Abstract
Mass cytometry is a novel platform for high-dimensional phenotypic and functional analysis of single cells. This system uses elemental metal isotopes conjugated to monoclonal antibodies to evaluate up to 42 parameters simultaneously on individual cells with minimal overlap between channels. The platform can be customized for analysis of both phenotypic and functional markers. Here, we will describe methods to stain, collect, and analyze intracellular functional markers and surface phenotypic markers on natural killer cells.
Keywords: CyTOF, Mass cytometry, Flow cytometry, Natural killer cell, Intracellular cytokine staining
1 Introduction
Mass cytometry is a next-generation flow cytometry platform with several technological advances that offer advantages over fluorescence-based flow cytometry when highly parametric analyses are required. Most notably, mass cytometry does not rely on detection of fluorescence, which requires compensation for spillover into adjacent channels. Instead, through the use of antibodies coupled to metal isotopes, mass cytometry can detect discrete isotope peaks without significant overlap [1, 2], thus ameliorating the need for compensation. In addition, more channels are available. The primary limiting factor is the chemistry required to conjugate the metals to the antibodies with high efficiency. Currently, mass cytometry allows for the detection of 42+ unique parameters rather than the 8–12 parameters that comprise a typical flow cytometry panel.
This increase in total parameters is critical for the study of natural killer (NK) cells, the diverse functional properties of which are influenced by combinatorial expression of multiple phenotypic markers [3, 4]. The functional properties of NK cells can be explored through multiple stimulation conditions including phorbol 12-myristate 13-acetate (PMA) + ionomycin, cytokines (such as IL-2, IL-12, IL-15, and/or IL-18), viral stimulation (such as HIV-1-infected or influenza-infected cells), or cell lines deficient in MHC-I expression (such as K562 or 721.221 cells). To analyze intracellular cytokines and chemokines, brefeldin A and monensin are added for the final 4 h of stimulation to maintain the cytokines intracellularly for detection [5]. Because CD107a (a lysosomal protein also known as LAMP-1) is briefly revealed at the cell surface during cytotoxic granule release, anti-CD107a antibodies added during stimulation serve as an indirect measure of cytotoxicity. Most antibodies used in mass cytometry bind directly to the target protein; however, in some cases, two-stage detection is more efficient. For optimal detection of CD107a by mass cytometry, anti-CD107a-APC is added during the stimulation, followed by isotope-conjugated anti-APC antibody as a surface stain.
Antibody–metal isotope pairs are available for purchase from Fluidigm (http://maxpar.fluidigm.com/product-catalog-metal.php). However, optimizing a panel that both explores the desired markers and accounts for isotope spillover and varying degrees of antibody signal intensity often requires a customized panel. Conjugation of antibodies and metal isotopes is an easily performed step that results in increased options for panel design, and has been previously described in detail [6]. Here we describe a method to profile both the phenotypic and functional characteristics of natural killer cells by conjugating antibodies to selected metal isotopes, surface and intracellular labeling of cells and analysis on a mass cytometer. The process of conjugating a custom panel for phenotypic analysis has been described previously; we will mention NK-cell-specific modifications here [3]. This chapter focuses on the addition of conditions for intracellular staining and functional assessment of NK cells, but can also be adapted for other cell types.
2 Materials
All reagents and containers must be free from heavy metal contaminants. No containers exposed to soap that may contain trace metal contaminants should be used. All buffers should be prepared and stored in disposable uncontaminated containers.
2.1 Antibody Conjugation
Maxpar® Metal labeling kits (Fluidigm Corporation).
Purified monoclonal antibodies (Fig. 1 and Table 1 provide examples), purified IgG or polyclonal. Must not have any carrier protein; otherwise, special order is required. Sodium azide is acceptable.
Centrifugal Filter Unit: 50 kDa Amicon Ultra—500 µL V bottom (Millipore) or 30 kDa Amicon Ultra 500 µL V bottom (Millipore).
Centrifugal Filter Unit: 3 kDa Amicon Ultra—500 µL V bottom (Millipore).
Aerosol Barrier (Filter) Pipette Tips.
Bond-Breaker™ TCEP Solution: 0.5 M TCEP (Tris(2-carboxyethyl)phosphine) (Pierce).
Microcentrifuge, ideally 2 units.
Heat block incubator or water bath at 37 °C.
PBS-based antibody stabilization solution (Candor Biosciences).
Nanodrop for protein quantification.
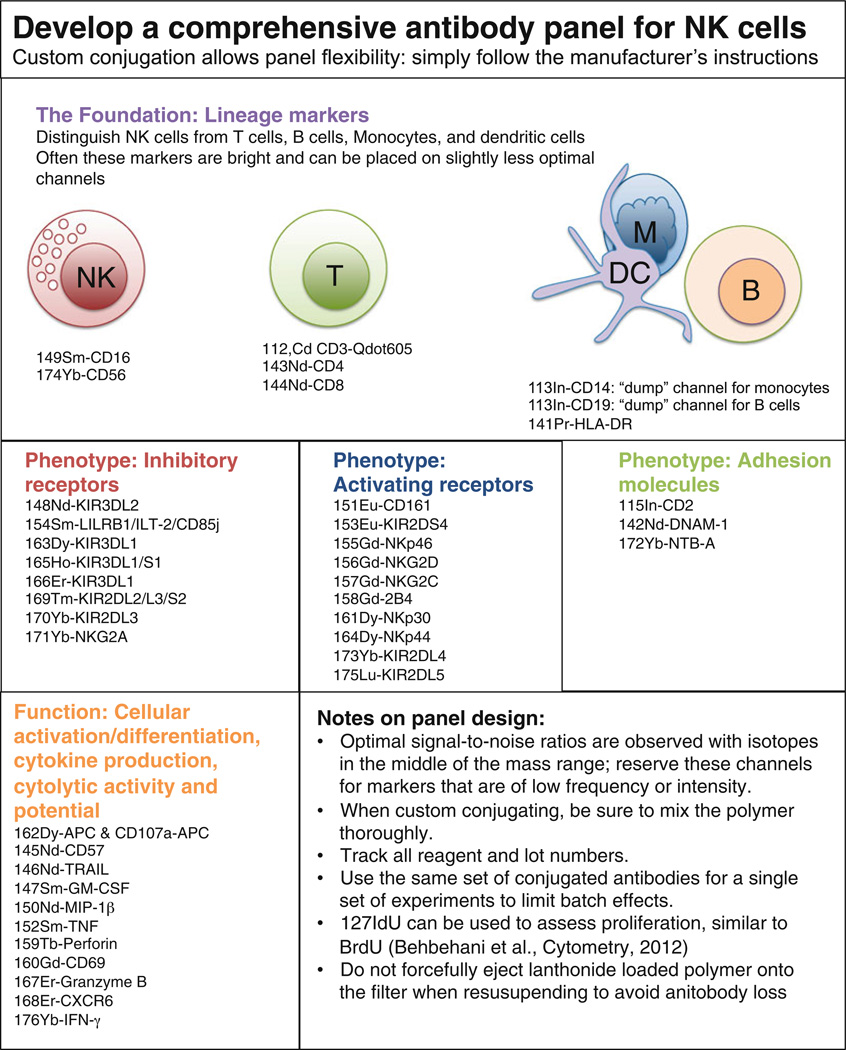
Fig. 1.
Schematic display of CyTOF panel development
Table 1.
Example of table to generate antibody surface and intracellular staining cocktails
| Isotope | Antibody | Clone | Supplier | Concentration (µg/mL) |
Working concentration (µg/mL) |
Volume to add a |
|---|---|---|---|---|---|---|
| Surface markers | ||||||
| 110,111,112,114Cd | CD3 | UCHT1 | Life Technologies (Qdot-605) | 100 tests/100 µL | 0.5 µL/test | 10.5 |
| 113In | CD14, CD19 | M5E2, HIB19 | Biolegend | 279, 342 | 2.5, 2.5 | 9.41, 7.68 |
| 115In | CD2 | RPA-2.10 | BD Biosciences | 302 | 0.625 | 2.17 |
| 141Pr | APC | APC003 | Biolegend | 252 | 5.0 | 20.83 |
| 142Nd | DNAM-1 | DX11 | BD Biosciences | 327 | 5.0 | 16.06 |
| 143Nd | CD4 | SK3 | Biolegend | 171 | 1.25 | 7.68 |
| 144Nd | CD8 | SK1 | Biolegend | 387 | 2.5 | 6.78 |
| 145Nd | CD57 | HCD57 | Biolegend | 630 | 0.625 | 1.04 |
| 146Nd | TRAIL | RIK-2 | Biolegend | 208 | 2.5 | 12.62 |
| 148Nd | KIR3DL2 | Lanier Lab | 266 | 1.25 | 4.93 | |
| 149Sm | CD16 | 3G8 | BD Biosciences | 385 | 1.25 | 3.41 |
| 151Eu | CD161 | DX12 | BD Biosciences | 319 | 2.5 | 8.23 |
| 153Eu | KIR2DS4 | FES172 | Beckman Coulter | 288 | 1.25 | 4.56 |
| 154Sm | ILT-2 | GHI/75 | Biolegend | 393 | 2.5 | 6.68 |
| 155Gd | NKp46 | 9E2/NKp46 | BD Biosciences | 385 | 1.25 | 3.41 |
| 156Gd | NKG2D | 1D11 | Biolegend | 257 | 5.0 | 20.43 |
| 157Gd | NKG2C | MAB1381 | R&D Systems | 138 | 2.5 | 19.02 |
| 158Gd | 2B4 | 2–69 | BD Biosciences | 324 | 2.5 | 8.10 |
| 161Dy | NKp30 | P30-15 | Biolegend | 286 | 2.5 | 9.18 |
| 163Dy | KIR3DL1 | DX9 | BD Biosciences | 415 | 2.5 | 6.33 |
| 164Dy | NKp44 | P44-8 | Biolegend | 293 | 1.25 | 4.48 |
| 165Ho | KIR3DL1/S1 | Z27.3.7 | Beckman Coulter | 328 | 1.25 | 4.00 |
| 166Er | KIR2DL1 | 143211 | R&D Systems | 313 | 1.25 | 4.19 |
| 168Er | CXCR6 | 56811 | R&D Systems | 396 | 5.0 | 13.26 |
| 169Tm | KIR2DL2/L3/S2 | GL183 | Beckman Coulter | 304 | 1.25 | 4.32 |
| 170Yb | KIR2DL3 | 180701 | R&D Systems | 273 | 2.5 | 9.62 |
| 171Yb | NKG2A | Z199 | Beckman Coulter | 428 | 2.5 | 6.13 |
| 172Yb | NTB-A | NT-7 | Biolegend | 94 | 1.25 | 13.96 |
| 173Yb | KIR2DL4 | 181703 | R&D Systems | 840 | 1.25 | 1.56 |
| 174Yb | CD56 | NCAM16.2 | BD Biosciences | 436 | 1.25 | 3.01 |
| 175Lu | KIR2DL5 | UP-R1 | Biolegend | 450 | 0.625 | 1.46 |
| #Vol CyFACS | 786.69 | |||||
| Intracellular markers | ||||||
| 147Nd | GM-CSF | BVD2-21C11 | Biolegend | 119 | 5.0 | 44.12 |
| 150Nd | MIP-1β | D21-1352 | BD Biosciences (Custom) | 551 | 5.0 | 9.53 |
| 152Sm | TNF-α | MAb11 | eBioscience | 454 | 5.0 | 11.56 |
| 159 Tb | Perforin | B-D48 | Abcam | 381 | 5.0 | 13.78 |
| 160Gd | CD69 | FN50 | Biolegend | 308 | 2.5 | 8.52 |
| 162Dy | HIVp24 | 38/5.4A | Abcam | 280 | 10.0 | 37.50 |
| 167Er | Granzyme B | 2CF/F5 | BD Biosciences | 442 | 5.0 | 11.88 |
| 176Yb | IFN-γ | 4S.B4 | eBioscience | 401 | 5.0 | 13.09 |
| #Vol CyFACS | 900.018 |
Volume to add = (Working Concentration/Concentration) × 50 × Number of Samples (in this case 20) + 1
Volume CyFACS = (50 × Number of Samples + 1) − Sum Total of the Volume of Antibodies
2.2 Mass Cytometry Labeling
Custom-conjugated antibodies or pre-conjugated antibodies purchased from Fluidigm (http://maxpar.fluidigm.com/product-catalog-metal.php).
Thermo Scientific™ Nunc™ 96 Deep Well Plates, Polystyrene. 96-well round bottom plates can be used for stimulation and incubation, but cells should be transferred to deep-well plates prior to surface staining.
MilliQ dH2O: No water should have contact with beakers or bottles washed with soap.
CyPBS: PBS without heavy metal contaminants, made from 10× PBS using MilliQ purified water, with no contact with glassware washed with soap.
CyFACS buffer: 0.1 % bovine serum albumin + 2 mM EDTA + 0.1 % sodium azide in CyPBS. Filter solution with a 0.2 µM filter.
Cisplatin Solution: Prepare 100 mM (Stock solution) in DMSO. Freshly prepare working solution of 10 mM cisplatin in PBS (100 µL cisplatin stock + 900 µL PBS).
Lysing Solution: Prepare 1× solution from 10× stock (BD FACS™ Lysing Solution, BD Biosciences) solution using MilliQ deionized water and store in a disposable plastic container.
0.1 µM spin filters (Millipore).
Permeabilization buffer: Prepare 1× permeabilization buffer from 10× stock solution (BD FACS™ Permeabilizing Solution 2, BD Biosciences) using MilliQ deionized water and store in a disposable plastic container.
Interchelator-PFA solution: Dilute Iradium-Interchelator solution (Fluidigm) 1:10,000 into 2 % paraformaldehyde solution. Prepare the 2 % Paraformaldehyde solution by diluting 16 % Stock Paraformaldehyde (Electron Microscopy Sciences) in CyPBS. Freshly prepare the Interchelator-PFA solution for each use.
Complete RPMI medium: RPMI-1640, 10 % Fetal Bovine Serum, 1× penicillin-streptomycin and 1× l-glutamine.
Refrigerated centrifuge equipped with rotor for spinning 96-well plates.
Aspirator with vacuum trap set-up.
2.3 Cell Stimulation
Cell Stimulation cocktail contains PMA and ionomycin (500×) (eBioscience): Freshly prepare 1× Working solution in complete RPMI medium (1:500 dilution).
CD107a-APC (Biolegend, Clone H4A3).
EDTA 0.5 M (Stock): prepare 20 mM working solution.
Monensin Solution (1000×) (eBioscience): Freshly prepare 1× Working solution in complete RPMI medium (1:1000 dilution).
Brefeldin A Solution (1000×) (eBioscience): Freshly prepare 1× Working solution in complete RPMI medium (1:1000 dilution).
2.4 Running CyTOF Mass Cytometry
Ice bucket.
Micropipettes.
Normalization beads (Fluidigm).
Filter-top tubes (BD Biosciences).
3 Methods
All steps may be completed at room temperature (RT) unless otherwise indicated.
3.1 Antibody Conjugation Using MaxPar Metal Labeling Kit
To conjugate antibodies simply follow manufacturer’s instructions provided with the MaxPar Metal Labeling Kit using the supplies listed above. An example of a customized panel to profile natural killer cell phenotype and function is shown in Fig. 1. The conjugation protocol has been described in depth previously [2–4]. Custom-conjugating antibodies gives more flexibility in panel design than would be available by purchasing pre-conjugated antibodies. The panel outlined in Fig. 1 contains receptors to identify major cell lineages (such as CD3, CD4, CD8, CD19, CD14, CD56) as well as markers for the major NK cell receptor families including the killer immunoglobulin-like receptors (e.g., KIR2DL1, KIR2DL2/L3/S2), Fc receptors (CD16), natural cytotoxicity receptors (NKp30, NKp44, NKp46), C-type lectins (NKG2A, NKG2C, NKG2D), and markers of maturity and differentiation (e.g. CD57). It also includes NK cell functional cytokines (IFN-γ, MIP-1β, TNF-α) as well as cytotoxicity markers (CD107a, Perforin, Granzyme B). In general, greater signal-to-noise ratios can be expected on isotopes in the middle of the mass range and markers that are of low frequency or low intensity are best reserved for these channels. Please see Notes 1–6 for additional suggestions regarding the conjugation of antibodies.
3.2 Surface and Intracellular Labeling of Cells for Mass Cytometry
Isolate human peripheral blood mononuclear cells by Ficoll gradient (see Note 7).
Wash the cells with complete RPMI medium and plate the cells at two million cells in 200 µL in a standard 96-well plate.
Add stimulation condition of choice; for example 1× Cell Stimulation cocktail (Subheading 2.3, item 1), 1× Brefeldin A and 1× Monensin. To assess degranulation include CD107a-APC at a dilution of 1:25 (25-fold dilution) in this Cell Stimulation cocktail. Incubate the cells at 37 °C for 4 h.
During the incubation, prepare the surface and intracellular antibody staining cocktails. This is best performed using a spreadsheet to calculate the quantities based on antibody titrations (Table 1).
At the end of the incubation add 20 µL of 20 mM EDTA to each well and mix by pipetting. Incubate for 10 min at room temperature.
Spin the plate at 750 × g for 3 min, aspirate the supernatant, and add 200 µL CyFACS. Transfer the cells to a deep 96-well plate.
Add 300 µL CyFACS to each well and spin the plate at 750 × g for 10 min. Aspirate the supernatant.
During centrifugation (step 7), dilute the 10 mM cisplatin working solution at a ratio of 1:200–1:50 (titration based on prior cisplatin batch optimization) in serum and antibiotic-free RPMI (see Notes 8 and 9).
Resuspend the cells in 400 µL of this diluted cisplatin.
Incubate cells for 1 min at RT.
Quench (see Note 9) with 400 µL of serum. Pipette to mix thoroughly.
Centrifuge plate at 750 × g for 10 min at RT. Aspirate the supernatant.
Add 500 µL of CyFACS buffer. Centrifuge plate at 750 × g for 10 min at RT. Aspirate the supernatant. After removing plate from the centrifuge, set centrifuge to 4 °C.
During centrifugation (step 13), centrifuge the surface antibody cocktail in a 0.1 µM Millipore spin filter for 3 min at 10,000 × g.
Resuspend pelleted cells in 50 µL of the antibody staining cocktail.
Incubate for 45 min on ice at 4 °C.
Add 500 µL CyFACS buffer to each well. Centrifuge plate at 750 × g for 10 min at 4 °C. Aspirate the supernatant.
Resuspend each well in 100 µL of 1× Lysing solution (Subheading 2.2, item 7) and incubate for 10 min at room temperature (see Note 10).
Add 500 µL CyFACS buffer to each well. Centrifuge at 750 × g for 10 min at 4 °C. Aspirate the supernatant.
Add 500 µL CyFACS buffer to each well. Centrifuge at 750 × g for 10 min at 4 °C. Aspirate the supernatant.
Resuspend cells in 200 µL of Permeabilization solution (Subheading 2.2, item 10). Incubate for 10 min at room temperature.
Add 500 µL CyFACS buffer to each well. Centrifuge at 750 × g for 10 min at 4 °C. Aspirate the supernatant.
During centrifugation (step 22), centrifuge the intracellular antibody cocktail in a 0.1 µM Millipore spin filter for 3 min at 10,000 × g.
Resuspend cells in 50 µL of the intracellular antibody staining cocktail.
Incubate for 45 min on ice at 4 °C.
Add 500 µL CyFACS buffer to each well, centrifuge at 750 × g for 10 min at 4 °C and aspirate the supernatant.
Repeat step 26 two additional times.
Resuspend cells thoroughly in 100 µL of freshly prepared Interchelator-PFA solution.
Incubate overnight at 4 °C.
The following day (the same day that the cells will be run on the mass cytometer), add 500 µL of CyPBS buffer to each well. Centrifuge at 1000 × g for 10 min at RT. Aspirate the supernatant, leaving 100 µL residual volume in the well.
Add 500 µL MilliQ water (metal-free) to each well. Centrifuge at 1000 × g for 10 min at RT. Aspirate the supernatant, leaving 100 µL residual volume in each well.
Repeat step 32 twice for a total of three washes in MilliQ water.
Resuspend cells in the residual 100 µL MilliQ water after the final wash.
Run on mass cytometer after resuspension in approximately 1 mL of MilliQ water with or without normalization beads immediately prior to the run. Pipette cells into a FACS tube through a cell-strainer cap to remove unwanted debris.
3.3 Running Samples on a CyTOF Mass Cytometer
Running samples on a CyTOF mass cytometer is normally performed by Flow Cytometry core services at most Institutions, to procure a detailed step-by-step protocol for operating CyTOF refer to the publication by Leipold M.D. et al. [7]. It is important to note that when looking for rare cell populations, higher numbers of total cells will need to be run through the mass cytometer. Although the maximum collection rate is estimated at 1000 cells/s, to ensure the avoidance of doublets and nebulizer clots, it is prudent to use low run speeds of 300 cells/s or lower. For assay normalization within and among runs, normalization beads should be used as described in [8]. It is helpful to count the cells in each well in order to appropriately estimate the ideal resuspension volume. Cells should be stored on ice while waiting to run on the mass cytometer.
3.4 Data Analysis
Due to the increase in potential marker combinations allowed for by mass cytometry, manual gating of all possible cellular subsets is not possible. In addition, this approach does not allow for the evaluation of unexpected cellular subsets of interest [2]. However, typical manual gating schema can also be used to major known cell subsets and functions, although there are important differences to consider when analyzing in FlowJo (see Notes 11 and 12 and Figs. 2 and 3). Boolean gating analysis can also be used to identify discreet cellular subsets as described by Horowitz et al. [4]. A number of analysis packages have now been developed in order to analyze CyTOF data in an unbiased manner. Spanning-tree progression analysis of density-normalized events (SPADE) is available through Cytobank and uses density-dependent down sampling, followed by agglomerative clustering, minimum spanning tree construction and upsampling to identify unique cellular subsets from CyTOF [9]. Citrus is an algorithm that identifies clusters of phenotypically similar cells in an unsupervised manner and identifies features (either functional or phenotypic) that are predictive of a selected group of samples with a specific endpoint or treatment relative to a control group [10]. Another tool to visualize these data in two dimensions is viSNE [11]. A complete step-wise protocol for analysis of CyTOF data is beyond the scope of this chapter, and we recommend that the user obtain training from their institution, Cytobank, or Fluidigm in the use of the various algorithms and analysis of these complex data sets and how to apply the programs to their specific research questions.
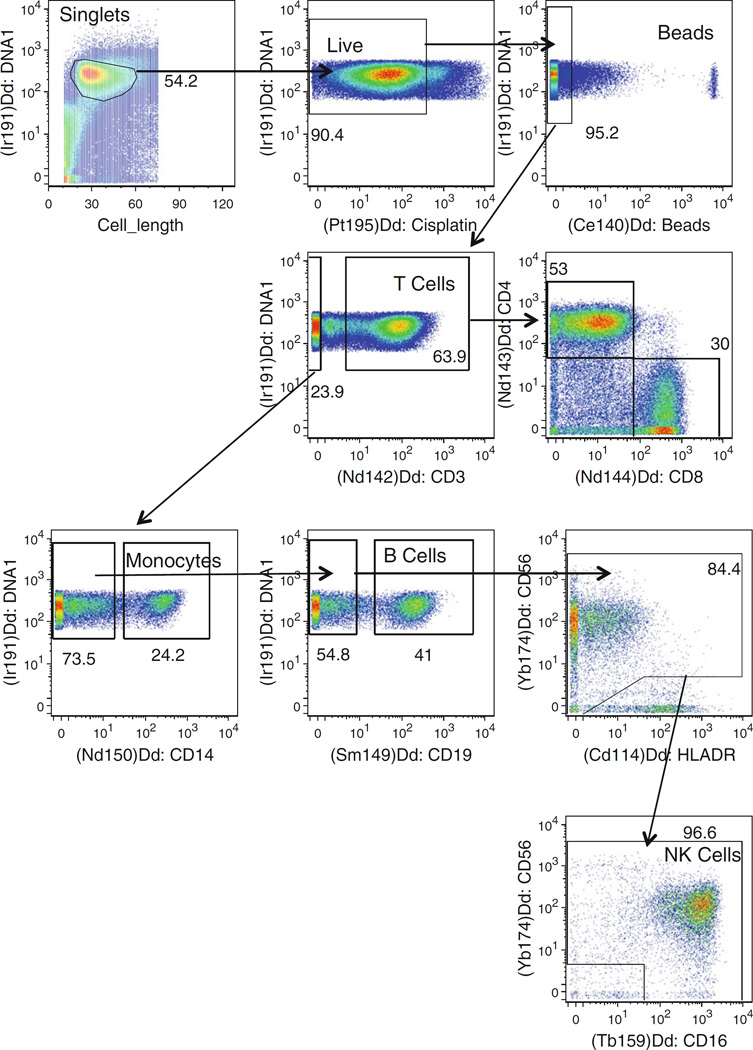
Fig. 2.
Gating strategy to identify NK cells and other major cell subsets by mass cytometry. The sequential gates to identify T-cell subsets, monocytes, B cells, and NK cells are shown
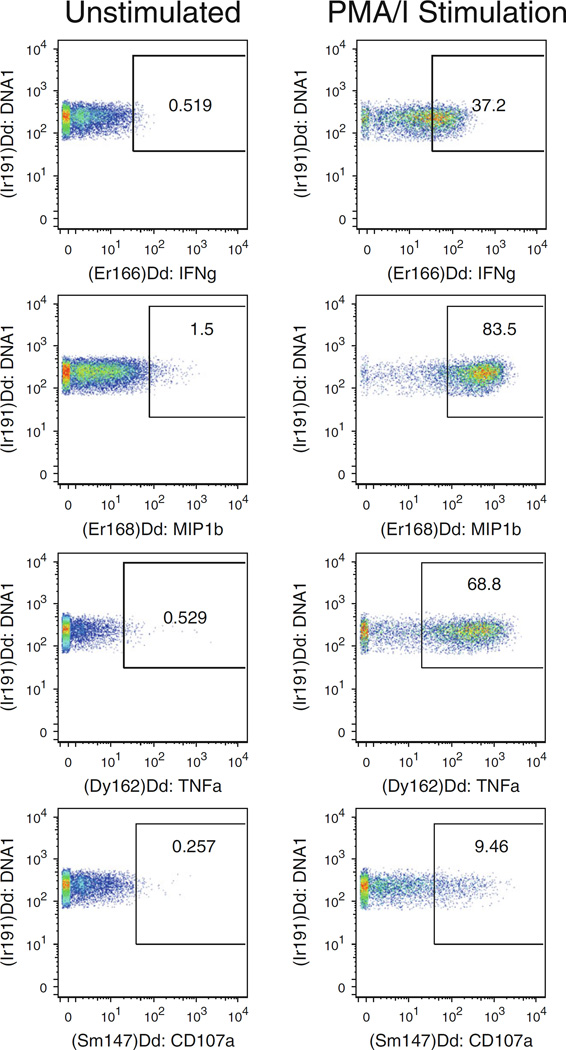
Fig. 3.
Representative CyTOF plots of functional markers on stimulated and unstimulated NK cells, gated as in Fig. 2
Footnotes
We have had success with IgM conjugations (e.g., CD57) although they are not technically supported by DVS/Fluidigm.
Qdot antibodies have a cadmium core that can be detected by mass cytometry on channels Cd111–Cd114. Bright antibodies (e.g. HLA-DR, CD3) are most effectively used on this channel or it can be designated as a dump channel.
Currently, Fluidigm does not sell kits to conjugate Gd157. Gd157 metals can be procured as 92 % + purity from Trace Sciences International, and conjugated with polymer from MaxPar kits.
Following conjugation, the total concentration of antibody should be determined by the concentration of A280 protein as measured by Nanodrop. The expected yield is approximately 2/3 of the initial antibody conjugated.
Antibodies should be titrated to determine optimal concentration. A typical starting point is 10 µg/mL, with five serial two-fold dilutions to test the range between 0.3125 and 10 µg/mL.
Based on the concentration of the antibody following concentration, and the optimal titration as determined empirically, the amount of antibody added per sample can be calculated in order to make the antibody staining cocktail. It is advisable to make extra staining cocktail. Example calculations for 20 samples are shown in Table 1.
Peripheral blood mononuclear cells can be used fresh or cryo-preserved and thawed prior to use. If the primary focus is NK cell responses, it is not necessary to rest the cells overnight prior to use.
Either DOTA-maleimide conjugated to a metal isotope [12] or cisplatin [13] can be used as a live-dead stain. Cisplatin use allows an additional antibody channel because its detection channel cannot be used in conjugations with other antibodies.
We have noted some batch-to-batch variability in the concentration of cisplatin required to see a clearly defined dead population. Validation can be performed with samples containing partially heat-killed cells. Cisplatin is a readily available platinum-based chemotherapeutic agent that passively accesses the cell interior of dead cells and rapidly reacts with protein nucleophiles such as R-SH or R-S-CH3 [10]. Cisplatin entry into cells is stopped through the addition of 100 % fetal bovine serum that reacts with residual extracellular cisplatin and prevents active transport into live cells.
It is acceptable to freeze the plate at −80 °C for up to 1 week after adding FACS Lyse working solution.
In order to analyze CyTOF data, settings on FlowJo need to be customized. In order to do so go to the “Preferences” tab, and under workspace select “CyTOF” in the lower right corner and set the value to 20,000. Select the “Define” button under the “Reading Digital Data Files” heading. Select “Side Scatter” and all fluorescence parameters to display with logarithmic staining. Select “Ignore Scaling suggested by the data file”. Add Time and Cell Length as parameters that should always be linear in the box on the top right. Set the Lowest Standard Log Conversion setting to 1 and the number of decades to display in log-converted data to 6 pulse area parameters and 6 pulse height parameters. Select the Enable Transformation box in addition to the Transform Height Parameters box. Set the number of decades to 5, additional negative display size to 0, and the width basis to −20.
References
- 1.Ornatsky O, Baranov VI, Bandura DR, et al. Multiple cellular antigen detection by ICP-MS. J Immunol Methods. 2006;308:68–76. doi: 10.1016/j.jim.2005.09.020. [DOI] [PubMed] [Google Scholar]
- 2.Bendall SC, Nolan GP, Roederer M, et al. A deep profiler’s guide to cytometry. Trends Immunol. 2012 doi: 10.1016/j.it.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strauss-Albee DM, Horowitz A, Parham P, et al. Coordinated regulation of NK receptor expression in the maturing human immune system. J Immunol. 2014;193:4871–4879. doi: 10.4049/jimmunol.1401821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Horowitz A, Strauss-Albee DM, Leipold M, et al. Genetic and environmental determinants of human NK cell diversity revealed by mass cytometry. Sci Transl Med. 2013;5 doi: 10.1126/scitranslmed.3006702. 208ra145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Newell EW, Sigal N, Bendall SC, et al. Cytometry by time-of-flight shows combinatorial cytokine expression and virus-specific cell niches within a continuum of CD8+ T cell phenotypes. Immunity. 2012;36:142–152. doi: 10.1016/j.immuni.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strauss-Albee DM, Blish CA. CyTOF: single cell mass cytometry for evaluation of complex innate cellular phenotypes. Experimental approaches for the investigation of innate immunity. 2016:27–39. [Google Scholar]
- 7.Leipold MD, Maecker HT. Mass cytometry: protocol for daily tuning and running cell samples on a CyTOF mass cytometer. JoVE. 2012 doi: 10.3791/4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finck R, Simonds EF, Jager A, et al. Normalization of mass cytometry data with bead standards. Cytometry. 2013;83A:483–494. doi: 10.1002/cyto.a.22271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qiu P, Simonds EF, Bendall SC, et al. An analysis extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat Biotechnol. 2011;29:886–891. doi: 10.1038/nbt.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bruggner RV, Bodenmiller B, Dill DL, et al. Automated identification of stratifying signatures in cellular subpopulations. Proc Natl Acad Sci U S A. 2014;111:E2770–E2777. doi: 10.1073/pnas.1408792111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amir ED, Davis KL, Tadmor MD, et al. visNe enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol. 2013;31:545–552. doi: 10.1038/nbt.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bendall SC, Simonds EF, Qiu P, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science (New York, NY) 2011;332:687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fienberg HG, Simonds EF, Fantl WJ, et al. A platinum-based covalent viability reagent for single-cell mass cytometry. Cytometry. 2012;81A:467–475. doi: 10.1002/cyto.a.22067. [DOI] [PMC free article] [PubMed] [Google Scholar]