Abstract
The opportunistic pathogen Pseudomonas aeruginosa produces colorful redox-active metabolites called phenazines, which underpin biofilm development, virulence and clinical outcomes. Though phenazines exist in many forms, the best studied is pyocyanin. Here, we describe pyocyanin demethylase (PodA), a hitherto uncharacterized protein that oxidizes the pyocyanin methyl group to formaldehyde and reduces the pyrazine ring via an unusual tautomerizing demethylation reaction. Treatment with PodA disrupts P. aeruginosa biofilm formation similarly to DNase, suggesting interference with the pyocyanin-dependent release of extracellular DNA into the matrix. PodA-dependent pyocyanin demethylation also restricts established biofilm aggregate populations experiencing anoxic conditions. Together, these results show that modulating extracellular redox-active metabolites can influence the fitness of biofilms.
Bacteria from phylogenetically diverse taxa secrete colorful redox-active metabolites, such as the well-studied phenazines produced by multiple species, including Pseudomonas aeruginosa (1, 2) (Fig. 1A,B). Phenazines can be toxic to other cells but also benefit their producers by facilitating extracellular electron transfer and survival in anoxic environments (3–6). These latter functions support the growth of antibiotic-resistant biofilms, and P. aeruginosa mutants that cannot make phenazines are defective in biofilm development (5). Accordingly, we reasoned that selectively manipulating phenazines might present a means to control biofilms. One way to influence extracellular metabolites is through active modification or degradation by other organisms (7–9). Recently, we described a group of mycobacteria that enzymatically degrade phenazines and identified genes that catalyze distinct steps in degradation (9). Here, we focus on the structure and function of a previously uncharacterized protein from Mycobacterium fortuitum encoded by MFORT_14352 (NCBI Accession number: EJZ13467) that catalyzes pyocyanin (PYO) degradation (9).
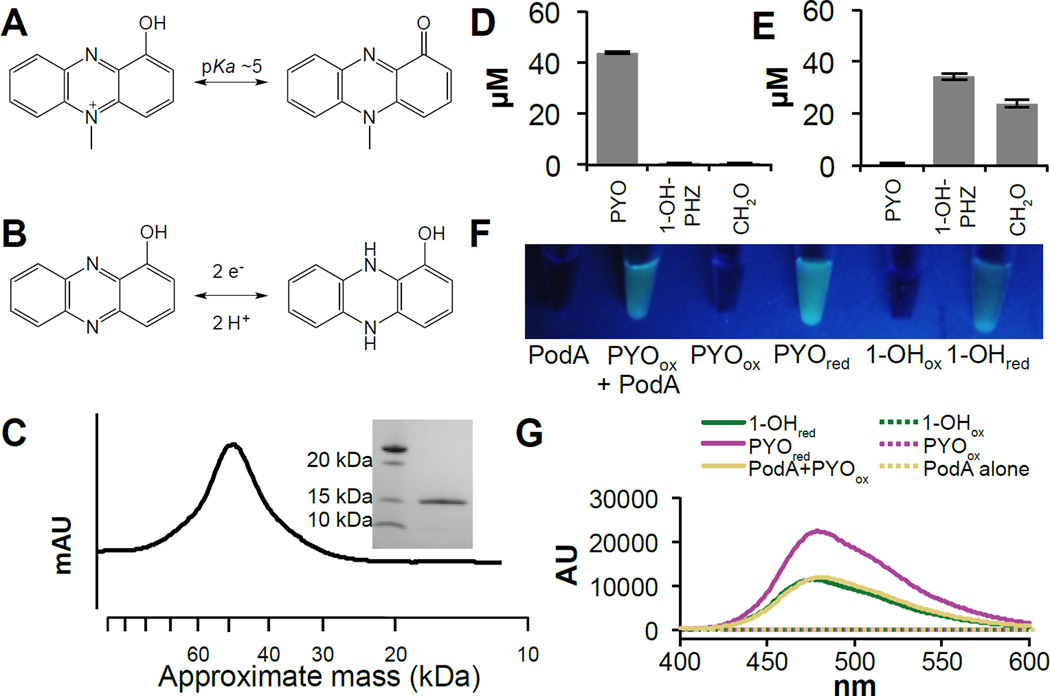
Figure 1. Biochemical analysis of the PodA reaction.
PodA catalyzes the demethylation of PYO (A) to reduced 1-OH-PHZ (B). (C) PodA30–162 purifies as a trimer by gel filtration chromatography (45.6 kDa). Inset is a denaturing gel demonstrating the size of monomeric PodA30–162. Incubations of PodA30–162 with PYO show the conversion of the starting material (D) to 1-OH-PHZ and formaldehyde (E). Data are average measurements from six reactions and error bars represent one standard deviation around the mean. Formaldehyde is derivatized to facilitate detection; the derivatization competes with other compounds in the mixture so stoichiometric production was not observed. (F) PodA30–162 is active under anoxic conditions, and the PYO containing reaction mixture fluoresces under UV illumination. This fluorescent product has an emission spectrum (250 nm excitation) consistent with a reduced phenazine (G). While both reduced PYO and 1-OH-PHZ have similar emission maxima (50 µM phenazine), the magnitude of the peak is consistent with the generation of reduced 1-OH-PHZ by PodA30–162.
To characterize its activity, we purified a heterologously expressed, truncated version of this protein (lacking a predicted N-terminal, membrane-spanning helix (10)), hereafter referred to as PodA30–162 (PYO demethylase), from Escherichia coli (Fig. 1C). PodA30–162 converts PYO to 1-hydroxyphenazine (1–OH-PHZ) and formaldehyde (Fig. 1D,E), indicating that it functions as a demethylase. Generally, enzyme-catalyzed N-demethylations proceed by oxidation of the methyl group to formaldehyde with electron transfer to a bound cofactor or iron-sulfur cluster (11, 12). Surprisingly, we found that PodA30–162 generates 1-OH-PHZ in the absence of either flavin or 2-oxoglutarate, which are required for most known N-demethylases (Fig. S1). Additionally, PodA30–162 functions under anoxic conditions suggesting a mechanism distinct from the oxygen-dependent Rieske-iron type demethylases (13). Kinetic analysis suggests that PodA is a high affinity PYO demethylase (Km < 1 µM and kcat = 1.20 ± 0.07 s−1) that operates over a wide regime of pH (<4.9 – 8.7) and salt concentrations (0 – 400 mM) (Fig. S2) with specificity for N-methylated phenazines (Fig S3). Under anoxic conditions, PodA30–162 catalyzes the formation of a reduced phenazine, suggesting that its substrate serves as the electron acceptor (Fig. 1F,G). This mechanism has not previously been observed for demethylases (11, 12). We propose the following reaction for PodA, wherein oxidized PYO (PYOox) is converted to reduced 1-OH-PHZ (1–OH-PHZred) (Fig. 1A,B):
C13H10N2O (PYOox) + H2O → C12H10N2O (1-OH-PHZred) + CH2O
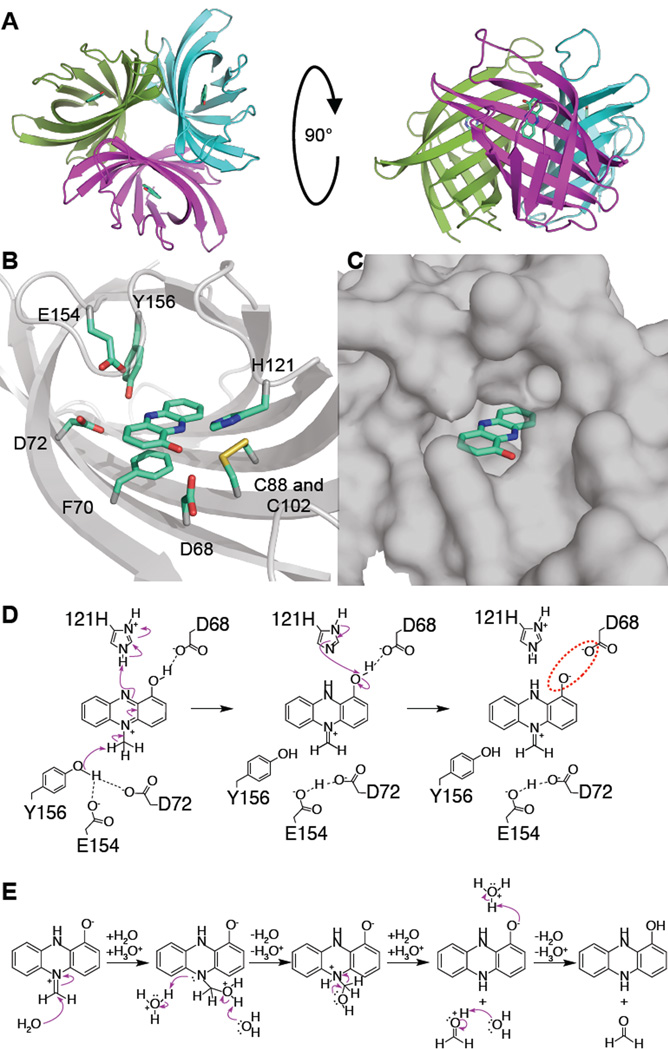
To test this model and better understand how PodA30–162 reduces its substrate, we solved the X-ray crystal structure at 1.8 Å resolution (Table S1) by molecular replacement using a search model generated by Robetta (14–16) (Fig. S4). PodA30–162 crystalized as a trimer in the asymmetric unit (Fig. 2A). Crystal formation occurred only in the presence of 1-OH-PHZ, which was visible in a putative solvent-exposed active site (Fig. 2B,C). We found no evidence of a bound cofactor or metals in the electron density of the active site (Fig. S5) further supporting the hypothesis that PodA catalyzes a novel demethylation reaction. Within the active site, there are several charged and polar residues (D68, D72, H121, E154, and Y156) and a nearby disulfide that are candidate catalytic residues (17); additionally, 1-OH-PHZ appears to bind via a π-π stacking interaction with F70 (Fig. 2B).
Figure 2. 1.8 Å crystal structure of PodA30–162.
(A) View of the PodA30–162 trimer with 1-OH-PHZ bound. The PodA active site (B) is solvent exposed (C) and contains charged and polar residues. There is a nearby disulfide ~3.5 Å from 1-OH-PHZ. (D) A proposed reaction mechanism based on the residues present in the active site. The model predicts that D72, E154 and Y156 are necessary for methyl deprotonation. H121 protonates the unmethylated N atom of the pyrazine ring, and D68 reprotonates H121. Formation of the negatively charged phenolate ion promotes product release due to an unfavorable electrostatic interaction (red dashed circle). D72 and E154 remain protonated, acidifying the active site to assist in formation of hydroxylated PYO (Fig. 1A) in the next catalytic cycle. (E) Hydrolysis of the product is spontaneous and occurs after release from the active site.
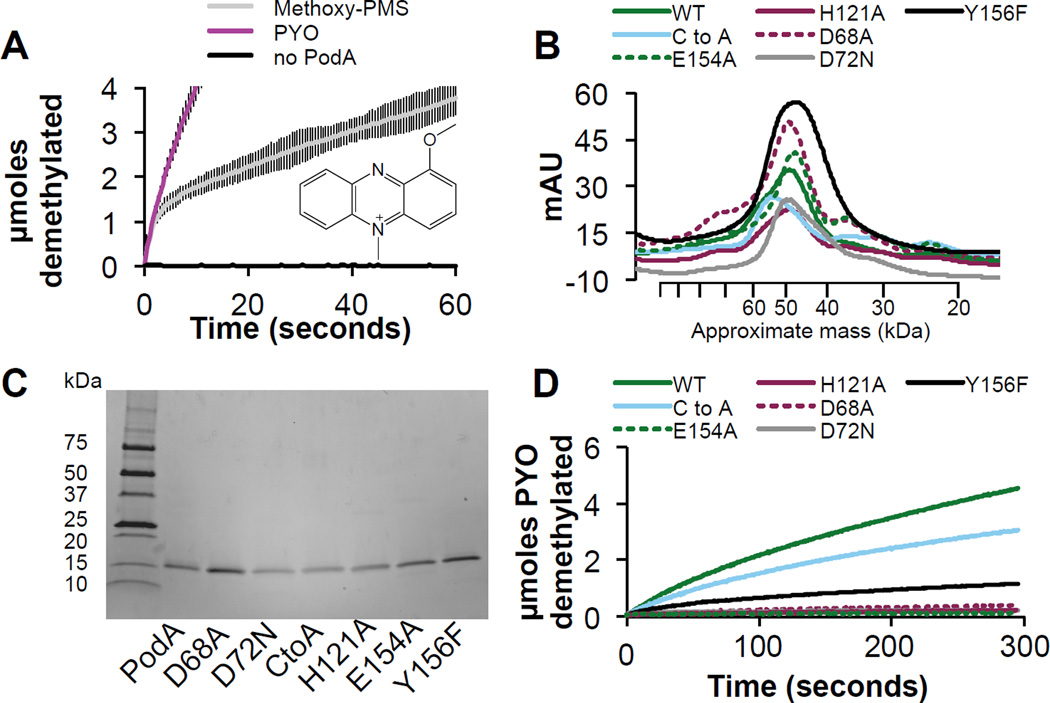
Based on the active site structure, we propose a mechanism relying on the presence of H121 functioning as an acid and D72, E154 and Y156 collectively functioning as a base (Fig. 2D). PYO binds in the phenol tautomeric form (Fig. 1A). Deprotonation of the methyl group results in iminium ion formation and concomitant reduction of the pyrazine ring, with the second nitrogen atom protonated by H121. H121 is then reprotonated by PYO’s hydroxyl group (pKa of His ~6 versus PYO ~5). The unfavorable interaction between the negatively charged phenolate ion and D68 drives product release. We hypothesize that PodA catalyzes the tautomerization of PYO to an iminium ion that is susceptible to hydrolysis outside of the enzyme: the structure indicates that the active site cannot accommodate both the substrate and a water molecule (Fig. 2B–E) (11). D72 and E154 remain protonated in this scenario, acidifying the active site, perhaps to ensure that PYO is in the hydroxylated form in subsequent reaction cycles (Fig. 1A). This scheme highlights the significance of the hydroxyl group of PYO to recharge PodA for subsequent reaction cycles and to drive product release. An alternative substrate, methoxyphenazine methosulfate (methoxy-PMS), which lacks the hydroxyl group, displays an initial burst of activity followed by a slower steady state (Fig. 3A), which is consistent with our model.
Figure 3. Molecular analysis of the PodA reaction mechanism.
(A) An alternative substrate — methoxy-PMS, inset — is demethylated by PodA30–162, but the reaction rate slows significantly after an initial burst, highlighting the importance of the hydroxyl group of PYO for catalysis and/or driving product release after deprotonation. Data are averages from triplicate measurements and error bars represent one standard deviation from the mean. (B) Residues in the PodA active site (Fig. 2B) were mutated and the resulting proteins purified; H121A, D68A, E154A, D72N, Y156F and the C88A, C102A (C to A) double mutant all purify as a trimer by gel filtration chromatography. (C) Mutant proteins were pure as assayed by reducing SDS-PAGE. (D) Activity of mutant proteins shows that the disulfide bond is not essential for activity. Y156F has ~25% wild type activity, and all other residues appear essential for catalysis.
To probe the proposed mechanism, we performed mutagenesis for each of the putative catalytic residues. H121A, E154A, D68A, and D72N mutants all formed a trimer (Fig. 3B,C) but had <10% wild type activity, consistent with the postulated catalytic mechanism (Fig. 3D). Y156F formed a trimer but retained ~25% wild type activity, consistent with this residue facilitating proton transfer to D72 and E154 but not being essential. A possible alternative mechanism could utilize the disulfide in the active site forming a covalent adduct with the phenazine, in analogy to some flavoproteins (18). However, our mutagenesis results indicate that the disulfide bond near the active site is not essential for catalytic activity although it may be important for structural stability (Fig. 3B–D) (Fig. S6).
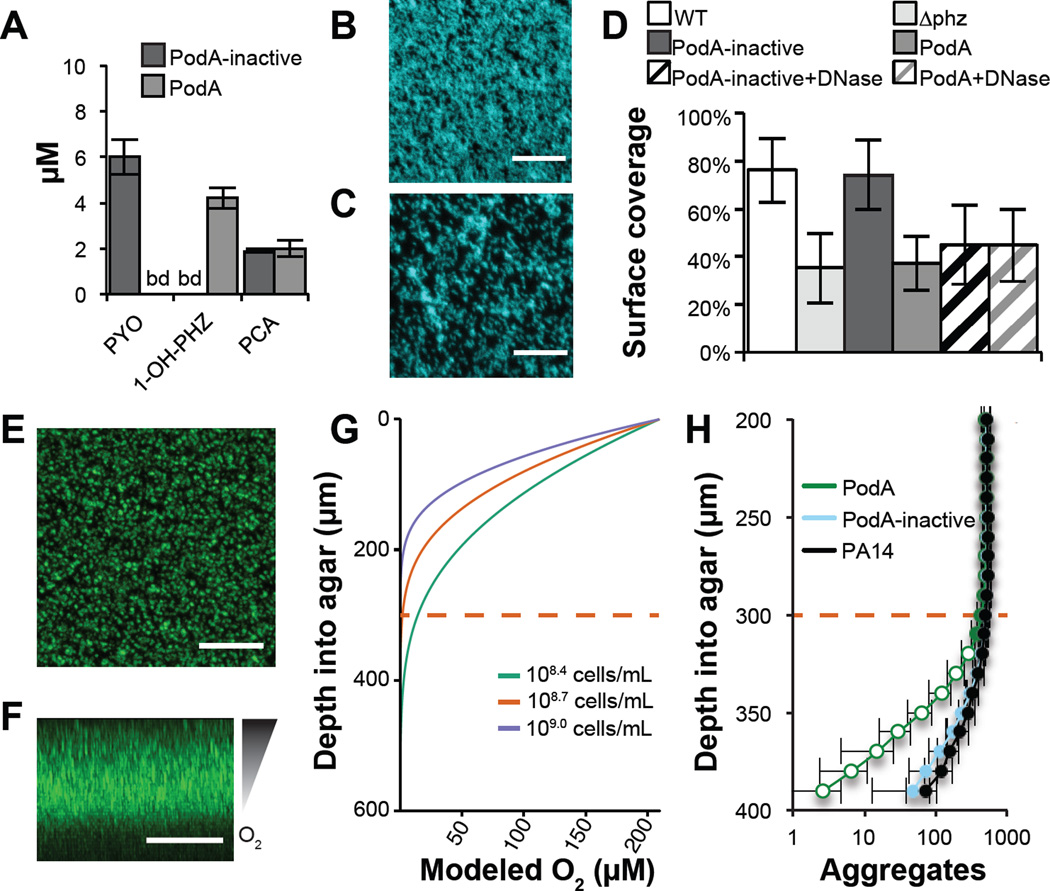
Because PodA requires only water and substrate for activity (Figs. 2D,E, S1, and S2), we hypothesized that it could degrade PYO during active production by P. aeruginosa (Fig. S7). PodA30–162 addition to P. aeruginosa planktonic culture results in the conversion of PYO to 1-OH-PHZ in both rich and minimal medium (Fig. S8). Encouraged by these findings, we assessed the impact of PodA on biofilm formation. Extracellular DNA (eDNA) comprises >50% of the P. aeruginosa biofilm matrix (19), and recently PYO was shown to drive eDNA release (20, 21) which is important early in biofilm development (19). We hypothesized that PodA30–162 might inhibit P. aeruginosa biofilms in part by attenuating DNA release, thereby removing an important matrix component and changing biofilm architecture. Because PYO does not completely block DNA release (20, 21), it is also possible that downstream PYO-DNA interactions may be important. We checked whether PodA30–162 could access PYO in the presence of DNA, as PYO is a known DNA intercalator (22); PodA30–162 catalyzed PYO demethylation in this context (Fig S8). We grew P. aeruginosa biofilms for 5 hours, staining them with DAPI to image biofilm structure. HPLC analysis of supernatants confirmed the conversion of PYO to 1-OH-PHZ by PodA30–162 in these cultures (Fig. 4A). Overall biofilm formation, as assayed by surface coverage (22), was reduced by PodA30–162 but not by the inactive PodA control (Fig. 4B–D), consistent with a role for PYO in early biofilm development. As previously shown, treatment with DNase independently inhibited biofilms (22). Interestingly, DNase or PodA30–162 treatment inhibited biofilms to the same extent, and dual treatment did not have an additive effect (Fig. 4D).
Figure 4. PodA30–162 inhibits biofilm formation and anoxic fitness of P. aeruginosa.
(A) Phenazines were measured by HPLC in biofilm supernatants after 5 hours of growth. PCA, phenazine-1-carboxylic acid. (B) P. aeruginosa forms a robust biofilm in the presence of inactivated PodA30–162 after 5 hours. (C) In the presence of PodA30–162, biofilm surface coverage was decreased. Surface coverage was 43.5 percent compared with 82.7 percent in the absence of PodA30–162 in the representative images shown. Scale Bars = 20 µm. (D) Surface coverage was lower in the presence of PodA30–162 (p < 10−6 vs. PodA-inactive, two-tailed Student’s t-test). DNase addition decreased surface coverage (p < 10−3 vs. PodA-inactive), but DNase and PodA30−162 combined did not have an additive effect (p > 0.05), consistent with an interaction between PYO and eDNA in supporting biofilms (22). Data are averages of 12 replicates taken from independent cultures. Error bars represent one standard deviation around the mean. ∆phz, PA14 mutant incapable of making phenazines. (E) Top down and (F) side views of P. aeruginosa grown embedded in 0.5% agar blocks for 27 hours. Scale bars = 200 µm. Oxygen depletion occurs at lower depths as a result of biological consumption outpacing diffusion from the surface, resulting in decreased biomass. (G) Oxygen diffusion model predicting the shape of the oxycline in agar blocks. Cell densities were estimated at 108.7 cells mL−1 based on aggregate number and volume. Modeling this concentration and 2-fold higher and lower densities suggests that oxygen depletion occurs ~300 µm ± 100 µm below the agar surface. Dashed red line indicates the approximate oxic-anoxic interface. (H) Biofilm aggregates detected at 10 µm increments below the agar surface. At depths near the oxic/anoxic interface (dashed red line), total biomass begins to decline. In assays treated for the last 5 hours with PodA30−162, there is an apparent biomass defect specifically at anoxic depths compared to untreated and inactive PodA treated controls, consistent with the importance of PYO for anoxic survival in P. aeruginosa. Data are averages of six independent experiments and error bars represent one standard deviation around the mean. Open symbols, p < 0.01, two-tailed Student’s t-test.
In addition to impacting early stages in biofilm development, phenazines can expand the habitable zone within established biofilms (5, 23). As P. aeruginosa biofilms mature, cells in deeper layers of the biofilm begin to experience oxygen limitation and redox stress (4, 23); these cells are believed to be slow growing and are highly resistant to antibiotics (24). Phenazines facilitate anoxic survival and alleviate redox stress (3, 25). Because PYO reacts with oxygen more efficiently than 1-OH-PHZ (26), we hypothesized that PodA activity might decrease anoxic fitness by disrupting PYO dependent electron shuttling to oxygen. To capture key features of in vivo biofilm aggregates (27, 28), cells were grown suspended in agar blocks at 37 °C for 22 hours to establish an oxygen gradient before a 5 hour treatment with PodA30–162 (Fig. 4E,F). Owing to constraints imposed by the incubator, we were unable to measure oxygen gradients directly, so we used a previously validated model to estimate the oxic-anoxic transition zone (29). Modeling our aggregate population indicated that anoxia occurs ~300 µm below the surface as a result of microbial consumption (Fig. 4G), consistent with what is known about oxygen diffusion into in vivo biofilms (29). We observed a decrease in detectable aggregates at depths 300–400 µm below the agar surface; additionally, cultures treated with PodA30–162 had a sharper decline in detectable aggregates below this depth (Fig. 4H). There was no significant difference in aggregate numbers above the predicted oxic-anoxic transition zone. How PYO demethylation restricts the aggregate population is unknown.
In conclusion, the discovery of a PYO demethylase that simultaneously catalyzes substrate reduction shows that redox-active pigments can participate in their own enzyme-catalyzed modification. Though PodA is the first member of a new class of tautomerizing demethylases that utilizes an oxidized substrate as the electron acceptor, this reaction is reminiscent of reduced flavin acting as the electron donor in its own destruction in vitamin B12 biosynthesis (30). It seems likely that the processing of other redox-active pigments and cofactors may operate by a similar mechanism where the redox activity of the substrate enables catalysis. That PodA can inhibit P. aeruginosa at different stages of biofilm development raises the possibility that selective degradation of extracellular electron shuttles may facilitate treatment of intractable infections.
Supplementary Material
Acknowledgments
The final model and native data set for PodA were submitted to the wwPDB under accession code 5K21. We thank Elena Perry, Lucas Meirelles, William DePas and Douglas Rees for assistance in experimental design and interpretation. KCC was supported by a Ruth L. Kirschstein National Research Service Award from the National Institutes of Health, National Institute of Allergy and Infectious Diseases, Grant no. F32AI112248. NRG was supported by the National Science Foundation Graduate Research Fellowship, Grant no. 1144469. This work was further supported by the Howard Hughes Medical Institute (HHMI), NIH (Grant 5R01HL117328-03) and the Molecular Observatory at the Beckman Institute, California Institute of Technology through the Gordon and Betty Moore Foundation and the Sanofi-Aventis Bioengineering Research Program at Caltech. Additional support was provided by the Stanford Synchrotron Radiation Lightsource, which is funded by the US DOE and NIH. SJC thanks St Hugh’s College, Oxford for research support.
Footnotes
Author contributions. KCC and DKN conceived the project. KCC, NRG, and DKN designed the experiments. KCC and NRG performed the experiments. KCC, NRG, SJC, and DKN analyzed and interpreted the results. KCC, SJC and DKN wrote the paper.
Supplementary Materials
Materials and Methods
Figures S1–S8
Tables S1, S2
References # 31–62
References and Notes
- 1.Turner JM, Messenger AJ. Occurrence, biochemistry and physiology of phenazine pigment production. Adv Microb Physiol. 1986;27:211–275. doi: 10.1016/s0065-2911(08)60306-9. [DOI] [PubMed] [Google Scholar]
- 2.Dietrich LE, Teal TK, Price-Whelan A, Newman DK. Redox-active antibiotics control gene expression and community behavior in divergent bacteria. Science. 2008;321:1203–1206. doi: 10.1126/science.1160619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Glasser NR, Kern SE, Newman DK. Phenazine redox cycling enhances anaerobic survival in Pseudomonas aeruginosa by facilitating generation of ATP and a proton-motive force. Mol Microbiol. 2014;92:399–412. doi: 10.1111/mmi.12566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Price-Whelan A, Dietrich LE, Newman DK. Pyocyanin alters redox homeostasis and carbon flux through central metabolic pathways in Pseudomonas aeruginosa PA14. J Bacteriol. 2007;189:6372–6381. doi: 10.1128/JB.00505-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramos I, Dietrich LE, Price-Whelan A, Newman DK. Phenazines affect biofilm formation by Pseudomonas aeruginosa in similar ways at various scales. Res Microbiol. 2010;161:187–191. doi: 10.1016/j.resmic.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y, et al. Phenazine-1-carboxylic acid promotes bacterial biofilm development via ferrous iron acquisition. J Bacteriol. 2011;193:3606–3617. doi: 10.1128/JB.00396-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moree WJ, et al. Interkingdom metabolic transformations captured by microbial imaging mass spectrometry. Proc Natl Acad Sci U S A. 2012;109:13811–13816. doi: 10.1073/pnas.1206855109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang ZJ, et al. Isolation, identification, and degradation characteristics of phenazine-1-carboxylic acid-degrading strain Sphingomonas sp. DP58. Curr Microbiol. 2007;55:284–287. doi: 10.1007/s00284-006-0522-7. [DOI] [PubMed] [Google Scholar]
- 9.Costa KC, Bergkessel M, Saunders S, Korlach J, Newman DK. Enzymatic degradation of phenazines can generate energy and protect sensitive organisms from toxicity. MBio. 2015;6:e01520–e01515. doi: 10.1128/mBio.01520-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 11.Anand R, Marmorstein R. Structure and mechanism of lysine-specific demethylase enzymes. J Biol Chem. 2007;282:35425–35429. doi: 10.1074/jbc.R700027200. [DOI] [PubMed] [Google Scholar]
- 12.Hagel JM, Facchini PJ. Biochemistry and occurrence of O-demethylation in plant metabolism. Front Physiol. 2010;1:14. doi: 10.3389/fphys.2010.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Summers RM, et al. Novel, highly specific N-demethylases enable bacteria to live on caffeine and related purine alkaloids. J Bacteriol. 2012;194:2041–2049. doi: 10.1128/JB.06637-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song Y, et al. High-resolution comparative modeling with RosettaCM. Structure. 2013;21:1735–1742. doi: 10.1016/j.str.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raman S, et al. Structure prediction for CASP8 with all-atom refinement using Rosetta. Proteins. 2009;77(Suppl 9):89–99. doi: 10.1002/prot.22540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim DE, Chivian D, Baker D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 2004;32:W526–W531. doi: 10.1093/nar/gkh468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gutteridge A, Thornton JM. Understanding nature's catalytic toolkit. Trends Biochem Sci. 2005;30:622–629. doi: 10.1016/j.tibs.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 18.Walsh CT, Wencewicz TA. Flavoenzymes: versatile catalysts in biosynthetic pathways. Nat Prod Rep. 2013;30:175–200. doi: 10.1039/c2np20069d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. Extracellular DNA required for bacterial biofilm formation. Science. 2002;295:1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- 20.Das T, Manefield M. Pyocyanin promotes extracellular DNA release in Pseudomonas aeruginosa. PLoS One. 2012;7:e46718. doi: 10.1371/journal.pone.0046718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Das T, Manefield M. Phenazine production enhances extracellular DNA release via hydrogen peroxide generation in Pseudomonas aeruginosa. Commun Integr Biol. 2013;6:e23570. doi: 10.4161/cib.23570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Das T, et al. Phenazine virulence factor binding to extracellular DNA is important for Pseudomonas aeruginosa biofilm formation. Sci Rep. 2015;5:8398. doi: 10.1038/srep08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dietrich LE, et al. Bacterial community morphogenesis is intimately linked to the intracellular redox state. J Bacteriol. 2013;195:1371–1380. doi: 10.1128/JB.02273-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Borriello G, et al. Oxygen limitation contributes to antibiotic tolerance of Pseudomonas aeruginosa in biofilms. Antimicrob Agents Chemother. 2004;48:2659–2664. doi: 10.1128/AAC.48.7.2659-2664.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Kern SE, Newman DK. Endogenous phenazine antibiotics promote anaerobic survival of Pseudomonas aeruginosa via extracellular electron transfer. J Bacteriol. 2010;192:365–369. doi: 10.1128/JB.01188-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, Newman DK. Redox reactions of phenazine antibiotics with ferric (hydr)oxides and molecular oxygen. Environ Sci Technol. 2008;42:2380–2386. doi: 10.1021/es702290a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bjarnsholt T, et al. The in vivo biofilm. Trends Microbiol. 2013;21:466–474. doi: 10.1016/j.tim.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 28.Kragh KN, et al. Role of Multicellular Aggregates in Biofilm Formation. MBio. 2016;7:e00237. doi: 10.1128/mBio.00237-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cowley ES, Kopf SH, LaRiviere A, Ziebis W, Newman DK. Pediatric cystic fibrosis sputum can be chemically dynamic, anoxic, and extremely reduced due to hydrogen sulfide formation. MBio. 2015;6:e00767. doi: 10.1128/mBio.00767-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taga ME, Larsen NA, Howard-Jones AR, Walsh CT, Walker GC. BluB cannibalizes flavin to form the lower ligand of vitamin B12. Nature. 2007;446:449–453. doi: 10.1038/nature05611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dietrich LE, Price-Whelan A, Petersen A, Whiteley M, Newman DK. The phenazine pyocyanin is a terminal signalling factor in the quorum sensing network of Pseudomonas aeruginosa. Mol Microbiol. 2006;61:1308–1321. doi: 10.1111/j.1365-2958.2006.05306.x. [DOI] [PubMed] [Google Scholar]
- 32.Rahme LG, et al. Common virulence factors for bacterial pathogenicity in plants and animals. Science. 1995;268:1899–1902. doi: 10.1126/science.7604262. [DOI] [PubMed] [Google Scholar]
- 33.Studier FW, Moffatt BA. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 34.DasGupta SK, Jain S, Kaushal D, Tyagi AK. Expression systems for study of mycobacterial gene regulation and development of recombinant BCG vaccines. Biochem Biophys Res Commun. 1998;246:797–804. doi: 10.1006/bbrc.1998.8724. [DOI] [PubMed] [Google Scholar]
- 35.Shih YP, Wu HC, Hu SM, Wang TF, Wang AH. Self-cleavage of fusion protein in vivo using TEV protease to yield native protein. Protein Sci. 2005;14:936–941. doi: 10.1110/ps.041129605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 37.Nash T. The colorimetric estimation of formaldehyde by means of the Hantzsch reaction. Biochem J. 1953;55:416–421. doi: 10.1042/bj0550416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones SB, Terry CM, Lister TE, Johnson DC. Determination of submicromolar concentrations of formaldehyde by liquid chromatography. Anal. Chem. 1999;71:4030–4033. [Google Scholar]
- 39.Sullivan NL, Tzeranis DS, Wang Y, So PT, Newman D. Quantifying the dynamics of bacterial secondary metabolites by spectral multiphoton microscopy. ACS Chem Biol. 2011;6:893–899. doi: 10.1021/cb200094w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mavrodi DV, et al. Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. J Bacteriol. 2001;183:6454–6465. doi: 10.1128/JB.183.21.6454-6465.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kemmer G, Keller S. Nonlinear least-squares data fitting in Excel spreadsheets. Nat Protoc. 2010;5:267–281. doi: 10.1038/nprot.2009.182. [DOI] [PubMed] [Google Scholar]
- 42.McIlwain H. The phenazine series. Part IV. Reactionsof alkyl phenazonium salts; the phenazyls. J Chem Soc. 1937:1704–1711. [Google Scholar]
- 43.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kabsch W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr D Biol Crystallogr. 2010;66:133–144. doi: 10.1107/S0907444909047374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Evans P. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 46.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Padilla JE, Yeates TO. A statistic for local intensity differences: robustness to anisotropy and pseudo-centering and utility for detecting twinning. Acta Crystallogr D Biol Crystallogr. 2003;59:1124–1130. doi: 10.1107/s0907444903007947. [DOI] [PubMed] [Google Scholar]
- 48.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Terwilliger TC, et al. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr D Biol Crystallogr. 2008;64:61–69. doi: 10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Headd JJ, et al. Use of knowledge-based restraints in phenix.refine to improve macromolecular refinement at low resolution. Acta Crystallogr D Biol Crystallogr. 2012;68:381–390. doi: 10.1107/S0907444911047834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Afonine PV, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Terwilliger TC, Adams PD, Moriarty NW, Cohn JD. Ligand identification using electron-density map correlations. Acta Crystallogr D Biol Crystallogr. 2007;63:101–107. doi: 10.1107/S0907444906046233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Terwilliger TC, Klei H, Adams PD, Moriarty NW, Cohn JD. Automated ligand fitting by core-fragment fitting and extension into density. Acta Crystallogr D Biol Crystallogr. 2006;62:915–922. doi: 10.1107/S0907444906017161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moriarty NW, Grosse-Kunstleve RW, Adams PD. electronic Ligand Builder and Optimization Workbench (eLBOW): a tool for ligand coordinate and restraint generation. Acta Crystallogr D Biol Crystallogr. 2009;65:1074–1080. doi: 10.1107/S0907444909029436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D Biol Crystallogr. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 59.Das T, Kutty SK, Kumar N, Manefield M. Pyocyanin facilitates extracellular DNA binding to Pseudomonas aeruginosa influencing cell surface properties and aggregation. PLoS One. 2013;8:e58299. doi: 10.1371/journal.pone.0058299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bolte S, Cordelieres FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- 62.Cohen D, et al. Oligoribonuclease is a central feature of cyclic diguanylate signaling in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2015;112:11359–11364. doi: 10.1073/pnas.1421450112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.