Screening of Arabidopsis mutants with altered seed mucilage allowed identification of UUAT1, a Golgi-localized protein that transports UDP-glucuronic acid and plays a role in the biosynthesis of pectin.
Abstract
UDP-glucuronic acid (UDP-GlcA) is the precursor of many plant cell wall polysaccharides and is required for production of seed mucilage. Following synthesis in the cytosol, it is transported into the lumen of the Golgi apparatus, where it is converted to UDP-galacturonic acid (UDP-GalA), UDP-arabinose, and UDP-xylose. To identify the Golgi-localized UDP-GlcA transporter, we screened Arabidopsis thaliana mutants in genes coding for putative nucleotide sugar transporters for altered seed mucilage, a structure rich in the GalA-containing polysaccharide rhamnogalacturonan I. As a result, we identified UUAT1, which encodes a Golgi-localized protein that transports UDP-GlcA and UDP-GalA in vitro. The seed coat of uuat1 mutants had less GalA, rhamnose, and xylose in the soluble mucilage, and the distal cell walls had decreased arabinan content. Cell walls of other organs and cells had lower arabinose levels in roots and pollen tubes, but no differences were observed in GalA or xylose contents. Furthermore, the GlcA content of glucuronoxylan in the stem was not affected in the mutant. Interestingly, the degree of homogalacturonan methylation increased in uuat1. These results suggest that this UDP-GlcA transporter plays a key role defining the seed mucilage sugar composition and that its absence produces pleiotropic effects in this component of the plant extracellular matrix.
INTRODUCTION
The plant cell wall is a complex and dynamic structure that is mainly composed of polysaccharides, with cellulose being a key component. The synthesis of noncellulosic polysaccharides (hemicellulose and pectin) occurs in the Golgi apparatus, where a number of glycosyltransferases (GTs) are located (Liepman et al., 2010; Scheible and Pauly, 2004). GTs transfer the sugar residue from an activated nucleotide donor, in the form of a UDP- or GDP-sugar, to a growing polysaccharide chain. Most GTs are type-II membrane-bound proteins with a catalytic domain facing the Golgi lumen (Sterling et al., 2001; Wulff et al., 2000; Scheible and Pauly, 2004). However, most nucleotide sugars utilized by GTs are produced in the cytosol (Bar-Peled and O’Neill, 2011; Bonin et al., 1997; Seifert, 2004). Therefore, the Golgi membrane is a physical barrier blocking access to the active GT site.
Nucleotide sugar transporters (NSTs) in the Golgi membrane overcome this topological problem and supply the substrates needed in the Golgi lumen for polysaccharide biosynthesis (Orellana et al., 2016; Reyes and Orellana, 2008; Temple et al., 2016). In Arabidopsis thaliana, the genes encoding for NSTs are similar to those encoding for plastidic triose phosphate translocators (TPTs); together, 44 NSTs and 7 TPTs form a gene family of 51 members (Knappe et al., 2003; Rautengarten et al., 2014). To date, a number of these NSTs from Arabidopsis have been functionally characterized, specifically transporters for GDP-mannose (GDP-Man), GDP-fucose (GDP-Fuc), UDP-galactose (UDP-Gal), UDP-glucose (UDP-Glc), UDP-rhamnose (UDP-Rha), and UDP-xylose (UDP-Xyl) (Bakker et al., 2005; Baldwin et al., 2001; Ebert et al., 2015; Handford et al., 2012, 2004; Norambuena et al., 2002, 2005; Rautengarten et al., 2014, 2016; Rollwitz et al., 2006).
Hemicellulose and pectins have diverse structures and sugar compositions, and several nucleotide sugars are required for their synthesis. UDP-glucuronic acid (UDP-GlcA) plays a critical role in noncellulosic polysaccharide synthesis, as it is the precursor for several nucleotide sugars involved in hemicellulose and pectin synthesis. These sugars include UDP-galacturonic acid (UDP-GalA), UDP-Xyl, UDP-arabinose (UDP-Ara), and UDP-apiose (UDP-Api) (Reboul et al., 2011). Therefore, UDP-GlcA is a major precursor required for hemicellulose and pectic polysaccharide synthesis. Substrate interconversion is required for polysaccharide biosynthesis, and in Arabidopsis leaves, polysaccharides containing sugars derived from UDP-GlcA account for nearly 50% of the cell wall biomass (Zablackis et al., 1995). The cytosol contains some of the enzymes for the interconversion of UDP-GlcA, such as soluble UDP-xylose synthase (UXS) (Harper and Bar-Peled, 2002; Pattathil et al., 2005; Kuang et al., 2016), UDP-apiose/UDP-xylose synthase (Guyett et al., 2009; Mølhøj et al., 2003), and UDP-arabinose mutase (Konishi et al., 2007; Rautengarten et al., 2011). There are also Golgi-localized interconverting enzymes. These include UDP-glucuronate epimerase, which converts UDP-GlcA into UDP-GalA (Gu and Bar-Peled, 2004; Mølhøj et al., 2004), membrane attached UXS (Harper and Bar-Peled, 2002; Kuang et al., 2016), and UDP-xylose-4-epimerase, which catalyzes UDP-Xyl to UDP-arabinopyranose (UDP-Arap) epimerization (Burget et al., 2003). All these enzymes are predicted type-II membrane proteins, and their catalytic domain faces the lumen. Therefore, the transport of UDP-GlcA into the Golgi lumen is a critical step for UDP-GalA biosynthesis, as well as for part of the luminal UDP-Xyl. Additionally, UDP-Arap produced from lumen-synthesized UDP-Xyl may rely on this transporter. UDP-GlcA transport is also important for glucuronoxylan biosynthesis because this polymer is synthesized in the Golgi lumen, where GlcA units are added to the xylan backbone. An NST that transports UDP-GlcA has been described in Caenorhabditis elegans, and mutations in the protein responsible for this activity lead to an abnormal development in this organism (Berninsone and Hirschberg, 2000). A Golgi-localized UDP-GlcA transporter is likely to play a critical role in plant cells by providing the substrate precursors needed for pectin and hemicellulose biosynthesis in the Golgi lumen. In this context, this transporter could play an important role in determining the content of plant cell wall sugars that are derived from UDP-GlcA.
To identify a UDP-GlcA transporter and analyze its role in defining cell wall composition, we took advantage of the fact that Arabidopsis seeds produce copious amounts of a pectin-rich substance that is referred to as seed coat mucilage. It is composed of gel-like molecules that are extruded from mature seeds following water imbibition (Saez-Aguayo et al., 2013; Western et al., 2000; Western, 2012; Young et al., 2008). Early visual examination of the mucilage provided evidence of its pectic nature and showed the presence of two distinct mucilage layers. Both layers contain large amounts of the GalA-containing polysaccharides rhamnogalacturonan I (RG-I) and homogalacturonan (HG). RG-I is mostly unbranched in the external layer, the soluble mucilage (SM), and it is branched, having arabinan and galactan side chains in the internal layer, the adherent mucilage (AM) (Macquet et al., 2007; Western et al., 2000; Willats et al., 2001). The AM also contains methylesterified HG and cellulose microfibrils (Macquet et al., 2007; Saez-Aguayo et al., 2013; Western et al., 2004; Willats et al., 2001). Because mucilage contains high pectin levels, changes in the pathway leading to the synthesis of UDP-GalA will alter RG-I or HG synthesis, affecting the seed mucilage formation and composition. Therefore, the seed coat NST expression analysis was combined with a mucilage release-screening assay of NST mutants to select any novel NSTs potentially involved in the RG-I or HG synthesis. We identified UUAT1 (UDP-URONIC ACID TRANSPORTER1), a gene encoding a protein that can transport UDP-GlcA and UDP-GalA in vitro. A knockout line lacking UUAT1 has less GalA and Rha in both AM and SM, and less Xyl in SM. Also, a decrease in arabinan content was observed in the seed coat. Analyses of UUAT1 expression in other organs and cells revealed differences in Ara content in uuat1 mutant versus wild-type tissue. Interestingly, besides changes in sugar content, a change in the HG methylation pattern was observed in the mucilage and more methyl groups were released from cell wall material from mucilage and stem, suggesting that HG methylation is also altered in some organs and indicating that pleiotropic changes might take place in the mutant cell wall. Our results suggest that UUAT1 transports UDP-GlcA in vivo. Furthermore, the loss of function of this transporter leads to changes in monosaccharide composition, in the cell wall, mainly in those sugars related to UDP-GlcA metabolism in the Golgi lumen. These results show the importance of the transport of UDP-GlcA in the biosynthesis of the plant cell wall.
RESULTS
Analysis of NSTs Expressed in Seed Coats and Identification of UUAT1
In silico data analyses revealed that 21 out of the 51 members of the NST/TPT Arabidopsis family (Rautengarten et al., 2014) are expressed in the seed coat during the developmental stages when mucilage is produced and accumulated in epidermal cells (Supplemental Figure 1) (Le et al., 2010; http://seedgenenetwork.net/arabidopsis). Of these 21 candidates, we disregarded those with reported functions (10 known NSTs) in the Arabidopsis UDP-rhamnose/UDP-galactose transporter (URGT) family (Rautengarten et al., 2014), UDP-galactose transporter 1 (UTR1), UTR2, and UTR3 (Norambuena et al., 2002, 2005; Reyes et al., 2006, 2010), and GalT1 (Bakker et al., 2005). Among the 11 target genes that were expressed throughout seed development and lack a known function (Supplemental Figure 1), At5g17630 was discarded from the analysis because as it belongs to the triose phosphate translocator clade. Of the 10 remaining genes, homozygous mutants could be obtained for seven of them, but only heterozygous mutants were obtained for the other three. Soluble mucilage content was assessed by measuring the uronic acid released following water imbibition. The results showed that a mutant allele of At5g04160, uuat1, exhibited the lowest level of mucilage uronic acid (Supplemental Figure 2). UUAT1 expression was also measured during seed development to confirm that it is expressed during the mucilage production stages (6 to 8 d after pollination [DAP]). Supplemental Figure 3 shows a peak in UUAT1 expression at 8 DAP, a pattern similar to the expression of genes involved in mucilage synthesis (Macquet et al., 2007; Saez-Aguayo et al., 2013; Rautengarten et al., 2014). UUAT1 encodes a polytopic transmembrane protein with 10 putative membrane spanning domains (Supplemental Figure 4) and belongs to a subclade composed of five paralogs with identities ranging from 81 to 49% (Supplemental Table 1). However, their expression levels are much lower than those of UUAT1 (Supplemental Figure 3).
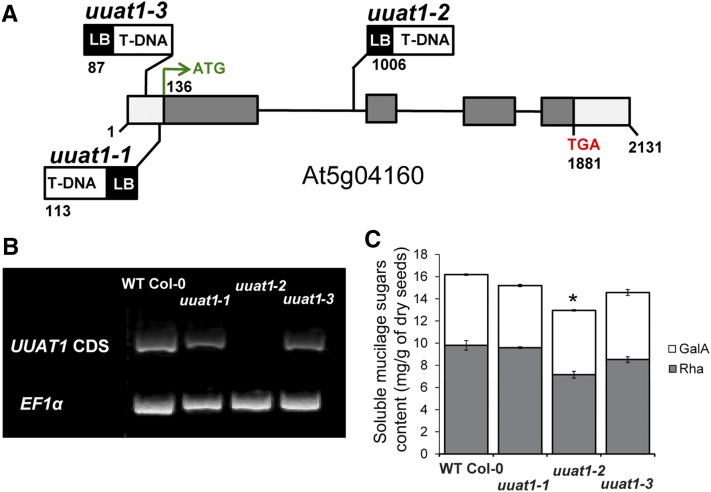
Given these results, we decided to focus on UUAT1 by analyzing its role in the biosynthesis of seed coat mucilage. Three T-DNA insertion lines were identified in the At5g04160 locus and were designated uuat1-1, uuat1-2, and uuat1-3 (Figure 1A). These mutant lines had a lower content of GalA and Rha residues in the SM fraction compared with the wild-type Col-0 plants (Figure 1C; Supplemental Table 2). When compared with the other two allelic lines, uuat1-2 exhibited the most pronounced decrease in both sugars. UUAT1 transcripts were undetectable in the uuat1-2 mutant line, whereas the other two lines (uuat1-1 and uuat1-3) exhibited some UUAT1 expression, albeit at lower levels than wild-type Col-0 (Figure 1B). Thus, we concluded that uuat1-2 had the strongest phenotype because it was a true knockout line, whereas the other alleles were knockdown lines and so the studies focused on the uuat1-2 allele. Molecular rescue of the uuat1-2 mutant confirmed that the absence of UUAT1 was responsible for the phenotypes observed in uuat1-2 (Supplemental Figure 5). The uuat1-2 line was transformed with a construct that contains the UUAT1 coding sequence (CDS) fused to a GFP tag and is driven by the UUAT1 endogenous promoter. Several independent transformants were obtained and the presence of the transgene was confirmed by RT-PCR (Supplemental Figure 5A). Wild-type ruthenium red staining of the AM and sugar content levels were observed in two independent transgenic lines, indicating that UUAT1-GFP had successfully rescued the mutant (Supplemental Figures 5B and 5C).
Figure 1.
Characterization of Mutants in UUAT1.
(A) Schematic representation of UUAT1 structure, as annotated in TAIR (http://www.arabidopsis.org/). The sites and orientations of the T-DNA insertions in allelic lines uuat1-1, uuat1-2, and uuat1-3 are indicated. Numbers indicate the positions (in base pairs) of the start and stop codons and the T-DNA insertion sites. White boxes, 5′- and 3′-untranslated regions; gray boxes, protein coding sequences; black lines, introns; LB, left border.
(B) Analysis of UUAT1 expression in T-DNA insertion lines. RT-PCR analyses were performed on RNAs isolated from wild-type Col-0, uuat1-1, uuat1-2, and uuat1-3 lines using specific primers for the full-length coding sequence of UUAT1. EF1α expression was used as a control.
(C) Measurement of galacturonic acid and rhamnose levels in soluble mucilage after 10 min of seed imbibition in water. Error bars represent se (n = 6) of three biological replicates. Asterisk indicates significant difference from the wild type using the t test, P < 0.05.
UUAT1 Is a UDP-Uronic Acid Transporter in the Golgi
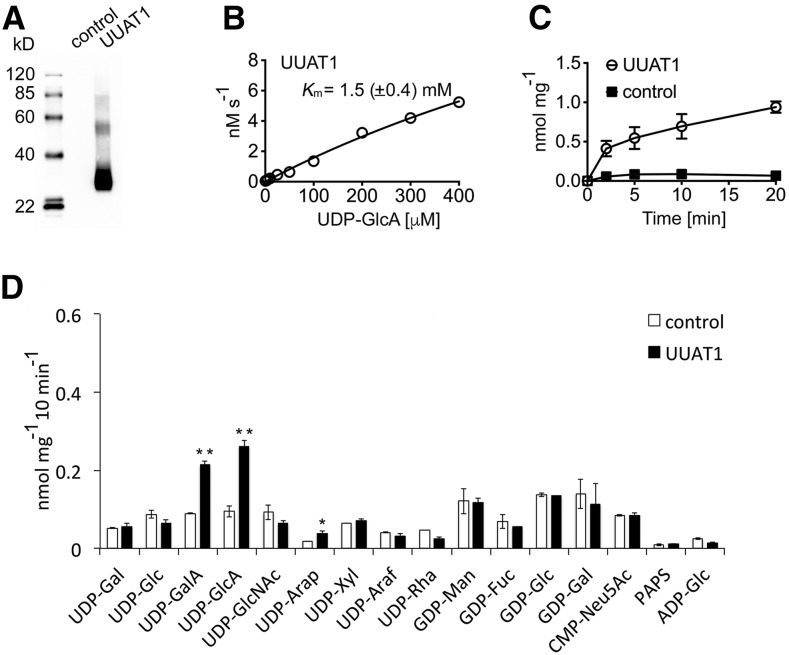
To determine the substrate specificity of UUAT1 in vitro, it was expressed heterologously in Saccharomyces cerevisiae (yeast), and transport assays were conducted as reported by Rautengarten et al. (2014). Transport assays were performed using the microsomal proteins reconstituted in proteoliposomes. An immunoblot analysis of the reconstituted protein confirmed the presence of UUAT1 in proteoliposomes (Figure 2A). Proteoliposomes were preloaded with UMP, GMP, CMP, or AMP and then incubated with a mixture of 15 nucleotides/nucleotide sugars to determine substrate specificity (Figure 2D; Supplemental Figure 6). Nontransported substrates were removed by gel filtration and the proteoliposome content analyzed with liquid chromatography-tandem mass spectrometry (LC-MS/MS). The substrate preference exhibited by UUAT1 could readily be assessed after LC-MS/MS analysis when compared with the empty vector control. UUAT1 demonstrated clear preferences for UDP-GlcA and UDP-GalA when proteoliposomes were preloaded with UMP (Figure 2D). No significant differences in transport activity between the control and UUAT1 were observed for any other nucleotide sugar apart from UDP-Arap, although this activity was much lower than that observed for the UDP-uronic acids (Figure 2D).
Figure 2.
UUAT1 Is a UDP-Uronic Acid Transporter.
(A) Immunoblot of the yeast microsomal fractions used to make the proteoliposomes. Total protein (2.5 µg) was probed with an anti-V5 tag antibody, and strong expression of UUAT1 (∼35 kD) was observed.
(B) Kinetics of UDP-GlcA transport at varying concentrations (0.5–400 µM) into proteoliposomes preloaded with UMP and then incubated for 2 min at 25°C.
(C) Time course for UDP-GlcA (50 µM) uptake into proteoliposomes preloaded with UMP and then incubated at 25°C. All values were normalized to the total protein content of the proteoliposome preparations and are means ± sd of four independent experiments.
(D) Quantification of nucleotide sugar uptake into proteoliposomes containing UUAT1 that were preloaded with UMP. Data are the means ± sd of four independent transport assays quantified by LC-MS/MS and normalized to the total protein content of the reconstituted proteoliposomes. The empty expression vector was used as a negative control. Significantly different values are marked with asterisks: *P < 0.05 and **P < 0.01; Student’s t test.
These substrate preferences were only specific when the proteoliposomes were preloaded with UMP and not with GMP, CMP, or AMP (Supplemental Figure 6). Given the additional negative charge present in UDP-uronic acids, UDP was also tested as a potential antiporter substrate, but no transport was observed (Supplemental Figure 6). Proteoliposomes preloaded with GMP could transport GDP-sugars and some lower activity was also observed when proteoliposomes were preloaded with AMP (Supplemental Figure 6), but this is likely to be the result of the endogenous transport activities of the yeast microsomal preparation, since the proteoliposome UUAT1 expression activity did not differ from the control, as has been observed previously (Ebert et al., 2015). UDP-GlcA transport by UUAT1 did not achieve saturation within the concentration range utilized in the assay (Figure 2B); however, transport was affected in a time dependent manner (Figure 2C). Analysis of transport rates indicated that UUAT1 has an apparent Km of 1.5 mM for UDP-GlcA (Figure 2B).
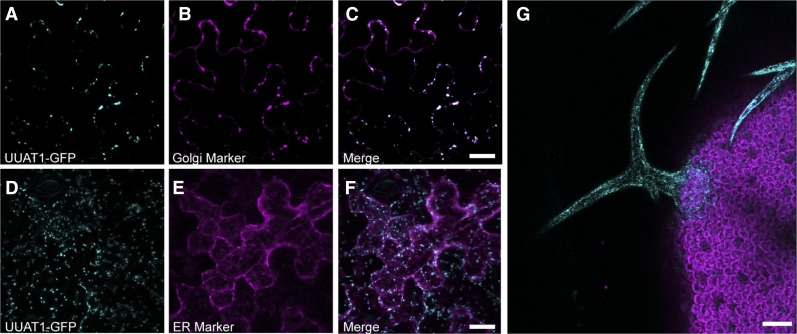
A C-terminal translational fusion with GFP was used to determine the subcellular localization of UUAT1 using confocal laser scanning microscopy on transient transformed epidermal cells (Figure 3). The UUAT1-GFP distribution pattern was compared with those obtained for the cis-Golgi marker α-mannnosidase-I (Saint-Jore-Dupas et al., 2006) and an endoplasmic reticulum (ER) marker (Nelson et al., 2007). The fluorescence signal obtained from the UUAT1-GFP protein colocalized with the punctate pattern obtained for the cis-Golgi marker α-Man-I but not with the ER marker (Figures 3A to 3F). To confirm the localization of UUAT1, trichomes of transgenic rescued plants expressing UUAT1-GFP under the endogenous promoter were analyzed, and they also showed motile structures exhibiting a punctate pattern, as has been described for Golgi resident proteins (Figure 3G) (Boevink et al., 1998). Taking these data together, these results indicate that UUAT1 is a Golgi-localized UDP-uronic acid transporter.
Figure 3.
UUAT1 Is Located in the Golgi Apparatus.
(A) to (F) Tobacco epidermal cells were cotransformed with agrobacteria carrying vectors containing Pro35S-UUAT1-GFP with the cis-Golgi marker α-Mannosidase-I-Cherry ([A] to [C]) or the ER marker wall-associated kinase-2-signal peptide-mCherry-HDEL ([D] to [F]). GFP labeling colocalized with the Golgi marker (C) but not with the ER marker (F). Bars = 5 μm.
(G) Subcellular localization of UUAT1-GFP in trichomes from uuat1-2 plants rescued with the ProUUAT1-UUAT1-GFP construct. Bar = 100 um.
Absence of UUAT1 Has Pleiotropic Effects on Seed Coat Cell Walls and Mucilage
In order to better understand the effects on the composition of cell wall polysaccharides caused by the absence of UUAT1, we analyzed the sugar content of the polysaccharides present in the seed mucilage (Table 1) in both wild-type Col-0 and uuat1-2 mutant plants. The SM, as expected, contained mostly GalA and Rha. However, decreases in both GalA (20%) and Rha (22%) were observed in the uuat1-2 mutant compared with the wild type (Table 1). Interestingly, despite the low Xyl levels in the sample, it displayed a similar reduction (21%). Furthermore, the seed+AM fraction from uuat1-2 showed a 5% decrease in GalA content along with a 9% reduction in Rha (Table 1).
Table 1. Sugar Composition of Seeds plus Adherent Mucilage (Seed+AM) and Extracted SM from Wild-Type Col-0 and uuat1-2 Plants.
| Structure | Sugar | Wild-Type Col-0 | uuat1-2 |
|---|---|---|---|
| Seed+AM | GalA | 20.67 (0.21)* | 19.68 (0.42)* |
| Rha | 21.77 (0.77)* | 20.02 (0.45)* | |
| Fuc | 1.74 (0.05) | 1.72 (0.03) | |
| Ara | 41.21 (1.59) | 39.62 (1.06) | |
| Xyl | 11.37 (0.56) | 11.45 (0.47) | |
| Man | 3.44 (0.12) | 3.69 (0.16) | |
| Gal | 30.23 (0.40) | 29.06 (1.13) | |
| Glc | 7.43 (0.26) | 8.18 (0.29) | |
| GlcA | 2.44 (0.08) | 2.55 (0.07) | |
| Total Seed+AM | 140.30 (2.80) | 135.96 (1.92) | |
| SM | GalA | 5.66 (0.21)* | 4.56 (0.22)* |
| Rha | 8.65 (0.49)* | 6.76 (0.43) * | |
| Ara | 0.08 (0.02) | 0.09 (0.01) | |
| Xyl | 0.43 (0.02)* | 0.34 (0.02)* | |
| Gal | 0.31 (0.04) | 0.26 (0.02) | |
| Total SM | 15.15 (0.64) | 11.99 (0.95) |
To analyze monosaccharide composition, a water-soluble extraction was used to isolate the SM fraction. The adherent mucilage cannot be detached from the seed and form the seed + soluble mucilage fraction (seed+AM). Sugar content was obtained using HPAEC-PAD from seed+AM and from SM. Values are in mg/g of dry seeds and are the means of three biological replicates. Standard errors are shown in parentheses for two technical replicates each. Asterisks indicate significant statistical differences using the Wilcoxon test (P < 0.05).
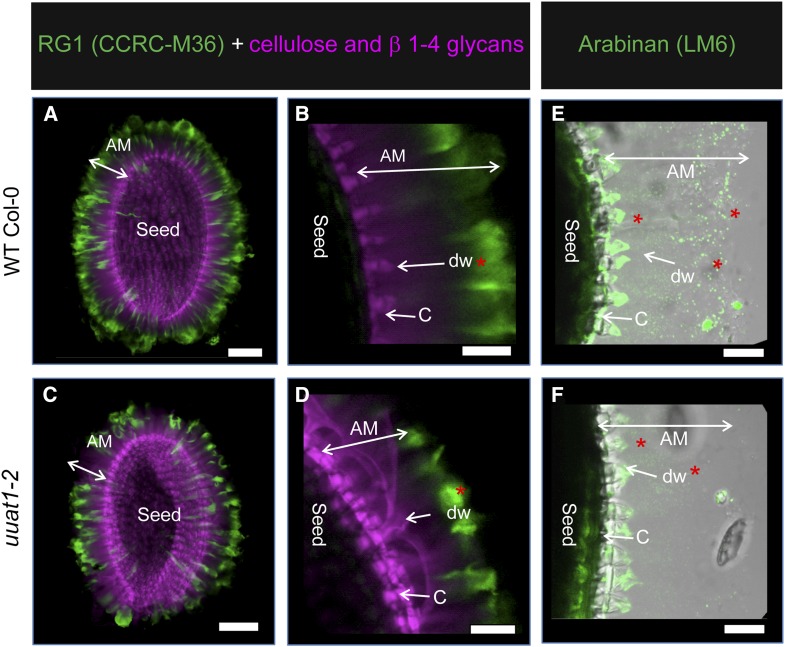
To further investigate the differences observed in the seed+AM fraction and to better understand which polysaccharides might be altered in the mutant, we performed whole-mount immunolabeling assays using antibodies against the epitopes present in cell wall polysaccharides (Figure 4). Based on the measured monosaccharide composition and considering the mucilage polysaccharide composition, the labeling was performed using the following antibodies: CCRC-M36 (anti-RG-I), LM6 (anti-arabinan), JIM7, and LM20 (both antimethylated HG) (Macquet et al., 2007; Verhertbruggen et al., 2009; Willats et al., 2001). In addition, to visualize the cell wall, we used calcofluor or propidium iodide, which stain β-1,4 glucans and polysaccharides. RG-I labeling was reduced in the AM of uuat1-2 seeds compared with wild-type Col-0 seeds (Figures 4A and 4C, green signal). Moreover, the calcofluor labeling showed a distal cell wall defect in the mutant when compared with wild-type Col-0 (Figures 4B and 4D, pink labeling), likely due to an abnormal cell wall rupture. The LM6 antibody showed less arabinan in the uuat1-2 AM compared with the wild-type Col-0, especially in the distal wall of the epidermal cells (Figures 4E and 4F). The lack of staining with the LM6 antibody was restored in the transgenic plants that expressed UUAT1-GFP with the endogenous promoter (Supplemental Figure 7), providing further evidence that this phenotype is due to the UUAT1 absence.
Figure 4.
The uuat1-2 Mutant Is Affected in Seed Mucilage Structure and Composition.
RG-I and arabinan labeling in adherent mucilage from wild-type Col-0 seeds and the uuat1-2 mutant line. Confocal microscopy optical section reconstruction of AM released from imbibed seeds. Asterisks represent differences in labeling. Bars = 100 μm in (A) and (C) and 50 µm in (B) to (F).
(A) to (D) CCRC-M36 antibody (green) was used to label RG-I domains, and calcofluor white was used to detect β-1,4-glucans (purple). (B) and (D) are magnifications of (A) and (C), showing greater detail of the AM, the distal cell wall (dw), and the columella cells (C).
(E) and (F) The LM6 antibody was used to label arabinan (green) in both wild-type Col-0 and uuat1-2 seeds.
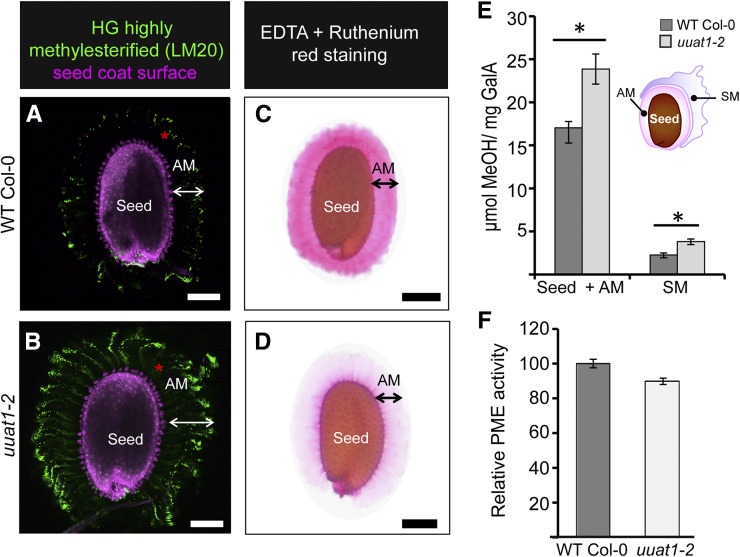
As the rupture of the distal cell wall upon water imbibition seemed to be altered, we reasoned that the cell wall stiffness might have changed. Because the degree of HG methylation affects the stiffness of the cell wall (Peaucelle et al., 2008), the LM20 and JIM7 antibodies were used to look at the highly methylesterified HG distribution (Figure 5; Supplemental Figure 8). An increase in LM20 labeling in AM was observed in the uuat1-2 mutant when compared with wild-type Col-0 (Figures 5A and 5B), suggesting the presence of HG with a higher degree of methyl esterification in the mutant. This result was also observed using the JIM7 antibody in the allelic lines uuat1-1 and uuat1-3 (Supplemental Figure 8). Analysis of the uuat1-2 lines expressing UUAT1 supported this observation by showing less labeling than in the mutant, but more labeling than in wild-type Col-0 when JIM7 and LM20 were used to assess the methylated HG content (Supplemental Figure 7). In addition, we performed ruthenium red staining in the presence of EDTA, a chelator that removes cations and increases the exposure of carboxylic groups, thus enhancing the ruthenium red staining (Figures 5C and 5D). Wild-type Col-0 seeds showed intense staining but mutant seeds exhibited a pale color, suggesting that fewer carboxylic groups were available for binding the dye (Figure 5D). To confirm changes in HG methylesterification in the uuat1-2 mutant, the contents of methyl groups present in the soluble mucilage and seed+AM fractions were determined by measuring the methanol released upon saponification. Both fractions displayed greater methanol release of 37 and 67%, respectively (Figure 5E). Finally, these HG methylesterification changes correlated with a 10% decrease in pectin methylesterase (PME) activity, measured in dry mutant seeds (Figure 5F). All these results provide strong evidence that UUAT1 absence leads to an increase of highly methylesterified HG epitopes in mucilage.
Figure 5.
The uuat1-2 Mutant Shows Increased Pectin Methylesterification and Has Reduced Pectin Methylesterase Activity in Seeds.
(A) and (B) Labeling of highly methylesterified HG in the adherent mucilage from seeds of wild-type Col-0 (A) and the uuat1-2 mutant line (B) imaged using confocal microscopy optical section reconstruction of AM released from imbibed seeds. The LM20 antibody (green) was used to label HG domains, and propidium iodide was used to stain the seed coat surface (pink). Bars = 100 μm.
(C) and (D) Appearance of seed adherent mucilage from wild-type Col-0 (C) and uuat1-2 (D) in the presence of a cation chelator. Seeds were stained with ruthenium red after 1 h of imbibition in 0.5 M EDTA. Bars = 100 μm.
(E) Degree of methylesterification in wild-type Col-0 and uuat1-2 in seed+AM and the soluble mucilage fractions. Error bars represent se (n = 16, from three biological replicates). ANOVA and Tukey tests were performed and compared with wild-type Col-0 (*P < 0.05).
(F) Seed PME activity. Total protein extracts from mature dry seeds of wild-type Col-0 and uuat1-2 were used to measure PME activity. The PME activity was normalized to the average wild-type activity (100). Error bars represent se (n = 16 for SM and n = 12 for seed+AM from three biological replicates).
UUAT1 Functions in Different Plant Organs
We next analyzed the UUAT1 expression pattern in organs such as roots, seedlings, rosette and cauline leaves, stems, flowers, siliques, and seeds at different development stages (Supplemental Figure 9). The UUAT1 transcript was detected by qPCR in all organs analyzed, with expression peaking in stems and flowers (Supplemental Figure 9A). The GUS reporter gene (Jefferson, 1989) was cloned under the control of the UUAT1 promoter and transformed into wild-type Col-0 plants to obtain spatial information regarding UUAT1 expression. Strong GUS activity was detected in roots, seedlings, trichomes, flowers, and developing seeds (Supplemental Figure 9B), confirming that UUAT1 is predominantly expressed in these organs.
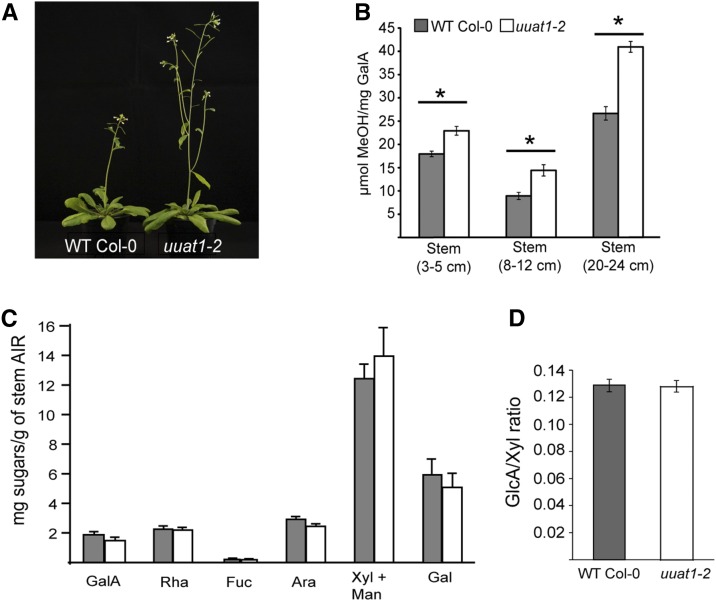
All obvious phenotypes in the uuat1-2 mutant were examined first to investigate whether the UUAT1 mutation leads to changes in plant development and/or cell wall composition in organs or tissues apart from the seed mucilage. Interestingly, the only change observed was in the primary stem, as uuat1-2 plants displayed an early elongation phenotype when compared with wild-type Col-0 plants (Figure 6A). However, this morphological difference disappeared once the plants reached a mature stage. No changes were observed in sugar composition of the stem cell wall for any of the sugars analyzed, including Xyl (Figure 6C), the most abundant sugar due to the presence of glucuronoxylan, one of the more abundant stem polymers. Because glucuronoxylan also contains GlcA in a given branching frequency, a carbohydrate gel electrophoresis (PACE) polysaccharide analysis (Mortimer et al., 2010) was used to quantify the oligosaccharides Xyl, Xyl2, and GlcAXyl4/[MeGlcA] Xyl4 released by xylanase GH11. No changes were found in the GlcA/Xyl ratio, suggesting that the glucuronoxylan structure is normal in the uuat1-2 mutant (Figure 6D). Additionally, because changes were observed in mucilage HG methylesterification, this modification was further analyzed in three development stages in stems. The methylesterification levels were observed to be significantly higher in all conditions in the uuat1-2 mutant when compared with wild-type Col-0 (Figure 6B).
Figure 6.
The uuat1-2 Mutant Line Displays an Early Stem Elongation Phenotype and an Increase in Methylesterification but Shows No Changes in Sugar Content in Stem Cell Walls.
(A) The uuat1-2 mutant displays early stem elongation when compared with the wild-type Col-0. Seven-week-old wild-type Col-0 and uuat1-2 plants show a pronounced difference in stem height. This difference disappears once the plants reach their adult state. This phenotype was observed in three biological replicates.
(B) Determination of the degree of methylesterification in stems. Error bars represent se (n = 8) from two biological replicates. Asterisks represent significant difference from wild-type Col-0 using ANOVA and Tukey tests (P < 0.05).
(C) High-performance anion exchange chromatography with pulsed amperometric detection (HPAEC-PAD) was used to quantify the cell wall extract monosaccharide composition from wild-type Col-0 and uuat1-2 stems (20–24 cm). Error bars represent se (n = 6) from three biological replicates.
(D) Ratio of GlcA/Xyl content of xylan products digested with GH11 xylanase. AIR material from basal stems of wild-type Col-0 and uuat1-2 was used to determine the frequency of GlcA branches on the xylan backbone using PACE. The quantity of each of the oligosaccharides released by GH11 xylanase [Xyl, (Xyl)2, GlcA-(Xyl)4/MeGlcA(Xyl)4] was calculated and the GlcA/Xyl ratio determined. Error bars represent se (n = 9) from three biological replicates.
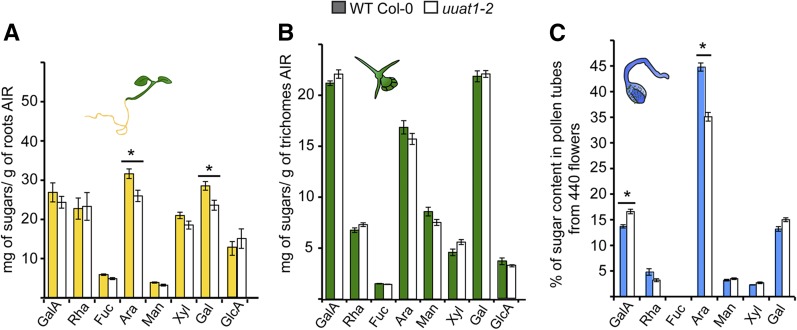
The alcohol-insoluble residue (AIR) sugar composition in uuat1-2 mutant roots, trichome, and pollen tube preparations was analyzed to uncover any possible changes in cell wall composition in other UUAT1-expressing organs (Figure 7). Ara levels were reduced in roots and pollen tubes. A slight decrease was observed in trichomes, but it was not significant. No decrease in GalA or Xyl was observed in any other of the tissues analyzed; thus, Ara was the only sugar whose content consistently decreased in the mutant, and this decrease was tissue specific. In conclusion, our results indicate that plants lacking UUAT1 show changes in the composition of cell wall monosaccharides derived from the metabolism of UDP-GlcA in the Golgi, with arabinose being the most affected. Furthermore, mutants in UUAT1 show enhanced levels of methylesterification in cell wall polysaccharides, a likely response to cope with changes in cell wall composition.
Figure 7.
Monosaccharide Composition of Different Organs, Tissues, or Cells from Wild-Type Col-0 and uuat1-2 Mutant Plants.
Quantification of the monosaccharide composition of the cell wall extracts from wild-type Col-0 and uuat1-2 mutant plants using HPAEC-PAD and gas chromatograph-flame ionization detection. Error bars represent se (n = 6) from three biological replicates. Asterisk indicates significant differences from the wild type using the Wilcoxon test (P < 0.05).
(A) Roots from 7-d-old plants.
(B) Trichomes from 14-d-old plants.
(C) Pollen tubes from 6-h-old plants.
DISCUSSION
UDP-GlcA is synthesized and utilized in the plant cell cytosol, but it is also required in the Golgi lumen for synthesis of UDP-GalA, UDP-Xyl, and UDP-Arap. Therefore, UDP-GlcA needs to be transported from the cytosol across the Golgi membrane into the Golgi lumen to be converted into these nucleotide sugars. Our work led to the identification of UUAT1, a protein that can transport UDP-GlcA, UDP-GalA, and low levels of UDP-Arap in vitro. However, UUAT1 is unlikely to transport significant amounts of UDP-GalA or UDP-Arap in vivo because both UDP-GlcA 4-epimerase and UDP-Xyl 4-epimerase, the enzymes involved in the synthesis of UDP-GalA and UDP-Arap, respectively, are located in the Golgi lumen (Burget et al., 2003; Gu and Bar-Peled, 2004; Mølhøj et al., 2004). A cytosolic salvage pathway for UDP-GalA has been reported (Yang et al., 2009) but requires release from the cell wall of GalA, which can then be converted into UDP-GalA by the enzymes GalA kinase and Sloppy, a promiscuous UDP-sugar pyrophosphorylase (Kotake et al., 2007). Therefore, this salvage pathway may be active only under certain circumstances and perhaps cytosolic UDP-GalA formed via this pathway could be transported by UUAT1. UDP-Arap can also be biosynthesized by UGE1 and UGE3 in the cytoplasm, using UDP-Xyl to form UDP-Arap. This would presumably be in addition to their roles in the UDP-Glc to UDP-Gal epimerization. However, mutations in these genes suggest that this cytosolic pathway is less important than that located in the Golgi (Rösti et al., 2007; Kotake et al., 2009; Rautengarten et al., 2011; Kotake et al., 2016). Furthermore, because UUAT1 did not exhibit in vitro transport activity for UDP-Araf (arabinose in its furanose form), we postulate that the main in vivo role for UUAT1 is to transport UDP-GlcA to the Golgi lumen. Finally, we believe the low UDP-Arap activity observed is not a function specific to UUAT1 because it is also observed in other nucleotide sugar transporters, such as the UDP-Rha/UDP-Gal transporters and the UDP-Xyl transporter (Ebert et al., 2015; Rautengarten et al., 2014)
UUAT1 is expressed in seed coat epidermal cells and knockout plants showed a number of mucilage-related phenotypes. The uuat1-2 mutant displayed reduced GalA and Rha content in both the SM and the seed+AM fraction. Thus, the GalA decrease could be explained by a lower UDP-GlcA transport rate to the Golgi lumen, the substrate required for the UDP-GalA synthesis. Thus, its reduced transport rate could lead to lower levels of this nucleotide sugar in the mutant. On the other hand, UUAT1 does not transport UDP-Rha, so the Rha decrease is likely due to an impairment in synthesis of the RG-I backbone, which is composed of repeating (GalA-Rha)n disaccharide units. Because one of the substrates (UDP-GalA) is reduced, it is likely that Rha incorporation has been also affected, leading to lower levels of this sugar in SM and the seed+AM fraction. Something similar occurs in mutants in URGT2, a UDP-Rha transporter that is also expressed in seed coat epidermal cells. SM in this mutant also exhibits lowered Rha and GalA, even though URGT2 does not transport UDP-uronic acids (Rautengarten et al., 2014). The GalA and Rha decrease observed in mucilage (a RG-I-enriched matrix) from UDP-Rha and UDP-GlcA transporter mutants suggests a coordination in the supply of both nucleotide sugars during RG-I biosynthesis.
The uuat1 mutants exhibit a decrease in arabinan in AM, as detected by LM6 antibody immunolabeling. Ara is an abundant sugar in the seed+AM fraction and can be present in other polysaccharides (Western et al., 2001), so changes in the total Ara content may not reveal the differences in a low-abundant Ara-containing polymer. The use of antibodies can detect precise changes in arabinan, which is present in the wild type and almost undetectable in the mutant, suggesting that UUAT1 plays an important role in providing the precursor for arabinan synthesis. The absence of UUAT1 also results in a reduction in Xyl in SM. However, this phenotype is different from the one observed for muci21 and irx14, xylosyltransferases mutants that show alterations in epidermal cell mucilage adhesion (Hu et al., 2016; Voiniciuc et al., 2015; Ralet et al., 2016). On the other hand, no Xyl changes were observed in the seed+AM fraction, suggesting a precise and discrete role for UUAT1 in Xyl-containing polymer synthesis.
The results show that both the SM and seed+AM matrices in the uuat1 mutants have lower levels of sugars (GalA, Xyl, and Ara) provided by UDP-sugars derived from the metabolism of UDP-GlcA in the Golgi. This supports the role of UUAT1 as an in vivo UDP-GlcA transporter. Because these sugars are not completely diminished, it suggests that other UDP-GlcA transporters are present in seed coat epidermal cells. Alternatively, some compensatory mechanisms, such as UDP-Xyl transport by specific transporters, could be activated (Ebert et al., 2015).
In addition to the changes in cell wall composition, our calcofluor staining studies revealed that distal cell walls of seed coat epidermal cells exhibited an abnormal rupture upon imbibition. Whether the changes observed in GalA, Rha, Xyl, and arabinan are responsible for this feature remains to be determined. However, an interesting observation was that the uuat1-2 mutant exhibits an increase in the level of HG methylation, a feature that was partially rescued in plants expressing UUAT1. The increase in labeling by the LM20 antibody in mucilage correlated with reduced ruthenium red staining, a dye that binds to the HG carboxyl groups, suggesting their blockage by methyl groups. In addition, more methyl groups were released from mucilage derived from uuat1-2 mutant seeds and lower PME activity was detected. This methylation increase may be the result of the adaptation that takes place in the uuat1-2 mutant to compensate for changes in its cell wall. Because the degree of HG methylation correlates to cell wall stiffness, methylation changes may contribute to the altered rupture of distal cell walls during seed imbibition. Regarding HG methylation and arabinan content, it is interesting to note that mutants containing lower HG methylation levels due to defective pectin methyltransferase activity exhibit an increase in Ara (Kim et al., 2015), which could account for higher arabinan levels. By contrast, uuat1-2 plants show a greater HG methylation and lower arabinan levels, suggesting that an inverse correlation may exist between arabinan content and the degree of HG methylation.
The expression pattern of UUAT1 indicates that it might have additional functional roles in other organs or cell types. Indeed, an evaluation of the cell wall sugar composition of other organs and cells from the uuat1-2 mutant showed lower levels of Ara in roots and pollen tubes, but not a decrease in Xyl or GalA. These results suggest that changes in the mutant plant cell wall composition are organ dependent. In this sense, Ara is the sugar most affected in the organs evaluated, suggesting that the supply of UDP-Ara for arabinosyltransferases is more affected in uuat1-2 plants. On the other hand, the levels of GalA and Xyl did not exhibit significant differences in the mutant, except in seeds. These results could be explained due to redundancy of paralog genes present in subclade V of the NST gene family (Rautengarten et al., 2014), which could supply UDP-GlcA for the UDP-GalA synthesis. This explanation is also valid for the absence of Xyl changes, but it is important to mention that UDP-Xyl levels in the Golgi lumen are also directly controlled by a UDP-Xyl transporter (UXT1), described recently by Ebert et al. (2015). Mutants in this transporter had decreased Xyl content in stems, and glucuronoxylan was strongly affected (Ebert et al., 2015), suggesting that the UDP-Xyl biosynthesized by cytosolic UXS and transported by UXT1 may be required for xylan synthesis. Furthermore, an Arabidopsis cytosolic UXS triple mutant was shown to have an irregular xylem phenotype, while the lumenal UXS triple mutant had no Xyl-associated phenotype (Kuang et al., 2016). These findings suggest that UUAT1 is less important in the synthesis of Xyl-containing polysaccharides. Furthermore, it is likely that the UDP-Xyl made in the cytosol is used to synthesize Xyl-containing polysaccharides, whereas the UDP-Xyl made in the Golgi lumen could be used for the synthesis of UDP-Arap in a tissue-specific manner. However, more data will be required to confirm this hypothesis. No changes were observed in the GlcA/Xyl ratio of glucuronoxylan, one of the main polymers containing GlcA, supporting the idea of redundancy in the UDP-GlcA transport.
Cell walls of the uuat1-2 mutant also exhibited other pleiotropic changes in sugar composition and HG methylation depending on the tissue analyzed. For instance, a Gal decrease was observed in roots, and a GalA increase occurred in pollen tubes. HGs can modulate cell wall stiffness in pollen tubes (Parre and Geitmann, 2005a), and callose content changes may also have an impact on the cell wall mechanics of pollen tubes (Parre and Geitmann, 2005b). Consequently, these changes may be a response to the Ara change observed in the uuat1-2 mutant. Stems also showed methylation increases at different development stages, which correlate with an early plant bolting phenotype. These pleiotropic events may correspond to adaptations of the mutant due to the absence of UUAT1 and are an indication of the plasticity displayed by the plant cell wall.
METHODS
Plant Material and Growth Conditions
Unless specified otherwise, Arabidopsis (Arabidopsis thaliana) and tobacco (Nicotiana benthamiana) plants were germinated and grown in a growth chamber using a long-day regime (16 h photoperiod); the light intensity was 100 μmol m−2 s−1 and the temperature 21°C. For seeds and aerial tissue collection, plants were grown in soil (Terracult blue substrate) supplemented with fertilizer (Basacote plus 6M; Compo Expert) in a relative humidity of 60%. The plants were grown in MS media (Duchefa) (2.155 g/L), 1% sucrose, and 0.8% agar to obtain root material. T-DNA insertion lines for UUAT1 (SALK_124146C/uuat1-1, SALK_105023C/uuat1-2, and SALK_048507/uuat1-3) were obtained from the ABRC (http://abrc.osu.edu/) using the SIGnAL Salk collection (Alonso et al., 2003). SALK_105023C was annotated as uuat1-2. Wild-type Col-0 and mutants were transformed using Agrobacterium tumefaciens (GV3101:pMP90) carrying the specified vectors, using the standard floral dip method (Clough and Bent, 1998).
Infiltration of Tobacco Leaves and Subcellular Localization of UUAT1-GFP
Six-week-old tobacco leaves were infiltrated with Agrobacterium strain GV3101:pMP90 as described (Batoko et al., 2000). Two independent transformations were undertaken for analyzing UUAT1 subcellular localization. ER localization was analyzed using UUAT1-GFP and the ER marker AtWAK-mCherry-HDEL (Wall Associated Kinase-2 carrying an ER retention signal). Golgi localization was analyzed by cotransformation of UUAT1-GFP and the Golgi-localized protein α-mannosidase I (α-mannosidase I-mCherry; Nelson et al., 2007). Fluorescent signals were analyzed 60 h after infiltration by confocal laser scanning microscopy using an Olympus Fluoview FV1000 spectral microscope.
GUS Staining
The histochemical localization of GUS activity was performed as described by Jefferson (1989). Tissues were imaged using an Olympus SZ61 stereoscopic microscope, and seeds were analyzed with an Olympus Fluoview FV1000 confocal microscope. Excitation at 488 nm and emission of 485 and 491 nm were used for the analysis of seed GUS staining by confocal microscopy (Truernit et al., 2008).
Ruthenium Red Staining
Mucilage released from mature dry seeds was stained either directly with 0.03% (w/v) ruthenium red or after imbibition in 0.5 M EDTA, pH 8.0, for 90 min. After EDTA treatment, seeds were stained for 2 min and observed with a light microscope (Olympus SZ61).
Genotyping
Genomic DNA was extracted from Arabidopsis 7-d-old cotyledons as described by Edwards et al. (1991). PCR was conducted to amplify the wild-type and mutant alleles using the primers described in Supplemental Table 3.
Cloning Procedures
The UUAT1 CDS without the stop codon was amplified from cDNA prepared from Arabidopsis leaf RNA, using the primers described in Supplemental Table 3. Resulting PCR products were introduced into the pENTR/D TOPO vector according to standard protocols (Life Technologies) to generate the entry clone pENTR-UUAT1. The C-terminal GFP fusion under the control of the cauliflower mosaic virus 35S promoter was generated by introducing the UUAT1 CDS from the entry clone into the gateway destination vector pK7FWG2.0 (Karimi et al., 2002) using LR clonase (Thermo Fisher Scientific). For the rescue construct, the intergenic region (653 bp) between At5g04170 and At5g04160 was defined as the UUAT1 promoter (pUUAT1) and was amplified from Arabidopsis genomic DNA using the primers described in Supplemental Table 3. Resultant PCR products were introduced into the pENTR 5-TOPO vector (Thermo Fisher Scientific) to generate the pENTR5-pUUAT1 entry clone. Both C-terminal GFP and HA fusions were obtained by recombining the entry clones pENTR-UUAT1 and pENTR5-pUUAT1 with destination vectors R4pGWB504 and R4pGWB513, respectively. For the transcriptional fusion of pUUAT1 to the GUS reporter gene, the entry clone pENTR5-pUUAT1 and the destination vector pKGWFS7 (Karimi et al., 2002) were recombined using LR clonase (Thermo Fisher Scientific).
Expression Analysis
Total RNA from stems, rosette and cauline leaves, and roots was extracted using Trizol (Thermo Fisher Scientific). For developing seeds, the pollination time in days after pollination was defined phenotypically as the time at which the flowers are just starting to open and the long stamens grow over the gynoecium, as previously described by Western et al. (2000). Seeds were dissected from approximately nine siliques in each DAP for further RNA extraction. RNA extractions were performed using an RNeasy Plus Micro Kit, according to the manufacturer’s instructions (Qiagen). One microgram of total RNA was used as a template for first-strand cDNA synthesis with an oligo(dT) primer and SuperScript II (Thermo Fisher Scientific), according to the manufacturer’s instructions. The primers described in Supplemental Table 3 were used to amplify PCR products from single-stranded cDNA in the wild-type and uuat1-2 samples for the CDS of UUAT1. The primers used to amplify EF1αA4 were those described by North et al. (2007). qPCR was performed using the Fast EvaGreen qPCR Master Mix kit (Mx3000P; Stratagene). Reactions contained 1 μL of 1:2 diluted cDNA in a total volume of 10 μL. Reactions were performed using primers that has been previously tested for their efficiency rates and sensitivity in a cDNA dilution series. The quantification and normalization procedures were done using the following equation, as described by Stratagene:
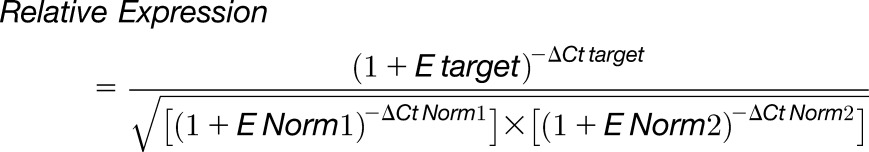 |
where E corresponds to the efficiency of amplification of target and reference genes, Ct is threshold cycle, and Norm1 and Norm2 refer to the references or normalizer genes. Primers for UUAT1, UUAT2, UUAT3, UUAT4, UUAT5 , EF1α (Hong et al., 2010), UBC9, and seed reference gene At4g12590 (Hong et al., 2010) were those described in Supplemental Table 3.
Analysis of in Vitro UUAT1 Transport
For heterologous expression, we used the uracil-auxotrophic Saccharomyces cerevisiae strain INVSc-1 (MATa his3D1 leu2 trp1-289 ura3-52 MAT his3D1 leu2 trp1-289 ura3-52; Thermo Fisher Scientific). The UUAT1 coding sequence was cloned onto the Gateway expression vector pYES-DEST52 (Thermo Fisher Scientific) and then introduced into the yeast strain with the S.c. EasyComp Transformation Kit (Thermo Fisher Scientific). The control was the yeast strain transformed with the empty vector pYES-DEST52. Microsomal fractions were obtained from 200-mL cultures grown at 30°C. Yeast cells were pelleted and spheroplasts produced in 10 mL resuspension buffer (50 mM potassium phosphate, pH 7.1, 1.4 M sorbitol, 10 mM NaN3, 40 mM 2-mercaptoethanol, and 6000 units of Lyticase [Sigma-Aldrich]) for 1 h at 37°C. Spheroplasts were harvested by centrifugation and washed with 0.8 M sorbitol, 10 mM triethanolamine/acetic acid, pH 7.2, and 1 mM EDTA. The spheroplasts were lysed with glass beads in 5 mL 0.8 M sorbitol, 10 mM triethanolamine/acetic acid, pH 7.2, 1 mM EDTA, protease inhibitor cocktail from Sigma-Aldrich, and 1 mM PMSF. Microsomes were isolated by sequential centrifugation (8000g for 10 min [F1] and 100,000g for 75 min [F2]). The F2 fraction was reconstituted in 10 mM Tricine-KOH, pH 7.5, 50 mM potassium gluconate, and 20% glycerol. Proteoliposomes were generated with acetone-washed soybean l-α-phosphatidylcholine (Avanti Polar Lipids) in reconstitution buffer (10 mM Tricine-KOH, pH 7.5, 50 mM potassium gluconate, and 20% glycerol). Reconstitution of microsomal membranes obtained from the UUAT1-expressing yeast or the control cells was undertaken using ∼400 µg microsomal protein in reconstitution buffer, lipid at a ratio of 13:1 (lipid:protein), 10 mM exchange substrate, and 50 mM octyl-β-glucoside. Unincorporated components were removed from reconstituted liposomes using Sephadex G50 columns (GE Healthcare). Aliquots (200 μL) were incubated with nucleotide sugar substrates at 25°C for the indicated times to assess transporter activities. Kinetic parameters were calculated with nonlinear regression using the Prism 7 application (GraphPad Software). Polyacrylamide gel electrophoresis was performed with 2.5 µg protein of proteoliposomes on a 7 to 15% SDS-PAGE gel. Immunoblotting was conducted with the anti-V5 antibody, using a 1:10,000 dilution (Thermo Fisher Scientific).
Nucleotide Sugar Quantification Using Tandem Mass Spectrometry
The transport assay reactions were purified using ENVI-Carb SPE columns (Sigma-Aldrich) and then lyophilized overnight, as outlined by Ito et al. (2014), and then analyzed by LC-MS/MS. Nucleotide sugars were separated using a Hypercarb column (150 mm × 1 mm, 5 μm) at a flow rate of 50 μL min−1 with an 1100 series HPLC system (Agilent Technologies) and a 4000 QTRAP LC-MS/MS system (Sciex) equipped with a Turbo ion spray ion source. Initial conditions were 95% buffer A (LC-MS grade water with 0.3% formic acid, pH 9.0, with ammonia) and 5% buffer B (100% acetonitrile) for 1 min followed by a gradient to 75% (A) in 20 min, then 50% (A) in 5 min before returning to 95% (A) in 5 min. The instrument was operated in negative ion mode, using the multiple reaction monitoring scan type. Declustering potential was −40, entrance potential was −10, and collision cell exit potential was −15. The ion spray voltage was set at −4200 V, source temperature (TEM) was 425°C, collision gas (CAD) was set to high, and source gas 1 (GS1) and 2 (GS2) were both set to 20. A time of 100 ms was applied for each transition, resulting in a duty cycle of 1.0501 s with both Q1 and Q3 resolution set to “unit.” All data were acquired using Analyst 1.6 Build 3773 (Sciex). Nucleotide sugars were quantified using MultiQuant 2.1 software (build 2.1.1296.02.1; Sciex) by integrating the signal peak areas of samples against a range of nucleotide sugar standards (2.5 to 20 pmol).
Seed Immunolabeling
Immunolabeling was performed with four monoclonal antibodies, CCRC-M36 (labels RG-I), LM6 (arabinan), JIM7 (highly methyl-esterified homogalacturonan), and LM20 (highly methyl-esterified homogalacturonan) (Saez-Aguayo et al., 2013). A double labeling with an antibody plus calcofluor white (0.01%) or propidium iodide (20 µg mL−1) was performed as indicated for each antibody to observe the seed surface and AM layer. Optical sections were obtained using an Olympus LX81 spectral confocal laser scanning microscope. A 488-nm argon laser line was used to excite Alexa Fluor 488, a 405-nm diode laser line was used to excite calcofluor white, and a 543-nm neon laser line was used to excite propidium iodide. Fluorescence emission was detected between 504 and 579 nm for Alexa Fluor 488, 412 and 490 nm for calcofluor white, and 550 nm and 725 nm for propidium iodide. For comparisons of the signal intensity within one experiment, the laser gain values were fixed.
Trichome Isolation
Trichome isolation was performed as described by Marks et al. (2008), with slight modifications. The aerial parts of 18-d-old seedlings were placed in a 50-mL tube with 15 mL of preheated (37°C) PBS (137 mM KCl, 10 mM K2HPO4, and 2 mM KH2PO4) containing 100 mM EGTA-KOH (pH 7.5) and 50 mg of glass beads (425 to 600 µm; Sigma-Aldrich). The plant material was then subjected to four cycles at maximum vortex speed for 30 s and on ice for 30 s. The trichomes were recovered using a nylon cell strainer (pore size 70 µm; BD Falcon) and resuspended in PBS buffer without EGTA.
AIR Preparation
Plant tissues were ground in liquid nitrogen and extracted twice in 80% ethanol with agitation for 1 h at room temperature followed by removal of lipids by washing twice with methanol:chloroform (1:1) and twice with acetone. The final AIR was dried overnight at room temperature. A sequential extraction procedure was used for determining the sugar composition of SM and seed+AM. Twenty milligrams of seeds was imbibed three times with 1 mL of water for 20 min; the SM was separated by 10 min of centrifugation at 12,000g, lyophilized, and resuspended in 300 μL of water before hydrolysis. The seed+AM fraction was lyophilized and AIR preparation was prepared as described above.
Acid Hydrolysis
Two milligrams of AIR was hydrolyzed for 20 min for soluble mucilage and 1 h for other tissues with 450 μL 2 M trifluoroacetic acid (TFA) at 121°C. TFA was evaporated at 60°C with nitrogen and the samples were washed twice in 250 μL of 100% isopropanol and dried in a SpeedVac. The suspension was clarified by passing through a syringe filter (pore size: 0.45 µm), transferred to a new tube, and used for HPAEC-PAD analysis as described below. Inositol was used as the internal control for TFA hydrolysis.
In Vitro Pollen Tube Growth and Cell Wall Extraction
Pollen was grown in vitro in a liquid medium according to the method described by Boavida and McCormick, (2007) and Dardelle et al. (2010). Forty freshly opened flowers were submerged in 1 mL of germination medium containing 5 mM CaCl2, 0.01% (w/v) H3BO3, 5 mM KCl, 1 mM MgSO4, and 10% (w/v) sucrose (pH 7.5), and tubes were shaken with a vortex to release the pollen grains from the anthers. Flowers were removed with a pair of tweezers, and the pollen suspension was then pelleted at 3200g for 6 min. New GM (250 µL) was added to the pellet, and pollen grains were grown in a growth chamber in the dark at 22°C for 6 h. Before any further manipulation, pollen germination and pollen tube growth were assessed with an inverted microscope. After 6 h, three volumes of 95% ethanol were added to the GM and stored at 4°C until use. Six-hour-old pollen tubes from 480 flowers were pooled, centrifuged at 5000g, and rinsed three times with 1 mL of 70% ethanol to remove salts and sucrose. The insoluble material was ground and treated three times with 70% ethanol at 70°C for 15 min followed by an incubation with 1 mL of a mixture of chloroform:methanol (1:1, v/v) for 15 min. After centrifugation (12,000g for 10 min), the remaining insoluble material was dried to yield the AIR fraction (∼1 mg). This experiment was performed three times.
Methylesterification Analysis of AIR Samples
The degree of methylesterification of wild-type Col-0 and uuat1-2 was analyzed in 2 mg of AIR preparations from roots or seeds with AM. For SM, 5 mg of seeds were imbibed in 200 μL of ultrapure water for 6 h, as described by Anthon and Barrett (2004). All experiments were done using three technical replicates and at least two biological replicates.
HPAEC-PAD
A Dionex ICS3000 ion chromatography system, equipped with a pulsed amperometric detector, a CarboPac PA1 (4 × 250 mm) analytical column, and a CarboPac PA1 (4 × 50 mm) guard column was used to quantify sugars. The separation of neutral sugars was performed at 40°C with a flow rate of 1 mL/min using an isocratic gradient of 20 mM NaOH for 20 min followed by a wash with 200 mM NaOH for 10 min. After every run, the column was equilibrated in 20 mM NaOH for 10 min. Separation of acidic sugars was performed using 150 mM NaOAc and 100 mM NaOH for 10 min at a flow rate of 1 mL/min at 40°C. Standard curves of neutral sugars (d-Fuc, l-Rha, l-Ara, d-Gal, d-Glc, d-Xyl, and d-Man) or acidic sugars (d-GalA and d-GlcA) were used for quantification.
Determination of Monosaccharide Composition of Pollen Tubes Using Gas Chromatography-Flame Ionization Detection
Samples were prepared as described by Dardelle et al. (2015). Approximately 0.5 mg of sample was hydrolyzed with 2 M TFA for 2 h at 110°C. Monosaccharides were then derivatized with 1 M methanol-HCl at 80°C overnight followed by a mixture of hexamethyldisiloxan:trimethyldisiloxan:pyridine (3:1:9) at 110°C for 20 min. After drying, derivatives were dissolved in 1 mL of cyclohexane and injected into the 3800 GC system equipped with a CP-Sil5-CB column. A temperature gradient from 120 to 160°C at 10°C min−1, 160 to 220°C at 1.5°C min−1, and 220 to 280°C at 20°C min−1 was used. Quantification was based on the internal standard and response factors previously determined for each monosaccharide.
Determination of PME Activity
Total protein extraction and PME activity assays were performed as described by Saez-Aguayo et al. (2013). Measurements of stained areas to determine PME activity were obtained using the ImageJ software (Abramoff et al., 2004)
Analysis of Stem Xylan Using PACE
AIR preparations and PACE were performed as described by Mortimer et al. (2010). One milligram of AIR from basal stems was incubated overnight in 0.1 M ammonium acetate buffer (pH 5.5) with an excess of Neocallimastix patriciarum Xyn11A xylanase at 21°C. Samples were derivatized with 8-aminonapthalene-1,3,6-trisulphonic acid (Invitrogen). After drying in vacuo, the samples were resuspended in 3 M urea (100 μL), of which 5 μL was loaded onto the gels. Samples were electrophoresed for 30 min at 200 V and then for 100 min at 1000 V. Gels were visualized using a Genebox (Syngene) equipped with a transilluminator with long-wave tubes emitting at 365 nm and a short-pass filter (500 to 600 nm). The quantity of each of the oligosaccharides released by Xyn11A [Xyl, (Xyl)2, GlcA-(Xyl)4/ Me-GlcA(Xyl)4] as well as the GlcA/Xyl ratio could be calculated by using the analytical software Genetools (Syngene). Results presented correspond to four biological replicates. The enzyme was a kind gift of Harry Gilbert (University of Newcastle, UK).
Accession Numbers
Nucleotide sequences for Arabidopsis UUAT1 have been deposited in GenBank (Benson et al., 2012) under accession numbers KT923621 (At5g04160, coding sequence) and KT923622 (At5g04160, promoter). T-DNA insertion lines in the At5g04160 locus were obtained from the ABRC: uuat1-1 (SALK_124146C), uuat1-2 (SALK_105023C), and uuat1-3 (SALK_048507).
Supplemental Data
Supplemental Figure 1. NST Genes Expressed during Seed Development.
Supplemental Figure 2. Uronic Acid Content in the Soluble Mucilage Fraction from Mutants in NST Genes.
Supplemental Figure 3. UUAT Expression in Developing Seeds.
Supplemental Figure 4. UUAT1 Hydropathy Plot.
Supplemental Figure 5. Rescue of the uuat1-2 Mutant Phenotype Using the UUAT1 ProUUAT1-UUAT1-GFP Construct.
Supplemental Figure 6. Exchange of Nucleotide Sugars with GMP, AMP, CMP, or UDP by UUAT1.
Supplemental Figure 7. Analyses of the Mucilage Phenotypes of uuat1-2 and the Rescued Lines Using Immunolocalization.
Supplemental Figure 8. Changes in Methylesterification Degree in uuat1 Allelic Mutants.
Supplemental Figure 9. UUAT1 Is Highly Expressed in Roots, Trichomes, Stems, and Seed Coat.
Supplemental Table 1. Percentage of Protein Identity among UUATs Family Members.
Supplemental Table 2. Sugar Composition of Soluble Mucilage from Wild-Type Col-0 and uuat1 Allelic Lines.
Supplemental Table 3. Sequences of Primers Used in This Study.
Supplementary Material
Acknowledgments
This work was supported by FONDECYT 11130498 (to F.C.R.), FONDECYT 3140415 (to S.S.-A.), FONDECYT 1151335, Fondo de Areas Prioritarias-Centro de Regulacion del Genoma-15090007, ECOS-CONICYT C14B02 and PFB-16 (to A.O.), and a CONICYT fellowship to H.T. Work conducted by the Joint BioEnergy Institute was supported by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research, through contract DE-AC02-05CH11231 between Lawrence Berkeley National Laboratory and the U. S. Department of Energy. J.L.H. is supported by an Australian Research Council Future Fellowship (FT130101165). Work conducted by Glyco-MEV (J.-C.M. and A.L.) was in part supported by the VASI research network from the Upper Normandy region and the French ministry of research. We thank Miriam Barros for her advice and expertise in confocal microscopy. We also thank Hernan Salinas and Alvaro Miquel for technical assistance with HPAEC analysis, and Flavien Dardelle and François Le Mauff for technical assistance with gas chromatography analyses of pollen tube cell walls.
AUTHOR CONTRIBUTIONS
S.S.-A., F.C.R., H.V.S., J.L.H., and A.O. designed the research. S.S.-A., C.R., B.E., D.S., T.E., H.T., O.S.-I., D.D., J.-P.P.-R., A.L., J.-C.M., and F.C.R. performed the experiments. S.S.-A., C.R., H.T., J.-C.M., P.D., J.L.H., H.V.S., F.C.R., and A.O. analyzed the data. S.S.-A., F.C.R., A.O., and H.T. wrote the article.
Glossary
- GT
glycosyltransferase
- NST
nucleotide sugar transporter
- TPT
triose phosphate translocator
- UDP-GlcA
UDP-glucuronic acid
- UXS
UDP-xylose synthase
- RG-I
rhamnogalacturonan I
- HG
homogalacturonan
- SM
soluble mucilage
- AM
adherent mucilage
- DAP
days after pollination
- CDS
coding sequence
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- ER
endoplasmic reticulum
- PME
pectin methylesterase
- PACE
carbohydrate gel electrophoresis
- AIR
alcohol-insoluble residue
- TFA
trifluoroacetic acid
- HPAEC-PAD
high-performance anion exchange chromatography with pulsed amperometric detection
Footnotes
Articles can be viewed without a subscription.
References
- Abramoff M.D., Magalhães P.J., Ram S.J. (2004). Image processing with ImageJ. Biophotonics International 11: 36–42. [Google Scholar]
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657. [DOI] [PubMed] [Google Scholar]
- Anthon G.E., Barrett D.M. (2004). Comparison of three colorimetric reagents in the determination of methanol with alcohol oxidase. Application to the assay of pectin methylesterase. J. Agric. Food Chem. 52: 3749–3753. [DOI] [PubMed] [Google Scholar]
- Bakker H., Routier F., Oelmann S., Jordi W., Lommen A., Gerardy-Schahn R., Bosch D. (2005). Molecular cloning of two Arabidopsis UDP-galactose transporters by complementation of a deficient Chinese hamster ovary cell line. Glycobiology 15: 193–201. [DOI] [PubMed] [Google Scholar]
- Baldwin T.C., Handford M.G., Yuseff M.I., Orellana A., Dupree P. (2001). Identification and characterization of GONST1, a golgi-localized GDP-mannose transporter in Arabidopsis. Plant Cell 13: 2283–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Peled M., O’Neill M.A. (2011). Plant nucleotide sugar formation, interconversion, and salvage by sugar recycling. Annu. Rev. Plant Biol. 62: 127–155. [DOI] [PubMed] [Google Scholar]
- Batoko H., Zheng H.-Q., Hawes C., Moore I. (2000). A rab1 GTPase is required for transport between the endoplasmic reticulum and Golgi apparatus and for normal Golgi movement in plants. Plant Cell 12: 2201–2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson D.A., Karsch-Mizrachi I., Clark K., Lipman D.J., Ostell J., Sayers E.W. (2012). GenBank. Nucleic Acids Res. 40: D48–D53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berninsone P.M., Hirschberg C.B. (2000). Nucleotide sugar transporters of the Golgi apparatus. Curr. Opin. Struct. Biol. 10: 542–547. [DOI] [PubMed] [Google Scholar]
- Boavida L.C., McCormick S. (2007). Temperature as a determinant factor for increased and reproducible in vitro pollen germination in Arabidopsis thaliana. Plant J. 52: 570–582. [DOI] [PubMed] [Google Scholar]
- Boevink P., Oparka K., Santa Cruz S., Martin B., Betteridge A., Hawes C. (1998). Stacks on tracks: the plant Golgi apparatus traffics on an actin/ER network. Plant J. 15: 441–447. [DOI] [PubMed] [Google Scholar]
- Bonin C.P., Potter I., Vanzin G.F., Reiter W.-D. (1997). The MUR1 gene of Arabidopsis thaliana encodes an isoform of GDP-D-mannose-4,6-dehydratase, catalyzing the first step in the de novo synthesis of GDP-L-fucose. Proc. Natl. Acad. Sci. USA 94: 2085–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burget E.G., Verma R., Mølhøj M., Reiter W.-D. (2003). The biosynthesis of L-arabinose in plants: molecular cloning and characterization of a Golgi-localized UDP-D-xylose 4-epimerase encoded by the MUR4 gene of Arabidopsis. Plant Cell 15: 523–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Dardelle F., Lehner A., Ramdani Y., Bardor M., Lerouge P., Driouich A., Mollet J.-C. (2010). Biochemical and immunocytological characterizations of Arabidopsis pollen tube cell wall. Plant Physiol. 153: 1563–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dardelle F., Le Mauff F., Lehner A., Loutelier-Bourhis C., Bardor M., Rihouey C., Causse M., Lerouge P., Driouich A., Mollet J.-C. (2015). Pollen tube cell walls of wild and domesticated tomatoes contain arabinosylated and fucosylated xyloglucan. Ann. Bot. (Lond.) 115: 55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert B., et al. (2015). Identification and characterization of a Golgi-localized UDP-Xylose transporter family from Arabidopsis. Plant Cell 27: 1218–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards K., Johnstone C., Thompson C. (1991). A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 19: 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X., Bar-Peled M. (2004). The biosynthesis of UDP-galacturonic acid in plants. Functional cloning and characterization of Arabidopsis UDP-D-glucuronic acid 4-epimerase. Plant Physiol. 136: 4256–4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyett P., Glushka J., Gu X., Bar-Peled M. (2009). Real-time NMR monitoring of intermediates and labile products of the bifunctional enzyme UDP-apiose/UDP-xylose synthase. Carbohydr. Res. 344: 1072–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handford M., Rodríguez-Furlán C., Marchant L., Segura M., Gómez D., Alvarez-Buylla E., Xiong G.-Y., Pauly M., Orellana A. (2012). Arabidopsis thaliana AtUTr7 encodes a golgi-localized UDP-glucose/UDP-galactose transporter that affects lateral root emergence. Mol. Plant 5: 1263–1280. [DOI] [PubMed] [Google Scholar]
- Handford M.G., Sicilia F., Brandizzi F., Chung J.H., Dupree P. (2004). Arabidopsis thaliana expresses multiple Golgi-localised nucleotide-sugar transporters related to GONST1. Mol. Genet. Genomics 272: 397–410. [DOI] [PubMed] [Google Scholar]
- Harper A.D., Bar-Peled M. (2002). Biosynthesis of UDP-xylose. Cloning and characterization of a novel Arabidopsis gene family, UXS, encoding soluble and putative membrane-bound UDP-glucuronic acid decarboxylase isoforms. Plant Physiol. 130: 2188–2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S.M., Bahn S.C., Lyu A., Jung H.S., Ahn J.H. (2010). Identification and testing of superior reference genes for a starting pool of transcript normalization in Arabidopsis. Plant Cell Physiol. 51: 1694–1706. [DOI] [PubMed] [Google Scholar]
- Hu R., Li J., Wang X., Zhao X., Yang X., Tang Q., He G., Zhou G., Kong Y. (2016). Xylan synthesized by Irregular Xylem 14 (IRX14) maintains the structure of seed coat mucilage in Arabidopsis. J. Exp. Bot. 67: 1243–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito N., Ito K., Ikebuchi Y., Kito T., Miyata H., Toyoda Y., Takada T., Hisaka A., Honma M., Oka A., Kusuhara H., Suzuki H. (2014). Organic cation transporter/solute carrier family 22a is involved in drug transfer into milk in mice. J. Pharm. Sci. 103: 3342–3348. [DOI] [PubMed] [Google Scholar]
- Jefferson R.A. (1989). The GUS reporter gene system. Nature 342: 837–838. [DOI] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195. [DOI] [PubMed] [Google Scholar]
- Kim S.-J., Held M.A., Zemelis S., Wilkerson C., Brandizzi F. (2015). CGR2 and CGR3 have critical overlapping roles in pectin methylesterification and plant growth in Arabidopsis thaliana. Plant J. 82: 208–220. [DOI] [PubMed] [Google Scholar]
- Knappe S., Flügge U.-I., Fischer K. (2003). Analysis of the plastidic phosphate translocator gene family in Arabidopsis and identification of new phosphate translocator-homologous transporters, classified by their putative substrate-binding site. Plant Physiol. 131: 1178–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi T., Takeda T., Miyazaki Y., Ohnishi-Kameyama M., Hayashi T., O’Neill M.A., Ishii T. (2007). A plant mutase that interconverts UDP-arabinofuranose and UDP-arabinopyranose. Glycobiology 17: 345–354. [DOI] [PubMed] [Google Scholar]
- Kotake T., Hojo S., Yamaguchi D., Aohara T., Konishi T., Tsumuraya Y. (2007). Properties and physiological functions of UDP-sugar pyrophosphorylase in Arabidopsis. Biosci. Biotechnol. Biochem. 71: 761–771. [DOI] [PubMed] [Google Scholar]
- Kotake T., Takata R., Verma R., Takaba M., Yamaguchi D., Orita T., Kaneko S., Matsuoka K., Koyama T., Reiter W.D., Tsumuraya Y. (2009). Bifunctional cytosolic UDP-glucose 4-epimerases catalyse the interconversion between UDP-D-xylose and UDP-L-arabinose in plants. Biochem. J. 424: 169–177. [DOI] [PubMed] [Google Scholar]
- Kotake T., Yamanashi Y., Imaizumi C., Tsumuraya Y. (2016). Metabolism of L-arabinose in plants. J. Plant Res. 129: 781–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang B., et al. (2016). The role of UDP-glucuronic acid decarboxylase(s) (UXS) in xylan biosynthesis in Arabidopsis. Mol. Plant 9: 1119–1131. [DOI] [PubMed] [Google Scholar]
- Le B.H., et al. (2010). Global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. Proc. Natl. Acad. Sci. USA 107: 8063–8070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liepman A.H., Wightman R., Geshi N., Turner S.R., Scheller H.V. (2010). Arabidopsis - a powerful model system for plant cell wall research. Plant J. 61: 1107–1121. [DOI] [PubMed] [Google Scholar]
- Macquet A., Ralet M.-C., Kronenberger J., Marion-Poll A., North H.M. (2007). In situ, chemical and macromolecular study of the composition of Arabidopsis thaliana seed coat mucilage. Plant Cell Physiol. 48: 984–999. [DOI] [PubMed] [Google Scholar]
- Marks M.D., Betancur L., Gilding E., Chen F., Bauer S., Wenger J.P., Dixon R.A., Haigler C.H. (2008). A new method for isolating large quantities of Arabidopsis trichomes for transcriptome, cell wall and other types of analyses. Plant J. 56: 483–492. [DOI] [PubMed] [Google Scholar]
- Mølhøj M., Verma R., Reiter W.D. (2003). The biosynthesis of the branched-chain sugar d-apiose in plants: functional cloning and characterization of a UDP-d-apiose/UDP-d-xylose synthase from Arabidopsis. Plant J. 35: 693–703. [DOI] [PubMed] [Google Scholar]
- Mølhøj M., Verma R., Reiter W.-D. (2004). The biosynthesis of D-Galacturonate in plants. functional cloning and characterization of a membrane-anchored UDP-D-Glucuronate 4-epimerase from Arabidopsis. Plant Physiol. 135: 1221–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer J.C., Miles G.P., Brown D.M., Zhang Z., Segura M.P., Weimar T., Yu X., Seffen K.A., Stephens E., Turner S.R., Dupree P. (2010). Absence of branches from xylan in Arabidopsis gux mutants reveals potential for simplification of lignocellulosic biomass. Proc. Natl. Acad. Sci. USA 107: 17409–17414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson B.K., Cai X., Nebenführ A. (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51: 1126–1136. [DOI] [PubMed] [Google Scholar]
- Norambuena L., Marchant L., Berninsone P., Hirschberg C.B., Silva H., Orellana A. (2002). Transport of UDP-galactose in plants. Identification and functional characterization of AtUTr1, an Arabidopsis thaliana UDP-galactos/UDP-glucose transporter. J. Biol. Chem. 277: 32923–32929. [DOI] [PubMed] [Google Scholar]
- Norambuena L., Nilo R., Handford M., Reyes F., Marchant L., Meisel L., Orellana A. (2005). AtUTr2 is an Arabidopsis thaliana nucleotide sugar transporter located in the Golgi apparatus capable of transporting UDP-galactose. Planta 222: 521–529. [DOI] [PubMed] [Google Scholar]
- North H.M., De Almeida A., Boutin J.-P., Frey A., To A., Botran L., Sotta B., Marion-Poll A. (2007). The Arabidopsis ABA-deficient mutant aba4 demonstrates that the major route for stress-induced ABA accumulation is via neoxanthin isomers. Plant J. 50: 810–824. [DOI] [PubMed] [Google Scholar]
- Orellana A., Moraga C., Araya M., Moreno A. (2016). Overview of nucleotide sugar transporter gene family functions across multiple species. J. Mol. Biol. 428: 3150–3165. [DOI] [PubMed] [Google Scholar]
- Parre E., Geitmann A. (2005a). Pectin and the role of the physical properties of the cell wall in pollen tube growth of Solanum chacoense. Planta 220: 582–592. [DOI] [PubMed] [Google Scholar]
- Parre E., Geitmann A. (2005b). More than a leak sealant. The mechanical properties of callose in pollen tubes. Plant Physiol. 137: 274–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattathil S., Harper A.D., Bar-Peled M. (2005). Biosynthesis of UDP-xylose: characterization of membrane-bound AtUxs2. Planta 221: 538–548. [DOI] [PubMed] [Google Scholar]
- Peaucelle A., Louvet R., Johansen J.N., Höfte H., Laufs P., Pelloux J., Mouille G. (2008). Arabidopsis phyllotaxis is controlled by the methyl-esterification status of cell-wall pectins. Curr. Biol. 18: 1943–1948. [DOI] [PubMed] [Google Scholar]
- Ralet M.C., Crépeau M.J., Vigouroux J., Tran J., Berger A., Sallé C., Granier F., Botran L., North H.M. (2016). Xylans provide the structural driving force for mucilage adhesion to the Arabidopsis seed coat. Plant Physiol. 171: 165–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rautengarten C., Ebert B., Herter T., Petzold C.J., Ishii T., Mukhopadhyay A., Usadel B., Scheller H.V. (2011). The interconversion of UDP-arabinopyranose and UDP-arabinofuranose is indispensable for plant development in Arabidopsis. Plant Cell 23: 1373–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rautengarten C., et al. (2014). The Golgi localized bifunctional UDP-rhamnose/UDP-galactose transporter family of Arabidopsis. Proc. Natl. Acad. Sci. USA 111: 11563–11568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rautengarten C., Ebert B., Liu L., Stonebloom S., Smith-Moritz A.M., Pauly M., Orellana A., Scheller H.V., Heazlewood J.L. (2016). The Arabidopsis Golgi-localized GDP-L-fucose transporter is required for plant development. Nat. Commun. 7: 12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reboul R., Geserick C., Pabst M., Frey B., Wittmann D., Lütz-Meindl U., Léonard R., Tenhaken R. (2011). Down-regulation of UDP-glucuronic acid biosynthesis leads to swollen plant cell walls and severe developmental defects associated with changes in pectic polysaccharides. J. Biol. Chem. 286: 39982–39992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes F., Orellana A. (2008). Golgi transporters: opening the gate to cell wall polysaccharide biosynthesis. Curr. Opin. Plant Biol. 11: 244–251. [DOI] [PubMed] [Google Scholar]
- Reyes F., Marchant L., Norambuena L., Nilo R., Silva H., Orellana A. (2006). AtUTr1, a UDP-glucose/UDP-galactose transporter from Arabidopsis thaliana, is located in the endoplasmic reticulum and up-regulated by the unfolded protein response. J. Biol. Chem. 281: 9145–9151. [DOI] [PubMed] [Google Scholar]
- Reyes F., León G., Donoso M., Brandizzí F., Weber A.P.M., Orellana A. (2010). The nucleotide sugar transporters AtUTr1 and AtUTr3 are required for the incorporation of UDP-glucose into the endoplasmic reticulum, are essential for pollen development and are needed for embryo sac progress in Arabidopsis thaliana. Plant J. 61: 423–435. [DOI] [PubMed] [Google Scholar]
- Rollwitz I., Santaella M., Hille D., Flügge U.-I., Fischer K. (2006). Characterization of AtNST-KT1, a novel UDP-galactose transporter from Arabidopsis thaliana. FEBS Lett. 580: 4246–4251. [DOI] [PubMed] [Google Scholar]
- Rösti J., Barton C.J., Albrecht S., Dupree P., Pauly M., Findlay K., Roberts K., Seifert G.J. (2007). UDP-glucose 4-epimerase isoforms UGE2 and UGE4 cooperate in providing UDP-galactose for cell wall biosynthesis and growth of Arabidopsis thaliana. Plant Cell 19: 1565–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez-Aguayo S., Ralet M.-C., Berger A., Botran L., Ropartz D., Marion-Poll A., North H.M. (2013). PECTIN METHYLESTERASE INHIBITOR6 promotes Arabidopsis mucilage release by limiting methylesterification of homogalacturonan in seed coat epidermal cells. Plant Cell 25: 308–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Jore-Dupas C., Nebenführ A., Boulaflous A., Follet-Gueye M.-L., Plasson C., Hawes C., Driouich A., Faye L., Gomord V. (2006). Plant N-glycan processing enzymes employ different targeting mechanisms for their spatial arrangement along the secretory pathway. Plant Cell 18: 3182–3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheible W.-R., Pauly M. (2004). Glycosyltransferases and cell wall biosynthesis: novel players and insights. Curr. Opin. Plant Biol. 7: 285–295. [DOI] [PubMed] [Google Scholar]
- Seifert G.J. (2004). Nucleotide sugar interconversions and cell wall biosynthesis: how to bring the inside to the outside. Curr. Opin. Plant Biol. 7: 277–284. [DOI] [PubMed] [Google Scholar]
- Sterling J.D., Quigley H.F., Orellana A., Mohnen D. (2001). The catalytic site of the pectin biosynthetic enzyme alpha-1,4-galacturonosyltransferase is located in the lumen of the Golgi. Plant Physiol. 127: 360–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truernit E., Bauby H., Dubreucq B., Grandjean O., Runions J., Barthélémy J., Palauqui J.-C. (2008). High-resolution whole-mount imaging of three-dimensional tissue organization and gene expression enables the study of phloem development and structure in Arabidopsis. Plant Cell 20: 1494–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temple H., Saez-Aguayo S., Reyes F.C., Orellana A. (2016). The inside and outside: topological issues in plant cell wall biosynthesis and the roles of nucleotide sugar transporters. Glycobiology 26: 913–925. [DOI] [PubMed] [Google Scholar]
- Verhertbruggen Y., Marcus S.E., Haeger A., Ordaz-Ortiz J.J., Knox J.P. (2009). An extended set of monoclonal antibodies to pectic homogalacturonan. Carbohydr. Res. 344: 1858–1862. [DOI] [PubMed] [Google Scholar]
- Voiniciuc C., Günl M., Schmidt M.H.-W., Usadel B. (2015). Highly branched xylan made by IRREGULAR XYLEM14 and MUCILAGE-RELATED21 links mucilage to Arabidopsis seeds. Plant Physiol. 169: 2481–2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Western T.L. (2012). The sticky tale of seed coat mucilages: production, genetics, and role in seed germination and dispersal. Seed Sci. Res. 22: 1–25. [Google Scholar]
- Western T.L., Burn J., Tan W.L., Skinner D.J., Martin-McCaffrey L., Moffatt B.A., Haughn G.W. (2001). Isolation and characterization of mutants defective in seed coat mucilage secretory cell development in Arabidopsis. Plant Physiol. 127: 998–1011. [PMC free article] [PubMed] [Google Scholar]
- Western T.L., Skinner D.J., Haughn G.W. (2000). Differentiation of mucilage secretory cells of the Arabidopsis seed coat. Plant Physiol. 122: 345–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Western T.L., Young D.S., Dean G.H., Tan W.L., Samuels A.L., Haughn G.W. (2004). MUCILAGE-MODIFIED4 encodes a putative pectin biosynthetic enzyme developmentally regulated by APETALA2, TRANSPARENT TESTA GLABRA1, and GLABRA2 in the Arabidopsis seed coat. Plant Physiol. 134: 296–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willats W.G., McCartney L., Knox J.P. (2001). In-situ analysis of pectic polysaccharides in seed mucilage and at the root surface of Arabidopsis thaliana. Planta 213: 37–44. [DOI] [PubMed] [Google Scholar]
- Wulff C., Norambuena L., Orellana A. (2000). GDP-fucose uptake into the Golgi apparatus during xyloglucan biosynthesis requires the activity of a transporter-like protein other than the UDP-glucose transporter. Plant Physiol. 122: 867–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T., Bar-Peled L., Gebhart L., Lee S.G., Bar-Peled M. (2009). Identification of galacturonic acid-1-phosphate kinase, a new member of the GHMP kinase superfamily in plants, and comparison with galactose-1-phosphate kinase. J. Biol. Chem. 284: 21526–21535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R.E., McFarlane H.E., Hahn M.G., Western T.L., Haughn G.W., Samuels A.L. (2008). Analysis of the Golgi apparatus in Arabidopsis seed coat cells during polarized secretion of pectin-rich mucilage. Plant Cell 20: 1623–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zablackis E., Huang J., Müller B., Darvill A.G., Albersheim P. (1995). Characterization of the cell-wall polysaccharides of Arabidopsis thaliana leaves. Plant Physiol. 107: 1129–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.