AtKNL2 possesses a conserved CENPC-k motif that is required for its centromeric localization and enables sequence-independent interaction with DNA in vitro but preferential binding with the centromeric repeat pAL1 in vivo.
Abstract
KINETOCHORE NULL2 (KNL2) is involved in recognition of centromeres and in centromeric localization of the centromere-specific histone cenH3. Our study revealed a cenH3 nucleosome binding CENPC-k motif at the C terminus of Arabidopsis thaliana KNL2, which is conserved among a wide spectrum of eukaryotes. Centromeric localization of KNL2 is abolished by deletion of the CENPC-k motif and by mutating single conserved amino acids, but can be restored by insertion of the corresponding motif of Arabidopsis CENP-C. We showed by electrophoretic mobility shift assay that the C terminus of KNL2 binds DNA sequence-independently and interacts with the centromeric transcripts in vitro. Chromatin immunoprecipitation with anti-KNL2 antibodies indicated that in vivo KNL2 is preferentially associated with the centromeric repeat pAL1. Complete deletion of the CENPC-k motif did not influence its ability to interact with DNA in vitro. Therefore, we suggest that KNL2 recognizes centromeric nucleosomes, similar to CENP-C, via the CENPC-k motif and binds adjoining DNA.
INTRODUCTION
Centromeres are the chromosomal positions at which the kinetochore complex assembles. The kinetochore complex is required for correct sister chromatid segregation, sister chromatid cohesion, chromosome movement, and cell cycle regulation (Henikoff and Dalal, 2005).
Normal kinetochore establishment depends on the centromeric histone H3 variant cenH3 originally described as human CENP-A (Earnshaw and Rothfield, 1985). CenH3 is an essential centromeric mark and substitutes for H3 histones at centromeric sites. The centromeric chromatin consists of blocks of H3 containing nucleosomes intermingled with cenH3 chromatin (Blower et al., 2002). In most plants and animals, centromeres are located within regions containing large numbers of tandem repeats of short sequences. CenH3-containing chromatin occupies only part of these repeats (Blower et al., 2002). In Arabidopsis thaliana, ∼15% of the 178-bp centromeric repeat pAL1 is associated with Arabidopsis cenH3 (Nagaki et al., 2003).
Thus, in most organisms, except Saccharomyces cerevisiae (Meluh et al., 1998), cenH3 localization is not solely determined by centromeric DNA sequences, but is rather regulated epigenetically. Some studies have proposed that cenH3 plays the role of epigenetic mark, since ectopically incorporated cenH3 (Lo et al., 2001; Nasuda et al., 2005), as well as tethering of cenH3 to noncentromeric loci (Mendiburo et al., 2011; Teo et al., 2013), leads to de novo kinetochore assembly at positions free of centromeric repeats. Incorporation of cenH3 is anticorrelated with other epigenetic marks such as DNA methylation and some histone modifications (Zhang et al., 2008). It was shown for Arabidopsis that the centromeric transcripts and corresponding small interfering RNAs might be involved in methylation of centromeric/pericentromeric DNA (Teixeira and Colot, 2010). Single-stranded centromeric transcripts can bind centromeric chromatin and may serve as a structural template to help in recruiting kinetochore proteins (Wong et al., 2007; Du et al., 2010).
The cenH3 deposition pathway includes three steps: (1) initiation, i.e., generation of the epigenetic context for cenH3 incorporation; (2) deposition; and (3) maintenance (reviewed in De Rop et al., 2012).
(1) In mammals, the Mis18 complex, which includes Mis18α, Mis18β, and Mis18BP1 (KNL2) proteins, plays a role in the initiation step and replenishment of cenH3. The human Mis18 complex is transiently present at centromeres after mitotic exit prior to cenH3 loading, and downregulation of Mis18 complex components results in a reduced amount of cenH3 at centromeres (Fujita et al., 2007). Fission yeast (Schizosaccharomyces pombe) Mis18 is also a part of the cenH3 assembly machinery (Hayashi et al., 2004). The Caenorhabditis elegans KNL2 and cenH3 colocalize at centromeres throughout the cell cycle and coordinate kinetochore assembly and chromosome segregation (Maddox et al., 2007). Arabidopsis KNL2 colocalizes with cenH3 and is associated with centromeres during all stages of the mitotic cell cycle, except from metaphase to mid-anaphase. Knockout of Arabidopsis KNL2 leads to anaphase bridges, reduced fertility, reduced levels of DNA methylation, and reduced transcription of the genes encoding cenH3 and histone modification proteins such as SUVH4 and SUVH9 (Lermontova et al., 2013).
Establishment of the epigenetic context for cenH3 loading requires other proteins. In mammalian cells, the kinetochore protein CENP-C recruits epigenetic factors such as DNA methyltransferases or KNL2 to centromeric and pericentromeric satellite sequences to promote epigenetic maintenance of CENP-A chromatin (Gopalakrishnan et al., 2009; Shono et al., 2015). CENP-C has been shown to interact directly with KNL2 (Moree et al., 2011; Dambacher et al., 2012) and with cenH3 nucleosomes (Westermann et al., 2003; Carroll et al., 2010). In vitro, CENP-C binds centromeric nucleosomes through the conserved CENPC motif (Kato et al., 2013). The loss of CENP-C leads to chromosome misalignment and segregation defects during mitosis and to increased transcription of centromeric repeats. CENP-I can also recruit KNL2 independently of CENP-C in HeLa cells (Shono et al., 2015).
(2) Deposition of cenH3 to centromeres depends on chaperones involved in nucleosome assembly. Two cenH3 chaperones have been identified: the Holliday Junction Recognition Protein (HJURP) and the Nuclear Autoantigenic Sperm Protein (Osakabe et al., 2010) in human and their respective homologs Scm3 and Sim3 in yeast (Pidoux et al., 2009). The Mis18 complex mediates the recruitment of HJURP to endogenous centromeres (Foltz et al., 2009; Barnhart et al., 2011).
(3) Centromeric chromatin maturation and maintenance of newly incorporated cenH3 have been less studied than the initiation and deposition steps.
Here, we showed that Arabidopsis KNL2 contains a conserved CENPC-like motif, designated CENPC-k, at its C terminus that is required for the centromeric localization of KNL2 in vivo. Mutagenesis of the conserved amino acids within the CENPC-k motif or complete deletion of the motif abolished centromeric localization of KNL2 in vivo. We also showed that substitution of the CENPC-k motif by the corresponding CENPC motif of Arabidopsis CENP-C restores centromere targeting of KNL2.
Additionally, we could demonstrate that the C terminus, but not the N terminus, of KNL2 interacts sequence-independently with DNA in vitro, but in vivo it associates preferentially with the centromeric repeat pAL1. Deletion of the CENPC-k motif did not influence the ability of KNL2 to interact with DNA in vitro.
RESULTS
Identification of a CENPC-Like Motif in KNL2
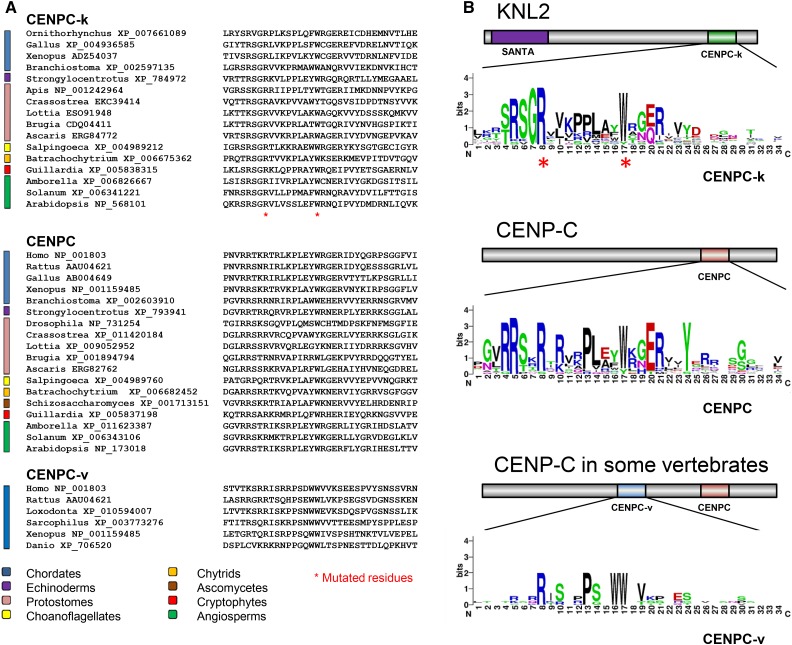
KNL2 was first described in nematodes and in vertebrates (Maddox et al., 2007), where it is characterized by both a C-terminal SANT/Myb domain (Boyer et al., 2002) and a unique N-terminal SANTA domain (Zhang et al., 2006). In Arabidopsis, KNL2 has a SANTA domain but lacks a SANT domain (Lermontova et al., 2013), suggesting that the SANTA domain is a conserved feature of KNL2 homologs. Recently, some insect proteins were reported to have both SANTA domains and a CENPC-like motif (Drinnenberg et al., 2014). Using BLASTP searches, we found that KNL2 homologs with a CENPC-like motif, hereafter referred to as a CENPC-k motif, are present in a wide spectrum of eukaryotes (Figure 1), including chytrids, cryptomonads, Arabidopsis, most invertebrates, and many vertebrates, but excluding therian mammals and Caenorhabditis elegans, the organisms in which KNL2 was first described (Maddox et al., 2007). This observation was recently independently made for vertebrate KNL2 proteins (Kral, 2016).
Figure 1.
CENPC-Like Motifs in KNL2 and CENP-C Homologs.
(A) Alignments of the three CENPC-like motifs present in KNL2 and CENP-C proteins of diverse eukaryotes. Asterisks indicate the conserved arginine (R) and tryptophan (W) residues that are found in all three alignments and that were mutated in this study.
(B) The same alignments presented in LOGO format (WebLogo; http://weblogo.berkeley.edu/logo.cgi) to more easily compare similarities and differences in the three motifs. Above each alignment is a domain map of the position of the CENPC-like motif in the KNL2 or CENP-C proteins.
The CENPC motif (Figure 1) is characteristic of the CENP-C protein (Brown, 1995; Talbert et al., 2004). The CENPC motif of rat CENP-C protein can bind directly to a chimeric H3/cenH3 nucleosome in vitro (Kato et al., 2013), suggesting that this motif binds to cenH3 nucleosomes in vivo. In some vertebrates, including therian mammals, Xenopus laevis (Kato et al., 2013), and teleost fishes (Kral, 2016), the CENP-C protein contains a second CENPC-like motif in the central region (here designated as the CENPC-v motif; Figure 1) that similarly binds H3/cenH3 hybrid nucleosomes (Kato et al., 2013). The presence of CENPC-k in KNL2 proteins suggests that it may be responsible for binding to cenH3 nucleosomes, similar to the CENPC and CENPC-v motifs of vertebrate CENP-Cs.
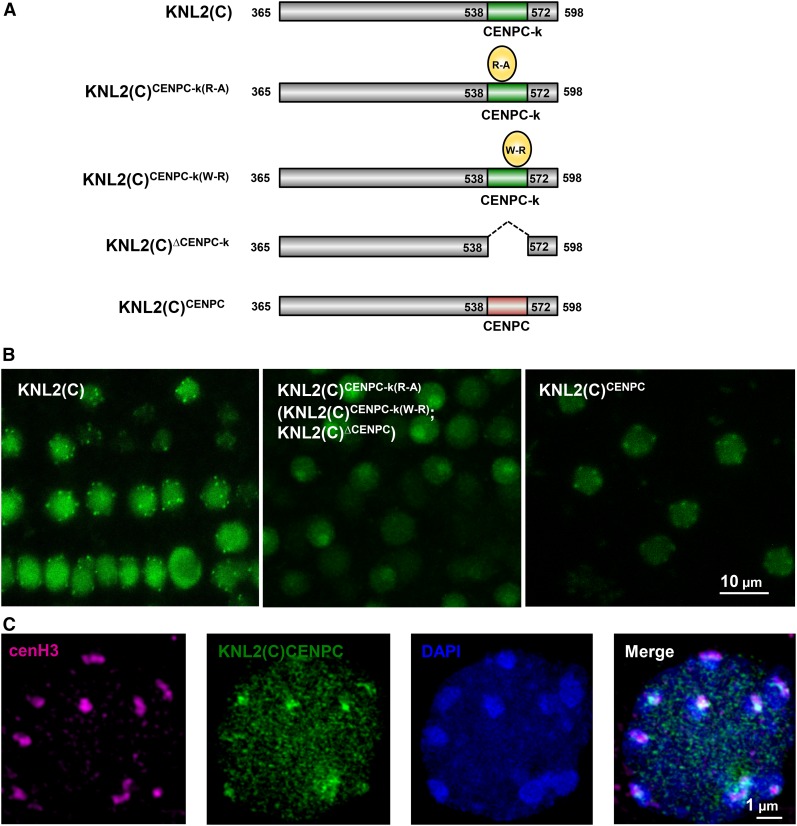
The CENPC-k Motif Is Required for the Centromeric Localization of KNL2 and Can Be Functionally Replaced by the CENPC Motif of CENP-C
Mutational analysis has identified critical residues of the CENPC motif (Heeger et al., 2005; Milks et al., 2009) or the CENPC-v motif (Song et al., 2002) that are essential for centromeric localization of CENP-C or for H3/cenH3 nucleosome binding (Kato et al., 2013). Two of these correspond to residues Arg-546 and Trp-555 of the Arabidopsis CENPC-k motif. The wild-type C-terminal part of KNL2 fused to EYFP can localize to centromeres (Lermontova et al., 2013). We therefore mutated Arg-546 and Trp-555 in this construct [KNL2(C)CENPC-k(R-A) and KNL2(C)CENPC-k(W-R)] (Figure 2A) to determine whether CENPC-k plays a similar role in KNL2 to the role of the CENPC and CENPC-v motifs in CENP-C. Additionally, we generated the construct with complete deletion of the CENPC-k motif [KNL2(C)∆CENPC-k] (Figure 2A). Analysis of transgenic Arabidopsis plants expressing these constructs showed that the mutagenized KNL2 variants are unable to localize to chromocenters/centromeric sites (Figure 2B). Fluorescence signals were detected in nucleoplasm and in nucleoli of all five independent transgenic lines analyzed for each construct. These results suggest that the CENPC-k motif of KNL2 in general, and the conserved Arg-546 and Trp-555 amino acids in particular, are required for in vivo localization of KNL2 at centromeres of Arabidopsis.
Figure 2.
Mutagenesis of Conserved Amino Acids at the CENPC-k Motif as Well as Its Complete Deletion Results in KNL2 Mislocalization That Can Be Restored by the CENPC Motif.
(A) Schematic view of EYFP-tagged KNL2 constructs: KNL2(C), KNL2(C)CENPC-k(R-A), KNL2(C)CENPC-k(W-R), KNL2(C)∆CENPC-k, and KNL2(C)CENPC.
(B) Subnuclear localization of KNL2(C) (left panel), KNL2(C)CENPC-k(R-A) (middle panel), and KNL2(C)CENPC (right panel) fused to EYFP in root tip nuclei of Arabidopsis transformed with corresponding constructs. KNL2(C)CENPC-k(W-R) and KNL2(C)∆CENPC-k proteins (indicated in brackets in the middle panel) showed the same localization pattern as KNL2(C)CENPC-k(R-A). The C-terminal part of KNL2 fused to EYFP showed nucleoplasmic and centromeric localization (Lermontova et al., 2013), while protein variants carrying the point mutations within the CENPC-k motif or lacking this motif completely lost an ability to localize at centromeres and were detected only at nucleoplasm and nucleolus. However, the C terminus of KNL2 with the substitution of CENPC-k by the CENPC motif was targeted to centromeres.
(C) Double immunostaining with anti-GFP [detecting the KNL2(C)CENPC EYFP fusion] and anti-cenH3 antibodies showed colocalization of both immunosignals at bright 4′,6-diamidino-2-phenylindole (DAPI)-stained chromocenters.
We addressed the question of whether replacement of CENPC-k by the CENPC motif of Arabidopsis CENP-C [KNL2(C)CENPC] will restore the ability of KNL2(C)∆CENPC-k to localize to centromeres. Analysis of transgenic Arabidopsis plants expressing the KNL2(C)CENPC construct has revealed that the CENPC motif is indeed able to target KNL2 to centromeres. Fluorescence signals were detected in the nucleoplasm and at centromeres of five independent transgenic lines similar to the KNL2(C) control (Figure 2B; Supplemental Figure 1). We showed previously that KNL2(C) fused with EYFP colocalizes with cenH3 at bright DAPI stained chromocenters (Lermontova et al., 2013) Double immunostaining with anti-GFP [detecting the KNL2(C)CENPC EYFP fusion] and anti-cenH3 antibodies showed the same colocalization (Figure 2C). These data suggest that CENPC-like motifs of KNL2 and of CENP-C proteins might play the same role in recognition of centromeric nucleosomes.
Additionally, leaves of Nicotiana benthamiana were transiently transformed by Agrobacterium tumefaciens with constructs expressing the wild-type C-terminal part of KNL2 (Lermontova et al., 2013), KNL2(C)∆CENPC-k, or KNL2(C)CENPC in a fusion with EYFP at their N or C termini, respectively. These constructs were expressed in N. benthamiana alone or in a combination with mCherry-cenH3. Supplemental Figure 2 indicates that in some cells the chimeric KNL2(C)CENPC protein localizes to centromeres and colocalizes with cenH3 similar to the KNL2(C) with the native CENPC-k motif, while KNL2(C)∆CENPC-k protein was detected only in the nucleoplasm and nucleolus. We assume that centromeric localization of the KNL2 variants and cenH3 can be seen only in mitotically active cells, since cenH3 (Lermontova et al., 2006) and very likely KNL2 can be loaded to centromeres only during the mitotic cell cycle. The proportion of cells showing colocalization varies depending on plant age and efficiency of double transformation. Our data demonstrate that Arabidopsis KNL2 can be targeted to centromeres of distantly related species such as N. benthamiana and that centromeric targeting requires presence of CENPC-k or CENPC motifs.
KNL2 Binds DNA Sequence-Independently in Vitro, but in Vivo It Is Associated with Centromeric Repeats
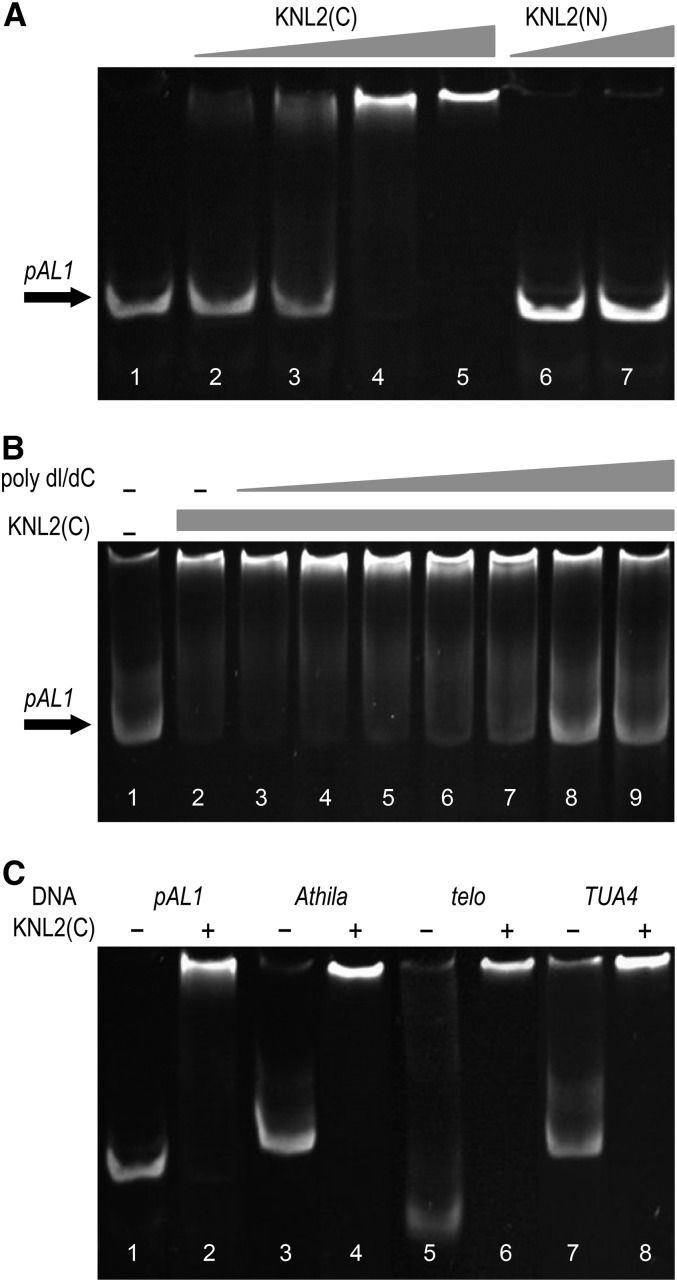
To test whether Arabidopsis KNL2 interacts with the centromeric DNA similar to the CENP-C protein of maize (Zea mays; Du et al., 2010), an electrophoretic mobility shift assay (EMSA) with recombinant KNL2 protein fragments and centromeric repeat pAL1 DNA was performed. The N-terminal part of KNL2 including the SANTA domain and the C terminus with the CENPC-k motif in fusion with a His-tag were separately expressed in Escherichia coli. Soluble proteins were purified under nondenaturation conditions and incubated with pAL1 (Figure 3A). The results showed that the band of pAL1 is shifted upwards only in the presence of recombinant C terminus, but not the N-terminal part of KNL2 (Figure 3A). The effect of KNL2(C) concentration on DNA binding was tested using constant amounts of pAL1 DNA and increased amounts of protein. The mobility of a portion of pAL1 DNA slightly shifted for DNA:protein ratios of 1:1 and 1:2, but with an increased amount of protein all pAL1 DNA shifted upwards. In the case of KNL2(N), no DNA binding was observed even when a high excess of protein (1:8 DNA:protein) was applied (Figure 3A). In addition to the nonradioactive EMSA, we employed the more commonly used radioactive method and produced similar results (Supplemental Figure 3).
Figure 3.
The C-Terminal Part of KNL2 Binds Centromeric and Noncentromeric DNA Sequences in Vitro.
(A) Nonradioactive EMSA with 4 pmol of the centromeric repeat pAL1 (all lanes) and 4 to 32 pmol of the recombinant C-terminal part of KNL2 (lanes 2–5) or 16 and 32 pmol of the recombinant N-terminal part of KNL2 (lanes 6 and 7), respectively, showed that the C terminus (lanes 4 and 5), but not the N terminus, of KNL2 interacts with the centromeric repeat pAL1 (n = 3).
(B) Nonradioactive EMSA with 4 pmol of pAL1 (all lanes) and 16 pmol of the C-terminal part of KNL2 (lanes 2–9). The unspecific competitor poly dI/dC was added in increasing concentrations of 0.1, 0.5, 1, 5, 10, 50, and 100 ng/μL (lanes 3–9). At an amount of 50 pmol poly dI/dC, the DNA binding of the C-terminal part of KNL2 and pAL1 get competed out (n = 2).
(C) Nonradioactive EMSA with 4 pmol of centromeric repeat pAL1 (lanes 1 and 2), transposable element Athila (lanes 3 and 4), telomeric repeat (lanes 5 and 6), euchromatic TUA4 sequence (lanes 7 and 8), and 16 pmol of the C-terminal part of KNL2 (even numbered lanes) shows that the C-terminal part of KNL2 interacts with centromeric and noncentromeric DNA (n = 3).
To test whether the C terminus of KNL2 interacts specifically with centromeric repeats, we performed a competition experiment in which poly(deoxyinosinic-deoxycytidylic) acid (poly dI/dC) was used. The DNA binding capability of the C-terminal KNL2 to pAL1 was abolished by 50 ng/μL poly dI/dC (Figure 3B). About 1 to 2.5 ng/μL poly dI/dC is usually used in EMSA to inhibit unspecific interactions (Smith and Humphries, 2009). Next, we analyzed the interaction of KNL2(C) with repetitive elements such as the centromeric transposable element Athila, the telomeric repeat and the coding region of tubulin TUA4 (Ay et al., 2009). The data showed that the C terminus of KNL2 binds all noncentromeric DNA sequences that were used (Figure 3C).
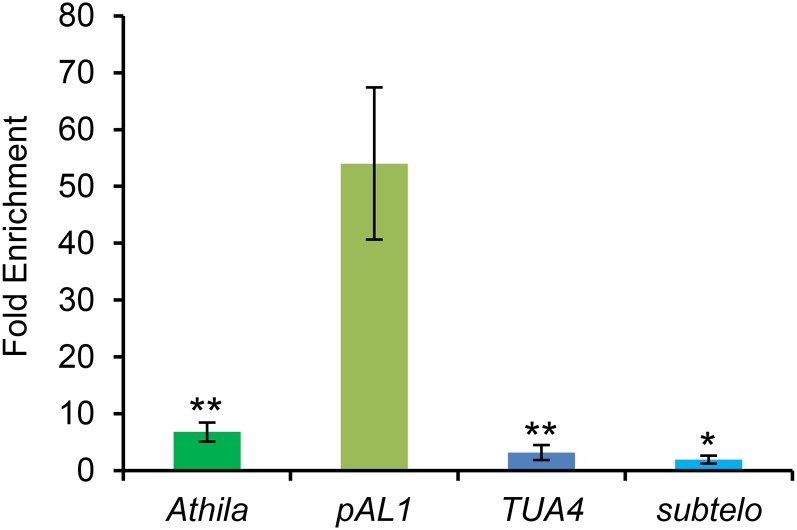
To analyze the interaction of KNL2 with DNA in vivo, we performed chromatin immunoprecipitation (ChIP). Antibodies against KNL2 were purified by affinity chromatography using CN-Br-Sepharose beads coupled with the KNL2(C) antigen and applied to chromatin isolated from seedlings of Arabidopsis wild-type plants (Figure 4). The results showed that in vivo, KNL2 is associated preferentially with the centromeric repeat pAL1 and to a much lower extent with other sequences.
Figure 4.
The ChIP Experiment with Anti-KNL2 Antibodies Showed That in Vivo KNL2 Is Mostly Associated with the Centromeric Repeat pAL1.
A ChIP experiment with sonicated chromatin isolated from 5 g of 8-d-old Arabidopsis wild-type seedlings was performed with anti-KNL2 antibodies (Lermontova et al., 2013). A subsequent qPCR on the KNL2 bound sequences performed with pAL1, Athila, TUA4 (nine biological replicates and three technical replicates), and subtelomeric (three biological and technical replicates) primers showed that KNL2 binds preferentially the centromeric sequence pAL1. Subtelomeric primers were chosen instead of telomeric ones to obtain more consistent CT values. Fold enrichment indicates the relative enrichment of the target sequence in the sample in comparison to the values of the nonantibody control. The bars indicate the sd. Asterisks indicate results of the t test between fold enrichments of pAL1 and respective noncentromeric sequences (double sided, type 2; *P < 0.5 and **P < 0.05).
The C Terminus of KNL2 Lacking the CENPC-k Motif Interacts with the Centromeric DNA in Vitro
We showed above that mutagenesis of conserved amino acids within the CENPC-k motif abolished centromeric localization of KNL2, indicating a role of the CENPC-k motif for the centromeric localization of KNL2 in vivo. Therefore, we addressed the question whether this motif is required for direct interaction of KNL2 with centromeric DNA in vitro.
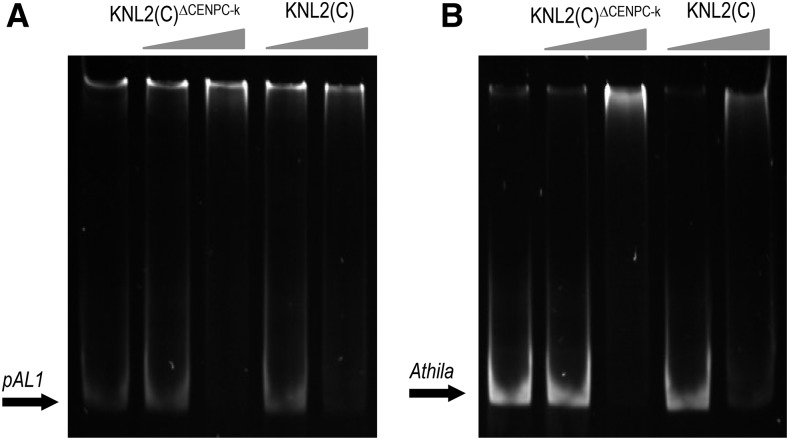
To test this hypothesis, the KNL2(C)∆CENPC-k fragment was subcloned into the pETG30A vector for expression of recombinant protein in E. coli. The purified KNL2(C)∆CENPC-k protein was used in EMSA with the centromeric repeat pAL1 and the centromeric transposable element Athila (Figure 5). The results showed that KNL2(C)∆CENPC-k binds pAL1 and Athila sequences with similar efficiency as the nonmutagenized variant (Figure 5). This suggests that the CENPC-k motif does not bind DNA directly, consistent with the proposed role of CENPC motifs in binding to the histones of the cenH3 nucleosome surface (Kato et al., 2013).
Figure 5.
The CENPC-k Motif Is Not Required for the Interaction of KNL2 with DNA in Vitro.
EMSA with KNL2(C)∆CENPC-k and the centromeric repeat pAL1 (A) or the transposable element Athila (B) showed that the absence of the CENPC-k motif does not influence the ability of KNL2(C) to interact with DNA. In both cases, KNL2(C) was used as a positive control (n = 2).
KNL2 Binds RNA in Vitro
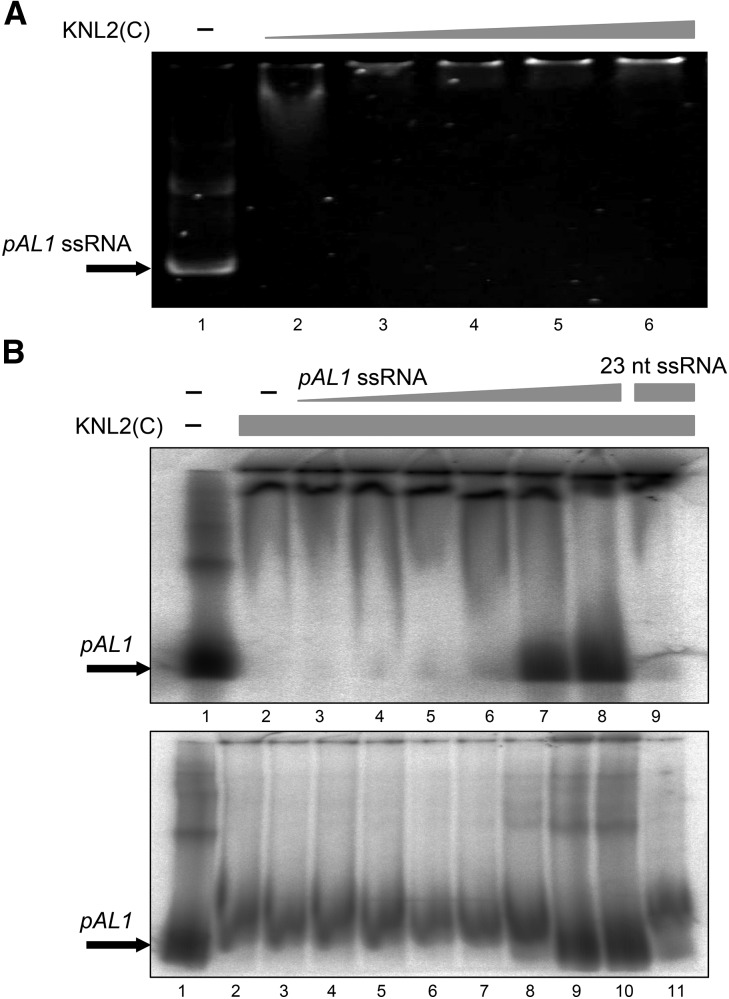
Previously, Du et al. (2010) showed that the CENP-C protein of maize binds not only DNA, but also RNA molecules. In a gel shift assay, DNA binding of CENP-C was promoted by small single-stranded RNAs (ssRNAs), while larger ssRNAs (>150 nucleotides) competed with the DNA. We have identified a 23-nucleotide ssRNA sequence (Supplemental Table 1) derived from the centromeric repeat pAL1 from small RNA-seq data of the wild type and tested whether this ssRNA interacts with KNL2 and influences its binding capability to pAL1 in EMSA. The selected ssRNA can bind to KNL2 (Supplemental Figure 4A) but had no effect on binding of pAL1 DNA by KNL2 (Supplemental Figure 4B). Additionally we generated the full-length forward and reverse centromeric transcripts (178-nucleotide pAL1 ssRNA; Supplemental Table 1) and used them in a gel shift assay with KNL2(C) or as competitor for the KNL2(C) pAL1 DNA interaction. We can demonstrate that centromeric transcripts bind KNL2(C) (Figure 6A) and compete out the pAL1 DNA interaction with KNL2(C) starting at a ratio of DNA:RNA 1:10 (Figure 6B).
Figure 6.
KNL2 DNA Binding Is Competed out by Centromeric pAL1 ssRNA Transcript.
(A) EMSA experiment with 8 pmol of pAL1 ssRNA (all lanes) and increased amounts of purified recombinant C-terminal KNL2 (8, 16, 20, 24, and 40 pmol in lanes 2–6, respectively; n = 3).
(B) Two radioactive EMSA experiments with 3.3 pmol labeled pAL1 (all lanes) and 26.6 pmol (upper gel) or 15 pmol (lower gel) purified recombinant C-terminal KNL2 (from lanes 2 to the rightmost). For the competition experiment, increasing amounts of pAL1 ssRNA forward transcript (upper gel, lanes 3–8: 1.5 and 3.3 pmol; 15, 33, 150, and 330 pmol; lower gel, lanes 3–10: 0.033 and 0.15 pmol; 0.33, 1.5, 3.3, 33, 150, and 330 pmol) or 330 pmol of 23-nucleotide ssRNA (last lane of each gel) were added. pAL1 ssRNA compete out the KNL2(C)-pAL1 DNA complex formation in contrast to the 23-nucleotide ssRNA. Forward and reverse pAL1 transcripts showed similar results (data not shown).
DISCUSSION
The Arabidopsis KNL2 Protein Possesses a CENPC-k Motif Required for Its Centromeric Localization
Based on sequence similarity, a conserved CENPC-k motif at the C terminus could be identified in Arabidopsis KNL2. Recently, Kral (2016) showed that the CENPC-k motif is present in KNL2 homologs of most vertebrates, but not of therian mammals. This motif can also be identified in KNL2 homologs across several eukaryotic kingdoms (Figure 1), indicating it is an ancient and conserved feature of KNL2. However, the functional importance of this motif in KNL2 homologs has remained elusive. The CENPC motif is a typical feature at the C terminus of CENP-C proteins, while the CENPC-v motif is also present at the central region of CENP-C in many vertebrates. In human cells, targeting of CENP-C to centromeric nucleosomes depends on either the CENPC motif or the CENPC-v motif (Lanini and McKeon, 1995; Yang et al., 1996; Song et al., 2002; Trazzi et al., 2002; Carroll et al., 2010). Kato et al. (2013) have demonstrated that mutagenesis of the conserved amino acids within both CENPC-v and CENPC motifs of rat CENP-C disturbed its binding to centromeric nucleosomes.
We showed that mutagenesis of the conserved amino acids Arg-546 and Trp-555 to Ala and Arg, respectively, within the CENPC-k motif, or complete deletion of this motif, abolished centromeric localization of Arabidopsis KNL2 (Figure 2; Supplemental Figure 1). However, the CENPC motif of the CENP-C protein restores the ability of KNL2(C)∆CENPC-k to localize at centromeres (Figure 2; Supplemental Figures 1 and 2). Therefore, we suggest that the CENPC-k motif of KNL2 is involved in binding centromeric nucleosomes, similar to the corresponding motif of the CENP-C protein. In view of the poor conservation of CENP-C and KNL2 proteins across eukaryotic kingdoms, outside of their CENPC-like motifs (Talbert et al., 2004; Maddox et al., 2007; Lermontova et al., 2013), it is possible that the CENPC-k motif of KNL2 or CENPC motif of CENP-C fused with any desired protein may lead to centromeric localization. Although this remains to be tested in further studies, this approach may provide an interesting tool for centromere research.
Arabidopsis KNL2 Binds DNA in a Sequence-Independent Manner and Interacts with Centromeric RNA Transcripts
Using EMSA experiments (Figure 3), we could demonstrate that the C terminus, but not the N terminus, of Arabidopsis KNL2 interacts with the centromeric DNA in vitro. In silico analysis of KNL2 did not reveal any recognized DNA binding family motifs, but predicted putative DNA/RNA binding regions around the CENPC-k motif (Supplemental Figure 5). The C terminus also showed an ability to interact with noncentromeric DNA in vitro, such as the transposable element Athila, the telomeric repeat, and the euchromatic tubulin gene region TUA4. However, according to our ChIP data (Figure 4), in vivo KNL2 is preferentially associated to the centromeric repeat pAL1. Since formaldehyde cross-linking, used for ChIP, can occur at the protein-DNA interface and reflect the in vivo binding of a specific factor to its cognate DNA sites (Toth and Biggin, 2000), we suggest that KNL2 interacts with the centromeric DNA directly.
Arabidopsis KNL2 may interact with DNA in a similar way to CENP-C proteins of animals and plants. Human CENP-C, which also recognizes DNA in a sequence-independent manner in vitro, preferentially interacts with the centromeric alpha-satellite DNA in vivo (Politi et al., 2002). Human CENP-C lacks a typical DNA binding domain, and its DNA binding regions are distributed within several broad domains that overlap the centromere-targeting CENPC and CENPC-v motifs (Sugimoto et al., 1994; Yang et al., 1996; Carroll et al., 2010).
Du et al. (2010) showed that the CENP-C protein of maize also does not contain a recognized DNA binding family motif and interacts with DNA via 122 amino acids encoded in exons 9 to 12, adjacent to the CENPC motif encoded in exon 13. Interaction with DNA via an adjacent dispersed DNA binding region may be a general feature of proteins with CENPC-like motifs.
Additionally, we found that the CENPC-k motif of Arabidopsis KNL2 is not required for its interaction with DNA, since the deletion of this motif did not influence the ability of KNL2 to interact with DNA in vitro (Figure 5). Conversely, a YFP-tagged CENP-C variant of maize with the deleted DNA binding region (exons 9–12), but containing the CENPC motif, showed only partially reduced centromeric localization in transient and stable transformation of maize (Du et al., 2010). We propose that targeting of KNL2 and CENP-C to centromeres mainly depends on their interaction with centromeric nucleosomes or other proteins via their CENPC-like motifs, while the interaction of both proteins with DNA via sequence-independent DNA binding regions allows a cooperative form of binding to centromeric chromatin that helps to anchor them at centromeric positions.
We also demonstrated that, similar to CENP-C of maize (Du et al., 2010), the C-terminal part of KNL2 interacts with ssRNAs such as the full-length centromeric pAL1 transcript (Figure 6) and a small 23-nucleotide ssRNA sequence derived from pAL1 (Supplemental Figure 4A). Although the precise role of centromeric transcripts remains unclear, it was suggested that they can bind centromeric chromatin and may serve as a structural template to help in recruiting kinetochore proteins (Wong et al., 2007; Du et al., 2010). The fact that in humans, a decrease in centromeric α-satellite transcription results in reduced level of CENP-C at centromeres and increased number of anaphases with lagging chromosomes (Chan et al., 2012) is supportive of this interpretation.
The Roles of KNL2 and CENP-C in Centromere Recognition and Kinetochore Assembly
Despite many common features, KNL2 and CENP-C are deployed differently in the cell cycle. In plants and mammals, CENP-C is present at centromeres throughout the cell cycle (Dawe et al., 1999; Sugimoto et al., 2000), while KNL2 of Arabidopsis and KNL2-2 of X. laevis are absent from centromeres from metaphase to anaphase (Lermontova et al., 2013; Moree et al., 2011), and human KNL2 is only transiently localized at centromeres after mitotic exit (Fujita et al., 2007). These differences reflect the role of CENP-C in kinetochore assembly and function, while KNL2 functions upstream of cenH3 loading and probably is not needed for the function of an established kinetochore.
Recruitment of vertebrate KNL2 to centromeres depends on CENP-C, which binds KNL2 through the C terminus containing the CENPC motif and cupin/dimerization domain (Moree et al., 2011; Dambacher et al., 2012). However, proper centromeric localization requires an interaction of KNL2 with CENP-C and Mis18α proteins (Stellfox et al., 2016). Kral (2016) has suggested that centromeric localization of mammalian KNL2, lacking the CENPC-k motif, might be more dependent on CENP-C than the localization of KNL2 homologs of other vertebrates. In X. laevis, a second mechanism of KNL2 loading during interphase has been invoked that is inhibited by CENP-C (Moree et al., 2011), possibly through competition for the same binding site on cenH3 nucleosomes. Therefore, it is likely that the mechanism of centromeric localization of KNL2 homologs of plants containing the CENPC-k motif is distinct from that of mammals and might be similar to that of other vertebrates. Given the broad phylogenetic distribution of the CENPC-k and CENPC motifs, such a mechanism may be common and ancestral for centromere maintenance.
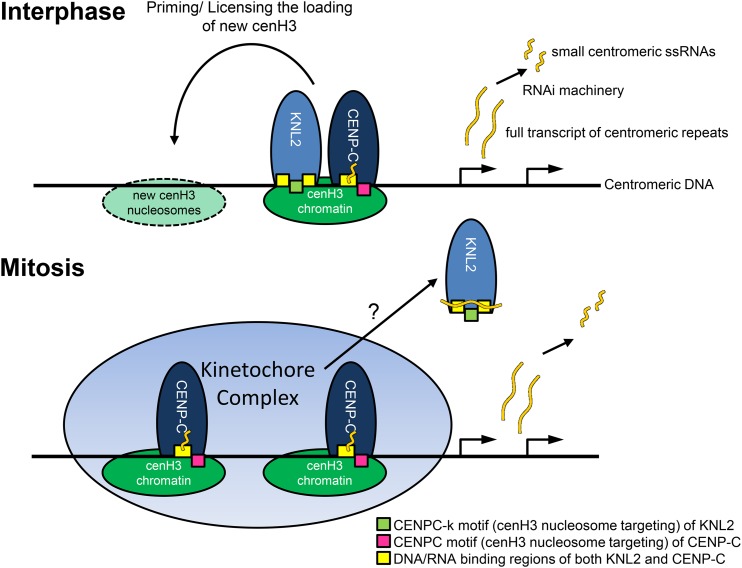
Here, we propose a model that describes the function and localization of KNL2 and CENPC at Arabidopsis centromeres (Figure 7). According to Du et al. (2010), binding of CENP-C to the centromeric DNA is stabilized by small ssRNAs; by contrast, we showed that binding of KNL2(C) to the pAL1 DNA is not influenced by small ssRNA. We demonstrated that full-length pAL1 transcripts compete with the pAL1 DNA for KNL2 binding. Therefore, we suggest that prior to mitosis, binding of KNL2 by long RNA transcripts might initiate release of KNL2 from centromeres, while ssRNAs stabilize CENP-C at the centromere, favoring the establishment of the kinetochore complex. We note that human CENP-C becomes stabilized at mitosis (Hemmerich et al., 2008).
Figure 7.
Model for the Localization and Function of the KNL2 and CENP-C Proteins of Plants in cenH3 Loading and Kinetochore Assembly.
Centromeric regions (black) of Arabidopsis chromosomes mainly consist of the tandem repeat pAL1 (178 bp) sequence packaged in cenH3 nucleosomes, which can be bound by KNL2 for new cenH3 deposition during G2 or by CENP-C for formation of the kinetochore. Centromeric DNA can be transcribed and processed into small RNAs. Centromeric transcripts compete with DNA for binding by KNL2 and can help KNL2 leave centromeric positions at early stages of mitosis, whereas ssRNA stabilizes CENP-C binding to centromeric DNA, helping to establish the mitotic kinetochore.
The sequence-independent DNA binding capability of KNL2 and CENP-C and their ability to recognize centromeric nucleosomes specifically via their CENPC-like motifs may shed some light on the establishment of new centromeres. CenH3 is commonly located at ectopic noncentromere sites in small amounts (Shang et al., 2013; Thakur et al., 2015), and if the normal centromere is disrupted, KNL2 and CENP-C may bind these cenH3 sites and the adjoining DNA to establish more cenH3 and a kinetochore by the priming activity of KNL2, CENP-C, or other proteins. Sequence-independent DNA binding capability gives KNL2, CENP-C, and cenH3 nucleosomes themselves the flexibility to support the establishment of rare de novo centromeres and to adapt to rapidly evolving centromeric satellites.
METHODS
Identification of KNL2 Homologs and CENPC-Like Motifs
KNL2 proteins were identified using the human, Arabidopsis thaliana, or other KNL2 proteins as query to search the NCBI protein database using BLASTP. Some searches were limited to specific taxa to assure broad phylogenetic representation. Putative KNL2 proteins were identified by homology to the SANTA domain. In searches where homology to the CENPC-k motif was not detected by BLASTP (for example using human KNL2 as query), the motif could be identified by visual inspection, except in therian mammals and Caenorhabditis species. Similar search strategies identified CENP-C proteins by homology to the CENPC motif (Talbert et al., 2004), with homology to the C-terminal cupin domain used as confirmation, except in plants and cryptomonads, which lack the cupin domain. CENPC-v motifs were identified by inspection or taken from Kato et al. (2013) or Kral (2016). Alignments were made using MUSCLE (http://www.ebi.ac.uk/Tools/msa/muscle/) and were trimmed and adjusted by hand. Logos were generated using the WebLogo website (http://weblogo.berkeley.edu/logo.cgi).
Plant Transformation and Plant Growth Conditions
Arabidopsis plants (accession Columbia-0) were transformed by the floral-dip method (Bechtold et al., 1993). Transgenic progenies were selected on Murashige and Skoog solid medium containing the relevant antibiotic. The presence of a transgene was confirmed by PCR. For each transformation at least 30 independent transgenic lines were generated and at least five independent lines were used for the in vivo localization studies.
Plants were germinated on Petri dishes under long-day conditions (16 h light [100 µmol m−2 s−1]/8 h dark, 20°C/18°C), grown for 4 weeks under short-day conditions (8 h light [100 µmol m−2 s−1]/16 h dark, 20°C/18°C), and then returned to long-day conditions.
Agrobacterium tumefaciens-mediated transient transformation of Nicotiana benthamiana plants was performed according to Walter et al. (2004). Epidermal cell layers of tobacco leaves were assayed for fluorescence 1 to 2 d after infiltration, at least 50 cells per plant were analyzed. For each construct transient transformation of N. benthamiana was repeated at least three times.
Site-Directed Mutagenesis and Plasmid Construction
The substitution of the two most conserved amino acids within the CENPC-k motif of KNL2, deletion of the CENPC-k motif and its substitution by the CENPC were performed by PCR using the Phusion site-directed mutagenesis kit (Thermo Fisher Scientific). Substitution of amino acids was performed by changing one nucleotide from each of the triplets encoding the conserved amino acids. A fusion construct KNL2(C)/pDONR221 (Lermontova et al., 2013) was PCR mutagenized using with the following primer pairs: KNL2_R_A_f and KNL2_R_A_r for the substitution of Arg by Ala, and KNL2_W_R_f and KNL2_W_R_r for the substitution of Trp by Arg (Supplemental Table 1). The CENPC-k motif of the KNL2(C)/pDONR221construct was deleted using PCR with KNL2_∆CENPC_f and KNL2_∆CENPC_r primers (Supplemental Table 1). The CENPC motif from Arabidopsis CENP-C was inserted into KNL2(C)∆CENPC-k/pDONR221 using two subsequent PCR reactions with CENP-Csubst_fr1 and CENP-Csubst_rev1; CENP-Csubst_fr2 and CENP-Csubst_rev2 primers, respectively (Supplemental Table 1). The mCherry-cenH3 fusion construct was generated by replacing the EYFP in the p35S:EYFP-cenH3 fusion construct (Lermontova et al., 2006) with mCherry. The resulting expression cassette, including 35S promoter, mCherry-cenH3, and Nos terminator, was subcloned into the pLH7000 vector containing the phosphinotricine resistance marker (www.dna-cloning-service.de) via the SfiI restriction site. All plasmid clones were prepared for sequence analysis using the Qiagen QIAprep kit according to the supplier’s instructions (Qiagen).
Preparation of DNA Samples and Competitors
Genomic DNA of Arabidopsis was extracted using Qiagen DNeasy plant mini kit. DNA fragments (centromeric repeat pAL1, transposable element Athila, subtelomeric repeat, and euchromatic TUA4 [tubulin gene] region) were amplified by PCR using primers listed in Supplemental Table 1. The PCR procedure was optimized for each target DNA sequence (Supplemental Table 2). DNA samples were purified by standard chloroform-phenol purification. For the radioactive EMSA, DNA samples were labeled with 5′-γ-ATP (32P) using T4 polynucleotide kinase according to the manufacturer’s protocol (Thermo Fisher Scientific).
Preparation of RNA Samples
The HPLC-purified 23-nucleotide ssRNA was ordered from Metabion. To produce full-length ssRNA of pAL1, pIDTBlue vectors (Integrated DNA Technologies) containing the forward or reverse sequences of pAL1 (Supplemental Table 1) was ordered. The plasmid was digested with PvuI according to the manufacturer’s protocol (Thermo Fisher Scientific/Fermentas).
The in vitro transcription was performed according to the “Molecular Cloning” protocol with minor changes (Green and Sambrook, 2012). The 5′ or 3′ overhangs were blunted using T4 DNA polymerase according to the manufacturer’s protocol (Thermo Fisher Scientific). For the in vitro transcription reaction, 15 units of T3 RNA polymerase (Thermo Fisher Scientific), 10 mM ribonucleoside triphosphate mix, and RiboLock (Thermo) as RNase inhibitor were used. The pIDTblue vector with reverse pAL1 served as template for the forward pAL1 ssRNA reaction, and for the reverse pAL1 ssRNA, the pIDTblue vector with forward pAL1 was used as template. The ssRNA was finally purified by ethanol precipitation.
Expression and Purification of Recombinant Protein
To produce recombinant N- and C-terminal fragments of the KNL2 protein in Escherichia coli for the EMSA experiments, the codon-usage optimized KNL2 cDNA clone in the pUC57 vector was generated by GenScript and used as a template for amplification of the cDNA fragments coding for the N- and C-terminal parts of KNL2 using primers listed in Supplemental Table 1. Both fragments were cloned into the Gateway-compatible pETG30A and pETG60A (EMBL) expression vectors and transformed into C43 (Lucigen) competent cells. Recombinant KNL2 fragments were expressed as histidine-fused proteins and purified on Ni2+-NTA agarose under nondenaturing conditions (Qiagen) according to the manufacturer’s protocol.
Nonradioactive EMSA
DNA and RNA samples were incubated with the N- or C-terminal parts of KNL2 at room temperature for 30 min in 20 μL EMSA buffer 1 (10 mM Tris, pH 7.5, 1 mM EDTA, 100 mM KCl, 0.1 mM DTT, 5% glycerol, and 0.01 mg/mL BSA). For competition experiments, 4 pmol of competitor DNA was added. DNA samples were subsequently loaded on an 8% native PAA gel and RNA on a 20% native PAA gel. Afterwards the DNA was stained by SYBR Safe (Thermo Fisher Scientific) and RNA by SYBR Gold (Thermo Fisher Scientific) for 30 min. If not stated otherwise, all EMSA experiments were repeated three times independently.
Radioactive EMSA
For the radioactive EMSA, radioactively labeled pAL1 DNA probe, KNL2 protein fragments, and, for certain experiments, RNA samples were incubated at room temperature for 30 min in a 10 μL of EMSA buffer 2 (10% glycerol, 25 mM HEPES, pH 7.6, 10 mM MgCl2, 0.2 mM CaCl2, 1 mM DTT, and 0.4 mM PMSF). After incubation, 2 μL of EMSA loading buffer (50% glycerol in EMSA buffer 2 with bromophenol blue) was added to each samples; subsequently, the samples were separated on a 5% native PAA gel and detected using the FLA 5100 phosphor imager (Fujifilm). If not stated otherwise, all EMSA experiments were repeated three times independently.
Immunostaining and Microscopy Analysis of Fluorescent Signals
Immunostaining of nuclei/chromosomes was performed as described previously (Lermontova et al., 2013). Arabidopsis seeds of lines harboring mutagenized KNL2(C)/pGWB641 variants or nonmutagenized control were germinated in agar medium in cover slip chambers (Nalge Nunc). Roots growing parallel to the cover slip bottom were analyzed in a LSM 510META confocal microscope (Carl Zeiss) using a 639 oil immersion objective (NA 1.4). EYFP was excited with a 488-nm laser line and fluorescence recorded with a 505- to 550-nm band-pass filter. Images were analyzed with the LSM software release 3.2.
ChIP
The ChIP was performed according Gendrel et al. (2005) with some modifications.
Step 1: Five grams of 8-d-old seedlings was used (biological replicate). Step 16: The sonication of the chromatin was performed using a Bioruptor disruptor (Diagenode) for six cycles with 30 s of high energy sonication and 30-s breaks at 4°C. Step 17: Twenty microliters of the sonicated plant chromatin was used as input control. Steps 19 to 22: Per one technical replicate or “nonantibody control,” 10 μL of Dynabead Protein A (Thermo Fisher Scientific) was washed two times with 1× PBS/0.1% Triton X-100 and one time with ChIP dilution buffer. Beads were applied in a magnetic rack (Qiagen) to separate beads from the supernatant. For the technical replicates, 50 μL ChIP dilution buffer and 50 μL of affinity chromatography-purified anti-KNL2 antibodies were added to the beads. Antibodies were purified using CN-Br-Sepharose coupled with the KNL2(C) antigen (Sigma-Aldrich protocol C9142 with minor changes). For the “nonantibody control,” 100 μL ChIP dilution buffer was added. Bead-antibody mixtures and controls were incubated at 4°C overnight. The diluted chromatin (biological replicate) was distributed equally to three technical replicates plus one “nonantibody control.” Step 27: Input samples were diluted up to 500 μL with elution buffer.
Quantitative Real-Time PCR
For quantitative real-time PCR, an iCycler iQ5 (Bio-Rad) and iQ SYBR Green Supermix (Bio-Rad) were used. Each ChIP sample (technical replicates, nonantibody controls, and input samples) was quantified three times and at least three independent biological replicates were used. Primers and procedure used for quantitative amplification of genomic regions after ChIP are defined in Supplemental Tables 1 and 2.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At KNL2 (At5g02520), At cenH3 (At1g01370), At CENP-C (At1g15660), pAL1 repeat (gi 164520998 and EU359556.1), Athila repeat (gi X81801), and tubulin (AT1G04820).
Supplemental Data
Supplemental Figure 1. Large-scale view and higher magnification of localization of KNL2(C), KNL2(C)CENPC-k(R-A), and KNL2(C)CENPC fused to EYFP in root tips of stable transformants of Arabidopsis.
Supplemental Figure 2. The CENPC motif of CENP-C protein restores the ability of KNL2(C)∆CENPC to localize at centromeres.
Supplemental Figure 3. The C-terminal part of KNL2 binds the centromeric pAL1 sequences also in radioactive EMSA.
Supplemental Figure 4. KNL2 DNA binding is not influenced by centromeric 23-nucleotide ssRNA.
Supplemental Figure 5. In silico analysis of KNL2 protein by BindN program (Wang and Brown, 2006) for DNA binding amino acid residues.
Supplemental Table 1. Primers used in this study.
Supplemental Table 2. PCR programs used for the amplification of DNA probes for EMSA.
Supplementary Material
Acknowledgments
We thank Christa Fricke, Joachim Bruder, Katrin Kumke, and Ulrike Gresch for technical assistance, Ingo Schubert and anonymous reviewers for critical reading of the article and helpful suggestions and discussions, Veit Schubert for help with super-resolution microscopy, and Karin Lipfert for help with preparation of figures. This work was supported by a grant from the Deutsche Forschungsgemeinschaft to I.L. (LE2299/1-2). D.D. is a postdoctoral fellow of the German Research Foundation (SFB) 648.
AUTHOR CONTRIBUTIONS
I.L., M.S., and P.T. designed the research. I.L., M.S., D.D., M.K., T.R., and U.C. performed research. I.L., M.S., P.T., D.D., M.K., and U.C. analyzed data. I.L., M.S., and P.T. wrote the article.
Glossary
- EMSA
electrophoretic mobility shift assay
- poly dI/dC
poly(deoxyinosinic-deoxycytidylic) acid
- ChIP
chromatin immunoprecipitation
- ssRNA
single-stranded RNA
Footnotes
Articles can be viewed without a subscription.
References
- Ay N., Irmler K., Fischer A., Uhlemann R., Reuter G., Humbeck K. (2009). Epigenetic programming via histone methylation at WRKY53 controls leaf senescence in Arabidopsis thaliana. Plant J. 58: 333–346. [DOI] [PubMed] [Google Scholar]
- Barnhart M.C., Kuich P.H.J.L., Stellfox M.E., Ward J.A., Bassett E.A., Black B.E., Foltz D.R. (2011). HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. J. Cell Biol. 194: 229–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold N., Ellis J., Pelletier G. (1993). In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana Plants. C. R. Acad. Sci. Life Sci. 316: 1194–1199. [Google Scholar]
- Blower M.D., Sullivan B.A., Karpen G.H. (2002). Conserved organization of centromeric chromatin in flies and humans. Dev. Cell 2: 319–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer L.A., Langer M.R., Crowley K.A., Tan S., Denu J.M., Peterson C.L. (2002). Essential role for the SANT domain in the functioning of multiple chromatin remodeling enzymes. Mol. Cell 10: 935–942. [DOI] [PubMed] [Google Scholar]
- Brown M.T. (1995). Sequence similarities between the yeast chromosome segregation protein Mif2 and the mammalian centromere protein CENP-C. Gene 160: 111–116. [DOI] [PubMed] [Google Scholar]
- Carroll C.W., Milks K.J., Straight A.F. (2010). Dual recognition of CENP-A nucleosomes is required for centromere assembly. J. Cell Biol. 189: 1143–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan F.L., Marshall O.J., Saffery R., Kim B.W., Earle E., Choo K.H., Wong L.H. (2012). Active transcription and essential role of RNA polymerase II at the centromere during mitosis. Proc. Natl. Acad. Sci. USA 109: 1979–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dambacher S., Deng W., Hahn M., Sadic D., Fröhlich J., Nuber A., Hoischen C., Diekmann S., Leonhardt H., Schotta G. (2012). CENP-C facilitates the recruitment of M18BP1 to centromeric chromatin. Nucleus 3: 101–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe R.K., Reed L.M., Yu H.G., Muszynski M.G., Hiatt E.N. (1999). A maize homolog of mammalian CENPC is a constitutive component of the inner kinetochore. Plant Cell 11: 1227–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rop V., Padeganeh A., Maddox P.S. (2012). CENP-A: the key player behind centromere identity, propagation, and kinetochore assembly. Chromosoma 121: 527–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drinnenberg I.A., deYoung D., Henikoff S., Malik H.S. (2014). Recurrent loss of CenH3 is associated with independent transitions to holocentricity in insects. eLife 3: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y., Topp C.N., Dawe R.K. (2010). DNA binding of centromere protein C (CENPC) is stabilized by single-stranded RNA. PLoS Genet. 6: e1000835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw W.C., Rothfield N. (1985). Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma 91: 313–321. [DOI] [PubMed] [Google Scholar]
- Foltz D.R., Jansen L.E., Bailey A.O., Yates J.R. III, Bassett E.A., Wood S., Black B.E., Cleveland D.W. (2009). Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell 137: 472–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y., Hayashi T., Kiyomitsu T., Toyoda Y., Kokubu A., Obuse C., Yanagida M. (2007). Priming of centromere for CENP-A recruitment by human hMis18alpha, hMis18beta, and M18BP1. Dev. Cell 12: 17–30. [DOI] [PubMed] [Google Scholar]
- Gendrel A.V., Lippman Z., Martienssen R., Colot V. (2005). Profiling histone modification patterns in plants using genomic tiling microarrays. Nat. Methods 2: 213–218. [DOI] [PubMed] [Google Scholar]
- Gopalakrishnan S., Sullivan B.A., Trazzi S., Della Valle G., Robertson K.D. (2009). DNMT3B interacts with constitutive centromere protein CENP-C to modulate DNA methylation and the histone code at centromeric regions. Hum. Mol. Genet. 18: 3178–3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M.R., Sambrook J. (2012). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; ). [Google Scholar]
- Hayashi T., Fujita Y., Iwasaki O., Adachi Y., Takahashi K., Yanagida M. (2004). Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell 118: 715–729. [DOI] [PubMed] [Google Scholar]
- Heeger S., Leismann O., Schittenhelm R., Schraidt O., Heidmann S., Lehner C.F. (2005). Genetic interactions of separase regulatory subunits reveal the diverged Drosophila Cenp-C homolog. Genes Dev. 19: 2041–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmerich P., Weidtkamp-Peters S., Hoischen C., Schmiedeberg L., Erliandri I., Diekmann S. (2008). Dynamics of inner kinetochore assembly and maintenance in living cells. J. Cell Biol. 180: 1101–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Dalal Y. (2005). Centromeric chromatin: what makes it unique? Curr. Opin. Genet. Dev. 15: 177–184. [DOI] [PubMed] [Google Scholar]
- Kato H., Jiang J., Zhou B.R., Rozendaal M., Feng H., Ghirlando R., Xiao T.S., Straight A.F., Bai Y. (2013). A conserved mechanism for centromeric nucleosome recognition by centromere protein CENP-C. Science 340: 1110–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kral L. (2016). Possible identification of CENP-C in fish and the presence of the CENP-C motif in M18BP1 of vertebrates. F1000 Res. 4: 474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanini L., McKeon F. (1995). Domains required for CENP-C assembly at the kinetochore. Mol. Biol. Cell 6: 1049–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lermontova I., Kuhlmann M., Friedel S., Rutten T., Heckmann S., Sandmann M., Demidov D., Schubert V., Schubert I. (2013). Arabidopsis kinetochore null2 is an upstream component for centromeric histone H3 variant cenH3 deposition at centromeres. Plant Cell 25: 3389–3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lermontova I., Schubert V., Fuchs J., Klatte S., Macas J., Schubert I. (2006). Loading of Arabidopsis centromeric histone CENH3 occurs mainly during G2 and requires the presence of the histone fold domain. Plant Cell 18: 2443–2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo A.W., Magliano D.J., Sibson M.C., Kalitsis P., Craig J.M., Choo K.H. (2001). A novel chromatin immunoprecipitation and array (CIA) analysis identifies a 460-kb CENP-A-binding neocentromere DNA. Genome Res. 11: 448–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox P.S., Hyndman F., Monen J., Oegema K., Desai A. (2007). Functional genomics identifies a Myb domain-containing protein family required for assembly of CENP-A chromatin. J. Cell Biol. 176: 757–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh P.B., Yang P., Glowczewski L., Koshland D., Smith M.M. (1998). Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell 94: 607–613. [DOI] [PubMed] [Google Scholar]
- Mendiburo M.J., Padeken J., Fülöp S., Schepers A., Heun P. (2011). Drosophila CENH3 is sufficient for centromere formation. Science 334: 686–690. [DOI] [PubMed] [Google Scholar]
- Milks K.J., Moree B., Straight A.F. (2009). Dissection of CENP-C-directed centromere and kinetochore assembly. Mol. Biol. Cell 20: 4246–4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moree B., Meyer C.B., Fuller C.J., Straight A.F. (2011). CENP-C recruits M18BP1 to centromeres to promote CENP-A chromatin assembly. J. Cell Biol. 194: 855–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaki K., Talbert P.B., Zhong C.X., Dawe R.K., Henikoff S., Jiang J. (2003). Chromatin immunoprecipitation reveals that the 180-bp satellite repeat is the key functional DNA element of Arabidopsis thaliana centromeres. Genetics 163: 1221–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasuda S., Hudakova S., Schubert I., Houben A., Endo T.R. (2005). Stable barley chromosomes without centromeric repeats. Proc. Natl. Acad. Sci. USA 102: 9842–9847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakabe A., Tachiwana H., Matsunaga T., Shiga T., Nozawa R.S., Obuse C., Kurumizaka H. (2010). Nucleosome formation activity of human somatic nuclear autoantigenic sperm protein (sNASP). J. Biol. Chem. 285: 11913–11921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidoux A.L., Choi E.S., Abbott J.K., Liu X., Kagansky A., Castillo A.G., Hamilton G.L., Richardson W., Rappsilber J., He X., Allshire R.C. (2009). Fission yeast Scm3: A CENP-A receptor required for integrity of subkinetochore chromatin. Mol. Cell 33: 299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politi V., Perini G., Trazzi S., Pliss A., Raska I., Earnshaw W.C., Della Valle G. (2002). CENP-C binds the alpha-satellite DNA in vivo at specific centromere domains. J. Cell Sci. 115: 2317–2327. [DOI] [PubMed] [Google Scholar]
- Shang W.H., et al. (2013). Chromosome engineering allows the efficient isolation of vertebrate neocentromeres. Dev. Cell 24: 635–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shono N., Ohzeki J., Otake K., Martins N.M., Nagase T., Kimura H., Larionov V., Earnshaw W.C., Masumoto H. (2015). CENP-C and CENP-I are key connecting factors for kinetochore and CENP-A assembly. J. Cell Sci. 128: 4572–4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A.J.P., Humphries S.E. (2009). Characterization of DNA-binding proteins using multiplexed competitor EMSA. J. Mol. Biol. 385: 714–717. [DOI] [PubMed] [Google Scholar]
- Song K., Gronemeyer B., Lu W., Eugster E., Tomkiel J.E. (2002). Mutational analysis of the central centromere targeting domain of human centromere protein C (CENP-C). Exp. Cell Res. 275: 81–91. [DOI] [PubMed] [Google Scholar]
- Stellfox M.E., Nardi I.K., Knippler C.M., Foltz D.R. (2016). Differential binding partners of the Mis18α/β YIPPEE domains regulate Mis18 complex recruitment to centromeres. Cell Reports 15: 2127–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K., Fukuda R., Himeno M. (2000). Centromere/kinetochore localization of human centromere protein A (CENP-A) exogenously expressed as a fusion to green fluorescent protein. Cell Struct. Funct. 25: 253–261. [DOI] [PubMed] [Google Scholar]
- Sugimoto K., Yata H., Muro Y., Himeno M. (1994). Human centromere protein C (CENP-C) is a DNA-binding protein which possesses a novel DNA-binding motif. J. Biochem. 116: 877–881. [DOI] [PubMed] [Google Scholar]
- Talbert P.B., Bryson T.D., Henikoff S. (2004). Adaptive evolution of centromere proteins in plants and animals. J. Biol. 3: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira F.K., Colot V. (2010). Repeat elements and the Arabidopsis DNA methylation landscape. Heredity (Edinb.) 105: 14–23. [DOI] [PubMed] [Google Scholar]
- Teo C.H., Lermontova I., Houben A., Mette M.F., Schubert I. (2013). De novo generation of plant centromeres at tandem repeats. Chromosoma 122: 233–241. [DOI] [PubMed] [Google Scholar]
- Thakur J., Talbert P.B., Henikoff S. (2015). Inner kinetochore protein interactions with regional centromeres of fission yeast. Genetics 201: 543–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth J., Biggin M.D. (2000). The specificity of protein-DNA crosslinking by formaldehyde: in vitro and in drosophila embryos. Nucleic Acids Res. 28: e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trazzi S., Bernardoni R., Diolaiti D., Politi V., Earnshaw W.C., Perini G., Della Valle G. (2002). In vivo functional dissection of human inner kinetochore protein CENP-C. J. Struct. Biol. 140: 39–48. [DOI] [PubMed] [Google Scholar]
- Walter M., Chaban C., Schütze K., Batistic O., Weckermann K., Näke C., Blazevic D., Grefen C., Schumacher K., Oecking C., Harter K., Kudla J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40: 428–438. [DOI] [PubMed] [Google Scholar]
- Westermann S., Cheeseman I.M., Anderson S., Yates J.R. III, Drubin D.G., Barnes G. (2003). Architecture of the budding yeast kinetochore reveals a conserved molecular core. J. Cell Biol. 163: 215–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong L.H., Brettingham-Moore K.H., Chan L., Quach J.M., Anderson M.A., Northrop E.L., Hannan R., Saffery R., Shaw M.L., Williams E., Choo K.H. (2007). Centromere RNA is a key component for the assembly of nucleoproteins at the nucleolus and centromere. Genome Res. 17: 1146–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C.H., Tomkiel J., Saitoh H., Johnson D.H., Earnshaw W.C. (1996). Identification of overlapping DNA-binding and centromere-targeting domains in the human kinetochore protein CENP-C. Mol. Cell. Biol. 16: 3576–3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Martyniuk C.J., Trudeau V.L. (2006). SANTA domain: a novel conserved protein module in Eukaryota with potential involvement in chromatin regulation. Bioinformatics 22: 2459–2462. [DOI] [PubMed] [Google Scholar]
- Zhang W., Lee H.R., Koo D.H., Jiang J. (2008). Epigenetic modification of centromeric chromatin: hypomethylation of DNA sequences in the CENH3-associated chromatin in Arabidopsis thaliana and maize. Plant Cell 20: 25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.