Abstract
In C. elegans, the anterior-posterior axis is established at the one-cell stage when the embryo polarizes along its long axis. One model suggests that a cue from the centrosome triggers symmetry breaking and is then dispensable for further steps in the process. In the absence of the initial centrosome cue, a redundant mechanism, reliant on the centrosome’s microtubules, can polarize the cell. Despite this model, data from multiple sources suggest that direct centrosome-contact with the cortex may play a role in ensuring robust polarization. Some of this past work includes analysis of pam-1 mutants, which lack a functional puromycin-sensitive aminopeptidase and have aberrant centrosome positioning and variable polarization defects. To better understand the role of centrosome dynamics in polarization, we looked in detail at centrosome behavior in relation to key polarity landmarks in pam-1 mutants as well as those lacking cortical flows. We provide evidence for a model in which sustained direct contact between the centrosome and the cortex acts to reinforce both the actomyosin and the microtubule-dependent pathways. This contact is necessary for polarization when flows are inhibited.
Keywords: C. elegans, polarity, anterior-posterior axis, centrosome, puromycin-sensitive aminopeptidase
Graphical abstract
Introduction
In Caenorhabditis elegans, the anterior-posterior axis is established shortly after fertilization through reorganization of the cortical cytoskeleton. A cue from the centrosome leads to symmetry breaking, when the first difference in cortical contractility along the axis is observed (Bienkowska and Cowan, 2012; Cheeks et al., 2004; Cowan and Hyman, 2004; Cuenca et al., 2003; Munro et al., 2004). Symmetry breaking begins as the smoothening of a small patch of the cortex near the centrosome to mark the future posterior pole. This comes about due to local inactivation of ECT-2 and the RHO-1 G protein and destabilization of the actin-myosin cytoskeleton (Jenkins et al., 2006; Tse et al., 2012). This symmetry breaking then leads to full polarity establishment, in which anteriorly-directed cortical flows move non-muscle myosin toward the anterior pole (Munro et al., 2004) and interior cytoplasm moves posteriorly, moving the centrosome into close contact with the posterior cortex (Hird and White, 1993). As a result, the entire posterior cortex becomes smooth while the anterior cortex exhibits continued contractility (Hird and White, 1993). At the boundary between these two domains is the pseudocleavage furrow, a transient ingression of the cell membrane that serves as a clear sign that polarization has taken place.
During symmetry breaking, the cortical PAR proteins begin to localize to distinct domains along the cortex, a localization that stabilizes during polarity establishment (Griffin, 2015; Rose and Kemphues, 1998). The PAR proteins PAR-3 and PAR-6 clear from a small cortical patch in the posterior and later become limited to the anterior half of the cortex. This allows posterior PAR proteins PAR-1 and PAR-2 to load to the posterior cortex, ultimately occupying the entire posterior half of the embryo (Cuenca et al., 2003). The PAR proteins then mediate further asymmetries in the embryo that result in an asymmetric first division.
How the centrosome initiates symmetry breaking and its role in polarity establishment has been the subject of numerous studies (Bienkowska and Cowan, 2012; Cowan and Hyman, 2004; Fortin et al., 2010; Goldstein and Hird, 1996; Hamill et al., 2002; Lyczak et al., 2006; Rappleye et al., 2002; Sadler and Shakes, 2000). Shortly after the completion of meiosis, the centrosome is positioned close to the future posterior pole where it acts to cue cell polarization. In experiments in which the centrosome was ablated at different time points, it was determined that the centrosome is needed for initiation but not maintenance of cell polarity. This role is independent of the centrosome’s role in microtubule nucleation (Cowan and Hyman, 2004; Sonneville and Gönczy, 2004; Tsai and Ahringer, 2007). However, the microtubules appear to be necessary for polarization in the absence of cortical flows (Motegi et al., 2011; Zonies et al., 2010). Thus, it has been proposed that there are two redundant mechanisms for initiating polarity – the first through an unknown centrosome cue to initiate flows and a second through microtubule loading of PAR-2 to the posterior cortex (Motegi and Seydoux, 2013).
The nature of the initial centrosome cue is unknown. Some work suggests that the centrosome can initiate AP polarity establishment from a distance. The centrosome is typically 2–7µm from the cell cortex at the time of polarity initiation (Bienkowska and Cowan, 2012). Distances further than this delay symmetry breaking both in wild-type and in some mutants, but do not prevent the process (Bienkowska and Cowan, 2012; McCloskey and Kemphues, 2012). Thus, a model in which initial centrosome distance from the cortex impacts polarity has been put forth. Once close enough to the cortex, the centrosome cues symmetry breaking, which is thought to be enough to lead to full polarization.
After symmetry breaking, flows of cytoplasm quickly move the centrosome toward a tight association with the posterior cortex. While this substantiates the idea that the centrosome may initiate polarity from anywhere in the cell, it fails to account for a potentially-continued role of the centrosome after initiation. In fact, research on centrosome positioning mutants has shown that distance from the cortex influences both the degree of success at polarity initiation and also maintenance of polarity. In embryos in which the centrosomes move prematurely from the cortex or spend less time in contact with the cortex, cell polarity phenotypes range from a complete lack of axis establishment to a failure to maintain polarization after initiation (Fortin et al., 2010; Lyczak et al., 2006; McCloskey and Kemphues, 2012). Similarly, when centrosomes are ablated after symmetry breaking, polarization is less robust, as posterior PAR domains are smaller (Cowan and Hyman, 2004). Thus, it is still an open question as to whether direct centrosome-cortical contact plays a role in axis polarization.
Little is known about the proteins that regulate centrosome positioning at the time of cell polarization. Initial proximity to the posterior cortex seems to depend on cytoplasmic microtubules (Bienkowska and Cowan, 2012). Factors that regulate protein turnover have also been implicated in this process. Mutations that disrupt the puromycin-sensitive aminopeptidase PAM-1 have aberrant centrosome movements that result in a failure to polarize (Fortin et al., 2010; Lyczak et al., 2006). This aminopeptidase is hypothesized to be involved in peptide activation or peptide degradation through the proteasome (Lyczak et al., 2006). Similarly, centrosomes are mislocalized when deubiquitylation machinery components (DUBs) are disrupted. math-33, usp-46, and usp-47 encode for deubiquitylation enzymes that when disrupted in isolation have no effect on polarity. However, in combination, these mutants show centrosome positioning defects that lead to polarity problems. Interestingly, math-33 mutants combined with RNAi for pam-1 show enhancement of the initial centrosome positioning defect (McCloskey and Kemphues, 2012). Mispositioning of the centrosomes is the cause of the polarization defects in these mutants, because when dynein is inactivated in pam-1 or DUB mutants, the centrosomes remain in tight association with the posterior cortex and these embryos exhibit proper polarization (Fortin et al., 2010; McCloskey and Kemphues, 2012). Thus, these proteins all appear to regulate polarity through centrosome positioning. However, it is not clear if PAM-1 works in a common pathway with DUBs or in two separate pathways, nor what the targets of these pathways are.
While distance from the cortex has been shown to dictate the extent of polarization (Bienkowska and Cowan, 2012; McCloskey and Kemphues, 2012), the timing of this contact, and the duration of this contact required for both initiation and maintenance of polarity, are not well understood. pam-1 mutants provide a system to better understand the relative importance of these interactions due to the variable expressivity of the phenotype. Centrosome dynamics vary among embryos both in proximity, timing of contact, and duration of contact. In addition, while some pam-1 mutant embryos show no signs of polarization, others polarize normally, or show initial polarization that is lost prior to the first cell division (Fortin et al., 2010; Lyczak et al., 2006). In this study, we have used this variability to better understand the relative importance of centrosome-cortical distance, timing, and duration in both the initiation and the sustained polarization of the anterior-posterior axis.
Here we provide evidence that sustained centrosome-cortical contact is necessary for proper polarization when cortical flows are compromised. By studying pam-1 mutants and those lacking cortical flows, we have found that direct contact between the centrosome and the cortex can reinforce both the actomyosin and the microtubule-dependent polarity establishment pathways.
Materials and Methods
Strain maintenance
C. elegans strains were propagated on NGM plates as previously described (Brenner, 1974). Worms were grown at 20°C or 15°C (pam-1 mutant strains). WT and pam-1 strains were shifted to 25°C for at least 5 hours prior to imaging. pam-1(or403) was used for all analysis (Lyczak et al., 2006). The following transgenic constructs were used: ruIs32 [pie-1::GFP::H2B + unc-119(+)] and ruIs57 [pie-1p::GFP::tubulin + unc-119(+)] (Praitis et al., 2001); ltIs37 [pie-1p::mCherry::his-58 (pAA64) + unc-119(+)]; xnls3 [par-6:PAR-6::GFP + unc-119 (+)] (Anderson et al., 2008); ddls6 [tbg-1::GFP + unc-119 (+)]; axls1327 [par-1::GFP]; ruls48[pAZ147 Ppie-1::tbb-2::GFP]. LP162, nmy-2(cp13[nmy-2::gfp + LoxP]) was used to image cortical activity.
RNAi
Bacteria expressing double stranded RNA were used and feeding RNAi was performed as described (Cockell et al., 2004; Fortin et al., 2010; Kamath et al., 2003; McCloskey and Kemphues, 2012). Feeding RNAi worms were shifted to 25°C before imaging. To ensure effective knock-down of nmy-2, loss of GFP in the LP162 strain was used as a control.
Imaging and analysis
Embryos were imaged on 3% agarose pads as previously described (Lyczak et al., 2006). Imaging was performed on a Nikon C1 confocal with EZC1 software or a Nikon C2 confocal with NIS-Elements AR 4.20 software. Z-stack time-lapse images were taken every 15 seconds with 1.5–2.0µm between Z frames (for centrosome analysis) or 5 µm between Z frames for NMY-2 analysis. Centrosome measurements from the cortex were gathered using NIS-Elements software and measurements were only made from time lapse sequences in which the centrosome appeared after the start of imaging and were measured in the X-Y plane. Contact times were made relative to nuclear envelope breakdown (NEBD). Nuclear envelope breakdown was defined as the first frame when a clear delineation between nuclear and cytoplasmic GFP was lost.
The extent of localization of PAR-1 at a landmark event of the cell cycle, nuclear envelope breakdown, was measured in the one-cell embryo using NIS-Elements. The length of the embryo was measured and the PAR-1 domain length was defined as the farthest anterior point along the cortex to which GFP::PAR-1 was observed. These two measurements were used to determine the percent embryo length of PAR-1 localization.
Statistical analysis
All graphs were generated using Excel and statistical significance was determined using a two-sided two-sample unequal variance t-test. Linear regression was completed using SAS 9.2.
Results and Discussion
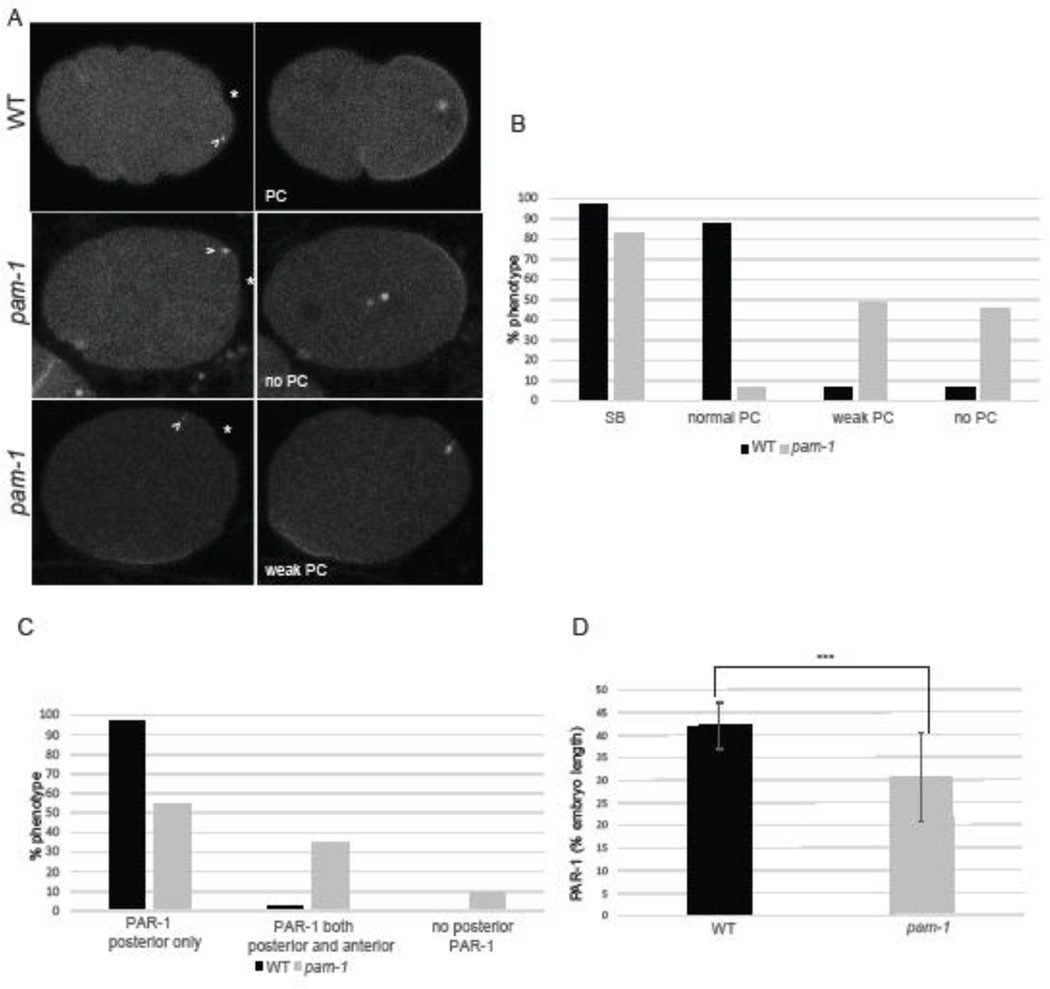
The role of the centrosome in polarity establishment is still poorly understood. The identity of the centrosome cue, the role of centrosome-cortical contact, and the timing of these events have not been fully dissected. To learn more about of the centrosome’s role in polarity establishment, we first compared a series of polarity markers, some not previously examined, in wild-type and pam-1 mutant embryos, so that they could be correlated with centrosome behavior. The initial sign of polarization is symmetry breaking, defined as the point where the centrosome initiates cortical smoothening in the posterior (Bienkowska and Cowan, 2012) (Figure 1A). In wild-type embryos, the initial symmetry breaking point then extends as the anterior and posterior cortex exhibit differences in contractility. The ultimate consequence of these differences is a deep pseudocleavage furrow that separates the anterior and posterior domains (Hird and White, 1993). In wild-type, nearly all embryos we examined exhibited normal symmetry breaking (n=31/32) followed by pseudocleavage (Figure 1 A–B). These two events were usually coupled in wild-type; the one embryo that did not exhibit symmetry breaking also did not have pseudocleavage, while a few other wild-type embryos had weak or absent pseudocleavage after symmetry breaking (Figure 1B).
Figure 1. pam-1 mutant embryos show defects in the establishment of polarity.
A) In wild-type embryos, the centrosome (arrowhead; γ-tub::GFP) appears close to the posterior cortex and triggers symmetry breaking (SB) (*). This is followed by pseudocleavage (PC) and localization of PAR-1 (GFP) to the posterior cortex. In pam-1 mutants, the centrosome is positioned similarly to wild-type at first and most embryos exhibit SB. However, most embryos exhibit no or weak PC and small or mislocalized PAR-1 domains. Posterior is to the right in all images B) The percentage of wild-type and pam-1 mutant embryos with different polarity phenotypes (WT=32; pam-1 =64). C) The percentage of wild-type (n=33) and pam-1 mutant (n=51) embryos with different PAR-1 localizations, as documented at the time of pronuclear envelope breakdown. D) pam-1 mutant embryos that localize PAR-1 posteriorly (n=46) exhibit smaller domains as compared to wild-type (n=32). Standard deviation is shown. *** p < 0.0001.
In pam-1 mutant embryos, these initial polarity landmarks were not always present, and were often decoupled. In comparison to wild-type, pam-1 mutant embryos exhibited symmetry breaking only 83% of time, while defects in pseudocleavage were more frequent (Figure 1 A–B; n=64) (Lyczak et al., 2006). Only 6% of pam-1 mutant embryos had normal pseudocleavage, while the remainder were split between those with weak pseudocleavage and those that lacked any signs of pseudocleavage. When pseudocleavage was looked at in relation to symmetry breaking, we noticed that nearly all embryos that lacked symmetry breaking (n=11) also lacked pseudocleavage (n=10/11), suggesting a link between the processes. However, when we looked at embryos that did exhibit the initial symmetry breaking event (n=53), we noted that only 7.5% of these embryos went on to exhibit normal pseudocleavage. The remaining embryos exhibited weak pseudocleavage (57%) or lacked pseudocleavage completely (36%). Thus, the initial symmetry breaking event did not always lead to full polarization. In wild-type, and in the current polarization model, full polarization follows after initiation from the centrosome (Bienkowska and Cowan, 2012; Cowan and Hyman, 2004). Our data suggests that PAM-1 is necessary for polarization following the initial symmetry breaking event.
In a previous study, we observed defects in PAR localization in pam-1 mutant embryos (Fortin et al., 2010; Lyczak et al., 2006). We wanted to quantify the extent of posterior PAR localization in pam-1 mutants as another measure of the extent of polarization. Using a PAR-1::GFP strain, we measured the extent of the PAR domains in wild-type and pam-1 mutants at the time of nuclear envelope breakdown (NEBD). All wild-type embryos, except the one that failed at symmetry breaking and pseudocleavage, exhibited normal PAR-1 posterior localization (Figure 1 A,C). This domain extended to about 42% of the embryo length at NEBD (Figure 1 D; n=32). In contrast, 10% of pam-1 mutant embryos exhibited no posterior PAR-1 localization at any time (Figure 1A, C). Some of these embryos had no cortical PAR-1 localization, while others had some anterior or lateral localization. While 90% of pam-1 mutant embryos had some posterior PAR-1 localization by NEBD, many had mislocalization prior to this time, or had additional anterior localization as well. About 20% of the embryos with eventual posterior localization had initial PAR-1 patches at the anterior to lateral cortex. In addition, 39% of all the posterior PAR-1 embryos had a second small domain of PAR-1 to the anterior, near the polar body (Figure 1C). We measured the domain of PAR-1 in pam-1 mutants where it localized to the posterior and found the domain to be substantially smaller than in wild-type, extending only 31% of the embryo length (Figure 1D; n=46; p<0.0001). Thus, even pam-1 mutant embryos that show signs of polarity have defects in the extent of polarization.
Initial centrosome distance from the cortex is similar in pam-1 mutant and wild-type embryos
Previous work has shown the importance of the initial centrosome distance from the cortex in the timing and establishment of polarity (Bienkowska and Cowan, 2012; McCloskey and Kemphues, 2012). Additionally, our work has shown that pam-1 mutant embryos exhibit aberrant centrosome positioning that causes the polarity defects (Fortin et al., 2010). Thus, we wanted to measure the initial position of the centrosome in relation to the cortex in both wild-type and pam-1 mutants and to correlate these distances with polarity landmarks. Centrosomes in wild-type embryos appeared at an average of 1.97 um from the cortex (Table 1 and Figure 2A), which was not significantly different from the centrosomes of pam-1 mutants, which initially appeared at an average of 1.71 um from the cortex (Table 1 and Figure 2A; p=0.59). This similarity between wild-type and pam-1 mutants in initial centrosome position separates pam-1 from other centrosome positioning mutants (McCloskey and Kemphues, 2012) and suggests that PAM-1 affects centrosome positioning after initial centrosome appearance.
Table 1.
Average distance of the centrosome from the cortex upon first appearance in relation to symmetry breaking (SB) and extent of pseudocleavage (PC).
| Centrosome- cortical distance (um) |
WT | pam-1 |
|---|---|---|
| All embryos | 1.97 ± 2.27 n=36 |
1.71 ± 2.11 n=52 p=0.59 |
|
Embryos with SB |
1.76 ± 1.90 n=35 |
1.63 ± 2.08 n=46 p=0.78 |
|
Embryos without SB |
9.49 n=1 |
2.32 ± 2.42 n=6 p=0.61 |
|
Embryos with normal PC |
1.66 ± 1.74 n=32 |
0.61 n=1 |
|
Embryos with weak PC |
3.96 ± 4.04 n=2 |
1.64 ± 1.88 n=28 p=0.97 |
|
Embryos without PC |
5.00 ± 6.36 n=2 |
1.85 ± 2.43 n=23 p=0.75 |
Distances were sorted into presence, absence, or weak pseudocleavage. Standard deviation is shown. No significant differences between pam-1 classes and wild-type (WT) embryos were found. P values were calculated by comparing WT all to pam-1 all; pam-1 SB classes to WT with SB; and pam-1 PC classes to WT with PC.
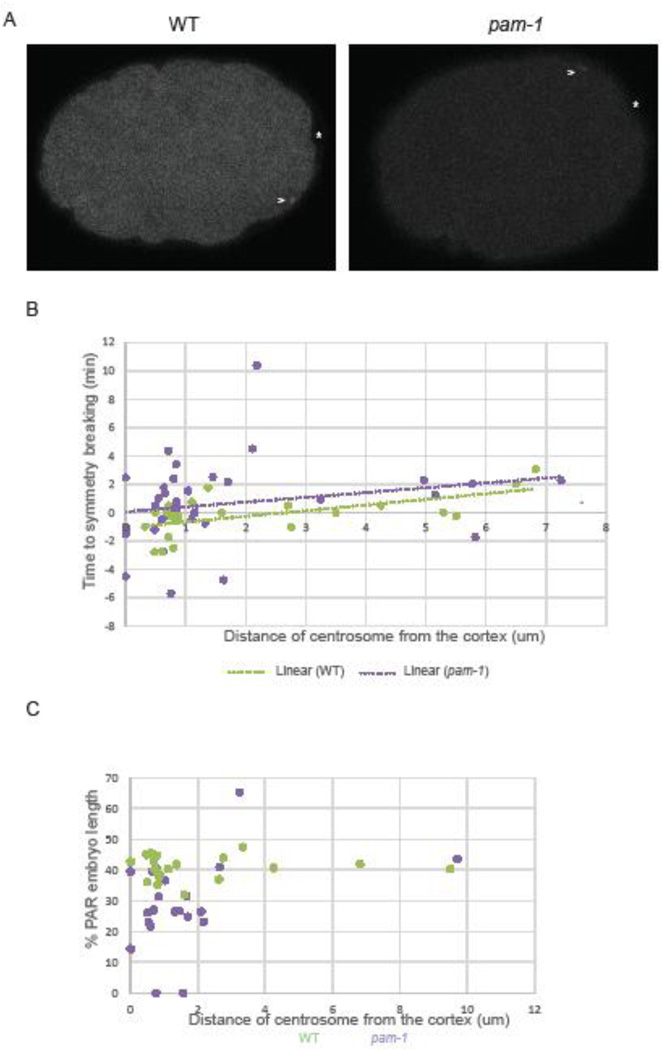
Figure 2. Polarity defects in pam-1 mutants are not caused by increased centrosome-cortical distance.
A) Centrosomes in both WT and pam-1 mutant embryos (arrowhead; γ-tub::GFP) appear near the cortex at the same relative distance and in most cases cue symmetry breaking (*). Posterior is to the right in all images. B) While initial centrosome distance correlates with increased time to symmetry breaking in WT embryos (n=23), this trend is not significant in pam-1 mutant embryos (n=36). C) When distance from the cortex is plotted against the extent of PAR-1 localization at NEBD, we see no correlation (WT n=23, pam-1 n=21). Each dot represents one embryo.
We then looked at the time required to break symmetry in these embryos from the first appearance of the centrosome. Overall, the time between centrosome appearance and symmetry breaking did not significantly differ between pam-1 mutant and wild-type embryos (Figure 2B; p=0.36). We used linear regression analysis to understand the correlation between centrosome distance from the cortex and the time required for symmetry breaking (Figure 2B). Prior studies have found that close proximity of the centrosome decreased the time that was required for symmetry breaking to occur, thus increasing the efficiency of polarity establishment (Bienkowska and Cowan, 2012). Despite much more variability in timing at the smaller distances, we found that for wild-type, the correlation of the variables were significant (R2 = 0.3485, P=0.0030, n=23), similar to past work (Bienkowska and Cowan, 2012). Smaller initial distances correlated with faster symmetry breaking. However, for pam-1 mutants, the correlation of the variables was not significant (R2 = 0.0478, P=0.2002, n=36). Despite this, the trend lines for wild-type and pam-1 mutants were similar (Figure 2B). We next looked at the centrosome distance in embryos that break symmetry and those that did not. When centrosome-cortical distance in pam-1 embryos with and without symmetry-breaking were compared to wild-type embryos with normal symmetry breaking, no significant difference was observed (Table 1; p=0.78, p=0.61). It is interesting to note however that the one wild-type embryo that failed to initiate symmetry breaking had a much larger centrosome-cortical distance. Thus, in pam-1 mutant embryos we could not find a connection between initial distance of the centrosome from the cortex and the time to symmetry breaking.
Next we examined pseudocleavage to see if centrosome-cortical distance affected this polarity landmark. When we compared pam-1 embryos with weak or absent pseudocleavage to wild-type embryos that exhibited normal pseudocleavage, the centrosome-cortical distance was not significantly different (Table 1; p=0.97, p=0.75). Thus, while the majority of pam-1 mutant embryos exhibited aberrant pseudocleavage, this was not correlated with differences in cortical-centrosome distance. Interestingly, while sample size was small, we did notice a trend in wild-type embryos with larger initial centrosome-cortex distances more likely to show aberrations in pseudocleavage (Table 1).
Finally, to see if variation in centrosome distance could impact the extent of localization of PAR-1, we conducted statistical analysis through correlation and linear regression of both wild-type and pam-1 data. For both wild-type and pam-1 data, we detected no significant correlation between centrosome distance from the cortex and the extent of PAR-1 localization (WT: R2 = 0.0016, P=0.8562, n=23, pam-1: R2 = 0.1316, P=0.1060, n=21). Wild-type embryos all showed consistent PAR-1 localization, while the variation in pam-1 mutant embryos did not relate to the centrosome distance (Figure 2C).
In the current model, close proximity of the centrosome to the cortex is thought to be sufficient to trigger symmetry breaking and axis polarization (Bienkowska and Cowan, 2012). Mutants in which the centrosomes appear farther from the cortex have delayed or absent polarization (McCloskey and Kemphues, 2012). However, our data with pam-1 mutants indicates that close centrosome-cortical proximity is not sufficient for polarization in these mutants. The initial distance of the centrosome to the cortex was similar in pam-1 mutants to wild-type (Figure 2), even though most pam-1 mutants had polarity defects (Figure 1). In addition, we found similarities in the centrosome-cortical distance in embryos that polarized well compared to those that polarized poorly. This similarity between wild-type and pam-1 mutants in initial centrosome position separates pam-1 from other centrosome positioning mutants (McCloskey and Kemphues, 2012) and suggests PAM-1 impacts the centrosome after its initial appearance.
Timing of centrosome contact at the cortex does not affect polarization
In wild-type, the centrosome cues symmetry breaking from a distance, but following this event, the centrosome is brought into tight association with the cortex. These events happen together in wild-type and have not been studied separately to determine if this later tight association is required for full polarization. In our previous work, we found that polarity could be restored to pam-1 mutants when centrosomes were in sustained contact with the cortex (Fortin et al., 2010). When we inactivated dynein heavy chain, the centrosome remained in contact with the cortex and polarized normally. Thus, we looked next at the timing and duration of direct centrosome-cortex interaction.
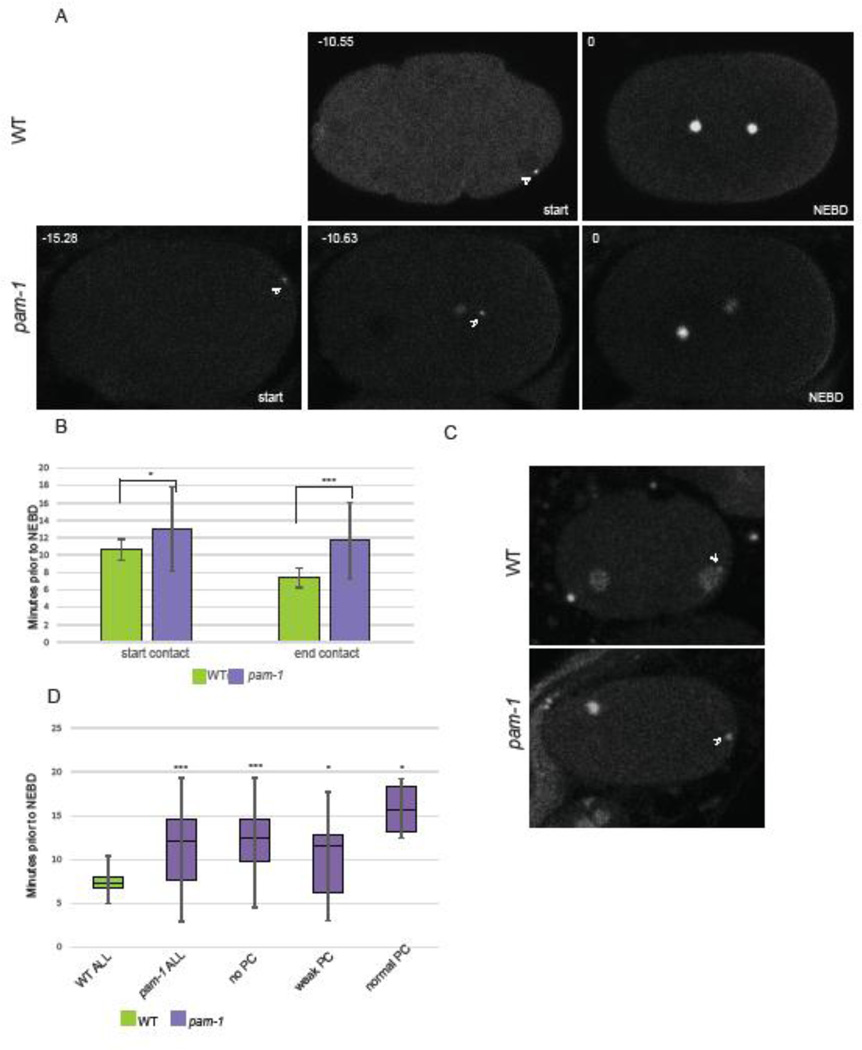
First, we examined the timing of the close contact during the embryonic cell cycle (Figure 3A). Since pam-1 mutants have a delayed meiotic exit (Lyczak et al., 2006), we wanted to see if centrosome contact was occurring at a time different from wild-type. This contact timing was defined as the time the centrosome initiated and terminated cortical contact relative to nuclear envelope breakdown (NEBD). We observed that wild-type embryos had centrosome-cortical contact beginning 10.59 minutes prior to NEBD and ending about 7.38 minutes prior to NEBD (Figure 3A–B; n=32 and n=39 respectively). In pam-1 mutant embryos where the centrosome contacted the cortex, it occurred earlier, starting 13 minutes prior to NEBD and ending 11.67 minutes prior to NEBD (Figure 3A–B; n=31 and 43 respectively). When we looked in strains carrying histone::GFP, to carefully discern the cell-cycle stage of the embryos, we noticed another difference between the two strains. In wild-type, the majority of embryos were in the pronuclear stage at the start of centrosome contact (n=19/22) while the remainder were in meiotic exit. By the end of centrosome contact, all wild-type embryos were in the pronuclear stage. However, in pam-1 mutants, only one exhibited contact during the pronuclear stage (n=14) while the remainder had centrosome contact during meiotic exit (Figure 3C). This change in timing is most likely due to the meiotic delay exhibited by pam-1 mutant embryos (Lyczak et al., 2006). Thus, relative to nuclear envelope break down, the centrosome left the cortex in pam-1 mutants before it started making contact in wild-type embryos.
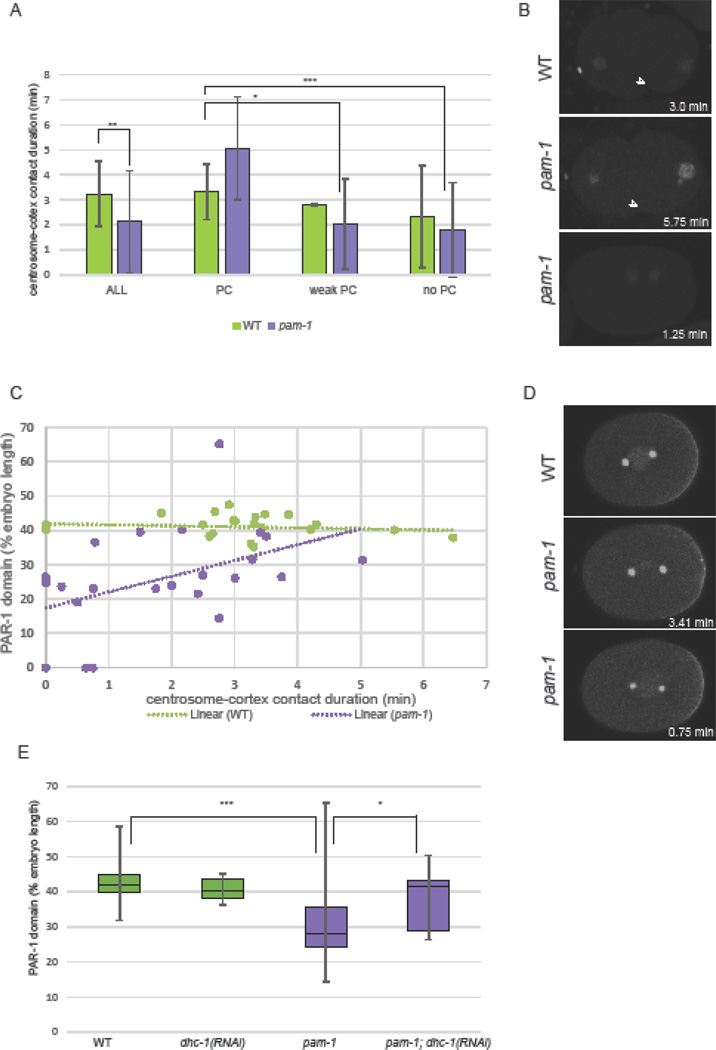
Figure 3. The timing of centrosome-cortical contact does not correlate with polarity defects.
A) In pam-1 mutants, the centrosome (γ-tub::GFP) contacts the cortex earlier in the cell cycle than WT. The times were measured back from nuclear envelope breakdown (NEBD). Images show the start of centrosome-cortical contact and a pam-1 mutant embryo at a time comparable to the start time for WT. Posterior is to the right in all images. B) Both the start and end of centrosome-cortical contact are significantly different. (WT n=39; pam-1 n=43) Standard deviations are shown. * p≤0.01; *** p≤0.0001. C) End of centrosome (arrowhead) contact times in wild-type occurred during the pronuclear phase as viewed by histone::GFP. In pam-1 mutants, end of centrosome contact usually occurred prior to the appearance of decondensed pronuclei. D) pam-1 mutant embryos were separated by extent of PC and end of centrosome contact was compared (no PC n=17; weak PC n=22; PC n=4). The mean in all three classes were different from WT. * p≤0.01; *** p≤0.0001. Box plots show range of data with the box representing the upper and lower quartile and the median. Whiskers show the max and min.
We next asked if the difference in centrosome-cortical contact timing correlated with the polarity defects. To do this, we separated pam-1 mutant embryos into three classes based on the extent of pseudocleavage (Figure 3D n=17, 22, 4). When embryos were separated, we noted that all pam-1 classes still had contact timing earlier in the cell cycle compared to wild-type (p≤0.0001; p≤0.01; p≤0.01). Both embryos that polarized well and those that polarized poorly exhibited earlier contact times. This lack of correlation suggests that the timing of contact is not a factor in polarization success. The earlier contact times in pam-1 embryos are most likely the result of the meiotic exit delays exhibited by pam-1 mutants. In our prior work, we showed that the meiotic exit delays are separable and not the cause of the polarity defects (Lyczak et al., 2006). When normal meiotic timing was restored through inactivation of cyclin B, embryos still failed to polarize. These results taken with our new data provide additional evidence that timing of centrosome-cortical contact is not a factor in polarization.
Duration of centrosome-cortical contact affects polarization
As centrosome distance and the timing of centrosome-cortical contact did not correlate with polarity defects in pam-1 mutant embryos, we hypothesized that centrosomes must contact the cortex for a specific amount of time to polarize the axis. Thus, we measured the duration of centrosome-cortical contact. In wild-type, the centrosome made contact with the cortex in 94.91% of the embryos examined (n=59). In contrast, the centrosome made contact with the cortex in 86.11% of pam-1 mutant embryos (n=72). We also found that the duration of centrosome-cortex was significantly shorter in pam-1 mutant embryos compared to wild-type (p<0.001; Figure 4A). While wild-type embryos had contact durations of 3.25 minutes on average, pam-1 embryos had contact durations of 2.14 minutes. Thus, centrosomes spend less time at the cortex in pam-1 mutant embryos compared to wild-type.
Figure 4. Short centrosome-cortical contact duration correlates with more severe polarity defects.
A) The centrosomes in pam-1 mutant embryos contact the cortex for a shorter time than WT. When separated by extent of PC, embryos with normal PC have contact times comparable to WT (p=0.20) Standard deviation is shown. p≤0.01; ** p≤0.001; ***p≤0.0001. (WT: n=59, 54, 2, 3; pam-1: n=72, 5, 28, 39) B) pam-1 mutants with longer contact times show normal pseudocleavage (arrowhead) as compared to those with shorter contact times. β-tub::GFP and histone::GFP – posterior to the right. C) Linear regression modeling finds a significant correlation between contact duration and the extent of posterior PAR-1 localization for pam-1 mutant embryos (n=23) but not WT (n=23). Each dot represents an embryo. D) Longer contact durations in pam-1 mutants (lower right of each picture) result in larger PAR-1 posterior domains and shorter contact durations result in smaller PAR-1 domains. Images at NEBD stage. γ-tub::GFP; PAR-1::GFP – posterior to the right. E) PAR-1 domains are smaller in pam-1 embryos (n=46) as compared to WT (n=32). When dhc-1 is inactivated, both WT (n=6) and pam-1 (n=14) embryos had PAR-1 domains similar to WT (p=0.48; p=0.29). * p≤0.05; *** p≤0.0001. Box plots show range of data with the box representing the upper and lower quartile and the median. Whiskers show the max and min.
As we did for other centrosome behaviors, we looked at the relationship between contact duration and symmetry breaking, pseudocleavage, and PAR-1 polarization. As wild-type and pam-1 mutant embryos exhibit no significant difference in the timing for symmetry breaking, and symmetry breaking occurs prior to centrosome-cortical contact, we asked if there was a relationship between symmetry breaking and subsequent centrosome-cortical contact. Symmetry breaking was scored for 49 pam-1 mutants and observed in all but eight. Of the eight that did not show symmetry breaking, most had centrosomes that never contacted the cortex (n=6/8) and the others had very short contact durations. On average, in pam-1 mutant embryos without symmetry breaking, the contact duration of the centrosome with the cortex was 0.14 ± 0.27 minutes as compared to 2.02± 1.65 minutes in the embryos that did show symmetry breaking (p<0.0001). Likewise, while only one wild-type embryo did not break symmetry, we noted that it also did not show any centrosome-cortical contact. Thus, it is possible that symmetry breaking is required for subsequent centrosome-cortical contact.
Next we divided our pam-1 mutant and wild-type embryos into those that exhibited normal pseudocleavage (n=5/72; n=54/59), those with weak pseudocleavage (n=28/72; n=2/59) and those without pseudocleavage (n=39/72; n=3/59) Figure 4A–B). When this was done, we saw that pam-1 embryos with strong pseudocleavage had duration times more similar to wild-type (p=0.19). In contrast, pam-1 embryos with either weak or absent pseudocleavage exhibited significantly shorter contact durations (p=0.002; p=0.0001). In addition, of the ten pam-1 mutants in which no centrosome contact was observed, seven exhibited no pseudocleavage and the others had very weak pseudocleavage. Thus, increased centrosome-cortical durations correlated with better polarization.
Similarly, we looked at the role of centrosome-cortex contact duration and the extent of PAR-1 localization to the posterior (Figure 4C–D). When we separated embryos into those with and without posterior PAR-1 localization, we found significantly shorter contact durations in those embryos without posterior PAR-1 (Table 2). We then looked at the size of the posterior PAR-1 domains. Using linear regression analysis, we found no association between contact duration and wild-type PAR domain size (R2 = 0.0165, P=0.5593, n=23). This was largely due to the striking consistency of PAR-1 domains among wild-type embryos, as well as consistency of centrosome-cortical contact durations in wild-type. In contrast, there was a significant association observed in pam-1 mutant embryos (R2 = 0.2014, P=0.0317, n=23). Shorter contact durations were strongly correlated with smaller posterior PAR-1 domains while longer durations had PAR-1 domains more similar to wild-type (Figure 4C–D). We hypothesized that PAR-1 domain size would be closer to wild-type, if we could increase the contact duration in pam-1 mutants. Previously we showed that inactivation of dynein heavy chain (dhc-1) in pam-1 mutants rescued polarity by preventing the centrosome from leaving the cortex (Fortin et al., 2010). Thus, we repeated this experiment and measured the PAR-1 domain size. We found that PAR domains were larger in pam-1; dhc-1(RNAi) embryos as compared to pam-1 alone (Figure 4E). In addition, both dhc-1(RNAi) and pam-1; dhc-1(RNAi) embryos had PAR-1 domain sizes similar to wild-type (Figure 4E; p=0.48 and p=0.29 respectively). Taken together, these data suggest that the polarity phenotypes in pam-1 embryos correlate with shortened centrosome-cortical contact and suggest that sustained centrosome-cortical contact is necessary for complete polarization in these mutants.
Table 2.
Duration of centrosome contact with the cortex correlated with PAR-1 localization.
| Duration of Centrosome-cortical contact (min) |
All | Posterior PAR-1 |
No posterior PAR-1 |
|---|---|---|---|
| pam-1 | 1.81 ± 1.43 n=26 |
2.10 ± 1.40 n=20 |
0.82 ± 1.13 n=6 p<0.0001 |
| nmy-2(RNAi) | 1.11 ± 1.30 n=16 |
1.57 ± 1.35 n=9 |
0.61 ± 1.03 n=7 p=0.10 |
Standard deviation is shown. p values shown compares two PAR classes for each strain.
Role of centrosome-cortical contact
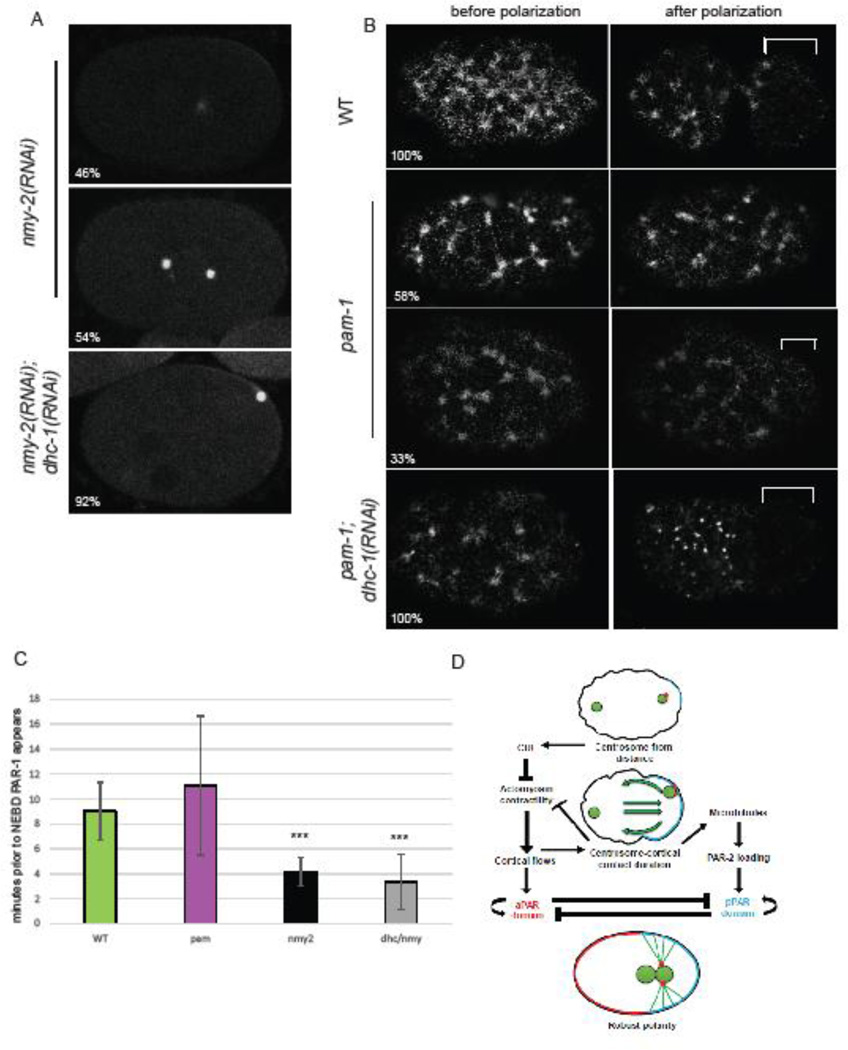
We next sought to examine how centrosome-cortex contact may influence the polarity cuing process. Does the centrosome contact reinforce the actomyosin flow pathway, the microtubule pathway, or both? To do this, we first asked if cortical flows are necessary to induce centrosome-cortical contact. We inactivated non-muscle myosin 2 (nmy-2), a protein previously shown to be necessary for cortical flows (Shelton et al., 1999). In all nmy-2(RNAi) embryos, we observed no difference in cortical activity along the axis or pseudocleavage, as previously reported (Figure 5A; n=16). We looked at centrosome positioning in these embryos and observed that centrosome-cortical contact occurred in only 50% of these embryos. While overall durations averaged 1.11 minutes (Table 2), when we averaged only embryos with centrosome contact, duration times were 2.23 ± 0.89 minutes. This duration, is similar to those seen in pam-1 mutants. Thus, while the centrosome can make contact with the cortex in the absence of flows, it does not occur with reproducibility. The similarity of centrosome-cortical timing between pam-1 and nmy-2(RNAi) embryos could mean that shortened durations in pam-1 mutants are a result of defects in cortical flow.
Figure 5. Centrosome-cortical contact enhances polarity through both the actomyosin-dependent and microtubule-dependent pathways.
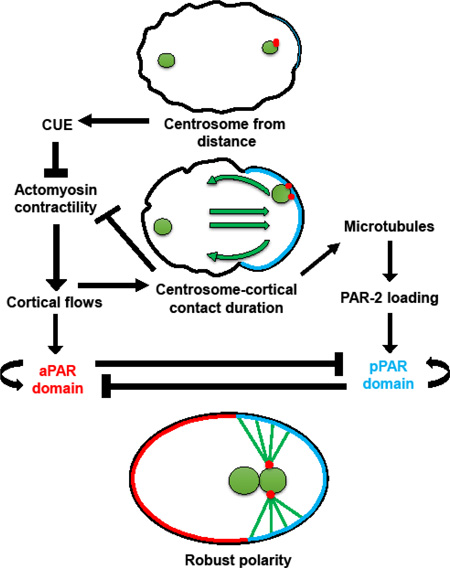
A) In embryos with no cortical flows, in the absence of NMY-2, by NEBD PAR-1 localizes to a small cortical patch 46% of the time. PAR-1::GFP and γ-tub::GFP. When dhc-1 is also inactivated, 92% of embryos now have PAR-1 localization at NEBD. B) pam-1 embryos have sparse NMY-2 foci in the cortex as compared to WT. While WT embryos show clearing of NMY-2 foci from the posterior after polarization, 58% of pam-1 mutants show no clearing and 33% show weak clearing. When dhc-1 is inactivated in pam-1 mutants, all embryos now show NMY-2 clearing from the posterior. NMY-2::GFP clearings shown with a bracket; posterior is to the right. C) PAR-1::GFP localization to the posterior occurs later when flows are absent. * p≤0.05; *** p≤0.0001 Standard deviation is shown. (WT: n=32; pam-1: n=44; nmy-2(RNAi): n=8; nmy-2, dhc-1(RNAi): n=10) D) A revision to the model of Motegi and Seydoux (2013). Centrosome-cortical contact enhances polarization through the actomyosin-dependent and the microtubule-dependent pathways. This contact is necessary when flows are weak or absent, but is redundant when cortical flows are strong.
To look at this in more detail, next we examined NMY-2 foci using NMY-2::GFP in pam-1 mutants. In wild-type, all embryos showed NMY-2 foci throughout the cortex prior to polarization, which cleared from the posterior during polarization (Figure 5C; n= 10; Munro et al., 2004). To have a comparison of myosin in embryos that fail to elicit cortical flows due to a centrosome defect, we looked at spd-5(RNAi) embryos. These embryos all showed normal NMY-2 foci that did not clear from the cortex (data not shown; n=9; Munro et al., 2004). When we examined pam-1 embryos, we noted that the NMY-2 foci appeared more sparsely arranged and larger, suggesting that PAM-1 may regulate the actomyosin cytoskeleton. As expected from the pam-1 pseudocleavage data (Figure 1B), we saw no posterior clearing or pseudocleavage in 58% of embryos, weak clearing and weak pseudocleavage in 33% of embryos and saw only one embryo with normal NMY-2 clearing and pseudocleavage (Figure 5C; n=12). Thus, it is possible that weak or absent flows in pam-1 mutants, may cause reduced centrosome-cortical contact. While this is likely, it does not explain how increased centrosome-cortical contact leads to better polarization outcomes.
How then does increased centrosome-cortical contact enhance polarization? Centrosome-cortical contact may enhance the microtubule-dependent polarity pathway, the actomyosin cortical flow pathway, or both. First we looked at the role of centrosome contact in embryos lacking the actomyosin pathway. Previously it was shown that in the absence of cortical flows, PAR-2 loads to the cortex through a microtubule dependent pathway, where it can then recruit PAR-1 (Motegi et al., 2011; Zonies et al., 2010). To address the role of centrosome-cortical contact in the absence of flows, we asked if PAR localization correlated with centrosome contact in nmy-2(RNAi) embryos. We found that only 56% of the nmy-2(RNAi) embryos had PAR localization at NEBD. We looked separately at embryos in which the centrosome contacted the cortex and those that did not and noticed a trend. While 75% of the embryos with centrosomecortical contact exhibited PAR localization, only 37.5% of embryos without centrosome-cortical contact did. Similarly, when we divided embryos by PAR localization, we noticed a trend toward shorter centrosome-cortex durations in embryos without PAR (Table 2; p=0.10). We next increased centrosome-cortical contact in embryos without flows, to see if increased contact enhanced polarity. To do this, we looked at embryos in which both nmy-2 and dhc-1 were inactivated. In these embryos, pseudocleavage failed due to the lack of flows, but the centrosome contacted the cortex in all embryos (n=12). Surprisingly, despite the lack of flows, PAR-1 localized to the cortex at NEBD in 92% of these embryos. This suggests that while flows may push the centrosome to the cortex in wild-type embryos, prolonged centrosome-cortical contact promotes PAR loading in the absence of cortical flows. Thus, our data suggests that centrosome-cortical contact can promote polarization through the microtubule-dependent pathway.
Because centrosome-cortical contact can enhance polarization through the microtubule pathway, next we asked if the centrosome-cortical contact in pam-1 mutant enhances polarity through only the microtubule-dependent polarization pathway or through the actomyosin pathway as well. First we compared the timing of PAR-1 loading to the cortex in pam-1 embryos as compared to embryos that lack flows (Figure 5B). In nmy-2(RNAi) embryos that localized PAR-1, PAR first became visible at the posterior about four minutes prior to NEBD, while in wild-type and pam-1 mutants, this occurred nine and eleven minutes prior to NEBD, respectively (Figure 5B). This delay in posterior PAR localization in nmy-2 embryos, was previously shown to be caused by reliance on the microtubule pathway (Motegi et al., 2011; Zonies et al., 2010) suggesting that pam-1 embryos may also use the flow dependent pathway. To see if centrosome-cortical contact in pam-1 embryos enhanced the flow-dependent polarity pathway we first looked at pseudocleavage, a visual sign of cortical flow. Strong pseudocleavage occurred in pam-1 embryos with longer duration times (Figure 4B). Even more striking, was the change in pseudocleavage in pam-1; dhc-1(RNAi) embryos. The embryos have increased centrosome-cortical contact but also all exhibit strong pseudocleavage and PAR polarity in all cases (Fortin et al., 2010 Figure 4E), suggesting enhanced flows. To see if NMY-2 clearing was restored when centrosome-cortical contact is enhanced, we looked at NMY-2::GFP in pam-1;dhc-1(RNAi) embryos. While NMY-2 foci were sparse and large similarly to pam-1 mutants alone, all embryos examined exhibited posterior NMY-2 clearing as well as pseudocleavage (Figure 5C; n=6). Thus, prolonged centrosome-cortical contact seems to enhance polarization through the actomyosin pathway as well as the microtubule-dependent pathway.
Conclusions
Here we provide evidence for a role of sustained centrosome-cortical contact in the polarization process. While this has been suggested from prior analysis of pam-1 mutants and other centrosome positioning mutants, a role for sustained contact has not previously been incorporated in the current model for axis establishment.
From our data, we propose a role for sustained centrosome-cortical contact in enhancement of both the actomyosin and the microtubule polarity establishment pathways (Figure 5D). The initial centrosome cue leads to local destabilization of the actomyosin cytoskeleton in the posterior, when still at a distance from the cortex (Bienkowska and Cowan, 2012; Cowan and Hyman, 2004; Munro et al., 2004). This initiates cortical flows that move the centrosome to the cortex in embryos where this has not happened on its own (Hird and White, 1993). Once at the cortex, the centrosome acts to reinforce polarity through interaction with both pathways. This reinforcement is redundant in wild-type embryos, where robust cortical flows are enough to fully polarize the embryo. However, our analysis reveals that centrosome-cortex contact and the duration of that contact are necessary when flows are reduced or inhibited. In the absence of flows, such as in nmy-2(RNAi) embryos, the centrosome cues polarity through the microtubule-dependent pathway. This PAR polarization is not always effective, and happens later than in wild-type embryos (Figure 5 and (Motegi et al., 2011; Zonies et al., 2010). We have shown that increased centrosome-cortical contact in these embryos increases the effectiveness of PAR polarization, but this polarization is still delayed, mostly likely due to reliance on the microtubule-dependent pathway. When actomyosin contractility is impaired but not absent, like in pam-1 embryos, centrosome-cortical contact can act both to enhance destabilization of posterior actomyosin as well as PAR localization. This can be seen in embryos lacking both pam-1 and dhc-1 and in enhanced psuedocleavage in embryos with longer contact durations. Hence, while centrosome-cortical contact is not required under normal cortical flow conditions, we propose a model where it can act to enhance the actomyosin-dependent as well as the microtubule-dependent polarity pathway in poor-flow conditions. Thus a continued role for the centrosome, after initial symmetry breaking is proposed (Figure 5D).
While we favor the model above, an alternative model is that centrosome-cortical contact only acts to enhance the microtubule-dependent polarity pathway. In this model, changes to the actomyosin cortex may occur secondarily to microtubule-dependent loading of PAR in pam-1 embryos. While the dynamics of the polarization in pam-1 embryos point to the first model, further experimentation will be needed to rule out this alternative possibility.
In addition to further insights into the role of the centrosome in polarization, our results also add to an understanding of the role of PAM-1 in polarization. While further research is necessary, the sparse and enlarged NMY-2 foci in pam-1 embryos suggest that PAM-1 regulates the actomyosin cytoskeleton. This may result in an inability to properly cue polarity through posterior destabilization of the cytoskeleton and subsequent cortical flows unless sustained centrosome-cortical contact follows. In both wild-type and pam-1 embryos, the centrosome is initially positioned close to the future posterior cortex (Bienkowska and Cowan, 2012; Cowan and Hyman, 2004). DUBs as well as the cytoplasmic microtubule network are needed for the initial positioning near the cortex and initial distance from the cortex influences the timing of symmetry breaking (Bienkowska and Cowan, 2012; McCloskey and Kemphues, 2012). Although centrosomes are in close proximity to the cortex in pam-1 mutant embryos, about 17% of the embryos do not break symmetry, indicating that close proximity is not sufficient for breaking symmetry in these mutants. Future studies will be needed to see if this is due to defects in the actomyosin cytoskeleton and its ability to respond to the centrosome cue. Following symmetry breaking, cytoplasmic flows ensure that the centrosome is positioned at the posterior cortex. The inability to elicit asymmetric actomyosin contractility may explain the centrosome positioning defects in pam-1 embryos. Despite the need for further research on PAM-1’s mechanism of action, this mutant has provided insights into the role of centrosome-cortical contact in ensuring polarization of the anterior-posterior axis.
Highlights.
-
◦
The centrosome is needed after the initial polarization cue.
-
◦
Sustained centrosome-cortical contact leads to full polarity.
-
◦
Contact enhances the actomyosin-dependent and microtubule-dependent polarity pathways.
Acknowledgments
This work was supported by NSF-RUI grant [IOS-0918950] and NIH grant [1 R15 GM110614-01]. Some of the strains were provided by the Caenorhabditis elegans Genetics Center, which is funded by the National Institutes of Health - National Center for Research Resources. We would like to thank B. Bowerman (University of Oregon) for some reagents and careful reading of the manuscript and P. Chi (Ursinus College) for reviewing our statistical analysis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson DC, Gill JS, Cinalli RM, Nance J. Polarization of the C. elegans embryo by RhoGAP-mediated exclusion of PAR-6 from cell contacts. Science. 2008;320:1771–1774. doi: 10.1126/science.1156063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienkowska D, Cowan CR. Centrosomes can initiate a polarity axis from any position within one-cell C. elegans embryos. Curr Biol. 2012;22:583–589. doi: 10.1016/j.cub.2012.01.064. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeks RJ, Canman JC, Gabriel WN, Meyer N, Strome S, Goldstein B. C. elegans PAR proteins function by mobilizing and stabilizing asymmetrically localized protein complexes. Curr Biol. 2004;14:851–862. doi: 10.1016/j.cub.2004.05.022. [DOI] [PubMed] [Google Scholar]
- Cockell MM, Baumer K, Gonczy P. lis-1 is required for dynein-dependent cell division processes in C. elegans embryos. J Cell Sci. 2004;117:4571–4582. doi: 10.1242/jcs.01344. [DOI] [PubMed] [Google Scholar]
- Cowan CR, Hyman AA. Centrosomes direct cell polarity independently of microtubule assembly in C. elegans embryos. Nature. 2004;431:92–96. doi: 10.1038/nature02825. [DOI] [PubMed] [Google Scholar]
- Cuenca AA, Schetter A, Aceto D, Kemphues K, Seydoux G. Polarization of the C. elegans zygote proceeds via distinct establishment and maintenance phases. Development. 2003;130:1255–1265. doi: 10.1242/dev.00284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortin SM, Marshall SL, Jaeger EC, Greene PE, Brady LK, Isaac RE, Schrandt JC, Brooks DR, Lyczak R. The PAM-1 aminopeptidase regulates centrosome positioning to ensure anterior-posterior axis specification in one-cell C. elegans embryos. Developmental biology. 2010;344:992–1000. doi: 10.1016/j.ydbio.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein B, Hird SN. Specification of the anteroposterior axis in Caenorhabditis elegans. Development. 1996;122:1467–1474. doi: 10.1242/dev.122.5.1467. [DOI] [PubMed] [Google Scholar]
- Griffin EE. Cytoplasmic localization and asymmetric division in the early embryo of Caenorhabditis elegans. Wiley interdisciplinary reviews. Developmental biology. 2015;4:267–282. doi: 10.1002/wdev.177. [DOI] [PubMed] [Google Scholar]
- Hamill DR, Severson AF, Carter JC, Bowerman B. Centrosome maturation and mitotic spindle assembly in C. elegans require SPD-5, a protein with multiple coiled-coil domains. Dev Cell. 2002;3:673–684. doi: 10.1016/s1534-5807(02)00327-1. [DOI] [PubMed] [Google Scholar]
- Hird SN, White JG. Cortical and cytoplasmic flow polarity in early embryonic cells of Caenorhabditis elegans. J Cell Biol. 1993;121:1343–1355. doi: 10.1083/jcb.121.6.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins N, Saam JR, Mango SE. CYK-4/GAP provides a localized cue to initiate anteroposterior polarity upon fertilization. Science. 2006;313:1298–1301. doi: 10.1126/science.1130291. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, Welchman DP, Zipperlen P, Ahringer J. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Lyczak R, Zweier L, Group T, Murrow MA, Snyder C, Kulovitz L, Beatty A, Smith K, Bowerman B. The puromycin-sensitive aminopeptidase PAM-1 is required for meiotic exit and anteroposterior polarity in the one-cell Caenorhabditis elegans embryo. Development. 2006;133:4281–4292. doi: 10.1242/dev.02615. [DOI] [PubMed] [Google Scholar]
- McCloskey RJ, Kemphues KJ. Deubiquitylation machinery is required for embryonic polarity in Caenorhabditis elegans. PLoS Genet. 2012;8:e1003092. doi: 10.1371/journal.pgen.1003092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motegi F, Seydoux G. The PAR network: redundancy and robustness in a symmetry-breaking system. Philos Trans R Soc Lond B Biol Sci. 2013;368:20130010. doi: 10.1098/rstb.2013.0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motegi F, Zonies S, Hao Y, Cuenca AA, Griffin E, Seydoux G. Microtubules induce self-organization of polarized PAR domains in Caenorhabditis elegans zygotes. Nat Cell Biol. 2011;13:1361–1367. doi: 10.1038/ncb2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro E, Nance J, Priess JR. Cortical flows powered by asymmetrical contraction transport PAR proteins to establish and maintain anterior-posterior polarity in the early C. elegans embryo. Dev Cell. 2004;7:413–424. doi: 10.1016/j.devcel.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Praitis V, Casey E, Collar D, Austin J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 2001;157:1217–1226. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappleye CA, Tagawa A, Lyczak R, Bowerman B, Aroian RV. The anaphase-promoting complex and separin are required for embryonic anterior-posterior axis formation. Dev Cell. 2002;2:195–206. doi: 10.1016/s1534-5807(02)00114-4. [DOI] [PubMed] [Google Scholar]
- Rose LS, Kemphues KJ. Early patterning of the C. elegans embryo. Annu Rev Genet. 1998;32:521–545. doi: 10.1146/annurev.genet.32.1.521. [DOI] [PubMed] [Google Scholar]
- Sadler PL, Shakes DC. Anucleate Caenorhabditis elegans sperm can crawl, fertilize oocytes and direct anterior-posterior polarization of the 1-cell embryo. Development. 2000;127:355–366. doi: 10.1242/dev.127.2.355. [DOI] [PubMed] [Google Scholar]
- Shelton CA, Carter JC, Ellis GC, Bowerman B. The nonmuscle myosin regulatory light chain gene mlc-4 is required for cytokinesis, anterior-posterior polarity, and body morphology during Caenorhabditis elegans embryogenesis. J Cell Biol. 1999;146:439–451. doi: 10.1083/jcb.146.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonneville R, Gönczy P. Zyg-11 and cul-2 regulate progression through meiosis II and polarity establishment in C. elegans. Development. 2004;131:3527–3543. doi: 10.1242/dev.01244. [DOI] [PubMed] [Google Scholar]
- Tsai MC, Ahringer J. Microtubules are involved in anterior-posterior axis formation in C. elegans embryos. J Cell Biol. 2007;179:397–402. doi: 10.1083/jcb.200708101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse YC, Werner M, Longhini KM, Labbe JC, Goldstein B, Glotzer M. RhoA activation during polarization and cytokinesis of the early Caenorhabditis elegans embryo is differentially dependent on NOP-1 and CYK-4. Mol Biol Cell. 2012;23:4020–4031. doi: 10.1091/mbc.E12-04-0268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zonies S, Motegi F, Hao Y, Seydoux G. Symmetry breaking and polarization of the C. elegans zygote by the polarity protein PAR-2. Development. 2010;137:1669–1677. doi: 10.1242/dev.045823. [DOI] [PMC free article] [PubMed] [Google Scholar]