Abstract
We have used a murine Achilles tendinopathy model to investigate whether tissue changes (such as collagen disorganization, chondroid metaplasia, and loss of tensile properties) which are broadly characteristic of human tendinopathies, are accompanied by changes in the expression of chromatin-modifying enzymes and the methylation status of promoter regions of tendon cell DNA. Tendinopathy was induced by two intra-tendinous TGF-β1 injections followed by cage activity or treadmill running for up to 28 days. Activation of DNA methyltransferases occurred at 3 days after the TGF-β1 injections and also at 14 days, but only with treadmill activity. Genome wide Methyl Mini-Seq™ analysis identified 19 genes with differentially methylated promoters, five of which perform functions with an apparent direct relevance to tendinopathy (Leprel2, Foxf1, Mmp25, Igfbp6 and Peg12). The functions of the genes identified included collagen fiber assembly and pericellular interactions, therefore their perturbation could play a role in the characteristic disorganization of fibers in affected tendons. We postulate that a study of the functional genomics of these genes in animal and human tendon could further delineate the pathogenesis of this multi-factorial complex disease.
Keywords: Methylome, Tendinopathy, Leprel2, Foxf1, Mmp25, Igfbp6, Peg12
Introduction
Tendon overuse, due to work and sports-related activities, is the initiating factor for tendinopathy in the majority of patients (1). Subsequent long-term impaired function due the chronicity of the disease can be dependent on age, genetic predisposition, co-morbidities (2), and adverse drug effects (3). A consistent feature of human and animal models of tendinopathy is the appearance of cell clusters in a chondroid matrix as defined by Alcian Blue or SafraninO positive staining (4–8). The accumulated sulfated glycosaminoglycans (largely aggrecan chondroitin sulfate) are correlated in humans to advanced clinical symptoms of pain, tenderness, and mechanical dysfunction (9). Our work on the pathogenic role of such chondroid deposits in murine Achilles tendinopathy showed that a single injection of transforming growth factor-β1 (TGF-β1) into the midportion of the tendon in adult male C57/Bl6 mice resulted in tendon swelling, hyperplastic aggrecan-rich deposits, collagen disorganization, and a loss of tensile properties. Mechanical stimulation of the impaired tendon via treadmill running was accompanied by elimination of chondroid deposits and recovery of mechanical properties (10). While this model appears very useful for studies on acute tendinopathy, it does not provide an opportunity to examine the pathways associated with chronic disease.
The pathway to chronicity favored by many in the field is that intra-tendinous stressors (metabolic or biomechanical) persist following an initiating injury, such as acute overload, and maintain the cellular activities responsible for chondroid deposition, collagen disorganization, and a loss of tissue material properties (11, 12). For example, overload of bovine flexor tendon generates areas of rounded cells, largely in the interfascicular matrix, accompanied by collagen disorganization and increased cellular staining for IL6 and MMP13 (13). As commonly seen in other connective tissues, such as cartilage (14) or skin (15), repair deficiencies in tendinopathy also appear to be associated with chronic turnover of pericellular components without a concomitant increase in the production of stable, long-lived matrix structures.
The cellular basis for repair of connective tissues includes proliferation and differentiation of endogenous multipotent cells (16) and in this regard it has been suggested, among many other approaches, that connective tissue growth factor stimulated recruitment of endogenous tendon CD146+ cells may be an effective first step (17). However, much as for any human complex disease, an understanding of the genomic and epigenomic regulation of the transcriptome will ultimately be needed to delineate the complex interplay between multipotent cells, matrix structure, and biomechanical factors. In relation to tendon biology, expression of G9a, a methyltransferase with specific affinity to the ninth lysine residue of histone three, was shown to regulate cellular differentiation in mouse tenocytes by expression of tenogenic transcription factors (18), including Scleraxis and Mohawk, which have been heavily studied as necessary modulators for proper tendon development (19). Additionally, the use of fluoroquinolones (linked to tendinopathy in human patients (20)) in human embryonic kidney cell cultures resulted in the inhibition of DNA methylases, collagen prolyl 4-hydroxylases, and hypoxia inducible factor 1a (also linked to tendinopathy (21)), suggesting a mechanistic epigenetic link to chronic tendinopathy (22).
While these recent studies suggest the important role of epigenetics in tendon biology, to our knowledge there no published studies on analyses for altered DNA methylation and histone modification in human tendinopathies or animal models of the disease. The current study was carried out, using a murine model of Achilles tendinopathy, to identify changes in expression of epigenetic modification enzymes, and to subsequently perform a genome-wide screening for changes in methylation of gene promoter regions associated with the development of the disease.
Methods
Murine model of Chronic Tendinopathy in Wild Type C57/Bl6 Mice
Mice for this study were 12 weeks old C57/Bl6 males, bred in-house from a colony established in 2007 using 2 breeding pairs obtained from Jackson Laboratories. At weaning, males were distributed randomly and maintained with free access to standard chow and water and with constant light cycles (12 hour light, 12 hours dark). All mice were maintained in the same groups for maturation to 12 weeks and randomly distributed to experimental protocols which were approved by the IACUC of Rush University.
We have previously published on the induction of tendinopathy in murine Achilles tendon by a single injection of TGF-β1 into the body of the tendon, and we noted that the early loss of biomechanical properties and matrix disorganization was largely reversed by extended treadmill running (10). To generate a model in which structural, biochemical, and cell biological changes become chronic, a more severe injury was generated by two consecutive injections of 100ng active TGF-β1 (human recombinant, Peprotech, Inc), 2 days apart under anesthesia with xylazine (5ug/g body weight) /ketamine (100ug/g body weight). This resulted in persistent histopathological changes up to 14 days with cage activity (CA), and worsening of the pathology with treadmill running (TM) to 14 days and 28 days. All injections were between 2pm and 4pm, and treadmill running was between 9am and 11am daily in a designated room as previously described (10). Mice were euthanized by CO2 asphyxiation and cervical dislocation between 9am and 10am and tissues collected immediately into RNALater (for RNA isolation) or proteinase K (for DNA isolation).
To control for potential epigenetic effects of anesthesia and needle injuries to the tendon body, we included the following experimental groups: mice (n=12) were anaesthetized as above on days 0 and 2 and sacrificed on day 3; mice (n=12) received anesthesia and needle injury (without TGF-β1) on days 0 and day 2. A summary of experimental groups and outcome measures is provided in Table S1
Histology
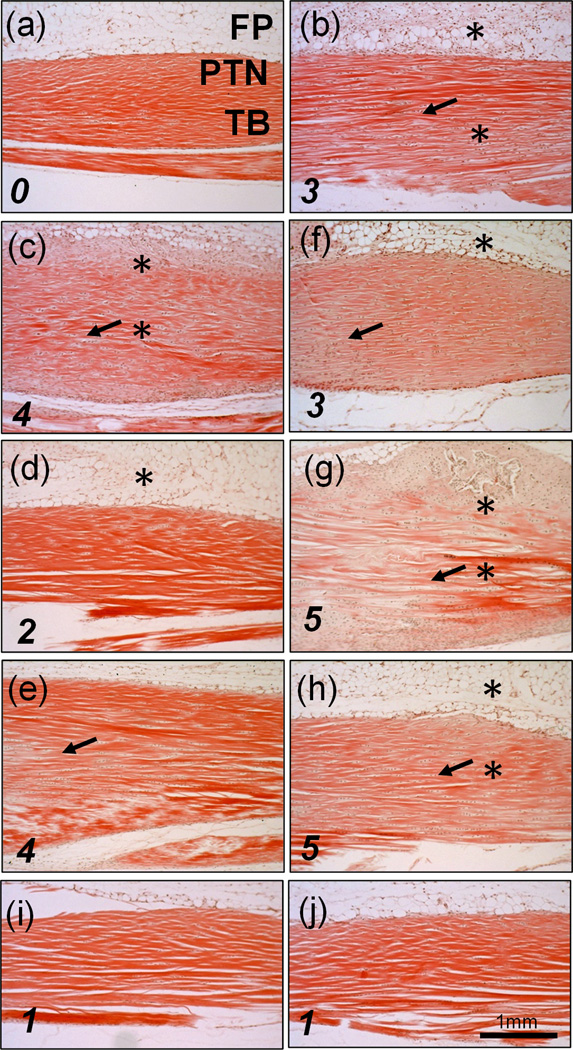
Histological evaluation was done as previously described (10). Briefly, after formalin fixation and decalcification, specimens were embedded in paraffin and 5µm thin sections were stained with Hematoxylin and Eosin and images taken at 40× magnification. Tendinopathy severity scores (0 to 5) were based on the degree of collagen disorganization observed by a blinded scorer in five 3.5mm long and 2.0mm wide longitudinal sections of each tendon. Typical scoring examples are provided in Figure 1.
Figure 1.
Hematoxylin and Eosin stained sections of Achilles tendons: (a) un-injured; (b) 3d acute TGF-β1 injury; (c) 14d; (d,e) 28d; (f) 14dTM, (g,h) 28dTM. (i) UI 14dTM, (j) UI 28dTM. Fat Pad (FP), Peritenon (PTN), tendon body (TB). * shows areas of hypercellularity and arrows denote regions of collagen disorganization. Tendinopathy severity scores, based on collagen disorganization are indicated in the bottom left hand corner (0= normal 5=severely disorganized)
Gene Expression Assays
Freshly harvested Achilles tendons were immediately isolated from all surrounding tissue with the peritenon left intact. RNA was isolated as previously described (10) from three separate pools of 12–24 combined Achilles tendons from un-injured (UI) mice and from each experimental and control group (Table S1). Briefly, tendon pools in RNALater were flash frozen with liquid N2, fragmented by hammer impact at −196 °C in a Bessman Tissue Pulverizer, recovered, and extracted in 1mL of Trizol by vortexing for 60 seconds. RNA was purified with an RNeasy MiniKit (Qiagen).Consistent with an anabolic response to injury, yields per tendon were: UI ~ 305 ng, post-injury 3d ~805 ng, post-injury 14d and 14dTM ~1800ng, and post-injury 28d and 28dTM ~ 800ng. A260:A280 for all preparations was >1.90. cDNA was synthesized with 0.5ug of mRNA (RT2 First Strand Kit, Qiagen). Chromatin modification enzyme transcript abundances were determined with SYBR qt-PCR array plates (PAMM-085Z, Qiagen) with 84 genes divided into the following functional groups: 6 DNA/Histone Demethylases (DHD), 3 DNA Methyltransferases (DM); 17 Histone Acetyltransferases (HA); 11 Histone Deacetylases (HD), 16 Histone Methyltransferases (HM), 7 Histone Phosphorylation (HP), 9 Histone Ubiquitinases (HU), and 15 SET Domain Proteins with HM activity (SET). The coefficient of variation for triplicate array assays was <3%. The same cDNA preparations were used for Taqman qt-PCR of single genes (Table S2), with primers from Thermo-Lifetech and the coefficient of variation was < 7%.
Changes in transcript abundance (ΔCt=Ct for transcript of interest minus Ct for the housekeeping gene B2m) were used to calculate the fold change (2^-ΔΔCt) from UI levels for each experimental group. A 1-way ANOVA with Tukey’s post-hoc test was conducted using GraphPad Prism 5 (La Jolla, CA) on the ΔCt values to determine the significance (p<0.05) in expression of genes in the injured relative to UI groups. Statistical evaluation was not possible on data from single tissue pool controls (anesthesia only, anesthesia with needle injury only, and treadmill only), although it should be recognized that the data obtained with any pool represents the average expression in 12–24 tendons.
Methyl-MiniSeq™ library construction
Libraries were prepared from 200–500 ng of genomic DNA (from single pools of 5–6 tendons per experimental group) digested with 60 units of TaqαI and 30 units of MspI (NEB) sequentially and then extracted with Zymo Research (ZR) DNA Clean & Concentrator™-5 kit (Cat#: D4003). Fragments were ligated to pre-annealed adapters containing 5’-methyl-cytosine instead of cytosine according to Illumina’s specified guidelines (www.illumina.com). Adaptor-ligated fragments of 150–250bp and 250–350bp in size were recovered from a 2.5% NuSieve 1:1 agarose gel (Zymoclean™ Gel DNA Recovery Kit, ZR Cat#: D4001). The fragments were then bisulfite-treated using the EZ DNA Methylation-Lightning™ Kit (ZR, Cat#: D5020). Preparative-scale PCR was performed and the resulting products were purified (DNA Clean & Concentrator™ - ZR, Cat#D4005) for sequencing on an Illumina HiSeq.
Methyl-MiniSeq™ Sequence alignments and data analysis
Sequence reads from bisulfite-treated EpiQuest libraries were identified using standard Illumina base-calling software and then analyzed using a Zymo Research proprietary analysis pipeline, which is written in Python and used Bismark with the murine mm10 reference genome (http://www.bioinformatics.babraham.ac.uk/projects/bismark/) to perform the alignment. Index files were constructed using the bismark_genome_preparation command and the entire reference genome. The --non_directional parameter was applied while running Bismark. All other parameters were set to default. Filled-in nucleotides were trimmed off during methylation calling. The methylation level of each sampled cytosine was estimated as the number of reads reporting a C, divided by the total number of reads reporting a C or T. Fisher’s exact test or t-test was performed for each CpG site which has at least five reads coverage, and promoter, gene body (intron and exon), and CpG island annotations were added for each CpG included in the comparison. For post-processing, CpG sites with methylation differentials greater than 80% (0.8) for any experimental group relative to UI were used. Data was further analyzed to identify site specific methylation events across groups and within CpG islands using the UCSC Genome Bioinformatics database (https://genome.ucsc.edu/).
Data Access
The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (Trella et al., 2016) and are accessible through GEO Series accession number GSE80396 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE80396).
Results
Histological Evaluation
Hematoxylin and Eosin staining was used to evaluate the collagen matrix and cellular responses during cage activity (CA) and treadmill activity (TM), subsequent to the two TGF-β1 injections into the Achilles tendon body (Figure 1; histopathology denoted by arrows for collagen disorganization (scoring shown in each panel) and asterisk for hypercellularity). The 3d acute response (1 day after second injection) consisted of cell proliferation in the body of the tendon, the peritenon, and the stroma of the adjoining adipose tissue, as well as localized collagen disorganization of the tendon at the injection sites (Figure 1b). In mice maintained with CA, hyper-cellularity, collagen disorganization, and chondroid deposits were still evident at 14 days (Figure 1c), but had largely dissipated by 28 days (Figure 1d), with small areas of increased cellularity seen in only one of the three mice examined (Figure 1e). When TM was applied after the injury, pathology was also evident at 14 days (Figure 1f), but had worsened by 28 days, with large areas of collagen disruption extending throughout the tendon body, and chondroid and occasional calcified deposits in between fiber bundles in the area surrounding the TGF-β1 injection sites (Figure 1g,h). Minimal changes in pathology were seen in UI mice with TM for 14 or 28 days (Figure 1i,j).
Expression of chromatin-modifying enzymes in the tendinopathy model
The intra-tendinous injections of TGF-β1 resulted in marked alterations in transcript abundance of genes involved in epigenetic modifications. Baseline expression levels of chromatin modification enzymes in UI mice can be seen in Table S3. In the 3d acute group (Table 1), 34 genes covering both histone and DNA modification enzymes showed significant (p<0.05) increases in expression and three histone-modifying enzymes (Kat2b, Hdac11, and Smyd1) showed a significant decrease. When mice were maintained with CA, the altered transcript levels were reversed to UI levels by 14d with the exception of Aurkb which remained 3.70-fold activated. At 28d, Aurkb returned to UI levels, however Kdm5c became significantly activated (2.44-fold).
Table 1.
Fold changes in transcript abundance of chromatin-modifying enzymes at 3 days
| Gene Group | Genes | 3d |
|---|---|---|
| DHD | Kdm1a | 2.00 (0.22) |
| DM | Dnmt1 | 5.24 (0.26) |
| Dnmt3b | 2.46 (0.56) | |
| HA | Ciita | 2.49 (0.25) |
| Csrp2bp | 2.12 (0.23) | |
| Esco2 | 26.51 (3.07) | |
| Hat1 | 3.31 (0.19) | |
| Kat2a | 3.10 (0.42) | |
| Kat2b | 0.57 (0.02) | |
| Kat7 | 1.74 (0.11) | |
| HD | Hdac1 | 2.25 (0.14) |
| Hdac11 | 0.27 (0.02) | |
| Hdac2 | 2.03 (0.18) | |
| Hdac3 | 1.95 (0.12) | |
| Hdac6 | 1.92 (0.16) | |
| HM | Carm1 | 2.61 (0.34) |
| Prmt1 | 3.81 (0.45) | |
| Prmt3 | 2.90 (0.06) | |
| Prmt5 | 2.69 (0.17) | |
| Prmt6 | 2.09 (0.30) | |
| Prmt7 | 2.08 (0.25) | |
| Setdb2 | 2.08 (0.24) | |
| Smyd1 | 0.36 (0.09) | |
| Suv39h1 | 3.57 (0.40) | |
| HP | Aurka | 33.57 (1.17) |
| Aurkb | 61.26 (2.30) | |
| Nek6 | 3.34 (0.29) | |
| Pak1 | 10.13 (0.84) | |
| HU | Dzip3 | 2.63 (0.22) |
| Rnf20 | 2.01 (0.10) | |
| Ube2a | 1.98 (0.17) | |
| Usp16 | 2.11 (0.28) | |
| Usp22 | 1.82 (0.31) | |
| SET | Setd1a | 1.60 (0.13) |
| Setd4 | 1.80 (0.31) | |
| Setd8 | 2.52 (0.31) | |
| Whsc1 | 5.03 (0.89) | |
All expression changes (average (standard deviation)) shown are significant (p<0.05) relative to un-injured levels.
Groups: DHD (DNA/Histone Demethylases), DM (DNA Methyltransferases), HA (Histone Acetyltransferases), HD (Histone Deacetylases), HM (Histone Methyltransferases), HP (Histone Phosphorylation), HU (Histone Ubiquitination), SET (SET Domain Proteins; Histone Methyltransferase Activity)
In contrast, when injured mice were exposed to TM (Table 2), mRNA abundance for six of the genes activated at day 3 (Dnmt3b, Ciita, Aurka, Aurkb, Pak1, and Whsc1) remained elevated at 14dTM, and Pak1 remained elevated at 28dTM. Furthermore, the transcript levels of two genes (Dnmt3a, Setd5) which were not altered by TGF-β1 injection at 3d, were increased at 14dTM. In addition, three genes (Hat1, Setd6, and Setd8) and two genes (Atf2 and Ehmt1) were decreased after 14dTM and 28dTM, respectively (all differences, p<0.05). Importantly, the expression of chromatin modification enzymes in control groups of UI mice with 14 or 28 days of TM (single pool of 15–17 tendons, no statistical test) showed no marked changes (fold change from UI <1.5-fold up or down) although a few genes showed changes which might indicate a response (Kdm1a and Atf2 (1.9-fold down at UI 28dTM), Kat6b (1.9-fold down at UI 28dTM), Smyd1 (2.1-fold up at UI 14TM and UI 28dTM) and Setd4 (2.3-fold up at UI 14dTM)). Overall, the lack of expression changes is consistent with the need for injections of TGF-β1 and TM to generate tendinopathy in this model.
Table 2.
Fold changes in transcript abundance of chromatin-modifying enzymes at 14 and 28 days with TM
| Gene Group | Genes | 14dTM | 28dTM |
|---|---|---|---|
| DHD | Kdm5c | 2.68 (0.16)* | 2.01 (0.67) |
| DM | Dnmt3a | 2.22 (0.34)* | 1.54 (0.52) |
| Dnmt3b | 2.05 (0.09)* | 1.61 (0.66) | |
| HA | Atf2 | 0.57 (0.18) | 0.52 (0.09)* |
| Ciita | 2.24 (0.35)* | 1.24 (0.35) | |
| Hat1 | 0.47 (0.04)* | 0.78 (0.11) | |
| HD | NONE | NONE | NONE |
| HM | Ehmt1 | 0.70 (0.05) | 0.57 (0.10)* |
| HP | Aurka | 2.91 (0.18)* | 1.23 (0.33) |
| Aurkb | 3.84 (0.55)* | 1.06 (0.04) | |
| Pak1 | 2.51 (0.09)* | 2.15 (0.35)* | |
| SET | Setd5 | 2.50 (0.35)* | 1.90 (0.61) |
| Setd6 | 0.66 (0.06)* | 0.89 (0.14) | |
| Setd8 | 0.46 (0.04)* | 0.59 (0.08) | |
| Whsc1 | 2.06 (0.18)* | 1.50 (0.13) | |
Values listed as average (standard deviation).
Significant (p<0.05) relative to uninjured levels.
Groups: DHD (DNA/Histone Demethylases), DM (DNA Methyltransferases), HA (Histone Acetyltransferases), HD (Histone Deacetylases), HM (Histone Methyltransferases), HP (Histone Phosphorylation), HU (Histone Ubiquitination), SET (SET Domain Proteins; Histone Methyltransferase Activity)
Notably, anesthesia alone or with needle injury appeared to affect expression levels at 3d (Table S4). 3dA alone resulted in decreased expression of epigenetic enzymes (with ten genes reduced >3-fold), whereas 3dAN enhanced expression of Esco2 (10.12-fold), Aurka (8.16-fold) and Aurkb (18.45-fold). Thus, since anesthesia alone appeared to have a broad inhibitory effect, it appears that only the transient high activation of Esco2 (26.51-fold), Aurka (33.57-fold) and Aurkb (61.26-fold) seen at 3d with the standard model (Table 1), can be partly explained by an effect of needle injury without TGF-β1.
Genome-wide alterations in the DNA Methylome
Changes in mRNA transcript abundance for DNA methylases and DNA/Histone demethylases (Tables 1 and 2) indicated that changes in the DNA methylome were likely to occur in this tendinopathy model. Therefore, a genome-wide DNA methylome analysis was conducted on tendons from each of the experimental groups (see Methods and Table S1). Following the acute TGF-β1 injury, 1058 individual CpG sites, with an additional 27 in CpG islands (CpG I), showed methylation differentials of > 80% (0.8) compared to UI levels. After 14 and 28 days, for both CA and TM, this was reduced to approximately 650 CpG sites with differential methylation (DM) (Table 3). The genome-wide and gene promoter region distribution of differentially hypomethylated and hypermethylated sites for each experimental group is given in Table S5. Most importantly for the present study, 3–5% of the DM sites for each experimental group were within promoter gene regions and a variable proportion of these were in CpG islands (Table 3). Overall, hypomethylation was the predominant response to the model tendinopathy, accounting for 74% (3d), 58% (14d), 87% (28d), 57% (14dTM), and 75% (28dTM) of all modified promoter CpG sites in these experimental groups.
Table 3.
Methylation of CpG sites of DNA in murine tendon for each experimental group
| Group | Genome-Wide | Non-Coding | Intron | Exon | Promoter | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| CpG I | Other | CpG I | Other | CpG I | Other | CpG I | Other | CpG I | Other | |
| 3d | 27 | 1058 | 6 | 539 | 9 | 412 | 13 | 99 | 4 | 31 |
| 14d | 9 | 622 | 0 | 347 | 4 | 232 | 5 | 29 | 0 | 26 |
| 28d | 17 | 620 | 0 | 338 | 4 | 230 | 14 | 47 | 9 | 23 |
| 14dTM | 19 | 693 | 1 | 357 | 2 | 280 | 15 | 45 | 4 | 26 |
| 28dTM | 17 | 609 | 0 | 333 | 2 | 230 | 11 | 37 | 5 | 27 |
All sites exhibited greater than 80% differential methylation compared to the same sites in un-injured tendons. Location of CpG sites (non-coding, intron, exon, and promoter) was determined using the UCSC database refseq gene track
Genes with differential promoter methylation and effects on transcript abundance
It has been proposed that DM of a CpG site within a promoter is most likely to affect transcription if it is located within a CpG island, or if it is one of multiple DM non-island sites (23). Table 4 lists the genes for each experimental group with CpG sites which fulfill these criteria, and their chromosomal location is given in Table S6. All genes of interest (except Foxr1) were identified in only one experimental group showing that differential hypomethylation of the promoter sites is transitory and therefore apparently specific to the time-course, activity level, and possibly pathology in this model. In addition, all sites (except for Sfi1) were found to be differentially hypomethylated, indicative of increased expression levels of all genes affected. Further, it is perhaps notable that the hypomethylation (>80%) of sites in the acute response at 3 days occurred in both island (4 sites in 4 genes) and non-island (7 sites in 3 genes) locations, whereas at later times (14dTM, 28d, and 28dTM), essentially all (12 of 13 sites) were in CpG islands, however the possible significance of this difference is not known.
Table 4.
Genes with differentially methylated promoter CpG sites for each experimental group
| Group | CpG I | Non CpG I | ||
|---|---|---|---|---|
| Gene | Methylation status | Gene | Methylation Status | |
| 3d | Baiap2l1 (1) | 80% HYPO | Cend1 (3) | 90% HYPO |
| Foxr1 (1) | 86% HYPO | Gm19557 (2) | 80% HYPO | |
| Grm4 (1) | 80% HYPO | Mirlet7c-2 (2) | 80% HYPO | |
| Rbmxl2 (1) | 80% HYPO | |||
| 14d | NONE | NONE | NONE | NONE |
| 28d | Gnas (2) | 91% HYPO | Hoxc4 (3) | 80% HYPO |
| Leprel2 (4) | 100% HYPO | |||
| Foxf1 (1) | 80% HYPO | |||
| Foxr1 (1) | 83% HYPO | |||
| Zrsr1 (1) | 84% HYPO | |||
| 14dTM | Igfbp6 (1) | 80% HYPO | NONE | NONE |
| Mmp25 (1) | 80% HYPO | |||
| Sfi1 (1) | 80% HYPER | |||
| 28dTM | Cnot7 (2) | 80% HYPO | NONE | NONE |
| Peg12 (1) | 80% HYPO | |||
| Shisa7 (1) | 96% HYPO | |||
| Usp9x (1) | 80% HYPO | |||
Numbers in parenthesis denote the number of affected sites within that gene promoter. Methylation status (%) represents the differential compared to un-injured mice with the average differential methylation given for multiple sites.
To examine the potential effect of these methylation changes on transcription, the fold-change in transcript levels (relative to UI) was determined for each gene in three tendon pools from the five experimental groups (Table 5). These assays indicated that transcription of most of these genes was modulated (primarily increased) to some degree at different stages of the model (for example, 20.9- & 6.1-fold in Peg 12, 4.5-fold in Cnot7, 4.4- & 3.8-fold in Usp9x, 3.9-fold in Leprel2, 3.6-& 3.8-fold in Mmp25 and 3.5-fold in Cend1), however the wide variability in transcript abundance between pools (details in Table S7) means that statistical significance was not achieved. Perhaps, the widely variable expression of these genes (coefficient of variation between 3 pools is ~12%) is indicative of short-term fluctuations in transcript abundance due to mRNA instability or multiple mechanisms of control. This idea is supported by the fact that the coefficient of variation was <7% for the assay of these genes in triplicates from a single RNA pool (data not shown).
Table 5.
Fold changes in transcript abundance for genes with differentially methylated promoter regions for each experimental group
| Gene | 3d | 14d | 28d | 14dTM | 28dTM | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| RNA | DM | RNA | DM | RNA | DM | RNA | DM | RNA | DM | |
| CpG I (Multiple Sites) | ||||||||||
| Cnot7 | 4.50 (2.28) |
- | 1.54 (0.88) |
- | 1.03 (0.15) |
- | 1.74 (1.00) |
- | 0.61 (0.56) |
HYPO |
| Gnas | 1.18 (0.63) |
- | 1.23 (0.51) |
- | 0.58 (0.12) |
HYPO | 1.36 (1.10) |
- | 0.93 (0.49) |
- |
| Leprel2 | 1.90 (0.10) |
- | 2.41 (1.18) |
- | 0.98 (0.36) |
HYPO | 3.87 (2.47) |
- | 1.29 (0.65) |
|
| CpG I | ||||||||||
| Usp9x | 4.39 (2.32) |
- | 2.45 (1.36) |
- | 0.61 (0.52) |
- | 3.76 (3.63) |
- | 1.22 (0.53) |
HYPO |
| Baiap2l1 | 2.94 (0.60) |
HYPO | 1.59 (0.45) |
- | 1.01 (0.49) |
- | 1.99 (1.55) |
- | 1.48 (0.81) |
- |
| Peg12 | 2.86 (1.23) |
- | 2.02 (1.55) |
- | 0.98 (0.35) |
- | 20.92 (23.1) |
- | 6.14 (10.2) |
HYPO |
| Mmp25 | 2.43 (0.31) |
- | 3.59 (1.55) |
- | 0.62 (0.24) |
- | 3.79 (2.80) |
HYPO | 1.10 (0.35) |
- |
| Sfi1 | 1.97 (0.37) |
- | 1.53 (1.13) |
- | 1.05 (0.39) |
- | 2.65 (1.46) |
HYPER | 1.66 (0.71) |
- |
| Foxf1 | 1.65 (0.58) |
- | 1.81 (2.27) |
- | 0.87 (0.16) |
HYPO | 3.05 (2.12) |
- | 1.55 (0.65) |
- |
| Zrsr1 | 0.60 (0.20) |
- | 1.13 (0.16) |
- | 0.99 (0.24) |
HYPO | 1.08 (0.47) |
- | 0.85 (0.45) |
- |
| Igfbp6 | 0.43 (0.13) |
- | 1.55 (0.40) |
- | 0.71 (0.07) |
- | 1.35 (1.25) |
HYPO | 0.98 (0.31) |
- |
| Rbmxl2 | 0.37 (0.13) |
HYPO | 0.95 (0.17) |
- | 1.06 (0.39) |
- | 1.00 (1.00) |
- | 0.89 (0.38) |
|
| Non CpG I (Multiple Sites) | ||||||||||
| Cend1 | 3.56 (0.90) |
HYPO | 1.85 (0.81) |
- | 0.65 (0.20) |
- | 2.69 (2.24) |
- | 1.29 (0.93) |
- |
| Hoxc4 | 1.47 (0.60) |
- | 1.30 (0.58) |
- | 0.88 (0.28) |
HYPO | 1.72 (1.21) |
- | 1.22 (0.53) |
- |
| Mirlet7c | 0.49 (0.19) |
HYPO | 1.59 (1.37) |
- | 0.99 (0.43) |
- | 0.76 (0.43) |
- | 0.93 (0.55) |
- |
Fold change (RNA column) values are listed as average (standard deviation) relative to un-injured levels and separated based on differential methylation of multiple sites in island or non-island locations and single sites in island locations.DM column denotes at what experimental time-point differential methylation occurred and whether site(s) were hypo- or hypermethylated related to un-injured levels.
Despite this variability, several potentially important associations between promoter methylation status and transcript abundance in the TGF-β1 injury model were evident (Table 5). For instance, hypomethylation of the Mmp25 promoter at 14dTM was accompanied by the predicted high transcript abundance, which was higher than at the 3d and 28dTM time-points at which the Mmp25 promoter was normally methylated. Conversely, hypermethylation of the Sfi1 promoter at 14dTM was accompanied by increased transcript abundance, suggesting that promoter methylation was not an important mechanism of control in this instance. This also appears to be the case for Igfbp6, where promoter hypomethylation at 14dTM was not associated with a significant change in transcripts. Hypomethylation at 3d of Cend1, Baiap2l1, Mirlet7c-2, and Rbmxl2 promoters was associated with the expected increase in abundance of transcripts for Cend1 and Baiap2l1, but there was a decreased abundance of Mirlet7c-2 and Rbmlx2 transcripts. Moreover, Cend1 and Baiap2l1 transcripts remained elevated in both 14d and 14dTM groups, despite normalization of the promoter methylation. Such results could be explained by particularly long half-lives for these mRNA species. For Leprel2, Hoxc4, and Foxf1 with promoter hypomethylation in the 28d group, the transcript abundance was unaltered relative to the UI group, however, for these genes the late hypomethylation may have resulted in increased transcription at >28d post-injury time-points, which was beyond the scope of the current study. It should also be noted that the increased transcript abundance for Leprel2 in the 3d and 14d groups, where promoter methylation was not changed relative to UI, suggests that expression of this gene (and others in Table 5) is controlled by factors other than promoter methylation. Also, for the three genes (Usp9x, Cnot7, and Peg12) showing >80% hypo-methylation at 28dTM, only Peg12 exhibited markedly increased transcript abundance at that time-point. Lastly, Foxr1, Grm4, and Shisa7 transcripts were below the level of detection at all time points, and Gm19557 (gene without any known function) was not assayed.
Discussion
The primary objective of this study was to use a murine Achilles tendinopathy model with acute, intermediate, and chronic phases to investigate whether tissue changes in the model (such as collagen disorganization, chondroid metaplasia, and loss of tensile properties (10)), which are broadly characteristic of human tendinopathies, are accompanied or preceded by changes in the methylation status of promoter regions of tendon DNA. Secondly, we aimed to determine whether promoter methylation status was predictive of changes in transcript abundance and whether the identity of the transcripts might provide novel insights into tendinopathic changes. In summary, 19 genes displayed differential promoter methylation in a treatment-group specific manner, and notably 18 of these genes involved hypomethylation. 12 of the identified genes also exhibited marked differences in mRNA transcript abundance between the groups. To investigate whether a putative change in the abundance of one or more of the 19 gene products might impact tendon function we undertook a limited literature search using the murine gene function on PubMed and identified studies relating the gene to multipotent mesenchymal cells, fibroblasts, tendon, and/or extracellular matrix. In this way we identified five genes (Leprel2, Foxf1, Mmp25, Igfbp6 and Peg12) which demonstrate strong relevance. Notably, three of the five exhibit functions closely related to collagen fiber organization.
Firstly, Leprel2 has the potential to profoundly affect collagen matrix organization in the tendon as it has a critically important and tissue-specific role in the unique assembly and packing of tendon collagen type I fibrils (24, 25). Thus, modulation of Leprel2 expression in the tendon could readily explain the fiber disorganization which is a hallmark of tendon pathologies, including tendinopathy. Secondly, Foxf1 induces integrin β3 expression, with high specificity in mouse embryonic fibroblasts (26). This finding appears to be important since this integrin was identified through the BORG database of genomic and biomedical knowledge as one of four strong candidate risk genes for human tendinopathy (which included COL11A2, ELN, ITGB3, and LOX) (27). Thirdly, proteomic analysis of pericellular proteins in developing tendons has shown that Mmp25 (like Leprel2 and Foxf1) is likely involved in collagen fibril and matrix assembly (28). Fourthly, a significance for Igfbp6 modulation in this model is suggested by its reported capacity to regulate cell proliferation in connective tissue disorders (29), and also by its sensitivity to hypoxia (30), which has often been implicated in tendinopathic change (21). Lastly, the dramatic increase in Peg12 expression in the 14dTM group might be related to its capacity to bind Gsk3 (31) and potentially reduce production of inflammatory cytokines in macrophage-type cells in response to toll-like receptors (32). Furthermore, in this regard it is interesting that analysis of the array data for chromatin-modifying enzymes shows that six of the genes most affected overall (Dnmt3b, Ciita, Aurka, Pak1, Hat1, and Atf2) have been implicated in the control of macrophage polarization in innate inflammation. While the role of inflammation in tendon injury is still heavily debated (33), recent evidence suggests that early macrophage involvement in ligament wound healing is necessary for maintenance of repair responses including angiogenesis, cell differentiation, and collagen production (34).
It should be noted that there was no clear relationship between methylation status and transcript abundance for many of the genes identified by this study. In this regard, recent reviews (35, 36) have underlined the complexity of the causes and effects of cytosine methylation on downstream transcription, so that the earlier supposition (37) that hypomethylation necessarily results in high transcription (and perhaps vice-versa) is, while possible in a context-dependent fashion, an over-simplification of the multiple possible effects of methylation levels. In addition, changes in transcript levels may partly or entirely result from other methods of regulation (e.g. enhancer methylation, transcription factor binding, and miRNA levels) which could contribute to the complexities of transcript abundance reported here.
In addition, cell populations from surrounding tissue (fat pad, synovial tissue, and peritenon) may be present in the samples at varying levels and potentially contribute to the variability exhibited. Due to the need for tissue pooling for mRNA assays and the limited numbers of mice used for this study (see Methods and Table S1), the n values for biological replicates was often not sufficient for robust statistical analysis. However, the post-hoc analysis presented here follows guidelines recently released by the American Statistical Association (38) for evaluating the biological significance (as opposed to statistical significance) of the type of data shown in this paper. Nonetheless, the illustration of transcript modulation for 15 of the genes identified by differential promoter methylation, make it likely that the activity of the protein products of these genes are involved to some degree in the pathogenesis of tendinopathy. In summary, despite these caveats, we postulate that a study of the molecular genomics of Leprel2, Foxf1, Mmp25, Igfbp6 and Peg12 in animal and human tendon could further delineate the pathogenesis of this multifactorial disease.
In conclusion, we wish to point out that in this report we have presented results based on a very limited mining of the data generated by genome wide methylome studies of this kind. Further ongoing work is aimed at utilizing all aspects of this data set (DM CpG sites throughout the genome) in pathway analysis, GO analysis, and identification of other potential disease associations relevant to tendinopathy. Furthermore, data has been deposited in NCBI’s Gene Expression Omnibus (Trella et al., 2016) and are publically accessible through GEO Series accession number GSE80396 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE80396).
Supplementary Material
Acknowledgments
Funding was obtained from the American Society of the Hand (RW), Arthritis Institute (AP), Katz Rubschlager Endowment for OA Research (AP), and NIH AR63144 (VMW). We would like to thank Ryan Sasada, Hunter Chung, and Keith Boohar of Zymo Research Inc.
Abbreviations
- CA
Cage Activity
- TM
Treadmill Running
- UI
Uninjured
- 3d
3 days post-injury
- 14d
14 days post-injury
- 28d
28d post-injury
- 14dTM
14 days post-injury with treadmill running
- 28dTM
28 days post-injury with treadmill running
- UI 14dTM
14d uninjured with treadmill running
- UI 28dTM
28 day uninjured with treadmill running
- TGF-β1
Transforming Growth Factor beta 1
- CpG I
CpG Island
- DM
differentially methylated
References
- 1.Abate M, Silbernagel KG, Siljeholm C, et al. Pathogenesis of tendinopathies: inflammation or degeneration? Arthritis research & therapy. 2009;11(3):235. doi: 10.1186/ar2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.September AV, Posthumus M, Collins M. Application of Genomics in the Prevention, Treatment and Management of Achilles Tendinopathy and Anterior Cruciate Ligament Ruptures. Recent Patents on DNA and Gene Sequences. 2012;6:212–223. doi: 10.2174/187221512802717358. [DOI] [PubMed] [Google Scholar]
- 3.Magnan B, Bondi M, Pierantoni S, Samaila E. The pathogenesis of Achilles tendinopathy: a systematic review. Foot Ankle Surg. 2014;20(3):154–159. doi: 10.1016/j.fas.2014.02.010. [DOI] [PubMed] [Google Scholar]
- 4.Han WM, Heo SJ, Driscoll TP, et al. Macro- to microscale strain transfer in fibrous tissues is heterogeneous and tissue-specific. Biophysical journal. 2013;105(3):807–817. doi: 10.1016/j.bpj.2013.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kannus P, Jozsa L. Histopathological changes preceding spontaneous rupture of a tendon. A controlled study of 891 patients. J Bone Joint Surg Am. 1991;73:1507–1525. [PubMed] [Google Scholar]
- 6.Corps AN, Robinson AH, Movin T, et al. Increased expression of aggrecan and biglycan mRNA in Achilles tendinopathy. Rheumatology. 2006;45(3):291–294. doi: 10.1093/rheumatology/kei152. [DOI] [PubMed] [Google Scholar]
- 7.de Mos M, Koevoet W, van Schie HT, et al. In vitro model to study chondrogenic differentiation in tendinopathy. The American journal of sports medicine. 2009;37(6):1214–1222. doi: 10.1177/0363546508331137. [DOI] [PubMed] [Google Scholar]
- 8.Khan M, Bonar F, Desmond PM, et al. Patellar tendinosis (jumper's knee): Findings at bistopathologic examination, US, MR lmaging. Musculoskelet Rad. 1996;200:821–827. doi: 10.1148/radiology.200.3.8756939. [DOI] [PubMed] [Google Scholar]
- 9.Attia M, Scott A, Carpentier G, et al. Greater glycosaminoglycan content in human patellar tendon biopsies is associated with more pain and a lower VISA score. British journal of sports medicine. 2014;48(6):469–475. doi: 10.1136/bjsports-2013-092633. [DOI] [PubMed] [Google Scholar]
- 10.Bell R, Li J, Gorski DJ, et al. Controlled treadmill exercise eliminates chondroid deposits and restores tensile properties in a new murine tendinopathy model. Journal of biomechanics. 2013;46(3):498–505. doi: 10.1016/j.jbiomech.2012.10.020. [DOI] [PubMed] [Google Scholar]
- 11.Jarvinen M, Jozsa L, Kannus P, et al. Histopathological findings in chronic tendon disorders. Scandinavian journal of medicine & science in sports. 1997;7(2):86–95. doi: 10.1111/j.1600-0838.1997.tb00124.x. [DOI] [PubMed] [Google Scholar]
- 12.Pingel J, Lu Y, Starborg T, et al. 3-D ultrastructure and collagen composition of healthy and overloaded human tendon: evidence of tenocyte and matrix buckling. J Anat. 2014;224(5):548–555. doi: 10.1111/joa.12164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spiesz EM, Thorpe CT, Chaudhry S, et al. Tendon extracellular matrix damage, degradation and inflammation in response to in vitro overload exercise. Journal of orthopaedic research : official publication of the Orthopaedic Research Society. 2015;33(6):889–897. doi: 10.1002/jor.22879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Plaas A, Osborn B, Yoshihara Y, et al. Aggrecanolysis in human osteoarthritis: confocal localization and biochemical characterization of ADAMTS5-hyaluronan complexes in articular cartilages. Osteoarthritis and cartilage / OARS, Osteoarthritis Research Society. 2007;15(7):719–734. doi: 10.1016/j.joca.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 15.Velasco J, Li J, DiPietro L, et al. Adamts5 deletion blocks murine dermal repair through CD44-mediated aggrecan accumulation and modulation of transforming growth factor beta1 (TGFbeta1) signaling. The Journal of biological chemistry. 2011;286(29):26016–26027. doi: 10.1074/jbc.M110.208694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maxson S, Lopez EA, Yoo D, et al. Concise review: role of mesenchymal stem cells in wound repair. Stem Cells Transl Med. 2012;1(2):142–149. doi: 10.5966/sctm.2011-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee CH, Lee FY, Tarafder S, et al. Harnessing endogenous stem/progenitor cells for tendon regeneration. The Journal of clinical investigation. 2015;125(7):2690–2701. doi: 10.1172/JCI81589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wada S, Ideno H, Shimada A, et al. H3K9MTase G9a is essential for the differentiation and growth of tenocytes in vitro. Histochem Cell Biol. 2015;144(1):13–20. doi: 10.1007/s00418-015-1318-2. [DOI] [PubMed] [Google Scholar]
- 19.Liu H, Zhu S, Zhang C, et al. Crucial transcription factors in tendon development and differentiation: their potential for tendon regeneration. Cell and tissue research. 2014;356(2):287–298. doi: 10.1007/s00441-014-1834-8. [DOI] [PubMed] [Google Scholar]
- 20.Kim GK. The Risk of Fluoroquinolone-induced Tendinopathy and Tendon Rupture: What Does The Clinician Need To Know? J Clin Aesthet Dermatol. 2010;3(4):49–54. [PMC free article] [PubMed] [Google Scholar]
- 21.Millar NL, Reilly JH, Kerr SC, et al. Hypoxia: a critical regulator of early human tendinopathy. Ann Rheum Dis. 2012;71(2):302–310. doi: 10.1136/ard.2011.154229. [DOI] [PubMed] [Google Scholar]
- 22.Badal S, Her YF, Maher LJ., 3rd Nonantibiotic Effects of Fluoroquinolones in Mammalian Cells. The Journal of biological chemistry. 2015;290(36):22287–22297. doi: 10.1074/jbc.M115.671222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schubeler D. Function and information content of DNA methylation. Nature. 2015;517(7534):321–326. doi: 10.1038/nature14192. [DOI] [PubMed] [Google Scholar]
- 24.Eyre DR, Weis M, Hudson DM, et al. A novel 3-hydroxyproline (3Hyp)-rich motif marks the triple-helical C terminus of tendon type I collagen. The Journal of biological chemistry. 2011;286(10):7732–7736. doi: 10.1074/jbc.C110.195768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hudson DM, Kim LS, Weis M, et al. Peptidyl 3-hydroxyproline binding properties of type I collagen suggest a function in fibril supramolecular assembly. Biochemistry. 2012;51(12):2417–2424. doi: 10.1021/bi2019139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malin D, Kim IM, Boetticher E, et al. Forkhead box F1 is essential for migration of mesenchymal cells and directly induces integrin-beta3 expression. Molecular and cellular biology. 2007;27(7):2486–2498. doi: 10.1128/MCB.01736-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saunders CJ, Jalali Sefid Dashti M, Gamieldien J. Semantic interrogation of a multi knowledge domain ontological model of tendinopathy identifies four strong candidate risk genes. Scientific reports. 2016;6:19820. doi: 10.1038/srep19820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith SM, Thomas CE, Birk DE. Pericellular proteins of the developing mouse tendon: a proteomic analysis. Connect Tissue Res. 2012;53(1):2–13. doi: 10.3109/03008207.2011.602766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raykha C, Crawford J, Gan BS, et al. IGF-II and IGFBP-6 regulate cellular contractility and proliferation in Dupuytren's disease. Biochimica et biophysica acta. 2013;1832(10):1511–1519. doi: 10.1016/j.bbadis.2013.04.018. [DOI] [PubMed] [Google Scholar]
- 30.Zhang C, Lu L, Li Y, et al. IGF binding protein-6 expression in vascular endothelial cells is induced by hypoxia and plays a negative role in tumor angiogenesis. Int J Cancer. 2012;130(9):2003–2012. doi: 10.1002/ijc.26201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bechard M, Trost R, Singh AM, Dalton S. Frat is a phosphatidylinositol 3-kinase/Akt-regulated determinant of glycogen synthase kinase 3beta subcellular localization in pluripotent cells. Molecular and cellular biology. 2012;32(2):288–296. doi: 10.1128/MCB.05372-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ko CY, Wang WL, Wang SM, et al. Glycogen synthase kinase-3beta-mediated CCAAT/enhancer-binding protein delta phosphorylation in astrocytes promotes migration and activation of microglia/macrophages. Neurobiol Aging. 2014;35(1):24–34. doi: 10.1016/j.neurobiolaging.2013.07.021. [DOI] [PubMed] [Google Scholar]
- 33.Dean BJ, Gettings P, Dakin SG, Carr AJ. Are inflammatory cells increased in painful human tendinopathy? A systematic review. British journal of sports medicine. 2016;50(4):216–220. doi: 10.1136/bjsports-2015-094754. [DOI] [PubMed] [Google Scholar]
- 34.Chamberlain CS, Leiferman EM, Frisch KE, et al. The influence of macrophage depletion on ligament healing. Connect Tissue Res. 2011;52(3):203–211. doi: 10.3109/03008207.2010.511355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dantas Machado AC, Zhou T, Rao S, et al. Evolving insights on how cytosine methylation affects protein-DNA binding. Brief Funct Genomics. 2015;14(1):61–73. doi: 10.1093/bfgp/elu040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baubec T, Schubeler D. Genomic patterns and context specific interpretation of DNA methylation. Curr Opin Genet Dev. 2014;25:85–92. doi: 10.1016/j.gde.2013.11.015. [DOI] [PubMed] [Google Scholar]
- 37.Jones PA, Laird PW. Cancer epigenetics comes of age. Nature genetics. 1999;21(2):163–167. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- 38.Wasserstein RL, Lazar NA. The ASA's statement on p-values: context, process, and purpose. The American Statistician. 2016 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.