The RSL4 (bHLH)-RHE (root hair-specific cis-element) regulatory module is conserved in tracheophytes for sustainable root hair growth.
Abstract
ROOT HAIR SPECIFIC (RHS) genes, which contain the root hair-specific cis-element (RHE) in their regulatory regions, function in root hair morphogenesis. Here, we demonstrate that an Arabidopsis thaliana basic helix-loop-helix transcription factor, ROOT HAIR DEFECTVE SIX-LIKE4 (RSL4), directly binds to the RHE in vitro and in vivo, upregulates RHS genes, and stimulates root hair formation in Arabidopsis. Orthologs of RSL4 from a eudicot (poplar [Populus trichocarpa]), a monocot (rice [Oryza sativa]), and a lycophyte (Selaginella moellendorffii) each restored root hair growth in the Arabidopsis rsl4 mutant. In addition, the rice and S. moellendorffii RSL4 orthologs bound to the RHE in in vitro and in vivo assays. The RSL4 orthologous genes contain RHEs in their promoter regions, and RSL4 was able to bind to its own RHEs in vivo and amplify its own expression. This process likely provides a positive feedback loop for sustainable root hair growth. When RSL4 and its orthologs were expressed in cells in non-root-hair positions, they induced ectopic root hair growth, indicating that these genes are sufficient to specify root hair formation. Our results suggest that RSL4 mediates root hair formation by regulating RHS genes and that this mechanism is conserved throughout the tracheophyte (vascular plant) lineage.
INTRODUCTION
Unicellular zygotes develop into multicellular organisms through the process of cell division and differentiation. The root hairs of vascular plants offer a useful model in which to study such cell differentiation. Each root hair is a tubular protrusion of a root epidermal cell that aids in the absorption of water and nutrients, anchorage to the soil, and interaction with soil microbes (Cutter, 1978). Root hairs show a linear developmental gradient along the longitudinal axis of the root from the meristem to the differentiation zone, beginning with determination of cell fate and followed by initiation, bulge formation, and, finally, tip growth (Grierson and Schiefelbein, 2008).
Three types of cell fate determination mechanisms in root epidermal cells determine whether a cell will become a root hair cell or a non-root-hair cell: random (type 1), asymmetrical cell division (type 2), and position dependent (type 3) (Dolan, 1996; Clowes, 2000; Schiefelbein, 2000). Among these, type 3 has been well characterized in Arabidopsis thaliana. In a non-root-hair cell position, a complex of WEREWOLF (WER; a MYB transcription factor [TF]), GLABRA3/ENHANCER OF GLABRA3 (GL3/EGL3; basic helix-loop-helix [bHLH] TFs), and TRANSPARENT TEST GLABRA (TTG; a WD40 protein) influences the expression of GL2 (a homeodomain TF) (Grierson and Schiefelbein 2008). GL2 directly suppresses the expression of ROOT HAIR DEFECTIVE6 (RHD6; a bHLH TF), RHD6-LIKE1 (RSL1), and RSL2, resulting in inhibition of root hair morphogenesis (Lin et al., 2015). At the root hair cell position, SCRAMBLED (a leucine-rich repeat receptor-like protein kinase) is likely to receive external signals from inner tissues (Kwak et al., 2005) and to suppress the expression of WER to inhibit formation of the complex with GL3/EGL3 and TTG, thus allowing the expression of RHD6 and subsequent root hair morphogenesis (Kwak and Schiefelbein, 2007; Grierson and Schiefelbein, 2008).
Root hair-specific morphogenetic genes are required for root hair morphogenesis and play roles in cell wall organization, vesicle secretion, signal transduction, and many other morphogenetic cellular processes (Grierson and Schiefelbein, 2008). In previous studies, we identified ROOT HAIR SPECIFIC (RHS) genes that are specifically expressed in root hair cells and are involved in root hair morphogenesis (Kim et al., 2006; Won et al., 2009). The identified RHS genes all contain the 16- or 17-bp root hair-specific cis-element (RHE) in their proximal promoter regions (Kim et al., 2006; Won et al., 2009). RHEs are functionally conserved in angiosperms to regulate root hair-specific gene expression despite differences in the upstream fate-determination type (Kim et al., 2006). This finding suggests that the RHE binding or RHS-regulating transcription factors are also functionally conserved in angiosperms.
In a previous study, RHD6 was shown to directly regulate RSL4 (a homolog of RHD6), but indirectly regulate EXPANSIN A7 (EXPA7; an RHS) (Yi et al., 2010). However, transcriptome analysis of wild-type, rsl4 mutant, and RSL4 overexpression Arabidopsis seedlings demonstrated that RSL4 upregulates the expression of many root hair-related genes, including RHS genes (Yi et al., 2010).
In this study, we conducted in vitro and in vivo binding assays to test the binding ability of RSL4 to the RHEs of RHS genes and examined the functional conservation of RSL4 in the tracheophyte (vascular plant) lineage by analyzing the function of orthologs of RSL4 from two eudicots, a monocot, and a lycophyte. Our results suggest that the RSL4-RHE regulatory module for root hair morphogenesis is conserved in tracheophytes.
RESULTS
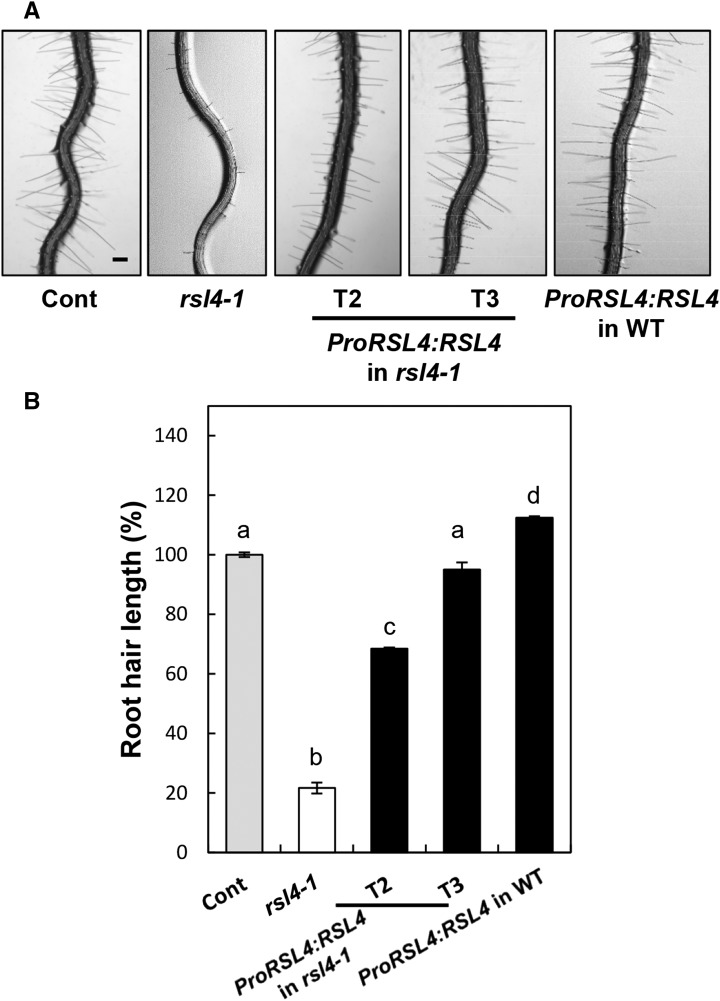
As previously reported (Yi et al., 2010), the loss-of-function rsl4-1 mutant seedling grew root hairs that were ∼20% the length of the control (Figure 1). Complementation of the rsl4-1 mutant with the RSL4:GFP transgene under the control of the RSL4 promoter (ProRSL4) restored the root hair growth to 68% of the length in the control in the T2 generation (heterozygotes plus homozygotes) and to 95% of the control in the T3 generation (homozygotes) (Figure 1; Supplemental Data Set 1). Overexpression of RSL4 with the ProRSL4:RSL4:GFP construct in the wild-type background increased root hair length to 112% of the control (Figure 1). Overexpression of RSL4 under the strong root hair-specific EXPA7 promoter (ProE7; Cho and Cosgrove, 2002; Kim et al., 2006) further increased root hair growth up to 131% of control length (Supplemental Figure 1 and Supplemental Data Set 1). Consistent with previous results, these data showed that RSL4 is a positive effector for root hair growth.
Figure 1.
RSL4 Is a Positive Regulator for Root Hair Growth.
(A) Root hair phenotypes of the control (Cont; ProE7:YFP), loss-of-function rsl4-1 mutant, RSL4-complemented T2 and T3 lines (ProRSL4:RSL4:GFP in the rsl4-1 background; T2, hetero- plus homozygotes for the transgene; T3, homozygote for the transgene), and T2 lines of ProRSL4:RSL4:GFP in the wild type (WT) background. Bars = 100 μm.
(B) Root hair lengths of the control, rsl4-1 mutant, complemented lines (ProRSL4:RSL4 in the rsl4-1 mutant background), and overexpression lines (ProRSL4:RSL4 in the wild-type background) as described in (A). Error bars indicate ±se from 162 to 4365 root hairs. Statistically significant differences are denoted with different letters (one-way ANOVA with Tukey’s unequal N HSD post hoc test, P < 0.05).
RSL4 Binds to the RHE of RHS Genes
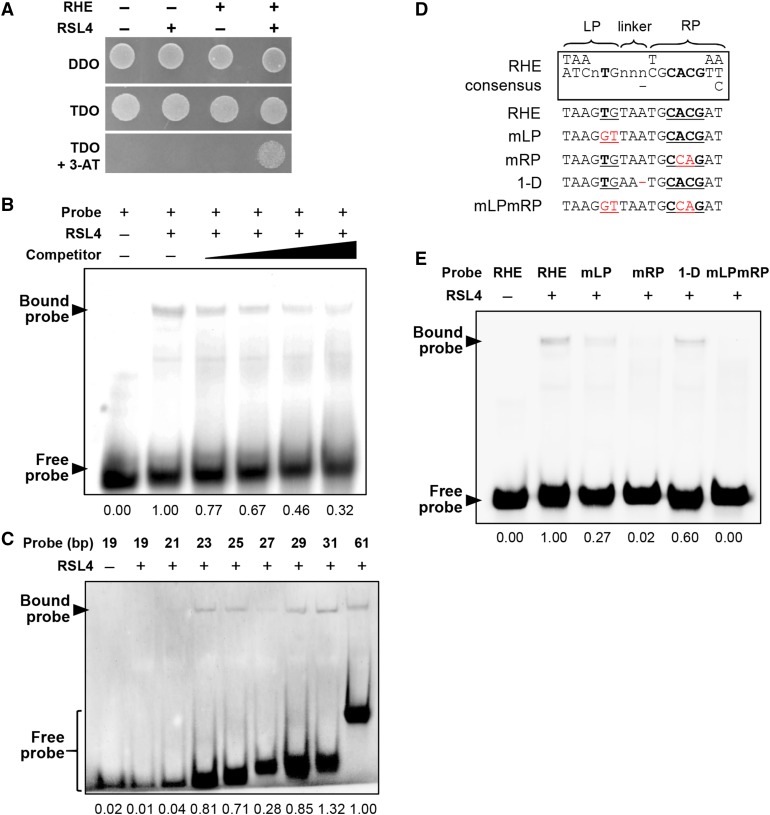
We performed several in vitro and in vivo binding assays to determine whether RSL4 binds to the RHE. In a yeast one-hybrid assay, using a tandem sequence of three copies of the RHE from EXPA7 (Kim et al., 2006; Supplemental Table 1) as bait and RSL4 as prey, yeast cells were able to grow in the presence of 3-amino-1,2,4-triazole in the triple dropout (TDO; -His/-Leu/-Trp) medium only with both the prey and the RHE bait (Figure 2A), suggesting that RSL4 binds to the RHE in yeast cells. In an electrophoresis mobility shift assay (EMSA) using the 61-bp promoter region, including a single RHE (Kim et al., 2006) from the Arabidopsis PROLINE RICH PROTEIN3 (PRP3) gene as the probe, the addition of heterologously expressed RSL4 proteins caused a typical mobility shift of the probe band, and the addition of the unlabeled competitor weakened the intensity of the shifted probe band (Figure 2B), indicating that RSL4 binds to the RHE in vitro. The core RHE motif is 16 or 17 bp (Kim et al., 2006); accordingly, we tested 19- to 61-bp probe sequences containing a 17-bp RHE at the center of each sequence. Substantial band intensity was maintained with the probes of 23-bp and longer, although the 27-bp probe showed a reduced band intensity (Figure 2C). However, probes shorter than 23 bp revealed almost no shifted band, indicating that at least a 2-bp margin at both ends of the RHE core is necessary for RSL4 binding.
Figure 2.
RSL4 Binds to the RHE.
(A) Yeast one-hybrid assay. The bait (RHE) is a tandem sequence of three copies of RHE, and the prey is RSL4. The yeast medium lacks Leu/Trp (DDO) or Leu/Trp/His (TDO). 3-AT, 3-amino-1,2,4-triazole.
(B) EMSA showing RSL4 binding to the RHE. GST-tagged RSL4 and the biotinylated RHE probe were used. The competitor is nonbiotinylated RHE of 20-, 30-, 50-, and 100-fold increases over the biotinylated probe amount.
(C) EMSA showing the effect of the RHE length on RSL4 binding. In each length (bp) of probe, the RHE core (17 bp) is located at the center of the probe.
(D) Sequences of mutated RHEs used for the EMSA in (E). Mutated nucleotides are marked in red. The line in the RHE consensus box indicates that the nucleotide in the position is missing. Bold letters depict highly conserved nucleotides. RHE consensus sequences are shown inside the box. RHE, wild-type RHE; mLP, RHE with mutated LP; mRP, RHE with mutated RP; 1-D, a 1-nucleotide deletion in the linker; mLPmRP, RHE with both mLR and mRP. RHE was from the Arabidopsis PRP3 gene.
(E) EMSA showing the effect of the RHE mutations on RSL4 binding. The RHE mutations are as described in (D). Numbers below the EMSA results ([B], [C], and [E]) indicate the relative band intensity of bound probe.
The RHE core consists of a 6-bp left part (LP) with a strictly conserved T nucleotide, a 2- to 3-bp linker, and an 8-bp right part (RP) with a strictly conserved CACG sequence, which generates a 16- or 17-bp functional RHE depending on RHE species (Kim et al., 2006; Figure 2D). Any single substitution in the strictly conserved nucleotides almost completely nullifies RHE-mediated root hair-specific gene expression, and a single nucleotide deletion and insertion mutation of a 3- and 2-bp linker, respectively, considerably compromises the function of the RHE (Kim et al., 2006). Using EMSA, we tested whether the effect of these RHE mutations on root hair-specific expression was caused by inhibition of RSL4 binding to the mutated RHEs. The mutation of the LP (mLP) considerably decreased RSL4 binding, while the RP mutation (mRP) showed an even greater inhibitory effect on RSL4 binding (Figure 2E). Mutations of both the LP and RP (mLPmRP) almost completely blocked RSL4 binding. A single nucleotide deletion in the linker (1-D) slightly decreased RSL4 binding. This EMSA result revealed a relationship between the binding of RSL4 to the RHE and RHE-mediated root hair-specific gene expression.
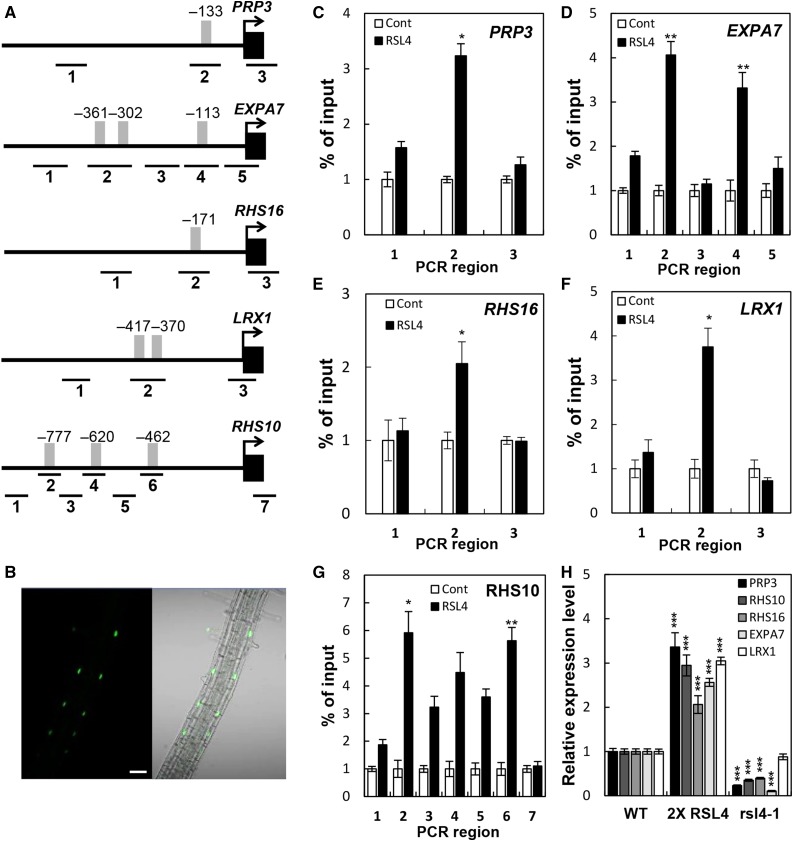
To investigate the binding ability of RSL4 to the RHE in vivo, we performed chromatin immunoprecipitation (ChIP) analyses with the promoter regions of five RHE-containing RHS genes (Kim et al., 2006; Won et al., 2009). These RHS genes contain one to three RHEs in their proximal promoter regions (Figure 3A). ChIP analyses were performed using RSL4:GFP-bound chromatin from the ProRSL4:RSL4:GFP transformant (Figure 3B), and ChIP-qPCR primers were designed to amplify the RHE or non-RHE regions as shown in Figure 3A. The ChIP data demonstrated that the percent of input of ChIP-qPCR was significantly higher in the RHE-containing regions than in the non-RHE regions (Figures 3C to 3G), suggesting that RSL4 preferentially binds to the RHE region in vivo.
Figure 3.
ChIP Analyses Showing RSL4 Binding to the RHE Region of RHS Gene Promoters.
(A) Locations of the RHE (gray bars) and ChIP-PCR regions (lines with #1–7) in the RHS gene promoters. Positions are relative to the start codon (+1).
(B) Confocal microscopy images showing RSL4:GFP (ProRSL4:RSL4:GFP in the wild-type background) signals in the nuclei of root hair cells. Bar = 50 μm.
(C) to (G) Enrichment fold of the ChIP-PCR fragment from the region shown in (A). The ChIP analysis was done with the wild type (Cont) and ProRSL4:RSL4:GFP (RSL4) transformant plants. Error bars indicate ±sd from three biological repeats. Values are relative to each Cont value and significantly different (*P < 0.05; **P < 0.005; Student’s t test) from the control value.
(H) RT-qPCR analysis of RHS transcripts from the wild type, ProRSL4:RSL4:GFP in the wild-type background (2× RSL4), and the rsl4-1 mutant. Error bars indicate ±sd from two biological repeats. The values are relative to the wild-type values and significantly different (***P < 0.0001; Student’s t test) from the wild-type values.
To examine whether the binding of RSL4 to the RHE induces RHS expression, RT-qPCR analysis was conducted on the five RHS genes used in the ChIP analysis in seedlings of the wild type, the wild-type background containing the ProRSL4:RSL4:GFP construct (which contains two copies of RSL4), and the rsl4-1 mutant. The expression of all five RHS genes was significantly increased by the introduction of an additional copy of RSL4 in the wild-type background containing the ProRSL4:RSL4:GFP construct and decreased in the rsl4-1 mutant compared with the levels in the wild type (Figure 3H), indicating that RSL4 binds to the RHE of RHS genes to activate their transcription.
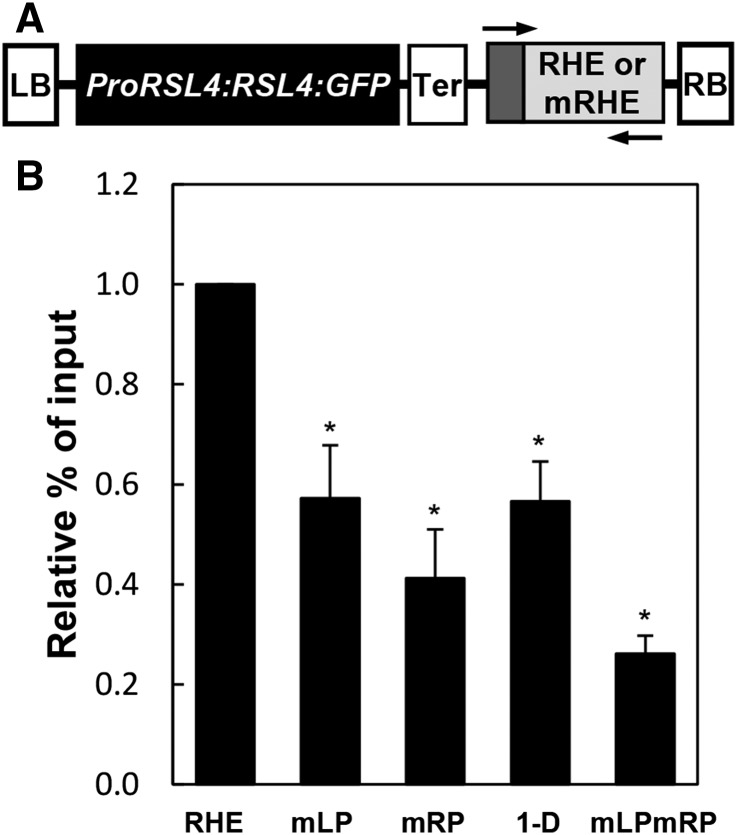
Next, ChIP analyses with mutated RHEs (mRHEs) were conducted to further investigate whether the RHE is a target of RSL4 in vivo. For these ChIP analyses, we designed five transgenic constructs that each contained ProRSL4:RSL4:GFP for RSL4:GFP immunoprecipitation, along with the wild-type RHE or one of four different mRHEs (mLP, mRP, 1-D, or mLPmRP, as described in Figure 2D) as an RSL4 binding target (Figure 4A). To distinguish these transgenic RHEs from the native RHE of PRP3 in the ChIP-qPCR analysis, a synthetic sequence for the annealing of the transgenic RHE-specific primer was added at the 5′ end of each transgenic RHE (Figure 4A). The ChIP analysis results showed that the binding capability of RSL4 was significantly decreased by the RHE mutations (Figure 4B). Furthermore, these results were consistent with the EMSA of mRHEs (Figure 2E), confirming that the RHE is the binding target of RSL4 both in vitro and in vivo.
Figure 4.
Effect of the RHE Mutations on RSL4 Binding in the ChIP Analysis.
(A) The transgenic construct of ProRSL4:RSL4:GFP with the RHE or mutated RHEs (mRHEs = mLP, mRP, 1-D, or mLPmRP for each independent construct as shown in Figure 2D) for the RSL4 binding ChIP analysis. The sequence of the RHE or mRHEs includes an artificial primer-annealing site (the dark gray border) for transgene-specific ChIP-PCR. LB, left border; Ter, transcription terminator; RB, right border of T-DNA. Arrows indicate the primers for ChIP-PCR.
(B) Relative percentage of input of the ChIP-PCR fragment from the RHE and mRHEs shown in (A). The ChIP analysis was conducted with ProRSL4:RSL4:GFP:RHE/mRHEs transformant plants. Error bars indicate ±se from three replicates. The values are relative to the RHE values and significantly different (*P < 0.005; Student’s t test) from the RHE values.
The Function of RSL4 Is Conserved in the Tracheophyte Lineage
Because the RHE motif is functionally conserved in angiosperms, the RHE binding transcription factor is likely to be conserved in angiosperms as well. To examine the conservation of RSL4 in the tracheophyte lineage, we characterized the RHEs from Selaginella moellendorffii (a lycophyte), identified RSL4 orthologs from S. moellendorffii and other angiosperms (a eudicot and a monocot), and examined the binding capacity of these RSL4 orthologs with the RHE in Arabidopsis and their function in root hair growth.
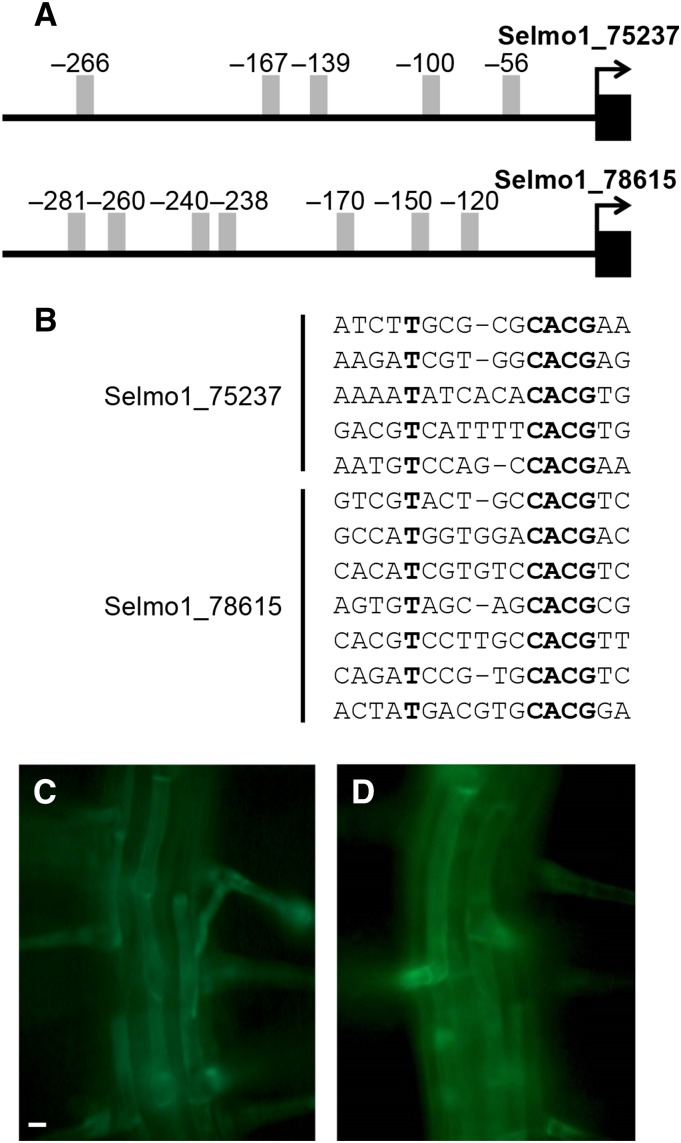
To identify the RHEs from S. moellendorffii, we first searched for the orthologs of Arabidopsis EXPA7 (RHE-containing root hair-specific EXPA gene) in S. moellendorffii (Cho and Cosgrove, 2002; Kim et al., 2006). Because EXPA7 is required for normal root hair growth in Arabidopsis (Lin et al., 2011) and its orthologs are well conserved throughout angiosperms (Kim et al., 2006), we presumed that root hair-specific orthologs of EXPA7 are involved in root hair growth in S. moellendorffii. A phylogenetic analysis showed that the S. moellendorffii genome includes two EXPAs that belong to the same clade as other EXPA7 orthologs from angiosperms (Supplemental Figure 2 and Supplemental Data Set 2). Their proximal promoter regions contained multiple RHEs: five RHEs in Selmo1_75237 and seven RHEs in Selmo1_78615 (Figures 5A and 2B). To determine whether the RHEs of the S. moellendorffii EXPA7 orthologs show root hair specificity, the RHE-containing promoter regions were fused with the minimal promoter region of Arabidopsis EXPA7 (mpE7, between −73 and +31 bp relative to the transcription initiation site) and the GFP reporter and introduced into Arabidopsis plants. The mpE7 region lacks the RHE but contains basic elements for transcription initiation, and the mpE7:GFP transformant itself is not able to express GFP (Kim et al., 2006). The RHE-containing promoter regions from both S. moellendorffii EXPAs directed root hair-enriched GFP expression in the Arabidopsis seedling root (Figures 5C and 5D). This result implied that S. moellendorffii contains the RHE binding RSL4-like transcription factor.
Figure 5.
S. moellendorffii RHEs Show Root Hair-Enriched Gene Expression in Arabidopsis.
(A) Promoter regions of two S. moellendorffii EXPA7-orthologous genes (Selmo1_75237 and Selmo1_78615) carrying multiple RHEs (gray bars). Numbers indicate relative positions to the start codon (+1).
(B) The RHE sequences of Selmo1_75237 and Selmo1_78615. Strictly conserved nucleotides are in bold.
(C) and (D) Root hair-enriched patterns of GFP expression driven by the RHEs of Selmo1_75237 (C) and Selmo1_78615 (D) in the Arabidopsis root. RHE regions of the S. moellendorffii genes are fused with the EXPA7 minimal promoter region to express GFP. Bar = 10 μm.
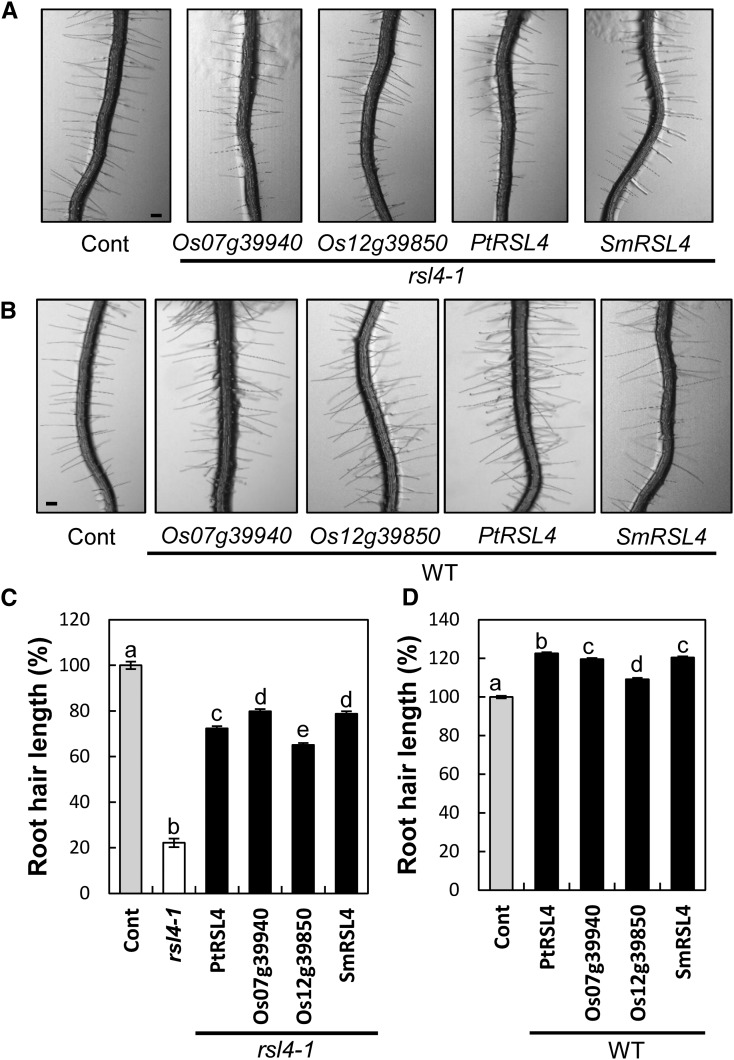
To demonstrate the conserved function of RSL4 in tracheophytes, we used phylogenetic analysis to identify the closest RSL4 homologs from poplar (Populus trichocarpa; POPTR 0002s12000.1.p, PtRSL4), rice (Oryza sativa; Os12g39850 and Os07g39940), and S. moellendorffii (Selmo1 419388, SmRSL4) (Supplemental Figure 3 and Supplemental Data Set 3). When these RSL4 homologs were expressed in the Arabidopsis rsl4-1 mutant background under the RSL4 promoter, root hair length was restored considerably (65–79% of the level of the wild type) (Figures 6A and 6C; Supplemental Data Set 1). When expressed in the wild-type background, root hair length was significantly increased (Figures 6B and 6D; Supplemental Data Set 1). When Os12g39850 was expressed under the strong root hair-specific promoter ProE7, one transgenic line showed a root hair length that was 193% that of the wild type (Supplemental Figure 4 and Supplemental Data Set 1). These results suggest that the RSL4 homologs from poplar, rice, and S. moellendorffii share a conserved function in mediating root hair growth.
Figure 6.
Tracheophyte RSL4 Homologs Enhance Root Hair Growth.
(A) and (B) Root images of control (Cont; ProE7:YFP) and transformants expressing RSL4 homologs of rice (ProRSL4:Os07g39940, Os07g39940; ProRSL4:Os12g39850, Os12g39850), poplar (ProRSL4:PtRSL4, PtRSL4), and S. moellendorffii (ProRSL4:SmRSL4, SmRSL4) in the rsl4-1 mutant (A) and wild-type (WT; [B]) backgrounds. Bars = 100 μm.
(C) Root hair length of the T3 homozygous lines expressing RSL4 homologs in the rsl4-1 mutant background. Error bars indicate ±se from 162 to 1169 root hairs (from two to five independent lines for transformants).
(D) Root hair length of the T2 lines expressing RSL4 homologs in the wild-type background. Error bars indicate ±se from 1851 to 3493 root hairs (from 6–10 independent lines for transformants). Statistically significant differences are denoted with different letters (one-way ANOVA with Tukey’s unequal N HSD post hoc test, P < 0.05) in (C) and (D).
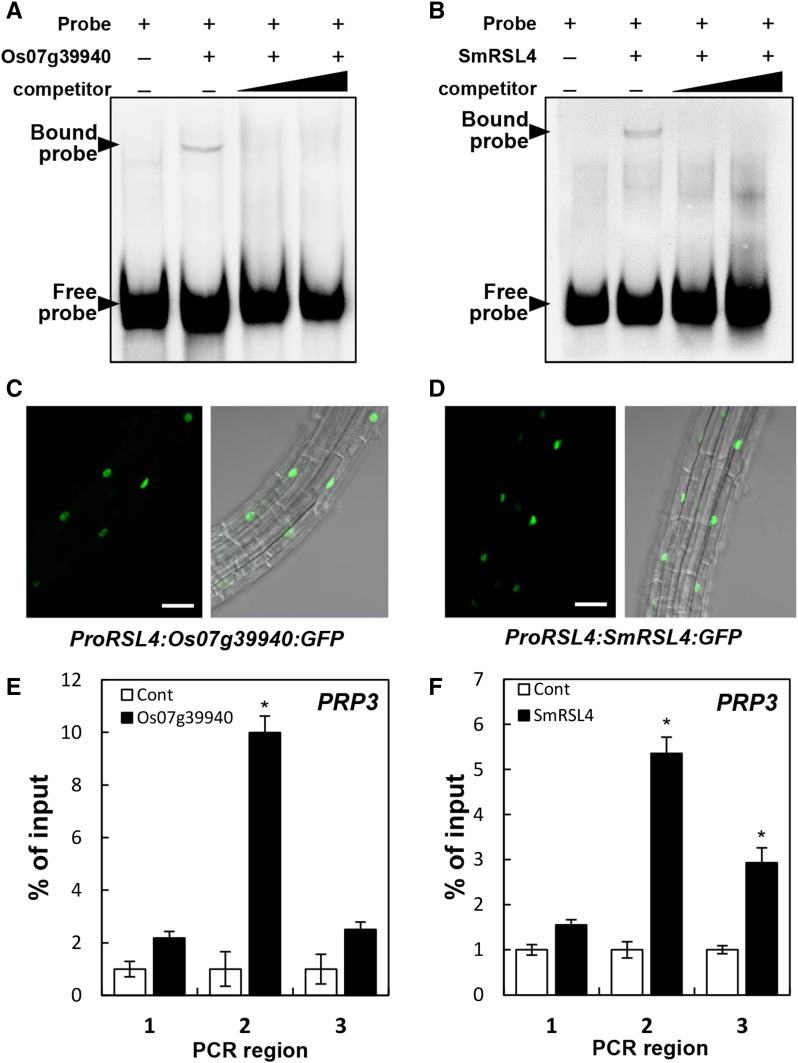
Next, we tested whether the RSL4 homologs bind to the RHE. In the EMSA, the RHE probe band was shifted by a rice RSL4 homolog (Os07g39940) and the S. moellendorffii homolog (SmRSL4), and the shifted band disappeared upon the addition of the unlabeled competitor RHE (Figures 7A and 7B), suggesting that these RSL4 homologs are able to bind to the RHE in vitro. To assess the binding of these RSL4 homologs to the RHE in vivo, we performed ChIP analysis on the RHE region of the Arabidopsis PRP3 promoter (Figure 3A) with the ProRSL4:RSL4-homolog:GFP transformants. The Os07g39940:GFP and SmRSL4:GFP fusion proteins targeted to the nuclei of root hair cells (Figures 7C and 7D). The ChIP-qPCR results showed that both Os07g39940 and SmRSL4 bound to the RHE region with high affinity (Figures 7E and 7F). These results suggest that the RSL4 homologs from the eudicot Arabidopsis, the monocot rice, and the lycophyte S. moellendorffii share a similar function in terms of their RHE binding and root hair-promoting capabilities.
Figure 7.
RSL4 Orthologs from Rice and S. moellendorffii Bind to the RHE in Vitro and in Vivo.
(A) and (B) EMSA showing the binding of Os07g39940 (A) and SmRSL4 (B) to the RHE. GST-tagged RSL4 orthologs and the biotinylated RHE probe were used. The competitor is nonbiotinylated RHE of 20- and 100-fold increases over the biotinylated probe amount.
(C) and (D) Confocal microscopy images showing fluorescent signals in the nuclei of root hair cells of plants expressing Os07g39940:GFP (ProRSL4:Os07g39940:GFP) in the wild-type background (C) and SmRSL4:GFP (ProRSL4:SmRSL4:GFP) in the wild-type background (D). Bars = 50 μm.
(E) and (F) Enrichment fold of the ChIP-PCR fragment from the region shown in Figure 3A. ChIP analysis was performed using wild-type (control [Cont]), ProRSL4:Os07g39940:GFP (Os07g39940, [E]), and ProRSL4:SmRSL4:GFP (SmRSL4, [F]) transformant plants. Error bars indicate ±se for two biological replicates. Values are relative to each Cont value and significantly different (*P < 0.05; Student’s t test) from the control value.
RSL4 Regulates Its Own Transcription in a Positive Feedback Loop
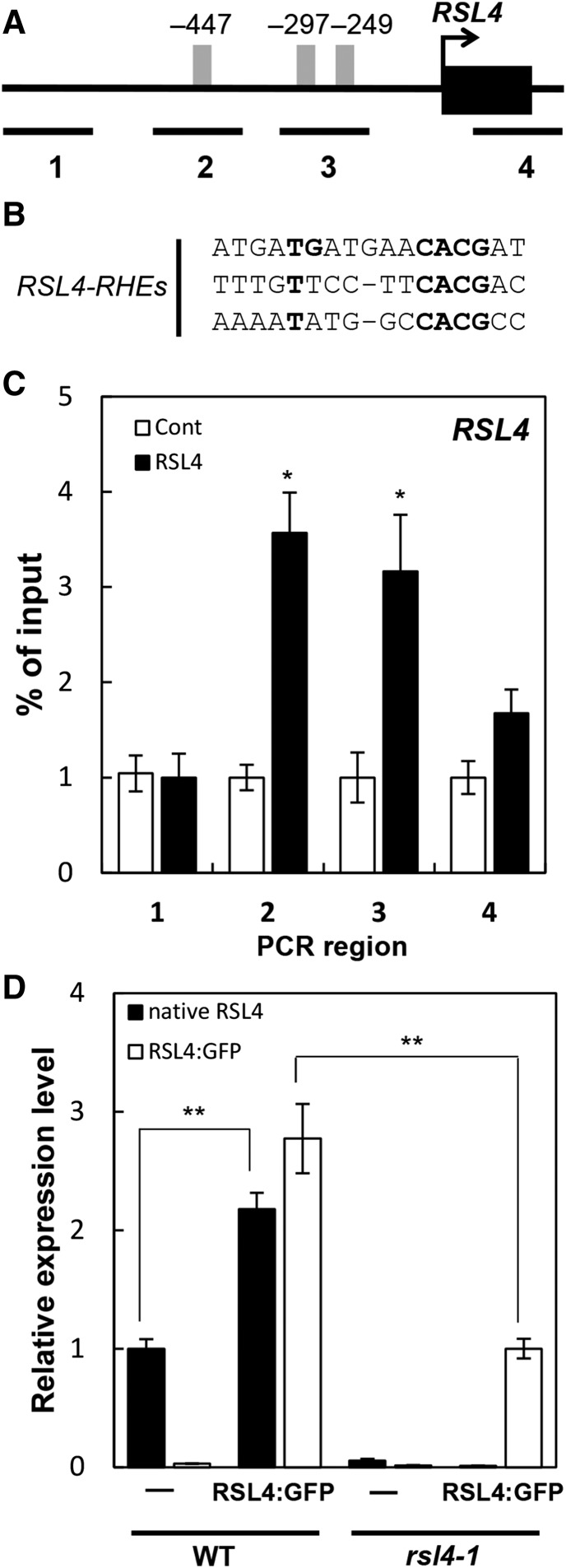
RSL4 itself has three RHE motifs in its proximal promoter region (Figures 8A and 8B). The promoters of the orthologous RSL4 genes from poplar, rice, and S. moellendorffii similarly contain one to five RHE motifs (Supplemental Figure 5). To test whether RSL4s positively regulate their own transcription, we conducted a ChIP analysis on the promoter of RSL4 with the ProRSL4:RSL4:GFP transformant. The binding of RSL4 to the RHE regions was significantly higher than the binding to the non-RHE regions (Figure 8C).
Figure 8.
RSL4 Binds to the RHEs of Its Own Gene Promoter and Amplifies Its Transcription.
(A) Locations of the RHE (gray bars) and ChIP-PCR regions (lines with #1–4) in the RSL4 promoter. Positions are relative to the start codon (+1).
(B) Sequences of three RHEs from the RSL4 promoter. Strictly conserved nucleotides are in bold.
(C) Enrichment fold of the ChIP-PCR fragment from the region shown in (A). The ChIP analysis was done with the wild type (Cont) and ProRSL4:RSL4:GFP (RSL4) transformant plants. Error bars indicate ±se for two biological replicates. Values are relative to each control value and significantly different (*P < 0.05; Student’s t test) from the control value.
(D) Transcript levels of native RSL4 and RSL4:GFP in the wild type, RSL4:GFP (ProRSL4:RSL4:GFP in the wild-type background), and the rsl4-1 mutant. Error bars indicate ±se from two biological repeats. The values are relative to the wild-type value and significantly different (**P < 0.001; Student’s t test) from the wild-type value.
To assess whether this binding represents a positive feedback loop, RSL4 transcript levels were compared between the wild type, the wild-type background containing the ProRSL4:RSL4:GFP construct, the rsl4-1 mutant, and the rsl4-1 mutant background containing the ProRSL4:RSL4:GFP construct by RT-qPCR analysis. If RSL4 exerted positive regulation on itself, the wild-type background containing the ProRSL4:RSL4:GFP construct would be expected to accumulate higher transcript levels of both the native RSL4 and the RSL4:GFP transgene than the wild-type plant and the rsl4-1 mutant background containing the ProRSL4:RSL4:GFP construct, respectively. Gene-specific primers were used in the RT-qPCR analysis to distinguish the native RSL4 gene from the RSL4:GFP transgene. Indeed, the RT-qPCR results showed that the wild-type background containing the ProRSL4:RSL4:GFP construct accumulated much higher levels of the native and transgenic RSL4 transcripts than did the wild-type plant and the rsl4-1 mutant containing the ProRSL4:RSL4:GFP construct, respectively (Figure 8D). This indicates that the transgenic RSL4:GFP further elevated the positive feedback regulation of the native RSL4 gene and the transgene itself, presumably by acting on the RHE motifs on the RSL4 promoter.
Expression of RSL4 Is Sufficient to Initiate Root Hair Growth in the Non-Root-Hair Cell Position and in the rhd6 Mutant
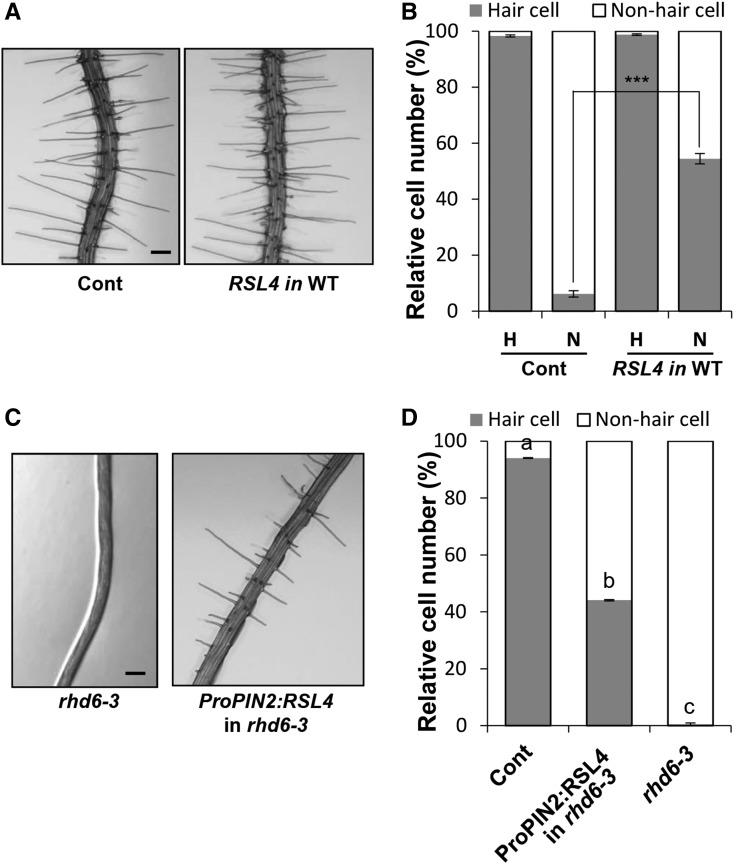
Our results demonstrated that RSL4 regulates RHE-containing RHS genes that are required for root hair formation. We tested whether the expression of RSL4 was sufficient to induce root hair growth in non-root-hair cells where RHD6 and its downstream processes for root hair morphogenesis are absent. The non-root-hair cell-specific GL2 promoter (ProGL2) was used to express RSL4 and its orthologs from rice and poplar in non-root-hair cells. Expression of RSL4 and its orthologs in non-root-hair cells considerably increased root hair numbers. Root hairs grew in only 6% of non-root-hair cells in the control plant, whereas the transformants expressing RSL4 and its orthologs grew root hairs in 54 to 74% of the non-root-hair cells (Figures 9A and 9B; Supplemental Figure 6 and Supplemental Data Set 1).
Figure 9.
RSL4 Induces Root Hair Growth in the Non-Root-Hair Cell Position and in the rhd6 Mutant.
(A) Root images of control (Cont; ProE7:YFP) and non-root-hair cells expressing transformants of RSL4 (ProGL2:RSL4, RSL4) in the wild-type (WT) background. Bars = 100 μm.
(B) Quantitative analysis for RSL4-induced root hair formation in the non-root-hair cell position. Relative number of root hair cells and non-root-hair cells of total counted epidermal cells in the root hair cell position (H) or in the non-root-hair cell position (N) was estimated. Error bars indicate ±se from 970 to 980 epidermal cells (from 20–21 roots for the transformants).
(C) Root images of the rhd6-3 mutant and ProPIN2:RSL4 in the rhd6-3 mutant background. The values are relative to the control value and are significantly different (***P < 0.0005; Student’s t test). Bars = 200 μm.
(D) Quantitative analysis of root hair formation in the root hair cell position as described in (B). Error bars indicate ±se from 600 to 1470 epidermal cells (from 12–26 roots for the transformants). Statistically significant differences are denoted with different letters (one-way ANOVA with Tukey’s unequal N HSD post hoc test, P < 0.05).
In addition, we tested whether the expression of RSL4 could restore root hair growth in the hairless rhd6-3 mutant background. RSL4 was expressed by the PIN2 promoter (ProPIN2) that is active in the meristem and elongation zones of the root epidermis. While the rhd6-3 mutant grew no root hairs, the introduction of ProPIN2:RSL4 in this mutant substantially restored root hair growth (Figures 9C and 9D; Supplemental Data Set 1). These results indicate that even when RHD6 and its downstream regulators (other than RSL4) are absent, RSL4 and its downstream targets (i.e., RHS genes) are able to initiate and maintain root hair morphogenesis.
DISCUSSION
The Structure of the RHE and RSL4 Binding
Our in vitro and in vivo binding assays showed that RSL4 and its orthologs from eudicot, monocot, and lycophyte plants directly bind to the RHE of RHS genes. The function of the RHE was originally determined by testing the effect of single nucleotide-level mutations of the RHE on root hair-specific gene expression and was supported by the comparative analysis of the RHEs from paralogs and orthologs of RHS genes (Kim et al., 2006; Won et al., 2009). Any single mutation of the strictly conserved nucleotides or change in linker length (2 to 3 bp or 3 to 2 bp; Figure 2D) drastically alters root hair-specific gene expression (Kim et al., 2006). Here, we found that the analogous mutations of the RHE reduced RSL4 binding both in vitro and in vivo, such that the mutation of the RP of the RHE reduced RSL4 binding to a greater extent than did the LP mutation, and the change in the linker length also substantially decreased RSL4 binding (Figures 2E and 4B). The similarity between the effects of these mutations on root hair growth (Kim et al., 2006) and on RSL4 binding strongly suggests that the functionality of RHEs is linked to the binding dynamics of the RSL4 transcription factor.
Because the functional parts of the RHE are separated by a linker, we questioned whether the RHE binds to two individual transcription factors and, if so, whether RSL4 solely binds to both the LP and RP or if additional transcription factors could be involved. The length of a transcription factor binding site varies from 5 to >30 bp but is typically ∼10 bp (Stewart et al., 2012). The 16- or 17-bp RHE core and the 23-bp minimum RHE length for RSL4 binding (Figure 2C) indicates that two transcription factor molecules could bind to the RHE. The structure of the RHE, i.e., two distinct motifs with a flexible linker between them, is similar to that of the nuclear receptor binding elements, where two homo- or heterodimeric nuclear receptor molecules bind to the element in different combinations depending on the length of the flexible linker between two 6-bp binding elements (Latchman, 2008). Because a mutation of either the LP or RP affected RSL4 binding, it is likely that RSL4 binds to both the LP and RP. This hypothesis is supported by the fact that the LP and RP are partially palindromic (Kim et al., 2006). However, because the RP mutation had a greater effect on RSL4 binding than did the LP mutation, RSL4 may preferentially bind to the RP. RSL4 may form a heterodimer with a close homolog, such as RSL5 (its closest homolog; Supplemental Figure 3). Both root hair growth and the expression of RSL4 target genes were further suppressed in the rsl2-1 rsl4-1 double mutant compared with the rsl4-1 single mutant (Yi et al., 2010), suggesting that RSL2 and RSL4 could possibly function together. However, because the rsl2-1 single mutation does not affect the expression of RSL4 target genes and has a weaker effect on root hair growth than does the rsl4-1 mutation (Yi et al., 2010), RSL4 is likely to perform the dominant role in RHE binding and regulation of RHS gene expression.
RSL4 Target Genes and Autoregulation of RSL4 Transcription
Two transcriptome analyses have been performed to investigate the RSL4 target genes. Yi et al. (2010) found 88 Arabidopsis gene loci that are upregulated by RSL4 overexpression and downregulated in the rsl4 mutant. Another recent analysis discovered 34 Arabidopsis genes that are induced by glucocorticoid-inducible RSL4 expression with cycloheximide in the rsl2 rsl4 double mutant (Vijayakumar et al., 2016). Twenty percent of the genes found by Yi et al. and none of those reported by Vijayakumar et al. represent known RHS genes. RHS genes were originally identified as containing the RHE and expressed only in root hair cells but not in other tissues and organs (Won et al., 2009). However, it is possible that RHE-containing genes can be expressed in other tissues (with other cis-elements). These genes, though not classified as RHS genes, may also be regulated by RSL4 through acting on the RHE. To determine if the genes found in the previously mentioned studies (88 by Yi et al. and 34 by Vijayakumar et al.) are RSL4 targets, we examined the 2.5-kb upstream region (from the start codon) of these genes for the presence of RHE (N4TN5-6CACGN2) motifs. We found that 113 of the 122 genes had at least one RHE (Supplemental Data Set 4), suggesting that these genes may be direct targets of RSL4. These 113 RHE-containing genes included an average of 3.7 RHEs per gene (Supplemental Table 2), and 81% of these genes had an RHE within −1.0 kb of the start codon (Supplemental Table 3). When the other RHS genes found by Won et al. (2009) are included, we now have at least 124 RHE-containing putative RSL4 target genes. Among these putative RSL4 target genes, the majority function in cell wall-related events (Supplemental Figure 7), pointing to cell wall dynamics as the major cellular process during root hair growth.
Interestingly, RSL4 and its orthologs contain RHEs (Figure 8A; Supplemental Figure 5), suggesting a direct positive loop for amplifying their own transcription. Our ChIP and RT-qPCR analyses demonstrated that RSL4 binds to its own RHEs and increases its transcript levels (Figure 8). Root hairs can grow more than 25 times the length of the root diameter in Arabidopsis (Yi et al., 2010); accordingly, root hair growth must require a sustainable mechanism to maintain this process. As RSL4 is the master regulator of various root hair RHS genes, the positive autoregulation of RSL4 and the resulting amplification of RHS gene products may be the critical factor for the sustainable growth of root hairs (Figure 10). In addition to the RHE-mediated positive feedback loop, the regulatory region of RSL4 may include the regulatory elements to accommodate RHD6- and auxin-mediated signaling, as RSL4 is a direct target of these two upstream factors (Yi et al., 2010). Our in silico analysis revealed that the promoter regions of RSL4 and its orthologs include multiple auxin response elements (Supplemental Figure 5).
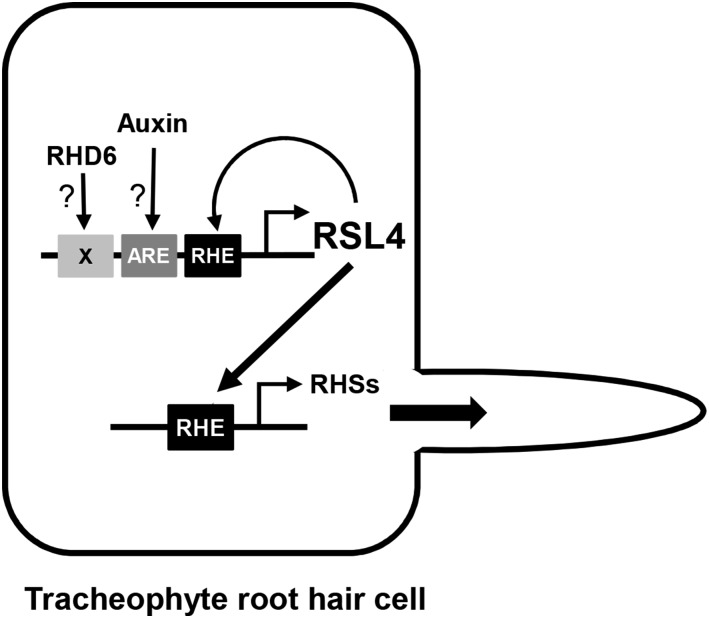
Figure 10.
Model Illustrating That Evolutionarily Conserved RSL4 Modulates Its Own Gene, Root Hair-Specific Genes, and Sustainable Root Hair Tip Growth.
RHD6 and auxin signaling directly regulate RSL4 by acting on the unidentified cis-element and auxin response element (ARE), respectively. RSL4 amplifies itself by binding to the RHE of its gene promoter. RSL4 directly modulates RHE-containing RHS genes to induce root hair formation and growth. Since the RHE and RSL4 are functionally conserved in angiosperms and a basal tracheophyte, this model is likely to be operational throughout vascular plants.
RSL transcription factors are divided into two classes, class I RSLs (RHD6 and RSL1) and class II RSLs (RSL2-5), where class I RSLs regulate class II RSLs (Pires et al., 2013). RSL4 is the direct target of RHD6 (Yi et al., 2010). Due to the lack of GL2 in root hair-forming cells, the gene-regulatory cascade, in which class I RSLs regulate class II RSLs that regulate RHSs and other root hair-forming genes, leads to root hair formation. Conversely, in non-root-hair cells, GL2 directly suppresses class I RSLs, RSL2, and other non-RSL-type bHLH transcription factor genes involved in root hair development (Lin et al., 2015), leading to no expression of RSL4 and RHS genes and no root hair formation. Although RSL4 is controlled by upstream RHD6, which modulates additional factors other than RSL4 for root hair formation, we tested whether RSL4 alone was sufficient to induce root hair formation. Our results showed that ectopic expression of RSL4 or its orthologs induced substantial root hair growth in non-root-hair cells and in the hairless rhd6 mutant (Figure 9). This suggests that expression of RSL4 is sufficient to induce root hair formation. Our result is contrasted with a recent report showing that expression of RHD6 in non-root-hair cells failed to induce root hair formation (Lin et al., 2015).
Conservation of the RSL4-RHE Regulatory Module for Root Hair Formation in Tracheophytes
The root hair is a basic cellular structure in vascular plants and is used as a model system to study the mechanisms for cell fate determination and cell morphogenesis. Although angiosperms, depending on the taxa, use different mechanisms to determine root hair cell or non-root-hair cell fate in the root epidermis, they share the common RHE in their RHS genes, and thus were hypothesized to share common transcription factors to regulate these RHS genes. In this study, we demonstrated that the RHE is the direct target of the bHLH transcription factor RSL4 and its angiosperm and lycophyte orthologs.
The roots of lycophytes and euphyllophytes (ferns and seed plants) evolved independently (Kenrick, 2002). Despite their different origins of the root, these two vascular plant groups share a common mechanism for determining root hair/non-root-hair cell fate; type 2 root hair cell fate determination occurs throughout the tracheophytes from lycophytes to angiosperms except for eudicots (Dolan, 1996; Clowes, 2000; Kim et al., 2006). Our results demonstrate that the RSL4-RHE regulatory module is also conserved between a lycophyte (S. moellendorffii) and angiosperms. Thus, our study is consistent with a recent report demonstrating the similarity in overall root developmental programs between these two vascular plant groups (Huang and Schiefelbein, 2015).
The RSL4-RHE regulatory module most likely evolved before the divergence of vascular plants, and as vascular plants evolved, different upstream fate-determining machineries evolved. The phylogenetic analysis of the RSL4 homologs from land plants revealed several RSL4-orthologous genes in a liverwort (Marchantia polymorpha) and a moss (Physcomitrella patens) (Supplemental Figure 3). Interestingly, the RSL4-orthologous genes from these bryophytes include multiple RHE-like motifs in their promoter regions as do the tracheophyte orthologs (Supplemental Figure 5), suggesting that the RSL4-RHE regulatory module may be found in bryophytes as well. The hierarchical relationship between class I and class II RSLs, where class I RSLs modulate class II RSLs, is conserved in both moss and Arabidopsis (Pires et al., 2013), and class I RSLs play a role in rhizoid development in liverwort and moss (Menand et al., 2007; Proust et al., 2016). Considering the morphological and functional similarity between root hairs and rhizoids, it is conceivable that the RSL4-RHE module might have emerged during early land plant evolution. Our work indicates that vascular plants repeatedly used the RSL4-RHE regulatory module as a toolkit for sustainable root hair growth, since the tip-growing cells of ancient plants had adopted this module.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana (Columbia) was used for transformation unless stated otherwise. Arabidopsis plants were transformed using Agrobacterium tumefaciens strain C58C1 (pMP90). Transformed plants were selected on hygromycin-containing plates (30 μg mL–1). All seeds were grown on agarose plates containing 4.3 g mL–1 Murashige and Skoog nutrient mix (Duchefa), 1% sucrose, 0.5 g mL–1 MES, pH 5.7, with KOH, and 0.8% agarose. Seeds were cold treated before germination at 23°C under a 16-h/8-h light/dark photoperiod. The light condition was 130 µmol m–2 s–1 with fluorescent light bulbs (FHF 32SS-EXD; Kumho Electric). For observation of root hairs, homozygous transformants were planted on antibiotic-free media, and T1 and T2 lines were planted on hygromycin containing media. Hygromycin did not significantly interfere with root hair development, as shown with the control ProE7:YFP transformants. Two control lines were adopted; the wild type for the rsl4-1 mutant analysis and ProE7:YFP (Lee and Cho, 2006; Ganguly et al., 2010) for transgenic analysis with hygromycin.
Observation of Root Hair Phenotypes
Root hair phenotypes were observed under a stereomicroscope (M205 FA; Leica). Root hair length was measured as described by Lee and Cho (2006) with modifications. Roots of 3-d-old seedlings were digitally photographed using the stereomicroscope at 40× magnification. The hair length of nine consecutive hairs protruding perpendicularly from each side of the root, for a total of 18 hairs from both sides of the root, was calculated using ImageJ 1.50b software (National Institutes of Health). To assess root hair cell distribution in the root epidermis, the ratio of root hair-containing cells in 10 consecutive epidermal cells from the H (root hair cell) position or from the N (non-root-hair cell) position was estimated from 15 to 20 roots of 4-d-old seedlings for each line.
Construction of Transgenes
The EXPA7 promoter (ProE7):GFP construct (Cho and Cosgrove, 2002; Kim et al., 2006) in the modified binary vector pCAMBIA1300-NOS (Lee et al., 2010) was used as the cloning vector to generate the GFP fusion proteins. The GL2 promoter (ProGL2; Lin and Schiefelbein, 2001) was used for non-root-hair cell-specific expression of RSL4 and its homologs. The PIN2 promoter (ProPIN2) from the ProPIN2:PIN2:GFP construct (Cho et al., 2007) was used to generate the ProPIN2:RSL4 transgene. All primers used for PCR are listed in Supplemental Table 1.
The RSL4 promoter region (ProRSL4, between −928 and +63 bp relative to the predicted transcription initiation site) was obtained by PCR using Arabidopsis genomic DNA as template and inserted into the HindIII/BamHI sites by replacing ProE7 of the ProE7:GFP construct. For the ProRSL4:RSL4:GFP, ProRSL4:Os07g39940:GFP, and ProRSL4:Selmo1_419388:GFP constructs, genomic fragments of RSL4 and Os07g39940 and the cDNA fragment of Selmo1_419388 were obtained by PCR using genomic DNA of Arabidopsis and rice (Oryza sativa) and cDNA of Selaginella moellendorffii as template and cloned into the XmaI/AvrII sites for RSL4 and the XmaI/MluI sites for Os07g39940 and Selmo1_419388 between the ProRSL4 and GFP regions of the ProRSL4:GFP construct. For the ProRSL4:Os12g39850 and ProRSL4:PtRSL4 constructs, genomic fragments were obtained by PCR using rice and poplar (Populus trichocarpa) genomic DNA, respectively, as template and were cloned into the PacI/XbaI sites downstream of ProRSL4 in the ProRSL4 construct.
For the ProRSL4:RSL4:GFP:RHE/mRHE constructs (Figure 4A), the RHE sequence of ProPRP3 was obtained by PCR using Arabidopsis genomic DNA as the template. For the ProRSL4:RSL4:GFP:RHE construct, the RHE sequence was cloned into the EcoRI site downstream of the NOS terminator in the ProRSL4:RSL4:GFP construct. For the ProRSL4:RSL4:GFP:mRHE construct, site-directed mutagenesis of the LP, RP, and linker of the RHE was performed by PCR. Primers used for site-directed mutagenesis of the mLP, mRP, 1-D, and mLPmRP are listed in Supplemental Table 1. For the ProRSL4:RSL4:GFP:mRHE construct, the PCR products of the mRHEs were cloned into the EcoRI site downstream of the NOS terminator.
The EXPA7 minimal promoter region (mpE7) (between −73 and +31 bp relative to the transcription initiation site) was obtained by PCR using Arabidopsis genomic DNA as the template and cloned into the SalI/BamHI sites upstream of GFP. The RHE-containing regions of Selmo1_75237 (−296 to approximately −42 bp relative to the start codon) and Selmo1_78615 (−313 to approximately −111 bp relative to the start codon) were obtained by PCR using S. moellendorffii genomic DNA as the template and cloned into the KpnI/SalI sites upstream of mpE7 in the mpE7:GFP construct.
The GL2 promoter region (ProGL2, −1951 to approximately +138 bp relative to the predicted transcription initiation site) was obtained by PCR using Arabidopsis genomic DNA as the template and cloned into the HindIII site. For the ProGL2:RSL4, ProGL2:Os12g39850, and ProGL2:PtRSL4 constructs, genomic fragments of RSL4, Os12g39850, and PtRSL4 were obtained by PCR and cloned into the BamHI/SpeI sites, the PacI/XbaI sites, and the PacI/XbaI sites downstream of ProGL2, respectively.
For the ProE7:RSL4, ProE7:Os12g39850, and ProPIN2:RSL4 constructs, genomic fragments of RSL4 and Os12g39850 were obtained by PCR and cloned into the BamHI/SpeI sites and the PacI/XbaI sites downstream of ProE7 or ProPIN2, respectively.
For the constructs used in the yeast one-hybrid assay, RSL4 cDNA was obtained by PCR from the Arabidopsis seedling cDNA library and cloned into the EcoRI/XhoI sites of the pGADT7-Rec prey vector (Clontech Laboratories). To make the bait construct (pHIS2:3X-E7-RHE), two complementary oligomer sequences of three tandem copies of RHE from the EXPA7 promoter (3X-E7-RHE; Supplemental Table 1) were synthesized, hybridized, and cloned into the EcoRI/SacI sites of the pHIS2 vector.
To express RSL4, Os07g39940, and Selmo1_419388 proteins in Escherichia coli for the EMSA, the cDNA sequences of RSL4, Os07g39940, and Selmo1_419388 were amplified by PCR from the Arabidopsis seedling cDNA library and cloned into the EcoRI/XhoI sites of the pGEX-4T-1 vector (GE Healthcare), generating fusion proteins with GST at the N termini of RSL4, Os07g39940, and Selmo1_419388 proteins.
All constructs were confirmed by nucleotide sequencing and introduced into Arabidopsis plants by the Agrobacterium-mediated floral dipping method. The transgene insertion in the Arabidopsis transformants was confirmed by PCR analysis using transgene-specific primers.
Yeast One-Hybrid Assay
The yeast one-hybrid assay was performed using the BD Matchmaker TM One-Hybrid Library Construction and Screening Kit accordingly to the manufacturer’s protocol. Yeast cells (Y187; Clonetech Laboratories) were cultured on the YPAD plate at 30°C for 1 to 2 d and resuspended in one-step buffer (50% PEG, 1 M LiAc, and 4 M DTT). Next, the plasmids, including the bait/prey constructs or the control construct, were added to the yeast solution and treated with heat shock at 42°C in a water bath for 45 min. The transformed yeasts cell suspension was dropped on SD (DDO for -Trp/-Leu, and TDO for -Trp/-Leu/-His with or without 3-amino-1,2,4-triazole) medium plates and cultured at 30°C for 5 to 7 d.
Preparation of Fusion Proteins and EMSA
To produce RSL4, Os07g39940, and Selmo1_419388 proteins for EMSA, the gene-containing vectors were transformed into E. coli strain BL21 and the transgene expression was induced with 0.25 mM isopropyl β-d-1-thiogalactopyranoside at 28°C for 4 h. The cells were harvested by centrifugation and lysed in B-PER Buffer (Thermo Scientific). The proteins in the lysate were bound to GSH resin (Elpisbio) at 4°C for 12 h and washed three times with TBS buffer. The bound proteins were eluted in elution buffer (15 mM GSH and 50 mM Tris, pH 6.17) at 4°C for 3 h. The eluted proteins were confirmed by SDS-PAGE separation and protein blot analysis using anti-GST antibody (GeneScript; catalog no. A00097; 1:10,000 dilution).
EMSA was performed using the LightShift Chemiluminescent EMSA kit (Thermo Scientific) following the manufacturer’s protocol. The assays were performed using recombinant proteins (GST-fused RSL4, Os07g39940, Selmo1_419388, and ARF5-DBD) and the 5′-biotinylated 61-bp RHE probe (aatgatggccaacatgtttttgaTCGTGCATTACACTTAGgtatgagaatctcaaacatac, the RHE core is shown in uppercases), unless stated otherwise, designed from the corresponding promoter region of Arabidopsis PRP3. The biotinylated probe was used at a concentration of 20 fmol/μL, and 20- to 100-fold of nonbiotinylated probe was used as the competitor. The protein was mixed with the probe or the probe and competitor in the binding buffer (10 mM Tris, pH 7.5, 50 mM KCl, and 1 mM DTT) with 50 ng μL–1 poly(dI-dC), 0.05% Nonidet P-40, 5 mM MgCl2, and 2.5% glycerol. The protein-probe complexes were resolved on an 8% polyacrylamide gel, transferred to a positively charged nylon membrane (Roche), and cross-linked by 254-nm UV for 8 min. The resulting bands were detected using the Chemiluminescent Nucleic Acid Detection Module Kit (Thermo Scientific).
Microscopy Observation of Fluorescent Proteins
Fluorescence signals of the GFP-fusion proteins from 4-d-old transgenic seedlings were observed using an LSM 510 confocal laser scanning microscope (Carl Zeiss).
ChIP and qPCR Analyses
ChIP analysis was performed as previously described (Gendrel et al., 2002; Haring et al., 2007). Four-day-old transgenic seedlings expressing the GFP-fusion proteins were vacuum-infiltrated in 1% formaldehyde solution for cross-linking. After quenching the cross-linking by adding glycine, the seedlings were ground in liquid nitrogen. The chromatin was isolated as described (Moehs et al., 1988), resuspended in nuclei lysis buffer (50 mM Tris-HCl, pH 8.0, 10 mM EDTA, and 1% SDS), and sonicated to obtain 0.5- to 1.0-kb fragments. The chromatin solution was precleared with salmon sperm DNA/Protein-A agarose beads (Millipore) at 4°C for 1 h and immunoprecipitated with anti-GFP antibody beads (MBL; code 598; 1:200 dilution) overnight. The immunocomplex was washed once with each of the following buffers: low-salt buffer (140 mM NaCl, 0.2% SDS, 0.5% Triton X-100, 2 mM EDTA, and 20 mM Tris-HCl, pH 8.0), high-salt buffer (500 mM NaCl, 0.2% SDS, 0.5% Triton X-100, 2 mM EDTA, and 20 mM Tris-HCl, pH 8.0), and LiCl wash buffer (0.25 M LiCl, 0.5% Nonidet P-40, 0.5% sodium deoxycholate, 1 mM EDTA, and 10 mM Tris-HCl, pH 8.0), and twice with TE buffer (10 mM Tris-HCl, pH 8.0, and 1 mM EDTA). Each wash buffer (1 mL) was added to the immunocomplex, mixed by rotating for 5 min, and centrifuged for 2 min at 5900g at 4°C. Chromatin was eluted from the beads by adding 300 μL elution buffer (1% SDS and 0.1 M NaHCO3) and incubated at 65°C for 15 min. After incubation, the beads were pelleted by a 1 min centrifugation at 16,100g at room temperature and the supernatant was collected. Cross-linking was reversed by adding 5 M NaCl (final 200 mM) for 7 h at 65°C and the resulting sample was treated with proteinase K (final concentration of 40 ng μL–1) to remove all the proteins. Antibody-untreated samples for estimating input DNA were also treated with the same processes. DNA from the reverse cross-linked samples was purified using the QIAquick PCR purification kit (Qiagen), and input DNA was estimated by PCR using ACTIN7 primers. The ChIP-qPCR analysis was done using the primer sets listed in Supplemental Table 1 and the amfiSure qGreen Q-PCR Master Mix (2X) without ROX (GenDepot) in the Chromo4 four-color real-time detector (Bio-Rad). The “% of input” value of each ChIP-qPCR fragment was calculated first by normalizing the fragment amount against the input value and then by normalizing the value from the transgenic plants against the value from the control plants. Each ChIP-qPCR reaction was performed in quadruplicate, and each experiment was repeated two to three times using chromatin samples prepared at different times.
RT-qPCR Analysis
Total RNA was isolated from the roots of 4-d-old seedlings using the RNeasy plant mini kit (Qiagen). cDNA was synthesized using TOPscript RT DryMIX (dT18; Enzynomics). RT-qPCR analyses were performed using amfiSure qGreen Q-PCR Master Mix (2X) without ROX (GenDepot) and Chromo4 four-color real-time detector (Bio-Rad). Gene-specific signals were normalized relative to ACTIN7 signals. Each RT-qPCR reaction was performed in triplicate, and each experiment was repeated two times using RNA samples prepared at different time points. Primers used are listed in Supplemental Table 1.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: AT1G27740 (RSL4), AT3G62680 (PRP3), AT1G12560 (EXPA7), AT4G29180 (RHS16), AT1G12040 (LRX1), AT1G70460 (RHS10), POPTR 0002s12000.1.p (PtRSL4), Selmo1_419388 (SmRSL4), and AT1G79840 (GL2).
Supplemental Data
Supplemental Figure 1. Root Hair-Specific Overexpression of RSL4 Enhances Root Hair Growth.
Supplemental Figure 2. A Phylogenetic Tree Showing EXPA7-Orthologous Sequences in Tracheophytes.
Supplemental Figure 3. Phylogenetic Relationship of RSL4 Homologs.
Supplemental Figure 4. The Effect of Overexpression of a Rice RSL4 Homolog (Os12g39850) on Root Hair Growth.
Supplemental Figure 5. Promoters of RSL4 and RSL4 Homologs Contain the RHE and Auxin Response Element.
Supplemental Figure 6. RSL4 Orthologs Induce Root Hair Growth in the Non-Root-Hair Cell Position and in the rhd6 Mutant.
Supplemental Figure 7. Functional Classification of 124 RHE-Containing RSL4 Target Genes.
Supplemental Table 1. The Primer List.
Supplemental Table 2. RHE Frequency.
Supplemental Table 3. RHE Distribution.
Supplemental Data Set 1. ANOVA Tables.
Supplemental Data Set 2. Alignments Used to Generate the Phylogeny Presented in Supplemental Figure 2.
Supplemental Data Set 3. Alignments Used to Generate the Phylogeny Presented in Supplemental Figure 3.
Supplemental Data Set 4. List of Putative RSL4 Target Genes Containing the RHE.
Supplementary Material
Acknowledgments
We thank Ji-Yun Kim and Woo-Rim Yang at Seoul National University for their kind advice with the ChIP analysis and Clint Chapple at Purdue University for kindly donating S. moellendorffii bulbils. This research was supported by grants from the Mid-career Researcher Program (2015002633) of the National Research Foundation and the Next-Generation BioGreen 21 program (The Agricultural Genome Center PJ011195) of the Rural Development Administration. H.-S.C. was partially supported by the Stadelmann-Lee Scholarship Fund, Seoul National University.
AUTHOR CONTRIBUTIONS
Y.H. performed most of the experiments. H.-S.C. performed the ChIP analysis on RSL4. H.-M.C. made the constructs for non-root-hair cell expression of RSL4 orthologs and PIN2-domain expression of RSL4 and analyzed their phenotypes. H.-T.C. designed the experiments. All authors contributed to analyzing the results and writing the manuscript.
Glossary
- TF
transcription factor
- bHLH
basic helix-loop-helix
- RHE
root hair-specific cis-element
- TDO
triple dropout
- EMSA
electrophoresis mobility shift assay
- LP
left part
- RP
right part
- ChIP
chromatin immunoprecipitation
Footnotes
Articles can be viewed without a subscription.
References
- Cho H.-T., Cosgrove D.J. (2002). Regulation of root hair initiation and expansin gene expression in Arabidopsis. Plant Cell 14: 3237–3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho M., Lee S.H., Cho H.-T. (2007). P-glycoprotein4 displays auxin efflux transporter-like action in Arabidopsis root hair cells and tobacco cells. Plant Cell 19: 3930–3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clowes F.A.L. (2000). Pattern in root meristem development in angiosperms. New Phytol. 146: 83–94. [Google Scholar]
- Cutter E.G. (1978). Plant Anatomy, 2nd ed. (Reading, MA: Addison-Wesley Publishing). [Google Scholar]
- Dolan L. (1996). Pattern in the root epidermis: An interplay of diffusible signals and cellular geometry. Ann. Bot. (Lond.) 77: 547–553. [Google Scholar]
- Ganguly A., Lee S.H., Cho M., Lee O.R., Yoo H., Cho H.-T. (2010). Differential auxin-transporting activities of PIN-FORMED proteins in Arabidopsis root hair cells. Plant Physiol. 153: 1046–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel A.-V., Lippman Z., Yordan C., Colot V., Martienssen R.A. (2002). Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science 297: 1871–1873. [DOI] [PubMed] [Google Scholar]
- Grierson C., Schiefelbein J. (2008). Genetics of Root Hair Formation. In Root Hairs, Emons A.M.C., Ketelaar T., eds (Berlin, Heidelberg, Germany: Springer Berlin Heidelberg; ), pp. 1–25. [Google Scholar]
- Haring M., Offermann S., Danker T., Horst I., Peterhansel C., Stam M. (2007). Chromatin immunoprecipitation: optimization, quantitative analysis and data normalization. Plant Methods 3: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Schiefelbein J. (2015). Conserved gene expression programs in developing roots from diverse plants. Plant Cell 27: 2119–2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenrick P. (2002). The origin of roots. In Plant Roots, 3rd ed., Waisel Y., Eshel A., Kafkafi U., eds (New York: Marcel Dekker; ), pp. 1–13. [Google Scholar]
- Kim D.W., Lee S.H., Choi S.B., Won S.K., Heo Y.K., Cho M., Park Y.I., Cho H.-T. (2006). Functional conservation of a root hair cell-specific cis-element in angiosperms with different root hair distribution patterns. Plant Cell 18: 2958–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak S.-H., Shen R., Schiefelbein J. (2005). Positional signaling mediated by a receptor-like kinase in Arabidopsis. Science 307: 1111–1113. [DOI] [PubMed] [Google Scholar]
- Kwak S.-H., Schiefelbein J. (2007). The role of the SCRAMBLED receptor-like kinase in patterning the Arabidopsis root epidermis. Dev. Biol. 302: 118–131.17027738 [Google Scholar]
- Latchman D.S. (2008). Eukaryotic Transcription Factors, 5th ed. (San Diego, CA: Academic Press; ). [Google Scholar]
- Lee O.R., Kim S.J., Kim H.J., Hong J.K., Ryu S.B., Lee S.H., Ganguly A., Cho H.-T. (2010). Phospholipase A(2) is required for PIN-FORMED protein trafficking to the plasma membrane in the Arabidopsis root. Plant Cell 22: 1812–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.H., Cho H.-T. (2006). PINOID positively regulates auxin efflux in Arabidopsis root hair cells and tobacco cells. Plant Cell 18: 1604–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C., Choi H.-S., Cho H.-T. (2011). Root hair-specific EXPANSIN A7 is required for root hair elongation in Arabidopsis. Mol. Cells 31: 393–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q., Ohashi Y., Kato M., Tsuge T., Gu H., Qu L.J., Aoyama T. (2015). GLABRA2 directly suppresses basic helix-loop-helix transcription factor genes with diverse functions in root hair development. Plant Cell 27: 2894–2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Schiefelbein J. (2001). Embryonic control of epidermal cell patterning in the root and hypocotyl of Arabidopsis. Development 128: 3697–3705. [DOI] [PubMed] [Google Scholar]
- Menand B., Yi K., Jouannic S., Hoffmann L., Ryan E., Linstead P., Schaefer D.G., Dolan L. (2007). An ancient mechanism controls the development of cells with a rooting function in land plants. Science 316: 1477–1480. [DOI] [PubMed] [Google Scholar]
- Moehs C.P., McElwain E.F., Spiker S. (1988). Chromosomal proteins of Arabidopsis thaliana. Plant Mol. Biol. 11: 507–515. [DOI] [PubMed] [Google Scholar]
- Pires N.D., Yi K., Breuninger H., Catarino B., Menand B., Dolan L. (2013). Recruitment and remodeling of an ancient gene regulatory network during land plant evolution. Proc. Natl. Acad. Sci. USA 110: 9571–9576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proust H., Honkanen S., Jones V.A.S., Morieri G., Prescott H., Kelly S., Ishizaki K., Kohchi T., Dolan L. (2016). RSL class I genes controlled the development of epidermal structures in the common ancestor of land plants. Curr. Biol. 26: 93–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefelbein J.W. (2000). Constructing a plant cell. The genetic control of root hair development. Plant Physiol. 124: 1525–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart A.J., Hannenhalli S., Plotkin J.B. (2012). Why transcription factor binding sites are ten nucleotides long. Genetics 192: 973–985. Erratum. Genetics 193: 1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayakumar P., Datta S., Dolan L. (2016). ROOT HAIR DEFECTIVE SIX-LIKE4 (RSL4) promotes root hair elongation by transcriptionally regulating the expression of genes required for cell growth. New Phytol. 212: 944–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won S.K., Lee Y.J., Lee H.Y., Heo Y.K., Cho M., Cho H.-T. (2009). Cis-element- and transcriptome-based screening of root hair-specific genes and their functional characterization in Arabidopsis. Plant Physiol. 150: 1459–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi K., Menand B., Bell E., Dolan L. (2010). A basic helix-loop-helix transcription factor controls cell growth and size in root hairs. Nat. Genet. 42: 264–267. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.