Abstract
Several studies have highlighted the changes in the gene expression due to the hypoxia response in fishes, but the systematic organization of the information and the analytical platform for such genes are lacking. In the present study, an attempt was made to develop a database of hypoxia responsive genes in fishes (HRGFish), integrated with analytical tools, using LAMPP technology. Genes reported in hypoxia response for fishes were compiled through literature survey and the database presently covers 818 gene sequences and 35 gene types from 38 fishes. The upstream fragments (3,000 bp), covered in this database, enables to compute CG dinucleotides frequencies, motif finding of the hypoxia response element, identification of CpG island and mapping with the reference promoter of zebrafish. The database also includes functional annotation of genes and provides tools for analyzing sequences and designing primers for selected gene fragments. This may be the first database on the hypoxia response genes in fishes that provides a workbench to the scientific community involved in studying the evolution and ecological adaptation of the fish species in relation to hypoxia.
Hypoxia, a condition where the deficient amount of oxygen reaches the body tissues, is a common abiotic stress in fishes that generally occurs due to the increase in the water temperature, presence of organic pollutants as well as aquatic flora, which consume ambient oxygen, and lead to rapid reduction of the oxygen tension in the water1. Hypoxia has a direct effect on the physiological and cellular function of fishes2. The rate of diffusion of the atmospheric oxygen in water becomes worse when the water surface is covered by ice, thick vegetation or coral reefs3,4. The consistent degradation of oxygen level in the tissues affects the regulation and expression of oxygen dependent genes. Further, the studies on the hypoxia tolerance have shown the imperative role in the rate of the oxygen diffusion and its concentration in tissue in the evolution and ecological context of the aquatic organisms, including fish, that force them for environmental adaptation in variety of ways5. A kind of environmental adaptation is considered to be the variability in hemoglobin structure and function to transport more oxygen to various tissues of the body6,7,8. An adaptation of alternative metabolic pathways of anaerobic energy production was also reported in the Carassius species of the family Cyprinidae during extreme hypoxia in which fermentation of the glucose produces ethanol and carbon dioxide9,10,11. Zebrafish undergoes evolutionary adaptation during embryogenesis and able to grow with enhanced hypoxia tolerance in the subsequent developmental stages12. The storage of large amounts of glycogen helps the organism to adapt for the long term survival in the hypoxic condition13. In fishes, evolutionary adaptation is not uniform and physiology of several species significantly alters under hypoxic conditions14,15,16,17.
Several approaches were conceded for the identification of the hypoxia responsive genes in fishes and their altered expression levels. Reports also revealed that the nuclear transcriptional factor is responsible for inducing gene expression in the hypoxic condition at the site, which is subsequently required for transcriptional activation of human EPO gene enhancer18. The discovery of hypoxia inducible factor (HIF) and its involvement in gene regulation during the hypoxic condition has been observed in many genes encoding EPO, VEGF glucose transporter-1 (GLUT-1), heme oxygenase (HO), transferrin, inducible nitric oxide synthase (iNOS), and the glycolytic enzyme aldolase A, enolase 1, lactate dehydrogenase A, phosphofructokinase L and phosphoglycerate kinase I19. HIF is a highly conserved heterodimer composed of an alpha and a beta subunit of protein chain that responds to the changes of the oxygen tension in the cellular environment. Three hypoxia inducible factors, each with two isoforms, viz. HIF-1α, HIF-1β, HIF-2α, HIF-2β, HIF-3α and HIF-3β, have been reported20. Rainbow trout was the leading fish in which expression and function of HIF-1α were studied21. The HIF-1 binds with a cis-regulatory DNA sequence, called hypoxia response element (HRE), in several genes reported for enhancing mRNA formation during the hypoxia22. Functional HREs are being reported in several mammalian genes23, while few studies have proposed functional HREs in fishes24,25.
Many genes characterized in response to the hypoxia in fishes are available in the public domain. Prior to this study, hardly any effort was made to compile and organize such genes in the form of a centralized digital resource for the fishes. To meet this, a database for hypoxia responsive genes in fish (HRGFish) was developed to collect, organize and manage the genes and gene information. HRGFish is a platform for analyzing genes and gene sets for evolutionary and diagnostic studies. The different sets of reported genes for hypoxia response were identified by screening the published research articles, and information including Gene Ontology (GO) terms were taken from on-line published sources. Linux Apache MySQL PHP and Perl (LAMPP) technology was used to create the database, design and connect the data-driven web pages integrated with browse, analysis and view functionalities. In silico approaches were applied to identify the upstream sequences, promoter regions and different sets of orthologous promoters from the genes in the database. An analysis of upstream region can be useful in understanding the gene regulation, regulatory elements, hypoxia response element (HRE) orientation, tolerance and the susceptibility of the species against hypoxia. The different types of analytical tools were included in the web interface of the database for analyzing the gene sequences and designing primer of the selected region. It is expected that the HRGFish resource might fill the knowledge gap in understanding the cellular adaptation in response to the hypoxia and the role of the intra- as well as inter-specific genetic variation in hypoxia tolerance.
Results
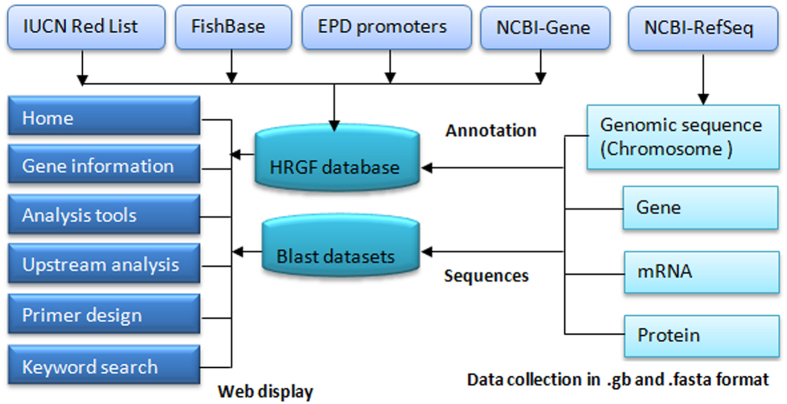
HRGFish presently covers a total of 818 gene sequences for 35 types of genes belonging to 38 fish species and is now freely accessible at URL: http://mail.nbfgr.res.in/HRGFish. The web based workbench of HRGFish includes menu items for retrieving, browsing and analyzing gene information and sequences. Tools for upstream and downstream analysis, primer design, sequence similarity and keyword searches have been integrated that enhance the utility and visibility of the database (Fig. 1).
Figure 1. Architecture and data flow diagram of HRGFish.
Browsing gene information
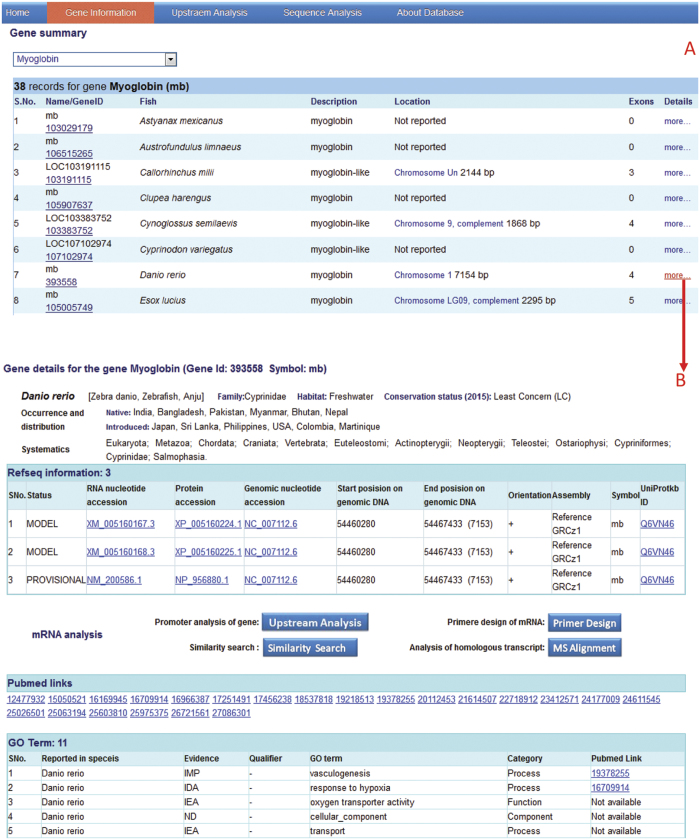
The web interface of HRGFish contains ‘Gene Information’ menu item that provides the ability to view the list of genes and the detailed information about the gene of interest. The selection of a gene from the list of genes displays all the orthologous genes in a table, presenting the gene name, description, chromosomal information, genomic accession, gene location, number of exons reported in the gene and the species name (Fig. 2A). Each row in the table of orthologous genes contains a hyperlink ‘more’ in the last column. A click event on this link opens up the ‘genedetails’ page. The ‘genedetails’ page provides more information about the gene (chromosome, mRNA, protein, GO and PubMed) and species (family, common name, synonyms, habitat, conservation status, occurrence, distribution and taxonomy) in different sections. The RefSeq gene information section presents IDs for mRNA variants, protein, genomic DNA, uniprot and localized region of genes and mRNAs in genomic DNA. The Pubmed provides a table of publications related to gene and the gene ontology section defines the go term of a gene like location, process and function of the gene product. All IDs presented on this page have links to their respective databases for cross validation of information. This page includes tools for different activities like: i) designing primer that uses gene/mRNA as a query of the selected region of the gene, ii) analyzing orthologous promoters of the gene, similarity analysis of the gene sequence of interest with sequences of the database, and iii) multiple sequence alignment analysis to observe the conserved regions across the species. The primer designing facility for selected exonic regions has been included for experimental validation by PCR (Fig. 2B).
Figure 2. Web interface of HRGFish database:
(A) display orthologous gene in fish species, and (B) details information about the gene and fish species.
Knowing about the database
The ‘About Database’ menu item in the web interface provides a graphical and textual description of tools included in the interfaces. This menu item also gives detailed information about the database and the tools to be used for browsing, retrieving and analyzing the information from the database.
Keyword search
The keyword search accepts keywords like species name, gene name, gene symbol, accession number, transcript ID and PubMed ID as input for retrieving records from the database. Different views for the different keywords have been prepared to display the information.
Upstream information
The ‘Upstream Analysis’ menu item included a drop down option ‘Pattern analysis’ in the web interface, which provides the ability to compute CG dinucleotide frequency, HRE motifs and observation of CpG islands in the upstream sequence of the selected gene. The sequence string and distribution of different motifs, like CG and HRE, are presented in graphical form, while CpG islands observation draws a graphical plot. The similarity search analysis of the upstream sequences with available reference promoters of the selected gene provides the common conserved patterns within the set of the orthologous promoters.
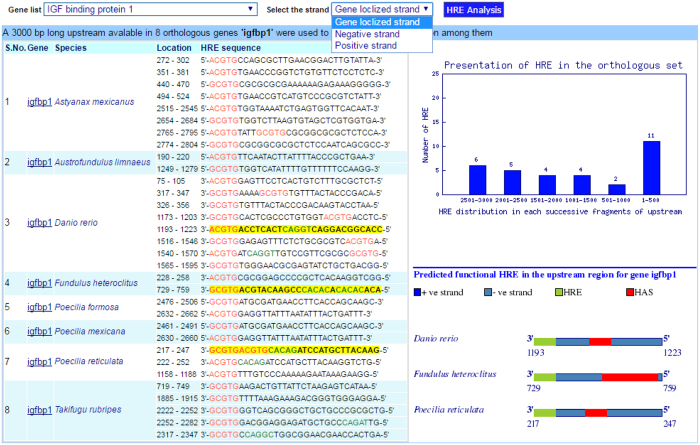
The ‘Upstream Analysis’ menu item also contains another drop-down option ‘HRE analysis’ which provides a separate interface for HRE analysis of all the orthologous genes. This module mines all HREs from the set of the orthologous upstream of a selected gene and the selected orientation and presents: (i) a table of all HREs and yellow highlighted predicted functional HRE, (ii) a graph of HRE distribution for each 500 nt long consecutive fragment, and (iii) a graphical plot of predicted functional HREs. This module facilitates the mining of canonical HRE from the sequence, including forward and reverse orientation in the set of the upstream of a selected gene (Fig. 3).
Figure 3. The HRE analysis page: Presenting all HRE in the orthologous set of IGFBP1 gene.
Bar diagram presents the frequency of HRE in each 500 nt long consecutive fragments of upstream, while graphical plots display the functional HRE in gene localized on the strand.
Primer design
The ‘Sequence Analysis’ menu item includes a submenu item ‘Primer design’ that uses a query interface. The query interface accepts input from the user and the result of query provides the details about the designed primers in a graphical format for both forward and reverse primer sets with a suitable length of flanking region. Additionally, the page presents information about the query specimen, selected subsequence and multiple primers with Tm values, GC content, start to end position and PCR product size. These designed primer sequences could be useful for analyzing and obtaining homologous promoters and exons of a gene across the species through PCR.
Sequence alignment
The ‘Sequence Analysis’ menu item in the HRGFish facilitates many options for analyzing the query sequences against the HRGFish database. The pairwise sequence similarity searching can be done through ‘Similarity search’ listed under ‘Sequence Analysis’ menu item and from the ‘genedetails’ page also. There are two methods to perform the alignment. One method is that the user selects any internal gene, mRNA and protein sequence as an internal query sequence to perform alignment with ‘HRGFish’ database. Another method is pairwise alignment that can be performed using the query sequence in FASTA format along with input parameters. Four input types are required for performing the alignment: (1) the selection of a target dataset as listed in Table 1, (2) the selection of a dataset-compatible Blast algorithm, (3) the selection of a sequence (DNA or protein) and (4) an input sequence in FASTA format. User input parameters such as maximum alignment number, identity percent cut off and word size can also be provided using Blastn, whereas Blastp and Blastx program use only the default parameters. The acceptance of these inputs through the submit button performs an alignment with the target sequences. If parameters are not selected correctly, the program terminates by giving a warning message. Thus, HRGFish platform provides the ability to the user for analyzing gene sequences, segments of a sequence, translated products and annotate genes in the newly reported fish genome.
Table 1. Different Blast-compatible datasets.
| Dataset name | Description | No. of sequences | Compatibility |
|---|---|---|---|
| upstrmdb | Upstream sequences of genes | 799 | Blastn |
| genedb | Genes sequences | 799 | Blastn |
| protdb | Protein with isoformes sequences | 1413 | Blastx, Blastp |
| mrnadb | Exonic sequences of mRNA and its varients | 1424 | Blastn |
| genomicmrnadb | Genomic mRNA sequences along with varients | 1414 | Blastn |
Apart from the different blast program for homologous sequence alignments, HRGFish also includes other useful programs under alignment analysis for specific search. The implemented program ‘Spidey’ provides the ability to the user to align mRNA sequence with the genomic sequence and supports to identify the transcribed regions and annotation information of a gene in the genomic sequences. The ORF finder program computes open reading frames for the user provided query mRNA as input. This program helps in the gene annotation process. In this way, these programs provide the ability to the user for gene identification and annotation obtained from exon-intron orientation in related species. The multiple sequence alignment program ‘ClustalW’ was implemented in HRGFish for analysis of conserved patterns among homologous mRNA sequences and obtaining orthologous clusters within a set of selected sequences.
Discussion
A number of hypoxia response genes have been characterized in fishes, but the attempts were lacking to compile and organize the information on a platform for collectively viewing and analyzing these data2. Hence, Hypoxia Responsive Genes in Fish (HRGFish) was developed to organize and manage the genes and their related information. The gene, upstream region and mRNA sequences were collected directly from the corresponding genomic sequences and reverse complement parsing methods were applied to the genes reported on the negative strand. Some analytical tools were implemented to analyze the gene, mRNA and the upstream sequences.
Orthology based promoter prediction and recognition of regulatory site in the upstream is an effective approach for promoter annotation26. The same approach was also applied here to display the orthologous set of promoter sequences across related species using reference promoter of the same gene of zebrafish. The localization of the binding sites for transcription factors provides support for promoter recognition in eukaryotes27. CpG islands are short interspersed DNA fragments enriched with dinucleotide motifs CG and found in or near the promoters of a gene and form transcription factor binding sites (TFBSs)28. In the present study, the observation of conserved motifs and analysis of CpG islands in the upstream region confirms the presence of the regulatory elements, like enhancer and promoter of a gene. In this way, the CpG island identification in the upstream region provides a support to identify the presence of TFBSs and annotation of the promoter regions.
The gene involved in the hypoxia response contains HRE elements for HIF-1 binding, which are upregulated during the hypoxia. HREs having functionally essential hypoxia binding site consists of the consensus sequence described in 5′ upstream enhancer and promoter regions and 3′ downstream of the 3′ UTR29. The functional HRE have been identified in several human genes that are associated with a HIF-1 binding site (5′-(A|G)CGTG-3′) and its HIF-1 ancillary sequence (HAS), 5′-CA(G|C)(A|G)(T|G|C)-3′, located at downstream23. But, reports on the functional HRE in fish was not available broadly. A novel HRE, 5′-GATGTG-3′, was reported inside the second introns in the lactate dehydrogenase-B gene in killifish25. The functional HRE in zebrafish have been reported for the IGFBP-1 genes along with HRE consensus sequence (5′-RCGTG-3′) and its HAS sequence (5′-CAGGT-3′) in the downstream spacing 15 nt30. The upstream sequences were taken for HRE analysis, and the approaches of Kimura’s and Kajimura’s were applied here for analyzing the upstream region of hypoxia response genes in fishes. An HRE sequence (5′-RCGTG-3′) and consensus HAS [5′-CA(G|C)(A|G)(T|G|C)-3′], located in a space of 7–15 nt downstream, were considered to determine the putative functional HRE from canonical sequences of upstream.
This platform has also a primer design module which may be used to design primers of the locating region of functional HRE on the upstream sequence for experimental validation.
The presence of HRE with an adjacent motif pattern of HAS in regulatory elements can be used as a marker to identify the gene induced by HIF-1 in hypoxic condition. The homology based annotation and findings of CpG islands and HRE pattern in the parsed upstream sequences support the existence of the promoter and enhancer regions.
The cross species PCR (cs-PCR) amplification was applied frequently to obtain common regions of a gene sequences among related species. Iyengar31 applied cs-PCR approach to amplify the region of interest into four species of non-human primates. The proposed work will help to design primers for amplification of the conserved exonic region of a gene to validate homologous gene in different fish species. Further, the reported genes for hypoxia response in zebrafish and other model fishes will help to identify the homologous genes in hypoxia susceptible and tolerant species using the cs-PCR, which may provide a greater insight into the stress management in the fishes.
Materials and Methods
Data collection and parsing
The peer-reviewed research articles were extracted from the NCBI’s PubMed using keywords, like hypoxia, anoxia etc., in fishes. The shortlisted articles from the search result were used for identifying the hypoxia responsive genes in fishes. Table 2 presents a list of genes identified from the shortlisted articles for hypoxia response in fishes. The information about the identified genes (Table 2) were collected from the Gene database32. For parsing and managing data in the HRGFish database, a Perl parsing program (GeneDB_Mapper.pl) was designed and implemented for parsing GeneID, gene ontology terms, PubMed ID and Accession ID of mRNA, protein and gene containing chromosome (genomic sequence) from the Gene database according to the database schema. The accession numbers for mRNA, protein and gene containing genomic sequences for the reported genes in the Gene database were collected and supplied in the Batch Entrez of NCBI33 and the data files in the FASTA format were downloaded. After that, another Perl script ‘GeneSeqParse.pl’ was designed and implemented for parsing gene and mRNA sequences by using their position of localization on the downloaded genomic sequences. An additional attempt was made to parse genomic mRNA sequences, because the downloaded mRNA from the curated database of NCBI had only coding regions (exonic) existing in the form of different variants in many genes which provided limited scope for analysis. On the other hand, the mRNA sequences with reported exons and introns were found much applicable for analysis and designing primers. Both types of mRNA sequences were taken into account for the study in different kind of analysis.
Table 2. Hypoxia response genes reported in fishes.
| S. No. | Gene name | Gene symbol | Pubmed IDs | Total genes in HRGFish |
|---|---|---|---|---|
| 1 | Aldolase A | aldoa | 8955077, 21266200 | 13 |
| 2 | Calcium/Calmodulin-dependent protein kinase | camk2g | 15821280, 25840431 | 9 |
| 3 | Ceruloplasmin | cp | 15741220 | 39 |
| 4 | Connective tissue growth factor | ctgf | 25455470 | 30 |
| 5 | CREB binding protein | cbp | 8917528, 17925579 | 23 |
| 6 | CREB regulated transcription coactivator 3 | crtc3 | 25840431 | 22 |
| 7 | E1A binding protein p300 | ep300 | 8917528 | 14 |
| 8 | Endothelial PAS domain protein 1 | epas1 | 25205386 | 31 |
| 9 | Enolase 1 | eno1 | 8955077, 21266200, 26762295 | 24 |
| 10 | Erythropoietin | epo | 21798198, 17579187 | 51 |
| 11 | Ferritin | fth1 | 11172064 | 42 |
| 12 | Fibrillarin | fbl | 17828398 | 27 |
| 13 | Glucose transporter 1 | slc2a1 | 10401038, 18941827, 12846834 | 4 |
| 14 | Glucose transporter 2 | slc2a2 | 19162213 | 22 |
| 15 | Glucose transporter 3 | slc2a3a | 16081168 | 1 |
| 16 | Glucose transporter 4 | slc2a4 | 16081168, 8941827 | 30 |
| 17 | Heme oxygenase-1 | hmox1 | 24780551 | 21 |
| 18 | Heme oxygenase-2 | hmox2 | 24780551 | 71 |
| 19 | Hemopexin | hpx | 11172064 | 20 |
| 20 | Hypoxia induced factor 1 alpha | hif1a | 11278461 | 35 |
| 21 | IGF binding protein 1 | igfbp1 | 16428465, 18769480, 21730259 21501614 | 9 |
| 22 | IGF binding protein 2 | igfbp2 | 22639285 | 25 |
| 23 | IGF binding protein 3 | igfbp3 | 18769480 | 9 |
| 24 | Lactate dehydrogenase B | ldhb | 19439190, 11441966 | 24 |
| 25 | Myoglobin | mb | 25026501, 25595439 | 38 |
| 26 | Noelin | noelin | 17828398 | 8 |
| 27 | Notch 2 | notch2 | 17828398 | 5 |
| 28 | P300-interacting transactivator | cited1 | 8917528, 20161383, 20547241 | 21 |
| 29 | Peroxisome proliferator activated receptor alpha | ppara | 25869933, 25205386, 25543049 | 27 |
| 30 | Titin | ttn | 11788825 | 12 |
| 31 | Transferrin | tf | 17646932, 11172064, 9242677 | 12 |
| 32 | Vascular endothelial growth factor A | vegfa | 22300081, 15177948 | 30 |
| 33 | Vascular endothelial growth factor B | vegfb | 22300081, 15177948 | 27 |
| 34 | Vascular endothelial growth factor C | vegfc | 22300081, 15177948 | 22 |
| 35 | VEGF receptor | flt1 | 20335444 | 20 |
| Total gene information | 818 |
Both Perl scripts support the inclusion of a new hypoxia responsive gene and gene information for updating HRGFish database. The gene information of the species was enriched with general information (taxonomy, habitat, occurrences, distribution) collected from FishBase34 and conservation status collected from IUCN35. The modus operandi applied in the Fish Karyome36 was used for integration of genomic data with other fisheries information. Figure 1 presents the complete architecture of the HRGFish database.
Identification of upstream elements
The upstream regions of different genes were collected. An orthologous cluster of upstream regions was formed and analyzed to deduce the conserved pattern of promoters. To carry out this work, genomic sequences of the reported gene for parsing the upstream sequence and a reference fish species, like zebrafish, in which promoter sequences are available, were essentially required. The EPD37 database was used to obtain promoter sequences of the zebrafish for the reported hypoxia responsive genes covered in HRGFish. Moreover, 3000 nt long upstream fragments located at −2900 to 100 position of the gene start site on genomic sequences were parsed. The upstream sequence of the selected genes was used to compute CpG (5′-C_phosphate_G-3′) island using CpG Ploter, HRE and HAS motif pattern. Further, alignment analysis of upstream sequences with reference promoters of zebrafish was done to deduce conserved patterns. These conserved patterns formed numerous clusters in the form of an orthologus set of promoter sequences for several genes.
Database development
MySQL, a relational database management system, was used to design and implement the database under the Linux operating platform on Intel Xeon based high performance computer server machine. Tables were designed and relationships between the tables were created using unique, primary and foreign keys. Tables ‘fishinfo’ and ‘taxonomy’ cover general information on the fish species and their systematics respectively. Tables ‘genesummary’, ‘gene2refseq’, ‘gene2go’ and ‘gene2pubmed’ contains data collected from Gene database. Tables ‘rna’ and ‘cds’ cover annotation information on genes. The FASTA format files of gene, mRNA and protein sequences were used by the program ‘formatdb’ of the Blast suite for building the local Blast-compatible target datasets to perform alignment between the query and the target sequences (Table 1). The Blast-compatible datasets were synchronized with the main database and getting updated automatically when the main database is updated.
Design and development of web interfaces
For web-based information delivery and analysis, user interactive web pages were designed and implemented using PHP (pre hypertext processor), Perl, DBI (database interface), CGI (common gateway interface), GD (graph display), Ajax, JavaScripts, CSS (cascading style sheets) and HTML technologies.
Implementation of tools for analysis
Different tools were implemented in the web interface to work with the HRGFish database. The web interface integrated with these tools provides the workbench for searching, browsing and analyzing sequences from the database (Fig. 1). The similarity search tools, like BLAST, Spidey38 and GetORF39, were implemented for analyzing the homologous sequences, CDS, mRNA and Open Reading frames in the gene sequences. ClustalW240 program was implemented for aligning a set of selected homologous sequences to analyze the conserved pattern among them. The primer341,42 program was implemented for designing primers using methodologies already described in FishMicrosat43 and FMiR44. The CpGplot39 was implemented to analyze the CpG island in the upstream region of the gene.
Conclusion
The primary goal of the HRGFish database is to serve as a repository of genes involved in hypoxia response in fishes and to provide a workbench for browsing information and analyzing the gene sequences. In addition, this platform provides the facility for primer designing for amplifying a selected region of a gene, which is applicable to identify the homologous sequence in other closely related fish species based on cross-species PCR amplification. HRGFish further highlights the annotation of the upstream sequences of genes for observation of HREs, TFBSs and other conserved elements of the promoter sequences which may play an important role in gene expression during hypoxia exposure. The findings of the HRE in the upstream regions of a gene confirm its expression that might be accelerated by a HIF transcription factor during hypoxia. Apart from that, the database contains taxonomy and general information of each fish species for categorization of the species and group-wise orthologous analysis of the promoter clusters. In future, the size of the database will grow by adding the records of new genes associated with hypoxia response. In this way, HRGFish is a prominent resource covering useful information and analysis tools for the cutting edge research areas related to the study of the hypoxia response and its applications in fishes, such as genetic relatedness among the species and genetic improvement programs of commercially important aquaculture species.
Additional Information
How to cite this article: Rashid, I. et al. HRGFish: A database of hypoxia responsive genes in fishes. Sci. Rep. 7, 42346; doi: 10.1038/srep42346 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Acknowledgments
Authors are thankful to the Director, ICAR-NBFGR, Lucknow, for providing necessary facilities to carry out this work. Further, authors are thankful to the CABin, ICAR-IASRI, New Delhi for financial assistance. Authors are also grateful to Dr. Satarudra Prakash Singh and Dr. Ajey Kumar Singh for their continued suggestions. We acknowledge Mrs. Rashmi Verma for her assistance in the literature survey.
Footnotes
The authors declare no competing financial interests.
Author Contributions I.R., N.S.N. and P.S. conceived the study and the conceptual design of the work. I.R. collected and compiled the data and programmed for data manipulation, designed, developed database and application modules for browsing and analyzing the data. A.K.P. tested the workflow model and application modules. R.K., B.K. and M.S. supported for fish biology and taxonomy information. I.R., A.K.P., N.S.N. and P.S. drafted the manuscript. All authors have read and approved the manuscript.
References
- Nilsson G. E. & Ostlund-Nilsson S. Hypoxia in paradise: widespread hypoxia tolerance in coral reef fishes. Proc Royal Soc London B-Biol Sci. 271, S30–S33 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Ju Z., Wells M. C. & Walter R. B. Genomic approaches in the identification of hypoxia biomarkers in model fish species. J Exp Mar Bio Ecol. 381, S180–S187 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poellinger L. & Johnson R. S. HIF-1 and hypoxic response: the plot thickens. Curr Opin Genet Dev. 14, 81–85 (2004). [DOI] [PubMed] [Google Scholar]
- Rosenberger A. E. & Chapman L. J. Hypoxic wetland tributaries as faunal refugia from an introduced predator. Ecol Freshwater Fish. 8, 22–34 (1999). [Google Scholar]
- Nikinmaa M. & Rees B. B. Oxygen-dependent gene expression in fishes. Am J Physiol Regul Integr Comp Physiol. 288, R1079–90 (2005). [DOI] [PubMed] [Google Scholar]
- Jensen F. B., Fago A. & Weber R. E. Hemoglobin structure and function. In: Fish Respiration, edited by Perry S. F. & Tufts B. L.San Diego: Academic 1–40 (1998). [Google Scholar]
- Jensen F. B. & Weber R. E. Proton and oxygen equilibria, their anion sensitivities and interrelationships in tench hemoglobin. Mol Physiol. 7, 41–50 (1985). [Google Scholar]
- Powers D. A. Structure, function, and molecular ecology of fish hemoglobins. Ann NY Acad Sci. 241, 472–490 (1974). [DOI] [PubMed] [Google Scholar]
- Holopainen I. J., Hyvärinen H. & Piironen J. Anaerobic wintering of crucian carp (Carassius carassius L.). II. Metabolic products. Comp Biochem Physiol. 83, 239–242 (1986). [DOI] [PubMed] [Google Scholar]
- Shoubridge E. A. & Hochachka P. W. The origin and significance of metabolic carbon dioxide production in anoxic goldfish. Mol Physiol. 1, 315–338 (1981). [Google Scholar]
- Van den Thillart G. Adaptations of fish energy metabolism to hypoxia and anoxia. Mol Physiol. 2, 49–61 (1982). [Google Scholar]
- Robertson C. E., Wright P. A., Köblitz L. & Bernier N. J. Hypoxia-inducible factor-1 mediates adaptive developmental plasticity of hypoxia tolerance in zebrafish, Danio rerio. Proc Biol Sci. 281, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vornanen M., Stecyk J. A. W. & Nilsson G. E. The anoxia-tolerant crucian carp (Carassius carassius L.). In Hypoxia (ed. Richards J. G., Farrell A. P. & Brauner C. J.). San Diego: Elsevier., 397–441 (2009). [Google Scholar]
- Stewart N. E., Shumway D. L. & Doudoroff, P. Influence of oxygen concentrations on the growth of juvenile larger mouth bass. Journal of Fisheries Research Board. Canada 24, 475–494 (1967). [Google Scholar]
- Thetmeyer H., Waller U., Black K. D., Inselmann S. & Rosenthal H. Growth of European sea bass (Dicentrarchus labrax L.) under hypoxic and oscillating oxygen conditions. Aquaculture 174, 355–367 (1999). [Google Scholar]
- Padmavathy P., Ramanathan N. & Francis T. Glucose, Lactate and Pyruvate Metabolism in Labeo rohita with Reference to Ambient oxygen. Asian Fisheries Science 16, 51–58 (2003). [Google Scholar]
- David M., Sangeetha J. & Harish E. R. Sodium cyanide induced alteration in the whole animal oxygen consumption and behavioural pattern of freshwater fish Labeo rohita. J Environ Biol. 36, 405–8 (2015). [PubMed] [Google Scholar]
- Semenza G. L. & Wang G. L. A nuclear factor induced by hypoxia Via denovo protein-synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Mol Cell Biol. 12, 5447–5454 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer N. V. et al. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1 alpha. Genes Dev. 12, 149–62 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- RytKönen K. T. et al. Subfunctionalization of cyprinid hypoxia-inducible factors for roles in development and oxygen sensing. Evolution 67, 873–882 (2013). [DOI] [PubMed] [Google Scholar]
- Soitamo A. J. Rabergh C. M., Gassmann M., Sistonen L. & Nikinmaa M. Characterization of a hypoxia-inducible factor (HIF-1alpha) from rainbow trout. Accumulation of protein occurs at normal venous oxygen tension. J Biol Chem. 276, 19699–705 (2001). [DOI] [PubMed] [Google Scholar]
- Wenger R. H., Stiehl D. P. & Camenisch G. Integration of oxygen signaling at the consensus HRE. Sci. STKE. 2005, re12 (2005). [DOI] [PubMed] [Google Scholar]
- Kimura H. et al. Identification of hypoxia-inducible factor 1 ancillary sequence and its function in vascular endothelial growth factor gene induction by hypoxia and nitric oxide. J Biol Chem. 276, 2292–8 (2001). [DOI] [PubMed] [Google Scholar]
- Rees B. B., Bowman J. A. & Schulte P. M. Structure and sequence conservation of a putative hypoxia response element in the lactate dehydrogenase-B gene of Fundulus. Biol Bull. 200, 247–51 (2001). [DOI] [PubMed] [Google Scholar]
- Rees B. B., Figueroa Y. G., Wiese T. E., Beckman B. S. & Schulte P. M. A novel hypoxia-response element in the lactate dehydrogenase-B gene of the killifish Fundulus heteroclitus. Comp Biochem Physiol A Mol Integr Physiol. 154, 70–7 (2009). [DOI] [PubMed] [Google Scholar]
- Barta E. et al. DoOP: Databases of Orthologous Promoters, collections of clusters of orthologous upstream sequences from chordates and plants. Nucleic Acids Res. 33, D86–90 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fickett J. W. & Hatzigeorgiou A. C. Eukaryotic promoter recognition. Genome Research 7, 861–878 (1997). [DOI] [PubMed] [Google Scholar]
- Deaton A. M. & Bird A. CpG islands and the regulation of transcription. Review. Genes Dev. 25, 1010–22 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fordel E., Geuens E., Dewilde S., De Coen W. & Moens L. Hypoxia/ischemia and the regulation of neuroglobin and cytoglobin expression. IUBMB Life 56, 681–7 (2004). [DOI] [PubMed] [Google Scholar]
- Kajimura S., Aida K. & Duan C. Understanding hypoxia-induced gene expression in early development: in vitro and in vivo analysis of hypoxia-inducible factor 1-regulated zebra fish insulin-like growth factor binding protein 1 gene expression. Mol Cell Biol. 26, 1142–55 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyengar S., Seaman M., Deinard A. S., Rosenbaum H. C. & Sirugo G. Analyses of cross species polymerase chain reaction products to infer the ancestral state of human polymorphisms. DNA Seq. 8, 317–27 (1998). [DOI] [PubMed] [Google Scholar]
- Brown G. R. et al. Gene: a gene-centered information resource at NCBI. Nucleic Acids Res. 43 (Database issue), D36–42 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCBI Resource Coordinators. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 43, D6–D17 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froese R. & Pauly D. FishBase. World Wide Web electronic publication. www.fishbase.org, version (April 2015) (2015).
- IUCN. Fish Conservation Status on the IUCN Red List. www.iucnredlist.org/freshwater (8 August 2014, date last accessed) (2014).
- Nagpure N. S. et al. Fish Karyome version 2.1: a chromosome database of fishes and other aquatic organisms. Database (Oxford) 15, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreos R., Ambrosini G., Cavin Périer R. & Bucher P. EPD and EPDnew, high-quality promoter resources in the next-generation sequencing era. Nucleic Acids Res. 41, D157–64 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheelan S. J., Church D. M. & Ostell J. M. Spidey: a tool for mRNA-to-genomic alignments. Genome Res. 11, 1952–7 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P., Longden I. & Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16, 276–277 (2000). [DOI] [PubMed] [Google Scholar]
- Larkin M. A. et al. Clustal W and ClustalX version 2.0. Bioinformatics 23, 2947–8 (2007). [DOI] [PubMed] [Google Scholar]
- Rozen S. & Skaletsky H. Primer3 on theWWWfor general users and for biologist programmers. Methods Mol Biol. 132, 65–386 (2000). [DOI] [PubMed] [Google Scholar]
- Untergasser A. et al. Primer3—new capabilities and interfaces. Nucleic Acids Res. 40, e115 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpure N. S. et al. FishMicrosat: a microsatellite database of commercially important fishes and shellfishes of the Indian subcontinent. BMC Genomics 14, 630 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpure N. S. et al. FMiR: A Curated Resource of Mitochondrial DNA Information for Fish. PLoS One 10, e0136711 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]