WIND1 directly activates the expression of another AP2/ERF transcription factor, ENHANCER OF SHOOT REGENERATION1, to promote callus formation and subsequent shoot regeneration.
Abstract
Many plant species display remarkable developmental plasticity and regenerate new organs after injury. Local signals produced by wounding are thought to trigger organ regeneration but molecular mechanisms underlying this control remain largely unknown. We previously identified an AP2/ERF transcription factor WOUND INDUCED DEDIFFERENTIATION1 (WIND1) as a central regulator of wound-induced cellular reprogramming in plants. In this study, we demonstrate that WIND1 promotes callus formation and shoot regeneration by upregulating the expression of the ENHANCER OF SHOOT REGENERATION1 (ESR1) gene, which encodes another AP2/ERF transcription factor in Arabidopsis thaliana. The esr1 mutants are defective in callus formation and shoot regeneration; conversely, its overexpression promotes both of these processes, indicating that ESR1 functions as a critical driver of cellular reprogramming. Our data show that WIND1 directly binds the vascular system-specific and wound-responsive cis-element-like motifs within the ESR1 promoter and activates its expression. The expression of ESR1 is strongly reduced in WIND1-SRDX dominant repressors, and ectopic overexpression of ESR1 bypasses defects in callus formation and shoot regeneration in WIND1-SRDX plants, supporting the notion that ESR1 acts downstream of WIND1. Together, our findings uncover a key molecular pathway that links wound signaling to shoot regeneration in plants.
INTRODUCTION
Many multicellular organisms regenerate their bodies after injury, and this regenerative capacity is vital for their survival after partial loss of their bodies. Plants, in particular, maintain high developmental plasticity during postembryonic development and display diverse forms of regeneration (Ikeuchi et al., 2016). One common example of plant regeneration is de novo organogenesis, i.e., the formation of new organs such as shoots and roots, from cut sites. This mode of regeneration has been widely used in agriculture as a tool, for instance, for propagation of elite cultivars and genetic engineering (Thorpe, 2007). As in animals, plant regeneration is initiated by at least two cellular mechanisms. One is by the reactivation of relatively undifferentiated cells existing in the somatic tissue and the other is by the reprogramming of mature somatic cells (Birnbaum and Sánchez Alvarado, 2008; Tanaka and Reddien, 2011; Ikeuchi et al., 2016). In some cases, these initiating cells directly regenerate new organs, but in other cases they first develop callus, a mass of dividing cells, from which new organs form (Hicks, 1994).
Molecular mechanisms underlying plant organ regeneration have been studied mostly in vitro where the balance between two plant hormones, auxin and cytokinin, determines the developmental fate of regenerating organs. Generally, a high ratio of auxin to cytokinin favors root regeneration, while a low ratio of auxin to cytokinin stimulates shoot regeneration (Skoog and Miller, 1957). Intermediate levels of auxin and cytokinin promote callus formation (Skoog and Miller, 1957). A protocol routinely used for Arabidopsis thaliana explants involves first incubation of a tissue fragment on auxin- and cytokinin-containing callus-inducing medium (CIM) to produce callus and subsequent transfer to cytokinin-rich shoot-inducing medium (SIM) and auxin-rich root-inducing medium to promote shoot and root regeneration, respectively (Valvekens et al., 1988). Accumulating evidence suggests that callus on CIM primarily derives from relatively undifferentiated pericycle cells through a genetic program underlying auxin-induced lateral root development (Che et al., 2007; Atta et al., 2009; Sugimoto et al., 2010). Accordingly, many regulators of lateral root development, including ABERRANT LATERAL ROOT4, AUXIN RESPONSE FACTOR7 (ARF7), ARF19, LATERAL ORGAN BOUNDARIES DOMAIN16 (LBD16), LBD17, LBD18, and LBD29, are required for callus formation on CIM (Sugimoto et al., 2010, Fan et al., 2012, Ikeuchi et al., 2013). A recent study has demonstrated that additional regulators, PLETHORA3 (PLT3), PLT5, and PLT7, are also needed to make CIM-induced callus pluripotent (Kareem et al., 2015). Key players acting downstream of PLT3, PLT5, and PLT7 to confer pluripotency are PLT1 and PLT2, which are also known for their role in root meristem development (Aida et al., 2004; Galinha et al., 2007). PLT3, PLT5, and PLT7, in addition, induce CUP SHAPED COTYLEDON1 (CUC1) and CUC2, important regulators of shoot meristem development during embryogenesis (Aida et al., 1997, 1999), presumably to introduce the potential to form shoots in the callus (Kareem et al., 2015). While CUC1 and CUC2 do not show an organized pattern of expression in CIM-induced callus, some root meristem regulators, such as WUSCHEL-RELATED HOMEOBOX5 (WOX5) and SCARECROW, display expression patterns similar to those observed in the root meristem (Gordon et al., 2007; Atta et al., 2009; Sugimoto et al., 2010). Thus, CIM-induced callus appears to represent a pluripotent cell mass that has characteristics more similar to root meristems (Ikeuchi et al., 2013).
Given that CIM-induced callus possesses root meristem-like properties, regenerating roots after transfer to root-inducing medium might be relatively straightforward, requiring further establishment of root meristem identity and execution of root developmental program by an auxin-induced transcriptional cascade (Ozawa et al., 1998; Che et al., 2002; Ikeuchi et al., 2016). By contrast, shoot regeneration on SIM ought to be more complex as it requires the conversion of root meristem fate into shoot meristem fate. What is central for the shoot meristem initiation is the activation of the key shoot stem cell regulator WUSCHEL (WUS) by cytokinin, which facilitates the partitioning of CIM-induced callus into WUS-expressing domains and CUC2-expressing domains (Che et al., 2006; Gordon et al., 2007; Chatfield et al., 2013). A cluster of CUC2-expressing cells continues to proliferate to form promeristems, in which polarized expression of the auxin transporter PIN-FORMED1 and another meristem regulator SHOOT MERISTEMLESS (STM) further organizes the formation of functional shoot meristem. Previous studies have also identified several other regulators, such as ENHANCER OF SHOOT REGENERATION1/DORNRÖSCHEN (ESR1/DRN), ESR2/DRN-LIKE (DRNL), and RAP2.6L, that contribute to shoot regeneration in vitro (Banno et al., 2001; Kirch et al., 2003; Che et al., 2006). An early study showed that overexpression of ESR1 promotes shoot regeneration without or at low doses of exogenous cytokinin (Banno et al., 2001). The esr1-1/drn-2 loss-of-function mutant, referred to as esr1-1 hereafter, however, does not display strong defects in shoot regeneration when cultured on CIM and SIM (Matsuo et al., 2011). By contrast, loss-of-function mutations in ESR2 and RAP2.6L cause clear defects in in vitro shoot regeneration (Matsuo et al., 2011; Che et al., 2006), suggesting that they play more profound roles.
We recently showed that wound stress provides another important cue for shoot regeneration, since intact plants cultured on CIM and SIM hardly regenerate shoots without wounding (Iwase et al. 2015). Wounding provokes various physiological responses, including rapid induction of reactive oxygen species, Ca2+ waves, and the production of stress-responsive hormones (Miller et al., 2009; Mousavi et al., 2013), but whether these early physiological responses direct cells for reprogramming is not established. A set of key regulators that are rapidly activated in response to wounding and have pivotal roles in wound-induced callus formation are a subfamily of AP2/ERF transcription factors, WOUND INDUCED DEDIFFERENTIATION1 (WIND1; aka RAP2.4), WIND2, WIND3, and WIND4 (Iwase et al., 2011a, 2011b). All WIND genes are induced by wounding and overexpression of each of them promotes callus formation (Iwase et al., 2011a, 2011b). Importantly, WIND1 substitutes the early wound response and confers pluripotency, since plants overexpressing WIND1 regenerate shoots on SIM without wound stress (Iwase et al., 2015). Conversely, dominant repression of WIND1 in WIND1-SRDX explants strongly blocks shoot regeneration, suggesting that WIND proteins function as key regulators of cellular reprogramming in response to wound stress (Iwase et al., 2015; Ikeuchi et al., 2016).
In this study, we set out to investigate how wound signaling links to regeneration at the molecular level. WIND proteins are likely to play key roles in this regulation and identification of genes directly targeted by WIND1 should help unveil how WIND1-mediated signaling controls the transcription of key regulators in regeneration. We provide both in vivo and in vitro evidence that WIND1 directly binds the ESR1 promoter and activates its expression. We also show that ESR1 functions downstream of WIND1 and facilitates both callus formation and shoot regeneration in response to wound stress. Our results thus uncover a key transcriptional mechanism that directly links the wound response to organ regeneration in plants.
RESULTS
Wound Stress Activates ESR1 Expression in a WIND1-Dependent Manner
Our previous microarray data showed that ESR1 expression is strongly upregulated in callus-overexpressing WIND1 under the control of the cauliflower mosaic virus 35S promoter (Iwase et al., 2011a). This is interesting, since ESR1 expression is restricted to the shoot apical meristem and leaf primordia during normal development (Kirch et al., 2003) and its overexpression confers increased shoot regenerative capacity in tissue culture (Banno et al., 2001). To validate this observation, we compared the level of ESR1 expression between 14-d-old wild-type seedlings and 35S:WIND1 callus. As shown in Supplemental Figure 1, our RT-qPCR analysis confirmed very low abundance of ESR1 transcripts in wild-type seedlings and a strong increase in 35S:WIND1 callus.
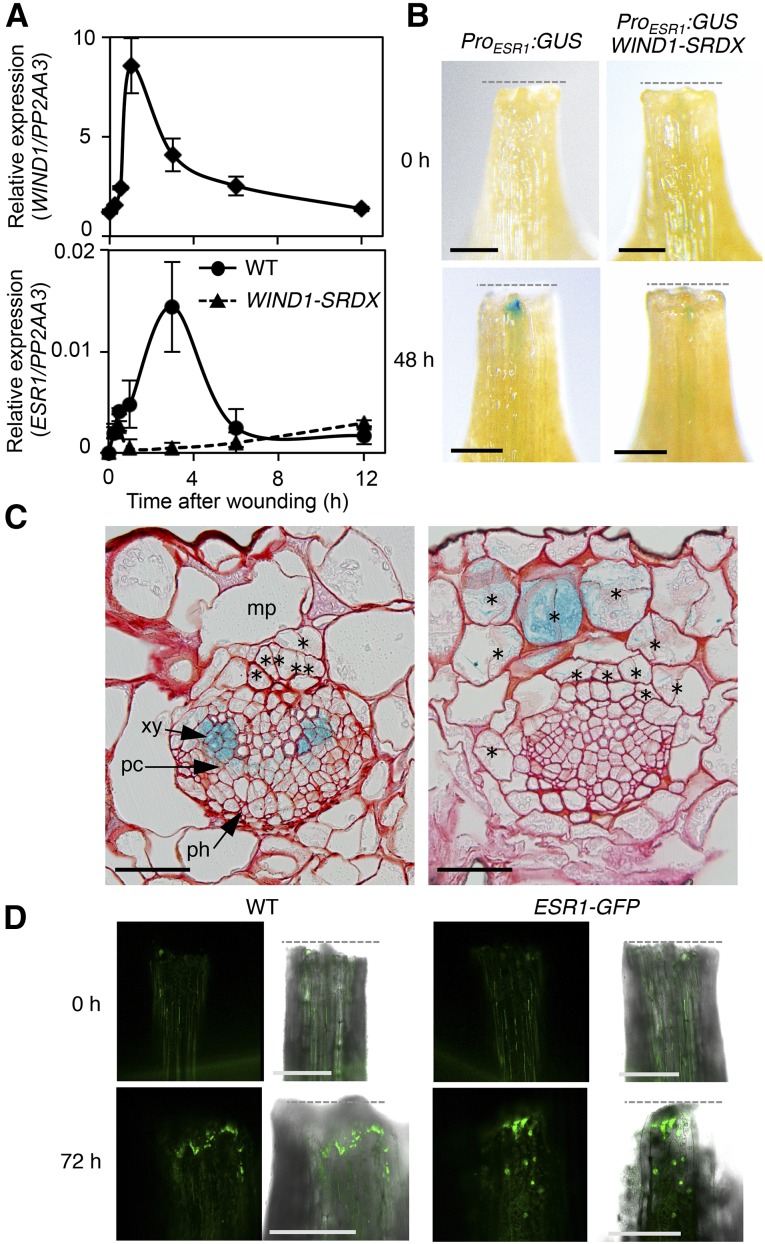
Given that the expression of the WIND1 gene is strongly activated by wounding (Iwase et al., 2011a), we then tested whether ESR1 is also upregulated in response to wound stress. Our time-course expression analysis using leaf explants dissected from 14-d-old wild-type seedlings revealed that WIND1 mRNA levels start to increase within 30 min after wounding and peak at 1 h (Figure 1A). Similarly, the level of ESR1 transcripts starts to increase within 30 min after wounding and peaks by 3 h (Figure 1A). ESR1 expression is barely detectable in wild-type root and hypocotyl explants, but we also detected an increase in the ESR1 expression in these organs after wounding (Supplemental Figures 2A and 2B). Importantly, wound-induced ESR1 activation is strongly suppressed in WIND1-SRDX explants (Figure 1A), suggesting that WIND1 is involved in ESR1 activation.
Figure 1.
Wounding Activates ESR1 Expression in a WIND1-Dependent Manner.
(A) RT-qPCR analysis of WIND1 (upper panel) and ESR1 (lower panel) expression after wounding. First and second leaves of 14-d-old wild-type seedlings were cut and leaf explants were cultured on phytohormone-free MS medium. WIND1 expression peaks at 1 h after wounding, and ESR1 expression peaks at 3 h. The induction of ESR1 is strongly suppressed in ProWIND1:WIND-SRDX (WIND1-SRDX) plants. Expression levels are normalized against those of PP2AA3. Data are mean ± se (n = 3, biological replicates).
(B) Induction of the ESR1 promoter activity at the wound sites of leaf petioles. Leaf explants of ProESR1:GUS and ProESR1:GUS WIND1-SRDX plants were cultured on MS medium. ESR1 activation is compromised in ProESR1:GUS WIND1-SRDX plants. Dashed lines mark wound sites. Representative images of petioles at 0 and 48 h after wounding are shown.
(C) Transverse section of ProESR1:GUS petioles close to wound sites at 48 h after wounding. GUS staining is found in several cell types, xylem parenchyma cells, procambium cells (left panel), and mesophyll cells (right panel) that have started to undergo cell division. Sections were counterstained by safranin O. Asterisks mark new division planes. mp, mesophyll cells; xy, xylem cells; pc, procambium cells; ph, phloem cells.
(D) Nuclear accumulation of ESR1-GFP fusion proteins within the epidermal cells near wound sites at 72 h after wounding. Note that wound stress produces strong green autofluorescence in both wild-type and ProESR1:ESR1-GFP (ESR1-GFP) plants, but these signals are found mostly at the cut edge or cytoplasm. Bars = 300 µm in (B) and (D) and 50 µm in (C).
We previously reported that the WIND1 induction by wounding is localized to wound sites (Iwase et al., 2011a). To explore the wound-induced expression of the ESR1 gene in planta, we examined the pattern of its promoter activity using ProDRN/ESR1:GUS lines, referred to hereafter as ProESR1:GUS, in which the expression of the GUS gene is driven by the promoter of ESR1 (Kirch et al., 2003). As expected, the promoter activity of ESR1 is not detected in intact leaf explants, but its activity is induced locally at wound sites (Figure 1B). We detected similar patterns of ESR1 promoter activity in wounded roots and hypocotyls (Supplemental Figures 2C and 2D). We also introduced the ProESR1:GUS construct into the WIND1-SRDX plants and found that WIND1 is required for the activation of the ESR1 promoter at wound sites (Figure 1B). Transverse sections of petioles close to wound sites showed that ESR1 promoter activity is often detected within the vasculature, i.e., in xylem parenchyma and procambium cells, but is also found in nonvascular cells, such as mesophyll, that have started to undergo cell division presumably to develop callus (Figure 1C). Liu et al. (2014) recently showed that wounding induces auxin accumulation at wound sites of Arabidopsis leaves. To uncouple the effect of wounding from auxin accumulation, we tested whether inhibition of auxin transport by N-1-naphthylphthalamic acid interferes with ESR1 activation at wound sites. As shown in Supplemental Figure 2E, 1 μM N-1-naphthylphthalamic acid does not block ESR1 expression at wound sites, suggesting that local auxin transport does not contribute to ESR1 activation at wound sites.
To further corroborate these observations, we generated ProESR1:ESR1-GFP transgenic plants (ESR1-GFP) in which we drove the expression of the ESR1-GFP fusion protein by the ESR1 promoter. We introduced this construct into the esr1-2/drn1 mutant, referred to hereafter as esr1-2 (Chandler et al., 2007; Matsuo et al., 2011), and confirmed that ESR1-GFP fusion proteins are functional by the complementation of cotyledon phenotypes in esr1-2 (Supplemental Figure 3A). As shown in Figure 1D, we detect clear accumulation of ESR1-GFP fusion proteins within the nuclei of cells close to wound sites. These observations demonstrate that the expression of the ESR1 gene is induced locally at wound sites and WIND1 is required for its activation.
WIND1 Directly Binds the ESR1 Promoter and Activates Its Expression
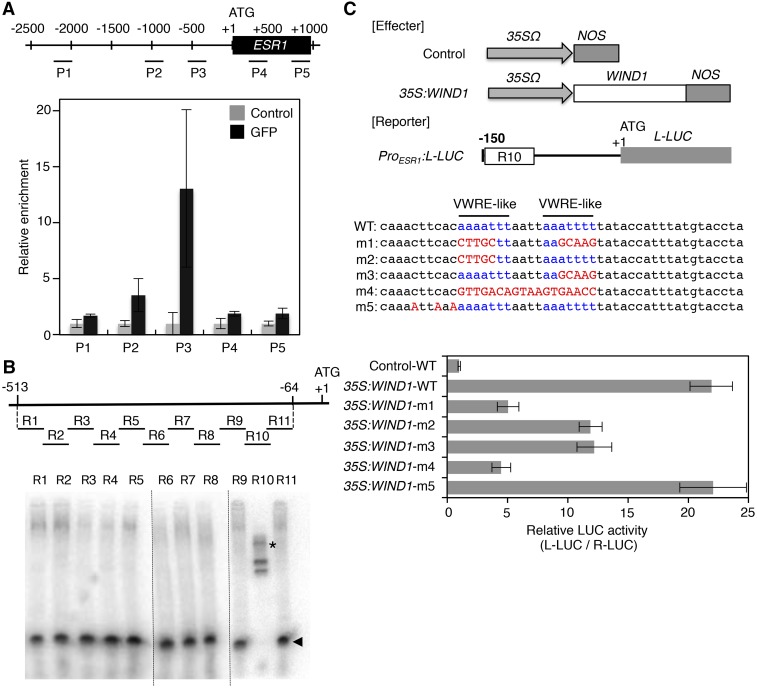
Having established that WIND1 is required for the induction of the ESR1 gene, we subsequently investigated whether WIND1 directly binds the ESR1 promoter in vivo. Using antibodies against GFP proteins, we immunoprecipitated WIND1-GFP proteins from root explants of ProWIND1:WIND1-GFP plants (Iwase et al., 2011a) and tested whether the chromatin of the ESR1 gene copurified with WIND1-GFP. Since we detected strong WIND1 expression in wound-induced callus (Iwase et al., 2011a), we used root explants cultured on Murashige and Skoog (MS) medium for 10 d, at which time they developed large callus at wound sites. As shown in Figure 2A, our chromatin immunoprecipitation coupled by quantitative PCR analysis detected a strong enrichment of the ESR1 promoter sequence using a pair of primers designed around 500 bp upstream of the translational start site. Using a particle bombardment-mediated transient expression assay, we subsequently investigated whether WIND1 could activate the ESR1 promoter in Arabidopsis MM2d culture cells. We fused a 1000-bp ESR1 promoter to the luciferase reporter gene and judged the promoter activation by the relative enzymatic activity of luciferase. As shown in Supplemental Figure 4A, ectopically expressed WIND1 displays strong transactivation activity for the 1000-bp ESR1 promoter, demonstrating that WIND1 can activate ESR1 in vivo. We also performed the transactivation assay using the ESR1 promoter truncated to 500, 250, 200, 150, and 100 bp from the translational start site and found that the 150-bp sequence upstream of the translational start site is sufficient for the ESR1 induction by WIND1.
Figure 2.
WIND1 Directly Binds the ESR1 Promoter and Activates Its Expression.
(A) Chromatin immunoprecipitation of WIND1-GFP fusion proteins on the ESR1 locus. Quantitative PCR analysis, using P1-P5 primers designed within the promoter and coding sequence of ESR1, shows the strongest enrichment of WIND1-GFP using P3 primers designed around −500 bp upstream of the translational start site. The black box represents the coding sequence of ESR1 and +1 ATG indicates the translational start site. Black lines mark the relative distance from the translational start site. Data are normalized against input DNA and shown as a relative enrichment of DNA immunoprecipitated with rabbit serum (control). Data are mean ± se (n = 3, technical replicates).
(B) EMSAs of MBP-WIND1-His6 protein’s binding to the ESR1 promoter in vitro. Upper panel shows the position of ∼50-bp DNA probes, designated as R1 to R11, that cover −513 to −64-bp nucleotides of the ESR1 promoter. Arrowheads and asterisks show free and shifted DNA probes, respectively. Dashed lines separate results obtained in three different experiments. Note that the band shifts only with the R10 probe, indicating that MBP-WIND1-His6 binds −153 to −104 bp upstream of the translational start site.
(C) WIND1-induced transient activation of the ESR1 expression in Arabidopsis culture cells. Upper panel shows the effector constructs, control and 35S:WIND1, and the reporter construct, ProESR1:L-LUC. For the effector constructs, gray arrows mark 35SΩ, the cauliflower mosaic virus 35S promoter with the tobacco mosaic virus omega translation amplification sequence, and gray boxes mark NOS, the Agrobacterium tumefaciens nopaline synthase transcriptional terminator. The white box marks the WIND1 coding sequence. For the reporter construct, the black bar represents the 150-bp promoter sequence of ESR1 with the R10 sequence marked with a white box. The gray box represents the coding sequence of L-LUC, encoding a firefly luciferase gene, and +1 ATG indicates the translational start site. The middle panel shows the wild-type R10 sequence with two VWRE-like motifs marked in blue and m1 to m5 mutations marked in red. Bottom panel shows the ESR1 promoter activity as judged by the L-LUC activity relative to R-LUC, Renilla luciferase. Cobombardment of 35S:WIND1 and ProESR1:L-LUC activates the ESR1 promoter. Note that abolishing each of the two VWRE-like motifs by m2 or m3 mutation results in reduced ESR1 induction and abolishing both motifs by m1 or m4 mutation has additive effects, indicating that both motifs are required for activation of ESR1 by WIND1. Data are mean ± se (n = 6, technical replicates).
To further characterize WIND1’s binding to the ESR1 promoter, we tested their direct interaction in vitro by an electrophoresis mobility shift assays (EMSAs). We expressed the WIND1 protein fused with maltose binding protein (MBP) and 6xHis-tag (His6) in Escherichia coli and purified the MBP-WIND1-His6 protein using the His-tag by affinity chromatography. We used the dehydration-responsive element (DRE; core sequence TACCGACAT) as a positive control, since WIND1 was previously shown to bind the DRE sequence (Lin et al., 2008). Our EMSA indeed showed that the MBP-WIND1-His6 protein strongly binds the DRE sequence in vitro, causing the DRE probe band to shift in the gel (Supplemental Figure 4B). Using this experimental setup, we also found that the MBP-WIND1-His6 protein, but not the MBP-GFP-His6 protein, binds the 513-bp sequence of the ESR1 promoter (Supplemental Figure 4C). To further narrow down WIND1’s binding site within the ESR1 promoter, we tested WIND1’s binding to 11 ∼50-bp DNA probes, designated as R1 to R11, that cover the −513 to −64 bp of the ESR1 promoter with a 10-bp overlap between each probe. As shown in Figure 2B and Supplemental Figure 4D, we found reproducible binding of MBP-WIND1-His6 protein specifically to the probe R10 that covers the −153- to −104-bp sequence of the ESR1 promoter.
The results from our transactivation assay and EMSA consistently suggest that the 150-bp ESR1 promoter is sufficient for WIND1’s binding and activation of the ESR1 gene. Interestingly, the R10 sequence where WIND1 likely binds within the 150-bp ESR1 promoter contains two vascular system-specific and wound-responsive cis-element (VWRE)-like motifs (core sequence AAATTT) previously implicated in wound-induced activation of gene expression in tobacco (Sasaki et al., 2002, 2006). We therefore asked whether these motifs are required for the activation of the ESR1 promoter by mutating either one or both of the VWRE-like motifs. Our transactivation activity assay revealed that the m1 and m4 mutations, abolishing both VWRE-like motifs, strongly hinder ESR1 induction by WIND1, whereas the m2 or m3 mutation, abolishing only the first or second VWRE-like motif, results in partial reduction in ESR1 activation (Figure 2C). By contrast, the m5 mutation, introduced outside the two VWRE-like motifs, does not alter ESR1 activation by WIND1 (Figure 2C). These results strongly suggest that WIND1 directly binds the two VWRE-like motifs within the ESR1 promoter to activate its expression.
ESR1 Promotes Callus Formation at Wound Sites
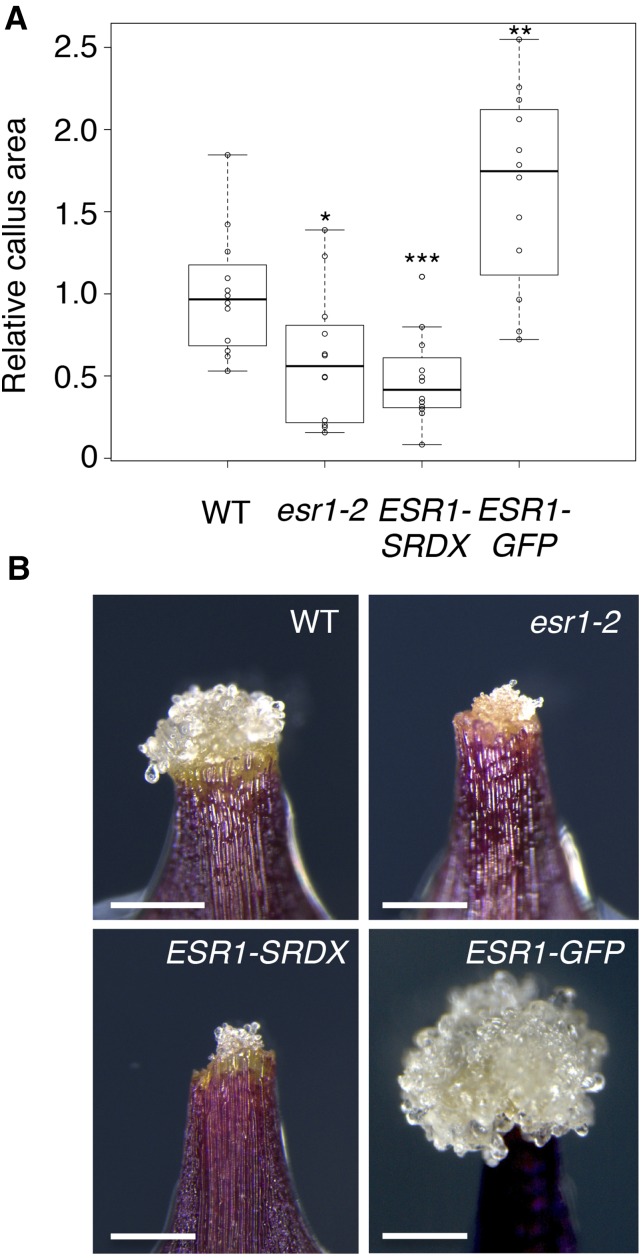
Given that WIND1 is required for callus induction at wound sites (Iwase et al., 2011a), we asked whether ESR1 also participates in wound-induced callus formation. Wild-type leaf explants cultured on MS medium develop a large mass of callus cells at wound sites (Figures 3A and 3B). By contrast, callus formation is severely compromised in esr1-2 leaf explants (Figures 3A and 3B). To further investigate the involvement of ESR1, we generated the ESR1-SRDX dominant repressor line in which we drove the expression of ESR1-SRDX chimeric proteins under the ESR1 promoter. We confirmed the cotyledon formation defects, previously described for esr1-2 mutants (Chandler et al., 2007), in the ESR1-SRDX plants. As expected, we also found clear defects in callus formation from leaf explants, demonstrating the requirement of ESR1 for wound-induced callus formation (Figures 3A and 3B). In addition, we found that ESR1-GFP plants develop larger callus at wound sites (Figures 3A and 3B). Our RT-qPCR analysis showed that, compared with the wild type, the level of ESR1 expression is at least 1.5-fold higher in ESR1-GFP plants after wounding (Supplemental Figure 3B), suggesting that the increased level of ESR1 expression promotes callus formation.
Figure 3.
ESR1 Promotes Callus Formation at Wound Sites.
(A) Callus formation at wound sites of wild-type, esr1-2, ProESR1:ESR1-SRDX (ESR1-SRDX), and ESR1-GFP leaf explants. Leaf explants were cultured on phytohormone-free MS medium, and callus phenotypes were scored at 8 d after wounding. Box plots represent the distribution of projected callus area (n = 12 per genotype). Statistical significance against the wild type was determined by a Student's t test (***P < 0.001, **P < 0.01, and *P < 0.1).
(B) Callus generated at wound sites of wild-type, esr1-2, ESR1-SRDX, and ESR1-GFP leaf explants. Representative images at 8 d after wounding are shown. Bars = 500 µm.
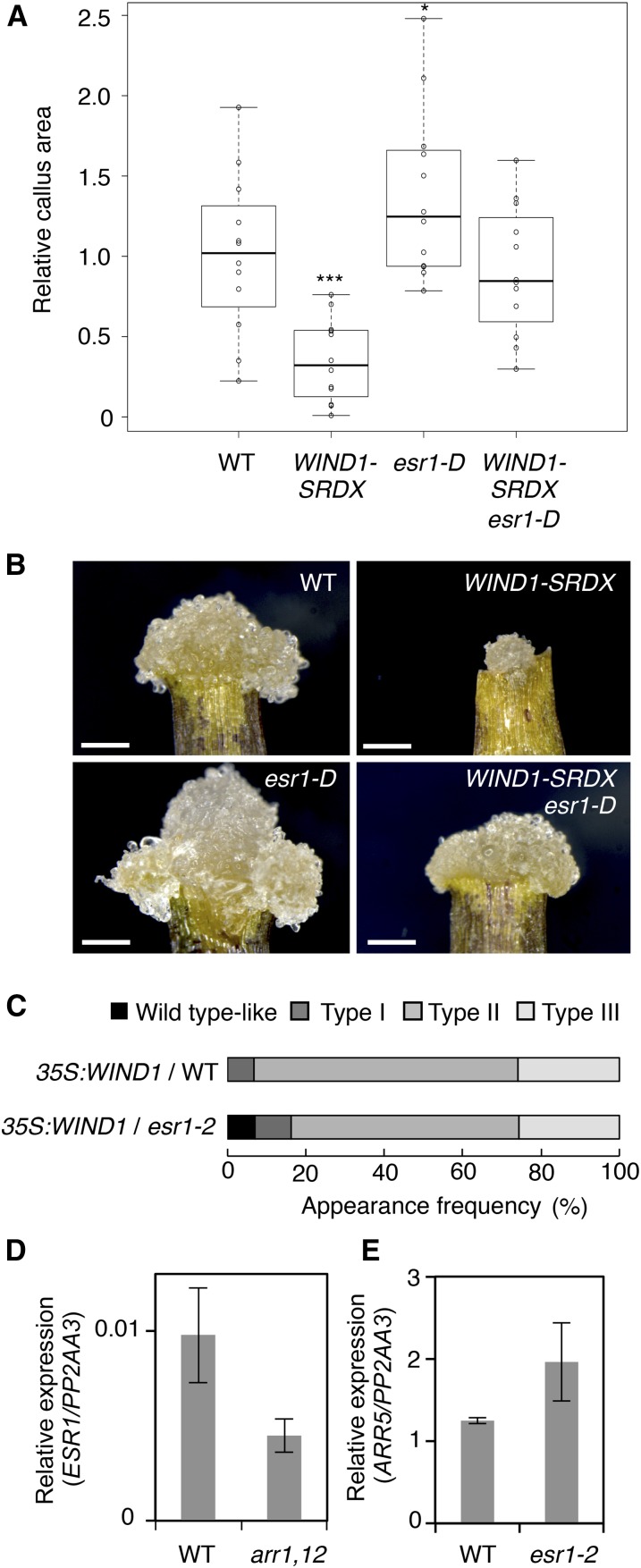
Our results so far are consistent with the hypothesis that WIND1 activates ESR1 expression to promote callus formation at wound sites. To validate this further, we generated double transgenic lines between WIND1-SRDX and esr1/drn-D gain-of-function mutants, referred to hereafter as esr1-D, in which the expression of ESR1 is ectopically activated by the 35S promoter (Kirch et al., 2003), and tested whether ectopic expression of ESR1 in WIND1-SRDX plants was sufficient to rescue defects in wound-induced callus formation. As shown in Figures 4A and 4B, WIND1-SRDX leaf explants, cultured on MS medium, display strong defects in callus formation, producing only a very small mass of cells at wound sites. Introduction of the esr1-D gain-of-function mutation into the WIND1-SRDX plants complements most of these callus formation defects, since leaf explants from WIND1-SRDX esr1-D double transgenic plants develop wild-type-like callus (Figures 4A and 4B). In addition, we examined whether the esr1-2 mutation blocked callus formation induced by WIND1 overexpression. As shown in Figure 4C, 35S:WIND1 T1 plants display weak, intermediate, and strong callus formation, which we previously classified as type I, type II, and type III plants, respectively (Iwase et al., 2011a). As expected, WIND1 overexpression in esr1-2 mutants causes milder callus phenotypes, producing ∼10% of wild-type-like T1 plants without any visible callus formation. These results therefore support that ESR1 functions downstream of WIND1 and promotes callus formation at wound sites.
Figure 4.
ESR1 Functions Downstream of WIND1 in Wound-Induced Callus Formation.
(A) Callus formation at wound sites of wild-type, ProWIND1:WIND1-SRDX (WIND1-SRDX), esr1-D, and WIND1-SRDX esr1-D leaf explants. Leaf explants were cultured on phytohormone-free MS medium, and callus phenotypes were scored at 8 d after wounding. Box plots represent the distribution of projected callus area (n = 12 per genotype). Statistical significance against wild-type was determined by a Student's t test (***P < 0.001 and *P < 0.1).
(B) Callus generated at wound sites of wild-type, WIND1-SRDX, esr1-D, and WIND1-SRDX esr1-D leaf explants. Note that ectopic induction of ESR1 rescues the callus formation deficiency in WIND1-SRDX explants. Representative images at 8 d after wounding are shown. Bars = 250 µm.
(C) The esr1-2 mutation partly suppresses WIND1-induced callus formation in T1 seedlings grown on MS medium. Phenotypic severity was scored according to Figure 2 in Iwase et al. (2011a). T1 plants showing weak, intermediate, and strong callus formation are classified as type I, type II, and type III plants, respectively (n = 104 for the wild type; n = 43 for esr1-2).
(D) The arr1 arr12 mutation partially suppresses the ESR1 expression after wounding.
(E) The esr1-2 mutation does not suppress the ARR5 expression after wounding. First and second leaves of 14-d-old wild-type, arr1 arr12, and esr1-2 seedlings were cut and leaf explants were cultured on phytohormone-free MS medium. Expression levels are normalized against those of the PP2AA3 gene. Data are mean ± se (n = 3, biological replicates).
We previously reported that WIND1 promotes callus formation via the B-type ARABIDOPSIS RESPONSE REGULATOR (ARR)-mediated cytokinin signaling pathway (Iwase et al., 2011a). To test the functional relationship between ESR1 and cytokinin signaling, we evaluated ESR1 expression in arr1 arr12 double mutants defective in B-type ARR signaling (Mason et al., 2005). Interestingly, our RT-qPCR analysis revealed that wound-induced activation of ESR1 is compromised in arr1 arr12 plants, while the esr1-2 mutation does not interfere with the expression of a cytokinin-responsive ARR5 gene (Argyros et al., 2008) (Figures 4D and 4E). Together, these results suggest that ESR1 functions downstream of B-type ARR-mediated cytokinin signaling.
ESR1 Promotes Shoot Regeneration at Wound Sites
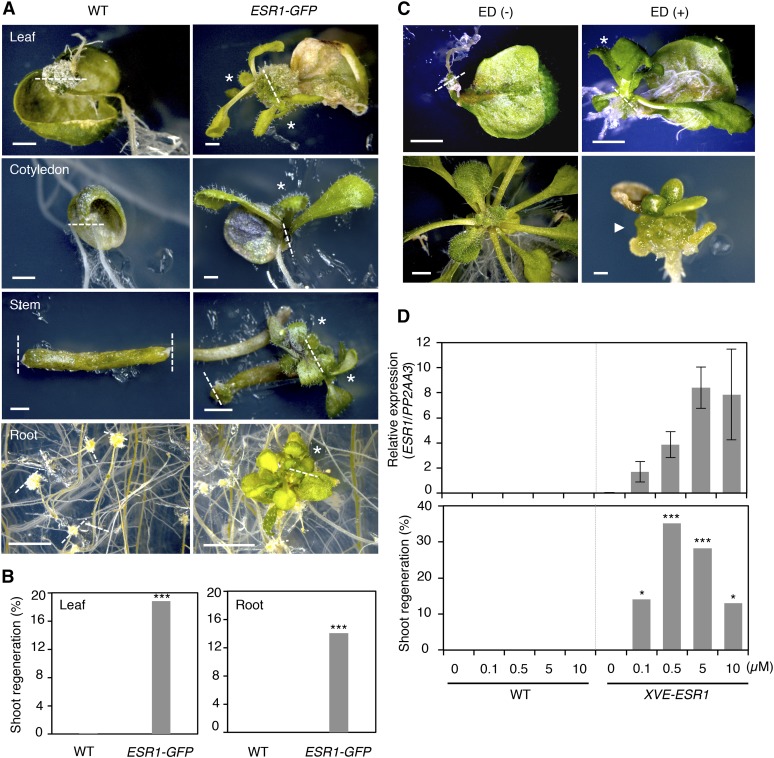
When Arabidopsis wild-type explants are cultured on MS medium without any exogenous plant hormones, they develop callus and often roots, but they hardly regenerate shoots from wound sites (Figure 5A). Unexpectedly, we noticed that leaf explants from ESR1-GFP plants regenerate shoots at wound sites (Figure 5A). Further investigation of this phenotype revealed that only 1 out of 646 (0.2%) leaf explants from wild-type plants regenerate shoots, whereas 31 out of 170 (18.2%) leaf explants from ESR1-GFP plants develop one or sometimes multiple shoots at wound sites (Figure 5B). Intriguingly, this phenotype is not limited to leaf explants, and we also observed shoot regeneration from various other organs such as cotyledons and inflorescence stems (Figure 5A). Similarly, root explants from wild-type plants develop callus at wound sites, but they never regenerate shoots under our culture condition (Figures 5A and 5B). By contrast, 89 out of 634 (14.0%) explants from ESR1-GFP plants develop shoots at wound sites (Figures 5A and 5B).
Figure 5.
ESR1 Promotes Shoot Regeneration at Wound Sites.
(A) Induction of shoot regeneration at wound sites of ESR1-GFP explants. Wild-type and ESR1-GFP explants were cultured on phytohormone-free MS medium for 50 d (leaves), 30 d (cotyledons and inflorescence stems), and 40 d (roots). Dashed lines represent wound sites and asterisks mark regenerating shoots.
(B) Quantitative analysis of shoot regeneration at wound sites. Leaf and root explants were cultured on phytohormone-free MS medium for 55 d. Data are shown as frequency (%) of explants regenerating shoots (n = 646 for wild-type leaves, 170 for ESR1-GFP leaves, 576 for wild-type roots, and 634 for ESR1-GFP roots). Statistical significance was determined by a proportion test (***P < 0.001).
(C) Induction of shoot regeneration at wound sites of XVE-ESR1 leaf explants. Top panel: XVE-ESR1 leaf explants were cultured on phytohormone-free MS medium in the absence (–) or presence (+) of 10 µM 17β-estradiol (ED). Bottom panel: Unwounded XVE-ESR1 plants develop callus in the presence of 10 µM ED. The dashed line represents wound sites and an asterisk marks regenerating shoots. An arrowhead marks callus developing from hypocotyls in intact XVE-ESR1 plants. Bars = 1 mm in (A) and (C).
(D) The frequency of shoot regeneration positively correlates with the level of ESR1 expression in XVE-ESR1 plants. ESR1 expression and shoot regeneration were quantified at 6 and 16 d, respectively, after the application of 0.1 to 10 μM β-estradiol. Expression levels are normalized against those of PP2AA3. Expression data are mean ± se (n = 3, biological replicates). Shoot regeneration is quantified as the frequency (%) of explants regenerating shoots (n = 50 per β-estradiol concentration). Statistical significance was determined by a proportion test (***P < 0.001 and *P < 0.1).
To further explore the causal relationship between ESR1 expression and shoot regeneration, we generated LexA-VP16-estrogen (XVE)-ESR1 transgenic plants in which we induced ESR1 expression by the application of 17β-estradiol (Figures 5C and 5D). Our RT-qPCR analysis confirmed that 0.1 to 10 μM 17β-estradiol induces ESR1 expression in a dose-dependent manner (Figure 5D). As expected, leaf explants from wild-type plants hardly develop shoots at wound sites in the presence of 17β-estradiol. In sharp contrast, leaf explants from up to 35% of XVE-ESR1 plants reproducibly regenerate shoots from wound sites, when cultured in the presence of 0.1 to 10 μM 17β-estradiol (Figures 5C and 5D). Interestingly, XVE:ESR1 plants regenerate shoots only upon wounding, and when they are grown without wound stress, they develop callus (Figures 5C). Nevertheless, these XVE:ESR1-derived calli are capable of regenerating shoots when transferred to SIM, indicating that they retain the potential to develop shoots (Supplemental Figure 5A). Together, these results demonstrate that the increased dosage of ESR1 strongly promotes shoot regeneration at wound sites.
The WIND1-ESR1 Pathway Is Required for Shoot Regeneration in Vitro
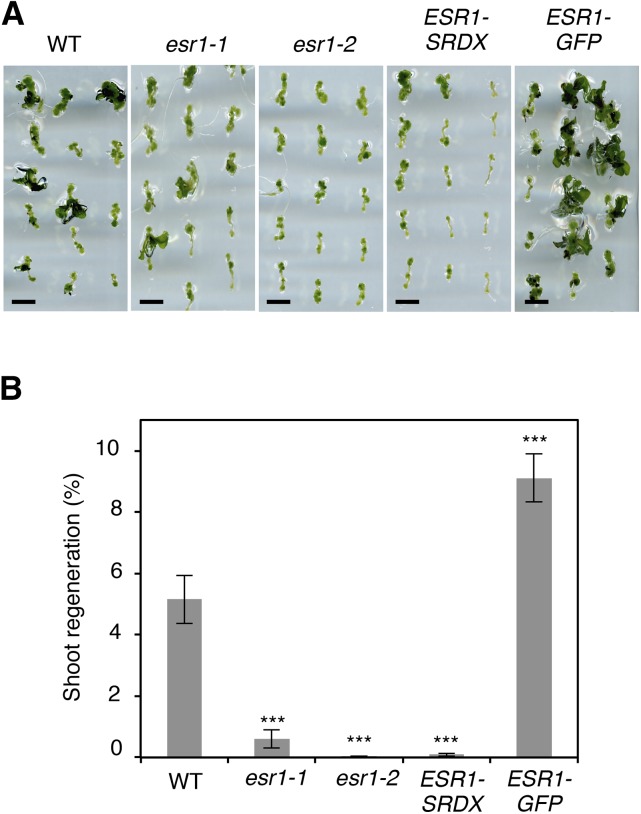
Having uncovered a clear enhancement of wound-induced shoot regeneration by ESR1, we reexamined the requirement of ESR1 for in vitro shoot regeneration. We incubated root explants of wild-type, esr1-1, and esr1-2 seedlings for 4 d on CIM and then transferred them to SIM to induce shoot regeneration. As shown in Figures 6A and 6B, both esr1-1 and esr1-2 display clear defects in shoot regeneration, producing fewer shoots compared with wild-type explants. Chandler et al. (2007) reported that the esr1-1 allele, carrying a dSpm transposon insertion immediately after the start codon, shows weaker cotyledon phenotypes than the esr1-2 allele, carrying the dSpm insertion in the central AP2 domain. We observed a similar trend for shoot regeneration phenotypes since esr1-1, but not esr1-2, explants occasionally develop shoots. In addition, we detected severe defects in shoot regeneration in ESR1-SRDX root explants (Figures 6A and 6B), confirming the requirement of ESR1 in in vitro shoot regeneration. Interestingly, we noticed that shoot regeneration is significantly enhanced in ESR1-GFP plants, forming nearly twice as many shoots from each root explant compared with the wild type (Figures 6A and 6B). By contrast, esr1-2, ESR1-SRDX, and ESR1-GFP root explants develop callus comparable to the wild type on CIM (Supplemental Figure 5B), suggesting that ESR1 does not play major roles in hormone-induced callus formation in vitro.
Figure 6.
ESR1 Promotes Shoot Regeneration in Vitro.
(A) Shoot regeneration of wild-type, esr1-1, esr1-2, ESR1-SRDX, and ESR1-GFP root explants in vitro. Root explants were cultured on CIM for 4 d and transferred to SIM. Representative images of root explants cultured on SIM for 18 d are shown. Bars = 5 mm.
(B) Quantitative analysis of shoot regeneration phenotypes. Regeneration phenotypes are scored as the number of regenerating shoots per explant. Data are mean ± se (n ≥ 30 per genotype). Statistical significance was determined by a Student’s t test (***P < 0.001).
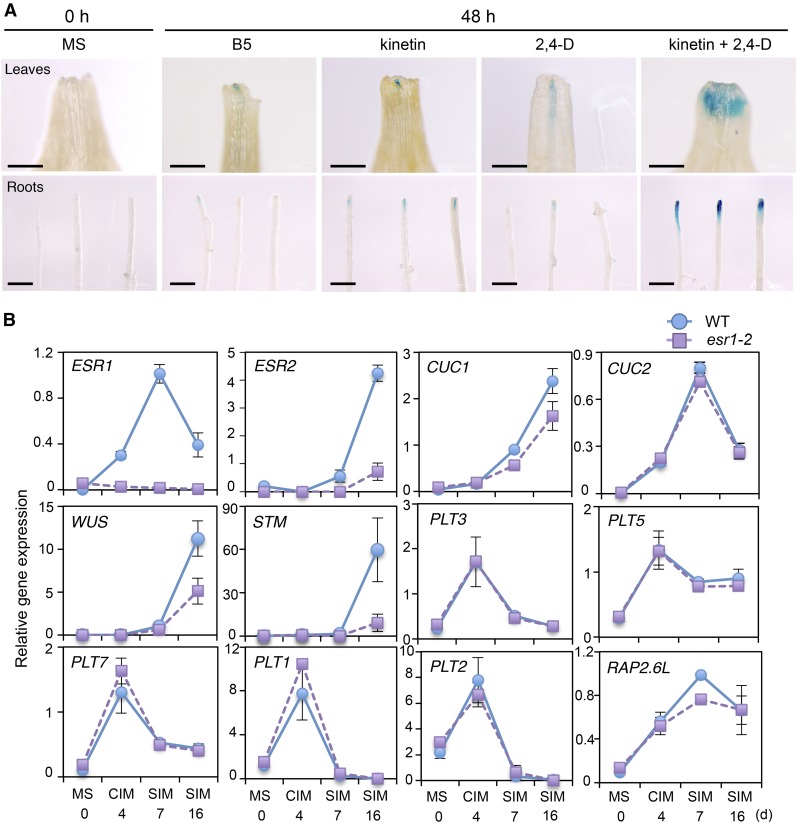
To investigate how ESR1 promotes shoot regeneration in vitro, we first examined ESR1 expression in explants cultured on CIM and SIM. Interestingly, incubation of petiole or root explants with kinetin and 2,4-D, cytokinin, and auxin in CIM strongly stimulates ESR1 promoter activity at wound sites, as shown by the enhanced GUS staining in ProESR1:GUS plants (Figure 7A). Importantly, the application of either kinetin or 2,4-D alone does not activate the ESR1 promoter in both petiole and root explants (Figure 7A), implying that cytokinin and auxin have synergistic effects on ESR1 activation. Our RT-qPCR analyses also confirmed that ESR1 expression is activated in root explants cultured for 4 d on CIM and also showed that its expression is enhanced, by more than 3-fold, after transfer to SIM (Figure 7B). Matsuo et al. (2011) previously showed that ESR2 is not expressed in Arabidopsis explants incubated on CIM and its expression is detected only after they start to form shoots on SIM. Our RT-qPCR analysis confirmed these results and further showed that the late activation of ESR2 expression is dependent on ESR1 (Figure 7B). Similarly, the expression of key shoot regulators, such as CUC1, WUS, STM, and RAP2.6L, is also increased after transfer to SIM (Gordon et al., 2007; Che et al., 2006; Chatfield et al., 2013), and their expression requires ESR1 to different degrees (Figure 7B). We also confirmed the previously reported upregulation of PLT1, 2, 3, 5, and 7 and CUC2 in explants cultured on CIM (Kareem et al., 2015) but found that none of their activation requires functional ESR1 (Figure 7B).
Figure 7.
ESR1 Is Required for the Transcriptional Activation of Shoot Regeneration Regulators in Vitro.
(A) Incubation of leaf (top panel) and root (bottom panel) explants on B5 medium containing both kinetin and 2,4-D strongly enhances the ESR1 promoter activity at wound sites. ProESR1:GUS explants were freshly prepared on MS medium and transferred to B5 medium with or without 0.1 mg/L kinetin and 0.5 mg/L 2,4-D. Representative images at 0 and 48 h after incubation are shown. Note that kinetin or 2,4-D alone does not activate the ESR1 promoter in both leaf and root explants. Bars = 500 µm.
(B) RT-qPCR analysis of ESR1 expression and other shoot regeneration regulators in wild-type and esr1-2 root explants cultured on CIM and SIM. Total RNA was extracted from root explants freshly prepared on MS medium (MS 0), cultured on CIM for 4 d (CIM 4), cultured on CIM for 4 d, and subsequently on SIM for 7 or 16 d (SIM 7 or SIM 16). Expression levels are normalized against those of PP2AA3. Data are mean ± se (n = 3, biological replicates).
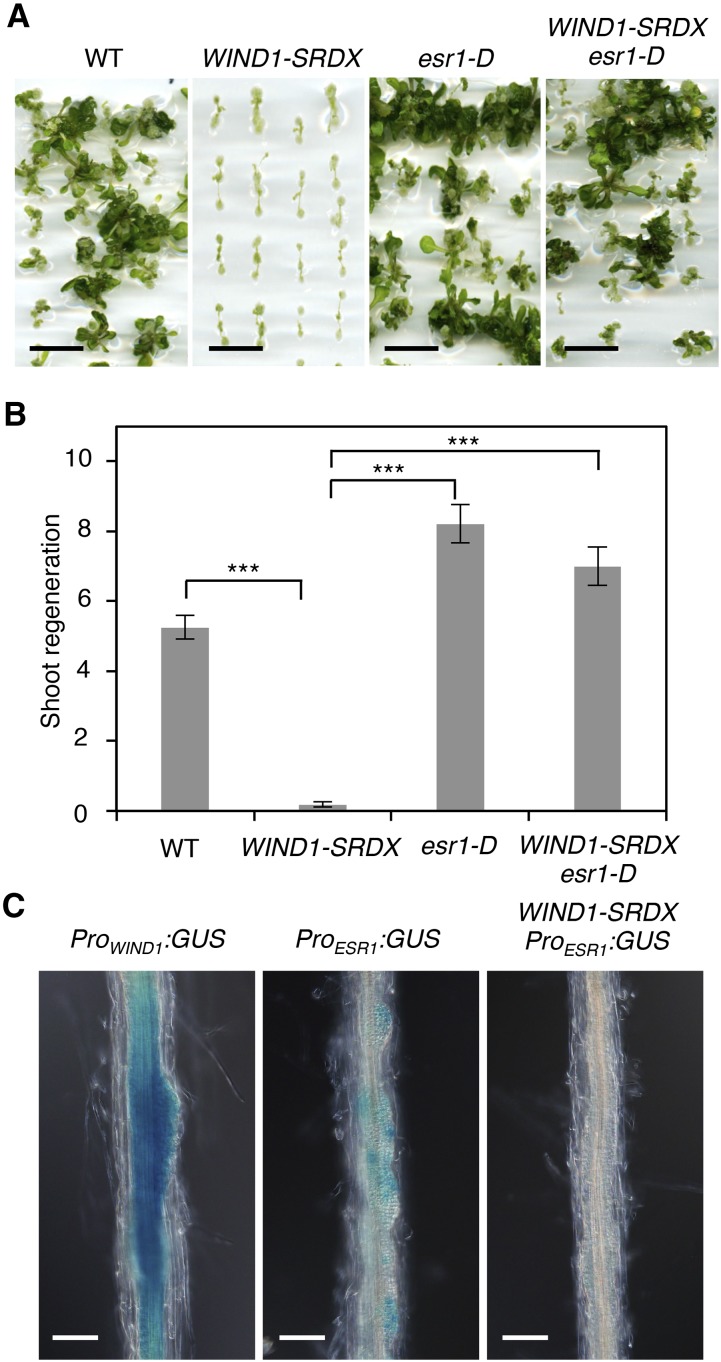
We previously demonstrated that WIND1-SRDX explants cultured on CIM and SIM are defective in shoot regeneration (Iwase et al., 2015; Figures 8A and 8B). Consistently, the WIND1 promoter is active in pericycle cells as well as callus cells derived from pericycle cells in root explants cultured on CIM and SIM (Iwase et al., 2011a; Figure 8C). To test whether ESR1 also acts downstream of WIND1 in this context, we examined the promoter activity of ESR1 in WIND1-SRDX root explants. As reported previously (Matsuo et al., 2011), ESR1 promoter activity is visible within callus of ProESR1:GUS root explants cultured on CIM and SIM (Figure 8C). By contrast, the ESR1 promoter activity is strongly suppressed in ProESR1:GUS WIND1-SRDX plants (Figure 8C), indicating the requirement of WIND1 for the activation of the ESR1 promoter. We also introduced the esr1-D construct into WIND1-SRDX plants and examined whether ectopic activation of ESR1 is sufficient to rescue the shoot regeneration phenotype in WIND1-SRDX explants. As expected, plants expressing both WIND1-SRDX and esr1-D show similar or slightly higher levels of shoot regeneration compared with the wild type (Figures 8A and 8B). These results thus demonstrate that ESR1 functions downstream of WIND1 and promotes shoot regeneration in vitro.
Figure 8.
ESR1 Functions Downstream of WIND1 in in Vitro Shoot Regeneration.
(A) Shoot regeneration of wild-type, WIND1-SRDX, esr1-D, and WIND1-SRDX esr1-D root explants in vitro. Root explants were cultured on CIM for 4 d and transferred to SIM. Representative images of root explants cultured on SIM for 21 d are shown.
(B) Quantitative analysis of shoot regeneration phenotypes. Regeneration phenotypes are scored as a number of regenerating shoots per explant. Data are mean ± se (n ≥ 50 per genotype). Statistical significance was determined by a Student’s t test (***P < 0.001). Note that ectopic induction of ESR1 is sufficient to rescue the shoot regeneration phenotypes in WIND1-SRDX explants.
(C) Induction of WIND1 and ESR1 promoter activities in callus developing from root explants on SIM. Root explants from ProWIND1:GUS, ProESR1:GUS, and ProESR1:GUS WIND1-SRDX plants were cultured on CIM for 4 d and transferred to SIM. Note that the promoter activity of ESR1 is strongly reduced by dominant repression of WIND1. Representative images at 1 d after transfer to SIM are shown. Bars = 500 µm in (A) and 100 µm in (C).
The WIND1-ESR1 Pathway Is Not Required for de Novo Root Regeneration at Wound Sites
Liu et al. (2014) recently showed that Arabidopsis leaf explants cultured on B5 medium are capable of developing new roots from wound sites. We found that Arabidopsis leaf explants cultured on MS medium also regenerate roots in the absence of a supply of exogenous phytohormones (Supplemental Figure 6A). Using this system, we asked whether WIND1 and ESR1 are also involved in root regeneration at wound sites. We typically observe >40% of wild-type leaf explants regenerating roots from wound sites (Supplemental Figure 6B). Interestingly, both WIND1-SRDX and ESR1-SRDX plants are capable of forming similar numbers of roots (Supplemental Figures 6A and 6B), suggesting that the WIND1-ESR1 pathway is not required for de novo root regeneration at wound sites.
DISCUSSION
In this study, we demonstrate that a wound-induced reprogramming regulator WIND1 directly activates ESR1 expression to promote callus formation and subsequent shoot regeneration at wound sites. Our data show that the level of ESR1 is a key determinant of shoot fate, since mild overexpression of ESR1 induces shoot formation at wound sites. WIND1 is also expressed in phytohormone-induced callus cells in explants cultured on CIM and SIM, and it is required for ESR1 activation and shoot regeneration in in vitro conditions. Our findings therefore uncovered an important transcriptional cascade underlying shoot regeneration in plants.
WIND1 as a Molecular Link between Wound Stress and Regeneration
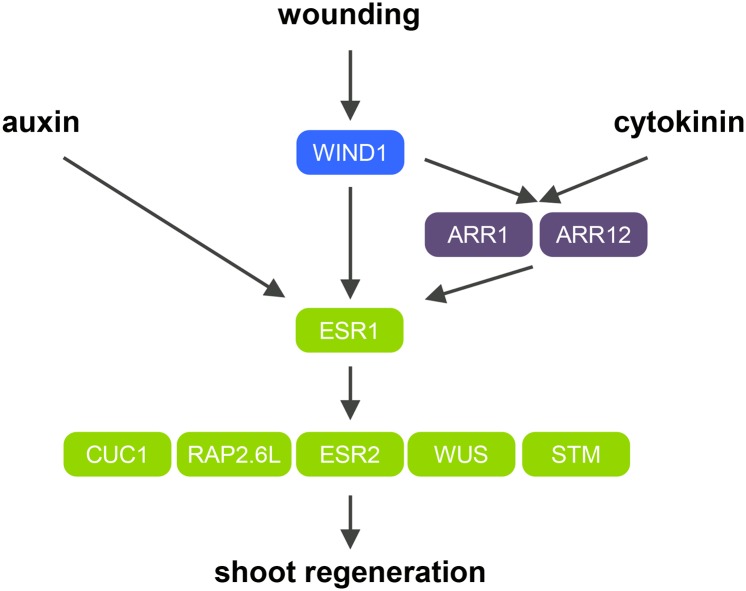
We previously showed that WIND1 promotes the reacquisition of competency for shoot regeneration (Iwase et al., 2015), but the precise molecular mechanisms underlying this control were not known. In this study, we demonstrate that a central role of WIND1 is to activate the expression of ESR1 to promote callus formation and shoot regeneration. Our data show that WIND1 first activates ESR1 expression, and culturing explants on auxin and cytokinin permits stronger ESR1 expression (Figure 9). It is interesting to note that the overexpression of ThWIND1-L, a WIND1 homolog from salt cress Thellungiella halophila, also induces ESR1 expression in Arabidopsis (Zhou et al., 2012), implying that WIND1-mediated ESR1 activation might be conserved at least among Brassicaceae plants.
Figure 9.
A Schematic Model Describing How the WIND1-ESR1 Pathway Promotes Shoot Regeneration in Arabidopsis.
Local wound stress induces WIND1 expression at wound sites. WIND1 directly binds the ESR1 promoter and activates its expression. WIND1 also activates ESR1 indirectly through enhancing the B-type ARR-mediated cytokinin signaling. ESR1 expression is synergistically enhanced by exogenously supplied auxin and cytokinin to further boost shoot regeneration in vitro. ESR1 is required for the upregulation of key shoot regulators such as CUC1, RAP2.6L, ESR2, WUS, and STM to promote shoot regeneration.
Our in vivo and in vitro data show that WIND1 directly binds the promoter of ESR1 using the two VWRE-like motifs (Figures 2B and 2C). The 14-bp VWRE motif (GAAAAGAAAATTTC) was first identified within the promoter of a wound-inducible peroxidase gene, tpoxN1, in tobacco (Nicotiana tabacum; Sasaki et al., 2006). A later study showed that two tobacco wound-responsive AP2/ERF family transcription factors, WRAF1 and WRAF2, bind the VWRE motif to induce the tpoxN1 gene after wounding and the core sequence (AAATTT) within the VWRE motif is essential for their binding (Sasaki et al., 2007). Interestingly, both WRAF1 and WRAF2 are induced within 30 min after wounding, preceding the accumulation of tpoxN1 transcripts after 1 h. We detect similar transcriptional changes for WIND1, starting within 30 min after wounding, followed by the peak accumulation of ESR1 transcripts after 1 h. The closest homolog of WRAF1 and WRAF2 in Arabidopsis is RAP2.6 (At1g43160), and its expression is also induced very rapidly after wounding (Kilian et al., 2007; Iwase et al., 2011a). It is thus plausible that one of the earliest wound-induced transcriptional changes is mediated by a set of wound-inducible AP2/ERF transcription factors and they activate target genes through binding the core VWRE motif within the target promoters. The physiological roles of RAP2.6 in the wound response have not been investigated so far, but it will be interesting to test whether RAP2.6 can also activate ESR1 using the VWRE-like motifs. In addition to ESR1, we found 228 other genes in Arabidopsis that carry two closely located VWRE-like motifs within the 1-kb promoter region, ∼10% of which are induced more than 2-fold by WIND1 overexpression (A. Iwase and B. Rymen, unpublished data). These genes are putative targets of WIND1, and future studies should investigate their functional relationships to WIND1. We should note that the core VWRE sequence does not resemble other GC-rich sequences such as DRE (Yamaguchi-Shinozaki and Shinozaki, 1994) and the GCC box (core sequence TAAGAGCCGCC) (Ohme-Takagi and Shinshi, 1995), recognized by other AP2/ERF transcription factors. While WRAF1 and WRAF2 do not recognize these GC-rich motifs, WIND1 recognizes both DRE and the GCC box at least in vitro (Lin et al., 2008; this study). It will be important to examine whether WIND1 binds both of these motifs in vivo and, if so, whether it activates different sets of genes, for instance, in a context-dependent manner.
We provide genetic evidence that WIND1 and ESR1 are not required for root regeneration from Arabidopsis leaf explants, at least under the condition used in this study. These results are in agreement with the finding that this form of root regeneration is driven by auxin accumulation at wound sites, which promotes the establishment of root cell fate through the activation of WOX11 and WOX12 (Liu et al., 2014). Intriguingly, Liu et al. (2014) showed that auxin-mediated callus formation (and subsequent root regeneration) derives primarily from leaf procambium cells, while our study suggests that wound-induced callus formation may originate from various cell types, including xylem parenchyma and mesophyll cells (Figure 1C). How these callus cells from different cellular origins contribute to shoot regeneration remains to be verified, but these observations together suggest that regeneration of roots and shoots from Arabidopsis leaf explants is operated by at least two distinct molecular mechanisms.
Activation of ESR1 Expression by Auxin and Cytokinin
How exogenously supplied auxin enhances ESR1 expression is not currently clear. Previous studies have shown that the auxin-inducible transcription factor ARF5/MONOPTEROS (MP) binds the ESR1/DRN promoter and activates its expression during embryonic development (Cole et al., 2009). The ESR1/DRN promoter possesses two canonical auxin-responsive elements where ARF5/MP binds in vivo, and mutations in these sequences alter auxin-induced expression of ESR1/DRN in developing embryos (Cole et al., 2009). Interestingly, the expression of ARF5/MP is strongly upregulated in explants cultured on CIM and SIM (Che et al., 2006), raising the possibility that ARF5/MP might participate in auxin-induced ESR1 expression in cultured explants. Alternatively, a recent study by Fan et al. (2012) has shown that ARF7 and ARF19 mediate auxin-induced callus formation by upregulating the expression of LBD16, LBD17, LBD18, and LBD29. It is thus possible that these ARFs bind the ESR1 promoter and activate its expression in parallel. We also do not rule out the possibility that ESR1 acts downstream of the ARF-LBD pathway, and future studies should clarify how these auxin-responsive transcriptional regulators directly or indirectly regulate ESR1 expression.
Our results show that cytokinin activates ESR1 expression at least partially through ARR1 and ARR12 (Figure 4D), demonstrating that ESR1 acts downstream of the B-type ARR-mediated pathway. How wound stress activates the B-type ARR pathway in a WIND-dependent manner is not currently known, but our data suggest that WIND1 may activate ESR1 both directly and indirectly via enhancing cytokinin signaling. In addition, Kareem et al. (2015) found that PLT3, PLT5, and PLT7 are the key regulators of in vitro shoot regeneration acting downstream of both auxin and cytokinin signaling. We showed that the expression of these PLT genes is not altered in esr1-2 mutants (Figure 7B), suggesting that they do not act downstream of ESR1. Instead, PLTs may function upstream of, or in parallel to, ESR1 and it will be interesting to investigate these functional relationships in future studies.
Role of ESR1 in Callus Formation and Shoot Regeneration
Our data suggest that ESR1 is induced immediately after wounding (Figure 1) and promotes callus formation at wound sites (Figure 3). We also show that the level of ESR1 expression is one key limiting factor for shoot regeneration, since mild overexpression of ESR1 dramatically improves shoot regeneration from wound sites (Figure 5). Intriguingly, XVE:ESR1 plants regenerate shoots only upon wounding and they develop callus without wound stress (Figure 5C). Banno et al. (2001) also reported callus formation in 35S:ESR1 plants and suggested that constitutive overexpression of ESR1 interferes with differentiation into shoot cells. Based on our observations, we hypothesize that ESR1 activation alone is not sufficient to fully establish the shoot fate and additional wound-induced events, such as induction and/or accumulation of some other signals, are also needed to confer shoot fate at wound sites.
Our data show that ESR1 is not essential for hormone-induced callus formation but that it plays pivotal roles in shoot regeneration in vitro (Supplemental Figure 5B; Figure 6). The cause for the apparent discrepancy between our results and a previous report (Matsuo et al., 2011) is not clear, but one possibility is that we employ slightly different culture conditions, such as long-day light condition as opposed to the continuous light conditions employed by Matsuo et al. (2011), and wild-type root explants appear to produce more shoots in our conditions. Matsuo and Banno (2008) reported that overexpression of ESR1-SRDX chimeric proteins blocks shoot regeneration, suggesting that ESR1, potentially together with other redundant transcriptional regulators, promotes shoot regeneration. Our observation further highlights the functional importance of ESR1, since loss of ESR1 in esr1 mutants or ESR1-SRDX expression by its own promoter is sufficient to cause severe regeneration defects (Figure 6). Since esr1 mutant calli turn green and develop some green foci (Figure 6), they might be able to develop shoot promeristems and/or shoot primordia, although they are severely impaired in shoot outgrowth. Shoot regeneration defects in esr1 mutants are accompanied by strong inhibition of key shoot meristem regulators, such as WUS and STM (Figure 7), further substantiating that these mutants fail to complete shoot regeneration. Given that the levels of PLT3, PTL5, and PLT7 expression are comparable between wild-type and esr1-2 mutants (Figure 7), esr1-2 calli likely retain reasonable levels of pluripotency, but they cannot progress through the shoot program without functional ESR1. A previous overexpression study suggested that ESR1 directly activates CUC1 in in vitro shoot regeneration (Matsuo et al., 2009). Our data show that ESR1 is required for CUC1 expression (Figure 7), supporting the notion that ESR1 functions upstream of CUC1. Kareem et al. (2015) showed that PLT3, PLT5, and PLT7 are required for CUC1 expression in vitro, but plt3 plt5 plt7 mutants still have some residual CUC1 expression on SIM. It is thus possible that these two pathways, governed by PLTs and ESR1, regulate CUC1 expression in parallel.
Our microarray data show that ESR2, a close homolog of Arabidopsis ESR1, is also upregulated in 35S:WIND1 callus (Iwase et al., 2011a), implying that WIND1 may also target ESR2 to promote shoot regeneration. Intriguingly, however, we do not detect any significant elevation of ESR2 expression after wound stress in any of the tissues examined, suggesting that wounding (or WIND1) does not directly induce ESR2 expression. Our data show that ESR2 expression is strongly dependent on ESR1 in in vitro conditions (Figure 7). Thus, the high levels of ESR1 in 35S:WIND1 callus may contribute to ESR2 induction. Matsuo et al. (2011) showed that ESR2 is expressed much later than ESR1 on SIM, and ESR1 indirectly activates ESR2 expression. These results also agree with the view that ESR2 acts downstream of ESR1 in in vitro shoot regeneration.
Together, this study has unveiled how a wound-induced transcriptional pathway integrates with signals mediated by externally supplied auxin and cytokinin to specify the developmental fate of regenerating organs. In nature, only a subset of plant species is capable of regenerating shoots from cut sites in the absence of exogenous hormones (Ikeuchi et al., 2016). Therefore, it will be interesting to explore whether the shoot regenerative potential of various plant species correlates with the inducibility of ESR1 after wounding. As WIND1 is likely to activate other developmental regulators, identifying additional downstream targets of WIND1 should further advance our understanding of how wound stress promotes regeneration at wound sites. Interestingly, many regulators acting during regeneration, including ESR1, are epigenetically silenced by Polycomb-mediated histone modification, but they are rapidly induced after wounding (Ikeuchi et al., 2015a, 2015b; A. Iwase and B. Rymen, unpublished data). Exploring how wound stress lifts the epigenetic repression of these regulators and allows their transcriptional induction will be another exciting challenge in future studies.
METHODS
Plant Materials, Growth Conditions, and Transformation
All plants used in this study were in the Col-0 background. The ProESR1/DRN:GUS, esr1-1/drn-2, esr1-2/drn-1, esr1/drn-D, ProWIND1:GUS, 35S:WIND1, ProWIND1:WIND1-GFP, ProWIND1:WIND1-SRDX, and arr1 arr12 plants were described previously (Kirch et al., 2003; Iwase et al., 2011a; Mason et al., 2005). Plants were grown at 22°C under long-day conditions with 16 h of white light (100 μmol m−2 s−1) and 8 h of darkness. For plant transformation, T-DNA vectors carrying an appropriate construct were introduced into Agrobacterium tumefaciens strain GV3101 by electroporation, and the resultant Agrobacterium was infiltrated into Arabidopsis thaliana by the floral dip method (Clough and Bent, 1998).
Callus Formation and Regeneration Assay
To induce callus or de novo root formation from petioles, first and second rosette leaves were cut with microscissors (Natsume Seisakusho; MB-50-15) and their explants were incubated on phytohormone-free MS medium supplemented with 1% sucrose and 0.6% Gellan gum (Gelzan; Sigma-Aldrich). To induce callus and shoot regeneration in vitro, root explants were first cultured on CIM to induce callus and then transferred to SIM to induce shoots (Valvekens et al., 1988). Wound-induced callus phenotypes were recorded at 8 d after wounding and CIM-induced callus phenotypes were recorded at 4 d after incubation on CIM. The projected area of callus was quantified by ImageJ.
Microscopy
GUS staining was performed as previously described (Kertbundit et al., 1991), and stained samples were observed using a Leica M165 C stereomicroscope. GFP signal was detected using a Leica TCS SP5 II confocal laser microscope. For Technovit sectioning, cut petioles of ProESR1:GUS plants were fixed in a FAA solution (formalin:acetic acid:70% ethanol, 1:1:18), dehydrated through a graded ethanol series, and embedded in Technovit 7100 (Heraeus Kulzer). Sections of 4-μm thickness were prepared with RM2135 (Leica), counterstained by safranin O, and observed under an Olympus BX51 microscope.
RNA Isolation and RT-qPCR Analysis
Total RNA was isolated with an RNeasy Plant Mini Kit (Qiagen) according to the manufacturer’s instructions and their cDNA was synthesized using a PrimeScript RT reagent kit with gDNA Eraser (TaKaRa). RT-qPCR analysis was performed using an Mx3000P qPCR system (Agilent Technologies) and Thunderbird SYBR qPCR mix (Toyobo). Three biological replicates were used for each treatment. The protein phosphatase 2A subunit A3 (PP2AA3) gene was used as a reference (Czechowski et al., 2005). A list of primers used for RT-qPCR is provided in Supplemental Table 1.
Plasmid Construction
To construct the ProESR1:ESR1-GFP and ProESR1:ESR1-SRDX vectors, genomic fragments containing the 2000-bp promoter sequence and ESR1 coding sequence were amplified by PCR and cloned into the pGFP_NOSG and pSRDX_NOSG vectors, respectively (Yoshida et al., 2013). The resulting ProESR1:ESR1-GFP and ProESR1:ESR1-SRDX fragments were subcloned into the pBCKH vector by Gateway LR Clonase II (Life technologies) for plant transformation (Mitsuda et al., 2006). To construct the ProESR1:LUC reporter vector, the 1000-bp promoter sequence of ESR1 was amplified by PCR and cloned into the pGEM-T Easy vector (Promega). The ProESR1:LUC vector with truncated ESR1 promoter was generated using primers that respectively amplify the 500-, 250-, 150-, and 100-bp sequence from the translational start site. The ProESR1:LUC vector with mutations in R10 were generated by introducing the respective mutations into the PCR primers. The sequence of a firefly luciferase (L-LUC) gene was PCR amplified from the GAL4GCC-LUC vector (Ohta et al., 2001) and inserted into the pGEM-T Easy vector together with a NOS terminator. To construct the pMGWA-WIND1 and pMGWA-GFP vectors, the coding sequence of WIND1 and GFP genes were PCR amplified without stop codons and cloned into the pDONR221 vector using Gateway BP Clonase II. The resultant plasmids were subsequently cloned into the pMGWA vector (Busso et al., 2005) using LR Clonase II (Life Technologies). To construct the pER8-ESR1 vector, the PCR-amplified coding sequence of ESR1 was cloned into the pER8 vector (Zuo et al., 2000). A list of primers used for PCR amplification is provided in Supplemental Table 1.
Transient Expression Assay
The Pro35S:WIND1 (Iwase et al., 2011a) and Pro35S:SG (Ohta et al., 2001) vectors were used as an effector and control, respectively. The ProESR1:L-LUC vector was used as a reporter, and the pPTRL vector, driving the expression of a luciferase gene from Renilla (R-LUC) by the 35S promoter (Fujimoto et al., 2000), was used as an internal control. Particle bombardment was performed using the Biolistic PDS-1000/He system (Bio-Rad), and luciferase assays were performed using the Dual-Luciferase Reporter Assay System (Promega) as previously reported (Hiratsu et al., 2002). Arabidopsis MM2d cultured cells (Menges and Murray, 2002) were used as host cells and luciferase activities were quantified using the Mithras LB940 Microplate Luminometer (Berthold Technologies).
Chromatin Immunoprecipitation
The chromatin immunoprecipitation experiment was performed following a previously reported protocol (Gendrel et al., 2005) with several modifications. Roots of 30-d-old Arabidopsis plants harboring ProWIND1:WIND1-GFP (Iwase et al., 2011a) were cut and their 5-mm explants were incubated on MS medium for 10 d to generate wound-induced callus. Approximately 1 g of fresh root explants was used as a starting material and WIND1-GFP proteins were immunoprecipitated using antibodies against GFP (Abcam; ab290). Sterile-filtered rabbit serum (Equitech-Bio; SR30) was used as a negative control.
EMSA
To express the MBP-WIND1-His6 and MBP-GFP-His6 proteins, Escherichia coli SoluBL21 cells (AMS Biotechnology) were transformed with pMGWA-WIND1 and pMGWA-GFP vectors. The resulting E. coli cells were grown at 37°C in Luria-Bertani medium containing 100 mg/L ampicillin until OD600 reached 0.6. The production of fusion proteins was induced at 18°C by adding 0.3 mM IPTG overnight. Cells were harvested by centrifugation and cell pellets were stored at −30°C until use. The cell pellets were resuspended in EMSA binding buffer (10 mM Tris-HCl, pH 7.5, 50 mM KCl, and 1 mM DTT) and lysed by sonication (Digital Sonifier 450D; Branson). After the addition of Triton X-100 to 0.2% (w/v), the cell slurry was incubated for 20 min at 4°C and clarified by centrifugation. The supernatant was passed through a column packed with Amylose resins (NEB) and eluted with EMSA binding buffer containing 10 mM maltose.
To generate the DNA probe, the 513-bp sequence of the ESR1 promoter was amplified with the set of biotinylated primers (Eurofins Genomics) listed in Supplemental Table 2 using the ESR1 promoter as template. The biotinylated PCR products were purified with the QIAquick gel extraction kit (Qiagen). Chemiluminescent EMSA was performed with a LightShift Chemiluminescent EMSA Kit (Thermo Scientific) according to the manufacturer’s instructions. The ∼50-bp oligo DNA probes were produced by annealing the complementary oligonucleotides listed in Supplemental Table 2. The double-stranded oligonucleotides were end-labeled with [γ-32P]ATP (Perkin-Elmer) using T4 Polynucleotide Kinase (NEB). The DNA binding reaction was performed for 20 min at room temperature in EMSA binding buffer containing 50 ng/μL poly(dI⋅dC) (Sigma-Aldrich) and 0.05% (w/v) Nonidet P-40. These reactions were resolved on 5% polyacrylamide/TBE gels (Bio-Rad) in half-strength TBE buffer (Bio-Rad), and the gels were dried with a HydroTech Gel Drying System (Bio-Rad). Radioactive probes were detected using a Typhoon FLA-7000 system (GE Healthcare).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative under the following accession numbers: WIND1 (At1g78080), ESR1 (At1g12980), ESR2 (At1g24590), ARR1 (At3g16857), ARR5 (At3g48100), ARR12 (At2g25180), CUC1 (At3g15170), CUC2 (At5g53950), WUS (At2g17950), STM (At1g62360), PLT1 (At3g20840), PLT2 (At1g51190), PLT3 (At5g10510), PLT5 (At5g57390), PLT7 (At5g65510), RAP2.6L (At5g13330), and PP2AA3 (At 1g13320).
Supplemental Data
Supplemental Figure 1. ESR1 is upregulated in callus induced by WIND1 overexpression.
Supplemental Figure 2. Wounding activates the ESR1 expression in roots and hypocotyls.
Supplemental Figure 3. Characterization of ProESR1:ESR1-GFP plants.
Supplemental Figure 4. WIND1 directly binds the ESR1 promoter in vitro.
Supplemental Figure 5. ESR1 overexpression induces callus with competency for shoot regeneration.
Supplemental Figure 6. WIND1 and ESR1 are not required for root regeneration from leaf explants.
Supplemental Table 1. A list of PCR primers used in this study.
Supplemental Table 2. A list of oligonucleotides used in EMSA.
Supplementary Material
Acknowledgments
We thank members of Sugimoto’s lab for discussions and comments on the manuscript. We thank Mariko Mouri, Chika Ikeda, Yasuko Yatomi, Mieko Ito, and Noriko Doi for their technical assistance. This work was supported by a grant from the Scientific Technique Research Promotion Program for Agriculture, Forestry, Fisheries, and Food Industry and by grants from the Ministry of Education, Culture, Sports, and Technology of Japan to A.I. (15K18565 and 15KK0265), M.I. (15K18564), and K.S. (26291064 and 15H05961). M.I. is supported by the RIKEN Special Postdoctoral Researcher Programme and the Naito Foundation.
AUTHOR CONTRIBUTIONS
A.I. and K.S. conceived the research and designed the experiments. A.I., H.H., M.I., M.O., and B.R. performed the experiments. M.N. and M.O.-T. provided the pGFP_NOSG vector, and S.K. provided the pMGWA-GFP vector. T.K., K.M., and E.G. helped with the chromatin immunoprecipiation experiment. A.I. and K.S. wrote the manuscript with input from all coauthors.
Glossary
- CIM
callus-inducing medium
- SIM
shoot-inducing medium
- MS
Murashige and Skoog
- EMSA
electrophoresis mobility shift assay
- DRE
dehydration-responsive element
- VWRE
vascular system-specific and wound-responsive cis-element
Footnotes
Articles can be viewed without a subscription.
References
- Aida M., Ishida T., Fukaki H., Fujisawa H., Tasaka M. (1997). Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell 9: 841–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida M., Ishida T., Tasaka M. (1999). Shoot apical meristem and cotyledon formation during Arabidopsis embryogenesis: interaction among the CUP-SHAPED COTYLEDON and SHOOT MERISTEMLESS genes. Development 126: 1563–1570. [DOI] [PubMed] [Google Scholar]
- Aida M., Beis D., Heidstra R., Willemsen V., Blilou I., Galinha C., Nussaume L., Noh Y.S., Amasino R., Scheres B. (2004). The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119: 109–120. [DOI] [PubMed] [Google Scholar]
- Argyros R.D., Mathews D.E., Chiang Y.H., Palmer C.M., Thibault D.M., Etheridge N., Argyros D.A., Mason M.G., Kieber J.J., Schaller G.E. (2008). Type B response regulators of Arabidopsis play key roles in cytokinin signaling and plant development. Plant Cell 20: 2102–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atta R., Laurens L., Boucheron-Dubuisson E., Guivarc’h A., Carnero E., Giraudat-Pautot V., Rech P., Chriqui D. (2009). Pluripotency of Arabidopsis xylem pericycle underlies shoot regeneration from root and hypocotyl explants grown in vitro. Plant J. 57: 626–644. [DOI] [PubMed] [Google Scholar]
- Banno H., Ikeda Y., Niu Q.W., Chua N.H. (2001). Overexpression of Arabidopsis ESR1 induces initiation of shoot regeneration. Plant Cell 13: 2609–2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum K.D., Sánchez Alvarado A. (2008). Slicing across kingdoms: regeneration in plants and animals. Cell 132: 697–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busso D., Delagoutte-Busso B., Moras D. (2005). Construction of a set Gateway-based destination vectors for high-throughput cloning and expression screening in Escherichia coli. Anal. Biochem. 343: 313–321. [DOI] [PubMed] [Google Scholar]
- Chandler J.W., Cole M., Flier A., Grewe B., Werr W. (2007). The AP2 transcription factors DORNROSCHEN and DORNROSCHEN-LIKE redundantly control Arabidopsis embryo patterning via interaction with PHAVOLUTA. Development 134: 1653–1662. [DOI] [PubMed] [Google Scholar]
- Chatfield S.P., Capron R., Severino A., Penttila P.-A., Alfred S., Nahal H., Provart N.J. (2013). Incipient stem cell niche conversion in tissue culture: using a systems approach to probe early events in WUSCHEL-dependent conversion of lateral root primordia into shoot meristems. Plant J. 73: 798–813. [DOI] [PubMed] [Google Scholar]
- Che P., Gingerich D.J., Lall S., Howell S.H. (2002). Global and hormone-induced gene expression changes during shoot development in Arabidopsis. Plant Cell 14: 2771–2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che P., Lall S., Nettleton D., Howell S.H. (2006). Gene expression programs during shoot, root, and callus development in Arabidopsis tissue culture. Plant Physiol. 141: 620–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che P., Lall S., Howell S.H. (2007). Developmental steps in acquiring competence for shoot development in Arabidopsis tissue culture. Planta 226: 1183–1194. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Cole M., Chandler J., Weijers D., Jacobs B., Comelli P., Werr W. (2009). DORNROSCHEN is a direct target of the auxin response factor MONOPTEROS in the Arabidopsis embryo. Development 136: 1643–1651. [DOI] [PubMed] [Google Scholar]
- Czechowski T., Stitt M., Altmann T., Udvardi M.K., Scheible W.R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139: 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan M., Xu C., Xu K., Hu Y. (2012). LATERAL ORGAN BOUNDARIES DOMAIN transcription factors direct callus formation in Arabidopsis regeneration. Cell Res. 22: 1169–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto S.Y., Ohta M., Usui A., Shinshi H., Ohme-Takagi M. (2000). Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 12: 393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galinha C., Hofhuis H., Luijten M., Willemsen V., Blilou I., Heidstra R., Scheres B. (2007). PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449: 1053–1057. [DOI] [PubMed] [Google Scholar]
- Gendrel A.-V., Lippman Z., Martienssen R., Colot V. (2005). Profiling histone modification patterns in plants using genomic tiling microarrays. Nat. Methods 2: 213–218. [DOI] [PubMed] [Google Scholar]
- Gordon S.P., Heisler M.G., Reddy G.V., Ohno C., Das P., Meyerowitz E.M. (2007). Pattern formation during de novo assembly of the Arabidopsis shoot meristem. Development 134: 3539–3548. [DOI] [PubMed] [Google Scholar]
- Hicks G.S. (1994). Shoot induction and organogenesis in vitro: A developmental perspective. In Vitro Cell Dev. Biol. 30: 10–15. [Google Scholar]
- Hiratsu K., Ohta M., Matsui K., Ohme-Takagi M. (2002). The SUPERMAN protein is an active repressor whose carboxy-terminal repression domain is required for the development of normal flowers. FEBS Lett. 514: 351–354. [DOI] [PubMed] [Google Scholar]
- Ikeuchi M., Sugimoto K., Iwase A. (2013). Plant callus: mechanisms of induction and repression. Plant Cell 25: 3159–3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi M., et al. (2015a). PRC2 represses dedifferentiation of mature somatic cells in Arabidopsis. Nat. Plants 1: 15089. [DOI] [PubMed] [Google Scholar]
- Ikeuchi M., Iwase A., Sugimoto K. (2015b). Control of plant cell differentiation by histone modification and DNA methylation. Curr. Opin. Plant Biol. 28: 60–67. [DOI] [PubMed] [Google Scholar]
- Ikeuchi M., Ogawa Y., Iwase A., Sugimoto K. (2016). Plant regeneration: cellular origins and molecular mechanisms. Development 143: 1442–1451. [DOI] [PubMed] [Google Scholar]
- Iwase A., Mitsuda N., Koyama T., Hiratsu K., Kojima M., Arai T., Inoue Y., Seki M., Sakakibara H., Sugimoto K., Ohme-Takagi M. (2011a). The AP2/ERF transcription factor WIND1 controls cell dedifferentiation in Arabidopsis. Curr. Biol. 21: 508–514. [DOI] [PubMed] [Google Scholar]
- Iwase A., Ohme-Takagi M., Sugimoto K. (2011b). WIND1: a key molecular switch for plant cell dedifferentiation. Plant Signal. Behav. 6: 1943–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase A., Mita K., Nonaka S., Ikeuchi M., Koizuka C., Ohnuma M., Ezura H., Imamura J., Sugimoto K. (2015). WIND1-based acquisition of regeneration competency in Arabidopsis and rapeseed. J. Plant Res. 128: 389–397. [DOI] [PubMed] [Google Scholar]
- Kareem A., Durgaprasad K., Sugimoto K., Du Y., Pulianmackal A.J., Trivedi Z.B., Abhayadev P.V., Pinon V., Meyerowitz E.M., Scheres B., Prasad K. (2015). PLETHORA genes control regeneration by a two-step mechanism. Curr. Biol. 25: 1017–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertbundit S., De Greve H., Deboeck F., Van Montagu M., Hernalsteens J.P. (1991). In vivo random beta-glucuronidase gene fusions in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 88: 5212–5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian J., Whitehead D., Horak J., Wanke D., Weinl S., Batistic O., D’Angelo C., Bornberg-Bauer E., Kudla J., Harter K. (2007). The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 50: 347–363. [DOI] [PubMed] [Google Scholar]
- Kirch T., Simon R., Grünewald M., Werr W. (2003). The DORNROSCHEN/ENHANCER OF SHOOT REGENERATION1 gene of Arabidopsis acts in the control of meristem ccll fate and lateral organ development. Plant Cell 15: 694–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R.-C., Park H.-J., Wang H.-Y. (2008). Role of Arabidopsis RAP2.4 in regulating light- and ethylene-mediated developmental processes and drought stress tolerance. Mol. Plant 1: 42–57. [DOI] [PubMed] [Google Scholar]
- Liu J., Sheng L., Xu Y., Li J., Yang Z., Huang H., Xu L. (2014). WOX11 and 12 are involved in the first-step cell fate transition during de novo root organogenesis in Arabidopsis. Plant Cell 26: 1081–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason M.G., Mathews D.E., Argyros D.A., Maxwell B.B., Kieber J.J., Alonso J.M., Ecker J.R., Schaller G.E. (2005). Multiple type-B response regulators mediate cytokinin signal transduction in Arabidopsis. Plant Cell 17: 3007–3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo N., Banno H. (2008). The Arabidopsis transcription factor ESR1 induces in vitro shoot regeneration through transcriptional activation. Plant Physiol. Biochem. 46: 1045–1050. [DOI] [PubMed] [Google Scholar]
- Matsuo N., Mase H., Makino M., Takahashi H., Banno H. (2009). Identification of ENHANCER OF SHOOT REGENERATION 1-upregulated genes during in vitro shoot regeneration. Plant Biotechnol. 26: 385–393. [Google Scholar]
- Matsuo N., Makino M., Banno H. (2011). Arabidopsis ENHANCER OF SHOOT REGENERATION (ESR)1 and ESR2 regulate in vitro shoot regeneration and their expressions are differentially regulated. Plant Sci. 181: 39–46. [DOI] [PubMed] [Google Scholar]
- Menges M., Murray J.A. (2002). Synchronous Arabidopsis suspension cultures for analysis of cell-cycle gene activity. Plant J. 30: 203–212. [DOI] [PubMed] [Google Scholar]
- Miller G., Schlauch K., Tam R., Cortes D., Torres M.A., Shulaev V., Dangl J.L., Mittler R. (2009). The plant NADPH oxidase RBOHD mediates rapid systemic signaling in response to diverse stimuli. Sci. Signal. 2: ra45. [DOI] [PubMed] [Google Scholar]
- Mitsuda N., Hiratsu K., Todaka D., Nakashima K., Yamaguchi-Shinozaki K., Ohme-Takagi M. (2006). Efficient production of male and female sterile plants by expression of a chimeric repressor in Arabidopsis and rice. Plant Biotechnol. J. 4: 325–332. [DOI] [PubMed] [Google Scholar]
- Mousavi S.A.R., Chauvin A., Pascaud F., Kellenberger S., Farmer E.E. (2013). GLUTAMATE RECEPTOR-LIKE genes mediate leaf-to-leaf wound signalling. Nature 500: 422–426. [DOI] [PubMed] [Google Scholar]
- Ohme-Takagi M., Shinshi H. (1995). Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 7: 173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M., Matsui K., Hiratsu K., Shinshi H., Ohme-Takagi M. (2001). Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13: 1959–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa S., Yasutani I., Fukuda H., Komamine A., Sugiyama M. (1998). Organogenic responses in tissue culture of srd mutants of Arabidopsis thaliana. Development 125: 135–142. [DOI] [PubMed] [Google Scholar]
- Sasaki K., Hiraga S., Ito H., Seo S., Matsui H., Ohashi Y. (2002). A wound-inducible tobacco peroxidase gene expresses preferentially in the vascular system. Plant Cell Physiol. 43: 108–117. [DOI] [PubMed] [Google Scholar]
- Sasaki K., Ito H., Mitsuhara I., Hiraga S., Seo S., Matsui H., Ohashi Y. (2006). A novel wound-responsive cis-element, VWRE, of the vascular system-specific expression of a tobacco peroxidase gene, tpoxN1. Plant Mol. Biol. 62: 753–768. [DOI] [PubMed] [Google Scholar]
- Sasaki K., Mitsuhara I., Seo S., Ito H., Matsui H., Ohashi Y. (2007). Two novel AP2/ERF domain proteins interact with cis-element VWRE for wound-induced expression of the tobacco tpoxN1 gene. Plant J. 50: 1079–1092. [DOI] [PubMed] [Google Scholar]
- Skoog F., Miller C.O. (1957). Chemical regulation of growth and organ formation in plant tissues cultured in vitro. Symp. Soc. Exp. Biol. 11: 118–130. [PubMed] [Google Scholar]
- Sugimoto K., Jiao Y., Meyerowitz E.M. (2010). Arabidopsis regeneration from multiple tissues occurs via a root development pathway. Dev. Cell 18: 463–471. [DOI] [PubMed] [Google Scholar]
- Tanaka E.M., Reddien P.W. (2011). The cellular basis for animal regeneration. Dev. Cell 21: 172–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorpe T.A. (2007). History of plant tissue culture. Mol. Biotechnol. 37: 169–180. [DOI] [PubMed] [Google Scholar]
- Valvekens D., Van Montagu M., Van Lijsebettens M. (1988). Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc. Natl. Acad. Sci. USA 85: 5536–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K., Shinozaki K. (1994). A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6: 251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K., Sakamoto S., Kawai T., Kobayashi Y., Sato K., Ichinose Y., Yaoi K., Akiyoshi-Endo M., Sato H., Takamizo T., Ohme-Takagi M., Mitsuda N. (2013). Engineering the Oryza sativa cell wall with rice NAC transcription factors regulating secondary wall formation. Front. Plant Sci. 4: 383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C., Guo J., Feng Z., Cui X., Zhu J. (2012). Molecular characterization of a novel AP2 transcription factor ThWIND1-L from Thellungiella halophila. Plant Cell Tissue Organ Cult. 110: 423–433. [Google Scholar]
- Zuo J., Niu Q.-W., Chua N.-H. (2000). Technical advance: An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 24: 265–273. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.