Abstract
Antimicrobial peptides (AMPs) are important components of the innate immune system that have been found to be effective against disease causing pathogens. Identification of AMPs through wet-lab experiment is expensive. Therefore, development of efficient computational tool is essential to identify the best candidate AMP prior to the in vitro experimentation. In this study, we made an attempt to develop a support vector machine (SVM) based computational approach for prediction of AMPs with improved accuracy. Initially, compositional, physico-chemical and structural features of the peptides were generated that were subsequently used as input in SVM for prediction of AMPs. The proposed approach achieved higher accuracy than several existing approaches, while compared using benchmark dataset. Based on the proposed approach, an online prediction server iAMPpred has also been developed to help the scientific community in predicting AMPs, which is freely accessible at http://cabgrid.res.in:8080/amppred/. The proposed approach is believed to supplement the tools and techniques that have been developed in the past for prediction of AMPs.
Antimicrobial peptides (AMPs) are important innate immune molecules, which have been found to be effective against several pathogenic micro-organisms like bacteria, virus, fungi, parasites etc1. AMP constitutes the first line of host defense against microbes2, where it causes the cell death of microbes either by disrupting its cell membrane or its intracellular functions3,4. Due to growing resistance of microbial pathogens against chemical antibiotics, AMPs have received attention as an alternative in recent years5. Specifically, due to the broad spectrum of activity and low propensity for developing resistance, AMPs are gaining popularity in clinical applications6.
Development of sequence-based computational tools can be helpful in designing the effective antimicrobial agents by identifying the best candidate AMP prior to the synthesis and testing against pathogens in wet-lab7. In this direction, computational tools like AntiBP1, AMPER8, CAMP3, AntiBP29, AVPpred10, ClassAMP11, iAMP-2L7 and EFC-FCBF12 have been developed for the prediction of AMPs. The binary (0, 1) and compositional features were used in AntiBP and AntiBP2 respectively to map the peptide sequences onto numeric feature vectors, where the numeric vectors were used as input in artificial neural network (ANN)13 and support vector machine (SVM)14 respectively for prediction of antibacterial peptides. In CAMP, random forest (RF)15, SVM and ANN supervised learning techniques were employed for prediction of AMPs, based on different physico-chemical (PHYC) features of peptides. In AVPpred, four different models viz., AVPmotif, AVPalign, AVMcompo and AVPphysico were developed for prediction of antiviral peptides only. The ClassAMP11 tool was developed for predicting the propensity of a peptide sequence as antibacterial, antiviral or antifungal peptide, by using SVM and RF machine learning techniques. In an another study, a two-level multi-class predictor was developed for identification of AMPs, based on Chou’s pseudo amino acid composition16 and fuzzy k-nearest neighbor7. Recently, Veltri et al.12 have developed a machine learning based computational approach for improved recognition of AMPs.
The above mentioned methods have their own advantages in generating knowledge for the prediction of AMPs. However, further improvement in prediction accuracy is required to minimize the number of false positives. In this study, we made an attempt to develop a computational approach for prediction of antibacterial, antiviral and antifungal peptides with higher accuracy. In this approach, combinations of compositional, PHYC and structural (STRL) features were used to map the peptide sequences onto numeric feature vectors, which were subsequently used as input in SVM for prediction. The proposed approach was found to perform better than several existing approaches for predicting AMPs, when comparison was made using bench mark dataset.
Material and Methods
As summarized and demonstrated by a series of recent publications17,18,19,20,21,22, in compliance with Chou’s 5-step rule23, to establish a really useful sequence-based statistical predictor for a biological system, the following five guidelines should be followed: (a) construct or select a valid benchmark dataset to train and test the predictor; (b) formulate the biological sequence samples with an effective mathematical expression that can truly reflect their intrinsic correlation with the target to be predicted; (c) introduce or develop a powerful algorithm (or engine) to operate the prediction; (d) properly perform cross-validation tests to objectively evaluate the anticipated accuracy of the predictor; (e) establish a user-friendly web-server for the predictor that is freely accessible to the public. In the following sections, we have described how to deal with these steps one-by-one.
Dataset
Positive
To construct the positive dataset, antibacterial, antiviral and antifungal peptide sequences were collected from publicly available databases (or datasets). Specifically, antibacterial peptides were collected from CAMP, APD324 and AntiBP2; antiviral peptides were collected from CAMP, APD3, LAMP25 and AVPpred; antifungal peptides were collected from CAMP, LAMP and APD3. The sequences having non-standard amino acids were then removed followed by removal of redundant sequences, similar to earlier studies7,12,26. Since AMPs are mostly 10–100 amino acids long1, sequences having less than 10 amino acids were also excluded from further analysis. A summary of the positive datasets is given in Table 1.
Table 1. Summary of the positive and negative datasets.
| Dataset | Bacterial | Viral | Fungal |
|---|---|---|---|
| Positive | CAMP3, APD324, AntiBP29 {3417} | CAMP, APD3, LAMP25, AVPpred10 {739} | CAMP, LAMP, APD3 {1496} |
| Negative | AntiBP2 {984} | AVPpred {893} | AntiBP2, AVPpred {1384} |
The value inside bracket {} is the number of sequences collected in that category.
Negative
The non-antibacterial and non-antiviral peptides were collected from AntiBP2 and AVPpred respectively. These non-antibacterial and non-antiviral peptides were respectively used as the negative dataset against the antibacterial and antiviral peptides. Further, these non-antibacterial and non-antiviral peptides were considered together as the negative dataset against the antifungal peptides. Similar to the positive dataset, sequences of the negative dataset were also processed. A summary of the negative datasets is also given in Table 1.
Feature generation
Since the peptide sequences are the strings of amino acids, they need to be mapped onto numeric feature vectors before being used as an input in supervised learning classifiers. In this study, three different categories of features i.e., compositional, PHYC and STRL were considered. In particular, 3 compositional (amino acid composition-AAC, pseudo amino acid composition-PAAC and normalized amino acid composition-NAAC), 3 PHYC (hydrophobicity, net-charge and iso-electric point) and 3 STRL (α-helix propensity, β-sheet propensity and turn propensity) features were considered (Table 2) for prediction of AMPs. The compositional and PHYC features were computed by using the “Peptide” package27 of R-software28, whereas the STRL features were computed by using the TANGO software29 available at http://tango.crg.es/. The TANGO server was first used by Torrent et al.30 for recognition of AMPs. Furthermore, to know the importance of each feature in predicting the antibacterial, antiviral and antifungal peptides, information gain was computed for all the 66 features [AAC (20) + PAAC (20) + NAAC (20) + PHYC (3) + STRL (3)]. To compute the information gain, the InfoGainAttributeEval function available in RWeka31 package was used.
Table 2. Summary of the feature sets.
| Feature category | Features in each category | #Features |
|---|---|---|
| Compositional | Amino acid composition (AAC) | 20 |
| Normalized AAC (NAAC) | 20 | |
| Structural (STRL) | Pseudo AAC (PAAC) | 20 |
| α-helix propensity | 1 | |
| β-sheet propensity | 1 | |
| Turn propensity | 1 | |
| Physico-chemical (PHYC) | Iso-electric point | 1 |
| Hydrophobicity | 1 | |
| Net-charge | 1 |
SVM-based prediction
We used SVM for prediction of AMPs because it is a non-parametric (does not make any assumption about the underlying probability distribution of the input dataset) and most widely used supervised learning technique in the field of bioinformatics, attributed to its sound statistical background32. The predictive ability of SVM, mainly depends upon the type of kernel function that maps the input data to a high-dimensional feature space, where the observations belong to different classes are linearly separable by a optimal separating hyper plane. In this work, the radial basis function (RBF) was used as kernel, due to its wide and successful application in most of the AMP prediction studies1,9,10,33. Further, in RBF kernel, default values of parameters gamma (gamma = 1/number of attributes) and cost (C = 1) were used to train and test the prediction model. The svm function available in the e1071 package34 of R-software was used to execute the SVM model. The scaling option was kept as TRUE in svm function, while training the model.
Performance evaluation
We considered different performance metrics viz., sensitivity (Sn), specificity (Sp), accuracy (Ac) and Matthew’s correlation coefficient (MCC) to evaluate the performance of the proposed approach. Since, the conventional formulae of these metrics are not quite intuitive, particularly MCC, Chen et al.35 derived a new set of equations for the above mentioned metrics based on the Chou’s symbols used in studying protein signal peptide cleavage sites36. The new formulae for these metrics are given in equation (1)
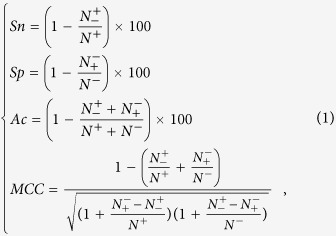 |
where 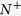 represents the total number of AMPs investigated,
represents the total number of AMPs investigated, 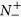 represents the number of AMPs incorrectly predicted as non-AMPs,
represents the number of AMPs incorrectly predicted as non-AMPs, 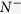 represents the total number of non-AMPs investigated and
represents the total number of non-AMPs investigated and 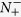 represents the number of non-AMPs incorrectly predicted as AMPs. The formulae given in equation (1) has made the meaning of Sn, Sp, Ac, and MCC much more intuitive and easier-to-understand, particularly for the meaning of MCC, as concurred by a series of studies published very recently19,20,37,38,39,40,41. The above formulae are valid only for the single-label systems, whereas for the multi-label systems, whose emergence has become more frequent in system biology42,43 and system medicine22,44,45, a different set of metrics is needed as elaborated in Chou46.
represents the number of non-AMPs incorrectly predicted as AMPs. The formulae given in equation (1) has made the meaning of Sn, Sp, Ac, and MCC much more intuitive and easier-to-understand, particularly for the meaning of MCC, as concurred by a series of studies published very recently19,20,37,38,39,40,41. The above formulae are valid only for the single-label systems, whereas for the multi-label systems, whose emergence has become more frequent in system biology42,43 and system medicine22,44,45, a different set of metrics is needed as elaborated in Chou46.
Training and validation
In an unbalanced dataset (i.e., the number of AMPs and non-AMPs are not same), machine learning based classifier may produce results biased towards the major class47 (having large number of sequences than the other class). Therefore, number of sequences of the major class was kept same as the number of sequences present in the minor class to train the prediction model effectively. Here, sequences of the major class were randomly drawn from the available sequences. Since one random set from major class may not be adequate to judge the generalized predictive ability of the classifier, one thousand random samples (drawn without replacement from major class) were used. Further, in each sample (consists of AMPs and non-AMPs) a 10-fold cross validation48 procedure was employed to assess performance of the predictor. Furthermore, to assess the impact of size (number of sequences) of dataset, three datasets with different sample sizes were used (Table 3).
Table 3. Number of sequences present (sample size) in three different datasets used for prediction of antibacterial, antiviral and antifungal peptides.
| Dataset | Bacterial | Viral | Fungal | |||
|---|---|---|---|---|---|---|
| #ABP | #nonABP | #AVP | #nonAVP | #AFP | #nonAFP | |
| 1st set | 100 | 100 | 100 | 100 | 100 | 100 |
| 2nd set | 500 | 500 | 500 | 500 | 500 | 500 |
| 3rd set | 983 | 983 | 738 | 738 | 1383 | 1383 |
#ABP: Number of antibacterial peptides, #nonABP: Number of non-antibacterial peptides, #AVP: Number of antiviral peptides, #nonAVP: Number of non-antiviral peptides, #AFP: Number of antifungal peptides, #nonAFP: Number of non-antifungal peptides. In all the cases the instances were randomly drawn (without replacement) from the available number of instances present in the respective classes.
Comparison with existing methods
Performance of the proposed approach was compared with that of latest AMP prediction tools viz., CAMP3, iAMP-2L7, EFC-FCBF12, EFC + 307-FCBF12. The comparison was made by using the Xiao et al. benchmark dataset7 (http://www.jci-bioinfo.cn/iAMP/data.html). In this dataset, the training set contains 770 antibacterial peptides and 2405 non-AMPs and the test set contains 920 AMPs and 920 non-AMPs. The same datasets have been used by Veltri et al.12 to evaluate the performance of EFC-FCBF and EFC + 307-FCBF approaches. Further, performances of the methods were compared in terms of area under receiving operating characteristics curve49 (AUC-ROC), area under precision-recall curve50 (AUC-PR) and MCC. For a binary classifier, recall is same as Sn (as defined in equation-1) and precision is defined as 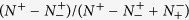 .
.
Development of prediction server
An online prediction server was also developed using hyper text markup language (HTML) and hypertext preprocessor (PHP), where a developed R-code was executed in the backend upon submission of peptide sequences in the FASTA format. The user can submit single or multiple sequences having only standard amino acid residues. This web server can be used to predict the probabilities with which a candidate peptide sequence can be classified into antiviral, antibacterial and antifungal categories.
Results
Performance analysis for predicting the antibacterial peptides
Three different sample sizes (100, 500, 983) were used for prediction of antibacterial peptides. Prediction accuracies for the sample size 983 are given in Table 4, whereas for the sample sizes 100 and 500 accuracies are provided in Supplementary Table S1. It is observed that the prediction accuracies are more precise (low standard error) for the sample size 983 as compared to that of sample sizes 100 and 500. Further, low prediction accuracies are observed with the compositional features alone, whereas 2–6%, ~1%, 2–4% and 4–5% increment in sensitivity, specificity, accuracy and MCC are observed respectively while the compositional, PHYC and STRL features are used together (Table 4 and Supplementary Table S1).
Table 4. Performance metrics of SVM in predicting antibacterial peptides for the sample size 983.
| Features | Performance metrics | |||
|---|---|---|---|---|
| Sn ± SE | Sp ± SE | Ac ± SE | MCC | |
| AAC + PAAC | 91.16 ± 0.71 | 93.41 ± 0.49 | 92.29 ± 0.36 | 0.85 ± 0.007 |
| AAC + NAAC | 91.29 ± 0.79 | 93.44 ± 0.49 | 92.37 ± 0.45 | 0.85 ± 0.009 |
| PAAC + NAAC | 91.29 ± 0.65 | 93.37 ± 0.51 | 92.33 ± 0.37 | 0.85 ± 0.007 |
| AAC + PAAC + NAAC | 91.35 ± 0.69 | 93.48 ± 0.52 | 92.41 ± 0.41 | 0.85 ± 0.008 |
| AAC + PAAC + PHYC + STRL | 93.81 ± 0.55 | 94.96 ± 0.40 | 94.39 ± 0.35 | 0.89 ± 0.007 |
| AAC + NAAC + PHYC + STRL | 93.87 ± 0.61 | 94.85 ± 0.39 | 94.36 ± 0.36 | 0.89 ± 0.007 |
| PAAC + NAAC + PHYC + STRL | 93.86 ± 0.65 | 94.91 ± 0.38 | 94.39 ± 0.35 | 0.89 ± 0.007 |
| AAC + PAAC + NAAC + PHYC + STRL | 93.85 ± 0.59 | 94.98 ± 0.36 | 94.69 ± 0.38 | 0.89 ± 0.008 |
SE: Standard Error.
Performance analysis for predicting the antiviral peptides
For the sample size 738, performance metrics of the proposed approach in predicting the antiviral peptides are given in Table 5, whereas for the sample sizes 100 and 500 accuracies are provided in Supplementary Table S2. It is seen that the prediction models based on the sample size 738 are more stable (low standard error) as compared to those based on sample sizes 100 and 500. Similar to antibacterial peptides, low prediction accuracies are also observed while only compositional features are used, whereas sensitivity, specificity, accuracy and MCC are observed to be increased by 1–3%, 1%, ~1% and 1–3% respectively while all the three features are accounted together (Table 5 and Supplementary Table S2). Besides, it is seen that the accuracies in predicting the antiviral peptides are low as compared to the antibacterial peptides.
Table 5. Performance metrics of SVM in predicting antiviral peptides for the sample size 738.
| Features | Performance metrics | |||
|---|---|---|---|---|
| Sn ± SE | Sp ± SE | Ac ± SE | MCC | |
| AAC + PAAC | 85.60 ± 0.56 | 90.72 ± 0.61 | 88.16 ± 0.38 | 0.76 ± 0.008 |
| AAC + NAAC | 85.42 ± 0.58 | 90.59 ± 0.69 | 88.00 ± 0.41 | 0.76 ± 0.008 |
| PAAC + NAAC | 85.47 ± 0.61 | 90.68 ± 0.59 | 88.08 ± 0.40 | 0.76 ± 0.008 |
| AAC + PAAC + NAAC | 85.49 ± 0.61 | 90.77 ± 0.62 | 88.13 ± 0.40 | 0.76 ± 0.008 |
| AAC + PAAC + PHYC + STRL | 88.67 ± 0.56 | 91.49 ± 0.68 | 90.08 ± 0.42 | 0.80 ± 0.008 |
| AAC + NAAC + PHYC + STRL | 88.46 ± 0.59 | 91.57 ± 0.64 | 90.01 ± 0.39 | 0.80 ± 0.008 |
| PAAC + NAAC + PHYC + STRL | 88.69 ± 0.59 | 91.49 ± 0.57 | 90.09 ± 0.34 | 0.80 ± 0.007 |
| AAC + PAAC + NAAC + PHYC + STRL | 88.65 ± 0.65 | 91.42 ± 0.67 | 90.08 ± 0.40 | 0.80 ± 0.008 |
SE: Standard Error.
Performance analysis for predicting the antifungal peptides
In case of antifungal peptides, prediction accuracies for the sample size 1383 are given in Table 6 and accuracies for the sample sizes 100 and 500 are provided in Supplementary Table S3. It is observed that the accuracies are more precise for the sample size 1383 as compared that of sample sizes 100 and 500. Similar to antibacterial and antiviral peptides, a decreasing trend in accuracies is observed for all the sample sizes, while PHYC and STRL features are not included in prediction. In particular, sensitivity, specificity, accuracy and MCC are increased by 1–2%, ~1%, ~1% and 1–2% respectively while compositional features are used along with the PHYC and STRL features (Table 6 & and Supplementary Table S3). Furthermore, the accuracies for predicting the antifungal peptides are found higher than that of antiviral peptides and lower than that of antibacterial peptides.
Table 6. Performance metrics of SVM in predicting antifungal peptides for the sample size 1383.
| Features | Performance metrics | |||
|---|---|---|---|---|
| Sn ± SE | Sp ± SE | Ac ± SE | MCC | |
| AAC + PAAC | 90.71 ± 0.29 | 93.14 ± 0.24 | 91.93 ± 0.16 | 0.84 ± 0.003 |
| AAC + NAAC | 90.82 ± 0.32 | 93.22 ± 0.25 | 92.02 ± 0.19 | 0.84 ± 0.004 |
| PAAC + NAAC | 90.76 ± 0.35 | 93.16 ± 0.25 | 91.96 ± 0.23 | 0.84 ± 0.005 |
| AAC + PAAC + NAAC | 90.77 ± 0.32 | 93.22 ± 0.21 | 92.00 ± 0.18 | 0.84 ± 0.004 |
| AAC + PAAC + PHYC + STRL | 92.33 ± 0.37 | 94.36 ± 0.22 | 93.35 ± 0.22 | 0.87 ± 0.004 |
| AAC + NAAC + PHYC + STRL | 92.32 ± 0.32 | 94.36 ± 0.23 | 93.34 ± 0.20 | 0.87 ± 0.004 |
| PAAC + NAAC + PHYC + STRL | 92.25 ± 0.29 | 94.38 ± 0.25 | 93.31 ± 0.17 | 0.87 ± 0.003 |
| AAC + PAAC + NAAC + PHYC + STRL | 92.30 ± 0.27 | 94.41 ± 0.25 | 93.35 ± 0.18 | 0.87 ± 0.004 |
SE: Standard Error.
Feature importance
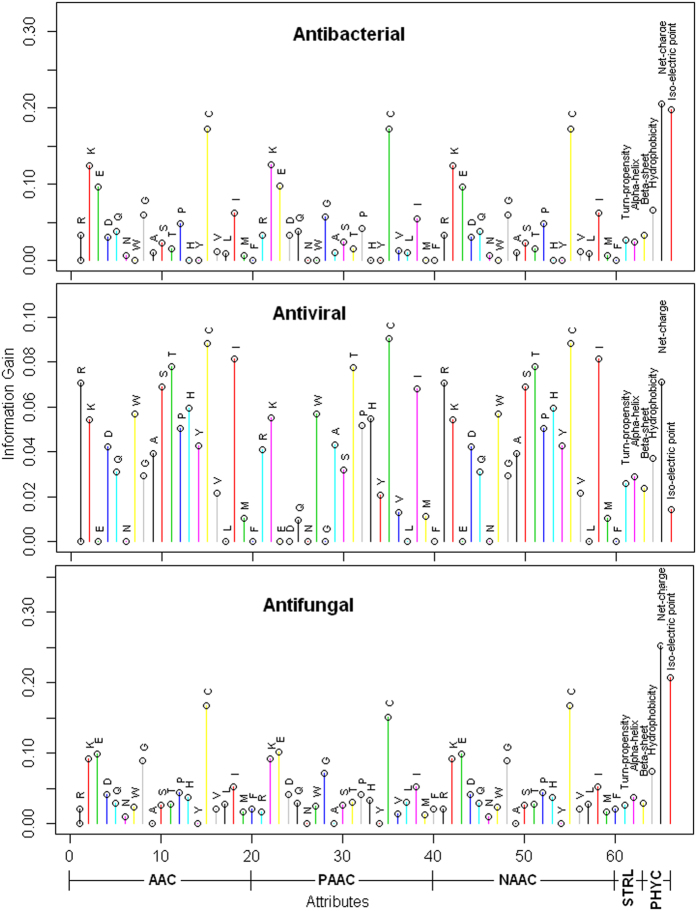
Based on top the model (AAC + PAAC + NAAC + STRL + PHYC), information gain for all the features was computed by using the largest sample size and are shown in Fig. 1. From the figure, it can be seen that the values of information gain are almost same for both the AAC and NAAC features. Further, it is observed that the information gain is highest for the feature net-charge followed by iso-electric point, while predicting the antibacterial and antifungal peptides. On the other hand, highest information gain is observed for the composition of amino acid C, while predicting the antiviral peptides. Furthermore, the STRL features are found less important (low information gain) than that of PHYC features and several compositional features. In particular, values of information gain are seen ≥0.05 for the amino acid compositions K, E. G, P, C and I in case of antibacterial and antifungal peptides, whereas it is ≥0.05 for the amino acid compositions R, K, W, S, T, P, H, C and I in case of antiviral peptides. Besides, values of information gain are observed close to zero for the amino acid compositions {N, W, V, L, M, F, H, Y}, {N, E, L, F} and {A, Y, N} in predicting the antibacterial, antiviral and antifungal peptides respectively. The values of information gain for other amino acids are observed to lie between 0 and 0.05.
Figure 1. Information gain for all the 66 features [AAC (20) + PAAC (20) + NAAC (20) + PHYC (3) + STRL (3)] in predicting antibacterial, antiviral and antifungal peptides.
Performance analysis for predicting the AMPs
For prediction of AMPs in general, positive dataset of AMPs was constructed by combining the antibacterial, antiviral and antifungal peptides, whereas negative dataset (non-AMP) was constructed by combining the non-antibacterial and non-antiviral peptides collected from AntiBP2 and AVPpred respectively. Besides, AMPs available in the LAMP were also included in the positive dataset. Finally, a dataset consisting of 5155 AMPs and 1384 non-AMPs was prepared. Similar to antibacterial, antiviral and antifungal, prediction of AMPs was also made with three different sample sizes i.e., 100, 500 and 1383. Moreover, the prediction was made only for the AAC + PAAC + PHYC + STRL and PAAC + NAAC + PHYC + STRL feature combinations, as little higher accuracies were obtained with these combinations in earlier predictions. The values of different performance metrics (averaged over 10-fold) are given in Table 7. From the table it is seen that the sensitivity, specificity and accuracy are > 90% for all the sample sizes. In addition, the performance of SVM with the above mentioned feature sets were also assessed by using Xiao benchmark training dataset, based on three different sample sizes (100, 500 and 769). The values of different performance metrics (averaged over 10-folds) are given in Table 8. From the table it is observed that the sensitivity, specificity and accuracy are ~94%, whereas for MCC it is ~88%. It is further seen that the prediction accuracies are more precise (low standard error) for the sample size 769.
Table 7. Accuracies of the proposed approach for the prediction of antimicrobial peptides.
| Feature | Sample size | Performance metrics |
|||
|---|---|---|---|---|---|
| Sn ± SE | Sp ± SE | Ac ± SE | MCC | ||
| AAC + PAAC + PHYC + STRL | 100 | 93.19 ± 2.32 | 95.13 ± 2.20 | 94.16 ± 1.56 | 0.88 ± 0.031 |
| 500 | 90.50 ± 1.30 | 93.68 ± 0.91 | 92.09 ± 0.73 | 0.84 ± 0.014 | |
| 1383 | 90.60 ± 0.66 | 92.98 ± 0.44 | 91.79 ± 0.39 | 0.84 ± 0.008 | |
| PAAC + NAAC + PHYC + STRL | 100 | 92.50 ± 2.62 | 95.39 ± 2.26 | 93.95 ± 1.66 | 0.88 ± 0.033 |
| 500 | 90.41 ± 1.31 | 93.77 ± 0.99 | 92.09 ± 0.75 | 0.84 ± 0.015 | |
| 1383 | 90.75 ± 0.82 | 92.94 ± 0.44 | 91.84 ± 0.40 | 0.84 ± 0.008 | |
SE: Standard Error.
Table 8. Prediction accuracies of the proposed approach in predicting the antimicrobial peptides using Xiao training dataset.
| Feature | Sample size | Performance metrics |
|||
|---|---|---|---|---|---|
| Sn ± SE | Sp ± SE | Ac ± SE | MCC | ||
| PAAC + NAAC + PHYC + STRL | 100 | 96.28 ± 1.76 | 95.58 ± 2.00 | 95.93 ± 1.30 | 0.91 ± 0.026 |
| 500 | 94.46 ± 0.72 | 93.83 ± 0.91 | 94.15 ± 0.53 | 0.88 ± 0.011 | |
| 769 | 94.10 ± 0.61 | 93.59 ± 0.81 | 93.84 ± 0.50 | 0.88 ± 0.010 | |
| AAC + NAAC + PHYC + STRL | 100 | 95.88 ± 1.95 | 95.57 ± 1.97 | 95.72 ± 1.35 | 0.91 ± 0.026 |
| 500 | 94.51 ± 0.81 | 93.73 ± 0.93 | 94.12 ± 0.62 | 0.88 ± 0.012 | |
| 769 | 94.08 ± 0.52 | 93.63 ± 0.83 | 93.85 ± 0.49 | 0.88 ± 0.009 | |
SE: Standard Error.
Comparative analysis
To further assess the predictive ability as compared to the existing approaches, performance of SVM with PAAC + NAAC + PHYC + STRL feature set (we call it iAMPpred) was compared with the performances of latest AMP prediction tools, by using Xiao benchmark dataset7. The results are given in Table 9. We observed that the accuracies of iAMPpred are much higher than that of all the four models of CAMP. In particular, it is observed that the AUC-ROC, AUC-PR and MCC values of iAMPpred are ~15%, ~20% and ~30% higher than all the four models of CAMP respectively. Though, iAMPpred and iAMP-2L performed at par in terms of MCC, AUC-ROC of iAMPpred is observed ~3% higher than that of iAMP-2L. Further, it is seen that the prediction accuracies (AUC-ROC, AUC-PR and MCC) of iAMPpred are also higher than that of EFC-FCBF and EFC + 307-FCBF (Table 9).
Table 9. Estimates of AUC-ROC, AUC-PR and MCC for different AMP prediction methods based on independent test dataset.
| Methods | AUC-ROC (%) | AUC-PR (%) | MCC |
|---|---|---|---|
| CAMP-SVM | 64 | 53 | 0.43 |
| CAMP-RF | 73 | 76 | 0.40 |
| CAMP-ANN | 80 | NA | 0.61 |
| CAMP-DA | 81 | 76 | 0.49 |
| iAMP-2L | 95 | NA | 0.90 |
| EFC-FCBF | 96 | 95 | 0.73 |
| EFC + 307-FCBF | 95 | 98 | 0.86 |
| iAMPpred | 98 | 99 | 0.91 |
Methods which provide continuous prediction values, we reported AUC-PR. Otherwise, “NA” is shown when methods only report a binary (AMP or nonAMP) prediction.
Comparison of iAMPpred with AntiBP2
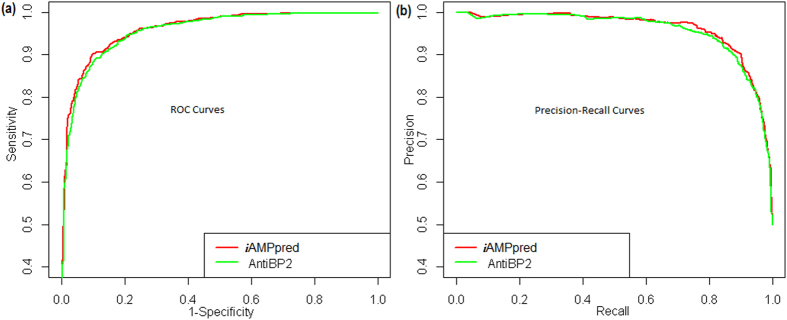
The performance of the iAMPpred was also compared with that of AntiBP2 (http://www.imtech.res.in/raghava/antibp2/) by considering the same dataset used in AntiBP2 that contains 999 antibacterial peptides and 999 non-antibacterial peptides. Since 5 sequences in the negative dataset were having non-standard amino acid residues they were excluded from the analysis, and the comparison was made using 999 positive and 994 negative sequences. The ROC and PR curves (averaged over 10-folds) are shown in Fig. 2. We observed that the areas covered under ROC and PR curves for iAMPpred are little higher than that of AntiBP2 respectively. This is in accordance with the results presented in Table 4 i.e., the values of performance metrics for PAAC + NAAC + PHYC + STRL feature set (feature set used in iAMPpred) are higher than that of AAC feature set (feature set used in AntiBP2).
Figure 2. ROC and PR curves of iAMPpred and AntiBP2 for the prediction of antibacterial peptides.
The performance of iAMPpred is found little higher than AntiBP2.
Comparison of iAMPpred with AVPpred
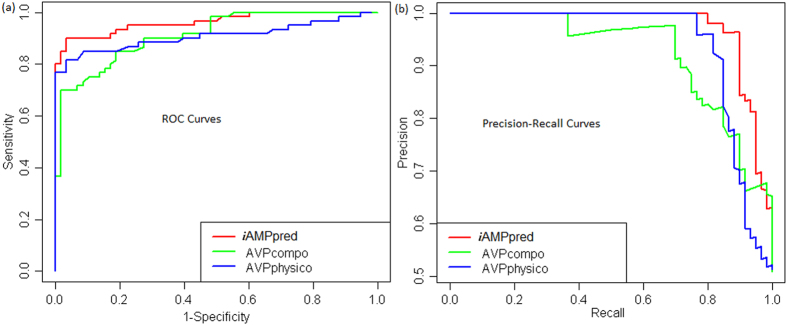
The performance of iAMPpred was further compared with that of AVPpred, by using training [T544(p) + 544(n)] and test [V60(p) + 60(n)] datasets available in AVPpred server (http://crdd.osdd.net/servers/avppred/collection.php?show=dataset). As the accuracies were reported to be higher for AVPcompo and AVPphysico models10, they were only considered for comparison. The ROC and PR curves for the test set are shown in Fig. 3. It is observed that the areas covered under both ROC and PR curves for iAMPpred are higher than that of both AVPcompo and AVPphysico models. Further, the AVPphysico model performed better than AVPcompo, which is similar to the observation made in Thakur et al.10.
Figure 3. ROC and PR curves of iAMPpred and AVPcompo, AVPphysico models of AVPpred for predicting the antiviral peptides.
The figure shows that the performance of iAMPpred is better than AVPcompo and AVPphysico models of AVPpred.
Performance analysis of ClassAMP
The performance of ClassAMP, which is meant for predicting the function type of AMPs, was also analyzed by using the Xiao testing dataset. Surprisingly, all the non-AMPs (920) were falsely predicted as AMPs (in any of the three classes) with more than 0.6 probabilities in case of SVM, whereas 915 were falsely predicted as AMPs while RF was used. On the other hand, only 34 and 8 AMPs were falsely predicted as non-AMPs in SVM and RF respectively. This implies that the ClassAMP might be biased towards predicting AMPs. Besides, the accuracies were found higher in iAMPpred as compared to that of ClassAMP in predicting the propensity of a peptide sequence as antibacterial, antiviral or antifungal peptides.
Analysis of organism-specific AMP prediction
Performance of iAMPpred was also assessed for prediction of AMPs specific to six different source organisms viz., plants, bacteria, cattle, insects, fishes and amphibians. AMPs for these organisms were collected from APD3 database (1348 AMPs from amphibians, 47 from cattle, 137 from fishes, 341 from insects and 216 from bacteria). The 920 non-AMPs of Xiao testing dataset was considered as the negative dataset against each of the positive datasets. The prediction accuracies in terms of different performance metrics (averaged over 10-fold cross validation) are given in Table 10. Highest accuracy in terms of MCC are observed for amphibians (0.97) followed by cattle (0.94), plants (0.93) and insects (0.92). Interestingly, accuracies for all the organisms are observed >96%, which suggests that the iAMPpred is also efficient in predicting the organism-specific AMPs.
Table 10. Performance metrics for iAMPpred in predicting organism-specific AMPs.
| Source Organism | Sn | Sp | Ac | MCC |
|---|---|---|---|---|
| Amphibian | 98.81 | 98.26 | 98.58 | 0.97 |
| Bacteria | 86.19 | 98.91 | 96.55 | 0.88 |
| Plant | 93.70 | 99.02 | 97.82 | 0.94 |
| Fish | 81.54 | 99.46 | 97.24 | 0.87 |
| Insect | 91.69 | 99.46 | 96.90 | 0.92 |
| Cattle | 98.33 | 99.89 | 98.44 | 0.94 |
Online prediction server: iAMPpred
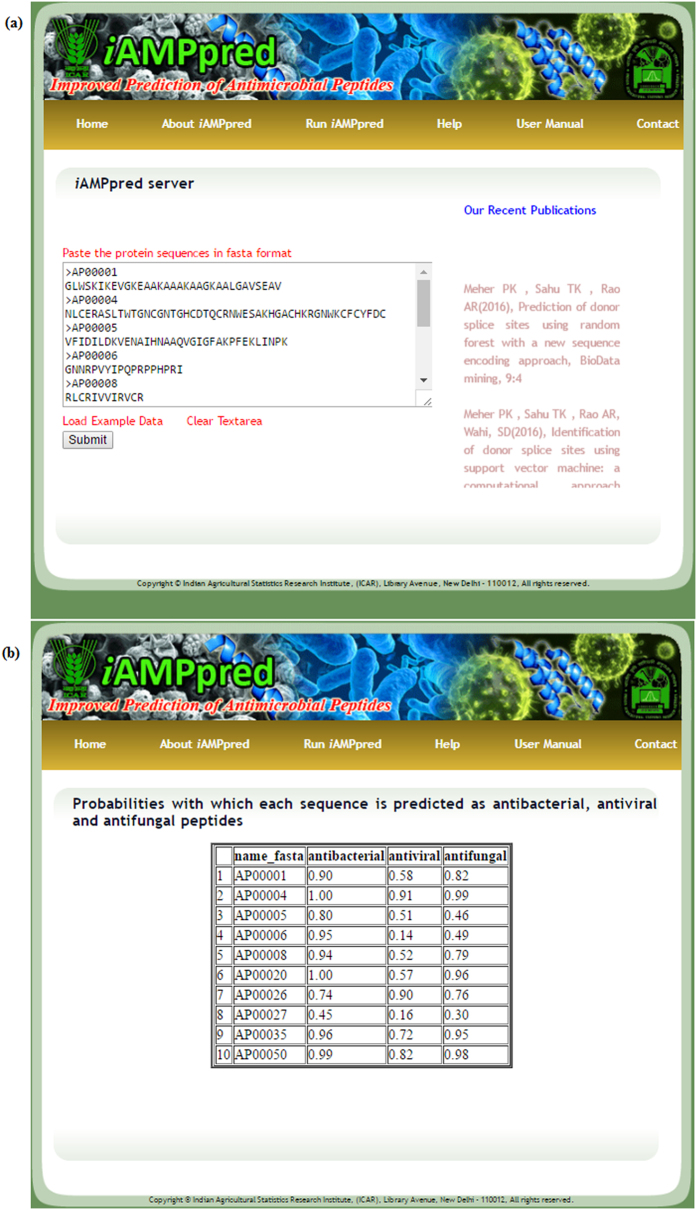
An online prediction server “iAMPpred” has been developed to predict the propensity of a peptide sequence as antibacterial, antiviral and antifungal peptides. Snapshots of the web pages showing the execution of iAMPpred for an example dataset along with the results are shown in Fig. 4. For user guidance with regard to feature generation, prediction method and input-output, links have been provided in the main menu. The sequences with probabilities of being antiviral, antibacterial and antifungal peptides are displayed in the result page. For reproducible research, links to download the trained datasets (http://cabgrid.res.in:8080/amppred/about.html) are also provided. The prediction server is freely accessible at http://cabgrid.res.in:8080/amppred.
Figure 4.
Snapshots of (a) server page of iAMPpred and (b) result page after execution of the program with an example dataset. The results are displayed in a tabular format showing the sequence identifier and the probabilities with which the sequences are predicted as antibacterial, antiviral and antifungal peptides.
Discussion
AMPs are natural antibiotics gaining attention as an alternative to the chemical antibiotics. Identification and designing of AMPs via wet lab experiments may be resource intensive. Thus, computational identification will supplement to the designing of new antimicrobial agents. This paper presents a SVM-based computational approach that can be used for predicting the effective AMPs with higher accuracy as compared to several existing approaches.
In this investigation, combinations of compositional, PHYC and STRL features were used to map the peptide sequences onto numeric feature vectors that were subsequently used as input in SVM for prediction of AMPs. Though, AAC9,10 and PAAC7,26 features have been used in earlier studies, the NAAC feature is used for the first time in our study for AMP prediction. Moreover, α-helix, β-sheet and turn propensity features were also used as they were reported to play an important role in discriminating the AMPs from non-AMPs30. Furthermore, Most of the earlier methods were evaluated based on a single dataset of AMPs, collected either from CAMP or APD/APD2 database. On the other hand, the sequences of AMPs used in this study were thought to be more representative as they were collected from several AMP databases. From information gain analysis, net-charge was found to be the most important feature followed by iso-electric point in predicting the antibacterial and antifungal peptides. On the other hand, the composition of amino acid C was observed to play the most important role in predicting the antiviral peptides. Further, the PHYC features were found to play a more important role than STRL features in predicting the antibacterial, antiviral and antifungal peptides. As far as the compositional features are concerned, amino acids K, P, C and I were found more important as compared to others in predicting the AMPs. On the other hand, the amino acid compositions {N, W, V, L, M, F, H, Y}, {N, E, L, F} and {A, Y, N} were found less important in predicting the antibacterial, antiviral and antifungal peptides respectively.
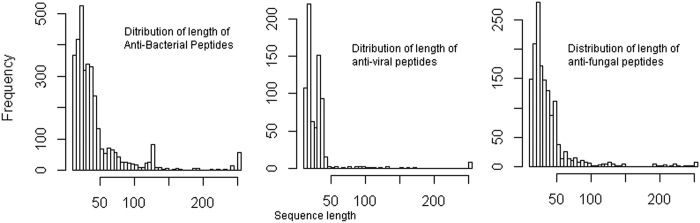
The prediction of antibacterial, antiviral and antifungal peptides was made by using three different sample sizes. Prediction accuracies were found to be more precise for the large sample sizes as compared to that of small sample sizes. Further, accuracies for predicting the antibacterial and antifungal peptides were found higher than that of antiviral peptides. This might be due to the longer sequence length (10–100 amino acids) of antibacterial and antifungal peptides and shorter sequence length (10–50 amino acids) of antiviral peptides (Fig. 5). Besides, PHYC and STRL determinants were found to play a more important role in the prediction of antibacterial peptides as compared to antiviral and antifungal peptides. Since the prediction accuracies (Sn, Sp, ACC) were also found to be higher (>90%) for prediction of AMPs in general (Table 7), the iAMPpred is believed to supplement the existing tools for predicting the antibacterial, antiviral and antifungal peptides independently as well as predicting the AMPs in general.
Figure 5. Distribution of length of the sequences in antibacterial, antiviral and antifungal peptides.
The antibacterial and antifungal peptides are > 50 amino acids long, whereas most of the antiviral peptides are < 50 amino acids long.
The performance of iAMPpred was also compared with that of several state-of-art AMPs prediction methods by using Xiao benchmark dataset. The iAMPpred was found to achieve higher accuracies than all the four models of CAMP, which might be due to the use of AAC and PHYC features in CAMP without STRL features. Moreover, the feature extraction in CAMP is based on the reduced alphabet due to which the information might be lost. The features employed in iAMP-2L is the correlated PAAC that constitutes a subset of iAMPpred feature set and this could be one of the reasons for the equivalent performance of iAMP-2L with iAMPpred. In EFC-FCBF, the evolutionary feature set was constructed and 40 informative features were selected by fast correlation based feature selection (FCBF)51 technique, which were then used as input in logistic classifier. The AUC-ROC and AUC-PR of EFC-FCBF were found closer to that of iAMPpred, which implies that the evolutionary features are also important in predicting AMPs. The EFC + 307-FCBF is an extension of EFC-FCBF, where 307 more PHYC features were used to train and test the model. Though the accuracy of this model was found at par with the iAMPpred, the number of features used in EFC + 307-FCBF (i.e., 347) is much larger than the number of features considered in iAMPpred (i.e., 46).
The performance of iAMPpred was also compared with the specific tools such as AntiBP2 and AVPpred meant for predicting antibacterial and antiviral peptides respectively. The accuracies of iAMPpred was found little higher than that of AntiBP2 but much higher than that of AVPpred. One of the possible reasons for this may be the non-consideration of NAAC, PAAC, STRL features in both AntiBP2 and AVPpred. The accuracy of iAMPpred was also found higher as compared to that of ClassAMP with Xiao testing dataset. Besides, iAMPpred achieved higher accuracies for organism-specific prediction of AMPs. The developed web server iAMPpred is believed to supplement the existing tools/techniques in predicting the AMPs.
Additional Information
How to cite this article: Meher, P. K. et al. Predicting antimicrobial peptides with improved accuracy by incorporating the compositional, physico-chemical and structural features into Chou’s general PseAAC. Sci. Rep. 7, 42362; doi: 10.1038/srep42362 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
The grant (Agril.Edn.4–1/2013-A&P dated 11.11.2014) received from Indian Council of Agriculture Research (ICAR) for Centre for Agricultural Bioinformatics (CABin) scheme of Indian Agricultural Statistics Research Institute (IASRI) is duly acknowledged. The authors also acknowledge the Division of Statistical Genetics for providing necessary support during the study. We are also highly thankful to the anonymous reviewers for their excellent suggestions.
Footnotes
The authors declare no competing financial interests.
Author Contributions Conceived and designed the study: P.K.M., A.R.R.; Collected and analyzed the sequence dataset: T.K.S., V.S.; Developed the prediction approach: P.K.M.; Developed the web server: T.K.S., P.K.M.; Drafted the manuscript: P.K.M., V.S., T.K.S. Corrected and refined the manuscript: P.K.M., T.K.S., A.R.R.; All authors read and approved the final manuscript.
References
- Lata S., Sharma B. K. & Raghava G. P. S. Analysis and prediction of antibacterial peptides. BMC Bioinform. 8, 263 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porto W. F., Souza V. A., Nolasco D. O. & Franco O. L. In silico identification of novel hevein-like peptide precursors. Peptides. 38, 127–136 (2012). [DOI] [PubMed] [Google Scholar]
- Yeaman M. R. & Yount N. Y. Mechanisms of antimicrobial peptide action and resistance. Pharmacol. 55, 27–55 (2003). [DOI] [PubMed] [Google Scholar]
- Brogden K. A. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 3, 238–250 (2005). [DOI] [PubMed] [Google Scholar]
- Thomas S., Karnik S., Barai R. S., Jayaraman V. K. & Thomas S. I. CAMP: a useful resource for research on antimicrobial peptides. Nucl. Acids. Res. 38 (Suppl 1), D774–D780 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marr A. K., Gooderham W. J. & Hancock R. E. W. Antibacterial peptides for therapeutic use: obstacles and realistic outlook. Curr. Opin. Pharmacol. 6, 468–472 (2006). [DOI] [PubMed] [Google Scholar]
- Xiao X., Wang P., Lin W. Z., Jia J. H. & Chou K. C. iAMP-2L: A two- level multi-labe classifier for identifying antimicrobial peptides and their functional types. Anal. Biochem. 436(2), 168–177 (2013). [DOI] [PubMed] [Google Scholar]
- Fjell C. D., Hancock R. E. & Cherkasov A. AMPer: a database and an automated discovery tool for antimicrobial peptides. Bioinform. 23(9), 1148–1155 (2007). [DOI] [PubMed] [Google Scholar]
- Lata S., Mishra N. K. & Raghava G. P. S. AntiBP2: improved version of antibacterial peptide prediction. BMC Bioinform. 11 (Suppl 1), S19 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur N., Qureshi A. & Kumar M. AVPpred: collection and prediction of highly effective antiviral peptides. Nucl. Acids. Res. 40, W199–204 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph S., Karnik S., Nilawe P., Jayaraman V. K. & Idicula-Thomas S. ClassAMP: a prediction tool for classification of antimicrobial peptides. IEEE/ACM Trans. Comput. Biol. Bioinform. 9(5), 1535–1538 (2012). [DOI] [PubMed] [Google Scholar]
- Veltri D., Shehu A. & Kamath U. Improving recognition of antimicrobial peptides and target selectivity through machine learning and genetic programming. IEEE/ACM Trans. Comput. Biol. Bioinform. x(x) (2015). [DOI] [PubMed] [Google Scholar]
- Haykin S. Neural Networks: a comprehensive foundation. Prentice Hall: Upper Saddle River, 1999. [Google Scholar]
- Vapnik V. N. Statistical learning theory. Wiley & Sons, New York, USA, 1998. [Google Scholar]
- Breiman L. Random Forests. Mach. Learn. 45(1), 5–32 (2001). [Google Scholar]
- Chou K. C. Prediction of protein cellular attributes using pseudo amino acid composition. Proteins. 43, 246–255 (2001). [DOI] [PubMed] [Google Scholar]
- Chen W., Ding H. & Feng P. iACP: a sequence-based tool for identifying anticancer peptides. Oncotarget. 7, 16895–16909 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia J., Liu Z. & Xiao X. pSuc-Lys: Predict lysine succinylation sites in proteins with PseAAC and ensemble random forest approach. J. Theor. Biol. 394, 223–230 (2016). [DOI] [PubMed] [Google Scholar]
- Jia J., Liu Z. & Xiao X. iCar-PseCp: identify carbonylation sites in proteins by Monto Carlo sampling and incorporating sequence coupled effects into general PseAAC. Oncotarget 7, 34558–34570 (2016a). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B. & Long R. iDHS-EL: Identifying DNase I hypersensi-tivesites by fusing three different modes of pseudo nucleotide composition into an en-semble learning framework. Bioinform. 32, 2411–2418 (2016). [DOI] [PubMed] [Google Scholar]
- Liu Z., Xiao X. & Yu D. J. pRNAm-PC: Predicting N-methyl-adenosine sites in RNA sequences via physical-chemical properties. Anal. Biochem. 497, 60–67 (2016). [DOI] [PubMed] [Google Scholar]
- Qiu W. R., Sun B. Q. & Xiao X. iPTM-mLys: identifying multiple lysine PTM sites and their different types. Bioinform. 32, 3116–3123 (2016). [DOI] [PubMed] [Google Scholar]
- Chou K. C. Some remarks on protein attribute prediction and pseudo amino acid composition (50th Anniversary Year Review). J. Theor. Biol. 273, 236–247 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Li X. & Wang Z. APD3: the antimicrobial peptide database as a tool for research and education. Nucl. Acids Res. 44(D1), D1087–1093 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Wu H., Lu H., Li G. & Huang Q. LAMP: A database linking antimicrobial peptides. PLoS ONE. 8(6), e66557 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, et al. Prediction of antimicrobial peptides based on sequence alignment and feature selection methods. PLoS ONE. 6(4), e18476 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio D., Rondon-Villarreal P. & Torres R. Peptides: A package for data mining of antimicrobial peptides. The R Journal. 7(1), 4–14 (2015). [Google Scholar]
- R Development Core Team. R: A language and environment for statistical computing. R foundation for statistical computing, Vienna, Austria, 2012. ISBN 3-900051-07-0, http://www.R-project.org/. [Google Scholar]
- Fernandez-Escamilla A. M., Rousseau F., Schymkowitz J. & Serrano L. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat. Biotech. 22(10), 1302–1306 (2004). [DOI] [PubMed] [Google Scholar]
- Torrent, et al. AMPA: An automated web server for prediction of protein antimicrobial regions. Bioinform. 28(1), 130–131 (2011). [DOI] [PubMed] [Google Scholar]
- Hornik K., Buchta C. & Zeileis A. Open-source machine learning: R meets Weka. Comput. Stat. 24(2), 225–232 (2009). [Google Scholar]
- Noble W. S. What is a support vector machine? Nat. Biotech. 24(12), 1565–1567 (2006). [DOI] [PubMed] [Google Scholar]
- Ng N. X., Rosdi B. A. & Shahrudin S. Prediction of antimicrobial peptides based on sequence alignment and support vector machine-pairwise algorithm utilizing LZ-complexity. BioMed Res. Int. 10.1155/2015/212715 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, et al. e1071: Misc functions of the Department of Statistics (e1071), TU Wien, R package version 1.6–1 (2012). [Google Scholar]
- Chen W., Feng P. M. & Lin H. iRSpot-PseDNC: identify recombination spots with pseudo dinucleotide composition. Nucl. Acids Res. 41, e68 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou K. C. Review: Prediction of protein signal sequences. Curr. Protein Pept. Sci. 3, 615–622 (2002). [DOI] [PubMed] [Google Scholar]
- Jia J., Zhang L. & Liu Z. pSumo-CD: Predicting sumoylation sites in proteins with covariance discriminant algorithm by incorporating sequence-coupled effects into general PseAAC. Bioinform. 32, 3133–3141 (2016b). [DOI] [PubMed] [Google Scholar]
- Liu B., Zhang D. & Xu R. Combining evolutionary information extracted from frequency profiles with sequence-based kernels for protein remote homology detection. Bioinform. 30, 472–479 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H., Deng E. Z. & Ding H. iPro54-PseKNC: a sequence-based predictor for identifying sigma-54 promoters in prokaryote with pseudo k-tuple nucleotide composition. Nucl. Acids Res. 42, 12961–12972 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S. H., Deng E. Z. & Xu L. Q. iNuc-PseKNC: a sequence-based predictor for predicting nucleosome positioning in genomes with pseudo k-tuple nucleotide composition. Bioinform. 30, 1522–1529 (2014). [DOI] [PubMed] [Google Scholar]
- Liu B., Fang L. & Long R. iEnhancer-2L: a two-layer predictor for identifying enhancers and their strength by pseudo k-tuple nucleotide composition. Bioinform. 32, 362–369 (2016a). [DOI] [PubMed] [Google Scholar]
- Lin W. Z., Fang J. A. & Xiao X. iLoc-Animal: A multi-label learning classifier for predicting subcellular localization of animal proteins. Mol. Biosyst. 9, 634–644 (2013). [DOI] [PubMed] [Google Scholar]
- Wu Z. C. & Xiao X. iLoc-Plant: a multi-label classifier for predicting the subcellular localization of plant proteins with both single and multiple sites. Mol. Biosyst. 7, 3287–3297 (2011). [DOI] [PubMed] [Google Scholar]
- Xiao X., Wang P. & Lin W. Z. iAMP-2L: A two-level multi-label classifier for identifying antimicrobial peptides and their functional types. Anal. Biochem. 436, 168–177 (2013). [DOI] [PubMed] [Google Scholar]
- Cheng X., Zhao S. G. & Xiao X. iATC-mISF: a multi-label classifier for predicting the classes of anatomical therapeutic chemicals. Bioinform. 2016, doi: 10.1093/bioinformatics/btw644. [DOI] [PubMed] [Google Scholar]
- Chou K. C. Some remarks on predicting multi-label attributes in molecular biosystems. Mol. Biosyst. 9, 1092–1100 (2013). [DOI] [PubMed] [Google Scholar]
- Meher P. K., Sahu T. K., Rao A. R. & Wahi S. D. A statistical approach for 5′ splice site prediction using short sequence motif and without encoding sequence data. BMC Bioinform. 15, 362 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson J., Salzberg S. & Fasman K. H. Finding genes in DNA with a hidden Markov model. J.Comput. Biol. 4, 127–141 (1992). [DOI] [PubMed] [Google Scholar]
- Fawcett T. Using rule sets to maximize ROC performance., Proc. Int’l Conf. Data Mining. 131–138 (2006). [Google Scholar]
- Davis J. & Goadrich M. The relationship between Precision-Recall and ROC curves. In: (ML’06): Proceedings of the 23rd international conference on machine learning. New York, USA, pp. 233–240 (2006). [Google Scholar]
- Yu L. & Liu H. Feature selection for high-dimensional data: A fast correlation-based filter solution. In ICML. 3, 856–863 (2003). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.