Summary
We now appreciate that the vascular endothelium plays a crucial role in regulating normal blood vessel physiology in the kidney. The gene products responsible are commonly expressed exclusively, or preferentially, in this cell type. However, despite the importance of regulated gene expression in the vascular endothelium, relatively little is known about the mechanisms that restrict endothelial-specific gene expression to this cell type. Even less is known about how gene expression might be restricted to endothelial cells of discrete regions of the kidney, such as the glomerulus or vasa recta. Although significant progress has been made toward understanding the regulation of endothelial genes through cis/trans paradigms, it has become apparent that additional mechanisms also must be operative. Classic models of transcription in vascular endothelial cells, specifically the cis/trans paradigm, have limitations. For instance, how does the environment have chronic effects on gene expression in endothelial cells after weeks or years? When an endothelial cell divides, how is this information transmitted to daughter cells? Chromatin-based mechanisms, including cell-specific DNA methylation patterns and post-translational histone modifications, recently were shown to play important roles in gene expression. This review investigates the involvement of epigenetic regulatory mechanisms in vascular endothelial cell-specific gene expression using endothelial nitric oxide synthase as a prototypical model.
Keywords: Cell-specific expression, endothelium, DNA methylation, histone post-translational modifications, noncoding RNAs
The endothelium forms the inner cellular lining of blood vessels and lymphatics and has a complex and important role in the health of patients with kidney disease. Endothelial dysfunction in both the renal and systemic vasculature can be the cause, or occur as a result, of renal dysfunction.1–5
At the heart of phenotype and function lies the molecular signature of the cell. Although endothelial cells (ECs) share functional and morphologic features, molecular studies have shown that many “endothelial” protein/messenger RNA markers are not uniform in the endothelium.6,7 In fact, gene expression profiling of 52 cultured human ECs showed distinct and characteristic tissue-specific profiles for cells from different regions.8 There are genes specific to arterial versus venous ECs, genes that are specific to macrovascular cells versus microvascular cells, and a broader correlation of tissue-specific endothelial gene expression with genes involved in body plan development. Functionally, these gene expression patterns reflect the numerous roles of ECs in normal physiology including regulation of vasomotor tone, hemostasis, cell and nutrient trafficking, permeability of vessels, and growth of new blood vessels.9,10
The expression of many of the key genes involved in these processes is unique, or highly restricted, to the endothelium. Yet, surprisingly, the molecular mechanisms underlying how endothelial-specific patterns of gene expression are established and maintained are poorly understood. In this review we examine the mechanisms responsible for endothelial-specific gene expression using endothelial nitric oxide synthase (eNOS) as a model.
VASCULAR ENDOTHELIUM IN THE KIDNEY
In support of its role in maintaining metabolic homeostasis, the kidney has a complex vascular network that exemplifies the heterogeneity in endothelial structure and function. Thus, the endothelium is differentially unique in the renal arteries, veins, glomerulus, peritubular capillary plexus, postcapillary venule, ascending vasa recta, and descending vasa recta. In glomerular capillary ECs, expression of the vascular endothelial growth factor receptors is important in the development of fenestrations and regulation of EC permeability.11–14 Glomerular capillary ECs also synthesize negatively charged molecules that form the glycocalyx, a membrane-associated cell surface layer of proteoglycans, glycosaminoglycans, glycoproteins, and glycolipids that participate in the charge selectivity of the glomerulus.15 In response to functional requirements and the microenvironment, eNOS is expressed more highly in the medullary vasa recta than in the glomerular and peritubular capillaries,16 whereas von Willebrand factor is expressed more robustly in arteries, veins, and peritubular capillaries versus glomerular capillaries.17
eNOS EXPRESSION IS IMPORTANT FOR NORMAL KIDNEY FUNCTION
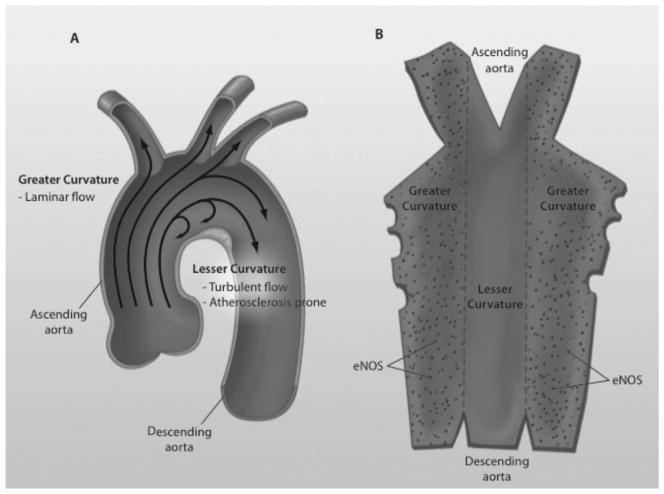
eNOS is a well-characterized endothelial-specific gene18,19 that is important in the control of blood vessel tone and remodeling,20–22 hemostasis,23 angiogenesis,24 and the mobilization of endothelial progenitor cells.25 The product of eNOS is nitric oxide (NO), which possesses vasodilatory, anti-inflammatory, and antithrombotic properties, among others.26–29 Dysregulated expression of eNOS is important in atherosclerosis,30 an important cause of morbidity and mortality in renal patients.31–34 Within a blood vessel wall, focal dynamic changes in eNOS expression are associated with the development of atherosclerotic lesions. Within the aortic arch, eNOS expression is high in the greater curvature, which in general is exposed to more uniform blood flow and has a low probability of developing atherosclerosis; conversely, eNOS expression is low in the lesser curvature, which is exposed to turbulent flow and has a higher probability of atherosclerosis (Fig. 1). This is in contrast to the expression patterns of the inflammatory gene p65, a component of nuclear factor κB, the expression of which is inverse compared with eNOS.35 Interestingly, the expression of eNOS in development has been linked to the initiation of flow within the circulation.36
Figure 1.
Blood flow, expression of eNOS, and atherosclerotic lesions in the aortic arch. (A) The lesser curvature of the aortic arch is exposed to turbulent flow and is more prone to atherosclerosis. The greater curvature, on the other hand, is exposed to laminar flow and is less prone to atherosclerosis. (B) The aorta is opened with a view of the endothelium. eNOS expression (dots) is noticeably decreased in the lesser curvature.
Within the kidney, eNOS knockout mice develop focal pathologic defects including glomerular hypoplasia and tubular cell death, leading to separation of glomeruli from tubules.37 Over time, the kidneys develop a thrombotic microangiopathy with endothelial cell loss, endothelial swelling, and intraluminal thrombi formation.38 In the setting of renal disease models including diabetic nephropathy and anti– glomerular basement membrane glomerulonephritis, deficiency of eNOS accelerates renal injury and the progression of disease.38–40 These data have suggested that eNOS may have a protective role in renal disease including diabetic nephropathy.41,42
The cell-specific expression of certain genes such as eNOS is critical to maintaining normal cellular function and thus normal organ function. A key question, then, is how the cell-specific expression of such genes is regulated in health, and dysregulated in disease.
MECHANISMS OF GENE REGULATION
The promise of the postgenome period argues that the key to human disease including kidney disease, lies within our DNA sequence. Given that the DNA sequence of every diploid human cell is the same, with the exception of T- and B-cell receptors in T and B cells, respectively, as well as somatic mutations, other factors must govern the difference between a glomerular endothelial cell and a renal artery vascular smooth muscle cell or a mesangial cell. How then are cell-specific patterns of gene expression established? How does the environment have chronic effects on gene expression in endothelial cells after weeks or years? When an endothelial cell divides, how is this information transmitted to daughter cells? The key question is how do cells with the same DNA sequence show heterogenous patterns of gene expression?
Until recently, the study of gene regulation has focused on the concept of specific transcription factors binding to canonical promoter elements to mediate transcriptional programs. These traditional cis/trans paradigms, however, do not fully explain characteristics of acquired renal diseases including differences between monozygotic twins, progression of severity over time, and relative late onset of disease.43 Rather, emerging evidence suggests that epigenetic mechanisms are implicated in the control of vascular endothelial gene expression in health and disease.44 It is now known that epigenetic mechanisms provide the key link between genes and the environment, play a fundamental role in the regulation of cell-specific gene expression, as well as the establishment and maintenance of cell fate. These insights led to the launch of the International Human Epigenome Consortium, whose goal is to map 1,000 reference epigenomes.45
DEFINITION OF EPIGENETICS
In its literal translation, “epi” means “above” and genetics refers to “the DNA code.” In the broadest sense, epigenetics can be defined as chromatin-based mechanisms important in the regulation of gene expression that do not involve changes in the DNA sequence per se.46 An important component of this definition is the concept that, in the absence of enzymatic modification, epigenetic marks are stable and heritable through cellular division, either mitosis or meiosis.
EPIGENETIC MECHANISMS
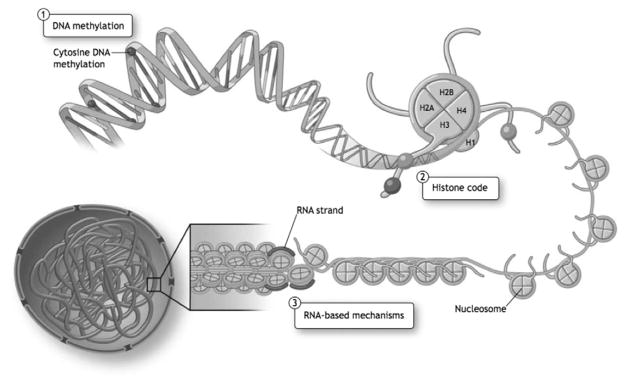
In mammalian systems, epigenetics encompasses three interconnected mechanisms that do not alter the double-stranded base pairing of DNA. The major epigenetic mechanisms are DNA methylation, histone density and post-translational modifications, and RNA-based mechanisms (Fig. 2).44
Figure 2.
Fundamental epigenetic mechanisms. Epigenetic mechanisms refer to three highly interrelated mechanisms that regulate gene expression, as follows: (1) DNA methylation refers to the covalent addition of a methyl group to the 5-position of cytosine in the context of CpG dinucleotides in mammals; (2) histone density and post-translational modifications affect chromatin structure: the fundamental repeating unit of chromatin is the nucleosome that is comprised of an octamer of two each of four core histone proteins; and (3) nuclear RNA-based mechanisms including long noncoding RNAs can direct chromatin structure. Reprinted with permission from Yan et al.43
DNA METHYLATION
DNA methylation occurs when a methyl group is added to the 5-position of cytosine to create 5-methyl-cytosine. In mammals, this occurs most commonly in the context of a CG dinucleotide, where a guanidine residue lies 3′ to a cytosine on a strand of DNA, and, when this occurs in promoter regions, generally is associated with gene repression. DNA methylation in this context plays a fundamental role in a variety of different processes such as genomic imprinting, X-chromosome inactivation, cancer, suppression of repetitive elements, and, more recently, cell-specific gene expression (reviewed by Bird47).
Although not well understood, methylated cytosines found in non-CG contexts also have been described in mammals.44,48 In human beings, with the exception of CpG islands, methylation occurs at 70% to 80% of all CpG dinucleotides in the human genome.49 CpG islands are regions in the genome spanning approximately 1 kb that have a high CpG content (relative to the overall C or G content), and generally are unmethylated. CpG islands span the promoter regions of 60% to 70% of all human genes and approximately 40% of tissue-specific genes (reviewed by Illingworth and Bird50).
DNA methylation is catalyzed by three DNA methyl-transferase (DNMT) enzymes: DNMT1, DNMT3a, and DNMT3b each catalyze the transfer of a methyl group of S-adenosylmethionine to the carbon 5 position of cytosine. DNMT1 is responsible for maintenance methylation through recognition of hemimethylated DNA during DNA replication, whereas DNMT3a and 3b are responsible for de novo DNA methylation during development. DNA methylation is fundamental for embryonic development because mice that lack DNMTs are embryonic lethal.51,52
There are three potential mechanisms by which DNA methylation leads to gene repression. First, 5-methyl-cytosine can sterically hinder transcription factor binding with their cis-elements (reviewed by Miranda and Jones53). Evidence for this repressive mechanism arises from studies showing that certain transcription factors such as c-myc,54 hypoxia inducible factor 1α,55 and the insulator protein CCCTC-binding factor (CTCF)56 are sensitive to DNA methylation. However, other transcription factors such as Sp157 are not affected by DNA methylation, indicating that there are other mechanisms at play. Second, DNA methylation can recruit methyl-binding proteins such as methyl CpG binding protein 1 and 2 (MeCP1, MeCP2),58 and methyl-CpG-binding domain protein 1–4 (MBD1–4),59 which in turn hinder other trans-factor binding. Finally, MeCP2 and MBD2 can recruit other chromatin-modifying co-repressor complexes such as histone deacetylases (HDACs),60,61 and histone demethylases53 to repress gene transcription further.
DNA DEMETHYLATION
The removal of the methyl group from cytosine, DNA demethylation, more recently has been believed to occur in a variety of different processes, such as development, tumorigenesis, neurogenesis, and the immune response. This process can occur via both passive/replication-dependent or active/replication-independent fashion. Passive DNA demethylation occurs when methyl groups are not added to the newly synthesized strand of DNA during replication whereas active replication involves the removal of the methyl group by putative demethylase enzymes (reviewed by Zhu62). An exciting new discovery involves the hydroxylation of 5-methyl-cytosine by tet methylcytosine dioxygenase (TET) family enzymes, creating 5-hydroxymethyl-cytosine. 5-Hydroxymethyl-cytosine may serve as an intermediate for DNA demethylation because it can be converted to cytosine.63,64
Intriguingly, and highly relevant to chronic kidney disease, Kim et al65 showed that parathyroid hormone could induce protein kinase C–mediated phosphorylation of MBD4, leading to conversion of 5-methyl-cytosine to cytosine and activation of cytochrome p450 27B1 (CYP27B1) during vitamin D biosynthesis. This is the key enzyme in the biosynthesis of vitamin D. Importantly, this report argues that phosphorylation of MBD4 allows it to recognize 5-methyl-cytosine in addition to its function as a DNA glycosylase.
HISTONE PROTEINS
In the nucleus, DNA does not exist on its own; rather, it is coiled around octamers of histone proteins to form nucleosomes. One nucleosome consists of 146 bp of DNA coiled around a histone octamer made up of two molecules each of histone H2A, H2B, H3, and H4. Adjacent nucleosome particles are connected via a shorter linker DNA sequence that variably can associate with histone H1. In heterochromatin, these repeating structures are tightly compacted with minimal linker DNA, whereas these structures are spaced more loosely in euchromatin, allowing for greater access to the DNA. This basic unit of chromatin then is compacted further into higher-order structures, ultimately forming a chromosome (reviewed by Misteli66).
Histone proteins are composed of a globular domain, as well as amino-terminal tails, which can undergo post-translational modifications that have a downstream effect on transcription (reviewed by Kouzarides67). These post-translational modifications (PTMs) include acetylation, methylation, phosphorylation, sumoylation, ubiquitination, and adenosine diphosphate ribosylation among others (reviewed by Kouzarides67 and Li et al68). The histone code hypothesis posits that the combination of these PTMs encodes regulatory information that is then “read” by proteins to generate a downstream response.69 Modifications such as the acetylation of H3 and H4, dimethylation or trimethylation of histone 3 lysine 4 (H3K4) at the promoter, as well as methylation of histone 3 lysine 36 (H3K36) in the gene body, are associated with active transcription. In contrast, histone 3 lysine 9 trimethylation (H3K9), histone 3 lysine 27 trimethylation (H3K27), and methylation of histone 4 lysine 20 (H4K20) at the promoter are repressive chromatin marks (reviewed by Li et al68) (Table 1). These modifications are dynamically regulated by opposing enzyme complexes that add, or remove, PTMs. Specifically, histone acetyltransferases add acetyl groups, whereas HDACs can remove acetyl groups (reviewed by Kouzarides67). Similarly, histone lysine methylation is regulated by histone methyltransferases such as the mixed lineage leukemia (MLL) family members, and demethylases such as the jumonji domain containing (JMJD) family members (reviewed by Li et al68).
Table 1.
Epigenetic Mechanisms and Gene Transcription
| Mechanism | Transcriptional Effect |
|---|---|
| DNA methylation (CpG dinucleotides) | ↓ |
| Histone posttranslational modifications | |
| Histone H3 | |
| Acetylation (K9, K14) | ↑ |
| Methylation | |
| K4 trimethylation | ↑ |
| K9 trimethylation | ↓ |
| K27 trimethylation | ↓ |
| K36 trimethylation | ↑ |
| K64 trimethylation | ↓ |
| Phosphorylation (S10) | ↑ |
| Histone H4 | |
| Acetylation (K5, K8, K12, K16) | ↑ |
| Methylation (K20) | ↓ |
| Long noncoding RNAs | |
| Xist, HOTAIR | ↓ |
Abbreviations: K, lysine; S, serine.
Aside from histone modifications that affect the electrostatic attraction of DNA with the core histone proteins, histone density is functionally relevant to transcription with lower histone density at the transcription start site generally associated with active transcription. Eviction of histones leading to lower histone density is mediated primarily through adenosine triphosphate–remodeling complexes such as Switch/Sucrose nonfermenting (SWI/SNF).70 Intriguingly, studies performed in human beings have shown H3/H4 eviction during the activation of vitamin D3–receptor–regulated genes.71 Relevant to this review, hypoxia represses transcription of the eNOS gene in vascular ECs, in part via regulating histone density.72
RNA-BASED MECHANISMS
The epigenetic marks described earlier are ubiquitous across cells types and the mechanisms that direct their specificity is not well described. Recent evidence suggests that nuclear RNA-based mechanisms can direct epigenetic memory.73 In particular, long noncoding RNAs (lncRNAs), to be distinguished from short non-coding RNAs such as microRNAs, can regulate groups of genes by binding to chromatin-modifying complexes such as the polycomb repressive complex 2 complex, which mediates H3K27 trimethylation, and mixed lineage leukemia 1, which mediates H3K4 trimethylation.73–75 These lncRNAs are defined as RNA molecules longer than 200 nt (reviewed by Mercer et al73), which do not code for protein, and function as an RNA molecule. Currently, thousands of lncRNAs have been annotated and play roles in a variety of cellular processes in development and mature cells.76,77 These lncRNAs were shown to have evolutionary conservation across 21 mammalian genomes, indicating their importance in higher-order mammals.76 In fact, some lncRNAs are transcribed from regions of the genome identified by genome-wide association studies to harbor disease-causing mutations including a region on chromosome 9 associated with coronary artery disease.78
One of the best studied long noncoding RNAs is X-inactivation–specific transcript, Xist, which is expressed by the X-chromosome inactivation center, and is responsible for inactivating one of the two female X-chromosomes in mammals (reviewed by Kanduri et al79). Furthermore, recent studies identified another ncRNA, HOX transcript antisense RNA (HOTAIR), which is transcribed from the HOXC locus and acts in trans to repress transcription in the HOXD locus through the interaction with the polycomb repressive complex 2.77 HOTAIR recently was implicated in breast cancer metastasis and mortality.80
Along with histone modifications, lncRNAs also have been shown to play a role in RNA-directed DNA methylation (reviewed by Payer and Lee81). Specifically, Xist antisense RNA, Tsix, negatively regulates Xist by recruiting DNMT3a to the Xist promoter of the active X chromosome, leading to DNA methylation and repression of Xist gene transcription.82 Collectively, these findings are beginning to uncover the fundamental role of lncRNAs in gene regulation.
REGULATION OF eNOS: CIS/TRANS PARADIGM
A simple model to account for the cell-restricted expression of endothelial genes such as eNOS is predicated on the classic cis/trans paradigm of gene expression. In this model, cis-DNA binding elements in the 5′-regulatory regions of genes specifically recruit trans-factors (or transcription factors). To account for cell-specific expression this model requires that cis-DNA binding elements be present in the 5′-regulatory regions of target genes and that the expression of the relevant trans-factors themselves be cell-restricted in expression. Although some cis-elements such as Sp-1, forkhead, and Ets elements are enriched in endothelial target gene promoters, these transcription factors are not.83,84 Indeed, there is no evidence for an endothelial master regulator, which is both necessary and sufficient for the expression of endothelial genes, unlike MyoD for skeletal muscle cells or peroxisome proliferator activated receptor-γ for adipocytes.85,86
REGULATION OF eNOS: EPIGENETICS
To elucidate the factors involved in eNOS expression, we performed transient transfection assays in endothelial cells (express eNOS) and vascular smooth muscle cells (VSMCs) (do not express eNOS) with an episomal eNOS core promoter-luciferase reporter construct.87 Intriguingly, both expressing and nonexpressing cell types displayed robust expression from the episomal constructs.87 These experiments provided us with strong evidence that nonexpressing cell types such as VSMCs contained the necessary trans-factors required for eNOS transcription. eNOS promoter-reporter transgenic mice containing the same eNOS promoter sequences, however, where the promoter-reporter construct is integrated into the genome, show an endothelial-restricted pattern of expression.88 These in vivo findings indicated that chromatin structure and epigenetics may play an important role in keeping eNOS switched off in nonexpressing cell types.
To determine if epigenetic mechanisms played a role in the endothelial-restricted pattern of eNOS, we first determined if DNA methylation played a role in eNOS regulation. By using sodium bisulfite genomic DNA sequencing to assay DNA methylation, we determined that there was a differentially methylated region in the core promoter (−361 to +3).87 Specifically, the eNOS promoter was hypomethylated in ECs, but was hypermethylated in nonexpressing cell types, such as human VSMCs and murine aortas.87 Furthermore, when treated with 5-azacytidine, a DNMT inhibitor, robust expression of eNOS in VSMCs was observed, indicating that promoter DNA methylation is involved directly in eNOS regulation.87 Moreover, the eNOS promoter in VSMCs (but not human umbilical vein endothelial cells [HUVECs]) was enriched in the methyl-binding protein MeCP2, a transcriptionally repressive mark.87 Taken together, these findings provide exciting evidence that DNA methylation plays a key role in the maintenance of cell-specific gene expression patterns.
Further, we showed that the eNOS core promoter in ECs was highly enriched in activating histone marks such as acetylated H3 and H4, and H3K4 dimethylation and trimethylation. These and other activating histone PTMs such as H4K12 acetylation and H3K9 acetylation comprise an active “histone code” at the eNOS promoter in ECs.89
Importantly, blocking histone deacetylase activity using trichostatin A (TSA) in nonexpressing cell types led to an increase in H3/H4 acetylation and an increase in steady-state eNOS messenger RNA levels, indicating that similar to DNA methylation, histone acetylation is fundamental for the regulation of eNOS.89 Taken together, these data provide evidence that the chromatin accessibility to ubiquitously expressed trans-factors governed through epigenetic regulation is critical for eNOS expression.
RNA-based mechanisms also play a role in eNOS regulation. We have found evidence of a natural anti-sense transcript, sONE, that regulates eNOS via post-transcriptional mechanisms. Interestingly, sONE participates both in the cell-specific expression of eNOS and in the hypoxia-mediated decrease in eNOS steady-state messenger RNA levels.90,91 Furthermore, a small, 27-nt RNA originating from the variable number tandem region of intron 4 can repress eNOS transcription in association with a reduction in activating epigenetic marks such as H3K9 and H4K12 acetylation at the promoter and an increase in repressive epigenetic marks such as DNA methylation at exon 3.92,93 Interestingly, copy number variation of the eNOS variable number tandem region is associated with increased risk of ischemic heart disease.94
EPIGENETICS, THE VASCULAR ENDOTHELIUM, AND KIDNEY DISEASE
Epigenetic mechanisms have become increasingly important as targets for both the treatment and pathogenesis of human diseases. Recently, we found that the HDAC inhibitor vorinostat decreased albuminuria and mesangial collagen IV deposition in a mouse model of diabetic nephropathy. Interestingly, these effects were found to be dependent on eNOS expression in the kidney.42 Another HDAC inhibitor, TSA, is being studied as a promising treatment for various cancers (reviewed by Carew et al95), as well as kidney diseases. TSA in combination with retinoic acid inhibited renal cell carcinoma proliferation.96 TSA also reduced cyst formation in polycystic kidney disease in mice,97,98 attenuated macrophage infiltration and fibrotic changes in tubulointerstitial injury,99 as well as inhibited renal fibroblast activation and tubular cell apoptosis in tubulointerstitial fibrosis.100 These findings suggest a prominent role for epigenetic mechanisms in the pathogenesis of kidney disease.
Intriguingly, recent studies have shown that DNA methylation of specific genes in cancer can itself cause hereditary nonpolyposis colorectal cancer (HNPCC), and that this modification can be inherited through the germline. In the absence of genetic mutation, allele-specific hypermethylation of the mismatch repair gene MutL homolog 1 (MLH1) led to gene silencing and subsequent HNPCC. These epimutations were germline because they were present in spermatozoa, as well as other somatic tissues such as buccal mucosa, hair follicles, and peripheral blood.101 Similarly, germline allelic-specific and mosaic hypermethylation of the mismatch repair gene mutS homolog 2 (MSH2) also was involved in HNPCC. Three siblings carried the germline epimutation and developed early onset HNPCC or endometrial cancers.102 Taken together these studies suggest that epimutations themselves are heritable, and can increase susceptibility to disease. Clearly, this will be an exciting area to explore.
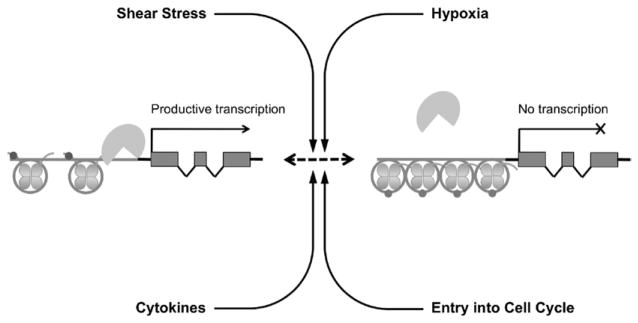
The role of epigenetic mechanisms is not limited to the constitutive expression of important endothelial genes such as eNOS as discussed earlier. The human inducible NOS (iNOS) gene is not expressed constitutively but is inducible only in certain cell types. Cell types such as ECs, which are strongly resistant to iNOS induction, have iNOS promoters with extensive DNA methylation whereas cell types in which iNOS is inducible have hypomethylated iNOS promoters.103 These mechanisms also are fundamental in mediating the endothelial cellular response to external stimuli such as cytokine stimulation, shear stress, and hypoxia,43 which are factors that play an important role in the pathogenesis of blood vessel diseases (Fig. 3).
Figure 3.
Epigenetics and environmental stimuli. Epigenetic pathways are important in the interface between genes and the environment. Important environmental stimuli such as entry into the cell cycle, shear stress, cytokines, and hypoxia are important in the epigenetic regulation of vascular endothelial gene expression in health and disease.
For instance, laminar shear stress can increase eNOS expression in part, via epigenetic alterations. Shear stress–mediated eNOS activation is mediated by increased p300 histone acetyltransferase activity, which increases acetylation of p65, as well as acetylation of histone H3 and H4 at the eNOS shear stress response element.104 Blocking p300 activity, on the other hand, inhibits eNOS activation.104 Importantly, we showed that eNOS expression is attenuated in regions of the mouse aorta predisposed to forming atherosclerotic lesions,35 which may occur through dysregulation of p300 activity. Furthermore, other studies have shown that shear stress induces the cAMP-response element-binding protein/CREB-binding protein (CREB/CBP) complex, which has histone acetyltransferase activity and can activate flow-responsive genes in HUVEC.105
With respect to hypoxia, our laboratory recently provided evidence that epigenetic mechanisms were important in the endothelial response to oxygen deprivation. Specifically, we found that hypoxia attenuated eNOS expression, concomitant with a decrease in histone acetylation, as well as H3K4 methylation.72 Intriguingly, this occurred as a result of hypoxia-induced eviction of acetylated histones at the eNOS promoter in ECs and reincorporation of histone proteins that lacked activating PTMs.72
SUMMARY
In this review, we have highlighted that the spatial and temporal regulation of gene expression patterns is critical to the normal functioning of our cells. We present epigenetics as a ubiquitous and powerful perspective from which to understand gene regulation, particularly in endothelial cells. Epigenetics is attractive as a model because it includes a mechanism by which the DNA sequence can interact with the environment and provides a therapeutic target for re-establishment of normal gene expression patterns in disease. To date, the study of epigenetics is in its infancy and the field is ripe for discoveries that have translational implications on vascular endothelial cells in kidney disease.
Acknowledgments
Financial support: Supported by a Career Investigator Award from the Heart and Stroke Foundation of Canada and by a grant from the Canadian Institute of Health Research (CIHR MOP 79475) (P.A.M.).
Footnotes
Conflict of interest statement: none.
References
- 1.Blum M, Yachnin T, Wollman Y, et al. Low nitric oxide production in patients with chronic renal failure. Nephron. 1998;79:265–8. doi: 10.1159/000045047. [DOI] [PubMed] [Google Scholar]
- 2.Kang DH, Kanellis J, Hugo C, et al. Role of the microvascular endothelium in progressive renal disease. J Am Soc Nephrol. 2002;13:806–16. doi: 10.1681/ASN.V133806. [DOI] [PubMed] [Google Scholar]
- 3.Petruzziello TN, Mawji IA, Khan M, Marsden PA. Verotoxin biology: molecular events in vascular endothelial injury. Kidney Int Suppl. 2009;112:S17–9. doi: 10.1038/ki.2008.612. [DOI] [PubMed] [Google Scholar]
- 4.Dworkin LD, Cooper CJ. Clinical practice. Renal-artery stenosis. N Engl J Med. 2009;361:1972–8. doi: 10.1056/NEJMcp0809200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lilien MR, Groothoff JW. Cardiovascular disease in children with CKD or ESRD. Nat Rev Nephrol. 2009;5:229–35. doi: 10.1038/nrneph.2009.10. [DOI] [PubMed] [Google Scholar]
- 6.DeLisser HM, Christofidou-Solomidou M, Strieter RM, et al. Involvement of endothelial PECAM-1/CD31 in angiogenesis. Am J Pathol. 1997;151:671–7. [PMC free article] [PubMed] [Google Scholar]
- 7.Aird WC. Spatial and temporal dynamics of the endothelium. J Thromb Haemost. 2005;3:1392–406. doi: 10.1111/j.1538-7836.2005.01328.x. [DOI] [PubMed] [Google Scholar]
- 8.Chi JT, Chang HY, Haraldsen G, et al. Endothelial cell diversity revealed by global expression profiling. Proc Natl Acad Sci U S A. 2003;100:10623–8. doi: 10.1073/pnas.1434429100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cines DB, Pollak ES, Buck CA, et al. Endothelial cells in physiology and in the pathophysiology of vascular disorders. Blood. 1998;91:3527–61. [PubMed] [Google Scholar]
- 10.Marsden PA, Brenner BM. Nitric oxide and endothelins: novel autocrine/paracrine regulators of the circulation. Semin Nephrol. 1991;11:169–85. [PubMed] [Google Scholar]
- 11.Aird WC. Phenotypic heterogeneity of the endothelium: I. Structure, function, and mechanisms. Circ Res. 2007;100:158–73. doi: 10.1161/01.RES.0000255691.76142.4a. [DOI] [PubMed] [Google Scholar]
- 12.Roberts WG, Palade GE. Increased microvascular permeability and endothelial fenestration induced by vascular endothelial growth factor. J Cell Sci. 1995;108:2369–79. doi: 10.1242/jcs.108.6.2369. [DOI] [PubMed] [Google Scholar]
- 13.Aird WC. Phenotypic heterogeneity of the endothelium: II. Representative vascular beds. Circ Res. 2007;100:174–90. doi: 10.1161/01.RES.0000255690.03436.ae. [DOI] [PubMed] [Google Scholar]
- 14.Satchell SC, Tasman CH, Singh A, et al. Conditionally immortalized human glomerular endothelial cells expressing fenestrations in response to VEGF. Kidney Int. 2006;69:1633–40. doi: 10.1038/sj.ki.5000277. [DOI] [PubMed] [Google Scholar]
- 15.Jeansson M, Haraldsson B. Morphological and functional evidence for an important role of the endothelial cell glycocalyx in the glomerular barrier. Am J Physiol Renal Physiol. 2006;290:F111–6. doi: 10.1152/ajprenal.00173.2005. [DOI] [PubMed] [Google Scholar]
- 16.Han KH, Lim JM, Kim WY, Kim H, Madsen KM, Kim J. Expression of endothelial nitric oxide synthase in developing rat kidney. Am J Physiol Renal Physiol. 2005;288:F694–702. doi: 10.1152/ajprenal.00085.2004. [DOI] [PubMed] [Google Scholar]
- 17.Pusztaszeri MP, Seelentag W, Bosman FT. Immunohistochemical expression of endothelial markers CD31, CD34, von Willebrand factor, and Fli-1 in normal human tissues. J Histochem Cytochem. 2006;54:385–95. doi: 10.1369/jhc.4A6514.2005. [DOI] [PubMed] [Google Scholar]
- 18.Marsden PA, Schappert KT, Chen HS, et al. Molecular cloning and characterization of human endothelial nitric oxide synthase. FEBS Lett. 1992;307:287–93. doi: 10.1016/0014-5793(92)80697-f. [DOI] [PubMed] [Google Scholar]
- 19.Gnanapandithen K, Chen Z, Kau CL, Gorczynski RM, Marsden PA. Cloning and characterization of murine endothelial constitutive nitric oxide synthase. Biochim Biophys Acta. 1996;1308:103–6. doi: 10.1016/0167-4781(96)00098-x. [DOI] [PubMed] [Google Scholar]
- 20.Huang PL, Huang Z, Mashimo H, et al. Hypertension in mice lacking the gene for endothelial nitric oxide synthase. Nature. 1995;377:239–42. doi: 10.1038/377239a0. [DOI] [PubMed] [Google Scholar]
- 21.Shesely EG, Maeda N, Kim HS, et al. Elevated blood pressures in mice lacking endothelial nitric oxide synthase. Proc Natl Acad Sci U S A. 1996;93:13176–81. doi: 10.1073/pnas.93.23.13176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rudic RD, Shesely EG, Maeda N, Smithies O, Segal SS, Sessa WC. Direct evidence for the importance of endothelium-derived nitric oxide in vascular remodeling. J Clin Invest. 1998;101:731–6. doi: 10.1172/JCI1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Freedman JE, Sauter R, Battinelli EM, et al. Deficient platelet-derived nitric oxide and enhanced hemostasis in mice lacking the NOSIII gene. Circ Res. 1999;84:1416–21. doi: 10.1161/01.res.84.12.1416. [DOI] [PubMed] [Google Scholar]
- 24.Lee PC, Salyapongse AN, Bragdon GA, et al. Impaired wound healing and angiogenesis in eNOS-deficient mice. Am J Physiol. 1999;277:H1600–8. doi: 10.1152/ajpheart.1999.277.4.H1600. [DOI] [PubMed] [Google Scholar]
- 25.Aicher A, Heeschen C, Mildner-Rihm C, et al. Essential role of endothelial nitric oxide synthase for mobilization of stem and progenitor cells. Nat Med. 2003;9:1370–6. doi: 10.1038/nm948. [DOI] [PubMed] [Google Scholar]
- 26.Katsuyama K, Shichiri M, Marumo F, Hirata Y. NO inhibits cytokine-induced iNOS expression and NF-kappaB activation by interfering with phosphorylation and degradation of IkappaB-alpha. Arterioscler Thromb Vasc Biol. 1998;18:1796–802. doi: 10.1161/01.atv.18.11.1796. [DOI] [PubMed] [Google Scholar]
- 27.Kubes P, Suzuki M, Granger DN. Nitric oxide: an endogenous modulator of leukocyte adhesion. Proc Natl Acad Sci U S A. 1991;88:4651–5. doi: 10.1073/pnas.88.11.4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.D’Souza FM, Sparks RL, Chen H, Kadowitz PJ, Jeter JR., Jr Mechanism of eNOS gene transfer inhibition of vascular smooth muscle cell proliferation. Am J Physiol Cell Physiol. 2003;284:C191–9. doi: 10.1152/ajpcell.00179.2002. [DOI] [PubMed] [Google Scholar]
- 29.Royston BD, Royston D, Pearson JD. Aprotinin inhibits platelet adhesion to endothelial cells. Blood Coagul Fibrinolysis. 1992;3:737–42. doi: 10.1097/00001721-199212000-00006. [DOI] [PubMed] [Google Scholar]
- 30.Wilcox JN, Subramanian RR, Sundell CL, et al. Expression of multiple isoforms of nitric oxide synthase in normal and atherosclerotic vessels. Arterioscler Thromb Vasc Biol. 1997;17:2479–88. doi: 10.1161/01.atv.17.11.2479. [DOI] [PubMed] [Google Scholar]
- 31.Culleton BF, Larson MG, Wilson PW, Evans JC, Parfrey PS, Levy D. Cardiovascular disease and mortality in a community-based cohort with mild renal insufficiency. Kidney Int. 1999;56:2214–9. doi: 10.1046/j.1523-1755.1999.00773.x. [DOI] [PubMed] [Google Scholar]
- 32.Fried LF, Shlipak MG, Crump C, et al. Renal insufficiency as a predictor of cardiovascular outcomes and mortality in elderly individuals. J Am Coll Cardiol. 2003;41:1364–72. doi: 10.1016/s0735-1097(03)00163-3. [DOI] [PubMed] [Google Scholar]
- 33.Go AS, Chertow GM, Fan D, McCulloch CE, Hsu CY. Chronic kidney disease and the risks of death, cardiovascular events, and hospitalization. N Engl J Med. 2004;351:1296–305. doi: 10.1056/NEJMoa041031. [DOI] [PubMed] [Google Scholar]
- 34.Young A, Garg AX. It’s about time: extending our understanding of cardiovascular risk from chronic kidney disease. J Am Soc Nephrol. 2009;20:2486–7. doi: 10.1681/ASN.2009101045. [DOI] [PubMed] [Google Scholar]
- 35.Won D, Zhu SN, Chen M, et al. Relative reduction of endothelial nitric-oxide synthase expression and transcription in atherosclerosis-prone regions of the mouse aorta and in an in vitro model of disturbed flow. Am J Pathol. 2007;171:1691–704. doi: 10.2353/ajpath.2007.060860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Teichert AM, Scott JA, Robb GB, et al. Endothelial nitric oxide synthase gene expression during murine embryogenesis: commencement of expression in the embryo occurs with the establishment of a unidirectional circulatory system. Circ Res. 2008;103:24–33. doi: 10.1161/CIRCRESAHA.107.168567. [DOI] [PubMed] [Google Scholar]
- 37.Forbes MS, Thornhill BA, Park MH, Chevalier RL. Lack of endothelial nitric-oxide synthase leads to progressive focal renal injury. Am J Pathol. 2007;170:87–99. doi: 10.2353/ajpath.2007.060610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakayama T, Sato W, Kosugi T, et al. Endothelial injury due to eNOS deficiency accelerates the progression of chronic renal disease in the mouse. Am J Physiol Renal Physiol. 2009;296:F317–27. doi: 10.1152/ajprenal.90450.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heeringa P, van Goor H, Itoh-Lindstrom Y, et al. Lack of endothelial nitric oxide synthase aggravates murine accelerated anti-glomerular basement membrane glomerulonephritis. Am J Pathol. 2000;156:879–88. doi: 10.1016/S0002-9440(10)64957-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakagawa T, Sato W, Glushakova O, et al. Diabetic endothelial nitric oxide synthase knockout mice develop advanced diabetic nephropathy. J Am Soc Nephrol. 2007;18:539–50. doi: 10.1681/ASN.2006050459. [DOI] [PubMed] [Google Scholar]
- 41.Wang CH, Li F, Hiller S, et al. A modest decrease in endothelial NOS in mice comparable to that associated with human NOS3 variants exacerbates diabetic nephropathy. Proc Natl Acad Sci U S A. 2011;108:2070–5. doi: 10.1073/pnas.1018766108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Advani A, Huang Q, Thai K, et al. Long-term administration of the histone deacetylase inhibitor vorinostat attenuates renal injury in experimental diabetes through an endothelial nitric oxide synthase-dependent mechanism. Am J Pathol. 2011;178:2205–14. doi: 10.1016/j.ajpath.2011.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yan MS, Matouk CC, Marsden PA. Epigenetics of the vascular endothelium. J Appl Physiol. 2010;109:916–26. doi: 10.1152/japplphysiol.00131.2010. [DOI] [PubMed] [Google Scholar]
- 44.Matouk CC, Marsden PA. Epigenetic regulation of vascular endothelial gene expression. Circ Res. 2008;102:873–87. doi: 10.1161/CIRCRESAHA.107.171025. [DOI] [PubMed] [Google Scholar]
- 45.Abbott A. Project set to map marks on genome. Nature. 2010;463:596–7. [PubMed] [Google Scholar]
- 46.Bird A. Perceptions of epigenetics. Nature. 2007;447:396–8. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- 47.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 48.Lister R, Pelizzola M, Dowen RH, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–22. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ehrlich M, Gama-Sosa MA, Huang LH, et al. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells. Nucleic Acids Res. 1982;10:2709–21. doi: 10.1093/nar/10.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Illingworth RS, Bird AP. CpG islands—‘a rough guide’. FEBS Lett. 2009;583:1713–20. doi: 10.1016/j.febslet.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 51.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–26. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 52.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–57. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 53.Miranda TB, Jones PA. DNA methylation: the nuts and bolts of repression. J Cell Physiol. 2007;213:384–90. doi: 10.1002/jcp.21224. [DOI] [PubMed] [Google Scholar]
- 54.Prendergast GC, Lawe D, Ziff EB. Association of Myn, the murine homolog of max, with c-Myc stimulates methylation-sensitive DNA binding and ras cotransformation. Cell. 1991;65:395–407. doi: 10.1016/0092-8674(91)90457-a. [DOI] [PubMed] [Google Scholar]
- 55.Wang GL, Semenza GL. Characterization of hypoxia-inducible factor 1 and regulation of DNA binding activity by hypoxia. J Biol Chem. 1993;268:21513–8. [PubMed] [Google Scholar]
- 56.Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–5. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 57.Harrington MA, Jones PA, Imagawa M, Karin M. Cytosine methylation does not affect binding of transcription factor Sp1. Proc Natl Acad Sci U S A. 1988;85:2066–70. doi: 10.1073/pnas.85.7.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lewis JD, Meehan RR, Henzel WJ, et al. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992;69:905–14. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- 59.Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol Cell Biol. 1998;18:6538–47. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jones PL, Veenstra GJ, Wade PA, et al. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–91. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 61.Ng HH, Zhang Y, Hendrich B, et al. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat Genet. 1999;23:58–61. doi: 10.1038/12659. [DOI] [PubMed] [Google Scholar]
- 62.Zhu JK. Active DNA demethylation mediated by DNA glycosylases. Annu Rev Genet. 2009;43:143–66. doi: 10.1146/annurev-genet-102108-134205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee DU, Agarwal S, Rao A. Th2 lineage commitment and efficient IL-4 production involves extended demethylation of the IL-4 gene. Immunity. 2002;16:649–60. doi: 10.1016/s1074-7613(02)00314-x. [DOI] [PubMed] [Google Scholar]
- 64.Tahiliani M, Koh KP, Shen Y, et al. Conversion of 5-methyl-cytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–5. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim MS, Kondo T, Takada I, et al. DNA demethylation in hormone-induced transcriptional derepression. Nature. 2009;461:1007–12. doi: 10.1038/nature08456. [DOI] [PubMed] [Google Scholar]
- 66.Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 67.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 68.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–19. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 69.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–5. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 70.Owen-Hughes T, Utley RT, Cote J, Peterson CL, Workman JL. Persistent site-specific remodeling of a nucleosome array by transient action of the SWI/SNF complex. Science. 1996;273:513–6. doi: 10.1126/science.273.5274.513. [DOI] [PubMed] [Google Scholar]
- 71.Ewing AK, Attner M, Chakravarti D. Novel regulatory role for human Acf1 in transcriptional repression of vitamin D3 receptor-regulated genes. Mol Endocrinol. 2007;21:1791–806. doi: 10.1210/me.2007-0095. [DOI] [PubMed] [Google Scholar]
- 72.Fish JE, Yan MS, Matouk CC, et al. Hypoxic repression of endothelial nitric-oxide synthase transcription is coupled with eviction of promoter histones. J Biol Chem. 2010;285:810–26. doi: 10.1074/jbc.M109.067868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–9. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 74.Khalil AM, Guttman M, Huarte M, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–72. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bertani S, Sauer S, Bolotin E, Sauer F. The noncoding RNA Mistral activates hoxa6 and hoxa7 expression and stem cell differentiation by recruiting MLL1 to chromatin. Mol Cell. 2011;43:1040–6. doi: 10.1016/j.molcel.2011.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 76.Guttman M, Amit I, Garber M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–7. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rinn JL, Kertesz M, Wang JK, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–23. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McPherson R, Pertsemlidis A, Kavaslar N, et al. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–91. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kanduri C, Whitehead J, Mohammad F. The long and the short of it: RNA-directed chromatin asymmetry in mammalian X-chromosome inactivation. FEBS Lett. 2009;583:857–64. doi: 10.1016/j.febslet.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 80.Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–6. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Payer B, Lee JT. X chromosome dosage compensation: how mammals keep the balance. Annu Rev Genet. 2008;42:733–72. doi: 10.1146/annurev.genet.42.110807.091711. [DOI] [PubMed] [Google Scholar]
- 82.Sun BK, Deaton AM, Lee JT. A transient heterochromatic state in Xist preempts X inactivation choice without RNA stabilization. Mol Cell. 2006;21:617–28. doi: 10.1016/j.molcel.2006.01.028. [DOI] [PubMed] [Google Scholar]
- 83.Fish JE, Marsden PA. Endothelial nitric oxide synthase: insight into cell-specific gene regulation in the vascular endothelium. Cell Mol Life Sci. 2006;63:144–62. doi: 10.1007/s00018-005-5421-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.De Val S, Black BL. Transcriptional control of endothelial cell development. Dev Cell. 2009;16:180–95. doi: 10.1016/j.devcel.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tapscott SJ, Davis RL, Thayer MJ, Cheng PF, Weintraub H, Lassar AB. MyoD1: a nuclear phosphoprotein requiring a Myc homology region to convert fibroblasts to myoblasts. Science. 1988;242:405–11. doi: 10.1126/science.3175662. [DOI] [PubMed] [Google Scholar]
- 86.Tontonoz P, Hu E, Spiegelman BM. Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell. 1994;79:1147–56. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 87.Chan Y, Fish JE, D’Abreo C, et al. The cell-specific expression of endothelial nitric-oxide synthase: a role for DNA methylation. J Biol Chem. 2004;279:35087–100. doi: 10.1074/jbc.M405063200. [DOI] [PubMed] [Google Scholar]
- 88.Teichert AM, Miller TL, Tai SC, et al. In vivo expression profile of an endothelial nitric oxide synthase promoter-reporter transgene. Am J Physiol Heart Circ Physiol. 2000;278:H1352–61. doi: 10.1152/ajpheart.2000.278.4.H1352. [DOI] [PubMed] [Google Scholar]
- 89.Fish JE, Matouk CC, Rachlis A, et al. The expression of endothelial nitric-oxide synthase is controlled by a cell-specific histone code. J Biol Chem. 2005;280:24824–38. doi: 10.1074/jbc.M502115200. [DOI] [PubMed] [Google Scholar]
- 90.Fish JE, Matouk CC, Yeboah E, et al. Hypoxia-inducible expression of a natural cis-antisense transcript inhibits endothelial nitric-oxide synthase. J Biol Chem. 2007;282:15652–66. doi: 10.1074/jbc.M608318200. [DOI] [PubMed] [Google Scholar]
- 91.Robb GB, Carson AR, Tai SC, et al. Post-transcriptional regulation of endothelial nitric-oxide synthase by an overlapping antisense mRNA transcript. J Biol Chem. 2004;279:37982–96. doi: 10.1074/jbc.M400271200. [DOI] [PubMed] [Google Scholar]
- 92.Zhang B, Ramesh G, Uematsu S, Akira S, Reeves WB. TLR4 signaling mediates inflammation and tissue injury in nephrotoxicity. J Am Soc Nephrol. 2008;19:923–32. doi: 10.1681/ASN.2007090982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang MX, Ou H, Shen YH, Wang J, Coselli J, Wang XL. Regulation of endothelial nitric oxide synthase by small RNA. Proc Natl Acad Sci U S A. 2005;102:16967–72. doi: 10.1073/pnas.0503853102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Casas JP, Bautista LE, Humphries SE, Hingorani AD. Endothelial nitric oxide synthase genotype and ischemic heart disease: meta-analysis of 26 studies involving 23028 subjects. Circulation. 2004;109:1359–65. doi: 10.1161/01.CIR.0000121357.76910.A3. [DOI] [PubMed] [Google Scholar]
- 95.Carew JS, Giles FJ, Nawrocki ST. Histone deacetylase inhibitors: mechanisms of cell death and promise in combination cancer therapy. Cancer Lett. 2008;269:7–17. doi: 10.1016/j.canlet.2008.03.037. [DOI] [PubMed] [Google Scholar]
- 96.Touma SE, Goldberg JS, Moench P, et al. Retinoic acid and the histone deacetylase inhibitor trichostatin a inhibit the proliferation of human renal cell carcinoma in a xenograft tumor model. Clin Cancer Res. 2005;11:3558–66. doi: 10.1158/1078-0432.CCR-04-1155. [DOI] [PubMed] [Google Scholar]
- 97.Xia S, Li X, Johnson T, Seidel C, Wallace DP, Li R. Polycystin-dependent fluid flow sensing targets histone deacetylase 5 to prevent the development of renal cysts. Development. 2010;137:1075–84. doi: 10.1242/dev.049437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cao Y, Semanchik N, Lee SH, et al. Chemical modifier screen identifies HDAC inhibitors as suppressors of PKD models. Proc Natl Acad Sci U S A. 2009;106:21819–24. doi: 10.1073/pnas.0911987106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Marumo T, Hishikawa K, Yoshikawa M, Hirahashi J, Kawachi S, Fujita T. Histone deacetylase modulates the proinflammatory and -fibrotic changes in tubulointerstitial injury. Am J Physiol Renal Physiol. 2010;298:F133–41. doi: 10.1152/ajprenal.00400.2009. [DOI] [PubMed] [Google Scholar]
- 100.Pang M, Kothapally J, Mao H, et al. Inhibition of histone deacetylase activity attenuates renal fibroblast activation and interstitial fibrosis in obstructive nephropathy. Am J Physiol Renal Physiol. 2009;297:F996–1005. doi: 10.1152/ajprenal.00282.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Suter CM, Martin DI, Ward RL. Germline epimutation of MLH1 in individuals with multiple cancers. Nat Genet. 2004;36:497–501. doi: 10.1038/ng1342. [DOI] [PubMed] [Google Scholar]
- 102.Chan TL, Yuen ST, Kong CK, et al. Heritable germline epimutation of MSH2 in a family with hereditary nonpolyposis colorectal cancer. Nat Genet. 2006;38:1178–83. doi: 10.1038/ng1866. [DOI] [PubMed] [Google Scholar]
- 103.Chan GC, Fish JE, Mawji IA, Leung DD, Rachlis AC, Marsden PA. Epigenetic basis for the transcriptional hyporesponsiveness of the human inducible nitric oxide synthase gene in vascular endothelial cells. J Immunol. 2005;175:3846–61. doi: 10.4049/jimmunol.175.6.3846. [DOI] [PubMed] [Google Scholar]
- 104.Chen W, Bacanamwo M, Harrison DG. Activation of p300 histone acetyltransferase activity is an early endothelial response to laminar shear stress and is essential for stimulation of endothelial nitric-oxide synthase mRNA transcription. J Biol Chem. 2008;283:16293–8. doi: 10.1074/jbc.M801803200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Illi B, Nanni S, Scopece A, et al. Shear stress-mediated chromatin remodeling provides molecular basis for flow-dependent regulation of gene expression. Circ Res. 2003;93:155–61. doi: 10.1161/01.RES.0000080933.82105.29. [DOI] [PubMed] [Google Scholar]