inversions and other complex rearrangements [5–8]. In this review, we bring together studies from multiple model systems to describe the unique attributes of MMEJ, compare it to other ‘alternative’ end-joining processes and highlight important unanswered questions regarding its cellular roles and regulation.
Is MMEJ an independent repair pathway?
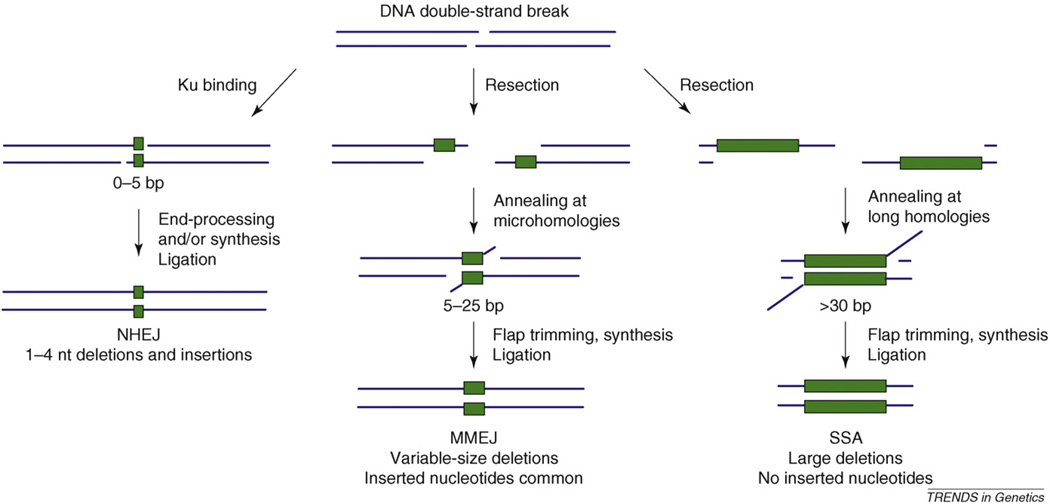
The ‘classical’ or ‘DNA-dependent protein kinase (DNA-PK) dependent’ end-joining pathway, referred to in this review as simply NHEJ, is remarkably flexible in its ability to repair a wide range of DSB substrates (for a review, see Ref. [9]). The core NHEJ factors, as defined in mammals, include the DNA-PK catalytic subunit (DNA-PKcs), the Ku70–Ku80 heterodimer and DNA ligase IV–XRCC4 (X-ray repair complementing defective repair in Chinese hamster cells 4). The Ku heterodimer binds to DNA ends and recruits several accessory factors, including the DNA-PKcs– Artemis nuclease and DNA polymerases μ and λ. These proteins process the broken ends in preparation for ligation by DNA ligase IV–XRCC4. Although both Saccharomyces cerevisiae and Schizosaccharomyces pombe lack DNA-PKcs and Artemis orthologs, the other core NHEJ components are conserved. During NHEJ, annealing of fully complementary single-stranded ends can result in accurate repair. However, most breaks created during normal cellular metabolism do not possess complementary ends and NHEJ frequently proceeds through annealing of short [four or fewer nucleotides (nts)] microhomologous sequences. Subsequent processing of DNA ends leads to small (1–4 nt) deletions and insertions in repair products (Figure 1).
Figure 1.
Comparison of NHEJ, MMEJ and SSA pathways in S. cerevisiae. During NHEJ repair of a DSB, the Ku70–Ku80 heterodimer binds, preventing DNA end resection. Repair proceeds by annealing at short microhomologies (green boxes), fill-in by Pol4 and ligation using DNA ligase IV, resulting in small deletion and insertion products. Both MMEJ and SSA require end resection or unwinding to reveal homologous sequences, although the length of homology required for MMEJ (5–25 bp) is shorter than for SSA. SSA and MMEJ also require 3′ flap cleavage before fill-in synthesis and ligation. Whereas MMEJ products can contain inserted nucleotides, these are never observed in SSA. Notably, although the three pathways share some common genetic requirements, overall they are distinct (Table 1).
The existence of homologous recombination-independent DSB repair pathway(s) that use five or more bases of microhomology during rejoining was demonstrated >20 years ago [10]. Subsequent studies with eukaryotic model systems supplied the first genetic evidence that the repair mechanism generating these events was distinct from NHEJ, although it occurred at a low frequency. Using plasmid rejoining assays (Box 1), two laboratories showed that repair in Ku80-deficient yeast or mice resulted in substantial increases in deletion size and extensive usage of microhomology at repair junctions [11,12]. Since these initial findings, recurrent observations of MMEJ have been reported, although relatively few studies have defined its genetic attributes.
Box 1. MMEJ assays.
Several different assay systems have been employed by researchers studying MMEJ-mediated DSB repair. The major techniques, together with advantages and potential pitfalls, are described here.
Plasmid end-joining assays
MMEJ was originally characterized by transformation of restriction enzyme-linearized plasmids into competent yeast cells and measurement of the percentage of cells that could form colonies on selective media [11,22,24]. Mutations in genes required for NHEJ severely reduced joining activity, but residual MMEJ-mediated repair occurred. Many researchers have subsequently refined the in vivo joining assays to augment MMEJ repair by using linearized plasmids with non-complementary or blunt ends. Plasmid end-joining assays have also been adapted for mammalian cells [12] and for in vitro studies with cell-free extracts [14].
Site-specific endonucleases
The topological and chromatin context of short linear DNA molecules (e.g linearized plasmids) might be different from breaks occurring on chromosomes. These issues have been addressed by using rare-cutting endonucleases (e.g. HO or I-SceI) to create dual, non-complementary breaks in a chromosomal context [26,85,86]. Endonuclease expression can be induced by changing temperature or by adding certain compounds to the growth medium, conferring two additional advantages: cell-cycle dependence of different repair pathways can be investigated and the repair products can be easily amplified by PCR for molecular analysis. Drosophila P-element transposons have also been exploited: P transposase produces DNA breaks with 17-nt non-complementary single-stranded overhangs. In HR-deficient flies, these breaks are readily repaired by DNA ligase IV-independent pathways [37], indicating that they are preferentially repaired by an alternative end-joining process.
Variations on the theme: microhomology-mediated recombination (MHMR)
Linearized plasmids that cannot autonomously replicate in yeast are, nonetheless, able to randomly integrate into the genome. The integration process, called microhomology-mediated recombination (MHMR), is similar to MMEJ in that it frequently uses stretches of microhomology longer than 4 bp and can result in large genomic deletions at integration sites [87]. MHMR is further promoted by agents that induce DSBs, including radiation, I-SceI endonuclease cutting and by restriction enzyme co-transformation. Although it is unknown if MMEJ is also induced by high levels of DNA damage, an increased number of broken DNA ends causes MMEJ to be favored over NHEJ [88]. DNA:protein ratios also have an effect, because more DNA ends favors Ku-independent and microhomology-dependent repair pathways [32,66]. These studies highlight the unanticipated effects that experimental design can have on the results of end-joining studies.
Accordingly, many ambiguities remain concerning the mechanism and purpose of MMEJ. For example, MMEJ has long been considered a ‘back-up’ mechanism to repair DNA breaks when NHEJ and other mechanisms fail [13,14]. Such a premise contributes to the idea that MMEJ might be irrelevant under normal physiological conditions and that, even if it occurs, no definitive genetic pathway can be assigned to it. However, recent studies have demonstrated that MMEJ in mammals is surprisingly robust, at least in certain contexts during immune system development. Class switch recombination (CSR), a process by which one class of antibody is converted to another in activated B lymphocytes, proceeds through the creation of DSBs in the immunoglobulin heavy-chain region. These breaks are normally repaired by NHEJ. However, three groups have now demonstrated that CSR in developing B cells that lack either DNA ligase IV or XRCC4 proceeds by end joining using substantial microhomology [15–17]. MMEJ can also operate efficiently during V(D)J recombination, a site-specific recombination reaction that generates diversity by rearranging V, D and J gene segments in T-cell receptor and antibody molecules. During V(D)J recombination, the Rag1–Rag2 (named for recombination activating gene) protein complex directs formation of DSBs which, like their counterparts in CSR, are usually repaired by NHEJ. Using Rag1 and Rag2 C-terminal truncation mutants, the Roth laboratory recently showed that MMEJ can partially rescue the end-joining defects of cells lacking either DNA-PKcs or XRCC4 [18]. Interestingly, they observed substantial MMEJ activity in wild-type cells expressing truncated Rag proteins, indicating that MMEJ can repair DSBs even when the NHEJ repair pathway is intact.
Whether MMEJ is simply a subclass of single-strand annealing (SSA), a pathway that creates deletions via annealing at directly repeated sequences of >30 bp [19], also remains controversial. In S. cerevisiae, Rad52 is required for efficient SSA. Therefore, the Rad52 dependence of certain, but not all, types of MMEJ events in S. pombe supports a mechanistic relationship between MMEJ and SSA [20]. However, as described later, there are distinct differences in the genetic requirements for MMEJ in comparison to SSA.
Finally, considerable confusion exists about the relationship between MMEJ and ‘alternative end joining’. Alternative end joining is loosely defined as the end-joining activities that operate when core NHEJ components are inactivated. By contrast, MMEJ refers to end-joining events that are mediated by substantial lengths of microhomology. Because MMEJ is active in the absence of certain core NHEJ factors (Ku70–Ku80 or DNA ligase IV), these two terms are currently used without clear distinction. However, not every alternative end-joining event is MMEJ and certain alternative end-joining events are clearly distinct from MMEJ, probably representing yet another end-joining pathway waiting to be defined.
Characterization of yeast MMEJ
Yeast model systems have served as the workhorse for end-joining studies and one of the first formal demonstrations of MMEJ came from assays investigating repair of restriction-enzyme-linearized plasmids that were transformed into Ku80-deficient S. cerevisiae [11] (Box 1). Yeast lacking other NHEJ-related proteins, including Lig4 or any member of the MRX complex (Mre11, Rad50 and Xrs2), also join linearized plasmids or broken chromosomes using MMEJ [21–24]. In S. cerevisiae, MMEJ becomes more efficient with blunt-ended breaks or incompatible overhangs, which are both poor NHEJ substrates. [11,21].
The length of microhomology required for MMEJ in a non-chromosomal context was characterized using oligonucleotide-mediated plasmid circularization, which assessed several parameters of DNA end sequences for joining ability and defined the relevant processing enzymes involved [25]. The results revealed three distinct subtypes of end-joining events: (i) those that depend on Ku70; (ii) those that require Rad52; and (iii) those independent of both Ku70 and Rad52. Ku70-dependent NHEJ predominates when short microhomologies of <5 bp are used for alignment, whereas annealing at homologies longer than 8 bp increasingly requires Rad52 for repair. Joints featuring 6–8 bases of complementary pairing require neither Ku70 nor Rad52, indicating that they represent true MMEJ in this system.
Recently, a comprehensive analysis of genes required for MMEJ was conducted using the yeast HO (homing) endonuclease (Box 1) to simultaneously create two DSBs with complementary or non-complementary ends at a chromosomal locus [26]. Surprisingly, the survival frequency with non-complementary ends was five times higher than with complementary ends, thereby demonstrating that MMEJ can operate efficiently in S. cerevisiae. The predominant MMEJ products that were recovered used eight to ten bases of microhomology and required both the MRX complex and the Rad1–Rad10 3′ flap endonuclease (Table 1). Repair was independent of both Rad52 and Ku70, thus, distinguishing it from SSA and NHEJ. A later study, using mutants of nearly all genes required for either SSA or NHEJ, revealed that S. cerevisiae Nej1 (NHEJ regulator 1), Srs2 (suppressor of Rad 6), Sae2 [homologous to mammalian CtBP-interacting protein (CtIP)] and Tel1 [a protein kinase homologous to mammalian ataxia telangiectasia mutated (ATM)] are involved in either the mechanics or regulation of MMEJ [27] (Table 1). In addition, DNA polymerases (Pols) previously implicated in NHEJ (e.g. Pol4), translesion synthesis (e.g. Rev3 and Rad30) and post-replication repair (e.g. Pol32) were also shown to be important in MMEJ. Together, these results indicate that MMEJ in S. cerevisiae is a molecular amalgam of SSA, NHEJ and other repair pathways. Like its sister pathway NHEJ, MMEJ is extremely versatile.
Table 1.
Proteins involved in the mechanism or regulation of MMEJa
| Protein | Organism in which studied | Proposed function in MMEJ | Involved in other repair pathways? | Refs |
|---|---|---|---|---|
| Mre11–Rad50– Xrs2b |
S. cerevisiae | Resection to expose microhomologies | NHEJ, HR | [26,27] |
| Exo1 | S. pombe, S. cerevisiae | Resection using 5′–3′ exonuclease activity | HR, SSA, MMR | [20,27] |
| Sae2c | S. cerevisiae | Promotion of resection | HR, SSA | [27] |
| Tel1d | S. cerevisiae | Phosphorylation of proteins important for resection |
HR | [27] |
| Srs2 | S. cerevisiae | Removal of Rad51 from single-stranded DNA | HR, SSA | [27] |
| Rad1–Rad10e | S. cerevisiae | Removal of 3′ flaps | SSA, NER, | [26,27,49] |
| Pol4 | S. cerevisiae, S. pombe | Short, fill-in synthesis | NHEJ | [20,27] |
| Pol η, Pol ζ | S. cerevisiae | Fill-in synthesis | TLS | [27] |
| Pol32 (pol δ subunit) |
S. cerevisiae | Synthesis to fill in single-strand gaps | HR | [27] |
| DNA ligase IV | S. cerevisiae | Ligation | NHEJ | [26] |
| DNA ligases I, III | Human and rodent cell lines | Ligation | HR, BER, NER | [50] |
| PARP-1 | Human and rodent cell lines | Synapsis of ends | BER | [52,54] |
| Hisrone H1 | Human cell lines | Unclear, stimulated or inhibited MMEJ in different studies |
N/A | [32,53] |
Abbreviations: BER, base excision repair; HR, homologous recombination; MMR, mismatch repair; NER, nucleotide excision repair; SSA, single-stand annealing; TLS, translesion synthesis.
NBS1 in mammals.
CtIP in mammals.
ATM in mammals.
XPF–ERCC1 in mammals.
In S. pombe, Decottingnies [20] found that circularization of PCR-derived extrachromosomal sequences containing at least 5 bp of microhomology requires Exo1, Rad22 (the Rad52 homolog) and Pol4, but is independent of Lig4 and Rad50. Additionally, Pms1 (elevated post-meiotic segregation 1) and Msh2 (MutS homolog 2) correct mismatches within the microhomologous tracts. The reasons for the discrepancy between MMEJ genetic requirements in budding versus fission yeast are not entirely clear, but might be related to differences in the experimental systems (i.e. intermolecular joining of chromosomal sequences versus intramolecular joining of extrachromosomal plasmids). Interestingly, in both budding and fission yeast, the Ku70–Ku80 heterodimer inhibits MMEJ and increases the extent of microhomology required for repair [20,27]. In ku70 mutants, accelerated single-stranded DNA formation accounts, in part, for increased MMEJ repair.
Alternative end joining in other eukaryotes – MMEJ or a separate repair mechanism?
Although joining of DNA ends using microhomologies was documented in mammals >20 years ago, the first clue that at least some of these events are distinct from NHEJ came from analyses of repair events in KU80−/− and XRCC4−/− cells [12,28,29]. Since then, most research in vertebrates has used biochemical assays that involve fractionating end-joining activities from crude cell extracts in NHEJ-deficient cell lines and identifying the enzymes responsible for the joining activities using simple biochemical means. Such approaches using cell-free extracts from Chinese hamster ovary cells, Xenopus laevis eggs and calf thymus have identified MMEJ-like activities that can rejoin linear DNA fragments [30–34].
Experiments conducted in human cell lines [35] and HeLa cell extracts [14] also provided evidence for a DNA-PKcs- and Ku-independent ‘alternative’ end-joining mechanism that is kinetically distinct from NHEJ, operating in the order of hours rather than minutes. Unlike MMEJ, this alternative end joining resulted in a large proportion of accurately repaired breaks, rarely resulted in deletions and frequently occurred with little or no complementary base pairing. Although these differences could, theoretically, result from an in vitro kinetic rejoining advantage afforded to the alternative-joining pathway [14], the results raise the important question of whether alternative end joining is functionally equivalent to MMEJ or whether it represents one or more distinct Ku-independent end-joining mechanisms that remain to be characterized. The latter hypothesis is bolstered by the recent observation that, whereas most breaks induced by the yeast site-specific homing endonuclease I-SceI in KU70−/− mammalian cells are repaired via MMEJ, some end-joining repair is microhomology-independent [36].
MMEJ-like repair events have also been reported in other model systems. DSB repair resulting from P transposable element excision in Drosophila is efficient in HR-deficient flies lacking DNA ligase IV, indicating the presence of a robust alternative end-joining pathway [37]. These repair events frequently use microhomologies but rarely create large deletions. A potential explanation for this occurrence lies in the nature of P-element-induced breaks, which possess 17-nt overhangs [38] with several regions of 3–8 nt microhomologies that can be used without a requirement for additional resection. By contrast, the repair of I-SceI-induced breaks in ku70 and lig4 mutant flies frequently results in large flanking deletions, although the usage of microhomologies during repair has not been thoroughly investigated [39].
Interestingly, a sizeable proportion of end-joining events in Drosophila involve de novo insertion of nucleotides at the junction [37]. Potential templates for these inserted nucleotides can frequently be identified in adjacent DNA, although the insertions are often imperfect copies of the flanking sequence. It is possible that these templated nucleotides (T nucleotides) might be used during repair as sources of microhomology when flanking microhomologous sequences are inadequate. Notably, similar inserted sequences are also observed after repair of ionizing-radiation-induced breaks in Arabidopsis thaliana [40] and I-SceI-induced breaks in tobacco plants [41,42].
A mechanistic model for MMEJ
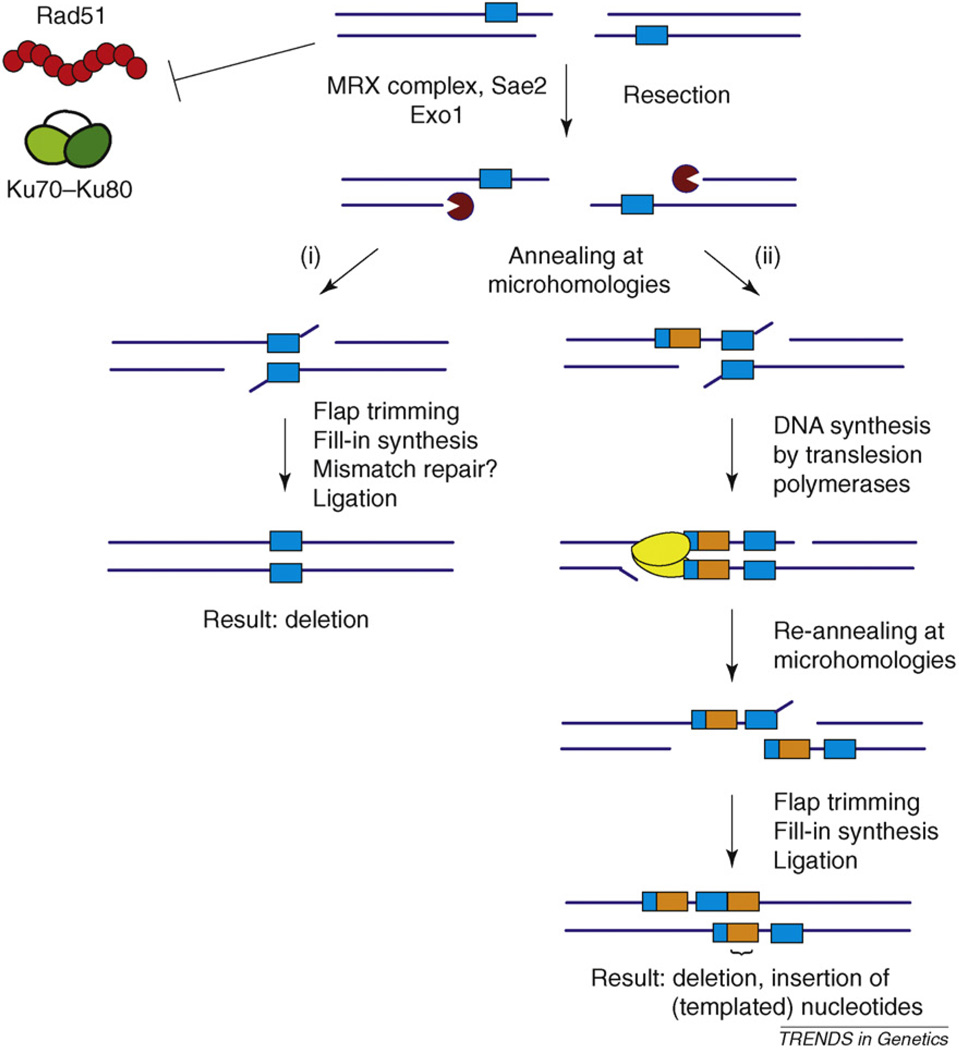
Based on studies from many organisms, we propose the following model, which predicts the intermediate steps of MMEJ and the biochemical activities of proteins involved in MMEJ (Table 1 and Figure 2).
Figure 2.
Model for MMEJ and alternative end-joining repair. During the initial stages of MMEJ, Ku70–Ku80 (green) and Rad51 (red), which inhibit MMEJ, are prevented from binding or are removed. This enables 5––3′ resection by the MRX complex, Sae2 and Exo1 (indicated by dark red partial circle) that reveals microhomologous sequences (blue boxes). These microhomologies transiently and dynamically anneal to each other. (i) In cases in which the annealing is stable, repair is completed by flap trimming, fill-in DNA synthesis and ligation, resulting in a deletion relative to the original sequence. Mismatch repair is not required for MMEJ, although it might have a supporting role. (ii) Alternatively, one or more translesion polymerases (yellow) can extend the annealed sequences (represented here by orange–blue boxes) using templated error-prone synthesis. Dissociation of the initial microhomologies and realignment at other microhomologous sequences, followed by flap trimming, fill-in DNA synthesis and ligation completes repair, resulting in a deletion plus insertion event. Many variations and iterations of (ii) can hypothetically occur, resulting in complex insertion–deletion junctions.
End resection
To expose proper microhomologies that mediate end annealing before joining, nucleolytic DNA end processing is probably required [27]. InS. cerevisiae, the MRX complex has been implicated in end processing, because Mre11 nuclease activity is essential for MMEJ [26,27]. In S. pombe, Exo1 is required for MMEJ and probably participates in the nucleolytic processing step [20]. This hypothesis is supported by observations that Exo1 overproduction can partially rescue the MMEJ defect of MRE11-deficient S. cerevisiae; moreover, the deletion of both MRE11 and EXO1 further reduces MMEJ frequency [27]. Recent findings indicate that mammalian CtIP, which is important for DNA end processing, can promote MMEJ-like repair [36]. The S. cerevisiae ATM and CtIP orthologues, Tel1 and Sae2, respectively, also stimulate end resection and promote MMEJ and HR, although they discourage NHEJ [27,43,44].
Studies in S. pombe have demonstrated that MMEJ and NHEJ might be cell-cycle regulated, such that NHEJ preferentially occurs at G1, whereas MMEJ predominates at S and G2 [20]. Substantial evidence shows that end resection is limited primarily to non-G1 cells because end resection requires Cdk1 cell-cycle-regulated kinase activity [45–47]. It is, thus, logical to propose that MMEJ occurs preferentially in non-G1 cells. Nevertheless, limited end resection occurs (albeit with slower kinetics) in G1-arrested cells and could be sufficient for most MMEJ processes [47,48]. In addition, DNA helicase activity might provide a mechanism to expose microhomologous sequences that are located far from the initial break.
Annealing of single-stranded DNA
The microhomology exposed by DNA resection or unwinding anneals to form a key MMEJ intermediate. In budding yeast, in which single-stranded DNA is coated with the Rad51 strand exchange protein, Srs2 promotes efficient MMEJ by displacing Rad51 [27]. During annealing, certain microhomologies seem to be preferentially used; for example, distinct 8–22 bp pair imperfect microhomologies are frequently used in both S. cerevisiae and Drosophila [6,26,37]. The ability of GC-rich microhomology to bypass the requirement of various canonical NHEJ factors during end joining indicates that at least one important attribute of microhomology is to provide base-pairing stability [25]. Nonetheless, why a given microhomology (sometimes possessing mismatches) is chosen over many others is currently unknown and is a crucial question for future research. Chromatin structure or other non-sequence-based features could potentially have important roles in determining how microhomology use is regulated.
Processing of DNA ends before ligation
After microhomology annealing, remaining non-complementary DNA segments form 3′ flaps that must be removed before ligation. Removal of 3′ flaps also assists in stable association of DNA ends and provides a proper substrate from which DNA synthesis can initiate for gap filling. Mirroring its role in the SSA pathway, the S. cerevisiae Rad1–Rad10 structure-specific endonuclease and its mammalian counterpart, xeroderma pigmentosum complementation group F (XPF)–excision repair cross complementation group 1 (ERCC1), has a key role in MMEJ by cleaving 3′ flaps from an annealed intermediate [26,27,49].
The presence of inserted nucleotides at many MMEJ junctions indicates that error-prone polymerases are frequently involved in the processing stages of repair. In both S. cerevisiae and S. pombe, Pol4 activity is necessary for normal levels of both MMEJ and NHEJ [20,27]. The translesion DNA polymerases Polη and Polζ also participate in MMEJ in S. cerevisiae [27]. Although Drosophila lacks a Pol4 orthologue, another translesion polymerase, such as Polθ, might be involved in T-nucleotide insertion during alternative end joining. One speculative model for the generation of T nucleotides involves an initial microhomology annealing step followed by extension by an error-prone polymerase, subsequent dissociation of the nascent DNA from its template and re-annealing at additional regions of microhomology (Figure 2). Such a model predicts that the protein–DNA complexes present during MMEJ are dynamic; moreover, it is consistent with MMEJ pathway flexibility. Interestingly, deletion of yeast POL32, a nonessential Polδ subunit, causes one of the most severe decreases in MMEJ repair frequency [27], thereby implicating Polδ in MMEJ-associated gap filling.
Ligation
Ligation of annealed and fully processed joints is achieved by two DNA ligases in yeast, Ligase I and Ligase IV (encoded by CDC9 and DNL4, respectively). Deletion of DNL4 or its accessory factor NEJ1 reduces MMEJ repair approximately by half but does not completely eliminate it, which is consistent with a role for DNA ligase I in MMEJ in yeast [26,27]. In human cells, DNA ligase I is also involved in MMEJ repair that uses 10 bp microhomologies [50]. Many eukaryotes possess a third ligase, DNA ligase IIIα, which, together with its partner XRCC1, seals single-strand breaks during base excision repair [51]. In principle, the MMEJ ligation step resembles the ligation of two single-strand break intermediates in close proximity [25]; thus, the participation of DNA ligase IIIα in MMEJ seems feasible. Indeed, multiple studies have provided substantial support for the involvement of DNA ligase IIIα and another base excision repair protein, poly(ADP-ribose) polymerase-1 (PARP-1), in both MMEJ and alternative end joining [50,52–54]. Intriguingly, DNA ligase IIIα is upregulated in chronic myeloid leukemia cell lines that are positive for the B cell receptor (BCR)–Abelson murine leukemia viral oncogene homolog (ABL) translocation [55]. Both Artemis and DNA ligase IV are also down-regulated in these cells, which is consistent with the observation that ~80% of DSBs in these cells are repaired using microhomologies.
In instances in which DNA ligase IIIα participates in the MMEJ ligation step, it remains unknown whether it functions independently or in concert with other protein partners (including XRCC1). As an XRCC1-deficient cell line shows no significant defect in rejoining efficiency or microhomology usage, DNA ligase IIIα might function independently of XRCC1 in MMEJ [56]. However, DNA ligase IIIα co-localizes with the Werner syndrome helicase-exonuclease (WRN) in nuclear foci at I-SceI-induced DSBs, indicating that it might partner with WRN and, possibly, other accessory proteins during MMEJ [55].
MMEJ regulation
Increasing evidence indicates that certain NHEJ-specific proteins might negatively regulate MMEJ. For example, the end-binding activity of the Ku heterodimer probably inhibits nucleolytic degradation, thus, rendering MMEJ more frequent in Ku-deficient cells [20,27,36,56]. A similar suppressive effect on MMEJ by DNA-PKcs occurs in vertebrates [57]. In addition, PARP-1 competes with Ku proteins for binding to broken DNA ends, perhaps promoting their synapsis, thereby directing breaks into an alternative end-joining pathway [54,58]. Ku proteins might also promote tethering of broken DNA ends [59]; this tethering might limit microhomology annealing after DNA cleavage. In cases in which NHEJ initiates, but is unable to proceed, Ku80 might be polyubiquitylated and, subsequently, dissociated from the break site [60], thereby enabling nucleases, helicases, polymerases and other MMEJ components to access the DNA ends.
Other proteins that regulate MMEJ prevalence have been identified, although their roles remain somewhat ambiguous. For example, histone H1 inhibited MMEJ repair in one study [32] but stimulated alternative end joining in a different system [53]. One explanation for the different results could be that the substrates used in the latter study did not require the use of large microhomologies for joining, thus, highlighting the need to distinguish between MMEJ and other alternative end-joining pathways. The Chk2 protein kinase and breast cancer 1 (BRCA1), a Chk2 substrate, also affect microhomology usage at DSB repair junctions and, thus, modulate MMEJ repair [61–63]. One likely explanation for these observations comes from new data indicating that a Brca1–CtIP–Mre11 complex facilitates DNA end resection to form single-stranded DNA [64].
Finally, interesting connections exist between MMEJ, mammalian DNA interstrand crosslink (ICL) repair pathways and the Fanconi anemia (FA) genes. FA is a hereditary disease caused by mutation of any one of thirteen FA genes and cell lines from FA patients are extremely sensitive to chemicals that induce ICLs (for a review, see Ref. [65]). Intriguingly, certain FA cell lines are deficient in DNA–PKcs-independent plasmid joining involving microhomologies [66], consistent with a role for FA genes in MMEJ. In addition, two Chinese hamster ovary cell lines that are sensitive to the crosslinking agent mitomycin C also demonstrate reduced MMEJ-mediated DSB repair [56]. Additional investigations into components that might be shared by both MMEJ and ICL repair pathways could provide new insights into the mechanisms and regulation of both.
Roles of MMEJ in chromosome instability and genetic variation
In mammals, MMEJ is frequently associated with chromosome rearrangements and cancer. Mice lacking both p53 and either DNA ligase IV, Ku70 or Ku80 develop pro-B cell lymphomas and accumulate nonreciprocal chromosome translocations, frequently involving multiple chromosomes. Sequencing of the translocation breakpoints provides evidence for 2–8 nt microhomologies at the junctions, indicating that MMEJ is probably responsible for at least some of these events [67–69]. Similarly, a large percentage of characterized chromosome translocation breakpoints from human cancers, including bladder cancers and leukemias, contain extensive microhomology that implicates MMEJ in their production [70–72] (Table 2). Interestingly, many translocations isolated from follicular and mantle cell lymphoma patients are associated with T-nucleotide insertions that frequently have mismatches relative to their presumptive templates, which are similar to those observed in alternative end joining in flies and plants (for a review, see Ref. [73]).
Table 2.
Table adapted, with permission, from Ref. [5].
Sequences surrounding t(9;22)(q34;q11) translocation breakpoints of BCR–ABL fusions from human leukemias.
Sequences that are physically present in each translocation are underlined. Each translocation is, therefore, composed of the 5′ BCR underlined sequence and the 3′ ABL underlined sequence. Breakpoints are designated by a forward slash (/). Microhomologous sequences flanking the translocation breakpoints are bolded and highlighted in yellow.
MMEJ also seems to be involved in telomere fusions in certain contexts in which telomere integrity is compromised. End-to-end fusion of shortened telomeres in A. thaliana lacking Ku or DNA ligase IV relies partially on Mre11 and is mediated by microhomology at the junctions [74]. In Drosophila, telomere fusions can also occur independently of both DNA ligase IV and Ku (for a review, see Ref. [75]), although, whether the fusions involve microhomologies is currently unknown. Recent findings show that mice with shortened telomeres, owing to telomerase inactivation, accumulate telomere fusions independently of DNA ligase IV [76]. Together, these studies indicate that the involvement of alternative end-joining pathways in telomere joining could be widespread.
Finally, MMEJ probably contributes to genetic variation within populations. Several genetic diseases are caused by genomic deletions with imperfect microhomologies present at their breakpoints (Table 3). In the human genome, extensive structural variations (SVs) exist that involve kilobase- to megabase-sized deletions, duplications and complex rearrangements [77]. Using massive paired-end mapping, Korbel et al. [77] mapped >1000 SVs and determined the breakpoint junctions of 188 simple insertions or deletions. >50% of the junctions seemed to have been created through end joining and a sizeable fraction of these involved joining at microhomologies of 5–8 nts, thus, indicating a potential role for MMEJ. Intriguingly, two recent studies demonstrated that an MMEJ-like process might also be involved in chemotherapeutic drug resistance in patients and cell lines harboring mutations in the breast cancer 2 (BRCA2) gene [78,79]. In many cases, inframe deletions flanking the original brca2 mutation occurred in the drug-resistant cells and these deletions encompassed microhomologies of five or more nucleotides [78]
Table 3.
MMEJ and human disease: sequences flanking deletion breakpoints from selected human diseasesa
| Breakpoint sequencesb | Gene | Disease | Refs |
|---|---|---|---|
| CFTR | Cystic fibrosis | [81] | |
| GALC | Krabbe | [82] | |
| RB | Retinoblastoma | [83] | |
| RB | Retinoblastoma | [83] | |
| BRCA2 | Pancreatic cancer cell line | [78] | |
| BRCA2 | Pancreatic cancer cell line | [78] | |
| Pancreatic cancer cell line | [78] | ||
| BRCA2 | Ovarian cancer | [78] |
Table adapted, with permission, from Ref. [5].
Sequences that are physically present on each deletion chromosome are underlined, whereas sequences at the 3′ end of each deletion are bracketed. Microhomologous sequences flanking the deletions are bolded and highlighted in yellow.
Concluding remarks and future perspectives
When MMEJ was initially characterized, it seemed to be a repair option that was used only when the dominant NHEJ pathway was compromised. However, recent investigations have demonstrated that MMEJ can operate even in the context of intact NHEJ and HR repair. This realization, combined with observations that MMEJ repair is often associated with genomic instability and cancer, prompted systematic studies that have begun to define its genetic underpinnings.
Notably, many questions remain unresolved (Box 2). First, the extent to which MMEJ contributes to overall DSB repair in NHEJ-proficient cells must be quantitated. Identifying the preferred MMEJ substrates should be helpful in such studies. MMEJ probably assumes a more important role in break repair if DNA ends are not readily compatible; such DNA ends are generated by ionizing radiation (for a review, see Ref. [80]) and during P-element transposition in Drosophila. In cases in which terminal microhomology is insufficient for MMEJ repair, embedded microhomologies internal to the break site could be used. Therefore, in vivo systems inducing DNA breaks that are preferentially and efficiently repaired by MMEJ or in vitro assays, which reconstitute MMEJ with a variety of substrates, will expedite its genetic and kinetic analysis. Given its dynamic nature, it seems likely that a core set of proteins carry out ‘simple’ MMEJ but that additional factors, especially the end processing enzymes, might modulate and regulate the reaction depending on the substrate that is presented [9]. It will be of particular interest to determine the origin of inserted nucleotides that are found at the junctions of many alternative end-joining products and to characterize the polymerase(s) responsible for their generation.
Box 2. MMEJ mysteries.
What is the relationship between MMEJ defined in yeast model systems and alternative end joining observed in multicellular eukaryotes?
Do multiple, distinct, NHEJ-independent alternative end-joining mechanisms exist?
Does MMEJ frequency differ between Ku-deficient and ligase IV-deficient cells?
Does MMEJ contribute substantially to double-strand break repair in non-lymphoid cells when NHEJ is functional?
Are there preferred substrates for MMEJ and how do these preferences affect its usage in different contexts?
What mechanism(s) are responsible for the generation and insertion of T-nucleotides observed in some alternative end-joining products?
Is MMEJ subject to regulation and, if so, how does this regulation ensure minimum genome instability?
Finally, whether the usage of MMEJ varies in different tissues and at different times during development needs to be determined. The prevalence of MMEJ involvement in bladder cancers and certain leukemias indicates potential bias towards MMEJ-mediated DSB repair in specific tissues, a hypothesis which has potential therapeutic implications. Furthermore, a more complete understanding of the cellular contexts which favor MMEJ might serve to improve cancer treatments, as demonstrated by the potential role of MMEJ in chemotherapy-induced brca2 mutation reversion [78,79]. These examples highlight the importance of clearly defining the roles and regulation of MMEJ in cancer and other genome destabilizing processes.
Acknowledgments
We thank F. Alt, J. Haber, G. Ira, T. Paull, T. Wilson, A. Yu and members of the Lee and McVey laboratories for insightful discussions that informed the review. We apologize to our colleagues whose work was not directly cited owing to space limitations. M.M. is supported by the Ellison Medical Foundation and by NSF grant MCB-0643253. S.E.L. is supported by NIH and the Texas Advanced Research Program and is a scholar of the Leukemia and Lymphoma Society.
Glossary
- Alternative end joining
end-joining repair in the absence of classical end-joining (NHEJ) repair factors such as DNA ligase IV and Ku70–Ku80. Because MMEJ is frequently observed in NHEJ-deficient cells and organisms, MMEJ probably constitutes one of several alternative end-joining mechanisms.
- DNA-PK-dependent non-homologous end joining (NHEJ)
also referred to as ‘classical end joining’. NHEJ relies on the Ku70–Ku80 and DNA ligase IV–XRCC4 heterodimers to repair DNA double-strand breaks. Repair can be accurate or it can result in small insertions or deletions at the junction site.
- Homologous recombination (HR)
repair that primes DNA synthesis by invading a homologous template to accurately restore DNA sequence at a double-strand break. Homologous sequences can be from a sister chromatid (after DNA replication), a homolog (in diploid organisms) or a duplicated sequence on the same or different chromosome. HR is essential for faithful meiotic chromosome segregation.
- Interstrand crosslinks (ICLs)
DNA lesions in which covalent bonds join two nucleotides on opposite strands of DNA. ICLs can be repaired by several different mechanisms in eukaryotes, which might involve HR and/or translesion polymerases.
- Microhomology-mediated end joining (MMEJ)
Ku-independent end joining mediated by base pairing between microhomologous sequences of approximately 5–25 nts. MMEJ always results in deletions and is frequently associated with chromosome translocations.
- Single-strand annealing (SSA)
a double-strand break repair pathway that proceeds by annealing of long (>30 nts) homologous sequences. Like MMEJ, it causes genomic deletions, but unlike MMEJ, SSA in yeast requires the Rad52 protein.
- Templated nucleotides (T nucleotides)
nucleotide sequences that are inserted at double-strand break repair junctions. If potential templates for the inserted sequence can be identified (frequently in nearby flanking sequence), the inserts are called T nucleotides. Inserted nucleotides for which templates cannot be identified are sometimes called ‘filler DNA’.
- Translesion polymerases
error-prone DNA polymerases that insert nucleotides across from lesions that block replicative polymerases. Each translesion polymerase has one or more ‘preferred’ lesions. Recent studies indicate that several translesion polymerases are involved in MMEJ in S. cerevisiae.
References
- 1.Keeney S, Neale MJ. Initiation of meiotic recombination by formation of DNA double-strand breaks: mechanism and regulation. Biochem. Soc. Trans. 2006;34:523–525. doi: 10.1042/BST0340523. [DOI] [PubMed] [Google Scholar]
- 2.Jung D, et al. Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Annu. Rev. Immunol. 2006;24:541–570. doi: 10.1146/annurev.immunol.23.021704.115830. [DOI] [PubMed] [Google Scholar]
- 3.Shrivastav M, et al. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008;18:134–147. doi: 10.1038/cr.2007.111. [DOI] [PubMed] [Google Scholar]
- 4.Nussenzweig A, Nussenzweig MC. A backup DNA repair pathway moves to the forefront. Cell. 2007;131:223–225. doi: 10.1016/j.cell.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 5.Chen C, et al. Chromosomal rearrangements occur in S. cerevisiae rfa1 mutator mutants due to mutagenic lesions processed by double-strand-break repair. Mol. Cell. 1998;2:9–22. doi: 10.1016/s1097-2765(00)80109-4. [DOI] [PubMed] [Google Scholar]
- 6.Yu X, Gabriel A. Ku-dependent and Ku-independent end-joining pathways lead to chromosomal rearrangements during double-strand break repair in Saccharomyces cerevisiae. Genetics. 2003;163:843–856. doi: 10.1093/genetics/163.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weinstock DM, et al. Formation of NHEJ-derived reciprocal chromosomal translocations does not require Ku70. Nat. Cell Biol. 2007;9:978–981. doi: 10.1038/ncb1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Welcker AJ, et al. Involvement of very short DNA tandem repeats and the influence of the RAD52 gene on the occurrence of deletions in Saccharomyces cerevisiae. Genetics. 2000;156:549–557. doi: 10.1093/genetics/156.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lieber MR, et al. Flexibility in the order of action and in the enzymology of the nuclease, polymerases, and ligase of vertebrate nonhomologous DNA end joining: relevance to cancer, aging, and the immune system. Cell Res. 2008;18:125–133. doi: 10.1038/cr.2007.108. [DOI] [PubMed] [Google Scholar]
- 10.Roth DB, Wilson JH. Nonhomologous recombination in mammalian cells: role for short sequence homologies in the joining reaction. Mol. Cell. Biol. 1986;6:4295–4304. doi: 10.1128/mcb.6.12.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boulton SJ, Jackson SP. Identification of a Saccharomyces cerevisiae Ku80 homologue: roles in DNA double strand break rejoining and in telomeric maintenance. Nucleic Acids Res. 1996;24:4639–4648. doi: 10.1093/nar/24.23.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liang F, et al. Chromosomal double-strand break repair in Ku80-deficient cells. Proc. Natl. Acad. Sci. U. S. A. 1996;93:8929–8933. doi: 10.1073/pnas.93.17.8929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lieber MR, et al. The mechanism of vertebrate nonhomologous DNA end joining and its role in V(D)J recombination. DNA Repair (Amst.) 2004;3:817–826. doi: 10.1016/j.dnarep.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 14.Wang H, et al. Biochemical evidence for Ku-independent backup pathways of NHEJ. Nucleic Acids Res. 2003;31:5377–5388. doi: 10.1093/nar/gkg728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pan-Hammarstrom Q, et al. Impact of DNA ligase IV on nonhomologous end joining pathways during class switch recombination in human cells. J. Exp. Med. 2005;201:189–194. doi: 10.1084/jem.20040772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soulas-Sprauel P, et al. Role for DNA repair factor XRCC4 in immunoglobulin class switch recombination. J. Exp. Med. 2007;204:1717–1727. doi: 10.1084/jem.20070255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan CT, et al. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 2007;449:478–482. doi: 10.1038/nature06020. [DOI] [PubMed] [Google Scholar]
- 18.Corneo B, et al. Rag mutations reveal robust alternative end joining. Nature. 2007;449:483–486. doi: 10.1038/nature06168. [DOI] [PubMed] [Google Scholar]
- 19.Sugawara N, et al. DNA length dependence of the single-strand annealing pathway and the role of Saccharomyces cerevisiae RAD59 in double-strand break repair. Mol. Cell. Biol. 2000;20:5300–5309. doi: 10.1128/mcb.20.14.5300-5309.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Decottignies A. Microhomology-mediated end joining in fission yeast is repressed by pku70 and relies on genes involved in homologous recombination. Genetics. 2007;176:1403–1415. doi: 10.1534/genetics.107.071621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang X, Paull TT. The Mre11/Rad50/Xrs2 complex and non-homologous end-joining of incompatible ends in S. cerevisiae. DNA Repair (Amst.) 2005;4:1281–1294. doi: 10.1016/j.dnarep.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 22.Wilson TE, et al. Yeast DNA ligase IV mediates nonhomologous DNA end joining. Nature. 1997;388:495–498. doi: 10.1038/41365. [DOI] [PubMed] [Google Scholar]
- 23.Moore JK, Haber JE. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol. Cell. Biol. 1996;16:2164–2173. doi: 10.1128/mcb.16.5.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schar P, et al. A newly identified DNA ligase of Saccharomyces cerevisiae involved in RAD52-independent repair of DNA double-strand breaks. Genes Dev. 1997;11:1912–1924. doi: 10.1101/gad.11.15.1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Daley JM, Wilson TE. Rejoining of DNA double-strand breaks as a function of overhang length. Mol. Cell. Biol. 2005;25:896–906. doi: 10.1128/MCB.25.3.896-906.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma JL, et al. Yeast Mre11 and Rad1 proteins define a Ku-independent mechanism to repair double-strand breaks lacking overlapping end sequences. Mol. Cell. Biol. 2003;23:8820–8828. doi: 10.1128/MCB.23.23.8820-8828.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee K, Lee SE. Saccharomyces cerevisiae Sae2- and Tel1-dependent single-strand DNA formation at DNA break promotes microhomology-mediated end joining. Genetics. 2007;176:2003–2014. doi: 10.1534/genetics.107.076539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bogue MA, et al. V(D)J recombination in Ku86-deficient mice: distinct effects on coding, signal, and hybrid joint formation. Immunity. 1997;7:37–47. doi: 10.1016/s1074-7613(00)80508-7. [DOI] [PubMed] [Google Scholar]
- 29.Kabotyanski EB, et al. Double-strand break repair in Ku86-and XRCC4-deficient cells. Nucleic Acids Res. 1998;26:5333–5342. doi: 10.1093/nar/26.23.5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuhfittig-Kulle S, et al. The mutagenic potential of nonhomologous end joining in the absence of the NHEJ core factors Ku70/80, DNA-PKcs and XRCC4-LigIV. Mutagenesis. 2007;22:217–233. doi: 10.1093/mutage/gem007. [DOI] [PubMed] [Google Scholar]
- 31.Feldmann E, et al. DNA double-strand break repair in cell-free extracts from Ku80-deficient cells: implications for Ku serving as an alignment factor in non-homologous DNA end joining. Nucleic Acids Res. 2000;28:2585–2596. doi: 10.1093/nar/28.13.2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang L, et al. Modulation of DNA end joining by nuclear proteins. J. Biol. Chem. 2005;280:31442–31449. doi: 10.1074/jbc.M503776200. [DOI] [PubMed] [Google Scholar]
- 33.Gottlich B, et al. Rejoining of DNA double-strand breaks in vitro by single-strand annealing. Eur. J. Biochem. 1998;258:387–395. doi: 10.1046/j.1432-1327.1998.2580387.x. [DOI] [PubMed] [Google Scholar]
- 34.Sandoval A, Labhart P. Joining of DNA ends bearing nonmatching 3′-overhangs. DNA Repair (Amst.) 2002;1:397–410. doi: 10.1016/s1568-7864(02)00013-7. [DOI] [PubMed] [Google Scholar]
- 35.DiBiase SJ, et al. DNA-dependent protein kinase stimulates an independently active, nonhomologous, end-joining apparatus. Cancer Res. 2000;60:1245–1253. [PubMed] [Google Scholar]
- 36.Bennardo N, et al. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 2008;4:e1000110. doi: 10.1371/journal.pgen.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McVey M, et al. End-joining repair of double-strand breaks in Drosophila melanogaster is largely DNA ligase IV independent. Genetics. 2004;168:2067–2076. doi: 10.1534/genetics.104.033902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beall EL, Rio DC. Drosophila P element transposase is a novel site-specific endonuclease. Genes Dev. 1997;11:2137–2151. doi: 10.1101/gad.11.16.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Preston CR, et al. Differential usage of alternative pathways of double-strand break repair in Drosophila. Genetics. 2006;172:1055–1068. doi: 10.1534/genetics.105.050138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shirley BW, et al. Effects of ionizing radiation on a plant genome: analysis of two Arabidopsis transparent testa mutations. Plant Cell. 1992;4:333–347. doi: 10.1105/tpc.4.3.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salomon S, Puchta H. Capture of genomic and T-DNA sequences during double-strand break repair in somatic plant cells. EMBO J. 1998;17:6086–6095. doi: 10.1093/emboj/17.20.6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gorbunova V, Levy AA. Non-homologous DNA end joining in plant cells is associated with deletions and filler DNA insertions. Nucleic Acids Res. 1997;25:4650–4657. doi: 10.1093/nar/25.22.4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mantiero D, et al. Dual role for Saccharomyces cerevisiae Tel1 in the checkpoint response to double-strand breaks. EMBO Rep. 2007;8:380–387. doi: 10.1038/sj.embor.7400911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clerici M, et al. The Saccharomyces cerevisiae Sae2 protein promotes resection and bridging of double strand break ends. J. Biol. Chem. 2005;280:38631–38638. doi: 10.1074/jbc.M508339200. [DOI] [PubMed] [Google Scholar]
- 45.Ira G, et al. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature. 2004;431:1011–1017. doi: 10.1038/nature02964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ferreira MG, Cooper JP. Two modes of DNA double-strand break repair are reciprocally regulated through the fission yeast cell cycle. Genes Dev. 2004;18:2249–2254. doi: 10.1101/gad.315804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aylon Y, et al. The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. EMBO J. 2004;23:4868–4875. doi: 10.1038/sj.emboj.7600469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu D, et al. Recruitment and dissociation of nonhomologous end joining proteins at a DNA double-strand break in Saccharomyces cerevisiae. Genetics. 2008;178:1237–1249. doi: 10.1534/genetics.107.083535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ahmad A, et al. ERCC1-XPF endonuclease facilitates DNA double-strand break repair. Mol. Cell. Biol. 2008;28:5082–5092. doi: 10.1128/MCB.00293-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liang L, et al. Human DNA ligases I and III, but not ligase IV, are required for microhomology-mediated end joining of DNA double-strand breaks. Nucleic Acids Res. 2008;36:3297–3310. doi: 10.1093/nar/gkn184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Caldecott KW, et al. An interaction between the mammalian DNA repair protein XRCC1 and DNA ligase III. Mol. Cell. Biol. 1994;14:68–76. doi: 10.1128/mcb.14.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Audebert M, et al. Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J. Biol. Chem. 2004;279:55117–55126. doi: 10.1074/jbc.M404524200. [DOI] [PubMed] [Google Scholar]
- 53.Rosidi B, et al. Histone H1 functions as a stimulatory factor in backup pathways of NHEJ. Nucleic Acids Res. 2008;36:1610–1623. doi: 10.1093/nar/gkn013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang M, et al. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res. 2006;34:6170–6182. doi: 10.1093/nar/gkl840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sallmyr A, et al. Up-regulation of WRN and DNA ligase IIIalpha in chronic myeloid leukemia: consequences for the repair of DNA double-strand breaks. Blood. 2008;112:1413–1423. doi: 10.1182/blood-2007-07-104257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Verkaik NS, et al. Different types of V(D)J recombination and end-joining defects in DNA double-strand break repair mutant mammalian cells. Eur. J. Immunol. 2002;32:701–709. doi: 10.1002/1521-4141(200203)32:3<701::AID-IMMU701>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 57.Perrault R, et al. Backup pathways of NHEJ are suppressed by DNA-PK. J. Cell. Biochem. 2004;92:781–794. doi: 10.1002/jcb.20104. [DOI] [PubMed] [Google Scholar]
- 58.Audebert M, et al. Effect of double-strand break DNA sequence on the PARP-1 NHEJ pathway. Biochem. Biophys. Res. Commun. 2008;369:982–988. doi: 10.1016/j.bbrc.2007.11.132. [DOI] [PubMed] [Google Scholar]
- 59.Soutoglou E, et al. Positional stability of single double-strand breaks in mammalian cells. Nat. Cell Biol. 2007;9:675–682. doi: 10.1038/ncb1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Postow L, et al. Ku80 removal from DNA through double strand break-induced ubiquitylation. J. Cell Biol. 2008;182:467–479. doi: 10.1083/jcb.200802146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang J, et al. Chk2 phosphorylation of BRCA1 regulates DNA double-strand break repair. Mol. Cell. Biol. 2004;24:708–718. doi: 10.1128/MCB.24.2.708-718.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhuang J, et al. Checkpoint kinase 2-mediated phosphorylation of BRCA1 regulates the fidelity of nonhomologous end-joining. Cancer Res. 2006;66:1401–1408. doi: 10.1158/0008-5472.CAN-05-3278. [DOI] [PubMed] [Google Scholar]
- 63.Zhong Q, et al. BRCA1 facilitates microhomology-mediated end joining of DNA double strand breaks. J. Biol. Chem. 2002;277:28641–28647. doi: 10.1074/jbc.M200748200. [DOI] [PubMed] [Google Scholar]
- 64.Chen L, et al. Cell cycle-dependent complex formation of BRCA1·CtIP·MRN is important for DNA double-strand break repair. J. Biol. Chem. 2008;283:7713–7720. doi: 10.1074/jbc.M710245200. [DOI] [PubMed] [Google Scholar]
- 65.Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat. Rev. Genet. 2007;8:735–748. doi: 10.1038/nrg2159. [DOI] [PubMed] [Google Scholar]
- 66.Lundberg R, et al. Deficient DNA end joining activity in extracts from fanconi anemia fibroblasts. J. Biol. Chem. 2001;276:9543–9549. doi: 10.1074/jbc.M008634200. [DOI] [PubMed] [Google Scholar]
- 67.Ferguson DO, et al. The nonhomologous end-joining pathway of DNA repair is required for genomic stability and the suppression of translocations. Proc. Natl. Acad. Sci. U. S. A. 2000;97:6630–6633. doi: 10.1073/pnas.110152897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Difilippantonio MJ, et al. DNA repair protein Ku80 suppresses chromosomal aberrations and malignant transformation. Nature. 2000;404:510–514. doi: 10.1038/35006670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhu C, et al. Unrepaired DNA breaks in p53-deficient cells lead to oncogenic gene amplification subsequent to translocations. Cell. 2002;109:811–821. doi: 10.1016/s0092-8674(02)00770-5. [DOI] [PubMed] [Google Scholar]
- 70.Bentley J, et al. DNA double strand break repair in human bladder cancer is error prone and involves microhomology-associated end-joining. Nucleic Acids Res. 2004;32:5249–5259. doi: 10.1093/nar/gkh842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang Y, Rowley JD. Chromatin structural elements and chromosomal translocations in leukemia. DNA Repair (Amst.) 2006;5:1282–1297. doi: 10.1016/j.dnarep.2006.05.020. [DOI] [PubMed] [Google Scholar]
- 72.Mattarucchi E, et al. Microhomologies and interspersed repeat elements at genomic breakpoints in chronic myeloid leukemia. Genes Chromosomes Cancer. 2008;47:625–632. doi: 10.1002/gcc.20568. [DOI] [PubMed] [Google Scholar]
- 73.Marculescu R, et al. Recombinase, chromosomal translocations and lymphoid neoplasia: targeting mistakes and repair failures. DNA Repair (Amst.) 2006;5:1246–1258. doi: 10.1016/j.dnarep.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 74.Heacock ML, et al. Telomere dynamics and fusion of critically shortened telomeres in plants lacking DNA ligase IV. Nucleic Acids Res. 2007;35:6490–6500. doi: 10.1093/nar/gkm472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rong YS. Telomere capping in Drosophila: dealing with chromosome ends that most resemble DNA breaks. Chromosoma. 2008;117:235–242. doi: 10.1007/s00412-007-0144-2. [DOI] [PubMed] [Google Scholar]
- 76.Maser RS, et al. DNA-dependent protein kinase catalytic subunit is not required for dysfunctional telomere fusion and checkpoint response in the telomerase-deficient mouse. Mol. Cell. Biol. 2007;27:2253–2265. doi: 10.1128/MCB.01354-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Korbel JO, et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318:420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Edwards SL, et al. Resistance to therapy caused by intragenic deletion in BRCA2. Nature. 2008;451:1111–1115. doi: 10.1038/nature06548. [DOI] [PubMed] [Google Scholar]
- 79.Sakai W, et al. Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature. 2008;451:1116–1120. doi: 10.1038/nature06633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jeggo P, Lobrich M. Radiation-induced DNA damage responses. Radiat. Prot. Dosimetry. 2006;122:124–127. doi: 10.1093/rpd/ncl495. [DOI] [PubMed] [Google Scholar]
- 81.Magnani C, et al. Short direct repeats at the breakpoints of a novel large deletion in the CFTR gene suggest a likely slipped mispairing mechanism. Hum. Genet. 1996;98:102–108. doi: 10.1007/s004390050167. [DOI] [PubMed] [Google Scholar]
- 82.Luzi P, et al. Characterization of the large deletion in the GALC gene found in patients with Krabbe disease. Hum. Mol. Genet. 1995;4:2335–2338. doi: 10.1093/hmg/4.12.2335. [DOI] [PubMed] [Google Scholar]
- 83.Canning S, Dryja TP. Short, direct repeats at the breakpoints of deletions of the retinoblastoma gene. Proc. Natl. Acad. Sci. U. S. A. 1989;86:5044–5048. doi: 10.1073/pnas.86.13.5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang JG, et al. Characterization of genomic BCR-ABL breakpoints in chronic myeloid leukaemia by PCR. Br. J. Haematol. 1995;90:138–146. doi: 10.1111/j.1365-2141.1995.tb03392.x. [DOI] [PubMed] [Google Scholar]
- 85.Guirouilh-Barbat J, et al. Impact of the KU80 pathway on NHEJ-induced genome rearrangements in mammalian cells. Mol. Cell. 2004;14:611–623. doi: 10.1016/j.molcel.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 86.Guirouilh-Barbat J, et al. Defects in XRCC4 and KU80 differentially affect the joining of distal nonhomologous ends. Proc. Natl. Acad. Sci. U. S. A. 2007;104:20902–20907. doi: 10.1073/pnas.0708541104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chan CY, et al. Ionizing radiation and restriction enzymes induce microhomology-mediated illegitimate recombination in Saccharomyces cerevisiae. Nucleic Acids Res. 2007;35:5051–5059. doi: 10.1093/nar/gkm442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Katsura Y, et al. Involvement of Ku80 in microhomology-mediated end joining for DNA double-strand breaks in vivo. DNA Repair (Amst.) 2007;6:639–648. doi: 10.1016/j.dnarep.2006.12.002. [DOI] [PubMed] [Google Scholar]