Clade E Growth-Regulating (EGR) phosphatases and Microtubule-Associated Stress Protein 1 (MASP1) modulate cell expansion and drought response via control of microtubule stability and organization.
Abstract
Plant growth is coordinated with environmental factors, including water availability during times of drought. Microtubules influence cell expansion; however, the mechanisms by which environmental signals impinge upon microtubule organization and whether microtubule-related factors limit growth during drought remains unclear. We found that three Clade E Growth-Regulating (EGR) Type 2C protein phosphatases act as negative growth regulators to restrain growth during drought. Quantitative phosphoproteomics indicated that EGRs target cytoskeleton and plasma membrane-associated proteins. Of these, Microtubule-Associated Stress Protein 1 (MASP1), an uncharacterized protein, increased in abundance during stress treatment and could bind, bundle, and stabilize microtubules in vitro. MASP1 overexpression enhanced growth, in vivo microtubule stability, and recovery of microtubule organization during drought acclimation. These MASP1 functions in vivo were dependent on phosphorylation of a single serine. For all EGR and MASP1 mutants and transgenic lines examined, enhanced microtubule recovery and stability were associated with increased growth during drought stress. The EGR-MASP1 system selectively regulates microtubule recovery and stability to adjust plant growth and cell expansion in response to changing environmental conditions. Modification of EGR-MASP1 signaling may be useful to circumvent negative growth regulation limiting plant productivity. EGRs are likely to regulate additional proteins involved in microtubule stability and stress signaling.
INTRODUCTION
Drought and reduced soil water potential (ψw) decrease plant growth by restricting both cell expansion and cell division (Skirycz and Inzé, 2010; Tardieu et al., 2011). Drought is thus a major constraint on plant productivity (Boyer, 1982; Claeys and Inzé, 2013). Active downregulation of growth, particularly shoot growth, during drought can be beneficial to plant survival by limiting the leaf area for transpiration and thus allowing conservation of soil water. However, such downregulation can also decrease biomass production and yield more than is needed under moderate severity drought. Understanding growth regulation in general, as well as identifying factors that detect changes in water status and limit growth during drought, has the potential to improve crop biomass, productivity, and yield stability (Claeys and Inzé, 2013). Regulation of growth under moderate drought is distinct from mechanisms determining survival of severe water deprivation where no growth occurs (Skirycz et al., 2011; Verslues, 2016).
A major facet of growth regulation is the control of cell expansion. The extent of cell expansion depends on solute deposition to generate turgor, cell wall properties, mechanical cues from neighboring cells, as well as many types of intra- and extracellular signaling (Hamant et al., 2008; Sampathkumar et al., 2014; Feng et al., 2016). Cell shape and expansion are greatly influenced by events at the plasma membrane-cell wall interface, including vesicle trafficking and cortical microtubule (MT)-mediated control of cell wall deposition (Landrein and Hamant, 2013; McMichael et al., 2013; Lei et al., 2014, Endler et al., 2015; Feng et al., 2016). Disruption of cortical MT organization by oryzalin disrupts directional (anisotropic) cell expansion and causes isotropic cell swelling (Baskin et al., 1994; Sugimoto et al., 2003). Many mutants with altered MT organization or stability are impaired in anisotropic growth and have developmental defects including spiral, twisting growth patterns (Thitamadee et al., 2002; Shoji et al., 2004; Bannigan et al., 2006; Ambrose et al., 2007; Korolev et al., 2007; X. Wang et al., 2007; Buschmann et al., 2009; Nakamura and Hashimoto, 2009; Hamada et al., 2013; Liu et al., 2016). Cortical MTs may themselves act as sensors or transducers of mechanical signals that regulate growth (Landrein and Hamant, 2013; Nick, 2013).
There is a rapid (1 h or less) loss of cortical MT organization upon exposure to salt or osmotic stress (Komis et al., 2001; Dhonukshe et al., 2003; Ban et al., 2013; Fujita et al., 2013; Endler et al., 2015). For osmotic stress, loss of MT organization is mediated in large part by phosphorylation of α-tubulin, which blocks the polymerization of α-β tubulin dimers (Ban et al., 2013; Fujita et al., 2013). As minus end depolymerization and MT catastrophe events continue even though polymerization is inhibited, the result is a loss of MT organization. Salt stress-induced loss of MT organization also involves proteasome-dependent degradation of the MT plus end binding protein Spiral 1 (SPR1; Wang et al., 2011). For salt stress, there is evidence that subsequent recovery of MT organization is essential for acclimation and resumption of growth (C. Wang et al., 2007; Wang et al., 2011), and this recovery involves the microtubule-associated proteins Companion of Cellulose Snynthase1 (CC1) and CC2 (Endler et al., 2015). For low ψw and osmotic stress, the role of MT recovery in stress acclimation and growth is less clear. Phosphorylation of α-tubulin was observed under lethal, or near lethal, plasmolyzing osmotic treatments, which precluded observations of longer term recovery and acclimation. Loss of MT organization induced by α-tubulin phosphorylation may mainly be a protection mechanism to prevent cellular damage (Ban et al., 2013). Treatment with high molecular weight polyethylene glycol (PEG), which reduces ψw and mimics many aspects of soil drying while avoiding plasmolysis, also causes an initial loss of MT organization followed by later recovery (Mei et al., 2012). This later MT recovery has been little investigated and the role of MT in growth and cell expansion during drought is uncertain.
The plant hormones ethylene and abscisic acid (ABA) are potent regulators of plant growth, drought response, and microtubule organization. Increased ethylene production or increased ethylene response inhibits anisotropic cell elongation and induces isotropic radial swelling of the hypocotyl and roots (Guzmán and Ecker, 1990; Pierik et al., 2007). The effect of ethylene on cell expansion involves rapid and extensive rearrangements of MT organization (Roberts et al., 1985; Pierik et al., 2007; Polko et al., 2012; Ma et al., 2016). Drought stress can lead to increased ethylene production that, in combination with ABA accumulation, regulates root and shoot growth during drought (Spollen et al., 2000; LeNoble et al., 2004). Exogenous ABA treatment can also induce MT reorganization (Seung et al., 2013; Takatani et al., 2015), although whether such MT reorganization is involved in ABA effects on growth during drought is not as clear.
Protein phosphorylation-dephosphorylation is important for drought and ABA signaling as well as MT organization. Pharmacological studies found that inhibitors of Type 1 and Type 2A (PP2A) protein phosphatases, as well as kinase inhibitors, caused root swelling (loss of anisotropic cell expansion) and MT disorganization (Baskin and Wilson, 1997). Also, some plant MT-associated proteins (MAPs) are known to be phosphorylated. For example, phosphorylation of MAP65 family proteins changes their MT binding properties (Komis et al., 2011). An important aspect of drought-related signaling is the activity of Type 2C protein phosphatases (PP2Cs), especially the Clade A PP2Cs whose activity is regulated by interaction with Pyrabactin Resistant-Like/Regulatory Component of Abscisic Acid Receptor (PYL/RCAR) ABA receptors (Cutler et al., 2010; Fuchs et al., 2013). Interaction with PYLs is specific to Clade A PP2Cs, which account for only nine of the 80 PP2Cs present in Arabidopsis (Arabidopsis thaliana; Fuchs et al., 2013; Sugimoto et al., 2014). For most of the other PP2Cs in Clades B to F, there is little information on their physiological function or on the phosphorylation sites they regulate. Untargeted “shotgun” phosphoproteomics has been used to identify phosphopeptides whose abundance is rapidly altered in response to osmotic stress or exogenous ABA (Kline et al., 2010; Stecker et al., 2014; Minkoff et al., 2015) and to identify targets of the SnRK2 kinases (Umezawa et al., 2013; Wang et al., 2013). Quantitative phosphoproteomics is a promising method to identify PP2C-regulated phosphoproteins; however, we are not aware of any studies that have attempted such an analysis.
A search in our laboratory for genes affecting drought-regulated traits, including proline accumulation and plant growth, led us to characterize the function of three Clade E PP2Cs that we found to regulate plant growth and MT organization. Phosphoproteomic analysis identified putative targets of these PP2Cs, including a new MT binding protein that accumulated during low ψw stress and promoted MT stability and growth in a phosphorylation-specific manner. Our results uncover cellular functions of uncharacterized Clade E PP2Cs, identify regulators of MT organization, and demonstrate the importance of MT organization and stability in continued plant growth during drought stress.
RESULTS
The EGR Clade E PP2Cs Are Negative Regulators of Growth during Drought Stress
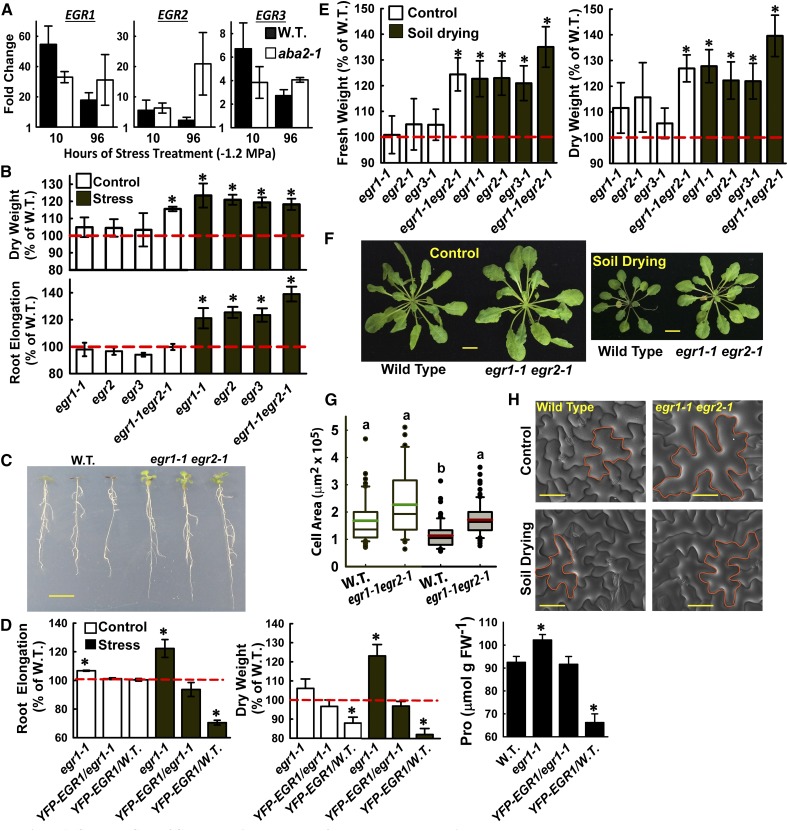
We hypothesized that some of the nearly 70 relatively uncharacterized Arabidopsis PP2Cs (Schweighofer et al., 2004; Fuchs et al., 2013) may function in drought signaling and identified three of the 13 Clade E PP2Cs as genes of particular interest (Supplemental Figure 1A; Schweighofer et al., 2004). These genes are hereafter referred to as Clade E-Growth-Regulating PP2C-1 (EGR1), EGR2, and EGR3. We focused on the EGRs in part because expression of EGR1 is correlated with the drought-regulated gene Δ1-pyrroline-5-carboxylate synthetase1 in the Plant Gene Expression Database (Horan et al., 2008) and EGR2 and EGR3 are close homologs of EGR1. EGR3 has also been recently identified as a putative leaf growth regulator by transcriptome analysis of stress- and circadian-regulated genes (Dubois et al., 2016). EGR expression was upregulated by low ψw in both the wild type and the ABA-deficient mutant aba2-1 (Figure 1A) but was less responsive to exogenous ABA (Supplemental Figures 1B and 1C), suggesting that EGR function may be distinct from ABA-regulated drought responses.
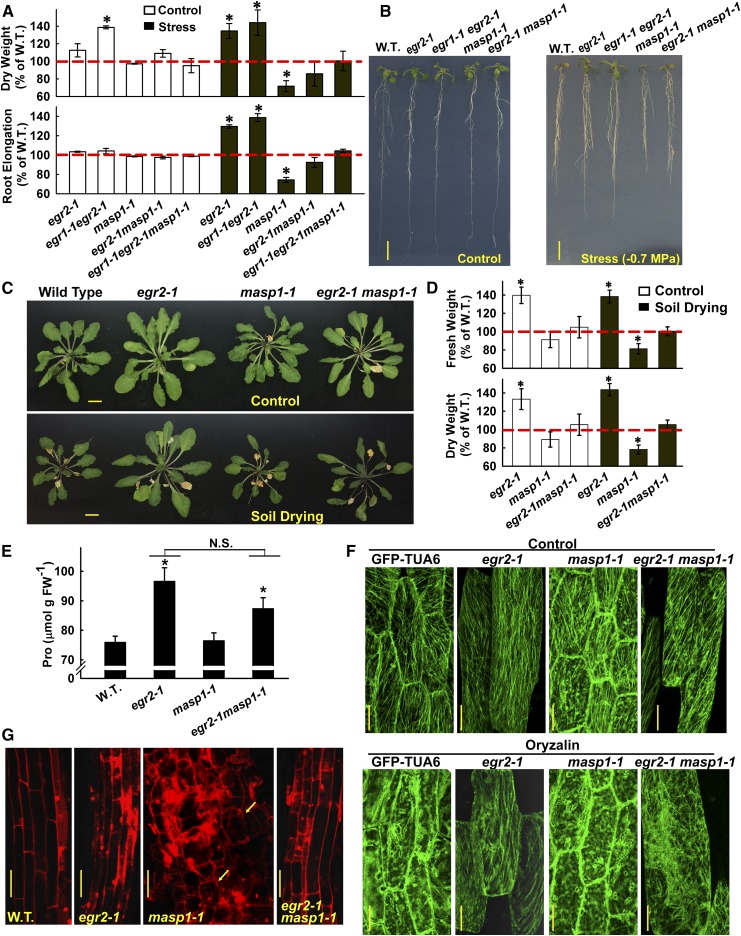
Figure 1.
Clade E EGR PP2Cs Are Negative Regulators of Growth and Cell Expansion.
(A) EGR expression at low ψw (−1.2 MPa) relative to the unstressed control for Col-0 (W.T.) and ABA-deficient mutant aba2-1. Data are means ± se, n = 6 from two independent experiments.
(B) Seedling dry weight (D.W.) and root elongation of the EGR mutant and overexpression lines in unstressed control conditions or 10 d after transfer to low ψw (−1.2 MPa). Data are relative to the wild type (mean ± se, n = 6 to 8 for seedling dry weight and n = 18 to 24 for root elongation, asterisk indicates significant difference compared with the wild type by one-sided t test [P ≤ 0.05]). Dashed red line indicates the wild-type level of growth (100%). Three or four seedlings were combined for each dry weigh measurement. egr2 and egr3 are combined data of two T-DNA alleles for each gene (Supplemental Figures 1A and 1D). Growth values of Col-0 wild type used for normalization are shown in Supplemental Figure 2.
(C) Representative seedlings of Col-0 wild type and egr1-1 egr2-1 after low ψw (−1.2 MPa) treatment. Five-day-old seedlings were transferred to low ψw and photographs taken 10 d later. Bar = 1 cm.
(D) Pro35S:YFP-EGR1 complements the increased growth and proline accumulation of egr1-1 and suppresses growth and proline in the wild type. Proline was measured 96 h after transfer to −1.2 MPa. Data are combined from two transgenic lines for both the egr1-1 and wild-type backgrounds and are means ± se (n = 4 to 12); asterisk indicates P ≤ 0.05 compared with the wild type. Dashed red line indicates the wild type level of growth (100%).
(E) Rosette fresh weight (F.W.) and dry weight for egr mutants in well-watered control plants or in plants subjected to partial soil drying. Data are expressed relative to the Col-0 wild type and are means ± se (n = 6 to 9) combined from two to three independent experiments. Two rosettes from plants grown in the same sector of the same pot were used for each fresh and dry weight measurement. Asterisks indicate significant difference compared with the wild type (P ≤ 0.05 by one-sided t test). Dashed red line indicates the wild-type level of growth (100%).
(F) Representative rosettes of the wild type and egr1-1 egr2-1 in control and soil drying treatments. Plants were 40 d old and were grown under short-day conditions. The soil drying treatment started at 18 d after planting and continued for 22 d with partial rewatering to control the extent of soil drying (see Methods for further details). Bars = 1 cm.
(G) Areas of epidermal pavement cells of leaf 6 from 38-d-old Col wild type and egr1-1 egr2-1 in control and soil drying treatments. Open boxes with green median lines show data for the unstressed control, and gray boxes with red median lines are stress treatment (black lines in each box indicate the mean, while box and whiskers indicate the 25 to 75 and 5 to 95 percentile ranges, respectively, and black circles show outlying data points). Data are means ± se (n = 40 to 80) combined from four to six plants. Lowercase letters above each box indicate significantly different groups (ANOVA on ranks, P ≤ 0.05; Supplemental Data Set 1).
(H) Representative scanning electron microscopy images with example epidermal pavement cells outlined in orange to illustrate the increased cell size in egr1-1 egr2-1, while retaining normal morphology. Bars = 50 μm.
We isolated T-DNA mutants for all three EGRs (Supplemental Figure 1D). Because EGR1 and EGR2 were strongly induced by stress and are the most closely related to each other in sequence (Supplemental Figure 1), we also generated an egr1-1 egr2-1 double mutant. Initial experiments showed that egr mutants had increased proline accumulation after transfer to PEG-infused agar plates adjusted to ψw representing conditions typical of mild to more severe, but not lethal, drought stress (−0.5 to −1.2 MPa; Supplemental Figure 1E). We further assayed seedling growth in unstressed control conditions (−0.25 MPa) and at low ψw (−1.2 MPa). Growth of all genotypes, including the wild type (Supplemental Figure 2), was inhibited by low ψw; however, egr mutants maintained higher root elongation and seedling dry weight (Figures 1B and 1C). egr1-1 egr2-1 also had significantly increased growth in the unstressed control condition (Figure 1B). For EGR2 and EGR3, two T-DNA alleles had essentially identical effects on growth (Figure 1B; egr2 and egr3 are combined data of the two alleles for each gene) and proline accumulation (Supplemental Figure 1E). We were unable to obtain multiple T-DNA alleles for EGR1; however, Pro35S:YFP-EGR1 complemented egr1-1 (Figure 1D). The complementation and similar phenotypes of all egr mutants indicated that the egr1-1 T-DNA insertion in the 5′ UTR of EGR1 likely blocks protein translation even though some mRNA is still produced in the mutant (Supplemental Figure 1D). Conversely, overexpression of EGR1 (in the Col-0 wild-type background) decreased growth and proline accumulation (Figure 1D). Together, these data indicate that EGRs act as negative regulators of several drought-related phenotypes.
To show that the egr growth phenotypes extended across different developmental stages and methods of low ψw imposition, 18-d-old wild-type and egr mutant plants were exposed to partial soil drying followed by quantification of rosette fresh and dry weight. The soil drying treatment, with partial rewatering to ensure that all plants were exposed to the same level of moderate soil drying stress, lasted 20 d and at the end of this period rosette fresh and dry weight of the stress-treated wild type was decreased by nearly 70% compared with the well-watered control. Because the degree of soil drying was maintained at a moderate level, there was little wilting or decrease in leaf relative water content for any of the genotypes assayed. Consistent with the PEG plate assays, egr mutants had increased rosette fresh and dry weight in the soil drying treatment. egr1-1 egr2-1 had the strongest effect and also had increased growth in the well-watered control (Figures 1E and 1F). Scanning electron microscopy showed that egr1-1 egr2-1 plants had a larger average size of epidermal pavement cells compared with the wild type under water stress conditions while still maintaining normal leaf morphology (Figures 1G and 1H; Supplemental Data Set 1). Thus, the increased growth of egr mutants was, at least in part, due to greater cell expansion and ability to maintain cell expansion despite water limitation. These data further demonstrated the role of EGRs in restricting growth.
Putative EGR Targets Identified by Quantitative Phosphoproteomics Are Enriched in Cytoskeleton- and Plasma Membrane-Associated Proteins
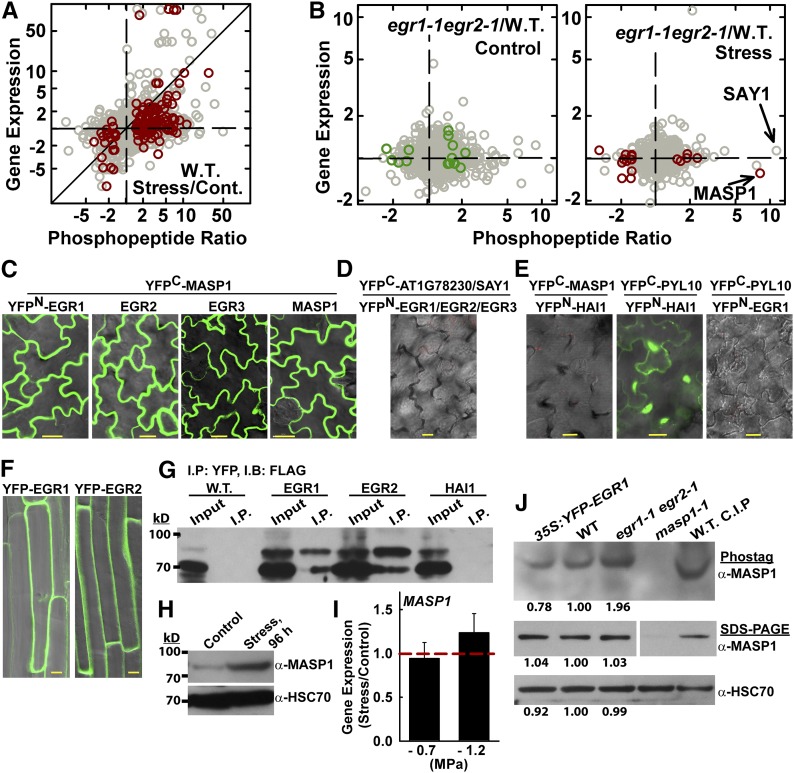
To identify targets of EGR regulation, we performed quantitative phosphoproteome and transcriptome analyses of the wild type and egr1-1 egr2-1 under control or low ψw (−1.2 MPa, 96 h) treatments. The gene expression data were mainly used as a companion for the phosphoproteomics data to compare proteins with stress-induced increase or decrease in phosphopeptide abundance to genes transcriptionally regulated by low ψw. Our phosphoproteomic analysis was unique compared with recent studies (Kline et al., 2010; Umezawa et al., 2013; Wang et al., 2013; Stecker et al., 2014; Minkoff et al., 2015) in that we used longer term (96 h; Supplemental Figure 3A) low ψw stress rather than short-term ABA or dehydration treatment. This was done primarily because EGR expression, as well as that of many other phosphatases and kinases (Bhaskara et al., 2012), was induced by such longer term stress treatment and because egr growth and proline phenotypes were observed in longer term low ψw treatment (Figure 1; Supplemental Figure 1). Our analysis identified more than 1500 phosphoproteins (Supplemental Figure 3B and Supplemental Data Sets 2 to 4), many of which were not previously listed in the PhosPhat database (Supplemental Figure 3C). In the wild type, 119 phosphopeptides were significantly more abundant at low ψw compared with the control condition, while 23 were significantly less abundant (Figure 2A; Supplemental Data Sets 5 and 6). Many of these proteins were from genes whose expression was unaffected by low ψw (Figure 2A; Supplemental Data Sets 5 and 6) and were not identified in previous phosphoproteomic studies (Supplemental Figure 3D and Supplemental Data Sets 4 and 7). Thus, our phosphoproteomic data revealed aspects of the low ψw response that could not be inferred from transcriptome data or previously existing phosphoproteomic data. However, it should be kept in mind that the longer term low ψw treatment used in our study allowed ample time for protein abundance to change. Thus, differences in phosphopeptide abundance could reflect a stress-induced difference in phosphorylation stoichiometry or a change in protein abundance. Both are of interest and distinct from transcriptional regulation; however, further experiments will be needed to distinguish between these two possibilities.
Figure 2.
Phosphoproteomics Analysis of Wild-Type and egr1-1 egr2-1 Plants Reveals Distinct Drought Effects on the Phosphoproteome and Identifies MASP1, an EGR-Interacting Phosphoprotein.
(A) Phosphopeptide abundance ratio versus gene expression for wild type (W.T.) stress versus control. Dark-red symbols indicate significant changes in phosphopeptide abundance (P ≤ 0.05 by one-sided t test and fold change ≥1.5). All other phosphopeptide data are plotted using gray symbols. Diagonal line indicates identical change in phosphopeptide abundance and gene expression. Data are fold change in phosphopeptide abundance (as indicated in the tick labels) plotted on a logarithmic scale. For the tick labels, ratios less than one were inverted and shown as negative fold change for clarity of presentation.
(B) Phosphopeptide abundance versus gene expression for egr1-1 egr2-1 versus the wild type in control and stress treatments. Format of data presentation is as described for (A). Dark red or green symbols indicate phosphopeptides with P ≤ 0.05 by one-sided t test and fold change ≥1.5 (with addition of MASP1, which has P = 0.07).
(C) BiFC interaction of MASP1 with EGRs and with itself in transient expression assays using intact Arabidopsis seedlings. Images of leaf epidermal pavement cells of unstressed seedlings are shown. Essentially identical results were seen in stress-treated (−1.2 MPa) seedlings. Bars = 20 μm.
(D) Representative image showing lack of interaction from BiFC analysis of EGRs with AT1G78320, a close homolog of MASP1, which lacks the MASP1 phosphorylation site (Supplemental Figures 3E and 3F and Supplemental Data Set 11) and SAY1 (Fig. 2B). Bars = 20 μm.
(E) Representative images showing the lack of BiFC fluorescence signal from the Clade A PP2C Highly ABA-Induced 1 (HAI1; AT5G59220) and MASP1 as well as EGR1 and PYL10 (identical results were seen for EGR2 and EGR3). The HAI1-PYL10 interaction was used as a positive control. Bars = 20 μm.
(F) Localization of YFP-EGR1 and YFP-EGR2 in stable transgenic plants. Cells in the root maturation zone of unstressed 11-d-old seedlings are shown. Similar localization was observed at low ψw. Bars = 10 μm.
(G) Coimmunoprecipitation of EGR1, EGR2, and MASP1. HAI1 and the uninfiltrated Avr-PTO line (W.T.) were used as a negative controls. YFP-tagged EGR1, EGR2, or HAI1 was transiently expressed along with FLAG-MASP1 and immunoprecipitation (I.P.) was performed with GFP-Trap resin to capture the YFP-tagged phosphatase. Immunoblot (I.B.) of the total protein extract (input) and immunoprecipitated proteins was performed using FLAG antisera. The experiment was repeated with similar results.
(H) Immunoblot using MASP1-specific antisera to detect endogenous MASP1 in wild-type seedlings under control or stress (−1.2 MPa, 96 h) conditions shows induction of MASP1 protein level at low ψw (100 μg of total protein was loaded per lane). Replicate blots were probed with HSC70 as a loading control.
(I) Quantitative RT-PCR analysis shows no increase of MASP1 expression in seedlings transferred to either −0.7 or −1.2 MPa low ψw stress for 96 h compared with unstressed plants. Data are means ± se (n = 6). Dashed red line indicates the level of expression in unstressed seedlings.
(J) Phos-tag gel analysis of MASP1 in stress-treated (−1.2 MPa, 96 h) seedlings. The identity of the phosphorylated MASP1 band was confirmed by analysis of masp1-1 and by treating the wild-type sample with calf intestinal phosphatase (C.I.P.). The same samples were also run on SDS-PAGE gels to assess MASP1 total protein level as well as HSC70 as a loading control (100 μg of protein was loaded per lane for both Phos-tag and regular SDS-PAGE). Band intensities relative to the wild type are shown for egr1-1 egr2-1 and Pro35S:YFP-EGR1 lines. For the MASP1 SDS-PAGE, all lanes are from the same blot, but intervening space was removed and part of the blot rotated to show the lanes in the same order as the other blots.
The egr1-1 egr2-1 phosphoproteomic data identified putative targets of EGR regulation that were consistent with EGR localization at the cell periphery and suggested a cytoskeleton-related function for the EGRs. Phosphopeptide abundances in egr1-1 egr2-1 were compared with those of the wild type for both the control and stress treatments (Supplemental Data Set 8). From this data set we identified 17 proteins with increased phosphopeptide abundance in egr1-1 egr2-1 and thus likely to represent targets of EGR regulation (Table 1, Figure 2B). Most of these proteins were encoded by genes whose expression was not affected by low ψw or egr1-1 egr2-1 (Figure 2B; Supplemental Data Set 8). Within these 17 proteins, there was a striking enrichment of cytoskeleton- and plasma membrane-associated proteins. This included the MT-associated proteins Basic Proline-rich Protein (BPP1), IQ Domain 32 (IQD32) (Hamada et al., 2013), and Nodulin/Glutamine Synthase-Like (NodGS) (Doskočilová et al., 2011) as well as the microfilament-associated protein Villin2 (VLN2) (van der Honing et al., 2012). Several sterile alpha motif (SAM) family proteins were also identified as putative MT-associated proteins (Hamada et al., 2013), suggesting that the SAM family member in Table 1 may also have MT-related function. Also, a rice (Oryza sativa) protein having Domain of Unknown Function (DUF) 966 similar to AT5G59790 enhanced stress tolerance and was localized around the cell periphery in a pattern similar to the EGRs (Luo et al., 2014); AT5G63640 may be involved in vesicle transport (ENTH/VHS/GAT family proteins are associated with vesicle transport, although no specific information exists for AT5G63640); and, AT3G01810, a protein of unknown function, has been previously identified as a plasma membrane phosphoprotein (Benschop et al., 2007). The match between localization of these proteins and EGR localization along the cell periphery (Figure 2F; see below) was a first indication that we had successfully identified EGR target proteins. There were also a number of other cytoskeleton- and trafficking-related proteins with marginally nonsignificant increases in phosphopeptide abundance in egr1-1 egr2-1 (Supplemental Data Set 8). These included MT-associated protein SUO (“shuttle” in Chinese; Hamada et al., 2013), microfilament-associated protein Altered Response to Gravity 1 (ARG1) (Harrison and Masson, 2008), as well as vesicle transport associated protein Stomatal Cytokinesis Defective 2 (SCD2) (McMichael et al., 2013). However, phosphopeptide abundances for these proteins were more variable (P = 0.06 or 0.09), and they must be considered with caution. There were also a number of proteins with significantly decreased phosphopeptide abundance in egr1-1 egr2-1 (Figure 2B; Supplemental Data Set 8). These may represent indirect effects of EGR signaling on phosphorylation or decreased abundance of these proteins in egr1-1 egr2-1.
Table 1. Proteins with Increased Phosphopeptide Abundance in egr1-1 egr2-1 Relative to the Wild Type.
| Protein ID | Phosphopeptide Ratio WT Stress/WT Control | P Value (WT) | Phosphopeptide Ratio egr1-1 egr2-1 Control/WT Control | P Value (Cont.) | Phosphopeptide Ratio egr1-1 egr2-1 Stress/WT Stress | P Value (Stress) | Description |
|---|---|---|---|---|---|---|---|
| Increased phosphopeptide abundance in egr1-1 egr2-1 in stress treatment | |||||||
| AT4G03260 | 4.4 | 0.13 | 1.8 | 0.20 | 8.2 | 0.07 | MASP1 |
| AT5G59790 | 1.0 | 1.00 | 1.2 | 0.70 | 2.3 | 0.02 | Unknown (DUF 966) |
| AT2G41740 | 1.5 | 0.58 | 1.5 | 0.45 | 1.9 | 0.04 | Villin 2 (VLN2) |
| AT1G65030 | 1.2 | 0.76 | 2.6 | 0.27 | 1.9 | 0.03 | DWD motif, part of CUL4-based E3 ubiquitin ligase |
| AT1G71710 | 1.1 | 0.93 | −1.4 | 0.58 | 1.7 | 0.01 | Inositol-polyphosphate 5-phosphatase |
| AT5G63640 | 1.1 | 0.30 | 1.1 | 0.73 | 1.6 | 0.03 | ENTH/VHS/GAT family |
| Increased phosphopeptide abundance in egr2-1 egr2-1 in control | |||||||
| AT1G70180 | 6.6 | 0.07 | 2.2 | 0.05 | −1.2 | 0.65 | Sterile alpha motif (SAM) |
| AT2G40070 | 2.1 | 0.29 | 2.0 | 0.01 | −1.3 | 0.41 | Basic proline-rich protein (BPP1) |
| AT3G53180 | 4.8 | 0.04 | 1.7 | 0.05 | 1.3 | 0.43 | Nodulin/Glutamine Synthase-Like (NodGS) |
| AT3G01810 | 1.4 | 0.56 | 1.7 | 0.02 | −1.4 | 0.27 | Unknown protein |
| AT2G32060 | 1.2 | 0.56 | 1.7 | 0.03 | 1.3 | 0.54 | Ribosomal protein L7Ae/L30e/S12e/Gadd45 |
| AT5G38600 | 1.1 | 0.89 | 1.6 | 0.01 | 1.0 | 0.83 | Proline-rich spliceosome-associated (PSP) protein |
| AT1G19870 | 1.6 | 0.10 | 1.6 | 0.04 | 1.1 | 0.75 | IQ-domain 32 (IQD32) |
| AT5G52310 | 14.1 | 0.07 | 1.5 | 0.05 | 1.3 | 0.43 | Responsive to Desiccation29A (RD29A) |
| AT3G46780 | 1.2 | 0.35 | 1.5 | 0.01 | 1.0 | 0.97 | Plastid transcriptionally active 16 (PTAC16) |
| AT3G47070 | −2.9 | 0.27 | 1.5 | 0.05 | 1.1 | 0.67 | Thylakoid soluble phosphoprotein TSP9 |
| AT1G17210 | 2.8 | 0.16 | 1.5 | 0.01 | 1.3 | 0.01 | IAP-like protein 1 (ILP1), phragmoplast associated |
A 1.5-fold increase and P value ≤ 0.05 (by one-sided t test) were the criteria to define putative EGR-regulated phosphoproteins (MASP1 is also included based on our further characterization of it). Significant fold increases are indicated by bold numbers. Additional data on phosphopeptide sequences as well as accompanying gene expression data can be found in Supplemental Data Set 8. WT, wild type.
The EGR transcriptome data showed a relatively limited effect of egr1-1 egr2-1 on gene expression (Supplemental Data Sets 9 and 10) compared with the extensive stress-induced transcriptional changes seen in the wild type and mutant of the Clade A PP2C HAI1 under the same stress treatment (Bhaskara et al., 2012). This seemed consistent with the cell periphery localization of EGRs and, along with the phosphoproteomic data, suggested that EGRs are not primarily regulators of transcription. Interestingly though, among the genes with altered expression in egr1-1 egr2-1, some were cytoskeleton- or trafficking-related, including a Spiral1-like gene (SPL4) as well as another ENTH/VHS family member and SNARE domain protein and two stress-related genes, C-repeat Binding Factor1 (CBF1) and Salt-Induced Serine Rich Protein (SIS1) and membrane signaling proteins.
MASP1 Is an EGR-Associated Phosphoprotein
Of the putative EGR targets (Table 1), AT4G03260, a protein of unknown function that we named Microtubule-Associated Stress Protein 1 (MASP1) stood out, as its phosphopeptide abundance (with phosphorylation at Ser-670) was dramatically increased in egr1-1 egr2-1 at low ψw (Table 1, Figure 2B; Supplemental Data Set 8). The same MASP1 phosphopeptide we identified was also found to be serine and threonine (Thr-673) phosphorylated in a previous study (Umezawa et al., 2013). However, the results of that study could not unambiguously assign the serine phosphorylation to Ser-668 or Ser-670 and did not find a change in MASP1 phosphopeptide abundance after 30 to 90 min of dehydration or ABA treatment. Although the current annotation of MASP1 lists it as a dynein light chain-like protein, its function has not been investigated and we found no similarity of MASP1 to dyneins or other MT binding proteins except for two short (five to six amino acids) regions of homology to Auxin-Induced in Root Cultures 9 (AIR9; Supplemental Figure 3E and Supplemental Data Set 11). The rest of the AIR9 protein is highly divergent from MASP1, and its reported function (Buschmann et al., 2006) differs substantially from our MASP1 results.
Bimolecular fluorescence complementation (BiFC) assays demonstrated that MASP1 associated with EGRs in a distinctive pattern around the cell periphery (Figure 2C), which matched EGR localization (Figure 2F) and the self-association of MASP1 (Figure 2C). As a control, the same BiFC analysis was performed using AT1G78230, which is the closest MASP1 homolog but lacks the MASP1 Ser-670 phosphorylation site (Supplemental Figures 3E and 3F). No interaction was observed between EGRs and AT1G78230 (Figure 2D). We also did not observe interaction of EGRs with SAY1, which had the largest fold increase in phosphopeptide abundance in egr1-1 egr2-1 (Figure 2B) but was variable and did not pass the statistical cutoff. MASP1 also did not interact with the clade A PP2C HAI1, while the known interaction of HAI1 with PYL10 was readily observed (Figure 2E). We also did not observe interaction of EGRs with PYL10. Consistent with the BiFC data, YFP-tagged EGR1 or EGR2 transiently expressed in Arabidopsis seedlings could coimmunoprecipitate FLAG-MASP1, while YFP-HAI1 could not (Figure 2G). Together these data demonstrated that the EGRs specifically interact with MASP1.
Immunoblotting using MASP1-specific antisera showed that MASP1 protein abundance was low and virtually undetectable in unstressed plants, but increased dramatically at low ψw (Figure 2H; Supplemental Figure 4C) despite MASP1 gene expression being unaffected by low ψw (Figure 2B; Supplemental Data Set 8). Quantitative RT-PCR analysis validated the microarray data and found no induction of MASP1 gene expression at low ψw (Figure 2I). Phos-tag gel analysis found that egr1-2 egr2-1 had increased abundance of phosphorylated MASP1, consistent with the phosphoproteomics data, albeit at a lesser fold change (Figure 2J). Conversely, EGR1 overexpression decreased the abundance of phosphorylated MASP1. MASP1 total protein level, detected by immunoblotting after conventional SDS-PAGE, did not differ between the wild type, egr1-1 egr2-1, and Pro35S:YFP-EGR1 (Figure 2J). This suggested that the increased phosphorylated MASP1 seen by Phos-tag gel analysis of egr1-1 egr2-1 was caused by reduced MASP1 dephosphorylation (or increased phosphorylation), while EGR1 overexpression led to increased MASP1 dephosphorylation (or reduced MASP1 phosphorylation). However, we were unable to reproducibly detect the nonphosphorylated band of MASP1 in plant extracts and thus could not quantitatively access the MASP1 phosphorylation stoichiometry. Note that the Phos-tag gel analysis was only conducted for samples from low ψw-treated plants as it was not possible to detect the low level of MASP1 expressed in unstressed plants. In the wild type, taking the immunoblot and proteomics data together, the strong increase in MASP1 protein abundance at low ψw indicated that the putative (P = 0.13) 4-fold stress-induced increase in MASP1 phosphopeptide abundance in the wild type (Table 1) may reflect increased MASP1 protein abundance (and could actually indicate dephosphorylation of MASP1 at low ψw, as MASP1 protein abundance increased by more than 4-fold). Together, the data indicated that EGRs affect the abundance of phosphorylated MASP1 and also showed that MASP1 is a low ψw-responsive protein.
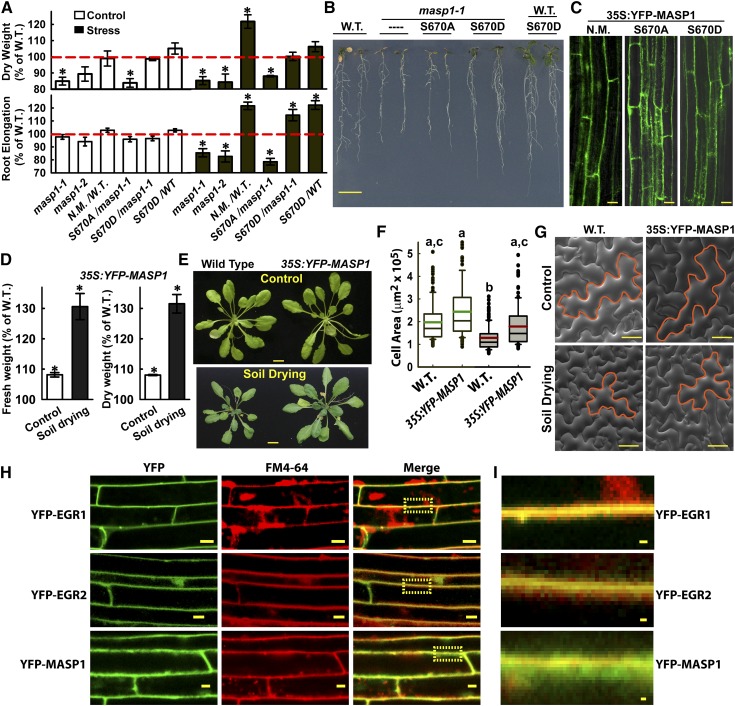
MASP1 Promotes Growth in a Manner That Depends upon Ser-670 Phosphorylation
Two T-DNA alleles of masp1 were isolated and shown to either lack MASP1 gene and protein expression (masp1-1) or express a truncated, presumably nonfunctional, MASP1 protein (masp1-2; Supplemental Figures 4A to 4C). Both alleles of masp1 had decreased growth (dry weight) in control and low ψw treatments and also had decreased root elongation at low ψw (Figures 3A and 3B). Conversely, MASP1 overexpression increased growth at low ψw (Figures 3A and 3B). Overexpression of phosphonull MASP1 (MASP1S670A) could not complement the reduced growth of masp1-1, while phosphomimic MASP1 (MASP1S670D) complemented masp1-1 and increased growth of the wild type at low ψw (Figures 3A and 3B). MASP1S670A and MASP1S670D were localized in the same pattern as nonmutated MASP1 and expressed at similar levels (Figure 3C). Overexpression of MASP1 was also highly effective at promoting growth during soil drying (Figures 3D and 3E). MASP1 overexpression increased leaf epidermal cell size (Figures 3F and 3G; Supplemental Data Set 12), similar to the phenotype of egr1-1 egr2-1 (Figures 1G and 1H).
Figure 3.
MASP1 Promotes Growth in a Phosphorylation-Dependent Manner and Is Localized in the Cell Cortex.
(A) Dry weight and root elongation of seedlings under control and stress (−1.2 MPa) conditions. Genotypes used were masp1-1 and masp1-2 as well as Pro35S:YFP-MASP1 expressed in the wild type (W.T.) or masp1-1. MASP1 alleles used in transgenic plants: N.M., no mutation wild-type MASP1; S670A, phosphonull MASP1; S670D, phosphomimic MASP1. Data are relative to the Col-0 wild type (mean ± se, n = 6 to 8, asterisk indicates significant difference compared with the wild type by one-sided t test [P ≤ 0.05]). Growth values of Col-0 wild type used for normalization are shown in Supplemental Figure 2. Dashed red line indicates the wild type level of growth (100%).
(B) Representative seedlings of Col-0 wild type (W.T.), masp1-1, masp1-1 transformed with MASP1S670A (phosphonull) or MASP1S670D (phosphomimic), and wild type transformed with MASP1S670D. Seedlings shown were subjected to −0.7 MPa low ψw treatment where MASP1-mediated effects on growth were similar or greater than the −1.2 MPa experiments shown in (A). Bar = 1 cm.
(C) Images of root cells showing that YFP-MASP1 unmutated (N.M.), phosphonull MASP1 (S670A), and phosphomimic MASP1 (S670D) all localized along the cell periphery and were expressed at a similar level. Bars = 20 μm.
(D) Relative rosette fresh weight and dry weight of Pro35S:YFP-MASP1 (unmutated, expressed in the Col-0 wild-type background) transgenic plants compared with the Col-0 wild type in control or soil drying treatments (mean ± se, n = 6 to 8, *P ≤ 0.05 by one-sided t test). For the Col-0 wild type, rosette fresh weight and dry weight were nearly 70% lower in the soil drying treatment relative to the well-watered control. The soil drying experiments and presentation of data are the same as described in Figures 1E and 1F.
(E) Representative rosettes of the wild type and Pro35S:YFP-MASP1 (unmutated, expressed in the Col-0 wild-type background) in the unstressed control and soil drying treatments. Bars = 1 cm.
(F) Areas of epidermal pavement cells of leaf 6 in Col wild type and Pro35S:YFP-MASP1 in control and soil drying treatments. Open boxes with green median lines show data for the unstressed control and gray boxes with red median lines are stress treatment (black lines in each box indicate the mean, while box and whiskers indicate the 25 to 75 and 5 to 95 percentile ranges, respectively, and black circles show outlying data points). Data are means ± se (n = 40 to 80) combined from four to six plants. Lowercase letters above each box indicate significantly different groups (ANOVA on ranks, P ≤ 0.05; Supplemental Data Set 12).
(G) Representative scanning electron microscopy images with example epidermal pavement cells outlined in orange to illustrate the increased size, but normal morphology, of Pro35S:YFP-MASP1 cells. Bars = 50 μm.
(H) Transgenic plants with stable expression of Pro35S:YFP-EGR1, EGR2, or MASP1 were incubated with FM4-64 to test for colocalization with the plasma membrane. Cells in the root elongation zone are shown. Yellow boxes in the merged images show the area that is enlarged in (I). Bars = 10 μm
(I) Enlargement of the yellow boxed areas in (H). Bars = 1 μm.
EGR localization and the EGR-MASP1 BiFC interaction were confined to the cell periphery, perhaps along the plasma membrane. However, we noted that the MASP1 localization pattern was more diffuse (cf. Figures 2F and 3C). Colocalization with FM4-64 was used to further compare the EGR and MASP1 localization and determine if they may be associated with the plasma membrane. EGR1 and EGR2 nearly completely colocalized with FM4-64 along the plasma membrane (Figures 3H and 3I), consistent with the presence of a hydrophobic N terminus in all three EGRs. MASP1 had partial colocalization with FM4-64 with a substantial portion of MASP1 being in the cell cortex just beneath the plasma membrane (Figures 3H and 3I). This was consistent with the interaction of EGRs and MASP1, but also indicated that MASP1 was not localized solely along the plasma membrane and may function in the cell cortex.
EGRs and Phosphorylated MASP1 Promote MT Recovery in Plants Acclimated to Low ψw
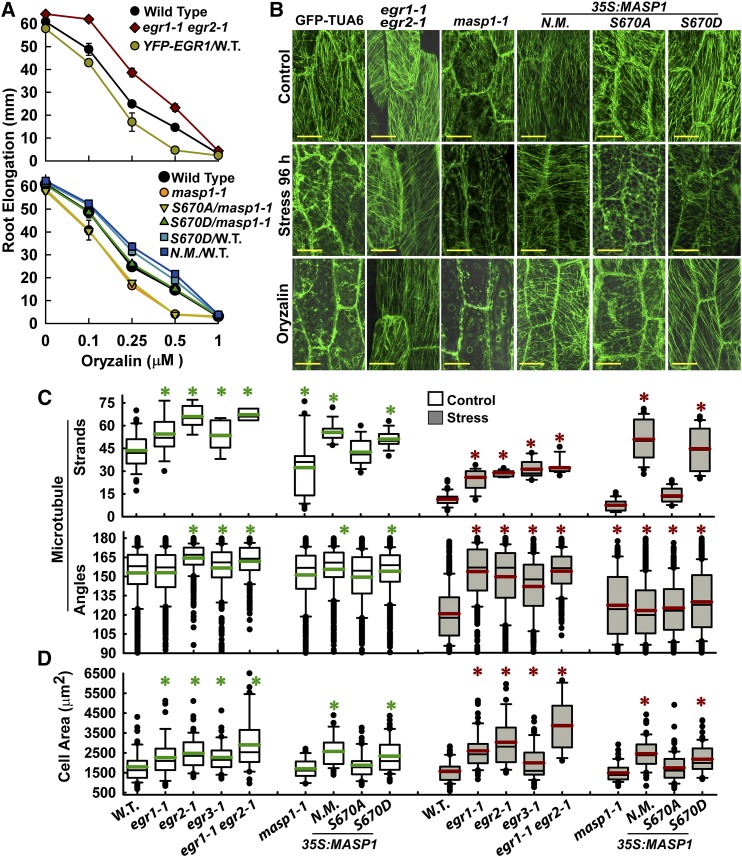
The enrichment of cytoskeleton-related proteins among putative EGR targets identified by phosphoproteomics (Table 1) indicated that EGRs and MASP1 may have roles in MT regulation. Consistent with this hypothesis, egr1-1 egr2-1 was more resistant to root growth inhibition by the MT-destabilizing drug oryzalin, while EGR1 overexpression decreased oryzalin resistance (Figure 4A). Conversely, masp1-1 was more sensitive to oryzalin than was the wild type and this sensitivity could be complemented by MASP1S670D phosphomimic but not MASP1S670A phosphonull. Overexpression of either unmutated MASP1 or MASP1S670D phosphomimic in the wild type increased oryzalin resistance (Figure 4A). Comparing the stress and oryzalin experiments, greater oryzalin resistance was consistently associated with enhanced growth at low ψw, while oryzalin sensitivity was associated with reduced growth at low ψw for all the EGR and MASP1 mutants and transgenic lines tested (cf. Figure 4A to Figures 1B, 1D, 3A, and 3B). This indicated that altered MT stability could be a key factor underlying the EGR and MASP1 growth phenotypes.
Figure 4.
egr Mutants and Plants Expressing MASP1 or Phosphomimic MASP1 Exhibit Enhanced Recovery of MT Organization at Low ψw and Also Have Increased Cell Size.
(A) Sensitivity of root elongation to various concentrations of oryzalin in egr1-1 egr2-1 and masp1 mutants as well as unmutated MASP1 (N.M.), MASP1 S670D (phosphomimic), and MASP1S670A (phosphonull) transformed into Col-0 wild type (W.T.) or masp1-1 (transgenic lines are the same as those shown in Figures 1 and 3). Four-day-old seedlings were transferred to plates containing the indicated oryzalin concentrations, and root elongation was measured over the subsequent 7 d. Data are means ± se, n = 6 to 25. All mutants or transgenic lines except for MASP1S670D/masp1-1 were significantly different from the wild type (P ≤ 0.05 by t test) in the 0.25 and 0.5 μM oryzalin treatments.
(B) Representative images of MT organization in hypocotyl cells at the base of the elongation zone visualized using GFP fused to Arabidopsis α-Tubulin6 (GFP-TUA6). Eleven-day-old seedlings were used for imaging. Stress = −1.2 MPa, 96 h. Oryzalin = 10 μM for 45 min. Bars = 20 μm.
(C) Quantification of microtubule strands per cell and microtubule angle distribution. Data are mean ± se, n = 10 to 50 for microtubule strands, n = 100 to 300 for angles, and asterisk indicates P ≤ 0.05 (by t test) compared with GFP-TUA6. Black lines in the boxes are the mean, and green or red lines are the median. Box and whiskers indicate the 25 to 75 and 5 to 95 percentile ranges, respectively, while black circles are outliers.
(D) Cell area of hypocotyl cells analyzed in (C). Data are means ± se, n = 20 to 60, asterisk indicates P ≤ 0.05 (by t test) compared with GFP-TUA6 (W.T.). The graph is formatted the same as in (C).
MT organization was directly examined using a GFP-TUA6 reporter line (Ueda et al., 1999), which was crossed with egr and masp1 mutants or directly transformed with MASP1 overexpression constructs. Expression of Pro35S:MASP1 in the GFP-TUA background led to the same stress growth phenotypes as in the wild-type Col-0 background (Supplemental Figure 4D). Consistent with previous studies (Mei et al., 2012; Fujita et al., 2013), we observed that transfer to low ψw caused disorganization of cortical MT arrays within a few hours (Supplemental Figure 5A), followed by partial recovery of MT organization at 72 h (Supplemental Figure 5A) and 96 h (Figure 4B). egr mutants were more resistant to loss of MT organization in the first few hours of stress treatment compared with the wild type; however, both egr mutants and the wild type had near complete loss of MT organization by 12 h after transfer to low ψw (Supplemental Figure 5A). As the plants acclimated to low ψw stress over longer times (72 and 96 h), it became clear that the egr mutants had more extensive MT recovery. This could be seen as both a greater number of visible MT strands as well as MT angle distribution closer to that observed in unstressed cells (Figures 4B and 4C; Supplemental Figure 5A). A similar, but less dramatic, effect of increased number of MT strands was observed in unstressed egr mutants (Figure 4C). Conversely, unstressed masp1-1 had less extensive cortical MT arrays and substantial cell-to-cell variability in MT number and arrangement (Figures 4B and 4C). After 96 h at low ψw, it was clear that masp1-1 was greatly impaired in its ability to recover MT organization (Figures 4B and 4C). Also, it was consistently observed that masp1-1 lacked thicker strands indicative of MT bundles, while egr1-1 egr2-1 had longer and thicker, bundle-like MT strands (Figure 4B).
The effect of MASP1 on MT organization depended on Ser-670 phosphorylation. Overexpression of unmutated MASP1 or MASP1S670D phosphomimic enhanced the recovery of MT strands after 96 h low ψw treatment (Figures 4B and 4C; Supplemental Figure 5B). The effect was similar to that of egr1-1 egr2-1 except that MASP1 overexpression did not cause the same change in MT angle distribution (Figure 4C). In contrast, MASP1S670A phosphonull had no significant effect on MT recovery at low ψw (Figure 4C). Consistent with the oryzalin root elongation assays, egr1-1 egr2-1 as well as overexpression of MASP1 or MASP1S670D phosphomimic stabilized MT against oryzalin-induced disorganization, while MASP1S670A phosphonull was less effective (Figure 4B).
As MT changes can affect cell expansion and anisotropic growth, hypocotyl cell size was also measured. Increased hypocotyl cell size was seen egr1-1 egr2-1 as well as in lines overexpressing unmutated MASP1 or MASP1S670D phosphomimic, but not MASP1S670A phosphonull (Figure 4D). This occurred while maintaining growth anisotropy (Supplemental Figure 6). Interestingly, we did not see any developmental defects in our mutants or transgenic lines. Propidium iodide (PI) staining of root cells confirmed that none of our lines had the spiral patterns of cell elongation seen in other mutants affecting MT organization (Supplemental Figure 7). In the PI staining, it could be seen that loss of MASP1 expression, and to a lesser extent EGR1 overexpression, led to isotropic swelling of cells in the elongation zone of plants at low ψw (Supplemental Figure 7). Expression of MASP1S670A phosphonull did partially complement the cell swelling of masp1-1 at low ψw but was not as effective as MASP1S670D phosphomimic.
EGRs Act Upstream of MASP1 in Growth Regulation
The similar but opposing growth phenotypes of egr and masp1 mutants as well as EGR effect on MASP1 phosphoprotein abundance indicated that EGRs may act upstream of MASP1 in the same pathway to control growth. To genetically test this hypothesis, we generated an egr2-1 masp1-1 double mutant. Growth assays indicated that egr2-1 masp1-1 was phenotypically similar to masp1-1, albeit with partial recovery of growth compared with masp1-1 (Figures 5A and 5B). Likewise, in soil drying experiments, egr2-1 masp1-1 had a partial recovery of growth but was overall similar in phenotype to masp1-1 (Figures 5C and 5D). Interestingly, proline accumulation in egr2-1 masp1-1 did not significantly differ from that of egr2-1 (Figure 5E). This was consistent with our finding that masp1-1 or MASP1 overexpression had no effect on proline accumulation at low ψw (Supplemental Figure 4E) and indicated that EGRs affect proline via a different pathway that does not involve MASP1. As the egr2-1 masp1-1 mutant was constructed in the GFP-TUA6 background, we also examined the MT response to oryzalin and found that egr2-1 masp1-1 had partial recovery of MT stability compared with egr2-1 (Figure 5F). Overall, these data were consistent with MASP1 acting downstream of and being required for EGR promotion of growth, while also showing that EGRs may regulate additional proteins involved in stress signaling and MT stability. It was also interesting that the cell swelling seen in masp1-1 at low ψw was largely recovered in egr2-1 masp1-1 (Figure 5G), likely because of the partial recovery of MT stability in egr2-1 masp1-1.
Figure 5.
MASP1 Is Epistatic to EGRs in Growth but Not Proline Accumulation.
(A) Analysis of dry weight (D.W.) and root elongation for seedlings under control and stress (−0.7 MPa) conditions for egr2-1, egr1-1 egr2-1, masp1-1, and egr2-1 masp1-1. Data are relative to the Col-0 wild type (W.T.; mean ± se, n = 6 to 8, asterisk indicates significant difference compared with the wild type by one-sided t test [P ≤ 0.05]) and are combined from two independent experiments. Typical growth values of Col-0 wild type used for normalization are shown in Supplemental Figure 2.
(B) Representative seedlings in the unstressed control (7 d after transfer) and at low ψw (−0.7 MPa, 10 d after transfer). Bars = 1 cm.
(C) Representative rosettes of Col-0 wild type, egr2-1, masp1-1, and egr2-1 masp1-1 in the unstressed control and soil drying treatments. The soil drying experiments and presentation of data are the same as described in Figures 1E and 1F. Bars = 1 cm.
(D) Relative rosette fresh weight and dry weight of egr2-1, masp1-1, and egr2-1 masp1-1 in control or soil drying treatments. Data are means ± se, n = 4 to 8, and asterisk indicates significant difference compared with the wild type by one-sided t test (P ≤ 0.05) combined from two independent experiments.
(E) Proline accumulation after 96 h at −1.2 MPa. Data are means ± se (n = 9 to 15) combined from two independent experiments. Asterisks indicate a significant difference compared with the wild type (P ≤ 0.05). N.S., not significant.
(F) Microtubule images for hypocotyl cells of untreated seedlings and seedlings treated with 10 μM oryzalin for 45 min. Bars = 20 μm.
(G) PI staining of cells in the root elongation zone for seedlings transferred to low ψw (−0.7 MPa) for 10 d. Arrows show examples where cell swelling can be most clearly seen in masp1-1. Bars = 50 μm.
Ethylene is an important regulator of growth during drought and can induce anisotropic cell swelling similar to that seen in masp1-1 (Baskin and Bivens, 1995). To test whether EGR-MASP1 phenotypes may be caused by altered ethylene signaling, we treated EGR and MASP1 mutant and overexpression lines with 1-aminocyclopropane-1-carboxylic acid (ACC) to increase ethylene production and aminoethoxyvinylglycine (AVG) to block ethylene production (Lewis et al., 2011). None of the mutants or transgenics had an altered response to ACC or AVG in the unstressed control except for masp1-1, where the 9% root growth inhibition seen in untreated plants was recovered by AVG treatment (Supplemental Figure 8). All of the mutants and overexpression lines had a similar response to ACC treatment at low ψw. At low ψw, AVG caused a 20 to 35% reduction in root elongation for all genotypes except the EGR1 overexpression line, which already grew less before AVG treatment and was less affected by AVG (Supplemental Figure 8). PI staining and observation of the root swelling phenotype showed that ACC application at low ψw induced root swelling in the wild-type, masp1-1, and EGR1 overexpression lines, but not in egr2-1 or egr2-1 masp1-1 (Supplemental Figure 9). In unstressed plants, this amount of ACC only induced root swelling in masp1-1. AVG suppressed root swelling at low ψw, but did not completely restore normal cell morphology (Supplemental Figure 9). The combined data indicate that any differences in response to ACC or AVG were a side effect of altered MT stability and that the effects of EGR-MASP1 on growth at low ψw were largely independent of ethylene. Particularly, the reduced MT stability of masp1-1 made it more susceptible to low ψw- or ACC-induced root swelling. AVG, or introduction of the egr2-1 mutation, alleviated masp1-1 root swelling (likely because of partial recovery of MT stability) but did not recover the reduced growth of masp1-1 at low ψw. Likewise, neither ACC nor AVG treatment suppressed the increased growth of egr1-1 egr2-1 to the wild-type level at low ψw.
MASP1 Directly Binds and Stabilizes MTs but Cannot Itself Promote MT Polymerization
The above data raised the question of whether the dramatic effect of MASP1 on MT stability could involve MASP1 binding to MT. MT cosedimentation assays using recombinant MASP1 and paclitaxel-stabilized MTs in vitro demonstrated direct, concentration-dependent MT binding (Figures 6A to 6C). No sedimentation of MASP1 was observed in controls lacking MTs (Figure 6B). Essentially identical results were observed for unmutated MASP1, MASP1S670D phosphomimic, and MASP1S670A phosphonull, thus demonstrating that MASP1 MT binding did not depend on Ser-670 phosphorylation. Similarly, the addition of MASP1 to paclitaxel-stabilized rhodamine-labeled MTs led to the formation of aggregates consistent with MT bundles and stabilized the MTs during cold treatment (Figure 6D). MASP1 MT bundling activity was also indicated by the thick strands seen in oryzalin-treated MASP1 and MASP1S670D phosphomimic-overexpressing plants (Figure 4B). Turbidity assays where MTs were polymerized in the presence of MASP1 showed a similar stabilizing effect of MASP1 regardless of Ser-670 phosphorylation (Figure 6E). However, in turbidity assays performed without the addition of glycerol to promote MT polymerization, MASP1 had no effect (Figure 6F). Together, these data indicate that MASP1 could bind and stabilize polymerized MTs, but could not itself induce polymerization.
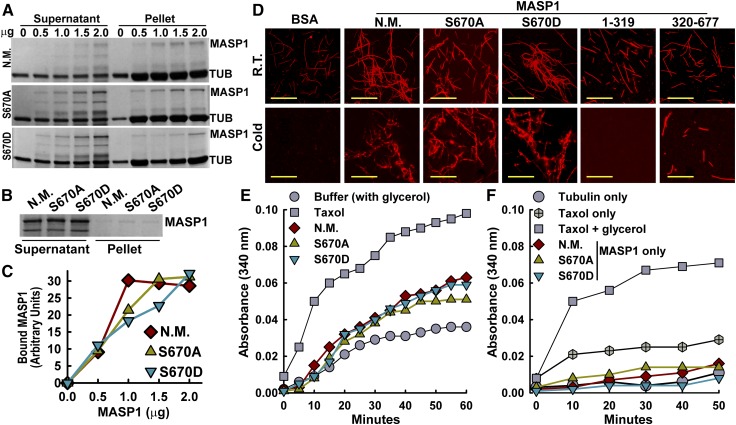
Figure 6.
MASP1 MT Interaction and MT Bundling.
(A) Cosedimentation MT binding assays using 0 to 2.0 mg of unmutated MASP1 (N.M.), MASP1S670A (phosphonull), or MASP1S670D (phosphomimic). TUB, tubulin. Note that the addition of even a small amount of MASP1 increased the amount of tubulin in the pellet fraction (to preserve MASP1 solubility, the final sedimentation was done at 10°C, which partially destabilized microtubules). Images shown are Coomassie-stained gels and are representative of two to three experiments.
(B) Minus MT control for the MASP1 MT binding experiments is shown in (A). Negligible MASP1 precipitation was seen in the absence of MTs. Gel shown is typical of several repeated experiments.
(C) Quantification of MASP1 sedimentation with microtubules. Data are means from two experiments.
(D) MASP1 MT bundling and stabilization in vitro visualized using rhodamine-labeled MTs. R.T., room temperature incubation of MTs with the indicated MASP1 alleles or truncations or BSA. Cold = 10°C, 45 min. Bars = 10 μm.
(E) Turbidity assay measuring MT polymerization in the presence of taxol, unmutated MASP1 (N.M.), MASP1S670A (phosphonull), or MASP1S670D (phosphomimic). Each type of MASP1 protein was present at 10 μM. All assays included glycerol to promote MT polymerization. Data shown are representative of three independent experiments.
(F) Turbidity assay performed in the same manner as in (E) except that glycerol was omitted for all reactions (except taxol + glycerol) to test whether MASP1 could promote MT polymerization.
MASP1 Structural Prediction and in Planta MT Association Further Indicate Its Function in MT Stability
Despite this demonstration of MT binding in vitro, we were unable to detect in planta MASP1 decoration of MTs in Pro35S:YFP-MASP1 transgenic plants. In transient expression assays, where higher local levels of MASP1 expression could be achieved, faint strands consistent with MT decoration could be observed (Figure 7A) and this pattern became more noticeable over longer periods of transient expression (Figure 7B). MT decoration was enhanced by oryzalin treatment for unmutated MASP1 and MASP1S670D phosphomimic, but not MASP1S670A phosphonull (Figure 7A). This indicated that high MASP1 expression may decrease MT dynamics, thus allowing MASP1 to accumulate on MTs. Oryzalin treatment exasperated this effect by blocking MT polymerization and the strong MT decoration again showed that high levels of unmutated or MASP1S670D phosphomimic, but not MASP1S670A phosphonull, could stabilize MTs in planta. Interestingly, phosphomimic MASP1, and to a lesser extent unmutated MASP1, was seen to cluster in foci along MTs (Figure 7A).
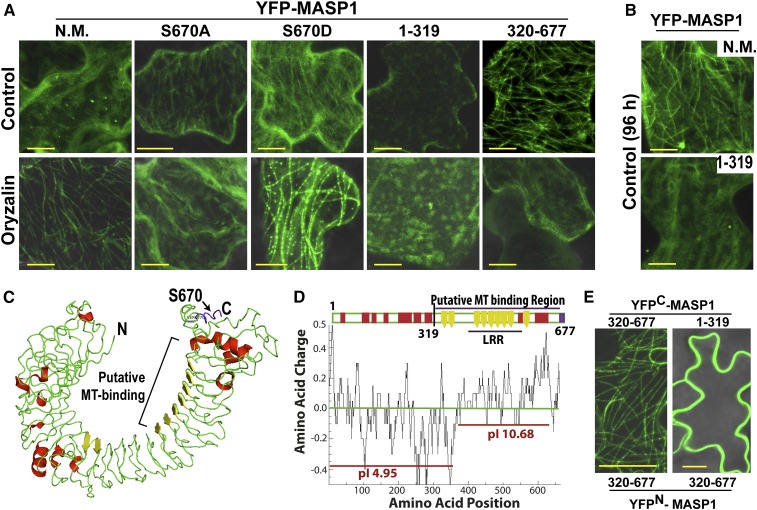
Figure 7.
MASP1 Structure and Localization Experiments Indicate That It Contains a Basic C-Terminal MT Binding Domain That Is Required and Sufficient to Decorate MTs in Planta.
(A) Localization and effect of oryzalin (10 μM) on transiently expressed YFP-MASP1 alleles and MASP1 truncations. Transient expression was done for 24 h and oryzalin treatment for 2.5 h for full-length unmutated MASP1 (N.M.), MASP1S670A (phosphonull), or MASP1S670D (phosphomimic) and 45 min for the MASP1 truncations (1 to 319 and 320 to 677). Bars = 10 μm.
(B) Transient expression of full-length MASP1 with no mutation (N.M.) and the N-terminal MASP1 fragment (amino acids 1 to 319) for 96 h. Bars = 10 μm.
(C) MASP1 structure predicted by I-Tasser (diagram generated by Pymol, nearly identical structure was predicted by Phyre2; Supplemental Figure 10A). Green = loop regions; red = helix, yellow = β-sheet. In addition, LRRsearch identified six LRR domains (Supplemental Figure 10B), which correspond to the β-sheet regions. The phosphopeptide containing Ser-670 identified in our phosphoproteomic analysis is shown in purple here and in Supplemental Figure 10B. MASP1 structure resembles typical LRR proteins, which form an α/β horseshoe fold; however, MASP1 does not have exterior helices after every turn of an interior β-sheet and thus has a less rigid secondary structure. The C-terminal β-sheet regions form a basic surface (D), which may directly contact MTs. The β-sheet LRR domains (yellow) may form multiple contact sites for lateral binding across the MTs. The internal diameter of the horseshoe structure (25 to 30 nm) is similar to the MT outer diameter (25 nm) and thus is compatible with lateral MT binding.
(D) Charge plot and pI analysis showing that the C-terminal half of MASP1, including the LRR rich region, which has basic pI consistent with direct MT binding, may be the site of direct MT binding. The charge distribution and less rigid structural fold of MASP1 (C) suggests that it binds the microtubule surface in a flexible manner leaving the two ends of the protein free for additional interactions. Charge plot was drawn using Emboss explorer (http://www.bioinformatics.nl/cgi-bin/emboss/charge) with a window size of 10 amino acids and pI of different sections of MASP1 calculated using the Compute pI/Mw tool (http://web.expasy.org/cgi-bin/compute_pi/pi_tool).
(E) BiFC assays of MASP1 C-terminal (320 to 677) and N-terminal (1 to 319) fragments. Bars = 20 μm.
Structural modeling showed that MASP1 forms an α/β horseshoe fold typical of leucine-rich repeat proteins (Figure 7C). The C-terminal portion of MASP1 has several leucine-rich repeat (LRR) domains, which line the inside of a hook-like structure the diameter of which (25 to 30 nm) is similar to the 25 nm MT outer diameter (Figure 7C; Supplemental Figures 10A and 10B). Also, the MASP1 predicted structure had mostly flexible loop regions (rather than helices) connecting the LRR domains. This indicated a relatively flexible structure consistent with MASP1 being able to stretch and bind laterally across several tubulin dimers as observed for mammalian TAU proteins (Butner and Kirschner, 1991). Charge plot and pI analysis showed that the C-terminal portion of MASP1 (amino acids 320 to 677) had a basic pI (Figure 7D) similar to other MT binding proteins (Hamada et al., 2013), further suggesting that this portion of MASP1 may directly contact MTs.
Consistent with this hypothesis, the MASP1 C-terminal fragment (amino acids 320 to 677), but not the N-terminal fragment (amino acids 1 to 319), decorated MTs in planta (Figure 7A). This localization pattern of the MASP1 C-terminal fragment was readily disrupted by oryzalin, indicating that it was associated with MTs but lacked the MT stabilization activity of full-length MASP1. Also, the MASP1 C-terminal fragment retained limited ability to stabilize, but not bundle, rhodamine-labeled MTs in vitro while the N-terminal MASP1 fragment had no effect (Figure 6D). MT decoration in vivo could also be observed in BiFC assays using the MASP1 C-terminal fragment for both halves of the BiFC interaction (Figure 7E. In this case, the BiFC signal may indicate dense MT binding that brings the split YFP halves in close enough proximity to generate a signal. In contrast, BiFC assay of the MASP1 N terminus with C terminus generated signal along the cell periphery (Figure 7D), suggesting that the N-terminal portion of MASP1 could sequester the C terminus away from MTs or otherwise obscure MT decoration. The predicted MASP1 structural analogs included several cell surface binding proteins (Supplemental Figure 10C), perhaps providing another clue that MASP1 may bind to other cellular structures in addition to its MT binding activity. Together, these data led to a model of EGR-MASP1 cellular function that includes EGR regulation of proteins along the plasma membrane and MASP1 MT binding and stabilization (Figure 8).
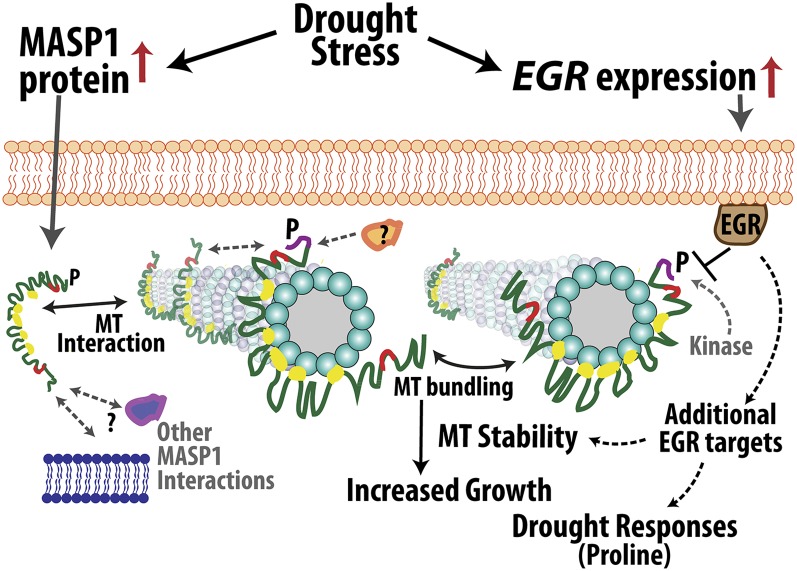
Figure 8.
Model of EGR-MASP1 Function.
EGRs and MASP1 physically associate but have opposing effects on plant growth, including effects on cell expansion. EGRs are negative growth regulators and increased EGR expression at low ψw restrains growth. MASP1 is a positive regulator of growth and unknown posttranscriptional mechanism(s) lead to increased MASP1 protein accumulation at low ψw. This increased MASP1 stabilizes MTs (as indicated by the arrows between MASP1 and MTs) and preserves the competence to grow under mild to moderate low ψw stress. The effects of MASP1 on growth are epistatic to those of EGRs, and MASP1 Ser-670 phosphorylation is required for its growth-promoting activity but not for MT binding [EGR attenuation of MASP1 phosphorylation is indicated by the T-bar between EGR and MASP1; MASP1 phosphorylation by unknown kinase(s) is indicated by gray dashed arrow]. MASP1 phosphorylation may affect other aspects of its function such as binding to additional MT-associated proteins or clustering at specific sites along MTs (indicated by gray dashed arrows and question mark). Functional analysis of MASP1 indicates that the C-terminal portion binds MTs, while the N-terminal portion is required for MT bundling and stabilization and may also act to sequester MASP1 away from MTs, possibly by binding to other structures in the cell cortex (binding events shown in this study are indicated by black double-sided arrows; possible association of MASP1 with other proteins or cellular structures is indicated by gray dashed double-sided arrows). By these or related mechanisms, MASP1 stabilizes and protects MT organization in a way that promotes cell expansion and continued growth. Additional EGR target proteins are likely to be involved in drought-related signaling, for example, in the regulation of proline accumulation or MASP1-independent effects on MTs.
DISCUSSION
Our results provide insight into plant growth regulation and drought response and also identify proteins influencing MT organization. EGR gene expression and MASP1 protein abundance are increased by low ψw (drought). Suppression of EGRs or ectopic expression of MASP1 increased growth and had an especially prominent effect on growth during drought stress. This occurred without causing the development defects often observed in MT-related mutants or in plants subjected to pharmacological treatments that alter MT stability (Thitamadee et al., 2002; Shoji et al., 2004; Bannigan et al., 2006; Ambrose et al., 2007; Korolev et al., 2007; X. Wang et al., 2007; Buschmann et al., 2009; Nakamura and Hashimoto, 2009; Hamada et al., 2013; Liu et al., 2016). While MASP1 stabilizes microtubule organization and promotes growth, EGRs act as a brake to slow growth and decrease MT stability. Thus, EGRs and MASP1 balance each other to selectively regulate MT organization and adjust plant growth in response to environmental stimuli (Figure 8). This balanced regulation is particularly relevant to drought, as inhibiting leaf growth can be a conservative strategy to prevent transpiration from outrunning the available water supply. Circumventing such negative growth regulation is a promising strategy to increase plant productivity in many drought scenarios; thus, EGRs and MASP1 are of interest for translational research to improve plant productivity during mild to moderate drought.
EGR-MASP1 Function in MT Regulation, Growth Regulation, and Drought Response
Analysis of egr2-1 masp1-1, as well as the EGR-MASP1 interaction and effect of egr mutants or EGR overexpression on the abundance of phosphorylated MASP1 (Figure 2), indicated that MASP1 acts downstream of EGRs in growth regulation. EGRs interacted with MASP1 along the cell periphery and attenuated the accumulation of phosphorylated MASP1. This could occur by direct EGR dephosphorylation of MASP1; however, we cannot rule out more indirect mechanisms, such as EGR-mediated inhibition of unidentified kinase(s) that phosphorylate MASP1. While MASP1 was critical for EGR-mediated growth promotion, our examination of several phenotypes also indicated a broader function of the EGRs. In particular, EGR-mediated regulation of proline accumulation was MASP1 independent and EGR2 also had some effect to enhance MT stability and alleviate root swelling independently of MASP1. An effect of EGR1 on inflorescence growth, as well as characterization of EGR1 phosphatase activity, has been reported (Sugimoto et al., 2014); however, there has been no reported mechanism of EGR action or analysis of EGR stress function. Likewise, our data show that MASP1 is a MT binding protein. Data presented here offer several intriguing possibilities for MASP1 function, which will require additional investigation. These include the function of MASP1 Ser-670 phosphorylation, effect of MASP1 on MT dynamics, and whether MASP1 affects trafficking or cell wall deposition as part of its growth regulation activity. To our knowledge, there is no previous functional information on MASP1 or any of its close homologs.
The data presented here are the most direct demonstration to date of the importance of MT in growth regulation during drought. Drought-related functions of MTs and MT binding proteins have been proposed as the rigid nature of cortical MTs make them particularly sensitive to mechanical perturbation (Landrein and Hamant, 2013; Nick, 2013), which may be involved in controlling cell morphology (Landrein and Hamant, 2013; Sampathkumar et al., 2014) and, possibly, drought sensing (Nick, 2013; Haswell and Verslues, 2015). However, drought-responsive proteins, which could allow stress signaling to communicate with the cytoskeleton (and vice versa), had not been identified. EGR-MASP1 function in MT regulation is distinct from phosphorylation of α-tubulin or degradation of SPR1, which lead to a rapid loss of MT organization in the initial hours of osmotic or salt stress (Wang et al., 2011; Fujita et al., 2013), but are of less clear relevance to MT recovery. A large part of the EGR-MASP1 effects on growth was due to changes in cell expansion. However, we cannot rule out additional effects of EGRs and MASP1 on cell division.
A Unique Phosphoproteomics Data Set to Understand Drought Signaling and EGR Function
Our phosphoproteomics data set identified a distinct and complementary set of phosphoproteins compared with previous stress phosphoproteomics experiments (Supplemental Figures 2C and 2D and Supplemental Data Sets 4 and 7) and the comparison to transcriptome data indicated that many of these were from genes not transcriptionally regulated by low ψw. These phosphoprotein abundance changes could represent either stress-induced changes in phosphorylation stoichiometry or posttranscriptional regulation of protein abundance. Both are of interest and difficult to access by other means. Overall, we propose that both our set of statistically significant phosphopeptide abundance changes (Supplemental Data Sets 5 and 6) as well as phosphorylation changes identified in multiple studies (Supplemental Data Sets 4 and 7) are a valuable resource for the study of phosphorylation-dephosphorylation mechanisms important for drought resistance.
Our phosphoproteomic analysis of egr1-1 egr2-1 identified a set of putative EGR targets enriched in cytoskeleton and plasma membrane-associated proteins. This result fits well with EGR localization and function to regulate MT organization and growth and demonstrated the utility of using phosphoproteomics to identify targets of phosphatase regulation. Our finding that MASP1 interacted with EGRs, affected a similar range of phenotypes, and acted genetically downstream of EGRs in growth regulation further validated the success of our phosphoproteomic analysis in discovering physiologically relevant EGR target proteins. At the same time, some of the effect of EGRs on MT stability, as well as EGR regulation of proline accumulation, was independent of MASP1. This indicated that other EGR targets are important for regulating cytoskeleton and drought phenotypes. This may involve additional proteins listed in Table 1. It should also be noted that the protein extraction method used here (9 M urea) does not efficiently solubilize very hydrophobic membrane proteins. It is possible that EGRs regulate the phosphorylation of additional membrane proteins and that strategies to enrich plasma membrane proteins (Zhang and Peck, 2011) would identify additional targets of EGR regulation.
Several MASP1 Features Distinguish It from Other MT Binding Proteins
This study uncovered several unique aspects of MASP1. One was that we could only observe MASP1 MT decoration under certain conditions or when using the C-terminal MASP1 fragment. These experimental observations, along with MASP1 localization and predicted structure, lead to the hypothesis that MASP1 may link MTs to structures, possibly including endomembrane structures, in the cell cortex (Figure 8). These possibilities will be of interest for further study as MT anchoring and connection to other cellular compartments is poorly understood in plants (Gardiner et al., 2001; Ambrose and Wasteneys, 2008; Brandizzi and Wasteneys, 2013). Several of our MASP1 observations are similar to models of MAP65 function (Smertenko et al., 2008; Zhang et al., 2012). However, MASP1 has no sequence or structural similarity to MAP65 family proteins. Some aspects of MASP1 function are also similar to the recently described Cellulose Synthase Microtubule Uncoupling (CMU) proteins (Liu et al., 2016) and CC proteins (Endler et al., 2015). CMU proteins bind MTs and are associated with membranes. However, loss of CMU function was not sufficient to detach MTs from the plasma membrane and the authors hypothesized that CMUs may be part of a protein complex that links MTs to the plasma membrane (Liu et al., 2016). CC proteins directly bound MTs and promoted MT recovery after salt stress. However, CC proteins seemed to only affect responses to salt stress but not osmotic stress (Endler et al., 2015; Wang et al., 2016). Neither CMUs nor CCs are structurally related to MASP1. It will be of interest for future studies to see if CC and CMU proteins have any functional interaction with MASP1.
A second unique aspect of MASP1 is that Ser-670 phosphorylation did not affect its MT binding and bundling activity in vitro but did determine its function in planta. MASP1 could directly bind MTs regardless of its Ser-670 phosphorylation status; however, Ser-670 phosphorylation was required for full activity of MASP1 in MT stabilization and growth promotion in planta. The in vitro data were broadly consistent with the physiological data, where MASP1S670A phosphonull retained a limited effect on cell length (Supplemental Figure 5), oryzalin sensitivity (Figure 4B), and root cell morphology (Supplemental Figure 6). Presumably, MASP1S670A binds MTs in vivo and has some stabilizing effect. At the same time, the inability of MASP1S670A phosphonull to complement masp1-1 and promote growth at low ψw indicated that Ser-670 phosphorylation affects aspects of MASP1 function other than its MT binding activity. It is possible that Ser-670 phosphorylation influences the interaction of MASP1 with other proteins involved in MT stabilization, influences recruitment of MASP1 to specific locations along MTs (as indicated by the foci of MASP1 S670D along MT; Figure 7A), or influences the association of MASP1 with specific structures in the cell cortex (Figure 8). MASP1 structure and the readily apparent MT decoration by the MASP1 C-terminal domain in planta indicate that it is the C-terminal portion of MASP1 that interacts directly with MTs; however, the C-terminal domain alone could not bundle MTs. The N-terminal domain alone did not decorate MTs (Figures 7A and 7B) and could also obscure the MT decoration of the C-terminal domain (Figure 7E). Thus, the N-terminal domain may be involved in interactions that sequester MASP1 away from MTs. Several aspects of these data, such as the basis for MASP1 clustering in foci along MTs and the identification of additional MASP1 interacting proteins, are worthy of further investigation and can provide clues to better understand the molecular basis of MASP1 effects on MT organization.
A third unique aspect of MASP1 (and the egr mutants) is that despite its MT stabilizing activity, mutation or ectopic expression of MASP1 produced none of the morphological defects seen in other MT-related mutants. This would explain why MASP1 was not found in genetic screens based on morphological traits. A final, and important, unique aspect of MASP1 is that its protein abundance was dramatically higher in plants acclimated to low ψw for 96 h even though MASP1 mRNA level was unchanged. The low abundance of MASP1 in unstressed plants could explain why MASP1 was not found in studies that purified MTs and identified MT interacting proteins (Korolev et al., 2005; Hamada et al., 2013) as none of these studies used stress-treated plants. Mechanisms controlling MASP1 protein abundance are also of interest for future research.
The predicted MASP1 structure of several putative MT contact points separated by flexible loops, as well as its MT stabilizing and bundling activity, is in many ways reminiscent of mammalian MT binding protein TAU and related proteins such as MAP2. TAU binds MT laterally across several tubulin dimers with the binding distributed across several sites each with relatively weak MT affinity (Butner and Kirschner, 1991). Such binding that links adjacent tubulin dimers across several protofilaments can inhibit MT depolymerization (Ichihara et al., 2001). TAU can also self-interact to link adjacent MTs (Ahmadzadeh et al., 2015). For MAP2, it was observed that sites of MAP2 clustering were associated with stopping of depolymerization and promotion of MT rescue (Ichihara et al., 2001). The effects of MASP1 on MT dynamics and whether it stabilizes MTs by affecting depolymerization is of interest for future studies. Interestingly, there are no plant orthologs of mammalian TAU or MAP2. Although basic MT components are conserved across eukaryotes, the unique aspects of the plant MT-plasma membrane-cell wall interface as well as the unique needs of plants to sense and respond to environmental stimuli have led to the evolution of a distinct repertoire of MT binding proteins and regulatory mechanisms. Our results indicate that EGRs and MASP1 are such unique plant MT-regulating proteins with an especially prominent role in the control of growth and cell expansion during drought stress.
METHODS
Mutants, Transgenic Plants, Stress Treatments, and Phenotype Assays
T-DNA insertion lines of Arabidopsis (Arabidopsis thaliana) were obtained from the ABRC. For construction of transgenic plants, the open reading frames of EGR1, EGR2, and MASP1 were amplified and cloned into entry vector pDONR207 or pDONR222 using the Gateway Cloning System (Life Technologies). Entry clones were further transferred to expression vectors pGWB442 (Nakagawa et al., 2007) and pEG104 (Earley et al., 2006) and then transferred to Agrobacterium tumefaciens strain GV3101 and floral dip Arabidopsis transformation was performed. The substitution of Ser-670 (codon AGT) to Asp (codon GAT) or Ala (codon GCT) to generate phosphomimic or phosphonull MASP1, respectively, was performed using site-directed mutagenic primers (Supplemental Data Set 13) and the entry vector pDONR222.2-MASP1 as a template. The PCR reaction was followed by incubation with DpnI at 37°C for 30 min to digest the template plasmid. The remaining PCR product (pDONR222.2-MASP1S670D or pDONR222.2-MASP1S670A) was gel purified, transformed into DH5α competent cells, and transformants selected on kanamycin (50 µg/mL) plates. Plasmids isolated from kanamycin-resistant colonies were sequenced to confirm the presence of the MASP1 mutation. Confirmed pDONR222.2-MASP1S670D or pDONR222.2-MASP1S670A was transferred by Gateway recombination to pGWB442 (for wild type and masp1-1 transformation) or pEG100 (for transformation of GFP-TUA6). Similarly, egr single mutants, egr1-1 egr2-1, and masp1-1 were introduced into GFP-TUA6 by crossing. The GFP-TUA6 transgenic line (Ueda et al., 1999) was obtained from the ABRC (stock number CS6551). Primers used for genotyping, cloning, site-directed mutagenesis, and RT-PCR are given in Supplemental Data Set 13.
Seedling growth and stress treatments on PEG-infused agar plates were performed as described previously (Verslues et al., 2006; Bhaskara et al., 2012). Five-day-old seedlings were transferred to fresh half-strength MS plates (control), PEG-infused plates (adjusted to different ψw severities as indicated in text or figures), or oryzalin-containing plates (oryzalin concentrations are given in text and figures). Growth was measured on the seventh day after transfer for control treatment and the tenth day after transfer for stress/oryzalin treatment. Dry weight was measured for groups of three to four seedlings after overnight drying in an incubator at 65°C. For proline measurement and immunoblotting (see below), seedlings were grown for 7 d and transferred to fresh half-strength MS plates and PEG-infused agar plates for 96 h. Proline content was measured by ninhydrin assay (Bates et al., 1973). For experiments involving ACC and AVG, seedlings were grown for 5 d on control media and then transferred to either control or PEG-infused plates (−0.7 MPa) with or without ACC (0.5 µM) or AVG (1 µM). Root growth measurement and PI staining of root elongation were performed after 7 d for control seedlings and after 10 d for stress-treated seedlings. For PI staining, seedlings were incubated in PI (10 µg/mL in water) for 1.5 to 2 min. Excess stain was removed by rinsing in water for 1 to 2 min and seedlings were examined by confocal microscopy (536-nm excitation and 617-nm emission) on a Zeiss LSM 510 Meta.
Soil drying experiments were conducted as previously reported (Bhaskara et al., 2015). A standard potting mix was combined with 25% Turface (Turface MVP; Profile Products). Four genotypes were planted in sectors (two plants per sector) of 8 × 8 × 10 cm (LxWxH) plastic pots to ensure that the different genotypes were exposed to the same extent of soil drying. Plants were grown in a short-day chamber (8 h light period, 25°C, light intensity of 100 to 120 μmol m−2 s−1 using mixed fluorescent and incandescent bulbs) and Hyponex nutrient solution (1 g L−1) supplied once per week. On the eighteenth day after planting, pots were watered to saturation, allowed to drain, and weighed. Water was withheld for 12 d (leading to a 50 to 60% reduction in pot weight), and each pot rewatered to 75% of the initial pot weight and then allowed to dry for another 8 to 10 d until the pot weight again reached 50 to 60% of the starting weight. At the end of the experiment, representative rosettes were photographed and the remaining rosettes weighed (fresh weight), rehydrated in cold water overnight, weighed again (hydrated weight), and then dehydrated overnight in an 60°C oven (dry weight). In some experiments, the sixth leaf was removed and analyzed by scanning electron microscopy (FEI Quanta 200 with a Quorum PP2000TR FEI cyro system).
Localization, Transient Expression, BiFC, and Coimmunoprecipitation
BiFC assays were performed using pSITE vectors (Martin et al., 2009), and transient expression was performed in 4-d-old Arabidopsis seedlings with dexamethasone-inducible AvrPto expression (ABRC stock number CS67140) using procedures adapted from those previously described (Tsuda et al., 2012). BiFC interaction was visualized 4 d after transfer of seedlings to either control or −1.2 MPa plates and 40 min after transfer to oryzalin plates using a Zeiss LSM 510 META microscope.
For coimmunoprecipitation assays of protein interaction, Pro35SYFP:EGR1, EGR2, or HAI1 (in vector pGWB442) was coinfiltrated with Pro35S:FLAG-MASP1 (in vector pGWB412) for transient expression in AvrPTO seedlings as described above for BiFC analysis. Approximately 1 g of infiltrated seedlings was collected after 4 d of infiltration. Seedlings were ground in liquid nitrogen and 500 μL of lysis buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 0.5% Triton X-100, 2× Roche EDTA-free protease inhibitor added along with 1 mM cross-linking reagent dithiobis-succinimidyl propionate). The lysate was placed on ice for 30 min with inverting tubes every 10 min and then Tris-HCl (pH 7.5) was added to the lysate to quench cross-linking. Lysate was further diluted to 1 mL using wash buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM EDTA, and 2× Roche protease inhibitor) and centrifuged at 20,000g for 10 min at 4°C. The clear supernatant was incubated with 50 μL of equilibrated GFP beads (Chromotek) for 4 h at 4°C with gentle agitation. Beads were collected by centrifugation at 2500g for 2 min and followed by washing two times with 500 μL of cold wash buffer. Beads were boiled for 10 min at 95°C in 2× SDS sample loading buffer, proteins were resolved by SDS-PAGE using an 8% separating gel, and immunoblots were performed using anti-GFP (Sigma-Aldrich) or anti-FLAG (Sigma-Aldrich) antibodies.
Subcellular localization of MASP1 and EGRs was analyzed in cells from the root elongation zone of T3 homozygous lines, and YFP was detected using excitation/emission wavelengths of 514/530 to 590 nm. For FM4-64 (Merck) colocalization, roots of 7-d-old seedlings were incubated with 2 μM FM4-64 for 1 min followed by a 1-min wash in distilled water. FM4-64 was detected with excitation/emission wavelengths of 488/575 to 610 nm.
Quantitative Phosphoproteomics Using I-Traq Labeling
Sample Preparation
Seven-day-old seedlings were transferred and grown on fresh MS agar plates or PEG-infused agar plates as described above to impose a low ψw stress of −1.2 MPa for 96 h. Seedlings were collected from three independent experiments (biological replicates) and the reproducibility of the stress treatment in each biological replicate was confirmed by measuring the proline content.
Sample preparation generally followed previous protocols (Wu et al., 2010) with some modifications. For protein extraction, samples from each biological replicate were ground in liquid nitrogen, further homogenized using cold trichloroacetic acid/acetone solution (10% trichloroacetic acid in acetone containing 0.07% 2-mercaptoethanol), and incubated for 1 h at −20°C. The homogenate was centrifuged at 17,000g for 15 min at 4°C. The protein pellet was washed by dissolving it in 10 volumes of ice-cold wash solution (0.07% 2-mercaptoethanol in acetone) and incubated at 20°C for 1 h. The protein pellet was washed twice using 10 volumes of ice cold wash solution and dried under a nitrogen stream. The dried protein pellets were redissolved in 3 to 4 mL of freshly prepared dissolution buffer (9 M urea and 25 mM Tris-HCl, pH 8.0) containing phosphatase inhibitors (Sigma Cocktail 2 and 3). After 1 h of precipitation at 4°C, the sample was again centrifuged at 20,000g for 15 min at 10°C and the supernatant collected in methanol-washed precooled centrifuge tubes. Protein concentration was measured using a Pierce BCA Protein Assay Kit.
Six milligrams of total protein was reduced with 10 mM DTT for 1 h at 37°C and alkylated with 25 mM iodoacetamide at room temperature for 30 min in the dark. Samples were quenched using DTT (5 mM final concentration), diluted to a concentration of 2 M urea using 25 mM Tris (pH 8.0), treated with benzonase (250 units/mL; Sigma-Aldrich; E8263), and incubated for 2 h at room temperature. Protein digestion was performed by first treating with Lys-C endopeptidase (1:200 [w/w]; Mass Spectrometry Grade; Wako) for 4 to 5 h at room temperature. The urea content of the sample was further diluted to <1 M using 25 mM Tris (pH 8.0), trypsin (1:50 [w/w]; Promega) was added, and samples were incubated overnight at room temperature. The tryptic peptides were acidified using 100% trifluoroacetic acid, desalted using a Sep-Pak C18 column (Waters; 200 mg, 3 mL bed volume), and eluted with 5 mL of 0.5% acetic acid in 50% acetonitrile. The eluate was dried in a speed vac. Phosphopeptide enrichment was performed using a Pierce TiO2 phosphopeptide enrichment and cleanup kit. Phosphopeptide labeling was done using an AB Sciex iTRAQ 8-plex reagent kit following the manufacturer’s instructions. In brief, the sample was dissolved in 30 μL of labeling buffer and iTRAQ reagents 113/117 for wild type control/stress and 116/121 for egr1-1 egr2-1 control/stress dissolved in 50 μL isopropanol. Ten microliters of 100% trifluoroacetic acid was then added and the samples were dried for 5 to 10 min in a speed vac to remove isopropanol. Samples labeled with individual iTRAQ tags were then combined in one tube, desalted using a Pierce graphite spin column and dried again. The desalted sample was resuspended in buffer A (10 mM KH2PO4 and 25% acetonitrile, pH 2.7 to 3.0) and fractionated on a strong cation exchange column (PolySULFOETHYL A column, 4.6 × 100 mm, 5 µm, 200Å; PolyLC) using an elution gradient from 0 to 50% buffer B (0.35 M KCl in buffer A; buffer B consisted of 7 mM KH2PO4, pH 2.65, and 30% acetonitrile). Samples were eluted over 20 min, and 10 fractions of 0.5 mL each were collected. Each fraction was desalted using a graphite spin column and dried under vacuum before proceeding to subsequent analysis.
LC-MS/MS Analysis and iTRAQ-Based Phosphopeptide Quantification
The strong cation exchange fractions were analyzed on a nanoUPLC system (nanoAcquity; Waters) coupled to an LTQ Orbitrap Velos hybrid mass spectrometer (Thermo Scientific). A C18 capillary column (75 μm × 250 mm, 1.7 μm, BEH130; Waters) was used to separate peptides with a 90-min linear gradient from 5 to 40% acetonitrile in 0.1% formic acid at a flow rate of 300 nL/min. The LTQ Orbitrap Velos MS was operated in data-dependent mode with the top 10 ions (charge states ≥2) from the MS survey scan selected for MS/MS (CID with multistage activation enabled and HCD) acquisitions. The parameters for the MS and MS/MS were set as follows: FT Orbitrap MS m/z range = 350 to 1600; resolving power = 60,000 with automatic gain control (AGC) target = 500,000; HCD: MSn AGC target = 50,000 with normalized collision energy = 45%, activation time = 10 ms, resolving power = 7500; CID: MSn AGC target = 5000 with normalized collision energy = 35%, activation Q = 0.25, activation time = 10 ms. The CID and HCD MS/MS were performed alternately for the same m/z precursor ion and had the following settings: minimum signal required = 2000, isolation width = 2; dynamic exclusion: repeat count = 1, repeat duration = 15 s, exclusion duration = 90 s. Neutral loss masses of phosphopeptides for multistage activation enabled CID were set at 97.98, 48.99, 32.66, and 24.49 for various charge states of the peptides. Peptide identification, phosphorylation, and quantification were performed using Proteome Discoverer software (v1.3; Thermo Fisher Scientific) with SEQUEST and Mascot (v2.3; Matrix Science) search engines. MS data were searched against TAIR10 protein sequence database with 27,416 sequence entries downloaded from The Arabidopsis Information Resource website (http://www.arabidopsis.org/). The parameters for database searches were set as follows: full trypsin digestion with 2 maximum missed cleavage sites, precursor mass tolerance = 10 ppm, fragment mass tolerance = 0.8 D (CID) and 50 mmu (HCD), dynamic modifications: oxidation (M) and phosphorylation (S, T, and Y), static modifications: carbamidomethyl (C), iTRAQ 8-plex (N terminus and K). The identified peptides were validated using the Percolator algorithm, which automatically conducted a decoy database search and rescored peptide spectrum matches using q-values and posterior error probabilities. All of the peptide spectrum matches were filtered with a q-value threshold of 0.01 (1% false discovery rate). Phosphopeptides were further validated using the phosphoRS algorithm with thresholds of pRS score ≥50 and pRS site probability ≥75%. For iTRAQ quantification, the ratios of iTRAQ reporter ion intensities in MS/MS spectra (m/z 113 and 117) from raw data sets were used to calculate the fold changes in phosphorylation between the control and treatment. Only unique unambiguous phosphopeptides with pRS site probabilities ≥75% were used for phosphoproteomic quantification (pRS probabilities for individual phosphopeptides are listed in Supplemental Data Sets 2 and 3). The phosphopeptide ratio was calculated from two or three biological replicates as indicated in Supplemental Data Set 4. Phosphopeptide abundances were statistically analyzed using a one-sided t test. Calculation of q-values indicated that only slightly more than 10% of phosphopeptides with P ≤ 0.05 would be expected to be false positives; thus, we used this as the main threshold for statistical significance. Phosphopeptide abundances were compared with gene expression data collected in samples from the same set of experiments. Microarray experiments were conducted as previously described (Bhaskara et al., 2012).
Phos-Tag-SDS-PAGE and Immunoblot
Protein Sample Preparation
Seedlings were ground in liquid nitrogen and further homogenized using cold extraction buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 1 mM DTT, 2× protease inhibitor [EDTA-free cocktail tablets; Roche], and 0.1% Triton X-100). The homogenate was centrifuged and protein concentration determined using a Pierce BCA protein assay kit. For in vitro phosphatase treatment, 50 μg protein was treated with 3 μg phosphatase (Sigma-Aldrich; P7640). The reaction was stopped by adding SDS loading buffer and heat inactivation at 70°C. Zn-Phos-tag SDS PAGE and immunoblot were performed as described previously (Kinoshita and Kinoshita-Kikuta, 2011) using conditions optimized for MASP1. Phos-tag gels were prepared using 5 mL of 4% stacking gel (1.785 mL of 1 M Bis-Tris, pH 6.8, 500 μL of 40% acrylamide, 5 μL TEMED, and 25 μL of 10% ammonium persulfate) and 20 mL of 7%, 50 μM Phos-tag resolving gel [7.14 mL of 1 M Bis-Tris, pH 6.8, 200 μL of 10 mM Zn(NO3)2⋅6H2O, 200 μL of 5 mM Phos-tag acrylamide (Phos-tag ligand, AAL-107), 10 μL TEMED, 3.5 mL of 40% acrylamide, and 100 μL 10% ammonium persulfate]. Protein samples (50 μg) with and without alkaline phosphatase treatment were loaded onto the gel and the gel was run at 20 V for 14 h at 4°C using 1× NuPAGE MOPS running buffer (50 mM MOPS, 50 mM Tris base, pH 7.8, 0.1% SDS, and freshly prepared 5 mM sodium bisulfite). The resolving gel was then soaked in 2 × 100 mL of NuPAGE transfer buffer (192 mM glycine, 25 mM Tris-base, pH 7.2, 5 mM sodium bisulfite, and 10% methanol containing 10 mM EDTA) twice for 30 min each time and washed once in transfer buffer without EDTA. The gel was further incubated in 100 mL NuPAGE transfer buffer containing 0.2% (w/v) SDS (two 20-min incubations). Protein on the gel was blotted on PVDF membrane by wet tank procedure at 30 V (at constant voltage) for 16 h at 4°C using NuPAGE transfer buffer containing 1 mM EDTA. MASP1 was detected using antisera obtained from AbMart (http://www.ab-mart.com/proteomics-monoclonal-antibodies_overview.html). Normal SDS-PAGE analysis and blotting (with HSC70 as a loading control) were conducted as previously described (Bhaskara et al., 2015).
In Vivo MT Analysis and Cell Size Measurement
MTs were observed in cells at the base of the hypocotyl elongation zone under confocal fluorescence microscopy. MT orientation was analyzed using the Fiji ImageJ package (http://fiji.sc/Fiji) essentially as described (Zhang et al., 2013). A vertical line was drawn that defined the long axis of the cell and the angle tool from ImageJ was used to measure the angle of each resolvable microtubule in that cell (i.e., bundles count as one microtubule for this analysis) with respect to the vertical line drawn. At least 15 to 20 cells were measured for each treatment. Cell length and width were measured by drawing a line through the longest and widest observable parts of the same cell that are used for measuring the MT number and angle. Cell area calculations assumed a rectangular cell shape (length × width) and anisotrophy was calculated as the ratio of cell length versus width.
Recombinant Protein Expression and Purification
For bacterial expression of His-tagged MASP1 proteins, constructs were cloned into the pET300NT destination vector and transformed into Escherichia coli strain BL21 (Rosetta DE3). Protein induction was done by incubating the culture overnight at 16°C with the addition of 0.1 M IPTG. Cells were harvested, resuspended in equilibration/wash buffer (50 mM HEPES, 300 mM KCl, 10% glycerol, 10 mM imidazole, and 1× Roche EDTA-free protease inhibitor cocktail, pH 7.4), and lysed using cell disruptor (Constant Systems). Clear lysate was applied to Mini Profinity IMAC cartridges using a BioLogic LP system (Bio-Rad). An imidazole gradient was used to elute the protein in elution buffer (50 mM HEPES, 300 mM KCl, 20% glycerol 300 mM imidazole, and 1× protease inhibitor at pH 7.4).
MT Cosedimentation Assay
One milligram of bovine brain tubulin was dissolved in 200 μL of ice cold GTB buffer (80 mM PIPES, pH 7.0, 2 mM MgCl2, and 0.5 mM EGTA) supplemented with 1 mM GTP. The protein concentration was 5 mg/mL and aliquots of 20 μL of tubulin stocks was snap-frozen in liquid nitrogen and stored at –70°C. For MT assembly, 20-μL aliquots of tubulin stock were defrosted by incubating for several minutes at room temperature and immediately transferred to ice. Tubulin polymerization was initiated by adding 2 μL of cushion buffer (80 mM PIPES, pH 7.0, 1 mM MgCl2, 1 mM EGTA, and 60% glycerol) followed by incubation at 35°C for 20 min. These assembled MTs were then diluted with 200 μL of 35°C prewarmed tubulin buffer and stabilized by adding taxol to a final concentration of 20 μM. This preparation yielded a population of stable MTs between 5 and 10 μm in length and at a concentration of ∼5.0 × 1011 MT/mL. This is equivalent to 5 μM tubulin dimer. For cosedimentation assays, MASP1 wild type and mutants were precentrifuged at 20,000g for 15 min at 4°C to remove any insoluble components. The specified concentrations of recombinant proteins were added to 20 μL of the above prepared prepolymerized MTs and the reaction was adjusted to a final volume of 50 μL with GTB buffer. The reaction was incubated for 30 min at room temperature and then centrifuged at 20,000g for 15 min at 4°C. Forty microliters of supernatant was transferred to a fresh tube and the remaining supernatant was carefully removed without disturbing the MT pellets. The MT pellets were then dissolved in 40 μL of tubulin buffer. Samples were added to 10 μL Laemmli sample buffer and heated at 95°C for 5 min before being resolved on 10% SDS-PAGE gels. Gels were stained with Coomassie Brilliant Blue R 250. In all the negative controls where there are no MTs or recombinant proteins, the reaction mixture was supplemented with GTB buffer.
Microtubule Bundling Assay
Porcine brain tubulin protein labeled with an activated ester of rhodamine [(5(and 6)-carboxytetramethylrhodamine succinimidyl ester) was obtained from Cytoskeleton. Labeled tubulin was reconstituted to 4 mg/mL with GTB buffer supplemented with 10% glycerol and 1 mM GTP. The reaction was incubated at 37°C for 20 min to polymerize the tubulin and form MTs. MTs were then incubated in the presence of 30 μM taxol at 37°C for 5 min to stabilize them. To test the MASP1 MT bundling activity, 6 μM of MASP1 alleles, MASP1 truncations, or BSA was added to 1 μL of MT stock and the final reaction volume was adjusted to 200 μL with prewarmed GTB buffer, supplemented with 20 μM taxol. For confocal microscopy observation, 5 μL of the mixture was mounted onto a glass slide beneath a cover slip and fluorescence was visualized using a filter set of 535-nm excitation and 585-nm emission.
To test MT stability under cold treatment, 6 μM protein was added to prepolymerized labeled microtubules and incubated at 10°C for 45 min.
Microtubule Polymerization Turbidity Assay
To test whether MASP1 promoted MT polymerization, polymerization assays were conducted using a low tubulin concentration (1 mg mL−1) in ice-cold GTB buffer supplemented with 1 mM GTP minus glycerol. Reactions were supplemented with taxol (5 to 10 μM) or MASP1 proteins. The reaction was started by the addition of 37°C prewarmed GTB buffer supplemented with 1 mM GTP. To test the MT stabilization effect of MASP1, other reactions were prepared in the same manner but with 5% glycerol to enhance MT polymerization. The reaction mixtures were then transferred to microtiter plates prewarmed to 37°C. MT polymerization was measured by recording absorbance at 340 nm every 5 or 10 min for 60 min. For the reactions with glycerol, three independent experiments were conducted with similar results and representative data are shown.
Structural Modeling and Analysis
For structure prediction and identification of structural orthologs, the MASP1 sequence was submitted to I-Tasser (Yang et al., 2015) and ModWeb (Pieper et al., 2014). The PyMOL visualization tool was used to generate the 3D representation of MASP1. LRR repeat regions were predicted using LRRsearch (Bej et al., 2014). The charge of individual amino acids was plotted using EMBOSS explorer and the pI value was calculated using the Compute pI/Mw tool. The phylogenetic tree of MASP1 and related proteins was constructed using phylogeny.fr (Dereeper et al., 2008).
Statistics
Quantitative data are reported as means ± se with data combined from two or more experiments and number of samples as indicated in the figure legends. Significant differences are based on one-sided t test (for phosphopeptide ratios or growth data reported relative to the wild type), t test, or ANOVA (performed in Sigma Plot 12). For phosphoproteomics data, q-values were calculated to estimate the false discovery rate as described above.
Accession Numbers
The complete egr1-1 egr2-1 microarray data are available in Gene Expression Omnibus (accession number GSE71237). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Vizcaíno et al., 2016) partner repository with the data set identifier PXD004869. Gene accession numbers are EGR1 (AT3G05640), EGR2 (At5G27930), EGR3 (At3G16800), and MASP1 (AT4G03260).
Supplemental Data
Supplemental Figure 1. Clade E Growth-Regulating (EGR) PP2C mutants, gene expression, and proline accumulation.
Supplemental Figure 2. Seedling growth measurements of Col-0 wild type in control as well as −0.7 and −1.2 MPa low ψw treatments.
Supplemental Figure 3. Phosphoproteomic analysis of drought acclimation identifies new aspects of the stress phosphoproteome including Microtubule Associated Stress Protien 1 (MASP1).
Supplemental Figure 4. MASP1 T-DNA lines, proline accumulation, and expression of MASP1 in the GFP-TUA6 background.
Supplemental Figure 5. Time course of MT structure in wild type (GFP-TUA6), egr1-1 egr2-1, and MASP1 overexpression lines after transfer from control (−0.25 MPa) to low ψw stress (−1.2 MPa).
Supplemental Figure 6. Cell size and anisotrophy of EGR and MASP1 mutant and overexpression lines.
Supplemental Figure 7. Propidium iodide staining of root cells of EGR and MASP1 mutant and overexpression lines.
Supplemental Figure 8. Effect of ACC and AVG on root elongation of EGR and MASP1 mutant and overexpression lines.
Supplemental Figure 9. Propidium iodide staining of EGR and MASP1 mutant and overexpression lines after ACC or AVG treatment at high or low ψw.
Supplemental Figure 10. MASP1 structure predicted by I-Tasser and Phyre2 and structural features putatively related to microtubule binding.
Supplemental Data Set 1. ANOVA table for the leaf epidermal cell size data shown in Figure 1G.
Supplemental Data Set 2. All phosphopeptides identified.
Supplemental Data Set 3. Nonredundant list of phosphopeptides and phosphoproteins identified.
Supplemental Data Set 4. Peptides found in two or three replicates of control and stress samples, phosphopeptide abundance ratios, and gene expression ratios between stress and control for wild type and comparison of phosphoproteins with increased or decreased phosphopeptide abundance in wild type to proteins found by Wang et al. (2013) and Umezawa et al. (2013).
Supplemental Data Set 5. Phosphopeptides significantly increased by low water potential stress in wild type based on P ≤ 0.05 and fold change >1.5-fold.
Supplemental Data Set 6. Peptides significantly decreased by low water potential stress in wild type based on P ≤ 0.05 and fold change >1.5-fold.
Supplemental Data Set 7. Comparison of proteins with increased and decreased phosphopeptide abundances identified in both our data and the data of Kline et al. (2010), Stecker et al. (2014), and Minkoff et al. (2015).
Supplemental Data Set 8. Phosphopeptides found in two or three replicates of wild type and egr1-1 egr2-1 under control and stress conditions along with expression of the corresponding gene in egr1-1 egr2-1 relative to wild type.
Supplemental Data Set 9. Genes up- or downregulated in egr1-1 egr2-1 seedlings relative to wild type under control (−0.25 MPa) conditions.
Supplemental Data Set 10. Genes up- or downregulated in egr1-1 egr2-1 compared with wild type at low ψw.
Supplemental Data Set 11. Protein sequences and alignments used to generate the phylogenetic tree of MASP1 and its closest homologs shown in Supplemental Figure 3E.
Supplemental Data Set 12. ANOVA table for the leaf epidermal cell size data shown in Figure 3F.
Supplemental Data Set 13. Primers used for cloning, genotyping, mutagenesis, and gene expression analysis.
Supplementary Material
Acknowledgments
We thank J.-Y. Huang and M.-J. Fang for microscopy assistance, W.-N. Jane for scanning electron microscopy analysis, R. Dixit (Washington University in St. Louis) for advice on microtubule analysis, T.E. Juenger (University of Texas-Austin) for statistical advice, the laboratory of M.-C. Shih (Agricultural Biotechnology Research Center, Academia Sinica) for assistance with ethylene inhibitors, T.-H. Yang and T.Z. Chang for laboratory assistance, and the Affymetrix Gene Expression Service Laboratory of Academia Sinica for microarray processing. This work was supported by the Taiwan Ministry of Science and Technology (NSC100-2311-B-001-008, NSC101-2311-B-001-012, and MoST103-2314-B-001-003 to P.E.V.) and funding from Academia Sinica.
AUTHOR CONTRIBUTIONS
P.E.V. conceived the research and designed experiments with G.B.B. G.B.B. performed all of the experiments, except the phosphoproteomics, and analyzed data. T.T.N. and G.B.B. performed the phosphoproteomic experiments. T.T.N. analyzed the data. T.-N.W. designed and supervised the phosphoproteomic experiments and analyzed the data. P.E.V. and G.B.B prepared the manuscript.
Glossary
- MT
microtubule
- ABA
abscisic acid
- BiFC
bimolecular fluorescence complementation
- PI
propidium iodide
- ACC
1-aminocyclopropane-1-carboxylic acid
- AVG
aminoethoxyvinylglycine
- LRR
leucine-rich repeat
- PEG
polyethylene glycol
- AGC
automatic gain control
References
- Ahmadzadeh H., Smith D.H., Shenoy V.B. (2015). Mechanical effects of dynamic binding between Tau proteins on microtubules during axonal injury. Biophys. J. 109: 2328–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose J.C., Wasteneys G.O. (2008). CLASP modulates microtubule-cortex interaction during self-organization of acentrosomal microtubules. Mol. Biol. Cell 19: 4730–4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose J.C., Shoji T., Kotzer A.M., Pighin J.A., Wasteneys G.O. (2007). The Arabidopsis CLASP gene encodes a microtubule-associated protein involved in cell expansion and division. Plant Cell 19: 2763–2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban Y., Kobayashi Y., Hara T., Hamada T., Hashimoto T., Takeda S., Hattori T. (2013). α-tubulin is rapidly phosphorylated in response to hyperosmotic stress in rice and Arabidopsis. Plant Cell Physiol. 54: 848–858. [DOI] [PubMed] [Google Scholar]
- Bannigan A., Wiedemeier A.M.D., Williamson R.E., Overall R.L., Baskin T.I. (2006). Cortical microtubule arrays lose uniform alignment between cells and are oryzalin resistant in the Arabidopsis mutant, radially swollen 6. Plant Cell Physiol. 47: 949–958. [DOI] [PubMed] [Google Scholar]
- Baskin T.I., Bivens N.J. (1995). Stimulation of radial expansion in arabidopsis roots by inhibitors of actomyosin and vesicle secretion but not by various inhibitors of metabolism. Planta 197: 514–521. [DOI] [PubMed] [Google Scholar]
- Baskin T.I., Wilson J.E. (1997). Inhibitors of protein kinases and phosphatases alter root morphology and disorganize cortical microtubules. Plant Physiol. 113: 493–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin T.I., Wilson J.E., Cork A., Williamson R.E. (1994). Morphology and microtubule organization in Arabidopsis roots exposed to oryzalin or taxol. Plant Cell Physiol. 35: 935–942. [PubMed] [Google Scholar]
- Bates L.S., Waldren R.P., Teare I.D. (1973). Rapid determination of free proline for water-stress studies. Plant Soil 39: 205–207. [Google Scholar]
- Bej A., Sahoo B.R., Swain B., Basu M., Jayasankar P., Samanta M. (2014). LRRsearch: An asynchronous server-based application for the prediction of leucine-rich repeat motifs and an integrative database of NOD-like receptors. Comput. Biol. Med. 53: 164–170. [DOI] [PubMed] [Google Scholar]
- Benschop J.J., Mohammed S., O’Flaherty M., Heck A.J.R., Slijper M., Menke F.L.H. (2007). Quantitative phosphoproteomics of early elicitor signaling in Arabidopsis. Mol. Cell. Proteomics 6: 1198–1214. [DOI] [PubMed] [Google Scholar]
- Bhaskara G.B., Nguyen T.T., Verslues P.E. (2012). Unique drought resistance functions of the highly ABA-induced clade A protein phosphatase 2Cs. Plant Physiol. 160: 379–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskara G.B., Yang T.-H., Verslues P.E. (2015). Dynamic proline metabolism: importance and regulation in water limited environments. Front. Plant Sci. 6: 484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer J.S. (1982). Plant productivity and environment. Science 218: 443–448. [DOI] [PubMed] [Google Scholar]
- Brandizzi F., Wasteneys G.O. (2013). Cytoskeleton-dependent endomembrane organization in plant cells: an emerging role for microtubules. Plant J. 75: 339–349. [DOI] [PubMed] [Google Scholar]
- Buschmann H., Hauptmann M., Niessing D., Lloyd C.W., Schäffner A.R. (2009). Helical growth of the Arabidopsis mutant tortifolia2 does not depend on cell division patterns but involves handed twisting of isolated cells. Plant Cell 21: 2090–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschmann H., Chan J., Sanchez-Pulido L., Andrade-Navarro M.A., Doonan J.H., Lloyd C.W. (2006). Microtubule-associated AIR9 recognizes the cortical division site at preprophase and cell-plate insertion. Curr. Biol. 16: 1938–1943. [DOI] [PubMed] [Google Scholar]
- Butner K.A., Kirschner M.W. (1991). Tau protein binds to microtubules through a flexible array of distributed weak sites. J. Cell Biol. 115: 717–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claeys H., Inzé D. (2013). The agony of choice: how plants balance growth and survival under water-limiting conditions. Plant Physiol. 162: 1768–1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler S.R., Rodriguez P.L., Finkelstein R.R., Abrams S.R. (2010). Abscisic acid: emergence of a core signaling network. Annu. Rev. Plant Biol. 61: 651–679. [DOI] [PubMed] [Google Scholar]
- Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., Chevenet F., Dufayard J.-F., Guindon S., Lefort V., Lescot M., Claverie J.-M., Gascuel O. (2008). Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36: W465–W469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhonukshe P., Laxalt A.M., Goedhart J., Gadella T.W.J., Munnik T. (2003). Phospholipase d activation correlates with microtubule reorganization in living plant cells. Plant Cell 15: 2666–2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doskočilová A., Plíhal O., Volc J., Chumová J., Kourová H., Halada P., Petrovská B., Binarová P. (2011). A nodulin/glutamine synthetase-like fusion protein is implicated in the regulation of root morphogenesis and in signalling triggered by flagellin. Planta 234: 459–476. [DOI] [PubMed] [Google Scholar]
- Dubois M., Claeys H., Van den Broeck L., Inzé D. (2016). Time of day determines Arabidopsis transcriptome and growth dynamics under mild drought. Plant Cell Environ. 10.1111/pce.12809. [DOI] [PubMed] [Google Scholar]
- Earley K.W., Haag J.R., Pontes O., Opper K., Juehne T., Song K., Pikaard C.S. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45: 616–629. [DOI] [PubMed] [Google Scholar]
- Endler A., Kesten C., Schneider R., Zhang Y., Ivakov A., Froehlich A., Funke N., Persson S. (2015). A mechanism for sustained cellulose synthesis during salt stress. Cell 162: 1353–1364. [DOI] [PubMed] [Google Scholar]
- Feng W., Lindner H., Robbins N.E. II, Dinneny J.R. (2016). Growing out of stress: The role of cell- and organ-scale growth control in plant water-stress responses. Plant Cell 28: 1769–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs S., Grill E., Meskiene I., Schweighofer A. (2013). Type 2C protein phosphatases in plants. FEBS J. 280: 681–693. [DOI] [PubMed] [Google Scholar]
- Fujita S., Pytela J., Hotta T., Kato T., Hamada T., Akamatsu R., Ishida Y., Kutsuna N., Hasezawa S., Nomura Y., Nakagami H., Hashimoto T. (2013). An atypical tubulin kinase mediates stress-induced microtubule depolymerization in Arabidopsis. Curr. Biol. 23: 1969–1978. [DOI] [PubMed] [Google Scholar]
- Gardiner J.C., Harper J.D.I., Weerakoon N.D., Collings D.A., Ritchie S., Gilroy S., Cyr R.J., Marc J. (2001). A 90-kD phospholipase D from tobacco binds to microtubules and the plasma membrane. Plant Cell 13: 2143–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzmán P., Ecker J.R. (1990). Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 2: 513–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada T., Nagasaki-Takeuchi N., Kato T., Fujiwara M., Sonobe S., Fukao Y., Hashimoto T. (2013). Purification and characterization of novel microtubule-associated proteins from Arabidopsis cell suspension cultures. Plant Physiol. 163: 1804–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamant O., Heisler M.G., Jönsson H., Krupinski P., Uyttewaal M., Bokov P., Corson F., Sahlin P., Boudaoud A., Meyerowitz E.M., Couder Y., Traas J. (2008). Developmental patterning by mechanical signals in Arabidopsis. Science 322: 1650–1655. [DOI] [PubMed] [Google Scholar]
- Harrison B., Masson P.H. (2008). ARG1 and ARL2 form an actin-based gravity-signaling chaperone complex in root statocytes? Plant Signal. Behav. 3: 650–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haswell E.S., Verslues P.E. (2015). The ongoing search for the molecular basis of plant osmosensing. J. Gen. Physiol. 145: 389–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horan K., Jang C., Bailey-Serres J., Mittler R., Shelton C., Harper J.F., Zhu J.K., Cushman J.C., Gollery M., Girke T. (2008). Annotating genes of known and unknown function by large-scale coexpression analysis. Plant Physiol. 147: 41–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichihara K., Kitazawa H., Iguchi Y., Hotani H., Itoh T.J. (2001). Visualization of the stop of microtubule depolymerization that occurs at the high-density region of microtubule-associated protein 2 (MAP2). J. Mol. Biol. 312: 107–118. [DOI] [PubMed] [Google Scholar]
- Kinoshita E., Kinoshita-Kikuta E. (2011). Improved Phos-tag SDS-PAGE under neutral pH conditions for advanced protein phosphorylation profiling. Proteomics 11: 319–323. [DOI] [PubMed] [Google Scholar]
- Kline K.G., Barrett-Wilt G.A., Sussman M.R. (2010). In planta changes in protein phosphorylation induced by the plant hormone abscisic acid. Proc. Natl. Acad. Sci. USA 107: 15986–15991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komis G., Apostolakos P., Galatis B. (2001). Altered patterns of tubulin polymerization in dividing leaf cells of Chlorophyton comosum after a hyperosmotic treatment. New Phytol. 149: 193–207. [DOI] [PubMed] [Google Scholar]
- Komis G., Illés P., Beck M., Šamaj J. (2011). Microtubules and mitogen-activated protein kinase signalling. Curr. Opin. Plant Biol. 14: 650–657. [DOI] [PubMed] [Google Scholar]
- Korolev A.V., Buschmann H., Doonan J.H., Lloyd C.W. (2007). AtMAP70-5, a divergent member of the MAP70 family of microtubule-associated proteins, is required for anisotropic cell growth in Arabidopsis. J. Cell Sci. 120: 2241–2247. [DOI] [PubMed] [Google Scholar]
- Korolev A.V., Chan J., Naldrett M.J., Doonan J.H., Lloyd C.W. (2005). Identification of a novel family of 70 kDa microtubule-associated proteins in Arabidopsis cells. Plant J. 42: 547–555. [DOI] [PubMed] [Google Scholar]
- Landrein B., Hamant O. (2013). How mechanical stress controls microtubule behavior and morphogenesis in plants: history, experiments and revisited theories. Plant J. 75: 324–338. [DOI] [PubMed] [Google Scholar]
- Lei L., Li S., Bashline L., Gu Y. (2014). Dissecting the molecular mechanism underlying the intimate relationship between cellulose microfibrils and cortical microtubules. Front. Plant Sci. 5: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeNoble M.E., Spollen W.G., Sharp R.E. (2004). Maintenance of shoot growth by endogenous ABA: genetic assessment of the involvement of ethylene suppression. J. Exp. Bot. 55: 237–245. [DOI] [PubMed] [Google Scholar]
- Lewis D.R., Negi S., Sukumar P., Muday G.K. (2011). Ethylene inhibits lateral root development, increases IAA transport and expression of PIN3 and PIN7 auxin efflux carriers. Development 138: 3485–3495. [DOI] [PubMed] [Google Scholar]
- Liu Z., Schneider R., Kesten C., Zhang Y., Somssich M., Zhang Y., Fernie A.R., Persson S. (2016). Cellulose-microtubule uncoupling proteins prevent lateral displacement of microtubules during cellulose synthesis in Arabidopsis. Dev. Cell 38: 305–315. [DOI] [PubMed] [Google Scholar]
- Luo C., Guo C., Wang W., Wang L., Chen L. (2014). Overexpression of a new stress-repressive gene OsDSR2 encoding a protein with a DUF966 domain increases salt and simulated drought stress sensitivities and reduces ABA sensitivity in rice. Plant Cell Rep. 33: 323–336. [DOI] [PubMed] [Google Scholar]
- Ma Q., Sun J., Mao T. (2016). Microtubule bundling plays a role in ethylene-mediated cortical microtubule reorientation in etiolated Arabidopsis hypocotyls. J. Cell Sci. 129: 2043–2051. [DOI] [PubMed] [Google Scholar]
- Martin K., Kopperud K., Chakrabarty R., Banerjee R., Brooks R., Goodin M.M. (2009). Transient expression in Nicotiana benthamiana fluorescent marker lines provides enhanced definition of protein localization, movement and interactions in planta. Plant J. 59: 150–162. [DOI] [PubMed] [Google Scholar]
- McMichael C.M., Reynolds G.D., Koch L.M., Wang C., Jiang N., Nadeau J., Sack F.D., Gelderman M.B., Pan J., Bednarek S.Y. (2013). Mediation of clathrin-dependent trafficking during cytokinesis and cell expansion by Arabidopsis stomatal cytokinesis defective proteins. Plant Cell 25: 3910–3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei Y., Gao H.B., Yuan M., Xue H.W. (2012). The Arabidopsis ARCP protein, CSI1, which is required for microtubule stability, is necessary for root and anther development. Plant Cell 24: 1066–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minkoff B.B., Stecker K.E., Sussman M.R. (2015). Rapid phosphoproteomic effects of abscisic acid (ABA) on wild-type and ABA receptor-deficient A. thaliana mutants. Mol. Cell. Proteomics 14: 1169–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T., et al. (2007). Improved Gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci. Biotechnol. Biochem. 71: 2095–2100. [DOI] [PubMed] [Google Scholar]
- Nakamura M., Hashimoto T. (2009). A mutation in the Arabidopsis gamma-tubulin-containing complex causes helical growth and abnormal microtubule branching. J. Cell Sci. 122: 2208–2217. [DOI] [PubMed] [Google Scholar]
- Nick P. (2013). Microtubules, signalling and abiotic stress. Plant J. 75: 309–323. [DOI] [PubMed] [Google Scholar]
- Pieper U., et al. (2014). ModBase, a database of annotated comparative protein structure models and associated resources. Nucleic Acids Res. 42: D336–D346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierik R., Sasidharan R., Voesenek L.A.C.J. (2007). Growth control by ethylene: Adjusting phenotypes to the environment. J. Plant Growth Regul. 26: 188–200. [Google Scholar]
- Polko J.K., van Zanten M., van Rooij J.A., Marée A.F.M., Voesenek L.A., Peeters A.J.M., Pierik R. (2012). Ethylene-induced differential petiole growth in Arabidopsis thaliana involves local microtubule reorientation and cell expansion. New Phytol. 193: 339–348. [DOI] [PubMed] [Google Scholar]
- Roberts I.N., Lloyd C.W., Roberts K. (1985). Ethylene-induced microtubule reorientations: mediation by helical arrays. Planta 164: 439–447. [DOI] [PubMed] [Google Scholar]
- Sampathkumar A., Yan A., Krupinski P., Meyerowitz E.M. (2014). Physical forces regulate plant development and morphogenesis. Curr. Biol. 24: R475–R483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweighofer A., Hirt H., Meskiene I. (2004). Plant PP2C phosphatases: emerging functions in stress signaling. Trends Plant Sci. 9: 236–243. [DOI] [PubMed] [Google Scholar]
- Seung D., Webster M.W., Wang R., Andreeva Z., Marc J. (2013). Dissecting the mechanism of abscisic acid-induced dynamic microtubule reorientation using live cell imaging. Funct. Plant Biol. 40: 224–236. [DOI] [PubMed] [Google Scholar]
- Shoji T., Narita N.N., Hayashi K., Asada J., Hamada T., Sonobe S., Nakajima K., Hashimoto T. (2004). Plant-specific microtubule-associated protein SPIRAL2 is required for anisotropic growth in Arabidopsis. Plant Physiol. 136: 3933–3944. Erratum. Plant Physiol. 137: 1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skirycz A., Inzé D. (2010). More from less: plant growth under limited water. Curr. Opin. Biotechnol. 21: 197–203. [DOI] [PubMed] [Google Scholar]
- Skirycz A., et al. (2011). Survival and growth of Arabidopsis plants given limited water are not equal. Nat. Biotechnol. 29: 212–214. [DOI] [PubMed] [Google Scholar]
- Smertenko A.P., Kaloriti D., Chang H.Y., Fiserova J., Opatrny Z., Hussey P.J. (2008). The C-terminal variable region specifies the dynamic properties of Arabidopsis microtubule-associated protein MAP65 isotypes. Plant Cell 20: 3346–3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spollen W.G., LeNoble M.E., Samuels T.D., Bernstein N., Sharp R.E. (2000). Abscisic acid accumulation maintains maize primary root elongation at low water potentials by restricting ethylene production. Plant Physiol. 122: 967–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecker K.E., Minkoff B.B., Sussman M.R. (2014). Phosphoproteomic analyses reveal early signaling events in the osmotic stress response. Plant Physiol. 165: 1171–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto H., Kondo S., Tanaka T., Imamura C., Muramoto N., Hattori E., Ogawa K., Mitsukawa N., Ohto C. (2014). Overexpression of a novel Arabidopsis PP2C isoform, AtPP2CF1, enhances plant biomass production by increasing inflorescence stem growth. J. Exp. Bot. 65: 5385–5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K., Himmelspach R., Williamson R.E., Wasteneys G.O. (2003). Mutation or drug-dependent microtubule disruption causes radial swelling without altering parallel cellulose microfibril deposition in Arabidopsis root cells. Plant Cell 15: 1414–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatani S., Hirayama T., Hashimoto T., Takahashi T., Motose H. (2015). Abscisic acid induces ectopic outgrowth in epidermal cells through cortical microtubule reorganization in Arabidopsis thaliana. Sci. Rep. 5: 11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardieu F., Granier C., Muller B. (2011). Water deficit and growth. Co-ordinating processes without an orchestrator? Curr. Opin. Plant Biol. 14: 283–289. [DOI] [PubMed] [Google Scholar]
- Thitamadee S., Tuchihara K., Hashimoto T. (2002). Microtubule basis for left-handed helical growth in Arabidopsis. Nature 417: 193–196. [DOI] [PubMed] [Google Scholar]
- Tsuda K., Qi Y., Nguyen V., Bethke G., Tsuda Y., Glazebrook J., Katagiri F. (2012). An efficient Agrobacterium-mediated transient transformation of Arabidopsis. Plant J. 69: 713–719. [DOI] [PubMed] [Google Scholar]
- Ueda K., Matsuyama T., Hashimoto T. (1999). Visualization of microtubules in living cells of transgenic Arabidopsis thaliana. Protoplasma 206: 201–206. [Google Scholar]
- Umezawa T., Sugiyama N., Takahashi F., Anderson J.C., Ishihama Y., Peck S.C., Shinozaki K. (2013). Genetics and phosphoproteomics reveal a protein phosphorylation network in the abscisic acid signaling pathway in Arabidopsis thaliana. Sci. Signal. 6: rs8. [DOI] [PubMed] [Google Scholar]
- van der Honing H.S., Kieft H., Emons A.M.C., Ketelaar T. (2012). Arabidopsis VILLIN2 and VILLIN3 are required for the generation of thick actin filament bundles and for directional organ growth. Plant Physiol. 158: 1426–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verslues P.E. (2016). Time to grow: factors that control plant growth during mild to moderate drought stress. Plant Cell Environ., doi/10.1111/pce.12827. [DOI] [PubMed] [Google Scholar]
- Verslues P.E., Agarwal M., Katiyar-Agarwal S., Zhu J., Zhu J.K. (2006). Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J. 45: 523–539. [DOI] [PubMed] [Google Scholar]
- Vizcaíno J.A., et al. (2016). 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44: D447–D456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Li J., Yuan M. (2007). Salt tolerance requires cortical microtubule reorganization in Arabidopsis. Plant Cell Physiol. 48: 1534–1547. [DOI] [PubMed] [Google Scholar]
- Wang P., Xue L., Batelli G., Lee S., Hou Y.J., Van Oosten M.J., Zhang H., Tao W.A., Zhu J.K. (2013). Quantitative phosphoproteomics identifies SnRK2 protein kinase substrates and reveals the effectors of abscisic acid action. Proc. Natl. Acad. Sci. USA 110: 11205–11210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Kurepa J., Hashimoto T., Smalle J.A. (2011). Salt stress-induced disassembly of Arabidopsis cortical microtubule arrays involves 26S proteasome-dependent degradation of SPIRAL1. Plant Cell 23: 3412–3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., McFarlane H.E., Persson S. (2016). The impact of abiotic factors on cellulose synthesis. J. Exp. Bot. 67: 543–552. [DOI] [PubMed] [Google Scholar]
- Wang X., Zhu L., Liu B., Wang C., Jin L., Zhao Q., Yuan M. (2007). Arabidopsis MICROTUBULE-ASSOCIATED PROTEIN18 functions in directional cell growth by destabilizing cortical microtubules. Plant Cell 19: 877–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Warren P., Shakey Q., Sousa E., Hill A., Ryan T.E., He T. (2010). Integrating titania enrichment, iTRAQ labeling, and Orbitrap CID-HCD for global identification and quantitative analysis of phosphopeptides. Proteomics 10: 2224–2234. [DOI] [PubMed] [Google Scholar]
- Yang J., Yan R., Roy A., Xu D., Poisson J., Zhang Y. (2015). The I-TASSER Suite: protein structure and function prediction. Nat. Methods 12: 7–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Fishel E., Bertroche T., Dixit R. (2013). Microtubule severing at crossover sites by katanin generates ordered cortical microtubule arrays in Arabidopsis. Curr. Biol. 23: 2191–2195. [DOI] [PubMed] [Google Scholar]
- Zhang Q., Lin F., Mao T., Nie J., Yan M., Yuan M., Zhang W. (2012). Phosphatidic acid regulates microtubule organization by interacting with MAP65-1 in response to salt stress in Arabidopsis. Plant Cell 24: 4555–4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z.J., Peck S.C. (2011). Simplified enrichment of plasma membrane proteins for proteomic analyses in Arabidopsis thaliana. Proteomics 11: 1780–1788. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.