Abstract
The circadian timing system consists on a distributed network of cellular clocks that together coordinate 24-h rhythms of physiology and behavior. Clock function and metabolism are tightly coupled, from the cellular to the organismal level. Genetic and non-genetic approaches in rodents have been employed to study circadian clock function in the living organism. Due to the ubiquitous expression of clock genes and the intricate interaction between the circadian system and energy metabolism, genetic approaches targeting specific tissue clocks have been used to assess their contribution in systemic metabolic processes. However, special requirements regarding specificity and efficiency have to be met to allow for valid conclusions from such studies. In this review, we provide a brief summary of different approaches developed for dissecting tissue clock function in the metabolic context in rodents, compare their strengths and weaknesses, and suggest new strategies in assessing tissue clock output and the consequences of circadian clock disruption in vivo.
Keywords: clock genes, metabolism, gene targeting, Bmal1, conditional knockout, CRE-loxP system
Introduction
A network of cellular clocks adapts physiology and behavior to a 24-h rhythmic environment. Epidemiological evidence suggests a strong association between circadian rhythm disruption—e.g., in shift workers—and metabolic disorders (1). The development of genetic rodent models has been essential in deciphering the mechanisms linking clock dysfunction and the pathogenesis of metabolic diseases, highlighting the therapeutic potential of circadian rhythm manipulation in this context. Here, we provide a brief overview of the metabolic consequences of circadian disruption derived from rodent model studies and discuss potential and pitfalls of emerging genetic techniques in studying clock-metabolism crosstalk.
Non-Genetic Circadian Disruption Models
Early approaches describing the interplay between circadian clock function and metabolism were based on models of non-genetic circadian disruption (Table 1). Lesion experiments identified the suprachiasmatic nucleus (SCN) as the central circadian pacemaker (2, 3). In rats, SCN lesions abolish plasma rhythms of glucose, insulin (4), and leptin (5). In terms of general energy homeostasis, there are differences between studies that range from slight weight reductions (6) to no effect (5) and marked obesity (7). In the latter study on mice, increased weight is accompanied by hepatic insulin resistance. In sum, these studies illustrate that the SCN regulates metabolic hormones and tissue physiology either directly or via its control on food intake rhythms.
Table 1.
Advantages and disadvantages of different clock targeting approaches in rodents.
| Paradigm | Advantages | Disadvantages |
|---|---|---|
| Non-genetic clock disruption |
|
|
| Classical (global) gene targeting |
|
|
| Conventional CRE-loxP gene targeting |
|
|
| Inducible CRE-loxP gene targeting |
|
|
| Chemogenetics |
|
|
| Optogenetics |
|
|
In an alternative paradigm, exposure to altered light regimens leads to disruption of behavioral circadian rhythms and energy metabolism. Mice exposed to bright day/dim night illumination lose their feeding rhythm along with reduced glucose tolerance and increased body weight (8). Constant light (LL)-exposed mice develop a higher body weight together with a profound loss in insulin sensitivity (7). In rats, LL exposure disrupts pancreatic beta cell clocks, correlating with reduced glucose-stimulated insulin secretion (9). In type-2 diabetes-prone rats, LL accelerates disease development due to a 50% reduction in beta cell mass (10). Most of these studies interpret their findings as a consequence of the disruptive effects of abnormal light exposure on SCN function. However, light can alter various centrally controlled functions such as mood and appetite independent of a clock effect (11, 12). Moreover, even though irregular light regimens are common for many animals and humans, there are only very few occasions where continuous light exposure above a certain threshold level may occur, making LL a highly artificial paradigm.
Several studies have outlined shift-working as a risk factor for the development of metabolic diseases (13), and various animal models have been developed to mimic this condition. Mice exposed to repeated 6-h advances of the light/dark (LD) cycle gain significantly more body weight than stable-LD controls (14). Interestingly, when mice are exposed to paradigms mimicking even more rapidly rotating weekly shift-work patterns, there is no or only a moderate effect on body weight (15). Sleep-restricting mice during their normal rest phase for 2 weeks leads to significant alterations in liver clock gene expression rhythms associated impaired pyruvate-stimulated gluconeogenesis, but no significant alterations in body weight (16). A similar approach in rats further yielded body weight increases and impaired glucose tolerance (17).
The mixed metabolic outcomes in the mentioned shift-work studies are likely directly related to the ability of the chosen paradigms to provoke food intake during the normal rest phase. Rest phase-restricted food access in mice promotes weight gain without increased energy intake (18). In line with that, while both the rapid-shift (15) and our sleep-restriction study in mice (16) showed no change in food intake rhythms, the 6-h shift model (14) and the rat sleep-restriction study (17) reported misaligned food intake. Considering the reciprocal interaction between energy intake and clock function, it has been speculated that epigenetic programming may be an important mechanism in the circadian regulation of energy metabolism—even across generations [reviewed in Ref. (19, 20)].
Metabolic Alterations in Conventional Clock Gene Mutant Mice
The development of clock gene mutant mouse models has provided a substantial tool to understand clock-metabolism interaction at the molecular level. Following the identification of circadian locomotor output cycles kaput (Clock) (21), further mammalian clock genes have been cloned and functionally characterized in corresponding mouse mutants (22–25). So far, brain and muscle ARNT-like 1 (BMAL1) has been identified as the only essential component of the molecular clockwork in mammals. Deletion of Bmal1 in mice abolishes behavioral circadian rhythms in constant environmental conditions (25). Gene expression profiling experiments show that the rate-limiting steps of various metabolic pathways are subject to circadian regulation (26). Bmal1 knockout (KO) mice show impaired glucose metabolism and insulin hypersensitivity (27, 28). At young age, they also gain weight more rapidly than wild-type littermates (28). Recently, an inducible global Bmal1 KO mouse model has been developed. Unexpectedly, adult-onset Bmal1 KOs do not suffer from many metabolic abnormalities described in the standard Bmal1 KOs (29).
Mice expressing a dominant-negative CLOCK variant (CLOCK-Δ19) on a C57BL/6J genetic background display altered 24-h feeding patterns (30). Clock-Δ19 mice are hyperphagic and show reduced energy expenditure. They also develop hyperglycemia, hyperlipidemia, hyperleptinemia, hypoinsulinemia, and increase in body weight and visceral adiposity under different diet conditions. These phenotypes may be partly explained by a reduced lipolytic capacity of white adipose tissue (31). By contrast, on an ICR genetic background, the same Clock-Δ19 mutation leads to a reduction in body weight and impaired dietary lipid absorption, suggesting that the genetic background influences clock-metabolism interaction (32).
Inconsistent findings were also reported for the metabolic consequences of mutations in another clock gene, Period (Per) 2. Per2Brdm1 mutant mice (24) show hyperphagy, diet-induced obesity (33), hyperinsulinemia with altered insulin sensitivity, hypoglycemia, and low fasting hepatic glycogen content (34). By contrast, Per2ldc mutants (35) show reduced adiposity, increased fatty acid oxidation, and hypotriglyceridemia (36), but are normoglycemic with improved clearance after glucose challenge (37). Of note, while genetic background may also play a role here, residual protein-coding transcripts have been detected in both mutants that may yield biologically active peptides (35, 38). Male, but not female, Per1/2/3 triple mutants become obese under an HFD (39). A Per3 KO alone has even stronger effects on male diet-induced weight gain, suggesting a genetic interaction of different Per genes in metabolic regulation (39).
Liver transcriptomic analyses from mice carrying Rev-Erbα loss- or gain-of-function alleles have identified it as a circadian regulator of cholesterol/lipid and bile acid homeostasis (40). In line with this, mice with adult-onset global loss of Rev-Erbα/β show deregulated glucose and lipid metabolism (41). Moreover, administration of REV-ERB agonists in mice induces body weight loss and decreased lipogenesis in liver and white adipose tissue, increased lipid and glucose oxidation in skeletal muscle, and elevated energy expenditure (42).
Together, these studies provide evidence that the circadian clock plays a fundamental role in energy homeostasis and that different clock components have specific functions in this context. However, because the mice used in these studies carry clock gene mutations in all tissues including the SCN, much like the non-genetic models discussed above, metabolic phenotypes may be confounded by systemic abnormalities such as altered sleep patterns, activity and feeding behaviors, or counteractive consequences of clock disruption in different tissues.
Tissue Clock Function in Metabolic Regulation
Several techniques have been developed to study the physiology of tissue circadian clocks (Table 1). In vitro experiments allow the study of tissue rhythms in the absence of external influences (43). However, such approaches have only limited potential for predicting the impact of tissue clock disruption on complex physiological processes such as energy metabolism. Transplantations have been used to study tissue clock function (44, 45). However, such techniques are highly invasive and only applicable for few tissues. Instead, conditional CRE-loxP-based gene targeting has been widely used for tissue-specific deletion of clock function in vivo (in most cases by targeting Bmal1) (46, 47). Here, we highlight some studies which have provided important insights into the contribution of different tissue clocks to metabolism.
Hepatocyte-specific deletion of Bmal1 (L-Bmal1 KO) improves glucose tolerance—the opposite effect of a global Bmal1 KO—which was attributed to reduced hepatic glucose export during the fasting phase via glucose transporter 2 (28). By contrast, mice with pancreatic beta cell-specific Bmal1 deletion (P-Bmal1 KOs) show hyperglycemia and impaired glucose tolerance (48) due to decreased insulin exocytosis (49). Further, while mice with muscle-specific deletion of Bmal1 (M-Bmal1 KO) show no significant change in systemic glucose regulation, impaired myocyte glucose uptake, and metabolism have been reported (50). Finally—and similar to global Bmal1 KOs and Clock-Δ19 mutants—mice with adipocyte-specific Bmal1 deletion (A-Bmal1 KOs) become obese (ca. 20% body weight) under HFD but not normal chow conditions. This phenotype correlates with misaligned food intake and blunted secretion rhythms of appetite-regulating polyunsaturated fatty acids from adipocytes (51).
Clock gene rhythms have been reported in central energy regulatory circuits (52). So far, few studies have addressed the biological function of specific brain clocks. With regard to metabolism, deletion of Bmal1 in steroidogenic neurons of the ventromedial hypothalamus reduces sympathetic activation of thermogenesis in brown adipose tissue (53).
These few examples make clear that tissue-specific dissection of clock gene function has provided a much more detailed picture on how circadian clocks affect energy metabolism in vivo. Nevertheless, conclusions drawn from these experiments are often confounded by intrinsic drawbacks of the classical CRE-loxP system which novel genetic tools may help to overcome.
Limitations of Classical Genetic Models in Chronobiology Research
Poor Specificity of the CRE Driver
In CRE-loxP-based gene targeting, the tissue-specific mutation is determined by the transcription dynamics of the CRE-expressing promoter. While this may not be such an issue for processes confined to specific tissues, for the ubiquitously active circadian system, this poses an important limitation: many allegedly tissue-specific promoters show varying amounts of off-target activity. For example, for two of the studies mentioned above, CRE drivers with off-target activities have been employed, namely Fabp4-Cre for A-Bmal1 and Pdx1-Cre for P-Bmal1 KOs. Critically, their expression in several metabolism-regulating circuits in the brain has been documented (54). Though the authors of these studies used different approaches to address this problem (such as supplementing in vitro/ex vivo data or repeating key experiments with another CRE driver), certain conclusions drawn from the in vivo metabolic experiments may still be ambiguous. Similarly, in the hypothalamic steroidogenic neuron Bmal1 KO study, the Sf1-Cre line used may very likely affect steroidogenic cells in other tissues (53).
Developmental Compensation
Some CRE drivers are developmentally active and clock genes are known to be involved in early developmental processes (55). Therefore, some phenotypes observed in adult mice might reflect changes in developmental programs while others may be masked by compensatory responses. A strategy circumventing this issue is to employ an inducible version of CRE (T2-CRE), which is activated only when animals receive tamoxifen (56). This approach has been used to globally delete Bmal1 and Rev-erbα/β (29, 41). In the M-Bmal1 KO study discussed above, a muscle strength phenotype was only observed in mutants with conventional, but not inducible CRE deletion (50).
Incomplete Recombination
Due to variations in expression levels or epigenetic effects, CRE-mediated recombination frequently does not occur in all cells of the same tissue. This issue is further exaggerated with the T2-CRE system. Thus, the lack of certain phenotypes in tissue-specific mutants may stem from non-recombined cells sufficient to maintain tissue function. In the circadian clock context, this issue becomes critical in tissues of strong intercellular coupling such as the SCN. Our own approach to target SCN pacemaker function using a Syt10-Cre-driver line revealed that only with the highest CRE dosage (Syt10Cre/Cre) and on a Bmal1flox/del background, recombination was sufficient to ablate behavioral rhythmicity (57).
The Non-Circadian Role of Individual Clock Genes
Many clock gene mutant studies fail to discern whether a phenotype is caused by altered clock rhythmicity or by loss of a specific clock component. For example, in the L-Bmal1 KO study, the mutation not only abolishes the transcription rhythm of Glut2, but also dramatically downregulates its overall expression (28). The multifaceted phenotype of Bmal1 KO mice suggests that this gene has important functions outside the circadian timekeeping system (58). Along this line, the monopolized use of Bmal1 targeting for tissue-specific clock deletion further exacerbates this issue since some of the reported phenotypes may be caused by a loss of Bmal1 rather than of the clockwork.
Emerging Genetic Techniques for Chronobiology Research
Some of the emerging novel genetic techniques may help to overcome the issues discussed above. Below we will highlight some methodologies that may help improving our understanding of the role of tissue clocks in complex physiological contexts.
Improving the Spatio-Temporal Resolution of Clock Gene Manipulation
Though pharmacologically inducible CRE systems bypass developmental effects, this strategy often compromises the recombination efficiency and complicates experimental designs (56). Moreover, the manipulation is irreversible, rendering it unsuitable for certain biological questions. As an alternative approach, chemogenetic manipulations such as tetracycline-based (i.e., Tet-ON/OFF) systems achieve anatomical specificity by expressing effectors under tissue-specific promoters, the activity of which depends on the presence/absence of an otherwise inert chemical (59, 60). These systems are reversible and have a very flexible space-time window for manipulations. For example, two complementary studies have established mouse lines expressing Clock-Δ19 and Rev-erbα as clock disruptors in brain and liver, respectively, in a tetracycline-dependent manner, to elucidate the relative contribution of central and peripheral clocks to physiological rhythms (61, 62). This revealed that circadian expression rhythms of most rhythmic transcripts in the liver depend on local oscillators whereas 10% of the rhythmic transcripts (including Per2) are sustained by systemic signals.
Optogenetic techniques have recently been introduced in chronobiology research. Photic stimulation on channelrhodopsin-2-expressing SCN neurons results in phase-resetting of SCN firing and behavioral rhythms (63). Other optogenetic systems allow for photo-switchable control of specific cellular and molecular processes (64). Of note, one strategy employs modified photo-sensitive plant cryptochrome 2 clock proteins from Arabidopsis (64), which would allow to directly affect molecular clock function by light. In addition to high spatial control (by directing light exposure), optogenetics may benefit chronobiology research because of their supreme temporal resolution, e.g., in phase-resetting experiments.
While standard viral transgene delivery approaches using short tissue-specific promoters often fall short to confer sufficient specificity (65), this is circumvented by combining virus injections with CRE-driver mouse lines targeting transgene expression to specific cells and tissues (66). Viral approaches further benefit from high spatio-temporal control through injection time/sites and viral capsid serotypes. They, however, often suffer from somewhat reduced penetrance and technical variability (67). Viruses can be used in conjugation with chemogenetic or optogenetic manipulations to interrogate the role of circadian clock in metabolic regulation and other physiological systems with unprecedented spatio-temporal resolution.
Perspectives for Developing Animal Models That Can Dissociate Circadian and Non-Circadian Functions of Clock Genes
Clock gene knockdown/KO experiments often cannot distinguish between circadian (i.e., timing related) and non-circadian effects of a given mutation. This may to some extent be addressed by comparing phenotypes between mutants of different clock genes. However, because of the interactive nature of clock genes this may often not yield further insights. As an alternative approach, the direct modulation of circadian period length without abolishing clock function itself in a tissue-specific manner is achievable via manipulating period-determining genes such as casein kinase I ϵ (44). To some extent, this and other similar approaches still inevitably change clock protein levels and, thus, pleiotropic clock gene output.
An ideal animal model would be one with altered phasing of clock gene expression rhythms but unaffected overall clock protein abundance. Generating such model has become possible with recent developments in genome editing such as CRISPR and TALEN (68, 69). CRISPR techniques have been used to “cure” retinal degeneration models in rats (70, 71). Together with the expanding knowledge of the role of the cis-regulatory elements of clock genes in determining the phase of expression rhythms, such as the identification of the phase-determining intronic enhancer of Cry1 (72), in vivo genome editing may allow to selectively manipulate clock gene phasing.
Conclusion
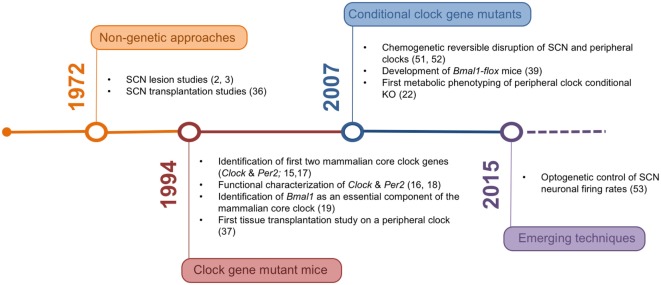
In the last decades, the tools for studying the mechanisms underlying organismal circadian timekeeping and its role in metabolic regulation have been constantly refined (Figure 1). The network structure of the circadian system represents an important challenge in this context, posing high demands on both tempo-spatial control and recombination efficiency in genetic experiments. Novel genetic approaches may help to overcome these issues and provide a clearer picture of the complex interaction of different tissue clocks in the regulation of energy metabolism.
Figure 1.
Timeline of the development of experimental rodent models and corresponding milestone papers in circadian tissue clock research.
Author Contributions
AT, MA, BL, and HO discussed the concept, complied the literature, and wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
This work was supported by grants from the Volkswagen Foundation (Lichtenberg Fellowship to HO), the German Research Association (DFG; GRK 1957 and SFBs 134 and 654), and an International Brain Research Organization (IBRO) fellowship to MA.
References
- 1.Gan Y, Yang C, Tong X, Sun H, Cong Y, Yin X, et al. Shift work and diabetes mellitus: a meta-analysis of observational studies. Occup Environ Med (2015) 72(1):72–8. 10.1136/oemed-2014-102150 [DOI] [PubMed] [Google Scholar]
- 2.Moore RY, Eichler VB. Loss of a circadian adrenal corticosterone rhythm following suprachiasmatic lesions in the rat. Brain Res (1972) 42(1):201–6. 10.1016/0006-8993(72)90054-6 [DOI] [PubMed] [Google Scholar]
- 3.Stephan FK, Zucker I. Circadian rhythms in drinking behavior and locomotor activity of rats are eliminated by hypothalamic lesions. Proc Natl Acad Sci U S A (1972) 69(6):1583–6. 10.1073/pnas.69.6.1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamamoto H, Nagai K, Nakagawa H. Role of SCN in daily rhythms of plasma glucose, FFA, insulin and glucagon. Chronobiol Int (1987) 4(4):483–91. 10.3109/07420528709078539 [DOI] [PubMed] [Google Scholar]
- 5.Kalsbeek A, Fliers E, Romijn JA, La Fleur SE, Wortel J, Bakker O, et al. The suprachiasmatic nucleus generates the diurnal changes in plasma leptin levels. Endocrinology (2001) 142(6):2677–85. 10.1210/endo.142.6.8197 [DOI] [PubMed] [Google Scholar]
- 6.Ishikawa K, Shimazu T. Circadian rhythm of liver glycogen metabolism in rats: effects of hypothalamic lesions. Am J Physiol (1980) 238(1):E21–5. [DOI] [PubMed] [Google Scholar]
- 7.Coomans CP, van den Berg SA, Lucassen EA, Houben T, Pronk AC, van der Spek RD, et al. The suprachiasmatic nucleus controls circadian energy metabolism and hepatic insulin sensitivity. Diabetes (2013) 62(4):1102–8. 10.2337/db12-0507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fonken LK, Workman JL, Walton JC, Weil ZM, Morris JS, Haim A, et al. Light at night increases body mass by shifting the time of food intake. Proc Natl Acad Sci U S A (2010) 107(43):18664–9. 10.1073/pnas.1008734107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qian J, Block GD, Colwell CS, Matveyenko AV. Consequences of exposure to light at night on the pancreatic islet circadian clock and function in rats. Diabetes (2013) 62(10):3469–78. 10.2337/db12-1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gale JE, Cox HI, Qian J, Block GD, Colwell CS, Matveyenko AV. Disruption of circadian rhythms accelerates development of diabetes through pancreatic beta-cell loss and dysfunction. J Biol Rhythms (2011) 26(5):423–33. 10.1177/0748730411416341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krysta K, Krzystanek M, Janas-Kozik M, Krupka-Matuszczyk I. Bright light therapy in the treatment of childhood and adolescence depression, antepartum depression, and eating disorders. J Neural Transm (Vienna) (2012) 119(10):1167–72. 10.1007/s00702-012-0863-2 [DOI] [PubMed] [Google Scholar]
- 12.Al-Karawi D, Jubair L. Bright light therapy for nonseasonal depression: meta-analysis of clinical trials. J Affect Disord (2016) 198:64–71. 10.1016/j.jad.2016.03.016 [DOI] [PubMed] [Google Scholar]
- 13.Boivin DB, Tremblay GM, James FO. Working on atypical schedules. Sleep Med (2007) 8(6):578–89. 10.1016/j.sleep.2007.03.015 [DOI] [PubMed] [Google Scholar]
- 14.Oike H, Sakurai M, Ippoushi K, Kobori M. Time-fixed feeding prevents obesity induced by chronic advances of light/dark cycles in mouse models of jet-lag/shift work. Biochem Biophys Res Commun (2015) 465(3):556–61. 10.1016/j.bbrc.2015.08.059 [DOI] [PubMed] [Google Scholar]
- 15.McGowan NM, Coogan AN. Circadian and behavioural responses to shift work-like schedules of light/dark in the mouse. J Mol Psychiatry (2013) 1(1):7. 10.1186/2049-9256-1-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barclay JL, Husse J, Bode B, Naujokat N, Meyer-Kovac J, Schmid SM, et al. Circadian desynchrony promotes metabolic disruption in a mouse model of shiftwork. PLoS One (2012) 7(5):e37150. 10.1371/journal.pone.0037150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salgado-Delgado RC, Saderi N, Basualdo Mdel C, Guerrero-Vargas NN, Escobar C, Buijs RM. Shift work or food intake during the rest phase promotes metabolic disruption and desynchrony of liver genes in male rats. PLoS One (2013) 8(4):e60052. 10.1371/journal.pone.0060052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arble DM, Bass J, Laposky AD, Vitaterna MH, Turek FW. Circadian timing of food intake contributes to weight gain. Obesity (Silver Spring) (2009) 17(11):2100–2. 10.1038/oby.2009.264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng D, Lazar MA. Clocks, metabolism, and the epigenome. Mol Cell (2012) 47(2):158–67. 10.1016/j.molcel.2012.06.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katada S, Imhof A, Sassone-Corsi P. Connecting threads: epigenetics and metabolism. Cell (2012) 148(1–2):24–8. 10.1016/j.cell.2012.01.001 [DOI] [PubMed] [Google Scholar]
- 21.Vitaterna MH, King DP, Chang AM, Kornhauser JM, Lowrey PL, McDonald JD, et al. Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science (1994) 264(5159):719–25. 10.1126/science.8171325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Antoch MP, Song EJ, Chang AM, Vitaterna MH, Zhao Y, Wilsbacher LD, et al. Functional identification of the mouse circadian Clock gene by transgenic BAC rescue. Cell (1997) 89(4):655–67. 10.1016/S0092-8674(00)80246-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun ZS, Albrecht U, Zhuchenko O, Bailey J, Eichele G, Lee CC. RIGUI, a putative mammalian ortholog of the Drosophila period gene. Cell (1997) 90(6):1003–11. 10.1016/S0092-8674(00)80366-9 [DOI] [PubMed] [Google Scholar]
- 24.Zheng B, Larkin DW, Albrecht U, Sun ZS, Sage M, Eichele G, et al. The mPer2 gene encodes a functional component of the mammalian circadian clock. Nature (1999) 400(6740):169–73. 10.1038/22118 [DOI] [PubMed] [Google Scholar]
- 25.Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, et al. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell (2000) 103(7):1009–17. 10.1016/S0092-8674(00)00205-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell (2002) 109(3):307–20. 10.1016/S0092-8674(02)00722-5 [DOI] [PubMed] [Google Scholar]
- 27.Rudic RD, McNamara P, Curtis AM, Boston RC, Panda S, Hogenesch JB, et al. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol (2004) 2(11):e377. 10.1371/journal.pbio.0020377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lamia KA, Storch KF, Weitz CJ. Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci U S A (2008) 105(39):15172–7. 10.1073/pnas.0806717105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang G, Chen L, Grant GR, Paschos G, Song WL, Musiek ES, et al. Timing of expression of the core clock gene Bmal1 influences its effects on aging and survival. Sci Transl Med (2016) 8(324):324ra316. 10.1126/scitranslmed.aad3305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science (2005) 308(5724):1043–5. 10.1126/science.1108750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shostak A, Meyer-Kovac J, Oster H. Circadian regulation of lipid mobilization in white adipose tissues. Diabetes (2013) 62(7):2195–203. 10.2337/db12-1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oishi K, Atsumi G, Sugiyama S, Kodomari I, Kasamatsu M, Machida K, et al. Disrupted fat absorption attenuates obesity induced by a high-fat diet in Clock mutant mice. FEBS Lett (2006) 580(1):127–30. 10.1016/j.febslet.2005.11.063 [DOI] [PubMed] [Google Scholar]
- 33.Yang S, Liu A, Weidenhammer A, Cooksey RC, McClain D, Kim MK, et al. The role of mPer2 clock gene in glucocorticoid and feeding rhythms. Endocrinology (2009) 150(5):2153–60. 10.1210/en.2008-0705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carvas JM, Vukolic A, Yepuri G, Xiong Y, Popp K, Schmutz I, et al. Period2 gene mutant mice show compromised insulin-mediated endothelial nitric oxide release and altered glucose homeostasis. Front Physiol (2012) 3:337. 10.3389/fphys.2012.00337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bae K, Jin X, Maywood ES, Hastings MH, Reppert SM, Weaver DR. Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron (2001) 30(2):525–36. 10.1016/S0896-6273(01)00302-6 [DOI] [PubMed] [Google Scholar]
- 36.Grimaldi B, Bellet MM, Katada S, Astarita G, Hirayama J, Amin RH, et al. PER2 controls lipid metabolism by direct regulation of PPARgamma. Cell Metab (2010) 12(5):509–20. 10.1016/j.cmet.2010.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bhatwadekar AD, Yan Y, Qi X, Thinschmidt JS, Neu MB, Li Calzi S, et al. Per2 mutation recapitulates the vascular phenotype of diabetes in the retina and bone marrow. Diabetes (2013) 62(1):273–82. 10.2337/db12-0172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng B, Albrecht U, Kaasik K, Sage M, Lu W, Vaishnav S, et al. Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell (2001) 105(5):683–94. 10.1016/S0092-8674(01)00380-4 [DOI] [PubMed] [Google Scholar]
- 39.Dallmann R, Weaver DR. Altered body mass regulation in male mPeriod mutant mice on high-fat diet. Chronobiol Int (2010) 27(6):1317–28. 10.3109/07420528.2010.489166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Le Martelot G, Claudel T, Gatfield D, Schaad O, Kornmann B, Lo Sasso G, et al. REV-ERBalpha participates in circadian SREBP signaling and bile acid homeostasis. PLoS Biol (2009) 7(9):e1000181. 10.1371/journal.pbio.1000181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cho H, Zhao X, Hatori M, Yu RT, Barish GD, Lam MT, et al. Regulation of circadian behaviour and metabolism by REV-ERB-alpha and REV-ERB-beta. Nature (2012) 485(7396):123–7. 10.1038/nature11048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Solt LA, Wang Y, Banerjee S, Hughes T, Kojetin DJ, Lundasen T, et al. Regulation of circadian behaviour and metabolism by synthetic REV-ERB agonists. Nature (2012) 485(7396):62–8. 10.1038/nature11030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamaguchi S, Isejima H, Matsuo T, Okura R, Yagita K, Kobayashi M, et al. Synchronization of cellular clocks in the suprachiasmatic nucleus. Science (2003) 302(5649):1408–12. 10.1126/science.1089287 [DOI] [PubMed] [Google Scholar]
- 44.Ralph MR, Foster RG, Davis FC, Menaker M. Transplanted suprachiasmatic nucleus determines circadian period. Science (1990) 247(4945):975–8. 10.1126/science.2305266 [DOI] [PubMed] [Google Scholar]
- 45.Oster H, Damerow S, Kiessling S, Jakubcakova V, Abraham D, Tian J, et al. The circadian rhythm of glucocorticoids is regulated by a gating mechanism residing in the adrenal cortical clock. Cell Metab (2006) 4(2):163–73. 10.1016/j.cmet.2006.07.002 [DOI] [PubMed] [Google Scholar]
- 46.Gu H, Marth JD, Orban PC, Mossmann H, Rajewsky K. Deletion of a DNA polymerase beta gene segment in T cells using cell type-specific gene targeting. Science (1994) 265(5168):103–6. 10.1126/science.8016642 [DOI] [PubMed] [Google Scholar]
- 47.Storch KF, Paz C, Signorovitch J, Raviola E, Pawlyk B, Li T, et al. Intrinsic circadian clock of the mammalian retina: importance for retinal processing of visual information. Cell (2007) 130(4):730–41. 10.1016/j.cell.2007.06.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marcheva B, Ramsey KM, Buhr ED, Kobayashi Y, Su H, Ko CH, et al. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature (2010) 466(7306):627–31. 10.1038/nature09253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perelis M, Marcheva B, Ramsey KM, Schipma MJ, Hutchison AL, Taguchi A, et al. Pancreatic beta cell enhancers regulate rhythmic transcription of genes controlling insulin secretion. Science (2015) 350(6261):aac4250. 10.1126/science.aac4250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dyar KA, Ciciliot S, Wright LE, Bienso RS, Tagliazucchi GM, Patel VR, et al. Muscle insulin sensitivity and glucose metabolism are controlled by the intrinsic muscle clock. Mol Metab (2014) 3(1):29–41. 10.1016/j.molmet.2013.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paschos GK, Ibrahim S, Song WL, Kunieda T, Grant G, Reyes TM, et al. Obesity in mice with adipocyte-specific deletion of clock component Arntl. Nat Med (2012) 18(12):1768–77. 10.1038/nm.2979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Piggins HD, Guilding C. The neural circadian system of mammals. Essays Biochem (2011) 49(1):1–17. 10.1042/bse0490001 [DOI] [PubMed] [Google Scholar]
- 53.Orozco-Solis R, Aguilar-Arnal L, Murakami M, Peruquetti R, Ramadori G, Coppari R, et al. The circadian clock in the ventromedial hypothalamus controls cyclic energy expenditure. Cell Metab (2016) 23(3):467–78. 10.1016/j.cmet.2016.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Harno E, Cottrell EC, White A. Metabolic pitfalls of CNS Cre-based technology. Cell Metab (2013) 18(1):21–8. 10.1016/j.cmet.2013.05.019 [DOI] [PubMed] [Google Scholar]
- 55.Landgraf D, Koch CE, Oster H. Embryonic development of circadian clocks in the mammalian suprachiasmatic nuclei. Front Neuroanat (2014) 8:143. 10.3389/fnana.2014.00143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leone DP, Genoud S, Atanasoski S, Grausenburger R, Berger P, Metzger D, et al. Tamoxifen-inducible glia-specific Cre mice for somatic mutagenesis in oligodendrocytes and Schwann cells. Mol Cell Neurosci (2003) 22(4):430–40. 10.1016/S1044-7431(03)00029-0 [DOI] [PubMed] [Google Scholar]
- 57.Husse J, Zhou X, Shostak A, Oster H, Eichele G. Synaptotagmin10-Cre, a driver to disrupt clock genes in the SCN. J Biol Rhythms (2011) 26(5):379–89. 10.1177/0748730411415363 [DOI] [PubMed] [Google Scholar]
- 58.Kondratov RV, Kondratova AA, Gorbacheva VY, Vykhovanets OV, Antoch MP. Early aging and age-related pathologies in mice deficient in BMAL1, the core componentof the circadian clock. Genes Dev (2006) 20(14):1868–73. 10.1101/gad.1432206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou X, Vink M, Klaver B, Berkhout B, Das AT. Optimization of the Tet-On system for regulated gene expression through viral evolution. Gene Ther (2006) 13(19):1382–90. 10.1038/sj.gt.3302780 [DOI] [PubMed] [Google Scholar]
- 60.Whissell PD, Tohyama S, Martin LJ. The use of DREADDs to deconstruct behavior. Front Genet (2016) 7:70. 10.3389/fgene.2016.00070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hong HK, Chong JL, Song W, Song EJ, Jyawook AA, Schook AC, et al. Inducible and reversible Clock gene expression in brain using the tTA system for the study of circadian behavior. PLoS Genet (2007) 3(2):e33. 10.1371/journal.pgen.0030033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol (2007) 5(2):e34. 10.1371/journal.pbio.0050034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jones JR, Tackenberg MC, McMahon DG. Manipulating circadian clock neuron firing rate resets molecular circadian rhythms and behavior. Nat Neurosci (2015) 18(3):373–5. 10.1038/nn.3937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tischer D, Weiner OD. Illuminating cell signalling with optogenetic tools. Nat Rev Mol Cell Biol (2014) 15(8):551–8. 10.1038/nrm3837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nathanson JL, Jappelli R, Scheeff ED, Manning G, Obata K, Brenner S, et al. Short promoters in viral vectors drive selective expression in mammalian inhibitory neurons, but do not restrict activity to specific inhibitory cell-types. Front Neural Circuits (2009) 3:19. 10.3389/neuro.04.019.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schnutgen F, Doerflinger N, Calleja C, Wendling O, Chambon P, Ghyselinck NB. A directional strategy for monitoring Cre-mediated recombination at the cellular level in the mouse. Nat Biotechnol (2003) 21(5):562–5. 10.1038/nbt811 [DOI] [PubMed] [Google Scholar]
- 67.Choudhury SR, Hudry E, Maguire CA, Sena-Esteves M, Breakefield XO, Grandi P. Viral vectors for therapy of neurologic diseases. Neuropharmacology (2016). 10.1016/j.neuropharm.2016.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Carlson DF, Fahrenkrug SC, Hackett PB. Targeting DNA With Fingers and TALENs. Mol Ther Nucleic Acids (2012) 1:e3. 10.1038/mtna.2011.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science (2014) 346(6213):1258096. 10.1126/science.1258096 [DOI] [PubMed] [Google Scholar]
- 70.Bakondi B, Lv W, Lu B, Jones MK, Tsai Y, Kim KJ, et al. In vivo CRISPR/Cas9 gene editing corrects retinal dystrophy in the S334ter-3 rat model of autosomal dominant retinitis pigmentosa. Mol Ther (2016) 24(3):556–63. 10.1038/mt.2015.220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Suzuki K, Tsunekawa Y, Hernandez-Benitez R, Wu J, Zhu J, Kim EJ, et al. In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature (2016) 540(7631):144–9. 10.1038/nature20565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ukai-Tadenuma M, Yamada RG, Xu H, Ripperger JA, Liu AC, Ueda HR. Delay in feedback repression by cryptochrome 1 is required for circadian clock function. Cell (2011) 144(2):268–81. 10.1016/j.cell.2010.12.019 [DOI] [PubMed] [Google Scholar]