Abstract
The contributions of phosphorylation-mediated signaling networks to colon cancer metastasis are poorly defined. To interrogate constitutive signaling alterations in cancer progression, the global phosphoproteomes of the patient-matched SW480 (primary colon tumor origin) and SW620 (lymph node metastasis) cell lines were compared with TiO2 and IMAC phosphopeptide enrichment followed by LC-MS/MS. Network analysis of the significantly altered phosphosites revealed differential regulation in cellular adhesion, mitosis, and mRNA translational machinery. mRNA biogenesis and splicing, transport through the nuclear pores, initiating translation, as well as mRNA stability and degradation were also affected. Though alterations in these processes have been associated with oncogenic transformation, control of mRNA stability has typically not been associated with cancer progression. Notably, the single phosphosite with the greatest relative change in SW620 was Ser2 on eukaryotic initiation factor 2 subunit 2 (eIF2S2), suggesting that SW620 cells translate faster or with greater efficiency than SW480 cells. These broad changes in the regulation of translation also occur without over-expression of eukaryotic initiation factor 4E (eIF4E). The findings suggest that metastatic cells exhibit constitutive changes to the phosphoproteome, and that mRNA stability and translational efficiency may be important targets of deregulation during cancer progression.
Keywords: colon cancer progression, phosphosite stoichiometry, signaling networks, IP-SRM, mRNA transport, mRNA translation, cellular adhesion, 4E-transporter (4E-T), IMAC, TiO2
INTRODUCTION
Colorectal cancer (CRC) deaths overwhelmingly result from metastasis rather than from locally confined tumors. Cancer progression is associated with invasive cellular behavior, migration, angiogenesis, and resistance to apoptosis [1]. All of these processes are regulated in part by protein phosphorylation. Significant crosstalk among these pathways and others facilitates both the acquisition of increasingly aggressive traits and resistance to drug treatments over time [2–6]. However, the relationships among these pathways and the molecular mechanisms of metastasis remain incompletely understood.
Because of the number of processes involved and the crosstalk among intracellular signaling pathways, the higher-order information provided by proteomics and phosphoproteomics is particularly useful for investigating the mechanisms of metastasis. A 2012 study by Wisniewski and coworkers compared the proteomes of formalin-fixed paraffin-embedded archived normal tissues and primary colon tumors from eight patients, as well as matched nodal metastases for seven of the patients [7]. While dramatic changes in the proteome were noted in the primary tumors compared to their matched normal tissues, the differences between the tumors and the metastases were much subtler. This nuanced discrepancy suggests that the metastatic proteomes may display more patient-to-patient variability, or that relatively few protein-level changes are needed for tumors to metastasize compared to the steps required for oncogenesis. On the other hand, post-translational modifications may be able to account for the phenotypic differences between Stage II and Stage III tumors that are not explained at the proteomic level.
One particularly valuable in vitro CRC model is the SW480/SW620 pair of cell lines, representing the chromosomal instability subtype of CRC that is commonly observed clinically [8, 9]. These patient-matched lines were derived from a primary adenocarcinoma and a lymph node metastasis, respectively [8]. They display differences in xenograft metastatic potential [10], migratory propensity, and drug sensitivity [11] that recapitulate behavior observed in vivo. Previous studies have compared the SW480 and SW620 cell lines at the proteome and secretome levels [12, 13], but there has been no report on the contributions of post-translational modifications to the metastatic phenotypes observed in SW620 cells relative to SW480 cells. Consequently, a comparison of the constitutive phosphorylation status of these two cell lines should provide a rich source of information about the unique behavior and vulnerabilities of metastatic colon cells.
To assess the relative constitutive phosphorylation in the two cell lines, we performed comparative phosphoproteomic analysis of these cells by liquid chromatography-mass spectrometry (LC-MS). Phosphopeptides from SILAC-labeled cell lysates were enriched with a combined IMAC (immobilized metal affinity chromatography) - TiO2 strategy and fractionated by high-pH reverse phase SPE (solid phase extraction) cartridges prior to LC-MS. Non-enriched peptides from the same lysates were similarly fractionated and analyzed by LC-MS to allow normalization of some phosphosites to protein expression changes. This discovery experiment identified altered signaling in adhesion, migration, mitosis, mRNA biogenesis, and regulation of translation. The network-level deregulation of mRNA processing is underscored by the finding that Ser2 of eIF2S2 exhibited the greatest phosphorylation increase in the SW620 cells. Therefore, SW620 cells appear to have a higher translational capacity than SW480 cells, and they accomplish this without overexpressing the mRNA 5’cap-binding protein eIF4E. Taken together, these data provide new evidence that metastatic cells deregulate mRNA stability and translational efficiency with constitutive changes to the phosphoproteome.
MATERIALS AND METHODS
Human and Animal Rights
This study used established human-derived cell lines, which do not require IRB approval or informed consent prior to new research.
Cell Culture and Harvest
Human colorectal cancer cell lines SW480 and SW620 were purchased from American Type Culture Collection (ATCC, Manassas, VA) and maintained in RPMI 1640 medium (Invitrogen, Gaithersburg, MD) supplemented with 10% fetal bovine serum (Thermo Scientific, Pittsburgh, PA) and 2mM L-glutamine (Invitrogen) and grown in 5% CO2 at 37°C. Cell lines were used within 3 months after receipt or resuscitation of frozen aliquots thawed from liquid nitrogen. The provider assured the authentication of these cell lines by cytogenetic analysis.
For stable isotope labeling of amino acids in cell culture (SILAC) experiments, the SW480 and SW620 cells were cultured in SILAC RPMI 1640 media containing 10% dialyzed fetal bovine serum plus 13C615N4-arginine and 13C615N2 -lysine (SILAC “Heavy”) or an equal concentration of the corresponding light isotopes (SILAC “Light”). Both cell lines were expanded for > 10 doublings. Cells were harvested at 60% confluency by scraping in ice-cold lysis buffer containing 8 M urea, 50 mM Tris HCl pH 8, 10 mM Na4(PO4)2, 1 mM phenylmethylsulfonylfluoride, 1mM each of the phosphatase inhibitors Na3VO3 and β-glycerophosphate, and 1 EDTA-free protease inhibitor tablet per 10 mL (Roche, Indianapolis, IN). Twelve 15 cm-dishes of each cell line were harvested to ensure sufficient material; four dishes mixed together were handled as a single biological/workflow replicate. For immunoprecipitation experiments, cells were grown in duplicate with reversed SILAC labels – one “light” replicate and one “heavy” replicate for each cell line. “Heavy” lysine in this experiment was the 13C6 isotope. These were harvested in in a lysis buffer containing 1% NP-40 (Tergitol) from Sigma (St. Louis, MO), 20 mm Tris pH 8.0, 137 mM NaCl, plus protease and phosphatase inhibitors as above.
Cell Lysis and Protein Digestion
Lysates were sonicated three times for 1 min each with a two-minute rest in between, then centrifuged at 4 °C for 10 minutes at 3000 x g. Protein concentration was determined in the supernatants by BCA assay (Thermo-Fisher Pierce, Waltham, MA). For discovery phosphoproteomics, 2 mg of lysates from each cell line were mixed 1:1, reduced with 5 mM DTT, and alkylated with 14 mM iodoacetamide. The alkylation reaction was quenched with an additional 5 mM DTT, then lysates were diluted with 50 mM ammonium bicarbonate (ABC) to <1.6 M urea. Bovine TPCK-treated trypsin (Sigma) was added at 1:50 enzyme: substrate (mg/mg) along with 5% trifluoroethanol prior to overnight incubation at 37°C. Digests were quenched with 0.4% trifluoroacetic acid (TFA). We set aside 500 µg aliquots for analysis of the non-enriched proteome and desalted the remaining 3.5 mg of peptides on 100 mg C18 Sep-Pak cartridges (Waters, Milford, MA) prior to phosphopeptide enrichment.
Phosphopeptide Enrichment and Fractionation
Phosphopeptides were enriched using both IMAC and TiO2 beads to enhance coverage, since the two chemistries have been reported to enrich complementary sub-proteomes [14, 15]. Digests were performed in triplicate on two occasions, once for each enrichment method. IMAC was performed three times sequentially, with the second and third enrichments mixed together, as described by us previously [15, 16]. Briefly, desalted peptides from 3.5mg of lysate were resuspended in loading buffer (40% ACN, 25mM formic acid in water) and mixed with 30uL of prepared PHOS-Select IMAC beads (Sigma). Beads and peptides were incubated for 1 hr with shaking at room temperature. The supernatant from the first IMAC was immediately added to fresh beads to begin the second IMAC enrichment. Similarly, the supernatant from the second IMAC was added to fresh beads to begin the third enrichment. Beads with bound peptides were rinsed three times in loading buffer, and then phosphopeptides were eluted by shaking 5 min in 50 mM K2HPO4, adjusted to pH10 with NH4OH. This elution step was repeated twice, and the combined eluates were acidified with formic acid (FA).
TiO2 was performed twice as previously reported [15, 17]. Bulk 5 µm Titanosphere particles were purchased from GL Sciences USA (Torrance, CA). Desalted peptides (from 3.5 mg of lysate) were resuspended in a solution of 65% ACN, 2% TFA, and saturated glutamic acid. Each replicate was mixed with 5.6 mg of TiO2 beads and shaken for 20 min at room temperature. Supernatants were removed and immediately added to fresh beads for the second enrichment. Beads with bound phosphopeptides were first washed in 65% ACN, 0.5% TFA for 20 min with shaking, then were washed twice in 65% ACN, 0.1% TFA for another 20 min with shaking. Phosphopeptides were eluted by shaking 20 min in 300 mM NH4OH/50% ACN, then twice with 500 mM NH4OH/60% ACN. The eluates from these three steps were combined and acidified with FA.
Hydrophilic-lipophilic balanced (HLB) reversed-phase Sep-Pak cartridges (Waters, Milford, MA) were used for high-pH reverse-phase fractionation of peptides. All solvents used for HLB fractionation contained aqueous 10mM ABC at pH 10, with acetonitrile added as indicated. Peptides were pH-adjusted by adding 100 mM ABC to a final concentration of 20 mM, then loaded onto cartridges with 1% ACN /99% 10 mM ABC and rinsed with the same buffer. Peptides were then eluted with 5%, 10%, 15%, 20%, 25%, 30%, 35%, and 80% ACN. For unmodified proteome analysis, a 500 µg aliquot of non-enriched peptide was prepared as eight fractions. The HLB fractions generated from each of the batches of IMAC and TiO2 enriched peptides (IMAC-1, IMAC-2&3, TiO2-1, and TiO2-2) were mixed to make four fractions, or 16 phosphopeptide fractions in total. HLB fraction #1 (5% ACN) was mixed with #5 (25% ACN), 10% ACN with 30% ACN, 15% ACN with 35% ACN, and 20% ACN with 80% ACN. After acidification with FA and vacuum drying to remove ACN, all fractions were desalted by ZipTip prior to LC-MS/MS analysis.
LC-MS/MS Analysis
LC-MS/MS analyses were performed with a 90 min analytical gradient on a 100 µm × 10 cm C18 column coupled to a Thermo Q-Exactive mass spectrometer. All analyses were performed in technical duplicate. Solvent A consisted of 0.1% FA in water and solvent B was 0.1% in ACN. The flow rate was kept at 1.2 µL/min with the column heated to 40 °C. Desalted phosphopeptide-enriched fractions were resuspended in 8 µL of 2% ACN with 0.1% FA and 2 µL were injected on-column. IMAC-enriched fractions were separated on a 2% to 30% ACN gradient. TiO2-enriched fractions were separated using a gradient from 2% to 25% ACN. Non-enriched samples were resuspended to 1 µg/µL after desalting; 1 µL was injected on-column. Gradients used for non-enriched HLB fractions varied by the fraction number. Fractions generated with 5%, 10%, 15%, and 20% ACN used a 2% to 30% ACN linear gradient; fractions generated with 25% ACN and 80% ran from 15% to 40% ACN in 85 min, and fractions generated with 30% and 35% ACN used a 2% to 40% ACN gradient. Unfractionated, non-enriched samples were analyzed with a 2–40% ACN gradient.
The mass spectrometer was operated in data-dependent MS/MS mode with a top-12 method. Dynamic exclusion was set to 30 s and charge 1+ ions were excluded as well. MS1 scans were collected from 400–2000 m/z with resolving power equal to 70,000 at 200 m/z. The MS1 AGC was set to 1×106. The isolation width was 2.0 m/z, and the normalized collision energy was set to 28%. The MS2 AGC was set to 2×104 with an intensity threshold of 8.3×103 and a maximum fill time of 120 ms. The resolving power of MS/MS scans was 17,500.
Data Analysis
LC-MS results were searched in MaxQuant 1.4.1.2 against the UniProt human database (version downloaded 4/2013, 89601 sequences) and a list of common contaminants. Cysteine carbamidomethylation was a fixed modification, and oxidized methionine was a variable modification. Phosphorylation of S, T, and Y residues were allowed for enriched fractions. Two missed cleavages were allowed and Trypsin/P was selected as the enzyme. The “first search” precursor mass tolerance was set to 20 ppm, “main search” tolerance was set to 10 ppm, and fragment mass tolerance was set to 50 ppm. The “first search” and “main search” precursor tolerances are parameters specialized to the MaxQuant software. SILAC ratios were calculated using the “highest change” setting. A 1% FDR was applied at the peptide and protein level.
“Heavy/Light” ratios were converted to SW620/SW480 ratios, then log2-transformed. Phosphosites were quantified at the peptide level, requiring at least two SILAC measurements per peptide. The phosphosite localization probability was calculated in MaxQuant. Only Class I phosphosites (those with 75% localization probability or better) were considered [18]. For non-enriched fractions, a minimum of two unique peptides were required for protein quantification. Phosphopeptide ratios were corrected for changes in protein abundance by subtracting the corresponding non-phosphorylated protein log2 -fold change. Altered regulation of phosphosites was determined using Bonferroni-corrected t-tests with α=0.05. The complete mass spectrometry dataset is available from the ProteomeXchange Consortium via the PRIDE partner repository under the dataset identifier PXD003708.
Network analysis was performed using the STRING database v.9.1 (string-db.org). Uniprot identifiers from 139 protein-abundance-corrected, significantly altered phosphoproteins were submitted for analysis, of which 124 contained a match in the database. Protein-protein interactions were mapped using the website’s “high confidence” setting, which represents 70% confidence or better in each interaction. For ease of visualization, disconnected protein nodes were excluded from the relevant figure. Gene Ontology (GO) term enrichment analysis was performed using STRING’s embedded tools. The human genome was set as the background dataset from which to measure enrichment. GO terms were considered significantly enriched against the background human genome with FDR-adjusted p-values >0.01.
Antibodies, Immunoprecipitation and Western Blots
Lysates were separated on SDS PAGE mini-gels, and proteins were transferred to nitrocellulose membranes for 1.5–2 h at 30 V. Rabbit polyclonal antibodies anti-4E-T (#ab55881) and anti-eIF4E (#ab1126) were purchased from Abcam (San Francisco, CA).The anti-4E-T antibody was either diluted 1:500 in 5% w/v milk in PBS and incubated for 1h at room temperature or used at 1:1000 overnight at 4 °C. Anti-eIF4E antibody was diluted 1:1000 in 5% w/v milk in PBS and incubated with membranes for 1h at room temperature. An anti-beta-actin antibody (Cell Signaling Technologies, Danvers, MA) was used at a 1:750 dilution in 5% BSA/TBST. Membranes were incubated with primary antibody for 1 hr at room temperature (or overnight at 4 °C for beta-actin), and then with HRP-linked anti-rabbit secondary antibody (Jackson Laboratories, Bar Harbor, ME) for 1 hr at room temperature. Bands were visualized with chemiluminescent substrate (Thermo-Fisher Pierce).
For immunoprecipitation (IP), 35 µL anti-4E-T antibody was incubated with 350 µg of mixed SILAC lysate for 4 hr at 4 °C. After this incubation, 175 µL Protein A-Dynabeads (Life Technologies, Grand Island, NY) were added and the mixtures were rotated 1 hr at 4° C. After washing three times in lysis buffer, the target protein was eluted twice from the beads by heating to 70 °C for 10 min in a buffer containing 1% SDS, 50 mM Tris pH 8, and 100 mM DTT. One-tenth of the eluate was retained for western blot confirmation of pull-down. Eluates were loaded directly onto Amicon 30k molecular weight cutoff spin filters (Millipore, Darmstadt, Germany) for FASP digestion [19]. Briefly, loaded samples were centrifuged at 14,000 xg and samples were alkylated with 50 mM iodoacetamide at room temperature for 20 minutes in the dark. Filters were then washed three times with 400uL 8M urea and then washed three times with 400uL 25 mM ammonium bicarbonate. Trypsin was mixed with 25 mM ammonium bicarbonate buffer and added to filters at a 1:30 enzyme:substrate ratio. Samples were digested on-filter for 12 hours and peptides were collected by centrifugation. The peptides were then desalted on StageTips for quantitative MRM analysis.
Targeted Phosphosite Stoichiometry Determination
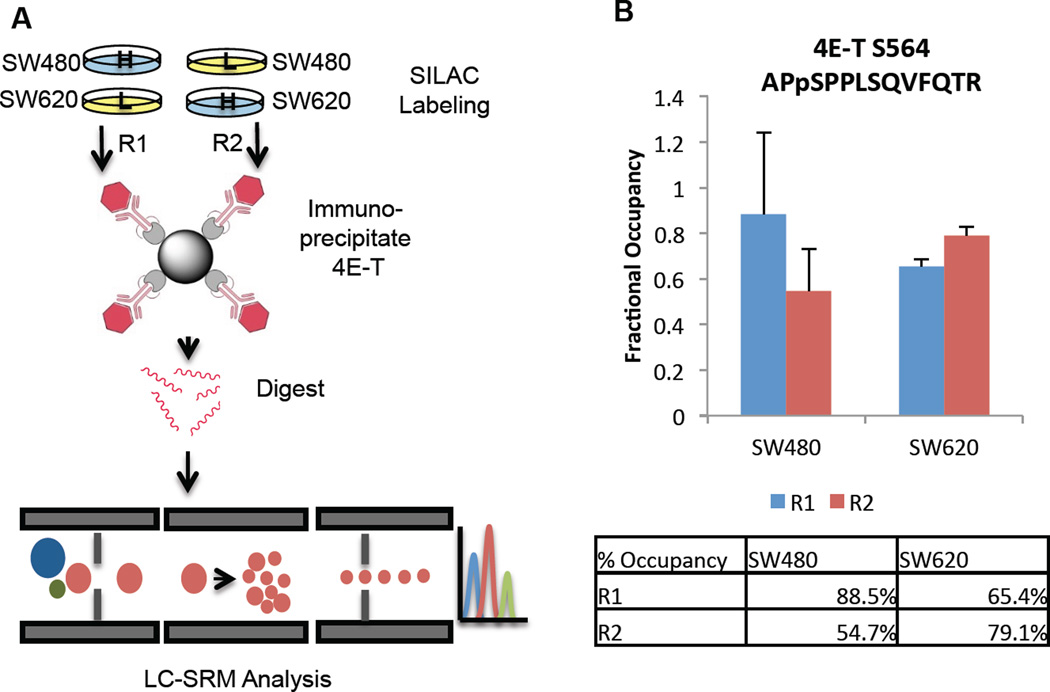
The phosphopeptide APpSPPLSQVFQTR on 4E-T (pSer564) was selected for targeted phosphosite stoichiometry determination. The corresponding non-phosphopeptide plus the proteotypic peptide ESSHSGVVLSVEEVEAGLK were also selected for monitoring. A complete table of transitions is given in ESM Table S1. Approximately 7 µg of desalted, dried peptides were resuspended in 15 µL of 5% ACN/0.1% FA and 2 µL were injected on column. Three replicate injections per biological sample were used. The 100 µm x 10 cm C18 column was operated at 500 nL/min with a gradient from 2–38% ACN in 60 min. A QTrap 5500 detected the peptides in MRM mode using 15 s dwell times. The fractional phosphorylation occupancy for each of the target sites was determined using the corresponding SILAC peptide as an internal standard. The phosphorylation rate can be calculated when SILAC ratios for the phosphopeptide, cognate nonphosphopeptide, and an unrelated peptide representing the total protein amounts are known [20]. The complete calculations are detailed in the Electronic Supplemental Information. In brief:
This can be calculated from the SILAC ratios as shown:
RESULTS & DISCUSSION
Combined IMAC/TiO2 Enrichment Strategy and Normalization to the Proteome Enables Detection of Widespread Changes to the Metastatic Phosphoproteome
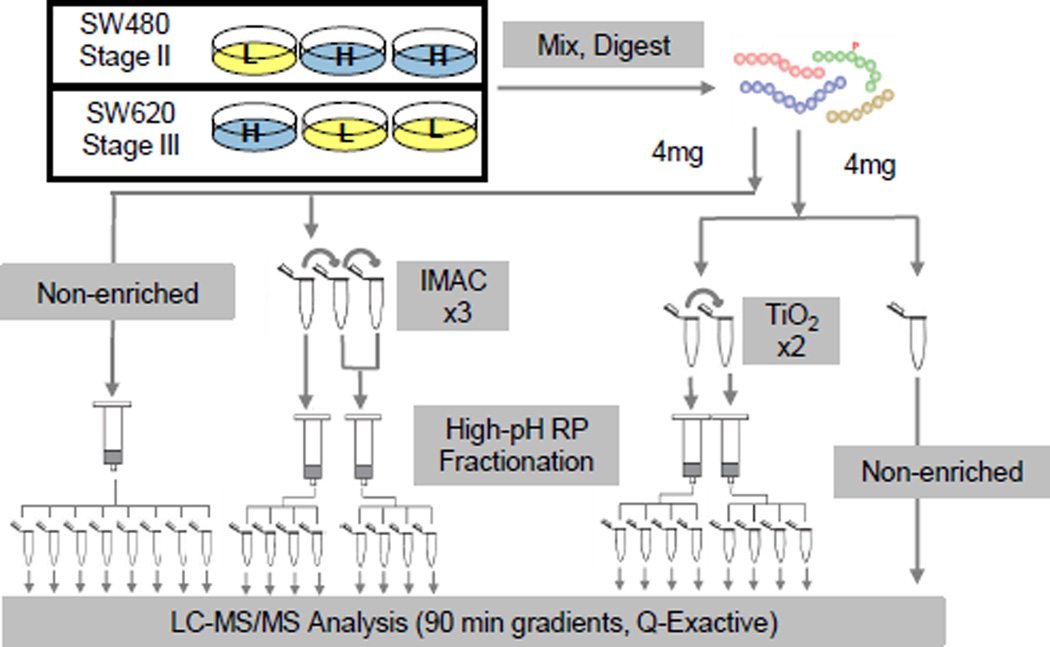
The constitutive phosphorylation differences between SW480 and SW620 were examined using comprehensive phosphoproteomic profiling. SILAC-labeled samples were mixed, digested and subjected to IMAC or TiO2 enrichment for phosphoproteomic analysis. A portion of the sample was not enriched for phosphopeptides and analyzed for protein abundance levels. A flow chart of the experimental design is shown in Figure 1.
Figure 1.
Preparation of the SW480 & SW620 phosphoproteome. Phosphoproteome and proteome workflow. Cell lines were SILAC labeled with heavy amino acids (blue, H) or light (yellow, L). Lysates were digested and non-enriched aliquots were reserved for proteome analysis. Phosphopeptides were enriched with either IMAC or TiO2 and then fractionated by high-pH reverse-phase (RP) chromatography. All fractions were desalted and analyzed by LC-MS/MS.
Combining IMAC and TiO2 enrichment strategies into one workflow allowed for more complete phosphoproteome coverage than either method alone. Our previous work demonstrated that multiple rounds of enrichment, performed sequentially, provided efficient enrichment and recovery of phosphopeptides [15, 16],. Three rounds of IMAC enrichment yielded 4,331 phosphopeptides, 30% of the total identifications. TiO2 was much more efficient, with 86% phosphopeptides in the first enrichment and 6,567 phosphopeptides identified overall from two enrichment steps. Of the confidently localized and quantified phosphosites, 56.6% were identified uniquely by one enrichment method or the other. For 23% of the quantified phosphopeptides, biological triplicate SILAC measurements were only possible when both enrichment chemistries were considered together.
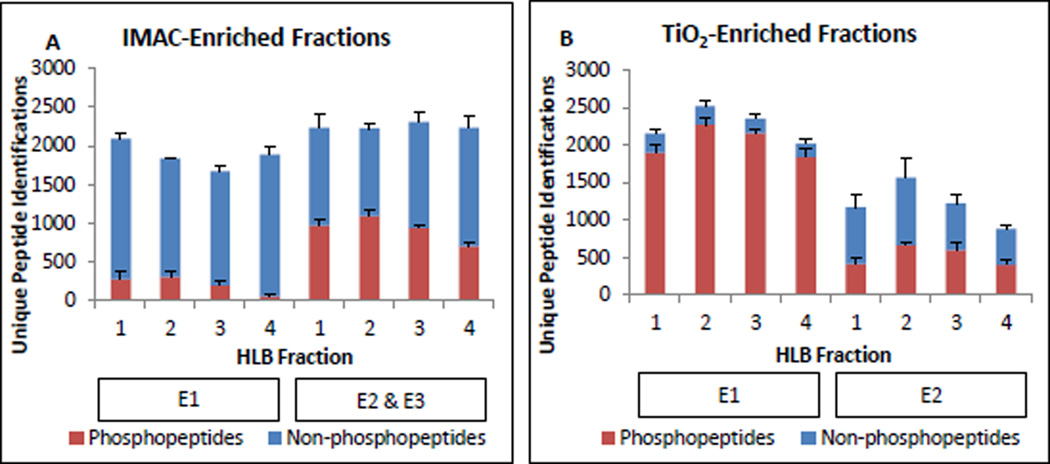
Unique peptide identifications in IMAC and TiO2-enriched fractions were plotted by high-pH reverse phase (HLB) fraction as shown in Figure 2A & B. Total peptide identifications were distributed equally across HLB fractions. This is especially visible in the IMAC-enriched fractions, where similar to 2000 unique peptides were identified in each fraction. Figure 2 also illustrates the relative contributions of each round of enrichment. The first-round TiO2 enrichment (E1) fractions contained nearly 90% phosphorylated peptides, with much lower specificity in the second round (E2). As fewer total peptides were detected in the E2 fractions, the data suggest E1 efficiently depletes most of the peptides that can bind specifically. With IMAC, the first round of enrichment recovered a tiny percentage of phosphopeptides, but the second and third rounds (combined) performed a bit better, with close to 50% specificity. This performance is not consistent with previous observations. After troubleshooting and purchasing of new resin, we concluded that this was due to batch-to-batch reproducibility problems with the product. In fact, the old beads outperformed fresh beads. These results show that this IMAC product requires multiple rounds of enrichment to achieve useful levels of phosphoproteome coverage and the optimized protocol is not consistently robust to manufacturer batch-to-batch variability. TiO2 chemistry provides the best phosphoproteome coverage in a single step, and adding a second round TiO2 enrichment enhances this coverage modestly, with 18% of TiO2 phosphopeptide identifications contributed by the second step.
Figure 2.
IMAC-HLB and TiO2-HLB workflow performance. All values shown are the average number of unique sequences identified per biological replicate and error bars display standard deviation. Phosphosite localization and SILAC quantification status were not considered at this stage. A. Unique peptide identifications by IMAC enrichment step (E1, E2&3) and HLB fraction number. Phosphopeptides are shown in red bars; non-phosphorylated peptides are in blue. B. TiO2-enriched peptide identifications by enrichment step (E1, E2) and HLB fraction number.
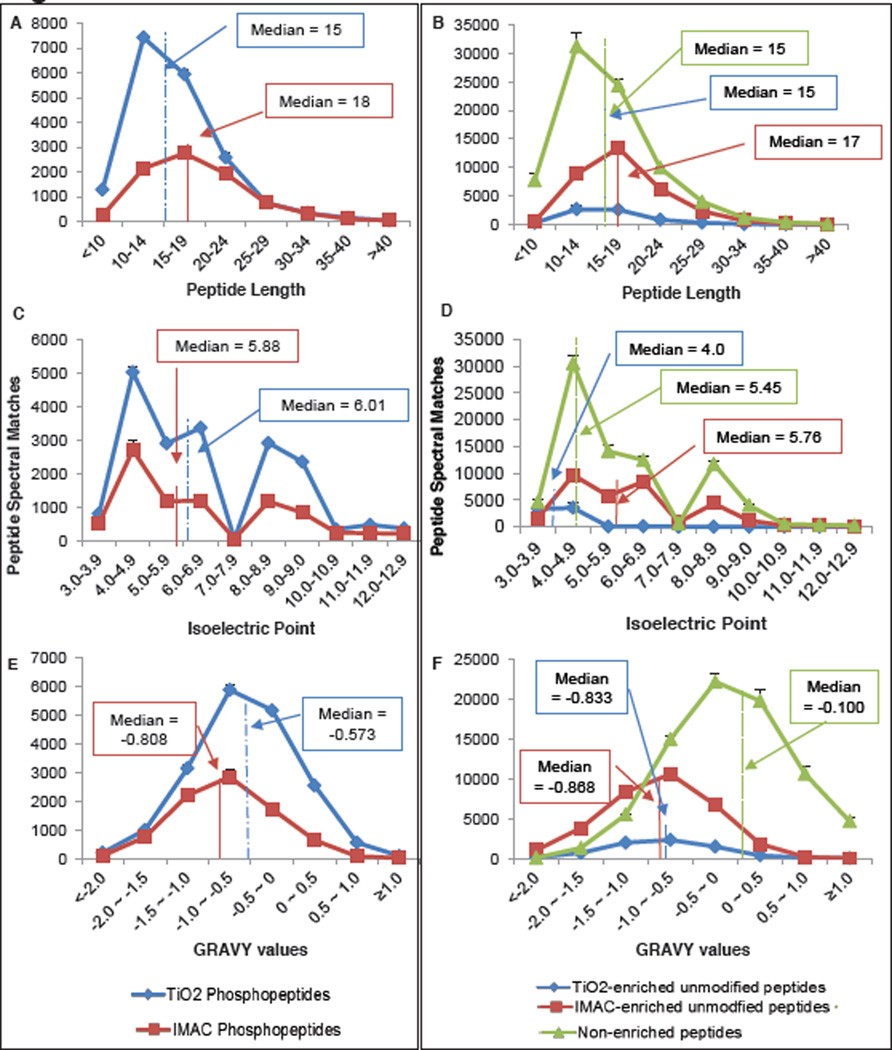
It has been reported by us and others that IMAC and TiO2 enrich distinct sub-proteomes [14, 15]. To explore this differential chemistry, we examined the length, isoelectric points (pIs), and grand average hydropathy (GRAVY) values of all the unique peptide sequences identified by IMAC enrichment, TiO2 enrichment, or without enrichment. The non-phosphorylated peptide spectral matches (PSMs) were aggregated separately from the phosphorylated PSMs for this analysis, which is summarized in Figure 3. IMAC- and TiO2-enriched phosphopeptides generally were of similar length, had similar pI distributions, and hydrophobicities (GRAVY values). The median phosphopeptide enriched by IMAC is slightly longer and more hydrophilic than that of TiO2, in agreement with our previous findings. However, the non-phosphorylated peptides reveal distinct patterns. Non-phosphorylated peptides found in TiO2 fractions were almost exclusively acidic, with pIs less than 5.0, whereas IMAC-enriched non-phosphorylated peptides followed the pI distribution of the unmodified tryptic proteome. Both IMAC and TIO2 were observed to enrich peptides with lower GRAVY values (more hydrophilic) than the tryptic proteome. Finally, the median non-phosphorylated peptide identified in IMAC fractions was just slightly longer than the median tryptic peptide identified in the unenriched proteome analysis. In fact, many additional missed-cleavage peptides were detected in the IMAC-enriched fractions in comparison to the TiO2-enriched or unenriched fractions (ESM Figure S1). In short, TiO2 specifically co-enriches acidic non-phosphorylated peptides alongside the target phosphopeptides, while contaminating unmodified peptides in IMAC fractions are truly nonspecific and largely resemble the general proteome.
Figure 3.
Properties of phosphopeptides and unmodified peptides identified after IMAC enrichment, TiO2 enrichment, or no enrichment. All values shown are the average number of unique sequences identified per biological replicate and error bars display standard deviation. Median values for each distribution are marked with solid, dotted, or dashed lines. A. Length of phosphorylated peptides. B. Length of unmodified peptides. C. Isoelectric point of phosphopeptides. D. Isoelectric point of unmodified peptides. E. Grand average hydropathy (GRAVY) values of phosphorylated peptides. F. GRAVY values of unmodified peptides.
Only a handful of other authors have used high pH RP chromatography in phosphoproteomic workflows, but it has performed favorably against traditional chromatography techniques so far [21–23]. These groups primarily used ammonium formate buffers at pH 10 with C18 columns. Song et.al showed that the orthogonality of high-pH RP as compared to traditional low-pH RP was improved by pooling non-adjacent fractions. This work was the first to demonstrate the RP-RP approach for phosphopeptide analysis [21]. Ficarro et.al. developed RP-RP and RP-SAX-RP workflows for phosphopeptide analysis that avoided the use of any specific phopshopeptide enrichment chemistry, such as IMAC or TiO2 [22]. This study found phosphopeptides, especially multiphosphopeptides, were weakly retained on C18 at high pH without the use of specialized ion-pairing reagents, such as tetrapropylammonium salts. As we did not examine the flow-through material from the HLB cartridges used in this study, we cannot rule out the loss of phosphopeptides from poor retention; however, this work and our previous work shows phosphopeptides eluting evenly across all fractions, from 5% ACN to 80% ACN. In a workflow similar to that described here, Batth et.al. fractionated peptides with high pH RP and then enriched phosphopeptides with one or two consecutive rounds of TiO2 [23]. They also noted that the C18 column degraded significantly with each use when ammonium formate was used as the solvent additive. This problem was solved by replacing the solvents with NH4OH. In contrast, the present work avoids this problem by using single-use HLB cartridges. To our knowledge, ours is the only workflow using benchtop SPE for RP fractionation. This strategy provides a simple, low-cost alternative for laboratories that do not have access to a dedicated HPLC system and fraction collector. The capability to fractionate multiple samples in parallel confers a significant time savings as well, especially as the number of samples increases.
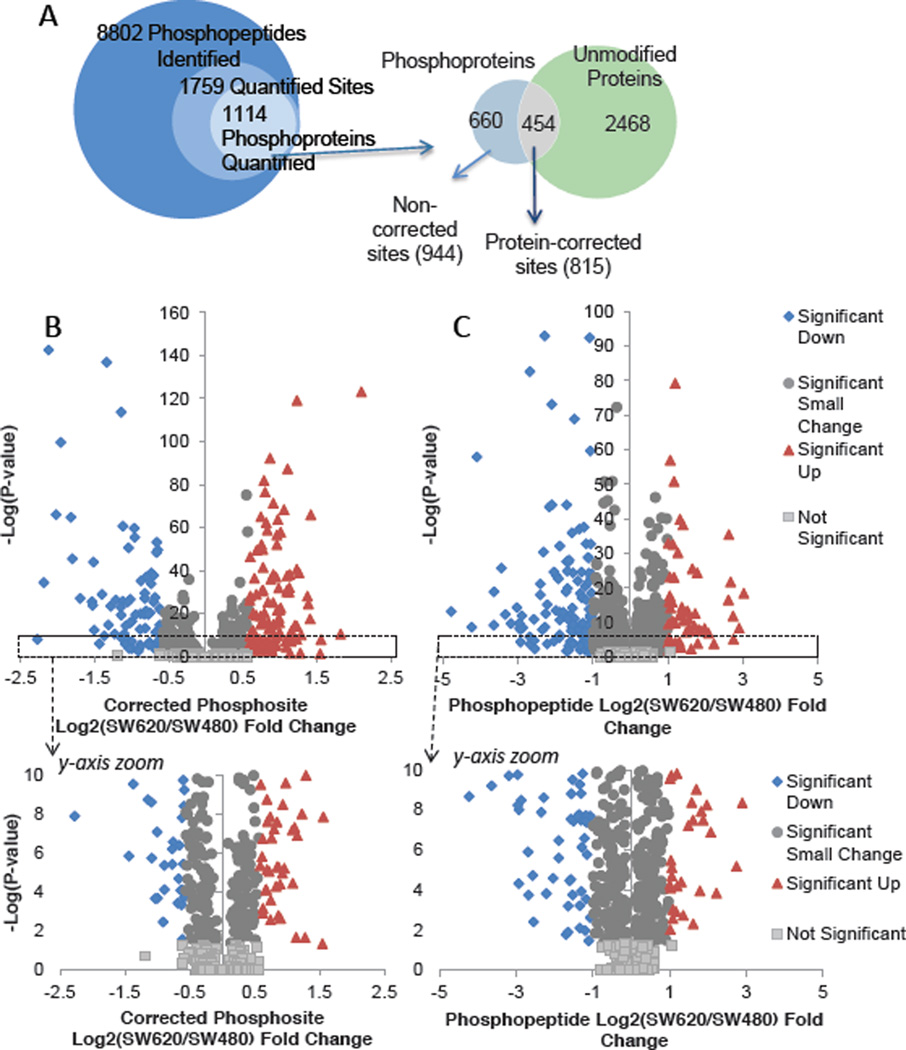
From 8,802 phosphopeptides identified, 1,759 distinct sites were quantified and localized in all three biological replicates. Good overlap of quantified phosphopeptides among the three biological replicates was obtained from both the IMAC and TiO2 workflows (ESM Figure S2). These 1759 quantified phosphosites mapped to 1,114 proteins (Figure 4A). Phosphosite SILAC ratios were corrected for differences in protein expression using values from the non-phosphorylated proteome. The utility of this strategy has been successfully demonstrated in yeast [24]. To maximize the number of corrected phosphosites calculated, protein-level data was included when at least two SILAC peptides were quantified in one or more biological replicates. Using 2922 protein groups meeting these criteria, 454 of the 1,114 phosphoproteins were normalized (Figure 4A). This included 815 of the 1,759 quantified phosphosites. From the phosphosites corrected for protein expression, a ±1.5 fold change cutoff, which corresponds to 0.586 on a log2 – scale, was applied after t-testing. This detected 108 phosphosites with increased phosphorylation in SW620 compared to SW480, and 86 sites decreased (Figure 4B). A more stringent 2-fold cutoff (1.0 on a log2 – scale) was applied to the non-corrected phosphosites, yielding 58 sites up-regulated and 114 sites down-regulated. Volcano plots of the corrected and non-corrected datasets are shown in Figure 4B. After the dataset was split, the corrected and the non-corrected phosphosites were analyzed as two distinct groups. Corrected phosphosite, non-corrected phosphosites, and protein-level quantification details are available in ESM Table S2–4.
Figure 4.
A. Total phosphopeptides identified and quantified, with the overlap between the phosphoproteome and unmodified proteome shown below. B., C. Phosphosite fold change vs. significance. Sites upregulated in the metastatic SW620 cells are shown in red triangles; those upregulated in the SW480 cells are shown in blue diamonds. Sites that changed less than the fold-change cutoff are shown in dark gray circles and nonsignificant sites are in light gray. (B) Protein abundance-corrected phosphosites are plotted by the log2(SW620/SW480) fold change. (C) Phosphosites for which protein abundance data was unavailable.
Gene Ontology and Network Analysis
Pathway and gene ontology analyses were performed using the significantly altered, corrected phosphosites. Both up- and down-regulated sites were analyzed together, as any given phosphosite may have either an “activating” or “inhibitory” role in regulating a protein’s known functions. Among the most significantly enriched Gene Ontology (GO) molecular functions were poly(A) RNA binding, kinase binding, and mRNA 5’ UTR binding (Table 1). Top cellular component terms included anchoring and adherens junctions, alongside nuclear components and the nuclear pore complex. Enriched biological processes included cellular component disassembly, mitotic cell cycle, mitotic nuclear envelope disassembly (all nuclear pore complex proteins), as well as RNA transport and establishment of RNA localization. The complete list of enriched GO biological processes, molecular functions, and cellular components are included in the ESM Table S5–7).
Table 1.
Selection of most significantly enriched Gene Ontology terms among protein-corrected phosphosites. Terms were ranked by significance and a maximum of ten terms are shown.
| Molecular Functions | Cellular Components | Biological Processes |
|---|---|---|
| poly(A) RNA binding RNA binding protein binding structural molecule activity kinase binding protein kinase binding enzyme binding mRNA 5’-UTR binding |
anchoring junction adherens junction extracellular vesicular exosome macromolecular complex cytosol intracellular non-membrane- bounded organelle nuclear part cytoskeleton nuclear pore nuclear lumen |
cellular component disassembly mitotic cell cycle mitotic nuclear envelope disassembly single-organism membrane organization RNA transport; establishment of RNA localization cell cycle nuclear envelope organization mRNA transport membrane organization cellular component disassembly involved in execution phase of apoptosis |
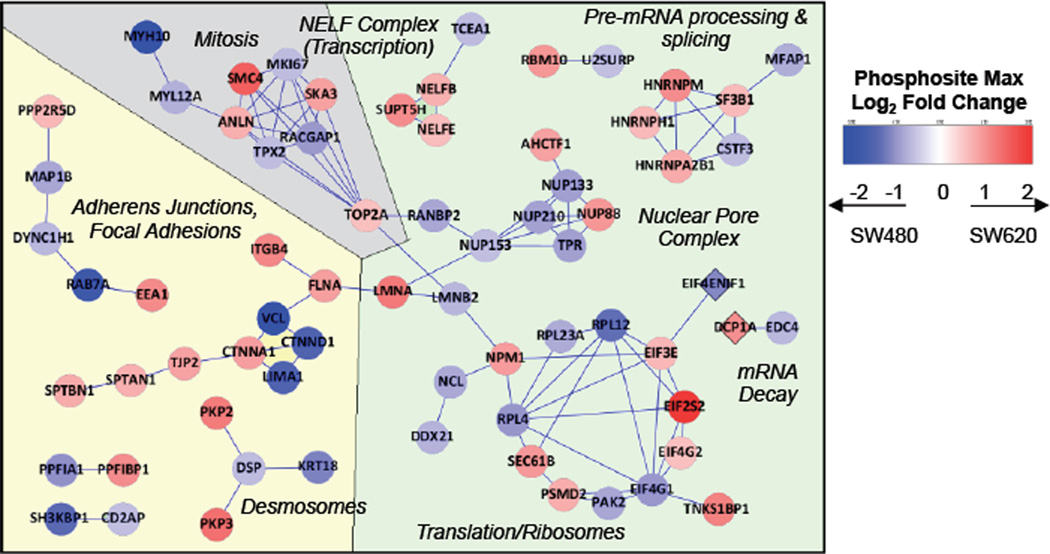
Plotting significantly altered phosphosites in STRING revealed several functional clusters that tended to align with GO annotation information [25] (Figure 5). One cluster centered around mitosis and functions related to the cell cycle, including such phosphoproteins as SMC4 (structural maintenance of chromosomes 4), TOP2A (topoisomerase IIA), SKA3 (spindle and kinetochore associated complex subunit 3), anillin, and MKI67. This perturbation in mitotic signaling is consistent with the more aggressive growth of the SW620 cell line. Likewise, since the SW620 cells are known to have greater migratory capacity, [10] it was expected that numerous phosphoproteins related to cytoskeletal organization, migration, and adhesion would be differentially regulated. Phosphoproteins in this cluster include delta-catenin, tight junction protein 2, zyxin, integrin beta 4, and desmoplakin.
Figure 5.
Network analysis of significant corrected phosphosites. Protein-expression corrected phosphosites that met significance criteria were compared against the STRING protein-protein interaction database. “High confidence” interactions are shown. The color of each node indicates the magnitude and direction of the SW620/SW480 fold change. Background colors highlight relationships observed among clusters: gray for mitosis-related phosphoproteins, yellow for cell-cell contacts and cytoskeleton-related clusters, and green for mRNA biogenesis-related clusters. EIF4ENIF1 (4E-T) and DCP1A, shown as diamonds, had missing protein-level information but had GO annotations that associated them with the mRNA – lifecycle-related clusters.
The remaining clusters included nuclear pore complex proteins, mRNA biogenesis and splicing factors, and translation initiation factors alongside ribosomal proteins. mRNA biogenesis, translation, and related processes are tightly regulated functions required by all cells, but a growing body of evidence reveals that their dysregulation contributes to carcinogenesis and to a lesser extent, metastasis [26, 27]. Human transcripts must compete for a limited pool of eIF4E (eukaryotic translation initiation factor 4E) cap-binding protein, which is one of several factors required for export to the cytosol through the nuclear pore complex. Overexpression of eIF4E contributes to oncogenic transformation in a wide array of cancers and plays a role in metastatic progression [28–30]. The regulatory functions of the phosphosites we observed on NUPs 88, 133, 153, and 210 are currently unknown, but multiple nucleoporins have been observed to modulate the transport and ultimate translational efficiency of specific cancer-related mRNA transcripts, as reviewed in recent manuscripts [31–33]. In short, there is a growing catalog of mechanisms that determine the translation rate of specific mRNA transcripts, and evidence that cancer cells hijack many of these mechanisms for survival [34]. There has been significantly less research tying these mechanisms to metastasis.
Our results are largely in agreement with previous proteomic studies of the SW480/SW620 cell lines. Ghosh and coworkers found that SW620 cells, as compared to SW480 cells, overexpressed several proteins involved in migration, adhesion, and cytoskeletal structure with novel roles in metastasis [12]. Our network analysis highlights these same processes and illustrates the role played by phosphorylation-mediated signaling. A comparison of the secretomes of these cell lines found trefoil factor 3 and growth/differentiation factor 15 upregulated in the SW620 cells [13]. Neither study specifically highlighted the role of mRNA biogenesis or decay; however, in our analysis, these processes were modulated exclusively at the phosphorylation site level rather than by protein abundance changes.
SW620 Cells Show Significant Phosphorylation Changes on Critical Cancer Proteins
Table 2 shows a selection of the phosphosites with the greatest fold-changes and significance levels. Many of these phosphoproteins have known roles in the functional clusters observed in the STRING analysis and in cancer progression. AHNAK and Septin 9 have been shown to be essential to pseudopod formation, migration, and EMT in metastatic cancer cell lines [35]. Vinculin, a component of focal adhesions, contains the single most down-regulated phosphosite in this study. Loss of vinculin expression has been shown to correlate with increased migration, invasiveness, and metastatic outcomes [36]. Both this work and that of others have reported that SW620 actually expresses more vinculin than SW480 [36], but the function of the phosphosite identified in this work is currently unknown. Nucleophosmin, which contains multiple highly up-regulated phosphosites, plays important roles in ribosome biogenesis, centrosome duplication, and apoptosis following DNA damage [37]. It has been reported to be overexpressed in metastatic colon cancers relative to primary tumors and normal tissues and to contribute to invasive behavior [38]. Pertaining to mRNA biogenesis and function, HNRPA2B1 [39], EIF4G1, EIF2S2, and ribosomal proteins L12 & L4 all contain highly significant phosphosites as well. Many of the phosphoproteins listed in this table and the specific sites identified would be worth further investigation to tease out the specific mechanistic roles of each site.
Table 2.
Most Significant Phosphosites Altered in SW620 and SW480.
| Gene Name | Protein Name |
Site | Phospho- peptide FC1 |
Protein FC1 |
Net Site FC1 | −log(P value) |
|---|---|---|---|---|---|---|
| AHNAK | Neuroblast differentiation- associated protein AHNAK |
S5731 | −0.643 | −2.133 | 1.489 | 227 |
| S5739 | −0.711 | 1.422 | 69 | |||
| S5841 | −1.074 | 1.059 | 71 | |||
| S216 | −1.301 | 0.832 | 62 | |||
| CLTC | Clathrin heavy chain 1 | T394 | −0.137 | −0.956 | 0.819 | 65 |
| EFHD2 | EF-hand domain-containing protein D2 |
S74 | 1.004 | 0.042 | 0.963 | 59 |
| EIF2S2 | Eukaryotic translation initiation factor 2 subunit 2 |
S2 | 1.660 | −0.445 | 2.105 | 126 |
| HIST1H1B | Histone H1.5 | S18 | 1.136 | −0.099 | 1.235 | 122 |
| S18 | 1.009 | 1.108 | 90 | |||
| HIST1H1E | Histone H1.4 | T18 | 0.986 | 0.012 | 0.975 | 67 |
| HNRNPA2B1 | Heterogeneous nuclear ribonucleoproteins A2/B1 |
S259 | 0.988 | 0.183 | 0.805 | 79 |
| ILF3 | Interleukin enhancer-binding factor 3 |
T596 | 1.234 | 0.221 | 1.013 | 61 |
| NPM1 | Nucleophosmin | S139 | 0.181 | −0.688 | 0.868 | 95 |
| S137 | 0.058 | 0.746 | 68 | |||
| PSMD2 | 26S proteasome non-ATPase regulatory subunit 2 |
S16 | −0.094 | −0.881 | 0.787 | 85 |
| SEPT9 | Septin-9 | S11 | 0.159 | −0.755 | 0.914 | 74 |
| CBFB | Core-binding factor, beta subunit |
S173 | 0.030 | 1.852 | −1.822 | 68 |
| EIF4G1 | Eukaryotic translation initiation factor 4 gamma 1 |
S1023 | −1.183 | −0.224 | −0.959 | 58 |
| KRT18 | Keratin, type I cytoskeletal 18 | S31 | −2.689 | −1.569 | −1.119 | 63 |
| S34 | −2.713 | −1.144 | 117 | |||
| PGAM1/4 | Phosphoglycerate mutase 1; Probable phosphoglycerate mutase 4 |
S118 | −1.547 | 0.414 | −1.961 | 102 |
| S118 | −1.610 | 0.414 | −2.024 | 69 | ||
| RPL12 | 60S ribosomal protein L12 | S38 | −1.342 | −0.002 | −1.340 | 140 |
| RPL4 | 60S ribosomal protein L4 | S295 | −0.942 | 0.024 | −0.966 | 63 |
| VCL | Vinculin | S290 | −1.632 | 0.494 | −2.126 | 145 |
FC, log2 fold change (SW620/SW480).
Besides highlighting particularly up- and down-regulated phosphosites, Table 2 also illustrates an interesting analytical observation. In certain cases, the large site fold changes are driven by phosphopeptide level changes, while others are driven by protein-level changes. Some phosphoproteins observed were extremely stable at the protein level but had highly significant phosphopeptide ratios, such as histones H1.4 and H1.5 and ribosomal proteins L4 & L12. In other cases, we observed modest phosphosite changes but dramatic protein differences. For example, Septin-9 pSer11 and nucleophosmin pSer137 and pSer139 have very modest phosphopeptide SW620/SW480 ratios, but exhibit dramatic protein-level changes which result in a significant corrected-phosphosite ratio. This situation implies that even though there are fewer copies of these proteins in SW620 cells, the fraction of copies that are phosphorylated at the indicated sites increased. Phosphosites like these whose occupancy changed in the opposite direction of protein abundance would not have been detected had the unmodified proteome not been analyzed.
Metastatic Cells Show Dysregulation in mRNA Biogenesis and Translation
The most significantly up-regulated phosphosite measured in SW620 as compared to SW480 was pSer-2 of eIF2S2. The eIF2a subunit uses a GTP to GDP hydrolysis to catalyze docking of the Met-tRNA to the ribosome pre-initiation complex in the final step of translation initiation. EIF2S2, or eIF2beta, participates in the exchange of the GDP for a fresh GTP. Blocking phosphorylation at Ser2 by mutation to alanine has been shown to cause a global translation slowdown [40]. A dramatic increase in phosphorylation at this site in the metastatic cell line suggests an increased rate of global translation in SW620s. Consistent with our observations, a 2006 microarray study of the SW480 and SW620 cell lines that used both total cellular mRNA and polysome profiling identified up-regulation of transcripts related to RNA transport, tRNA metabolism, and translational machinery [41]. Other critical transcripts pertained to cell cycle, viability, and apoptosis. The authors found over 80% of the inter-cell line variation was represented exclusively at the polysome level, suggesting that adaptations required for metastasis were better achieved by modulating the translational efficiency of particular transcripts rather than simply changing the copy number.
The cap-binding protein eIF4E has previously been reported to be overexpressed in many cancers and has been tied to oncogenesis and progression [29, 42, 43]. To evaluate whether oncogenic over-expression of eIF4E might account for the general over-activation of translation in SW620, the abundance level of the protein was examined. The data demonstrated, however, that this “limiting reagent” of translation is just slightly more abundant in SW480 than SW620, with an average log2 ratio = −0.46 (3 replicates). Western blots against eIF4E carried out on the original SILAC samples obtained a similar result (Figure 6). Therefore, the observed global mRNA regulatory perturbations occur despite fewer copies of eIF4E being available to the SW620 cells.
Figure 6.
Immunoblots against 4E-T, DCP1A, and eIF4E in the SILAC-labeled lysates used for proteome and phosphoproteome analysis. Beta-actin served as the loading control. Triplicate lanes of each biological replicate lysate were prepared across two membranes; a representative image is shown.
The ATCC reports that both SW480 and SW620 have driver KRAS, HRAS, and NRAS mutations. Ras signaling through ERK, PI3K/AKT, and mTORC2 is known to activate cap-dependent translation, especially by acting upon the eIF4E-eIF4G-eIF4A-mRNA cap complex [43]. Thus, upregulated translation would be expected to be a natural consequence of mutant Ras. However, since SW480 and SW620 both show constitutive Ras activation, this pathway is not sufficient to explain the deregulation of translation specific to the metastatic cells. Despite the extensive research that has been done to understand the regulation of translation, the stimuli that are responsible for the difference between SW480 and SW620’s phosphorylation in this network are unclear.
mRNA Degradation Phosphoproteins 4E-T and DCP1A
Having observed broad phosphorylation-level regulatory alterations to mRNA biogenesis and translational circuits, the next question was whether any of the non-corrected phosphopeptides had GO annotations to connect them to this network. This analysis yielded three such phosphopeptides: APpSPPLSQVFQTR (Ser564) of eukaryotic translation initiation factor 4E transporter (4E-T; also EIF4ENIF1 or eIF4E nuclear import factor 1) as well as LRLpTPQHDQIQTQPLGK (Thr401) and KASpSPpSPLTIGTPESQR (Ser522/523, Ser525) of mRNA decapping protein 1A (DCP1A) (ESM Table S-8). In the latter peptide, pSer525 was localized with 98% confidence but the second phosphorylation could be attached at either 522 or 523. The 4E-T peptide containing pSer564 was found to be about twice as concentrated in SW480 as in SW620; the two DCP1A phosphopeptides were two-fold up-regulated in the SW620 cell line in the discovery experiment. Since no unmodified peptides for these proteins were identified in our proteome analysis, these phosphopeptides could not be normalized for protein abundance.
4E-T has been shown to recruit mRNA cap-binding protein eIF4E to processing bodies (or P-bodies), where the mRNA is either sequestered or degraded. A recent report showed JNK phosphorylates 4E-T at sites distinct from Ser564 upon an increase in reactive oxygen species, which leads to an increase in P-body size [44]. The function of pSer-564 has not yet been reported, nor have there been any previous reports of 4E-T function changing in correlation to metastasis.
DCP1A acts as part of a larger decapping complex to remove the protective 5’ cap from mRNA marked for degradation. It operates primarily in the cytoplasm and has been shown to interact with 4E-T in processing-bodies (P-bodies) under stress –triggered JNK signaling [45]. DCP1a has also been reported to be a target of TGF-B signaling [46]. The specific roles of pSer525 and pThr401 are uncharacterized, however, the C-terminal domains containing these sites (380–470, 470–582) have been shown to contribute to PB localization [47]. The protein is also known to become hyperphosphorylated during M phase, including at Ser522/523. Alanine mutants of these two sites have been shown to interfere with PB localization but do not completely abolish it. In contrast to its well-recognized role in mRNA decay, DCP1A has not previously been linked to cancer progression.
It was unclear whether the apparent phosphosite changes on 4E-T and DCP1A were artifacts of protein abundance changes or if they represent true differences in site stoichiometry. Westerns indicate that 4E-T is expressed at similar levels in the two cell lines (Figure 6), so we proceeded to measure the absolute site stoichiometry at Ser564 by SRM. 4E-T was prepared for SRM analysis by immunoprecipitating in duplicate from pooled SILAC SW480 and SW620 lysates (Figure 7A). An immunoblot showed the target protein was pulled down efficiently (ESM Figure S1). SRM revealed that 4E-T Ser564 was phosphorylated 55–88% in SW480 and 65–79% in SW620; there was no significant change between cell lines. Technical variation was relatively low, with coefficients of variation between 4–10% (ESM Table S9). The phosphopeptide SW620/SW620 ratios measured by SRM trended in the same direction as those measured in the discovery experiment, but with a smaller magnitude (ESM Table S9). Thus, a modest decrease in protein copy number best explains the differences in phosphorylation initially observed between the cell lines. While this site does not correlate to metastatic status, the high fractional occupancy at this site does suggest a critical function for pSer564 in logarithmically growing cells. Furthermore, finding the site occupancy unchanged between cell lines reiterates the importance of considering protein abundance changes in phosphoproteomic experiments.
Figure 7.
A. IP-MRM workflow. SW480 and SW620 cells were SILAC-labeled and a reversed label was employed in a duplicate experiment, carried out in parallel. B. 4E-T pS564 site stoichiometry as measured by IP-MRM.
As with 4E-T, DCP1A was examined first by immunoblot, which revealed no cell line-dependent difference in protein abundance. However, DCP1A from SW620 cells exhibited a gel mobility shift which is consistent with hyperphosphorylation. This high degree of modification prevented the selection of tryptic proteotypic peptides suitable for SRM analysis; therefore, DCP1A was not amenable to targeted site stoichiometry analysis. When hyperphosphorylation is present or suspected, it becomes especially crucial to avoid the selection of peptides containing serine and threonine. DCP1A contains just two such tryptic peptides, and neither of these was detectable in immunoprecipitates.
The IP-SRM analyses of 4ET and DCP1A illustrate the ongoing challenges for targeted phosphoprotein analysis, and more broadly, for targeted analysis of any PTM. Many phosphoproteins of biological interest are found at relatively low abundance in vivo and thus require immunoprecipitation or other enrichment from tissue or cellular extracts. While a targeted analysis meant to simply measure total protein concentration can select among the peptides most amenable to SRM analysis, this choice does not exist for a phosphosite target. If multiple phosphorylation isomers are possible for the peptide(s) of interest, these must all be surveyed. Site stoichiometry determination adds an extra challenge. The occupancy of some sites may be highly variable in vivo. When the occupancy is either very high or very low, either the phosphopeptide or its counterpart non-phosphopeptide must necessarily exist at low abundance. All these factors must be taken into consideration during targeted assay design. Fundamentally, adequate sensitivity for all relevant peptide species is the greatest obstacle to meaningful assay results.
CONCLUSION
This is the first comparison of the constitutive phosphoproteome of the colon cancer Stage II and Stage III cell lines, SW480 and SW620. With 815 protein-normalized phosphosites quantified, along with many novel phosphosites detected as well, this work should provide a valuable resource to other researchers. Comprehensive phosphopeptide profiling shows that the metastatic cell line is distinguished from the primary tumor line by differential regulation in cellular adhesion, cytoskeletal structure, mitosis, mRNA transport, and translation. Since many of the sites detected in this analysis are not functionally characterized, additional work will be required to tease out the specific relationships between the driver kinases, the phosphorylation of translation proteins, and metastatic phenotypes. Further investigation will be required to reveal how regulation of translation contributes to the metastatic phenotypes that SW620 displays, as well as to generalize these findings to other cell lines.
In summary, this work illustrates the breadth of deregulation in the constitutive phosphoproteome of metastatic cells, especially in the mRNA translation machinery. Our findings suggest there is much left to investigate regarding the selective advantage that metastatic cells gain from translation deregulation. Likewise, it remains unclear which signaling stimuli are responsible for the level of deregulation that is observed. More straightforward explanations for translation perturbations, like overexpression of eIF4E or activated Ras, are not enough to differentiate the two isogenic cell lines, so there may be uncharacterized factors that are influencing translation in these cell lines. Finally, this work suggests that metastatic cells may be uniquely susceptible to translation inhibitors. Further work will be required to test whether this hypothesis will be borne out.
Acknowledgments
The work was funded by Walther Cancer Foundation and the Notre Dame Harper Cancer Research Institute for postdoctoral funding for XY and salary support for ABH. This report was supported by the National Science Foundation (CAREER Award, CHE-1351595) and the National Institutes of Health (1R01GM110406-01) for AS and ABH. The work was also funded by Department of Defense Visionary Postdoctoral Fellowship Award to XY (USAMRAA W81XWH-12-1-0412). We thank the staff from the Mass Spectrometry and Proteomics Facility at the University of Notre Dame, especially Dr. Matthew Champion, for their helpful discussions and advice, and for their help maintaining the mass spectrometry equipment.
Footnotes
COMPLIANCE WITH ETHICAL STANDARDS
The authors declare that they have no conflicting interests.
ELECTRONIC SUPPLEMENTARY MATERIAL
In the ESM_1.xlsx are Table S1: SRM transitions for 4E-T and Tables S2–4: Complete LC-MS results for Corrected Phosphosites, Non-Corrected Phosphosites, and Proteins.
The file “ESM_2.pdf” contains: Figure S1 (missed cleavages), Figure S2 (Biological replicate reproducibility), Tables S5–7 (Detailed GO annotations), Table S8 (Phosphopeptides selected for further analysis, Figure S3 (4E-T pulldown), and Table S9 (4E-T IP-SRM measured peptide ratios).
REFERENCES
- 1.Valastyan S, Weinberg RA. Tumor metastasis: Molecular insights and evolving paradigms. Cell. 2011;147:275–292. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hsu PP, Kang SA, Rameseder J, Zhang Y, Ottina KA, Lim D, Peterson TR, Choi Y, Gray NS, Yaffe MB, Marto JA, Sabatini DM. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science. 2011;332:1317–1322. doi: 10.1126/science.1199498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McCubrey Ja, Steelman LS, Chappell WH, Abrams SL, Wong EWT, Chang F, Lehmann B, Terrian DM, Milella M, Tafuri A, Stivala F, Libra M, Basecke J, Evangelisti C, Martelli AM, Franklin Ra. Roles of the Raf/MEK/ERK pathway in cell growth, malignant transformation and drug resistance. Biochim Biophys Acta - Mol Cell Res. 2007;1773:1263–1284. doi: 10.1016/j.bbamcr.2006.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Longley DB, Johnston PG. Molecular mechanisms of drug resistance. J Pathol. 2005;205:275–292. doi: 10.1002/path.1706. [DOI] [PubMed] [Google Scholar]
- 5.Misale S, Yaeger R, Hobor S, Scala E, Janakiraman M, Liska D, Valtorta E, Schiavo R, Buscarino M, Siravegna G, Bencardino K, Cercek A, Chen C-T, Veronese S, Zanon C, Sartore-Bianchi A, Gambacorta M, Gallicchio M, Vakiani E, Boscaro V, Medico E, Weiser M, Siena S, Di Nicolantonio F, Solit D, Bardelli A. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012;486:532–536. doi: 10.1038/nature11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Voulgari A, Pintzas A. Epithelial-mesenchymal transition in cancer metastasis: Mechanisms, markers and strategies to overcome drug resistance in the clinic. Biochim Biophys Acta - Rev Cancer. 2009;1796:75–90. doi: 10.1016/j.bbcan.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 7.Wiśniewski JR, Ostasiewicz P, Duś K, Zielińska DF, Gnad F, Mann M. Extensive quantitative remodeling of the proteome between normal colon tissue and adenocarcinoma. Mol Syst Biol. 2012;8:611. doi: 10.1038/msb.2012.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leibovitz A, Stinson JC, McCombs WB, McCoy CE, Mazur KC, Mabry ND. Classification of human colorectal adenocarcinoma cell lines. Cancer Res. 1976;36:4562–4569. [PubMed] [Google Scholar]
- 9.The Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hewitt RE, McMarlin a, Kleiner D, Wersto R, Martin P, Tsokos M, Stamp GW, Stetler-Stevenson WG, Tsoskas M. Validation of a model of colon cancer progression. J Pathol. 2000;192:446–454. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH775>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 11.Bauer KM, Lambert PA, Hummon AB. Comparative label-free LC-MS/MS analysis of colorectal adenocarcinoma and metastatic cells treated with 5-fluorouracil. Proteomics. 2012;12:1928–1937. doi: 10.1002/pmic.201200041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghosh D, Yu H, Tan XF, Lim TK, Zubaidah RM, Tan HT, Chung MCM, Lin Q. Identification of key players for colorectal cancer metastasis by iTRAQ quantitative proteomics profiling of isogenic SW480 and SW620 cell lines. J Proteome Res. 2011;10:4373–4387. doi: 10.1021/pr2005617. [DOI] [PubMed] [Google Scholar]
- 13.Xue H, Lu B, Zhang J, Wu M, Huang Q, Wu Q, Sheng H, Wu D, Hu J, Lai M. Identification of Serum Biomarkers for Colorectal Cancer Metastasis Using a Differential Secretome Approach. J Proteome Res. 2010;9:545–555. doi: 10.1021/pr9008817. [DOI] [PubMed] [Google Scholar]
- 14.Bodenmiller B, Mueller LN, Mueller M, Domon B, Aebersold R. Reproducible isolation of distinct, overlapping segments of the phosphoproteome. Nat Methods. 2007;4:231–237. doi: 10.1038/nmeth1005. [DOI] [PubMed] [Google Scholar]
- 15.Yue X, Schunter A, Hummon AB. Comparing multistep immobilized metal affinity chromatography and multistep TiO2 methods for phosphopeptide enrichment. Anal Chem. 2015;87:8837–8844. doi: 10.1021/acs.analchem.5b01833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yue X, Hummon AB. Combination of multistep IMAC enrichment with high-pH reverse phase separation for in-depth phosphoproteomic profiling. J Proteome Res. 2013:4176–4186. doi: 10.1021/pr4005234. [DOI] [PubMed] [Google Scholar]
- 17.Li Q, Ning Z, Tang J, Nie S, Zeng R. Effect of peptide-to-TiO2 beads ratio on phosphopeptide enrichment selectivity. J Proteome Res. 2009;8:5375–5381. doi: 10.1021/pr900659n. [DOI] [PubMed] [Google Scholar]
- 18.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 19.Wiśniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 20.Olsen JV, Vermeulen M, Santamaria A, Kumar C, Miller ML, Jensen LJ, Gnad F, Cox J, Jensen TS, Nigg Ea, Brunak S, Mann M. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal. 2010;3:ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- 21.Song C, Ye M, Han G, Jiang X, Wang F. Reversed-Phase-Reversed-Phase Liquid Chromatography Approach with High Orthogonality for Multidimensional Separation of Phosphopeptides. Anal Chem. 2010;82:53–56. doi: 10.1021/ac9023044. [DOI] [PubMed] [Google Scholar]
- 22.Ficarro SB, Zhang Y, Carrasco-Alfonso MJ, Garg B, Adelmant G, Webber JT, Luckey CJ, Marto JA. Online Nanoflow Multidimensional Fractionation for High Efficiency Phosphopeptide Analysis. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.O111.011064. O111.011064-O111.011064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Batth TS, Francavilla C, Olsen JV. Off-line high-pH reversed-phase fractionation for in-depth phosphoproteomics. J Proteome Res. 2014;13:6176–6186. doi: 10.1021/pr500893m. [DOI] [PubMed] [Google Scholar]
- 24.Wu R, Dephoure N, Haas W, Huttlin EL, Zhai B, Sowa ME, Gygi SP. Correct interpretation of comprehensive phosphorylation dynamics requires normalization by protein expression changes. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M111.009654. M111.009654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, Jensen LJ. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parsyan A, Hernández G, Meterissian S. Translation initiation in colorectal cancer. Cancer Metastasis Rev. 2012;31:387–395. doi: 10.1007/s10555-012-9349-9. [DOI] [PubMed] [Google Scholar]
- 27.Blagden SP, Willis AE. The biological and therapeutic relevance of mRNA translation in cancer. Nat Rev Clin Oncol. 2011;8:280–291. doi: 10.1038/nrclinonc.2011.16. [DOI] [PubMed] [Google Scholar]
- 28.Culjkovic B, Topisirovic I, Borden KLB. Controlling gene expression through RNA regulons: The role of the eukaryotic translation initiation factor eIF4E. Cell Cycle. 2007;6:65–69. doi: 10.4161/cc.6.1.3688. [DOI] [PubMed] [Google Scholar]
- 29.Robichaud N, Del Rincon SV, Huor B, Alain T, Petruccelli La, Hearnden J, Goncalves C, Grotegut S, Spruck CH, Furic L, Larsson O, Muller WJ, Miller WH, Sonenberg N. Phosphorylation of eIF4E promotes EMT and metastasis via translational control of SNAIL and MMP-3. Oncogene. 2015;34:2032–2042. doi: 10.1038/onc.2014.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pettersson F, del Rincon SV, Emond A, Huor B, Ngan E, Ng J, Dobocan MC, Siegel PM, Miller WH. Genetic and pharmacologic inhibition of eIF4E reduces breast cancer cell migration, invasion and metastasis. Cancer Res. 2015;75:1102–1113. doi: 10.1158/0008-5472.CAN-14-1996. [DOI] [PubMed] [Google Scholar]
- 31.Culjkovic-Kraljacic B, Borden KLB. Aiding and abetting cancer: MRNA export and the nuclear pore. Trends Cell Biol. 2013;23:328–335. doi: 10.1016/j.tcb.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Köhler A, Hurt E. Gene Regulation by Nucleoporins and Links to Cancer. Mol Cell. 2010;38:6–15. doi: 10.1016/j.molcel.2010.01.040. [DOI] [PubMed] [Google Scholar]
- 33.Strambio-De-Castillia C, Niepel M, Rout MP. The nuclear pore complex: bridging nuclear transport and gene regulation. Nat Rev Mol Cell Biol. 2010;11:490–501. doi: 10.1038/nrm2928. [DOI] [PubMed] [Google Scholar]
- 34.Loreni F, Mancino M, Biffo S. Translation factors and ribosomal proteins control tumor onset and progression: how? Oncogene. 2014;33:2145–2156. doi: 10.1038/onc.2013.153. [DOI] [PubMed] [Google Scholar]
- 35.Shankar J, Messenberg A, Chan J, Underhill TM, Foster LJ, Nabi IR. Pseudopodial actin dynamics control epithelial-mesenchymal transition in metastatic cancer cells. Cancer Res. 2010;70:3780–3790. doi: 10.1158/0008-5472.CAN-09-4439. [DOI] [PubMed] [Google Scholar]
- 36.Li T, Guo H, Song Y, Zhao X, Shi Y, Lu Y, Hu S, Nie Y, Fan D, Wu K. Loss of vinculin and membrane-bound beta-catenin promotes metastasis and predicts poor prognosis in colorectal cancer. Mol Cancer. 2014:1–15. doi: 10.1186/1476-4598-13-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grisendi S, Mecucci C, Falini B, Pandolfi PP. Nucleophosmin and cancer. Nat Rev Cancer. 2006;6:493–505. doi: 10.1038/nrc1885. [DOI] [PubMed] [Google Scholar]
- 38.Liu Y, Zhang F, Zhang X, Qi L, Yang L, Guo H, Zhang N. Expression of Nucleophosmin/NPM1 correlates with migration and invasiveness of colon cancer cells. J Biomed Sci. 2012;19:53. doi: 10.1186/1423-0127-19-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wojtuszkiewicz A, Assaraf YG, Maas MJ, Kaspers GJ, Jansen G, Cloos J. Pre-mRNA splicing in cancer: the relevance in oncogenesis, treatment and drug resistance. Expert Opin Drug Metab Toxicol. 2015;11:673–689. doi: 10.1517/17425255.2015.993316. [DOI] [PubMed] [Google Scholar]
- 40.Llorens F, Duarri A, Sarró E, Roher N, Plana M, Itarte E. The N-terminal domain of the human eIF2beta subunit and the CK2 phosphorylation sites are required for its function. Biochem J. 2006;394:227–236. doi: 10.1042/BJ20050605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Provenzani A, Fronza R, Loreni F, Pascale A, Amadio M, Quattrone A. Global alterations in mRNA polysomal recruitment in a cell model of colorectal cancer progression to metastasis. Carcinogenesis. 2006;27:1323–1333. doi: 10.1093/carcin/bgi377. [DOI] [PubMed] [Google Scholar]
- 42.Graff JR, Konicek BW, Carter JH, Marcusson EG. Targeting the Eukaryotic Translation Initiation Factor 4E for Cancer Therapy. Cancer Res. 2008;68:631–634. doi: 10.1158/0008-5472.CAN-07-5635. [DOI] [PubMed] [Google Scholar]
- 43.Pelletier J, Graff J, Ruggero D, Sonenberg N. Targeting the eIF4F Translation Initiation Complex: A Critical Nexus for Cancer Development. Cancer Res. 2015;75:250–263. doi: 10.1158/0008-5472.CAN-14-2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cargnello M, Tcherkezian J, Dorn JF, Huttlin EL, Maddox PS, Gygi SP, Roux PP. Phosphorylation of the eukaryotic translation initiation factor 4E-transporter (4E-T) by c-Jun N-terminal kinase promotes stress-dependent P-body assembly. Mol Cell Biol. 2012;32:4572–4584. doi: 10.1128/MCB.00544-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ferraiuolo MA, Basak S, Dostie J, Murray EL, Schoenberg DR, Sonenberg N. A role for the eIF4E-binding protein 4E-T in P-body formation and mRNA decay. J Cell Biol. 2005;170:913–924. doi: 10.1083/jcb.200504039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bai R-Y, Koester C, Ouyang T, Hahn SA, Hammerschmidt M, Peschel C, Duyster J. SMIF, a Smad4-interacting protein that functions as a co-activator in TGFbeta signalling. Nat Cell Biol. 2002;4:181–190. doi: 10.1038/ncb753. [DOI] [PubMed] [Google Scholar]
- 47.Aizer A, Kafri P, Kalo A, Shav-Tal Y. The P body protein Dcp1a is hyper-phosphorylated during mitosis. PLoS One. 2013;8:e49783. doi: 10.1371/journal.pone.0049783. [DOI] [PMC free article] [PubMed] [Google Scholar]