SUMMARY
Long noncoding RNAs (lncRNAs) affect gene regulation through structural and regulatory interactions with associated proteins. The Polycomb complex often binds to lncRNAs in eukaryotes and a lncRNA, COLDAIR, associates with Polycomb to mediate silencing of the floral repressor, FLOWERING LOCUS C (FLC) during the process of vernalization in Arabidopsis. Here we identified an additional Polycomb-binding lncRNA, COLDWRAP. COLDWRAP is derived from the repressed promoter of FLC and is necessary for the establishment of stable repressed state of FLC by vernalization. Both COLDAIR and COLDWRAP are required to form a repressive intragenic chromatin loop at the FLC locus by vernalization. Our results indicate that vernalization–mediated Polycomb silencing is coordinated by lncRNAs in a cooperative manner to form a stable repressive chromatin structure.
eTOC Blurb
Flowering control occurs in part through repression of FLOWERING LOCUS C (FLC). Kim and Sung identify a FLC promoter-derived noncoding RNA (COLDWRAP) that is induced by vernalization and functions with the lncRNA COLDAIR to retain Polycomb at the FLC promoter through the formation of a repressive intragenic chromatin loop.
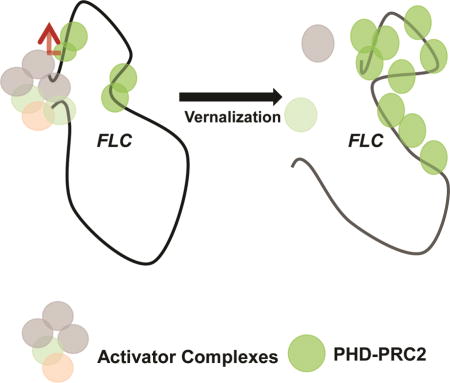
INTRODUCTION
Environmentally induced regulation of gene expression is critical for the proper development of eukaryotic organisms. Given their sessile nature, plants have evolved intricate regulatory systems to provide developmental plasticity to respond to changing environments. One such example is the floral transition. Flowering is one of major developmental changes in the life cycle of flowering plants. A number of studies revealed that sophisticated regulatory networks control the onset of floral transition in response to both endogenous and environmental conditions (Amasino and Michaels, 2010; Andres and Coupland, 2012; Kim et al., 2009). To increase the survival of progeny, flowering occurs only under proper environmental conditions. Seasonal change is a major environmental variable that greatly influences the floral transition. Two major environmental stimuli, photoperiod and winter cold, are critical for plants to recognize the seasonal change. Inductive photoperiods provoke a cascade of gene expression changes that results in the activation of floral integrators. Unlike photoperiod, winter cold does not immediately induce flowering. Rather, winter cold renders plants to be competent to flower under inductive photoperiods, through a process known as vernalization. Because there is a temporal separation between cold stimuli and its biological response, flowering, many vernalization systems have evolved to establish mitotically stable epigenetic changes at critical flowering loci (Bastow et al., 2004; Sung and Amasino, 2004).
One major epigenetic change caused by vernalization is the silencing of the floral repressor, FLOWERING LOCUS C (FLC). This silencing of FLC is mediated by an evolutionarily conserved Polycomb Repression Complex 2 (PRC2) (De Lucia et al., 2008; Gendall et al., 2001; Kim and Sung, 2013). A prolonged exposure to cold triggers the increased enrichment of PRC2 and increased levels of a repressive histone mark, Histone H3 Lys 27 trimethylation (H3K27me3), at FLC chromatin (De Lucia et al., 2008; Kim and Sung, 2013). The PRC2-mediated H3K27me3 histone mark is stably maintained at FLC chromatin even after plants are returned to warm temperature, ensuring the competence achieved by vernalization is stable. During and after vernalization, PRC2 spreads throughout FLC chromatin including the promoter region (Angel et al., 2011; Finnegan and Dennis, 2007; Gendall et al., 2001).
Long noncoding RNAs (lncRNAs) are emerging as key regulators for recruitment of Polycomb complexes (Brockdorff, 2013; Kaneko et al., 2014; Kaneko et al., 2010; Rinn et al., 2007; Zhao et al., 2010). In Arabidopsis, an lncRNA, COLDAIR, is necessary for increased occupancy of PRC2 at FLC chromatin during vernalization through its direct association with PRC2 (Heo and Sung, 2011). COLDAIR is derived from the 1st intron of FLC and reaches its maximum level of expression at around 20 days of cold exposure and decreases to the levels comparable to pre-vernalization condition, as the FLC locus is silenced (Heo and Sung, 2011). A number of PRC2-associated lncRNAs have been identified in mammals, including promoter-derived lncRNAs (Kanhere et al., 2010; Negishi et al., 2014). Previous studies suggested that the proximal promoter region of FLC is necessary for a proper vernalization response (Coustham et al., 2012; Helliwell et al., 2011; Sheldon et al., 2009), although it is not clear how the proximal promoter region of FLC contributes to Polycomb-mediated silencing of FLC.
Here, we identify an additional lncRNA from the proximal promoter of FLC and find that the formation of a repressive chromatin loop by lncRNAs is critical to maintain the Polycomb-mediated silencing of FLC by vernalization.
RESULTS
Promoter-derived noncoding RNA, COLDWRAP, from FLC locus in Arabidopsis
To examine the dynamics of PRC2-RNA interaction over the course of vernalization at FLC, we used an RNA immuno-precipitation (RIP) using the polyclonal antibody against CLF (CLF, Arabidopsis homolog of human EZH2; (Hyun et al., 2013)) followed by tiled RT-PCR approach (Figures S1A and S1B). Using RIP followed by RT-PCR, we observed transiently increased association of COLDAIR with PRC2 during the course of vernalization as expected (Figures S1A and B). We also observed additional overlapping RNA transcripts from the promoter region of FLC that co-precipitate with PRC2 and named this transcript COLDWRAP (Cold of Winter-induced noncoding RNA from the Promoter) (Figures 1A, 1B, S1A and S1B). During cold exposure, COLDAIR associates transiently with PRC2, whereas COLDWRAP remains associated throughout cold exposure (Figures 1A, S1A and S1B). COLDAIR transcription transiently increases during cold exposure (Figure 1B), whereas COLDWRAP transcription persists during and even after cold exposure (Figure 1A).
Figure 1.
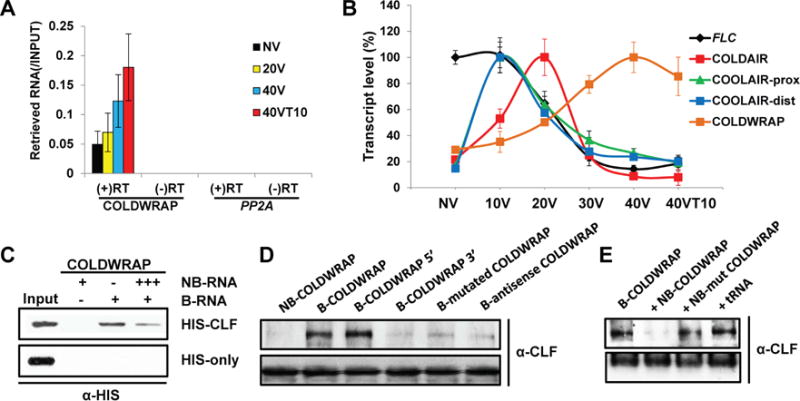
Identification of COLDWRAP. (A) RIP using polyclonal CLF antibody retrieves COLDWRAP RNA. (+) RT: with reverse transcription, (−) RT: without reverse transcription. (b) Expression patterns of FLC, COOLAIR (proximal and distal), COLDAIR, and COLDWRAP transcripts during the course of vernalization. (A, B) Data (mean ± SD of quantitative PCR; biological replicates n = 3). NV, non-vernalized. 10V, 10 days of vernalization. 20V, 20 days of vernalization. 40V, 40 days of vernalization. 40VT10, 40 days of vernalization followed by 10 days of normal growth temperature. (C) In vitro RNA-binding assays. C-terminal His-tagged CLF recombinant protein binds to in vitro transcribed (IVT)-biotinylated COLDWRAP RNA. (D) RNA-binding assay using IVT-biotinylated RNAs and CLF polyclonal antibody. (E) RNA-binding assay using non-biotinylated competitive RNAs. (C–E) NB: non-biotinylated, B: biotinylated. Also see Supplementary Figures S1 and S2.
COLDWRAP is transcribed in a sense direction relative to FLC mRNA and its major transcription starts at 225 bp upstream from the FLC mRNA transcription start site (Figure S1C). Similar to COLDAIR, COLDWRAP has a 5′ cap (Figures S1D–S1F), but the majority of the transcripts appears not to be polyadenylated (Figure S1G). COLDWRAP is ~ 316 bases long and overlaps with 5′ end of FLC mRNA as determined by 3′ tiled RT-PCR (Figures S1C and S1E). To rule out the possibility of COLDWRAP being an alternative FLC mRNA transcript or unspliced FLC transcripts, we performed a series of RT-PCR assays spanning various regions of FLC (Figures S2A–S2C). COLDWRAP is not co-amplified with either FLC mRNA or unspliced FLC (Figure S2C). In addition, COLDWRAP and FLC mRNA exhibit opposite expression patterns during vernalization, further confirming the independent transcription of COLDWRAP (Figures S2B, S2C–S2E).
COLDWRAP physically associates with PRC2
COLDWRAP interacts with recombinant CURLY LEAF (HIS-CLF) protein in vitro (Fig. 1C). To address the specificity of the interaction between COLDWRAP and the native PRC2 complex, we used nuclear extracts from wild-type plants and polyclonal CLF antibody to perform in vitro RNA-binding assays (Figures 1D and 1E). We detected significant binding of native CLF to the sense strand of COLDWRAP but weakly to the antisense strand of COLDWRAP, confirming the specific interaction of COLDWRAP with native PRC2 (Figure 1D). We further dissected COLDWRAP regions to identify an RNA motif of COLDWRAP that is responsible for its PRC2 association. The 5′ half of COLDWRAP is sufficient to pull-down PRC2 in vitro, whereas the 3′ half of COLDWRAP does not bind to CLF-containing PRC2 (Figure 1D). From prediction (Gruber et al., 2008) of the secondary structure of COLDWRAP (Figure S2F), we identified several stable secondary structures of the sort that have been implicated in RNA-protein interactions (Kanhere et al., 2010; Zhao et al. 2010; Mercer and Mattick, 2013; Somarowthu et al., 2015). Point mutations that disrupted the predicted secondary structures of COLDWRAP RNA (Figure S2F) reduced the level of precipitated PRC2 in in vitro RNA-binding assays (Figure 1D). In addition, the mutated form of COLDWRAP failed to compete with the wild-type COLDWRAP for the binding of PRC2 in the in vitro binding assays (Figure 1E). This is not due to altered stability of the mutated RNA, as there is no significant difference in the stability of in vitro transcribed RNAs (Figure S2G). Therefore, the secondary structures in the 5′ half of COLDWRAP appear to be critical for the proper association of PRC2 with COLDWRAP.
Biological roles of COLDWRAP in the PRC2-mediated FLC silencing by vernalization
To address the biological function of COLDWRAP in the vernalization response, we took the RNAi knockdown approach. More than 70% of the primary RNAi-containing transgenic plants show a reduced vernalization response (Figure S3). Two stable COLDWRAP knockdown lines with significantly reduced levels of COLDWRAP during vernalization were chosen for further analysis (Figures S3A–C). The levels of other ncRNAs from FLC, COLDAIR and COOLAIR (Heo and Sung, 2011; Swiezewski et al., 2009), are not largely affected in these COLDWRAP knockdown lines (Figures S3D and S3E). The reduced vernalization response observed in COLDWRAP knockdown lines is consistent with increased levels of FLC expression (Figure S3F).
To evaluate the effect of COLDWRAP on the repression of FLC by vernalization, we determined the relative enrichment of PRC2 at FLC over the course of vernalization by chromatin immunoprecipitation (ChIP) assay using anti-CLF antibodies (Figure S3G). The reduced enrichment of PRC2 is observed at the promoter regions of FLC (Figure S3G) in COLDWRAP RNAi lines compared to the wild type. Consistent with the reduced occupancy of PRC2 at FLC, the accumulation of H3K27me3 at regions of FLC is substantially reduced in COLDWRAP RNAi lines, particularly at the promoter regions of FLC (Figures S3H and S3I).
Although knockdown approaches have been widely used to address biological significance of lncRNAs (Heo and Sung, 2011; Orom and Shiekhattar, 2011; Tsai et al., 2010; Willingham et al., 2005), knockdown approaches have limitations, including weaker than expected phenotypes. Because COLDWRAP is transcribed from the proximal promoter of FLC, using a traditional insertion mutant would not be informative. To overcome the disadvantage of knockdown approach and the lack of insertion mutant, we utilized a deletion allele of flc (flc-2). The flc-2 mutant harbors a deletion that encompasses the entire COLDWRAP and COLDAIR regions (Michaels and Amasino, 1999). We created a mutant form of COLDWRAP RNA that does not precipitate PRC2 in vitro (Figures 1D and 1E). The same mutations were introduced into the full-length FLC genomic construct that is sufficient to complement flc mutants ((Michaels and Amasino, 1999); Figures 2 and S4). The mutated construct was introduced in flc-2 background to create the “functional-null” allele of COLDWRAP (mutant COLDWRAP) (Figure S4A). FLC transgenic lines are often subject to the dosage effect and result in a rather wide-range of flowering times (Coustham et al., 2012; Michaels and Amasino, 1999). Therefore, we generated a large number of primary transgenic plants with the wild-type FLC genomic construct for comparison (Figures 2A and 2B). About 43% of primary transgenic lines (57 out of 131) carrying the mutant COLDWRAP show very late flowering after vernalization (rosette leaf numbers > 80) (Figures 2A–B, S4B and S4C). On the other hand, we did not observe this extreme late-flowering phenotype in primary transgenic lines carrying the wild-type allele of COLDWRAP after vernalization (Figures 2A–D).
Figure 2.
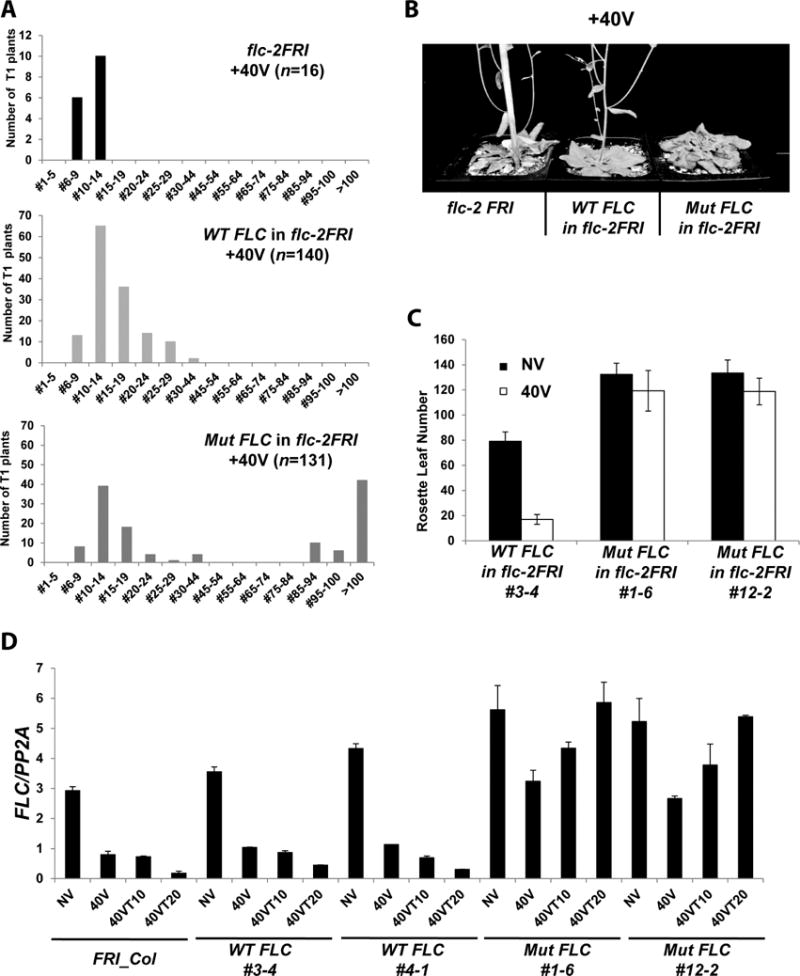
COLDWRAP is necessary for vernalization response. (A) Flowering times of flc-2FRI (parental; top), the primary transgenic lines carrying the wild-type FLC transgene in flc-2FRI after vernalization (middle) and the primary transgenic lines carrying the mutant COLDWRAP in flc-2FRI after vernalization (bottom). X-axis; range of rosette leaf numbers. (B) Representative flowering behaviors of flc-2 FRI, flc-2 FRI transformed with the wild-type FLC transgene, and flc-2 FRI transformed with the mutant COLDWRAP after 40 days of vernalization. (C) Flowering times of a representative transgenic line carrying the wild-type FLC transgene, and two representative lines carrying the mutant COLDWRAP. (D) Changes in FLC mRNA during the course of vernalization in the wild type (FRI_Col) and two representative transgenic lines carrying the wild-type FLC transgene (WT FLC #3-4 and #4-1) and two representative transgenic lines carrying the mutant COLDWRAP (Mut FLC #1-6 and #12-2). Data (relative levels; mean ± SD of quantitative RT-PCR; biological replicates n = 3). Also see Supplementary Figures S3 and S4.
Due to the dosage effect of FLC on flowering time (Michaels and Amasino, 1999), FLC transgene may result in variations in flowering time depending on its insertion location and the relative strength of transgene expression. Therefore, we examined the level of FLC expression of randomly selected T2-pools of transgenic plants of early-flowering and late-flowering cohorts among wild-type FLC (WT_FLC) and mutant COLDWRAP (Mut_FLC) transgenic lines (Fig. S4J). The result indicated that the early-flowering cohort of mutant COLDWRAP might be due to the integration of FLC transgene into the genome where the behavior of the endogenous FLC locus cannot fully be recapitulated (i.e. the low level of FLC expression was observed without vernalization), an indicative of the transgene effect. Therefore, we chose several representative WT_FLC and Mut_FLC transgenic lines from the ones that show significant level of FLC expression without vernalization for further study.
Effects of the mutant COLDWRAP on the PRC2-mediated FLC silencing by vernalization
The mutant COLDWRAP does not impair the FLC expression prior to vernalization, as determined by their flowering time and the level of FLC expression (Figure 2). The level of FLC mRNA decreases during cold exposure in all genotypes compared to the levels of non-vernalized plants (Figure 2D). Two representative transgenic lines (WT_FLC) carrying the wild-type FLC and non-transgenic FRI-Col maintained their repressed levels of FLC after vernalization (Figure 2D). On the contrary, the repression of FLC is not stable in the transgenic lines carrying the mutant COLDWRAP (Mut_FLC) after vernalization (Figure 2D). Unstable repression of FLC by vernalization is commonly observed in Polycomb mutants (Bastow et al., 2004; Helliwell et al., 2011; Heo and Sung, 2011; Sung and Amasino, 2004; Sung et al., 2006a), consistent with a model in which the mutant COLDWRAP compromises Polycomb function at FLC. Expression of FLOWERING LOCUS T (FT), a downstream target of FLC, remains at low levels even after vernalization, consistent with late flowering phenotype of mutant COLDWRAP lines compared to wild type (Figure S4D). The transgenic lines carrying the mutant COLDWRAP flower later than either the transgenic lines carrying the wild-type COLDWRAP or non-transgenic lines without vernalization (Figure 2C). The level of FLC mRNA in the transgenic lines carrying mutant COLDWRAP is higher than that of wild type (FRI_Col) plants or transgenic plants carrying wild-type FLC (WT_FLC) plants without vernalization (Figure 2D), which is consistent of the involvement of Polycomb in the regulation of FLC without vernalization (De Lucia et al., 2008; Kim and Sung, 2013). Expressions of five FLC-related genes are not affected in the mutant COLDWRAP lines compared to the wild-type plants (Figure S4E), and there is no COLDWRAP-like sequences found in five FLC-related gene loci. Therefore, COLDWRAP specifically affects the regulation of FLC.
The enrichment of CLF and H3K27me3 at FLC by vernalization also decreases in the transgenic lines carrying the mutant COLDWRAP, consistent with the requirement of COLDWRAP in Polycomb-mediated silencing (Figures 3A and 3B). We also detected reduced levels of COLDWRAP precipitated with PRC2 in RIP assays using anti-CLF antibody only in transgenic lines carrying the mutant COLDWRAP (Figure 3C). This is in agreement with in vitro data that show reduced levels of precipitation of PRC2 with mutated form of COLDWRAP (Figure 1D). The reduced levels of precipitated COLDWRAP is not due to the reduction in the level of COLDWRAP expression, as COLDWRAP is expressed at a comparable level in COLDWRAP mutant lines (Figure S4F). Therefore, the mutant COLDWRAP affects vernalization through its physical association with PRC2. The mutant COLDWRAP also causes reduced precipitation of COLDAIR (Figure 3C), indicating the cooperative role of the two lncRNAs in Polycomb-mediated FLC silencing by vernalization.
Figure 3.
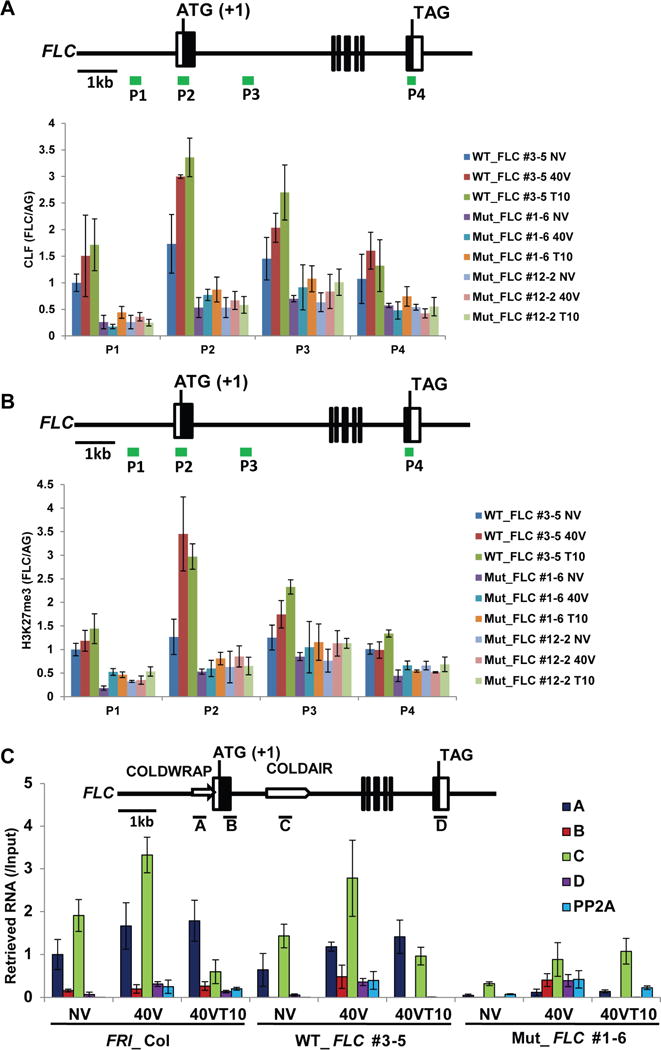
COLDWRAP is necessary for vernalization-mediated H3K27me3 enrichment at FLC chromatin. (A) Changes in occupancy of CLF at FLC chromatin during the course of vernalization. (B) Changes in enrichment of H3K27me3 at FLC chromatin during the course of vernalization. (C) Relative fold changes of RNA retrieved by RIP using anti-CLF antibody followed by quantitative RT-PCR in wild type (FRI_Col), a representative transgenic line carrying the wild-type FLC transgene (WT_FLC #3-5), and a representative transgenic line carrying the mutant COLDWRAP (Mut_FLC #1–6). (A–C) Data (mean ± SD of quantitative PCR; biological replicates n=3). Also see Supplementary Figures S3 and S4.
Restoration of vernalization response by in trans expression of COLDWRAP
To address whether FLC-derived lncRNAs function as a nascent transcript (in cis) or in trans, we introduced the CaMV35S-driven COLDWRAP transgene into the mutant COLDWRAP lines and the wild type. Primary transgenic plants do not show significant difference in flowering times compared to non-transgenic lines without vernalization (Figures 4A, S4G and S4H). After 40 days of cold treatment, most primary transgenic plants exhibit a normal vernalization response (Figures 4A–C). This is not due to the inactivation of FLC itself, as non-vernalized transgenic plants show late flowering in the absence of vernalization treatment (Figures 4A–C). The level of FLC mRNA is stably repressed by vernalization in mutant COLDWRAP primary transgenic plants carrying 35S:COLDWRAP (Mut_FLC #1-6+35S:COLDWRAP) compared to that of mutant COLDWRAP (Mut_FLC #1-6) lines (Figure 4D). This result indicates that the exogenous expression of COLDWRAP restores the vernalization response and, therefore, that COLDWRAP is able to act in trans to mediate the repression of FLC by vernalization.
Figure 4.
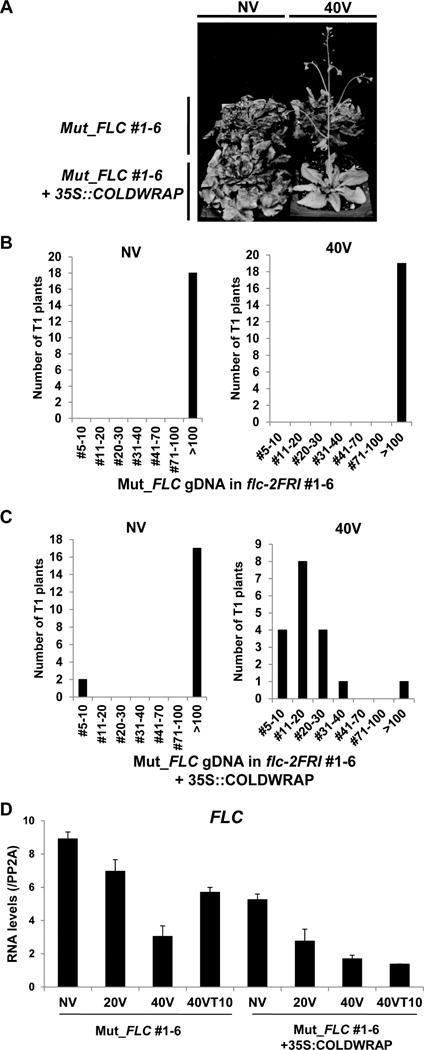
Restoration of vernalization response of transgenic lines carrying the mutant COLDWRAP mutant by COLDWRAP. (A) Representative flowering behaviors of transgenic lines carrying the mutant COLDWRAP and the mutant COLDWRAP, complemented with 35S::COLDWRAP without (NV) and with (40V) vernalization. (B) Flowering times of a representative transgenic line carrying the mutant COLDWRAP (#1-6) without (NV) and with (40V) vernalization (n = 19). (C) Flowering times of the primary transgenic lines carrying the mutant COLDWRAP complemented with the 35S::COLDWRAP (n = 18). (D) Levels of FLC mRNA during the course of vernalization between Mut_FLC gDNA in flc-2FRI (#1-6) and Mut_FLC gDNA in flc-2FRI+35S:COLDWRAP T1 primary transgenic lines. Data (relative levels; mean ± SD of quantitative RT-PCR; n =3). Also see Supplementary Figure S4.
Formation of repressive intragenic gene loop at FLC locus by vernalization
Chromatin conformation changes have been implicated in Polycomb-mediated gene silencing (Lanzuolo and Orlando, 2012; Lanzuolo et al., 2007; Simon and Kingston, 2009) and some lncRNAs have been shown to be involved in coordinating chromatin conformation changes (Orom and Shiekhattar, 2011). To examine chromatin conformation at FLC by vernalization, we employed a chromatin conformation capture (3C) assay (Figures 5 and S5). We observed a strong chromatin loop formation between the promoter and 3′ end of the first intron (downstream region from the COLDAIR transcribed site) (Figures S5A). We tested a series of primers that cover the entire region of FLC. Among tested promoter and the 5′end regions of FLC, the COLDWRAP transcribed region (F4) shows the most strong chromatin loop formation with 3′ end of the first intron of FLC (Figures S5A). The gene loop is present prior to vernalization and is gradually enhanced by vernalizing cold, indicating that chromatin loop function is part of the system required for stable maintenance of FLC repression by vernalization (Figures 5A–D and S5B). In COLDWRAP knockdown lines, the extent of chromatin loop formation is significantly reduced (Figures 5A–D, and S5B). In addition, the formation of this vernalization-mediated chromatin loop is impaired in transgenic lines carrying the mutant COLDWRAP, but not in transgenic lines carrying the wild-type COLDWRAP (Figure 5E), further demonstrating that COLDWRAP is required for the formation of the chromatin loop between the promoter region and the 3′end of the first intron. An additional chromatin loop exists between the promoter of FLC and the downstream of 3′ end of FLC locus (Figures 5A–C, and S5B). Unlike the loop between the promoter and the intron, this loop disappears during vernalization, implying it has a role in favoring the active state of FLC (Crevillen et al., 2013; Zhu et al., 2015).
Figure 5.
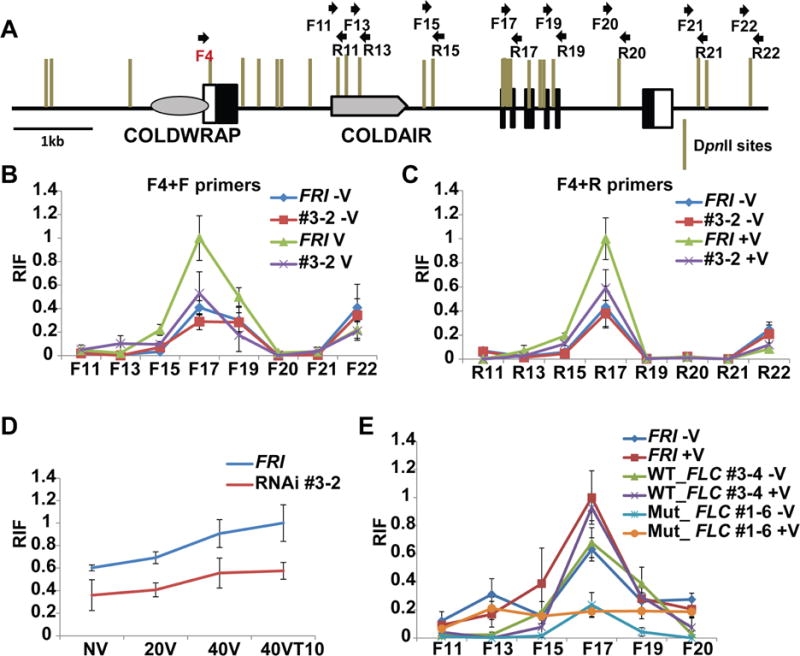
Chromatin conformation capture (3C) assays (A) Relative positions of primer sets used for 3C assays. (B) Relative Interaction Frequency (RIF) in 3C assay between F4 and a series of F primer regions of FLC in the wild type (FRI) and the COLDWRAP RNAi line (#3-2). (C) RIF in 3C assay between F4 and a series of R primer regions of FLC in the wild type (FRI) and the COLDWRAP RNAi line (#3-2). (D) RIF in 3C assay between F4 and R17 regions of FLC in the wild type (FRI) and the COLDWRAP RNAi line (#3-2) during the course of vernalization. (E) RIF in 3C assay between F4 and a series of F primer regions of FLC in the wild type (FRI), the transgenic line carrying the wild-type FLC transgene (WT_FLC #3-4) and the transgenic line carrying the mutant COLDWRAP (Mut_FLC #1-6). (B~E) Maximum interaction frequency is set as 1 and relative fold changes are shown. −V, without vernalization. +V, with vernalization. NV, non-vernalized. 10V, 10 days of vernalization. 20V, 20 days of vernalization. 40V, 40 days of vernalization. 40VT10, 40 days of vernalization followed by 10 days of normal growth temperature. Data (mean ± SD of quantitative 3C; biological replicates n=3). Also see Supplementary Figure S5.
PRC2 mediates the formation of repressive intragenic gene loop at FLC locus by vernalization
PRC2 that is specifically involved in vernalization-mediated FLC silencing in Arabidopsis includes at least two Plant-HomeoDomain (PHD) finger proteins, VERNALIZATION INSENSITIVE 3 (VIN3) and VIN3-LIKE 1 (VIL1), and the PRC2 complex containing these proteins is often referred to as PHD-PRC2 complex (De Lucia et al., 2008). Other core components of PRC2 are involved in a range of developmental programs, which often impedes the interpretation of flowering phenotypes when they are mutated (i.e. early developmental defects; (Hennig and Derkacheva, 2009; Kohler et al., 2003; Schubert et al., 2005)). VIN3 and VIL1, however, appear to be specific to the regulation of FLC and FLC-related loci, and mutants in them do not show other pleiotropic phenotypes (Greb et al., 2007; Kim and Sung, 2013; Sung and Amasino, 2004; Sung et al., 2006b). Therefore, we evaluated the role of PRC2 in the formation of intragenic repressive chromatin loop at FLC using vin3 and vil1 mutants. The level of chromatin loop formation within FLC is significantly compromised in both vin3-1 FRI and vil1-1 FRI mutants compared to the wild-type plants (Figures 6A and 6B). This result indicates that VIN3/VIL1-containing PRC2 contributes to the formation of intragenic repressive chromatin loop at FLC locus in response to vernalization.
Figure 6.
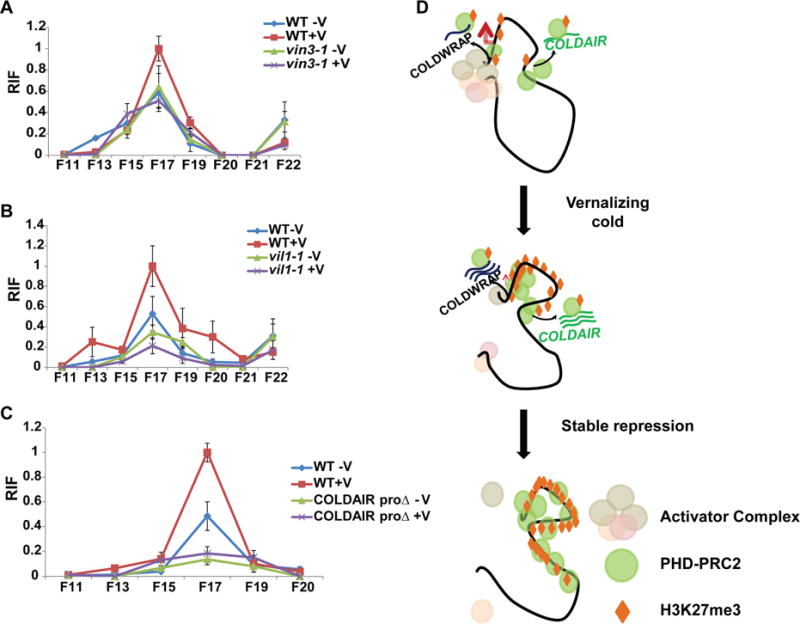
The formation of a repressive chromatin loop at the FLC locus. (A) RIF in 3C assay between F4 and a series of F primer regions of FLC in the wild type (FRI) and vin3 mutant (vin3-1FRI). (B) RIF in 3C assay between F4 and a series of F primer regions of FLC in the wild type (FRI) and vil1 mutant (vil1-1FRI). (C) RIF in 3C assay between F4 and a series of F primer regions of FLC in the wild type (WT) and a representative COLDAIR promoter deletion line (COLDAIR proΔ gFLC). (A~C) Maximum interaction frequency is set as 1 and relative fold changes are shown. −V, without vernalization. +V, with vernalization. (D) A proposed model for Polycomb-mediated FLC repression by two ncRNAs during the course of vernalization. During early cold, COLDAIR functions to recruit PRC2 at the first intron region of FLC. Subsequently, COLDWRAP helps PRC2 spread to the promoter region, which results in spreading of repressive histone mark, H3K27me3. This spreading is achieved at least in part by the formation of an intragenic chromatin loop between the COLDAIR-transcribed region and the promoter of FLC. Also see Supplementary Figure S6.
Previously, another lncRNA, COLDAIR, was shown to be involved in the PRC2-mediated silencing of FLC by vernalization through its role in the recruitment of PRC2 to FLC. To address the role of COLDAIR in the formation of intragenic repressive chromatin loop at FLC, we created a null allele of COLDAIR in flc-2 mutant background (COLDAIR_proΔ gFLC; Figure S6). We used a genomic construct of FLC in which the intrinsic promoter of COLDAIR is deleted (Figure S6A). After vernalization, the majority of primary transgenic lines show significantly late flowering compared to wild type (Figures S6B and S6C). Expression of COLDAIR is not detectable and the de-repression of FLC by vernalization is observed in stable COLDAIR_proΔ gFLC transgenic lines (Figure S6D and S6E). Furthermore, the vernalization response is restored in COLDAIR_proΔ gFLC transgenic lines by introducing the wild-type copy of COLDAIR (Figure S6F). Therefore, we used COLDAIR_proΔ gFLC transgenic lines to evaluate the contribution of COLDAIR to the formation of intragenic repressive chromatin loop at FLC (Figure 6C). Vernalization-mediated chromatin loop formation is severely impaired in COLDAIR_proΔ gFLC transgenic lines (Figure 6C). Thus, our results show that the formation of an intragenic chromatin loop at FLC requires the association of PRC2 at FLC that is accomplished by two lncRNAs and the intragenic repressive chromatin loop is involved in establishing stable Polycomb-mediated silencing of FLC by vernalization (Figure 6D).
DISCUSSION
There is growing evidence that various long noncoding RNAs are engaged in the control of gene expression in eukaryotes (Mercer and Mattick, 2013; Rinn and Chang, 2012; Vance and Ponting, 2014; Wilusz et al., 2009; Yang et al., 2014; Zhao et al., 2008). However, there remains much to learn about the mechanisms by which long noncoding RNAs control target gene expression. Our study shows that two long noncoding RNAs coordinate with a histone H3K27 methyltransferase complex, PRC2, to mediate vernalization-mediated silencing of FLC through the formation of intragenic gene loop within the FLC locus. The involvement of multiple lncRNAs in the Polycomb silencing demonstrates diverse roles of lncRNAs in the regulation of gene expression in eukaryotes.
Our study indicates that putative secondary structures in the 5′ half of COLDWRAP is necessary for the association with PRC2 in vitro and in vivo. A long noncoding RNA, HOTAIR, forms modular structures that are implicated in its function through the association of various protein components (Somarowthu et al., 2015; Tsai et al., 2010). Predicted secondary structures of COLDWRAP suggest that secondary structures may constitute a structural module in COLDWRAP that is necessary for the physical interaction with PRC2. The binding specificity of an RNA motif with PRC2 is questionable as PRC2 appears to bind promiscuously to many different RNAs in in vitro assays (Davidovich et al., 2013). However, mutated COLDWRAP impairs the PRC2 binding in vitro (Fig. 1D, E) and in vivo (Fig. 3C) and our data show that structural integrity of noncoding RNAs is necessary for the proper function of lncRNA-PRC2 in vivo. Therefore, our results support the biological significance of the secondary structure of lncRNAs in Polycomb-mediated silencing. Comprehensive transcriptome data from 998 ecotypes of Arabidopsis are available (Kawakatsu et al., 2016). It is interesting to note that we observed the COLDWRAP transcript reads from about 12% (123 ecotypes out of 998 ecotypes) of the RNA-Seq data, suggestive of the variations among natural accessions of Arabidopsis in the transcription of COLDWRAP. Whether these variations contribute to the vernalization response remains to be addressed. In addition, it will be interesting to determine whether natural variations in the secondary structures of COLDWRAP may play roles in fine-tuning the vernalization response among natural accessions.
Although there are clear associations of COLDAIR and COLDWRAP with PRC2 during vernalization, it remains to be investigated how PRC2 is recruited to FLC in a locus-specific manner by lncRNAs. One possible model involves co-transcriptional association of lncRNA and PRC2. In this model, lncRNAs become associated with PRC2 as they are transcribed. However, both COLDWRAP and COLDAIR are capable of restoring a vernalization response when they are expressed in trans (Fig. 4A–C and S6F). Therefore, co-transcriptional association is not necessary for COLDWRAP and COLDAIR activity. An alternative model, consistent with the trans effect, predicts the presence of direct DNA-RNA interaction between target loci and lncRNAs. HOTAIR’s target loci share a sequence motif that is preserved in HOTAIR itself (Chu et al., 2011) and likewise COLDAIR and CODWRAP share sequence with FLC, suggesting that the DNA-RNA interaction may mediate the recruitment of PRC2 to certain targets. Whether a direct DNA-RNA interaction is actually needed for COLDWRAP and COLDAIR effects on FLC remains to be established.
A number of proteins have been identified as lncRNA binding proteins (Chu et al., 2011; Chu et al., 2015; Jeon and Lee, 2011; Sarma et al., 2014; Tsai et al., 2010). Some of these lncRNA-interacting proteins are involved in the target specificity. For example, a transcriptional repressor protein, Yin and Yang 1 (YY1), is capable of binding to both DNA and RNA through different motifs and acts to specifically tether Xist lncRNA onto X chromosome (Jeon and Lee, 2011). This exemplifies an alternative targeting model in which a protein component bridges between lncRNA and target DNA. Some lncRNAs also serve as scaffold molecules to harbor multiple protein complexes and deliver them to target loci (Batista and Chang, 2013; Tsai et al., 2010). HOTAIR binds to both E(z), a H3K27 methyltransferase, and LSD1, a H3K4 demethylase (Tsai et al., 2010). Whether COLDWRAP and/or COLDAIR is able to bind to additional proteins to target PRC2 to FLC and/or to mediate other chromatin-modifying activities through its direct association with other chromatin-remodeling complexes remains to be addressed. The constitutive expression of COLDWRAP driven by 35S promoter is not sufficient to trigger the silencing of FLC in the absence of vernalization (Figure S4), indicating that other vernalization-triggered events (i.e. the induction of VIN3, COLDAIR and COOLAIR) are also required to repress FLC by vernalization.
The spatial organization of the genome and higher-order chromosome structures in the nucleus is important for gene expression and genome function in higher eukaryotes (Dixon et al., 2012; Gibcus and Dekker, 2013; Jin et al., 2013; Van Bortle and Corces, 2012). Some reports examined Arabidopsis chromatin conformation using genome-wide Hi-C technique (Feng et al., 2014; Grob et al., 2013). These studies showed that the Arabidopsis genome includes relatively short-range genomic interactions, compared to those of mammalian genomes which have many long-range chromosomal interactions. Short-range chromatin loop formation is closely related to control of gene expressions in Arabidopsis (Ariel et al., 2014; Cao et al., 2014; Crevillen et al., 2013; Wang et al., 2015). For example, a chromatin loop between a distal enhancer motif and a proximal promoter part of the floral activator gene, FLOWERING LOCUS T (FT) are required to its gene activation in a photoperiod-dependent manner (Cao et al., 2014). The formation of an intragenic repressive gene loop at the FLC locus by vernalization requires both COLDWRAP and COLDAIR lncRNAs. In addition, the loop is formed between the promoter region (where COLDWRAP originates) and the 1st intron (where COLDAIR originates). This implies that two lncRNAs, COLDWRAP and COLDAIR, paly cooperative roles in the formation of chromatin loop to establish stably repressed chromatin at the FLC locus by vernalization. Promoter-derived noncoding RNAs from Polycomb target loci have been observed in other organisms (Enderle et al., 2011; Kanhere et al., 2010). Therefore, it is likely that these dynamic changes of topological genomic structure directly influence the transcriptional outputs in response to developmental or environmental cues in eukaryotes. Our study indicates that “intragenic” loop formation could be a general mechanism of gene repression.
We have shown that both COLDAIR and CODWRAP lncRNAs are required to establish the silencing of FLC by vernalization. COLDWRAP apparently affects the formation of COLDAIR-PRC2 RNP complex (Figure 3C), suggesting the presence of a positive feedback between COLDWRAP and COLDAIR in establishing repressive chromatin at FLC locus. This cooperative nature of two lncRNAs may serve as a safety lock to ensure that the silencing of FLC occurs only in response to vernalization. Also, this intra-locus loop within FLC might function to prevent re-formation of promoter-3′UTR loop after vernalization (Crevillen et al., 2013) which is considered to enhance transcription of FLC by recycling RNA Pol II from the end of the gene back to the promoter (Laine et al., 2009; Tan-Wong et al., 2009).
In conclusion, we show that vernalization-mediated FLC silencing includes an elegant step-wise system involving multiple lncRNAs by which a seasonal cue (exposure to the prolonged cold of winter) results in the epigenetic repression of a target gene and the initiation of an important developmental program. Thus vernalization transpires when two lncRNAs cooperate to translate a seasonal cue (cold of winter) into repression of the target gene FLC and subsequent release of the flowering program.
STAR★Methods
KEY RESOURCE TABLE
Separate file submitted.
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to the corresponding author, Dr. Sibum Sung (sbsung@austin.utexas.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All Arabidopsis plants are in the Columbia FRIsf2 background. Seeds were germinated on agar plates, following stratifications at 4°C for 2 days, and grown for 10 days under short day (SD) (8h light: 16h dark at 22°C). Seedlings were transplanted to soil or harvested for experiments. Vernalization treatment was performed as previously described (Kim et al., 2010). Non-vernalized seedlings were grown for 11 days after germination under short day condition. For 40VT0 and 40VT10 plants, seedlings were grown for 7 days and 4 days, respectively, after seed germination and then seedlings were subject to cold treatment. Samples were harvested for experiments right after the cold treatment (40V) or grown for 10 days more (40VT10) under SD.
For 8kb genomic FLC construct, genomic DNA fragment that contains about 1.7-kb promoter, coding and 0.7-kb 3′ part sequence of FLC was amplified by PCR amplification. Genomic FLC fragment was clonedinto pPZP211 binary vector. Using the wild-type genomic FLC construct as a template, mutated form of genomic FLC fragment was amplified by site-directed mutagenesis method using pairs of mutagenic primers and flanking primers and then cloned into pPZP211 vector. Both of WT and mutated form of genomic FLC constructs were transformed into Agrobacterium tumefaciens GV3101 strain and transformed into flc-2;FRI-Col plants. Full length of COLDWRAP and COLDAIR are cloned into pENTR vector (Invitrogen) and then transferred into pEARLEY100 vector using LR clonase (Thermo Fisher Scientific). Cloned sequence was confirmed by sequencing and used for transformation to create the mutant COLDWRAP or COLDAIR transgenic lines. For COLDWRAP knockdown lines, 94 bp of 5′ COLDWRAP was amplified and subcloned into pENTR directional TOPO vector (Invitrogen) followed by LR reaction (Invitrogen) into the RNAi destination vector pB7GWIWG2(II) using LR clonase (Invitrogen). The construct was transformed into Agrobacterium tumefaciens strain GV3101 and transformed into FRI-Col wild-type plants by Agrobacterium-mediated floral dip transformation. All transgenic lines were selected on selective antibiotic marker-containing medium, and were confirmed by PCR.
METHOD DETAILS
Tiling RNA-IP assay around FLC genomic region
Whole seedlings were harvested and treated with formaldehyde-containing buffer (0.4 M sucrose, 10 mM Tris (pH 8.0), 1 mM EDTA, 1 mM PMSF, 1% formaldehyde) for cross-linking. Plants tissues were dried and ground in liquid nitrogen, then re-suspended in nuclei isolation buffer (0.25M sucrose, 5mM PIPES (pH 8.0), 5mM MgCl2, 85mM KCl, 15mM NaCl, 1% Triton X-100, RNaseOUT (40 U/ml, Thermo Fisher Scientific), 1mM PMSF, 1 tablet of Roche protease inhibitors cocktail), and placed on ice for 15 minutes. The crude nuclei fraction was centrifuged top speed for 10 minutes at 4°C. Each pellet was re-suspended in lysis buffer (50mM HEPES (pH 7.5), 150mM NaCl, 1% Triton X-100, 1mM EDTA, 0.1% Na deoxycholate, 1% SDS, 1mM PMSF, RNaseOUT (40 U/ml, Thermo Fisher Scientific), 1 tablet of Roche protease inhibitors cocktail) and was sonicated five times, each 15 seconds. Sonicated lysates were centrifuged with top speed for 15 minutes at 4°C. Each supernatant was transferred to new tubes and added with 50 ul of protein A agarose beads (Roche, IN, USA) and rotated for 30 minutes at 4°C at for pre-clearing. Potein A agarose beads were removed by spin-down and supernatants were transferred to new tubes. Each supernatant was volumed up to 1 ml with dilution buffer (1.1% Triton X-100, 1.2mM EDTA, 16.7mM Tris-HCl (pH8.0), 167mM NaCl, RNaseOUT (40 U/ml, Thermo Fisher Scientific), 1 tablet of Roche protease inhibitors cocktail) and then 10% per sample was preserved as an input RNA sample and frozen at −80°C. Anti-CLF antibody was added to each sample tube and each tube was rotated overnight at 4°C to form immune complexes. Next day, each immune complex was sequentially washed using low-salt, high-salt, LiCl and TE buffer (pH 8.0). After washing, each sample was added by 100 ul of elution buffer (1% SDS, 0.1M NaHCO3, RNaseOUT (40 U/ml, Invitrogen)) and incubated for 10 minutes at room temperature. Elution step was repeated once. Eluted samples were treated with 20 ug of proteinase K and incubated for 1 hour at 42°C. For reverse-crosslinking, each sample was treated with 10 ul of 5M NaCl and incubated for 1 hour at 65°C. RNA was purified using TRIzol (Invitrogen) according to manufacturer’s instruction. After treatment of RQ1 DNase (Promega) for 25 minutes at 37°C, RNA were used for first strand cDNA syntheses using random primers (Gene Link) with M-MLV System (Promega). To exclude the possibility of amplification of contaminated DNA, we performed the same RT reaction without M-MLV reverse transcriptase. This sample was used as a negative control for further qRT-PCR analysis.
RNA expression analysis
Total RNAs were extracted from whole seedling plants using TRIzol (Invitrogen). DNase I enzyme (Invitrogen) was treated for 30 minutes at 37°C to eliminate contaminated genomic DNA from total RNAs. Five micrograms of total RNA were used for synthesis of first strand cDNA using random primers and then used for real-time qRT-PCR analyses using ViiA 7 real time system (Life Technologies). Reactions were carried out in a total volume of 10 ul with Maxima SYBR green master mix (Thermo Fisher Scientific, USA). Primer sequences used in quantitative RT-PCR analyses are listed in Supplementary Table 1.
5′ Rapid Amplification of cDNA ends (RACE)
We used 5′RACE System kit (Invitrogen, Carlsbad, CA, USA) to determine the 5′end of COLDWRAP. With purified total RNAs from 40 days-vernalized seedlings, COLDWRAP forward or reverse primers (Supplementary Table 1) were designed and used to synthesize first strand cDNA by using the M-MLV System (Promega) and further procedures were performed according to the manufacturer’s instructions. The PCR reaction using primer set of AUAP/COLDWRAP reverse primer only produced a single PCR band (Supplementary Fig. 2C) and this PCR product was cloned into pCR2.1_TOPO TA cloning kit (Thermo Fisher Scientific) and sequenced.
Determination of 3′end of COLDWRAP by RNA blot analysis and tiling 5′-RACE PCR
To determine the approximate size of COLDWRAP, we carried out the RNA blot analysis. 40 ug of total RNA from non-vernalized and vernalized seedlings were used for RNA blotting. Full or the 5′ part of COLDWRAP (157 bp) was amplified and cloned into pBluscript SK vector. Orientation of inserted DNA was confirmed by sequencing using T7 promoter primer. For synthesis of single-stranded antisense probe for COLDWRAP, plasmid was digested by SacI restriction enzyme to produce linear plasmid DNA and then eluted form gel using Gel extraction kit (Qiagen, USA). Each 262 bases of sense FLC 7th exon sequence and 208 bases of sense PP2A sequence was amplified and cloned into pENTR vector (Thermo Fisher Scientific). For synthesis of single-stranded antisense probe for each FLC and PP2A, each pENTR clone was digested by NotI restriction enzyme to produce linear plasmid DNA and then eluted from gel using Gel extraction kit (Qiagen, USA). Each linearized plasmid DNA was used to produce radioactive-labeled (with α-P32 UTP) antisense strand-specific RNA by using T7 RNA polymerase (Roche, IN, USA) to detect sense FLC and COLDWRAP transcript in total RNA blot using Riboprobe System (Promega). A PP2A (At1g13320) was used as a loading control. To determine the 3′end of COLDWRAP, we employed tiling 5′RACE PCR method as described previously (Heo and Sung, 2011). In brief, a series of tiling reverse primers were designed around predicted 3′ end of COLDWRAP (Supplementary Fig. S2B) and used for the synthesis of first strand cDNA. These first strand cDNA products were used as a template to amplify COLDWRAP using primer combination of AAP/COLDWRAP-specific C2-2 reverse primer (Supplementary Fig. S2B; Supplementary Table S1) and further used for second PCR amplification with primer combination of AUAP/COLDWRAP-specific C2 reverse primer (Supplementary Fig. S2D).
Chromatin Immunoprecipitation (ChIP) assay
Whole seedling plants were harvested and cross-linked in 1% formaldehyde-containing buffer. Cross-linked seedling plants were ground in liquid nitrogen and used for further ChIP experiments as previously described (Kim et al., 2010). To measure specificity of CLF antibody, we performed ChIP assay between WT and clf-29 mutants. Enrichment level at FLC, AG, and FUS3 (At3g26790) loci were quantified by qPCR, normalized to the reference gene, PP2A (At1g13320) (Figure S1H). Information for primer sequences used in ChIP-qPCR analyses were shown in Supplemental Data Table 1.
RNA pull-down assay using in vitro transcribed biotinylated RNA
RNA pull-down assay was conducted as previously described (Heo and Sung, 2011). In short, Biotin-labeled RNAs were in vitro transcribed using the Biotin RNA Labeling Mix (Roche, IN, USA) and T7 RNA polymerase (Roche, IN, USA), and then treated with RNase-free DNaseI enzyme (Thermo Fisher Scientific). Transcribed RNAs were purified using RNeasy Mini Kit (Qiagen) and three micrograms of biotin-labeled RNAs were used for binding assay with His-tagged recombinant CLF protein or nuclear extract from FRI-Col wild type plants. Ten micrograms of non-biotinylated RNAs were used for the competition assay. After 6 hours incubation at 4°C in pull-down buffer [50mM Tris-Cl (pH 7.5), 100mM NaCl, 2mM DTT, 0.05% NP-40, RNase-OUT RNAse inhibitor (40U/mL; Thermo Fisher Scientific) and a protease inhibitor tablet (Roche, IN, USA)], streptavidin agarose beads (Roche, IN, USA) was added to each binding reaction and further incubated for overnight at 4°C. Next day, protein A-agarose beads were added into samples and rotated for additional 2 hours at 4°C. Then, beads were washed at 4°C briefly five times using pull-down buffer and boiled in SDS loading buffer, and then loaded onto SDS-PAGE gel for further Western blot analysis using anti-His (Santa Cruz Biotech) or anti-CLF antibodies.
Chromosome Conformation Capture (3C) assay
3C assays were performed according to Louwers et al. (Louwers et al., 2009) with minor modifications. Arabidopsis whole seedlings were cross-linked with 1% formaldehyde buffer at 4°C for 30 minutes. Seedlings were dried and ground in liquid nitrogen. Nuclei were isolated and treated with 0.3% SDS at 65°C for 40 minutes. SDS was sequestered with 1% Triton X-100. Digestions were performed overnight at 37°C with 400 U of DpnII restriction enzyme (NEB). Restriction enzymes were inactivated by addition of 1.6% SDS and incubation at 65°C for 10 minutes and then, 2% Triton X-100 was added to sequester SDS. Ligations were performed at 22°C overnight in 4-ml volume using 200 U of T4 DNA ligase (NEB). Reverse crosslinking was performed for 6 hours at 65°C. After Proteinase K (NEB) treatment, DNA was purified by phenol/chloroform/isoamyl-alcohol (25:24:1) extraction and ethanol precipitation. Quantitative PCR reaction (qPCR) was performed using Maxima SYBR green master mix (Thermo Scientific) according to the manufacturer’s instructions. Quantitative PCR reaction was performed on ViiA™ 7 Real-Time PCR System (Life Technologies). Primer sequences used in 3C-qPCR analyses are listed in Supplementary Table 1.
QUANTIFICATION AND STATISTICAL ANALYSIS
Measurement of flowering time
Flowering time was measured as a rosette leaf number at a bolting stage under the long-day condition. Numbers of plants used for measurement of flowering time were indicated in Figure legends (Fig. 2, 4, Fig S3, S4, S6)
RNA-IP followed by quantitative RT-PCR analysis
Relative amounts of retrieved RNA were calculated using double delta Ct method by comparing relative level to input RNA. Quantitative RT-PCR reactions were performed using ViiA 7 real time system (Life technologies, USA) with 33 cycles. Number of biological replicates used for qRT-PCR were indicated in Figure legends (Figure S1A~B). The mean and SD of these sampled calculations are shown. Primer sets spanning FLC genomic regions used in previous study were used for quantitative tiling RT-PCR analysis (Supplementary Table 1).
Quantitative RT-PCR analysis
Relative transcript level of each gene was calculated using double delta Ct method by comparing to relative expression level of PP2A reference gene (Czechowski et al., 2005). For COLDAIR, total RNA were used for first strand cDNA syntheses using random primers (Gene Link) with M-MLV System (Promega) and subsequently combination of anchor primers, AAP and AUAP (Invitrogen) and gene-specific reverse primers (COLD1R and COLD2R) were used for detection (Fig. S3D). Number of biological replicates used for qRT-PCR were indicated in Figure legends (Fig 1–4, Fig S2~S4, Fig S6). The mean and SD of these sampled calculations were shown in Figures.
ChIP analysis
Quantitative PCR reaction (qPCR) was performed and calculated using double delta Ct method by comparing relative level to input DNA. ChIP data for FLC were quantified by qPCR, normalized to the internal reference gene (AG, At4g18960) and then relative fold changes were represented. Quantitative PCR reaction was performed in a 384-well PCR plate of ViiA 7 Real-Time PCR System (Life Technologies). Numbers of biological replicates used for ChIP-qPCR were indicated in Figure legends (Fig. 3, S3). The mean and SD of these sampled calculations were shown in Figures.
3C-qPCR analysis
Relative interaction frequencies (Hagege et al., 2007) were calculated by quantitative PCR reaction. In brief, cycle threshold (Ct) value was determined for the each 3C sample with primer combination between anchor primers (4F) and the rest of FLC primers (series of F primers or R primers which span FLC region (Supplementary Table 1). Then, we also determined the Ct value of PP2A loading control (LC), a PP2A primer set which does not span DpnII restriction sites (Supplementary Fig. S8D) to correct variations in DNA concentration among 3C samples. Thus, ratios of Ct value of 3C product to Ct value of PP2A (LC) of all samples were calculated. Next, we corrected primer efficiencies for primer pairs by using control template DNA (CTD) as described in (Crevillen et al., 2013). The CTD was obtained by digesting subcloned FLC genomic region (FLC-pPZP211 plasmid) DNA with DpnII restriction enzyme (NEB), and subsequent random ligation with T4 DNA ligase (NEB). FLC-pPZP211 plasmid is about 20-kb long, and contains whole FLC genomic sequence (about 8kb). We measured Ct value of CTD for each primer pair and then used to normalize ratio of Ct value of all 3C samples. Finally, relative interaction frequency was calculated and shown as proportion of each fragment to value of highest peak fragments (i.e. F4+F17 fragment in Fig. 5B or F4+R15 fragment in Supplementary Fig S8A) in 3C assays. Numbers of biological replicates used for qRT-PCR were indicated in Figure legends (Fig. 5, 6, S5). The mean and SD of these sampled calculations were shown in Figures.
Supplementary Material
Highlights.
A long noncoding RNA, COLDWRAP, is transcribed from the repressed FLC promoter
COLDWRAP is necessary for vernalization-mediated FLC silencing
FLC silencing by vernalization includes the formation of an intragenic chromatin loop
lncRNAs and PRC2 are necessary for the formation of an intragenic chromatin loop
Acknowledgments
This work was funded by a grant from the National Institutes of Health (GM100108) and in part by a grant from the Next-Generation BioGreen 21 Program (PJ011106; RDA, Republic of Korea) to S. S. We thank Dr. Enamul Huq for helpful comments on the manuscript. Extended Data contains additional data.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
D.-H. K. conducted the experiments, D.-H. K. and S. S. designed the experiments and wrote the paper.
References
- Amasino RM, Michaels SD. The timing of flowering. Plant Physiol. 2010;154:516–520. doi: 10.1104/pp.110.161653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres F, Coupland G. The genetic basis of flowering responses to season al cues. Nat Rev Genet. 2012;13:627–639. doi: 10.1038/nrg3291. [DOI] [PubMed] [Google Scholar]
- Angel A, Song J, Dean C, Howard M. A Polycomb-based switch underlying quantitative epigenetic memory. Nature. 2011;476:105–108. doi: 10.1038/nature10241. [DOI] [PubMed] [Google Scholar]
- Ariel F, Jegu T, Latrasse D, Romero-Barrios N, Christ A, Benhamed M, Crespi M. Noncoding transcription by alternative RNA polymerases dynamically regulates an auxin-driven chromatin loop. Mol Cell. 2014;55:383–396. doi: 10.1016/j.molcel.2014.06.011. [DOI] [PubMed] [Google Scholar]
- Bastow R, Mylne JS, Lister C, Lippman Z, Martienssen RA, Dean C. Vernalization requires epigenetic silencing of FLC by histone methylation. Nature. 2004;427:164–167. doi: 10.1038/nature02269. [DOI] [PubMed] [Google Scholar]
- Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockdorff N. Noncoding RNA and Polycomb recruitment. Rna. 2013;19:429–442. doi: 10.1261/rna.037598.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S, Kumimoto RW, Gnesutta N, Calogero AM, Mantovani R, Holt BF., 3rd A distal CCAAT/NUCLEAR FACTOR Y complex promotes chromatin looping at the FLOWERING LOCUS T promoter and regulates the timing of flowering in Arabidopsis. Plant Cell. 2014;26:1009–1017. doi: 10.1105/tpc.113.120352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44:667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C, Zhang QC, da Rocha ST, Flynn RA, Bharadwaj M, Calabrese JM, Magnuson T, Heard E, Chang HY. Systematic discovery of Xist RNA binding proteins. Cell. 2015;161:404–416. doi: 10.1016/j.cell.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coustham V, Li P, Strange A, Lister C, Song J, Dean C. Quantitative modulation of polycomb silencing underlies natural variation in vernalization. Science. 2012;337:584–587. doi: 10.1126/science.1221881. [DOI] [PubMed] [Google Scholar]
- Crevillen P, Sonmez C, Wu Z, Dean C. A gene loop containing the floral repressor FLC is disrupted in the early phase of vernalization. EMBO J. 2013;32:140–148. doi: 10.1038/emboj.2012.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005;139:5–17. doi: 10.1104/pp.105.063743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidovich C, Zheng L, Goodrich KJ, Cech TR. Promiscuous RNA binding by Polycomb repressive complex 2. Nat Struct Mol Biol. 2013;20:1250–1257. doi: 10.1038/nsmb.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lucia F, Crevillen P, Jones AM, Greb T, Dean C. A PHD-polycomb repressive complex 2 triggers the epigenetic silencing of FLC during vernalization. Proc Natl Acad Sci U S A. 2008;105:16831–16836. doi: 10.1073/pnas.0808687105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enderle D, Beisel C, Stadler MB, Gerstung M, Athri P, Paro R. Polycomb preferentially targets stalled promoters of coding and noncoding transcripts. Genome Res. 2011;21:216–226. doi: 10.1101/gr.114348.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Cokus SJ, Schubert V, Zhai J, Pellegrini M, Jacobsen SE. Genome-wide Hi-C analyses in wild-type and mutants reveal high-resolution chromatin interactions in Arabidopsis. Mol Cell. 2014;55:694–707. doi: 10.1016/j.molcel.2014.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan EJ, Dennis ES. Vernalization-induced trimethylation of histone H3 lysine 27 at FLC is not maintained in mitotically quiescent cells. Curr Biol. 2007;17:1978–1983. doi: 10.1016/j.cub.2007.10.026. [DOI] [PubMed] [Google Scholar]
- Gendall AR, Levy YY, Wilson A, Dean C. The VERNALIZATION 2 gene mediates the epigenetic regulation of vernalization in Arabidopsis. Cell. 2001;107:525–535. doi: 10.1016/s0092-8674(01)00573-6. [DOI] [PubMed] [Google Scholar]
- Gibcus JH, Dekker J. The hierarchy of the 3D genome. Mol Cell. 2013;49:773–782. doi: 10.1016/j.molcel.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greb T, Mylne JS, Crevillen P, Geraldo N, An H, Gendall AR, Dean C. The PHD finger protein VRN5 functions in the epigenetic silencing of Arabidopsis FLC. Curr Biol. 2007;17:73–78. doi: 10.1016/j.cub.2006.11.052. [DOI] [PubMed] [Google Scholar]
- Grob S, Schmid MW, Luedtke NW, Wicker T, Grossniklaus U. Characterization of chromosomal architecture in Arabidopsis by chromosome conformation capture. Genome Biol. 2013;14:R129. doi: 10.1186/gb-2013-14-11-r129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber AR, Lorenz R, Bernhart SH, Neubock R, Hofacker IL. The Vienna RNA websuite. Nucleic Acids Res. 2008;36:W70–74. doi: 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagege H, Klous P, Braem C, Splinter E, Dekker J, Cathala G, de Laat W, Forne T. Quantitative analysis of chromosome conformation capture assays (3C-q PCR) Nat Protoc. 2007;2:1722–1733. doi: 10.1038/nprot.2007.243. [DOI] [PubMed] [Google Scholar]
- Helliwell CA, Robertson M, Finnegan EJ, Buzas DM, Dennis ES. Vernalization-Repression of Arabidopsis FLC Requires Promoter Sequences but Not Antisense Transcripts. PLoS One. 2011;6:e21513. doi: 10.1371/journal.pone.0021513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig L, Derkacheva M. Diversity of Polycomb group complexes in plants: same rules, different players? Trends Genet. 2009;25:414–423. doi: 10.1016/j.tig.2009.07.002. [DOI] [PubMed] [Google Scholar]
- Heo JB, Sung S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science. 2011;331:76–79. doi: 10.1126/science.1197349. [DOI] [PubMed] [Google Scholar]
- Hyun Y, Yun H, Park K, Ohr H, Lee O, Kim DH, Sung S, Choi Y. The catalytic subunit of Arabidopsis DNA polymerase alpha ensures stable maintenance of histone modification. Development. 2013;140:156–166. doi: 10.1242/dev.084624. [DOI] [PubMed] [Google Scholar]
- Jeon Y, Lee JT. YY1 tethers Xist RNA to the inactive X nucleation center. Cell. 2011;146:119–133. doi: 10.1016/j.cell.2011.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin F, Li Y, Dixon JR, Selvaraj S, Ye Z, Lee AY, Yen CA, Schmitt AD, Espinoza CA, Ren B. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503:290–294. doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S, Bonasio R, Saldana-Meyer R, Yoshida T, Son J, Nishino K, Umezawa A, Reinberg D. Interactions between JARID2 and noncoding RNAs regulate PRC2 recruitment to chromatin. Mol Cell. 2014;53:290–300. doi: 10.1016/j.molcel.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S, Li G, Son J, Xu CF, Margueron R, Neubert TA, Reinberg D. Phosphorylation of the PRC2 component Ezh2 is cell cycle-regulated and up-regulate s its binding to ncRNA. Genes Dev. 2010;24:2615–2620. doi: 10.1101/gad.1983810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanhere A, Viiri K, Araujo CC, Rasaiyaah J, Bouwman RD, Whyte WA, Pereira CF, Brookes E, Walker K, Bell GW, et al. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Mol C ell. 2010;38:675–688. doi: 10.1016/j.molcel.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakatsu T, Huang SS, Jupe F, Sasaki E, Schmitz RJ, Urich MA, Castanon R, Nery JR, Barragan C, He Y, et al. Epigenomic Diversity in a Global Collection of Arabidopsis thaliana Accessions. Cell. 2016;166:492–505. doi: 10.1016/j.cell.2016.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Doyle MR, Sung S, Amasino RM. Vernalization: winter and the timing of flowering in plants. Annu Rev Cell Dev Biol. 2009;25:277–299. doi: 10.1146/annurev.cellbio.042308.113411. [DOI] [PubMed] [Google Scholar]
- Kim DH, Sung S. Coordination of the Vernalization Response through a VIN3 and FLC Gene Family Regulatory Network in Arabidopsis. Plant Cell. 2013;25:454–469. doi: 10.1105/tpc.112.104760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Zografos BR, Sung S. Vernalization-mediated VIN3 Induction Overcomes the LIKE-HETEROCHROMATIN PROTEIN1/POLYCOMB REPRESSION COMPLEX2-mediated epigenetic repression. Plant Physiol. 2010;154:949–957. doi: 10.1104/pp.110.161083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler C, Hennig L, Spillane C, Pien S, Gruissem W, Grossniklaus U. The Polycomb-group protein MEDEA regulates seed development by controlling expression of the MADS-box gene PHERES1. Genes Dev. 2003;17:1540–1553. doi: 10.1101/gad.257403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laine JP, Singh BN, Krishnamurthy S, Hampsey M. A physiological role for gene loops in yeast. Genes Dev. 2009;23:2604–2609. doi: 10.1101/gad.1823609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzuolo C, Orlando V. Memories from the polycomb group proteins. Annu Rev Genet. 2012;46:561–589. doi: 10.1146/annurev-genet-110711-155603. [DOI] [PubMed] [Google Scholar]
- Lanzuolo C, Roure V, Dekker J, Bantignies F, Orlando V. Polycomb response elements mediate the formation of chromosome higher-order structures in the bithorax complex. Nat Cell Biol. 2007;9:1167–1174. doi: 10.1038/ncb1637. [DOI] [PubMed] [Google Scholar]
- Louwers M, Splinter E, van Driel R, de Laat W, Stam M. Studying physical chromatin interactions in plants using Chromosome Conformation Capture (3C) Nat Protoc. 2009;4:1216–1229. doi: 10.1038/nprot.2009.113. [DOI] [PubMed] [Google Scholar]
- Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. 2013;20:300–307. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell. 1999;11:949–956. doi: 10.1105/tpc.11.5.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negishi M, Wongpalee SP, Sarkar S, Park J, Lee KY, Shibata Y, Reon BJ, Abounader R, Suzuki Y, Sugano S, et al. A new lncRNA, APTR, associates with and represses the CDKN1A/p21 promoter by recruiting polycomb proteins. PLoS One. 2014;9:e95216. doi: 10.1371/journal.pone.0095216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orom UA, Shiekhattar R. Long non-coding RNAs and enhancers. Curr Opin Genet Dev. 2011;21:194–198. doi: 10.1016/j.gde.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annual review of biochemistry. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma K, Cifuentes-Rojas C, Ergun A, Del Rosario A, Jeon Y, White F, Sadreyev R, Lee JT. ATRX directs binding of PRC2 to Xist RNA and Polycomb targets. Cell. 2014;159:869–883. doi: 10.1016/j.cell.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert D, Clarenz O, Goodrich J. Epigenetic control of plant development by Polycomb-group proteins. Curr Opin Plant Biol. 2005;8:553–561. doi: 10.1016/j.pbi.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Sheldon CC, Jean Finnegan E, James Peacock W, Dennis ES. Mechanisms of gene repression by vernalization in Arabidopsis. Plant J. 2009 doi: 10.1111/j.1365-313X.2009.03883.x. [DOI] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- Somarowthu S, Legiewicz M, Chillon I, Marcia M, Liu F, Pyle AM. HOTAIR forms an intricate and modular secondary structure. Mol Cell. 2015;58:353–361. doi: 10.1016/j.molcel.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung S, Amasino RM. Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature. 2004;427:159–164. doi: 10.1038/nature02195. [DOI] [PubMed] [Google Scholar]
- Sung S, He Y, Eshoo TW, Tamada Y, Johnson L, Nakahigashi K, Goto K, Jacobsen SE, Amasino RM. Epigenetic maintenance of the vernalized state in Arabidopsis thaliana requires LIKE HETEROCHROMATIN PROTEIN 1. Nat Genet. 2006a;38:706–710. doi: 10.1038/ng1795. [DOI] [PubMed] [Google Scholar]
- Sung S, Schmitz RJ, Amasino RM. APHD finger protein involved in both the vernalization and photoperiod pathways in Arabidopsis. Genes Dev. 2006b;20:3244–3248. doi: 10.1101/gad.1493306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiezewski S, Liu F, Magusin A, Dean C. Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature. 2009;462:799–802. doi: 10.1038/nature08618. [DOI] [PubMed] [Google Scholar]
- Tan-Wong SM, Wijayatilake HD, Proudfoot NJ. Gene loops function to maintain transcriptional memory through interaction with the nuclear pore complex. Genes Dev. 2009;23:2610–2624. doi: 10.1101/gad.1823209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bortle K, Corces VG. Nuclear organization and genome function. Annu Rev Cell Dev Biol. 2012;28:163–187. doi: 10.1146/annurev-cellbio-101011-155824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance KW, Ponting CP. Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends Genet. 2014;30:348–355. doi: 10.1016/j.tig.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Xia H, Zhang Y, Zhao S, Zhao C, Hou L, Li C, Li A, Ma C, Wang X. Genome-wide high-resolution mapping of DNA methylation identifies epigenetic variation across embryo and endosperm in Maize (Zea may) BMC genomics. 2015;16:21. doi: 10.1186/s12864-014-1204-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, Aza-Blanc P, Hogenesch JB, Schultz PG. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science. 2005;309:1570–1573. doi: 10.1126/science.1115901. [DOI] [PubMed] [Google Scholar]
- Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Froberg JE, Lee JT. Long noncoding RNAs: fresh perspectives into the RNA world. Trends Biochem Sci. 2014;39:35–43. doi: 10.1016/j.tibs.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, Song JJ, Kingston RE, Borowsky M, Lee JT. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell. 2010;40:939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D, Rosa S, Dean C. Nuclear organization changes and the epigenetic silencing of FLC during vernalization. J Mol Biol. 2015;427:659–669. doi: 10.1016/j.jmb.2014.08.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


