Abstract
Epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs) such as gefitinib are one of gold standard treatment options for nonsmall-cell lung cancer (NSCLC) patients, which eventually fail due to the acquired resistance and relapse because of the development of secondary activating mutations such as T790M in EGFR. Predicting chemo-responsiveness of cancer patients provides a major challenge in chemotherapy. The goal of the present study is to determine whether phospholipid signatures of tumor extracellular vesicles (EV) are associated with gefitinib-resistance of NSCLC. A sophisticated MS-based shotgun lipidomic assays were performed for in-depth analysis of the lipidomes of gefitinib-resistant (PC9R) and responsive (PC9) NSCLC cells and their shed EV from these cell lines (PC9EV or PC9REV). Lipid MALDI-MS analysis showed that EV phospholipid composition was significantly distinct in PC9R, compared to PC9 cells. Following statistical analyses has identified 35 (20 positive and 15 negative ion mode) differentially regulated lipids, which are significantly over- or underexpressed in PC9R EV, compared to PC9 EV (p value < 0.01, fold change > 1.5). Our phospholipid signatures suggest that EV associates with drug sensitivity, which is worthy of additional investigation to assess chemoresistance in patients with NSCLC treated with anti-EGFR TKIs.
Keywords: Extracellular vesicles, Lipidopmics, MALDI-MS, Nonsmall-cell-lung cancer, Phospholipids
1 Introduction
Lung cancer is the leading cause of cancer-related mortality in United States, and nonsmall cell lung cancer (NSCLC) represents over 85% of lung cancer, with less than 10% five-year survival rate [1]. Most of NSCLC patients are diagnosed as stage 3b or stage 4. In patients with over stage 3, cancers cannot be removed by surgery, thus most of NSCLC patients are treated with combined chemotherapy and radiotherapy. More than 60% of NSCLC showed overexpression of epidermal growth factor receptor (EGFR), a transmembrane protein with tyrosine kinase activity required for signal transduction [2]. Thus, EGFR has become a therapeutic target for NSCLC, and various EGFR tyrosine kinase inhibitors (TKIs) have been developed and tested in preclinical and clinical settings [1, 3]. Approximately 15% of NSCLC patients in United States have mutations to the EGFR [4], and they showed better therapeutic effects, suggesting that EGFR mutations predict whether treatment including TKIs may help patients [5].
Our laboratory has demonstrated that tumor cells shed extracellular vesicles (EV) into the extracellular space in previous publications [6–10]. EV have been reported not only as a biological cargo, but also as a potential biomarker, since they have found to contain various bioactive biomolecules such as nucleic acids (DNA, mRNAs, miRNAs, and small non-coding RNAs), oncoproteins, and metabolites [11–13]. Since these bioactive EV can be isolated from various biological specimens including urine and serum, EV have been considered as potential noninvasive biomarkers assessing tumor progression and drug response [14, 15]. Recently, we could identify gefitinib (an ATP competitive inhibitor of EGFR tyrosine kinase)-resistance associated proteome in EV [16]. These results suggested that EV could be a therapeutic biomarker for predicting clinical response to EGFR targeting drugs.
Phospholipid metabolism is regulated by extracellular stimuli such as various growth factors, inflammation-associated cytokines, oncogenes, and hypoxia conditions, and plays a key role in cell motility, invasion and metastasis of tumors [17–24]. Given the previous reports showing that metabolic perturbation of phospholipids was associated with various cancer types [25–27], the composition of phospholipids may be critical for deciding fate of tumor cells. Alteration of levels of phospholipid metabolites such as phosphatidylcholine (PC), and phosphatidylethanolamine has been often considered as biochemical indicators of tumor progression or drug response [28–31].
Although composition and abundance of phospholipid species may be also important for biological roles of EV, such as maintenance of cellular membrane structure and signal transduction via EV, global lipidomics analysis of EV remain elusive. Recently, our laboratory has developed and established the MALDI-TOF-based MS analysis method, which is a feasible approach to identify specific lipid profile of various biological samples. We have applied this method to profile phospholipids in ovarian cancer tissues and rat brain tissues [17, 32]. Moreover, our phospholipid-based imaging MS could elucidate the spatial localization of phospholipids throughout the entire brain or tumor tissues, and reveal their membrane structures. More recently, MALDI-MS-based lipid profiling classified subsets of cancer types including clear cell renal carcinoma, cholangiocarcinomas, pancreatic cancers, HER2-positive breast cancer, and ovarian cancer patients [18, 33–35]. In our previously reported lipid profile, phospholipids such as PC{34:1} [M+H]+ (m/z = 760.6), PC{34:1} [M+K]+(m/z = 798.6), and PC{32:0} [M+H]+ (m/z = 734.6) were identified as unique lipid signature of breast cancer compared to normal tissues [18], suggesting that lipid profiles correlate with clinicopathological features of breast cancers.
In this paper, we present the first EV phospholipid profile associated with NSCLC with resistance to gefitinib. Our MALDI-TOF-MS/MS analysis detected total 67 significantly differential phospholipids between PC9 and PC9R cells including phosphatidylcholines (PC), lysophosphatidylcholines (LPC), sphingomyelines (SM), phosphatidylglycerols (PG), and phosphatidylinositols (PI), lysophosphatidylinositols (LPI). EV phospholipid composition was significantly distinct in gefitinib resistant NSCLC, compared to parent cells. Our findings suggest that a transition to an acquired resistant phenotype may accompany alteration of phospholipids reside in EV, and that these alterations of phospholipid composition might be relevant to gefitinib resistance as a novel predictive biomarker for response to gefitinib.
2 Materials and methods
2.1 Cell culture
The PC9R cell line was established as a gefitinib-resistant clone from parent PC9 cells. Briefly, to establish a gefitinib-resistant subline, the PC9 parent cells were selected by incubation in medium containing gefitinib in a stepwise manner, which was started from 0.01 to 2 μM of gefitinib for several months. PC9 and PC9R cells were cultured under a humidified atmosphere of 5% CO2 at 37 °C in RPMI-1640 medium (Sigma, St Louis, MO, USA) supplemented with 10% FBS, 100 μg streptomycin, and 100 units/mL penicillin (Invitrogen, Carlsbad, CA). The cells were then harvested by trypsinization and subcultured at a density of 6 × 103 cells/cm2. The media were changed after one day of subculture during culture, and cell cultures were passaged again at 70–80% confluence.
2.2 Isolation of EV from conditioned medium
To obtain purified EV, PC9, and PC9R cells were washed with PBS twice, and then incubated in serum-free medium for additional 24 h. Conditioned media were harvested and centrifuged at 500 × g for 15 min, followed by another centrifugation at 800 × g for 15 min to remove cell debris. After Supernatant concentration using Stirred Ultrafiltration Cells 8400 (Millipore, USA), it was loaded onto 0.8 and 2.7 M sucrose cushions in 20 mM HEPES/150 mM NaCl buffer (pH 7.2) and subsequently ultracentrifuged at 100 000 g for 2 h at 4 °C. To purify EV, sequential ultracentrifugation on a sucrose cushion and OptiPrep (50% iodixanol, Axis-Shield PoC AS, Oslos, Norway) density gradient were performed repeatedly [36]. Sequentially, fractions containing EV were added onto 0.8 and 2.7 M sucrose cushions after dilution in sucrose dilution buffer [20 mM HEPES/150 mM NaCl, pH 7.2], before ultra-centrifugation at 200 000 g for 70 min at 4 °C. The interface between the 0.8 M and 2.7 M sucrose cushion was harvested to apply for step-density gradient ultracentrifugation (2.5 mL of 5% OptiPrep, 3 mL of 20% OptiPrep and 4.8 mL of 30% OptiPrep). Ten fractions of equal volume were collected from the top of the tube. The 3rd fraction (1.4 mL) containing EV was transferred to 9 mL PBS followed by ultra-centrifugation at 100 000 g for 1 h at 4 °C to collect EV pellet for further study. The EV purified fractions were stored at −80 °C until start extraction of lipid [18, 19].
2.3 Transmission electron microscopy
For the direct visualization of EV in the transmission electron microscope, the purified EV were placed on to glow-discharged carbon-coated copper grids. After 2 min incubation at room temperature, EV were removed by blotting with a filter paper and negative-stained with a drop of 1% aqueous uranyl acetate. The grid was washed and dried. Negatively stained EV were visualized with a JEM 1400 (Jeol, Japan) at an acceleration voltage of 100 kV.
2.4 Immunoblot analysis
Isolated EV from PC9 or PC9R were lysed and 25 μg of EV lysates were analyzed by Western blotting using the following antibodies: rabbit polyclonal (Abcam, 1:1000 dilution), rabbit polyclonal CD9 (Abcam, 1:1000 dilution); and rabbit monoclonal CD63 (Abcam, 1:1000 dilution).
2.5 Phospholipid extraction of EV
Total 20 μg EV were used for each lipid analysis. EV phospholipids were extracted by Bligh & dyer method [37]. Briefly, EV were directly transferred into 3 mL of CHCl3:ethanol (1:2, v/v) in 15 mL conical glass tube. Each sample was vortexed and sonicated for 10 min, followed by cooling on ice for 10 min. The samples were centrifuged at 2500 g for 10 min after vortex. The bottom organic phase containing the phospholipids was dried in a speed vacuum. All experiments were run in triplicate [18, 19].
2.6 Lipid MALDI-MS analysis
For phospholipid MALDI-MS analysis, 10 μL of lipid samples diluted in methanol/chloroform (70/30, v/v) were mixed with 10 μL of the binary matrix solution (7 mg each of 2, 5-dihydroxybenzoic acid (DHB) and α-cyano-4-hydroxycinnamic acid (CHCA) in 1 mL of 70% methanol plus 0.1% TFA) and 10 αL of 9-aminoacridine (10 mg/mL; dissolved in isopropanol/acetonitrile (60/40, v/v)) for negative mode analysis [32, 38]. Each sample was deposited on a 384 target plate (BrukerDaltonics, Bremen, Germany) in ten replicates (positive mode), five replicates (negative mode) followed by evaporation in desiccator for homogeneous crystallization in order to get iterative results. MALDI-MS analysis was performed using an Ultraflex III TOF/TOF mass spectrometer (BrukerDaltonics, Bremen, Germany) equipped with a 200-Hz smartbeam laser as an ionization source. All of the spectra were acquired with delay: 180 ns; ion source 1: voltage, 25 kV, ion source 2: voltage, 21.65 kV; lens voltage: 9.2 kV and optimized for 500–1200 Da with an average of 1000 shots. Before each data acquisition, an external calibration was conducted using lipid-mixed calibration standards with m/z ranges of 674–834 Da (positive ion mode) and 564–906 Da (negative ion mode). MALDI-LIFT (MS/MS) analysis was directly performed on the sample spot after MALDI-MS to confirm lipids structure. We precede the lipid identification using Lipidomics Gateway (http://www.lipidmaps.org) based on major fragment ions of ms/ms spectrum after performing manual mono isotope selection. <LIFT mode condition> CID mode = false; PCIS mass limit = 2–4 Da; Ion source voltage 1/2 = 8kV/7.1 kV; LENS voltage = 3.6 kV; LIFT voltage 1/2 = 19 kV/4.3 kV in positive mode. CID mode = false; = PCIS mass limit = 2–4 = Da; Ion source voltage 1/2 = 8 kV/7.1 kV; LENS voltage = 3.6 kV; LIFT voltage 1/2 = 19 kV/4.2 kV in negative mode.
2.7 Preprocessing of MALDI MS data
All preprocessing steps were performed using MALDIquant package [39] in R. The intensities of the features in each single spectrum were transformed to square root scale for variance stabilization and smoothed with moving average algorithm. The spectrum background was estimated with statistics-sensitive nonlinear iterative peak-clipping algorithm and used for baseline correction. To enable a quantitative comparison across multiple spectra, the peak intensities were normalized using probabilistic quotient normalization method [40]. Each spectrum was normalized by a reference spectrum that is a median spectrum of all samples. Briefly, the normalization procedure was as follows: (1) normalization of all spectra to equal TIC, (2) calculation of the reference spectrum that is a median spectrum of all samples, (3) for each spectrum, calculation of the quotients of the intensities of the spectrum with those of the reference spectrum, (4) calculation of the median of these quotients, and (5) division of all intensities of the spectrum by the median of quotients calculated at step 4. The features with a S/N of higher than three were considered as peaks. The peaks belonging to the same mass were aligned by the statistical regression-based approach using the identification of landmark peaks and the estimation of a nonlinear warping function.
2.8 Statistical analysis of MALDI MS data
For comparison of multiple spectra from different samples, all the following statistical analyses were performed using MetaboAnalyst 2.0, a web-based software for quantitative data analysis [41]. First, missing values in the data obtained from a preprocessing procedure were replaced by half of the minimum positive value in the data. Then the data were normalized to have the same total intensity for all spectra. The intensity values of each peak across multiple spectra were mean-centered and divided by the standard deviation. Principal component analysis (PCA) was performed to show the variance and differences among samples from PC9 and PC9R. The differentially regulated phospholipids (DRL) between PC9 and PC9R were identified using the following criteria: (1) p values from t-test are less than 0.01; (2) absolute fold changes are larger than 1.5. To show the relationship between samples and features, hierarchical clustering of DRLs was performed using “Euclidean distances” and “ward” linkage. The Pearson’s correlation coefficients between spectra were calculated and clustered to identify peaks that are correlated.
2.9 Quantitative confirmation of target lipids using Triple Quadrupole LC-MS
To further validate differential expression of the lipids identified by MALDI-MS analysis, we performed LC-MS analysis using triple quadrupole, which outperforms MALDI-TOF in sensitivity, specificity, and dynamic range. The quantification of target lipids (protonated form) in EV samples was performed by 6490 Accurate-Mass Triple Quadrupole (QqQ) LC-MS coupled to a 1200 series HPLC system (Agilent Technologies, Wilmington, DE, USA) with a Hypersil GOLD column (2.1 × 100 mm id; 1.9 μm, Thermo science). This approach provides high sensitivity by iFunnel technology consisting of three components: Agilent Jet Stream technology, a hexabore capillary, and a dual ion funnel. We described detail information in the Supplementary Materials and Methods.
3 Results
3.1 MALDI-MS analysis identified lipidomes
Previous studies from our laboratory and others suggest the alteration of phospholipid metabolism is associated with cancer progression or subpopulation [19, 20, 26–28, 42]. Here we attempted to test whether alteration in phospholipid composition in EV shed from NSCLC cells with acquired resistance to gefitinib (PC9R) from sensitive control cells (PC9). Good signature predicting resistance will suggest that phospholipid profile of shed EV into biofluids (e.g. urine or plasma) using MS-based analysis may be used for monitoring on drug resistance after gefitinib-based chemotherapy in clinic.
In order to interrogate global alteration of phospholipids, EV shed from PC9 and PC9R cells were isolated (Fig. 1A). Electron microscopic images of shed EV from PC9 or PC9R were shown in Fig. 1B, and several EV-specific protein markers, CD81, CD9, and CD63, were analyzed by Western blot analysis (Fig. 1C). MS-based lipidomic analysis was performed (biological triplicates and ten technical replicates), generating a total of 67 lipidome profiles for PC9_cell (whole cell lysates of PC9, n = 30 in positive, n = 15 in negative), PC9R_cell (whole cell lysates of PC9R, n= 30 in positive, n = 15 in negative), PC9_EV (EV isolated from conditioned medium of PC9 culture, n = 30 in positive, n = 15 in negative) or PC9R_EV (EV isolated from conditioned medium of PC9R, n = 30 in positive, n = 15 in negative). We have obtained mass spectra by MALDI-MS-based analysis exhibited profile of positive- and negative-ion reflector modes. MALDI-TOF Spectrum process, data processing and statistical analysis for this analysis were shown in workflow in Fig. 1A.
Figure 1.
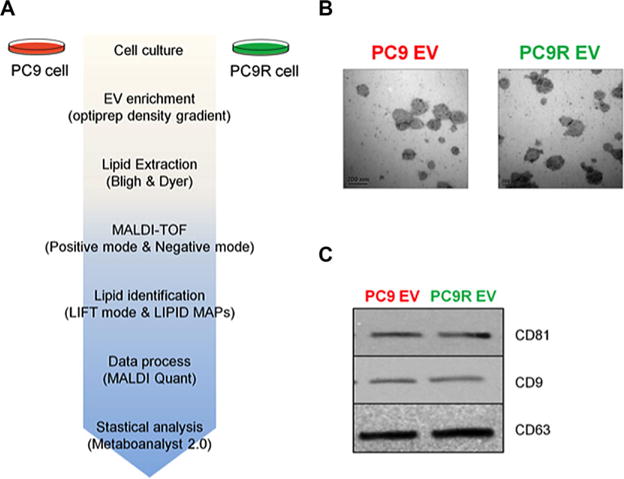
Schematic diagram of lipidomic analysis and characteristics of the isolated EV. (A) Whole cells and EV derived from PC9 and PC9R were prepared for lipidomics analysis. Lipids were isolated from each sample and analyzed using MALDI-MS as described in Materials and Methods. MALDI-TOF spectra were processed using MALDI Quant and R package. Datasets were analyzed using MetaboAnalyst. (B) Electron microscopy showed that purified EV varied in size from 40 to 500 nm. The EVs were not contaminated by cellular debris or protein aggregates. Scale bar = 200 nm. (C) Western blot analysis showing the EV markers in both EV samples.
Representative average mass spectra for lipids obtained from EV of PC9 (PC9_EV, red) or PC9R (PC9R_EV, green) were shown in positive- (Fig. 2A) and negative- (Fig. 2B) ion modes. In positive mode, most of the phospholipid spectra appeared on m/z 500–900; however, only m/z 912.7 was detected above m/z 900 with low intensity and some of the major peaks, such as m/z 969.8, 995.8, 997.8, 1013.9 were identified triglyceride (data not shown). In negative mode, most phospholipid peaks were detected on the vicinity of m/z 700–900 (Fig. 2B). All 30 and 15 spectra obtained in positive and negative modes were consistent and showed clear distinction between PC9 and PC9R (Fig. 2C and 2D). Chromatograms of PC9_cell, PC9R_cell, PC9_EV, or PC9R_EV and the combined spectra of all samples were shown in Supporting Information Fig. 1.
Figure 2.
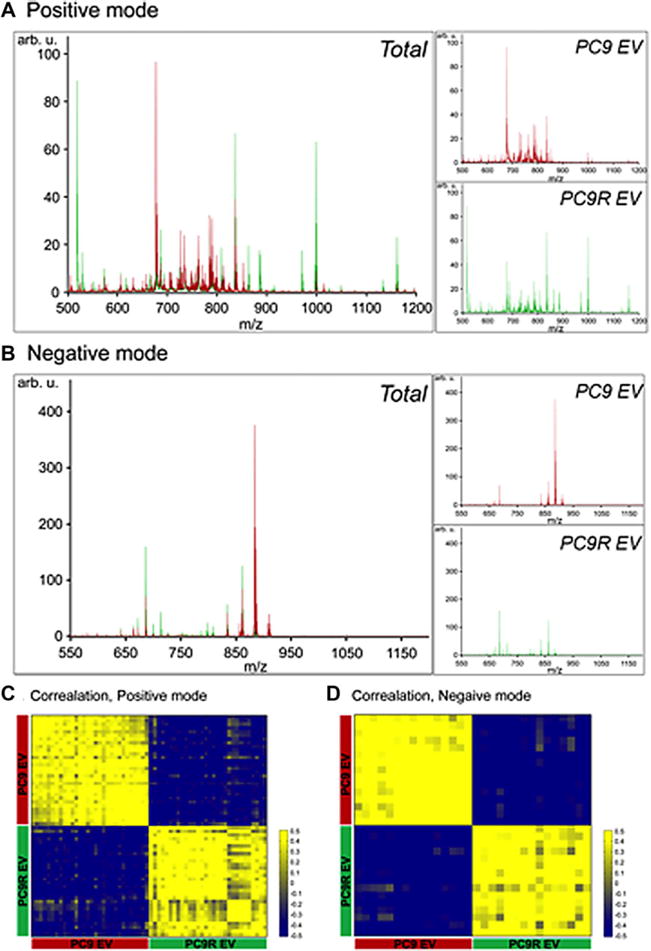
Lipidomic analysis of EV from PC9 (PC9_EV) and PC9R cells (PC9R_EV). (A) Average chromatogram of PC9_EV (in red) or PC9R_EV (in green) obtained with positive ion mode. (B) Average chromatogram of PC9_EV (in red) or PC9R_EV (in green) obtained with negative ion mode. (C) Heatmap of correlation between 30 spectra obtained from positive ion mode. Yellow and blue denote positive and negative correlation between spectra, respectively. (D) Heatmap of correlation between 15 spectra obtained from negative ion mode. Yellow and blue denote positive and negative correlation between spectra, respectively.
Figure 3 shows representative fragmentation spectra obtained from positive and negative ionization modes by LIFT mode. In positive mode, m/z 782.6 has a particular head group containing choline combined with phosphate (m/z 184), sodium adducted phosphate (m/z 147), and other specific fragment ion peaks of the phospholipids (Fig. 3A). The m/z 782.6 can be identified simultaneously as sodium adducted form and protonated form because it has two forms; (1) major m/z 147 peak and (2) small but detectable m/z 184 peak. As described in previous literature [43–45], m/z 782.6 was annotated in two ways. Our group also has used ms/ms spectra of PC {34:1} [M+Na]+ as a reference in a published paper [46]. So, when ms/ms spectra have m/z 184 and m/z 147 at the same time, we annotated them as sodium adducted form and protonated form, respectively. The fragmentation spectra of m/z 861.6 in negative mode were also shown (Fig. 3B). The fragmentation spectra contain m/z 241 (head group of phosphatidylinositol) and fatty acid chains (m/z 283; C18:0, m/z 279; C18:2). All the other identified phospholipids were confirmed by fragment ions (LIFT mode) in the same ways (data not shown).
Figure 3.
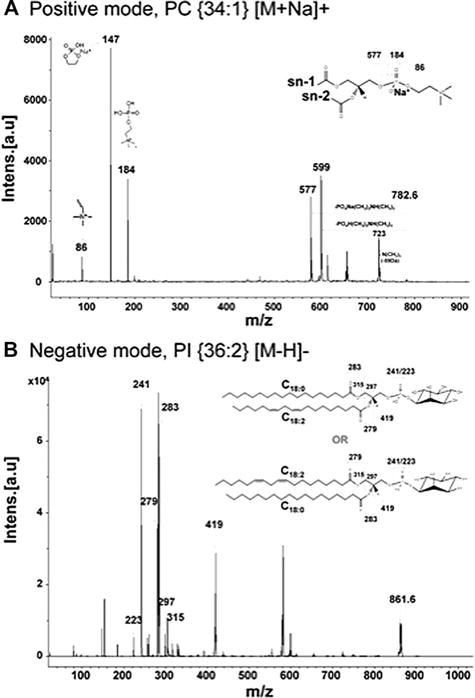
Fragmentation spectra using the LIFT technique for representative phospholipid species. (A) PC{34:1}[M+Na]+ in positive ion mode and (B) PI{18:0/18:2}[M-H]− in negative ion mode.
An unsupervised multivariate data analysis technique, PCA analysis was performed on the MS spectral data of the EV phospholipids in positive and negative mode (Figs. 4A and 5A). Each sphere represented a single spectrum, and spheres were shown closer than others when profile of lipids was similar. PCA analysis showed that composition of EV phospholipids was more distinctive than those of whole cells, showing a remarkable discrimination of certain phospholipids in EV (Supporting Information Fig. 2A). Phospholipids of two cell lines, PC9R and PC9, were less clearly differentiated. PC9R_EV (light blue) exhibited a clear differentiation from PC9_EV (green) with a good separation and dispersion both in positive (Supporting Information Fig. 2A) and negative modes (data not shown), suggesting that EV can be effective biomarker discriminating gefitinib sensitive versus resistant NSCLC. Clustering using Euclidean distance measure and ward clustering algorithm linkage demonstrated the greater correlations in DRL between PC9_cells and PC9R_cell, or PC9_EV and PC9R_EV (Supporting Information Fig. 2B), than those between cell and EV.
Figure 4.

Differentially regulated phospholipids (DRL) identified in positive mode. (A) Principal component analysis; A principal component analysis plot for phospholipids mass spectrum of PC9_EV (shown in red), PC9R_EV (shown in green). (B) Volcano plot showing relative intensities of DRL in PC9R_EV compared to PC9_EV, which were measured by MALDI-TOF-MS in positive ion mode (fold change > 1.5, p value < 0.01)
Figure 5.

DRL in EV identified by MS in negative ion mode. (A) Score plot showing that a segregation of signatures between PC9_EV and PC9R_EV. (B) Volcano Plot including DRL in PC9R_EV compared to PC9_EV, which were determined in negative mode (fold change > 1.5, p value < 0.01).
3.2 MS-based lipid profiling can classify gefitinib-resistant NSCLC from responsive tumor cells
Total 48 peaks including m/z 884.7 (p value, 6.0448 10−47), m/z 832.7 (p value, 5.16 × 10−47), or m/z 728.5 (p value, 1.28 × 10−47), were obtained in positive (p) ion mode MS. Under the strict selection criteria (fold change > 1.5, p value < 0.01), our positive ion mode lipidome quantification using MetaboAnalyst 2.0 could identify 20 DRL in total. Main lipid and PL classes identified in a positive mode were indicated by their m/z ratios in Table 1. Phosphatidylcholines (PC), lysophosphatidylcholines (LPC), and sphingomyelines (SM) were identified in positive ion mode. Seventeen phospholipids containing PC{42:6} [M+Na]+, PC{40:7} [M+H]+, and PC{32:3} [M+H]+ were in PC9R_EV, while 31 phospholipids containing SM{d40:2} [M+H]+, PC{38:5} [M+H]+, and PC{34:2} [M+H]+ were significantly underexpressed in PC9R_EV, compared to PC9_EV (Table 1). Ratios of PC9R_EV/PC9_EV and p values were also shown in Table 1 (positive mode). PC and SM, which are major constituents of cell membranes, were prominent in positive mode. In negative ion mode, we also found that 19 phospholipids which expressed differently in PC9R_EV or PC9_EV (Table 2). PI and PG were identified mainly and ten phospholipids containing PI, PG class were overexpressed in PC9R_EV, while nine phospholipids containing only PI class were underexpressed in PC9R_EV, compared to PC9_EV. Fifteen phospholipids were shown significant difference between PC9_EV and PC9R_EV in negative mode (fold change > 1.5, p value < 0.01). PI, an acidic phospholipid that plays a key role in EGFR signaling pathway, was detected in negative ionization mode. PI has both neutral and anionic structures, which indicate an arrangement of hydroxyl groups on the inisitol head group in order to maximize hydrogen-bonding interactions. Furthermore, both structures demonstrate strong interaction between the two of the inisitol hydroxyls and the phosphate moiety. These interactions are especially critical in the stabilization of [PI–H]− with simultaneous hydrogen bonds to both charge bearing oxygens [47].
Table 1.
Identification and quantification of MS peaks in EV from PC9 or PC9R cells in positive ion mode.
| m/z | pa) | Ratiob) | Assignment |
|---|---|---|---|
| Overexpressed in extracellular vesicles of PC9R cell | |||
| 754.5 | 2.00 × 10−1 | 1.03 | PC {34:4} [M+H]+ |
| 720.6 | 5.23 × 10−3 | 1.05 | PC{O-32:0}[M+H]+ |
| 706.6 | 1.72 × 10−13 | 1.2 | PC {30:0} [M+H]+ |
| 756.5 | 1.64 × 10−10 | 1.22 | PC {34:3} [M+H]+ |
| 742.5 | 1.24 × 10−15 | 1.36 | PC {O-34:3} [M+H]+/PC {P-34:2} [M+H]+ |
| 750.6 | 3.40 × 10−21 | 1.41 | PC {34:6} [M+H]+ |
| 834.7 | 4.97 × 10−20 | 1.45 | PC {40:6} [M+H]+ |
| 692.6 | 2.14 × 10−31 | 1.56 | PC{O-30:0}[M+H]+ |
| 804.6 | 2.05 × 10−19 | 1.61 | PC {38:7} [M+H]+ |
| 722.5 | 1.50 × 10−32 | 1.69 | PC {30:3} [M+Na]+ |
| 550.5 | 1.84 × 10−20 | 2.39 | LPC {20:1} [M+H]+ |
| 806.7 | 1.16 × 10−37 | 2.4 | PC {36:3} [M+Na]+/PC {38:6} [M+H]+ |
| 770.6 | 6.76 × 10−49 | 6.06 | PC {O-36:3} [M+H]+/PC {P-36:2} [M+H]+ |
| 912.7 | 7.03 × 10−35 | 6.96 | PC {44:6} [M+Na]+ |
| 728.5 | 1.28 × 10−44 | 9.25 | PC {32:3} [M+H]+ |
| 832.7 | 5.16 × 10−47 | 11.76 | PC {40:7} [M+H]+ |
| 884.7 | 6.0448 × 10−47 | 13.75 | PC {42:6} [M+Na]+ |
| Underexpressed in extracellular vesicles of PC9R cell | |||
| 732.6 | 9.18 × 10−1 | 0.99 | PC {32:1} [M+H]+ |
| 746.5 | 6.04 × 10−1 | 0.99 | PC {O-34:1} [M+H]+/PC {P-34:0} [M+H]+ |
| 782.6 | 2.58 × 10−1 | 0.97 | PC {34:1} [M+Na]+/PC {36:4} [M+H]+ |
| 718.7 | 3.15 × 10−3 | 0.93 | PC {O-32:1} [M+H]+/PC {P-32:0} [M+H]+ |
| 704.6 | 4.04 × 10−4 | 0.93 | PC {30:1} [M+H]+ |
| 820.6 | 9.04 × 10−3 | 0.91 | PC {40:5} [M+H]+ |
| 768.6 | 4.89 × 10−9 | 0.88 | PC {O-36:4} [M+H]+/PC {P-36:3} [M+H]+ |
| 780.6 | 3.33 × 10−7 | 0.88 | PC {36:5} [M+H]+ |
| 760.6 | 1.08 × 10−6 | 0.85 | PC {34:1} [M+H]+ |
| 787.8 | 9.40 × 10−18 | 0.82 | SM{d40:1}[M+H]+ |
| 703.6 | 1.1716 × 10−18 | 0.75 | SM {d34:1} [M+H]+ |
| 725.5 | 1.70 × 10−19 | 0.75 | SM {d34:1} [M+Na]+ |
| 522.3 | 8.13 × 10−9 | 0.74 | LPC {18:1} [M+H]+ |
| 810.6 | 2.10 × 10−25 | 0.73 | PC {36:1} [M+Na]+/PC {38:4} [M+H]+ |
| 675.6 | 1.42 × 10−26 | 0.73 | SM {d32:1} [M+H]+ |
| 774.6 | 1.12 × 10−27 | 0.71 | PC {34:5} [M+Na]+ |
| 812.6 | 1.14 × 10−22 | 0.71 | PC {38:3} [M+H]+ |
| 798.6 | 1.56 × 10−19 | 0.71 | PC {34:1} [M+K]+ |
| 759.7 | 1.41 × 10−23 | 0.7 | PC {38:1} [M+H]+ |
| 794.6 | 1.42 × 10−18 | 0.69 | PC {O-38:5} [M+H]+/PC {P-38:4} [M+H]+ |
| 518.3 | 6.30 × 10−15 | 0.68 | LPC {18:3} [M+H]+ |
| 504.5 | 1.12 × 10−16 | 0.62 | LPC {14:1} [M+K]+ |
| 649.5 | 2.01 × 10−9 | 0.55 | SM {d30:0} [M+H]+ |
| 546.4 | 1.16 × 10−24 | 0.55 | LPC {20:3} [M+H]+ |
| 562.4 | 5.08 × 10−23 | 0.54 | LPC {18:0} [M+K]+ |
| 796.6 | 1.85 × 10−30 | 0.53 | PC {O-38:4} [M+H]+/PC {P-38:3} [M+H]+ |
| 572.6 | 5.08 × 10−21 | 0.51 | LPC {22:4} [M+H]+ |
| 544.3 | 1.07 × 10−49 | 0.28 | LPC {20:4} [M+H]+ |
| 758.6 | 3.07 × 10−47 | 0.19 | PC {34:2} [M+H]+ |
| 808.6 | 4.13 × 10−54 | 0.15 | PC {38:5} [M+H]+ |
| 785.7 | 3.58 × 10−59 | 0.08 | SM{d40:2}[M+H]+ |
Peak position was indicated by the m/z ratio. MS peaks were assigned using lipid databases, Lipid Search or LIPID Metabolites and Pathways Strategy (LIPID MAPS) database (www.lipidmaps.org).
p-value <0.01, fold change>1.5.
Ratio, extracellular vesicles of PC9R cell/PC9 cell ratio.
P-, plasmenyl; O-, plasmanyl.
PC, phosphatidylcholines.
P-LPC, lysophosphatidylcholines.
P-SM, sphingomyelines.
Table 2.
Identification and quantification of MS peaks in EV from PC9 or PC9R cells in negative ion mode.
| m/z | pa) | Ratiob) | Assignment |
|---|---|---|---|
| Overexpressed in extracellular vesicles of PC9R cell | |||
| 599.3 | 0.00030159 | 1.2213 | LPI {18:0} [M-H]− |
| 571.3 | 4.7026 × 10−5 | 1.2858 | LPI {16:0} [M-H]− |
| 819.6 | 2.97 × 10−12 | 1.6714 | PI {O-34:2} [M-H]−/PI {P-34:1} [M-H]− |
| 677.5 | 1.92 × 10−8 | 1.8357 | PG {O-30:1} [M-H]−/PG {P-30:0} [M-H]− |
| 747.5 | 7.70 × 10−13 | 1.9846 | PG {34:1} [M-H]− |
| 833.6 | 3.02 × 10−27 | 2.079 | PI {34:2} [M-H]− |
| 861.6 | 6.28 × 10−25 | 2.1993 | PI {36:2} [M-H]− |
| 805.5 | 3.39 × 10−20 | 2.995 | PI {32:2} [M-H]− |
| 807.6 | 7.69 × 10−27 | 4.0045 | PG {O-40:6} [M-H]−/PG {P-40:5} [M-H]− |
| 809.6 | 3.36 × 10−18 | 4.4588 | PI {32:1} [M-H]− |
| Underexpressed in extracellular vesicles of PC9R cell | |||
| 859.5 | 0.67216 | 0.99218 | PI {36:3} [M-H]− |
| 597.3 | 1.8187 × 10−8 | 0.69517 | LPI {18:1} [M-H]− |
| 857.5 | 1.06 × 10−18 | 0.5215 | PI {36:4} [M-H]− |
| 883.5 | 2.89 × 10−24 | 0.45728 | PI {38:5} [M-H]− |
| 907.6 | 5.10 × 10−22 | 0.42615 | PI {40:7} [M-H]− |
| 911.6 | 3.45 × 10−11 | 0.36459 | PI {40:5} [M-H]− |
| 881.5 | 4.92 × 10−15 | 0.3427 | PI {38:6} [M-H]− |
| 909.6 | 3.62 × 10−27 | 0.3302 | PI {40:6} [M-H]− |
| 855.5 | 3.06 × 10−27 | 0.32854 | PI {36:5} [M-H]− |
Peak position was indicated by the m/z ratio. MS peaks were assigned using lipid databases, Lipid Search or LIPID Metabolites and Pathways Strategy (LIPID MAPS) database.
p-value<0.01, fold change>1.5.
Ratio, extracellular vesicles of PC9 cell/PC9R cell ratio.
P-, plasmenyl; O-, plasmanyl.
PI, phosphatidylinositols.
PG, phosphatidylglycerols.
3.2 Identification of phospholipids enriched in EV derived from the gefitinib-resistant NSCLC
Score plot shown in Fig. 4A clearly demonstrated that phospholipids of PC9R_EV are significantly different from those of PC9_EV. We could identify the phospholipids, which were differentially expressed in PC9R_EV (Fig. 4B). We next assessed how much EVs isolated from PC9R are differentially expressed compared to EV from PC9 using volcano plots. Volcano plots revealed that the DRL were largely classified into over- or underexpressed clusters with differential expression patterns in PC9_EV, and PC9R_EV (Fig. 4B). Annotated DRL (pink dots) with a fold change > 1.5 and p value < 0.01 were shown in the volcano plot. A heatmap shown in positive mode (Supporting Information Fig. 3A) indicated that up-(red) or downregulated DRL (green) in PC9R_EV in comparison to PC9_EV, or PC9_EV compared to PC9R_EV, revealing a clear distinction between PC9_EV versus PC9R_EV in both mode. We could identify the differentially expressed phospholipids in negative mode (Fig. 5). A volcano plot suggested that PG{O-40:6} [M-H]−/PG{P-40:5} [M-H]−, PI{32:2} [M-H]−, and PI{32:1} [M-H]− were significantly overexpressed in PC9R_EV (Fig. 5B). A heatmap in Supporting Information Fig. 3B also suggest phospholipids, which were differently expressed in PC9R_EV, compared to PC9_EV.
3.3 Quantitative confirmation of target lipids using Triple Quadrupole LC-MS
First, six lipid standards were used to optimize the MRM conditions of PC, PG, PI, LPC, LPI, and SM. To construct the transition (precursor m/z (Q1) > product m/z (Q3)), the adducted ions of each lipid were confirmed by MS scan. As a result, in the positive ion mode of ESI, each lipid was detected with several adducted ions, such as a hydrogen ion (H+; m/z 1.01) and an ammonium ion (NH4+; m/z 18.03). The Q3 of transition and MS/MS collision energy were also optimized by the automatic tool. The optimized MRM conditions of six lipids were listed in Supporting Information Table 1. Among differentially expressed lipids (**p < 0.01) from MALDI-TOF, protonated lipids in positive mode and deprotonated lipids in negative mode were analyzed using QqQ LC-MS. Consequently, nine lipids containing PC, LPC, SM, PI have similar results of increase and decrease aspect. PC {40:7} (**p < 0.05), and LPC {20:1} (*p < 0.1) were increased in PC9R EV. PC {34:2}(**p < 0.05), PC {38:5} (*p < 0.1), SM {d40:2}(**p < 0.05), LPC {20:4}(**p < 0.05), LPC {22:4}(*p < 0.1), PI {36:4} (**p < 0.05), and PI {36:5}(**p < 0.05) were decreased in PC9R EV (Supporting Information Fig. 4).
4 Discussion
Predicting of drug responses remains as a major challenge in NSCLC cancer patients. In our previous study, we showed that EV isolated from gefitinib resistant NSCLC cell line, PC9R, are bioactive by demonstrating that treatment of EV shed from PC9R (PC9R_EV) suppressed sensitivity to cisplatin-induced apoptosis of recipient PC9 parental cells. We found mTOR signaling pathway as key molecules transferred through EV cargo leading to an enhanced drug resistance [16]. Pharmaceutical inhibition of mTOR could resensitize the gefitinib-resistance PC9R cells, resulting in increased cell apoptosis in response to gefitinib. In present study we identified a “shotgun” lipidomics profile using MALDI-TOF MS on the whole cell lysates and EV shed from in both positive- and negative-ion modes. This represents the most comprehensive “global” lipidome analysis of shed EV, based on our knowledge, performed to date assessing differences between resistant versus sensitive NSCLC.
For the rapid and sensitive monitoring of the molecular compositions and abundances of individual phospholipid species, we and other groups have developed a comprehensive lipidome analysis method based on MS and/or MS/MS. Although there are several caveats associated with the use of MS methods for lipidome analysis mainly due to structural diversity and complexity of the lipid mixtures, monitoring of phospholipid composition in EV shed from tumor cells has a benefit since it may suggest characteristics of cancer cells. In order to enhance diagnostic accuracy, a histology-guided phospholipid profiling approach may be attempted. Our MS analysis revealed that the PC content including PC{42:6} [M+Na]+, PC{40:7} [M+H]+, PC{32:3} [M+H]+, PC{44:6} [M+Na]+, and PC{O-36:3} [M+H]+/PC{P-36:2} [M+H]+, increased in PC9R_EV in comparison of PC9_EV. Negative ion mode MS revealed that various PI and PG were up- or downregulated in PC9R_EV. PI{32:1} [M-H]−, PG{O-40:6} [M-H]−/PG{P-40:5} [M-H]−, and PG{32:2} [M-H]− were significantly upregulated. SM, including SM{d30:0} [M+H]+ and SM{d40:2} [M+H]+, were significantly decreased in PC9R_EV. It is notable that nine phospholipids such as PC {40:7} [M+H]+ and SM {d40:2} were detected in QqQ LC-MS as well as MALDI-TOF analyses, and showed a significantly consistent pattern of changes in both analyses.
Sphingomyelin (SM) is common source of several phospholipid metabolites, including ceramide and sphingosine-1-phosophate (S-1-P) [48–50]. Ceramides, bioactive signaling sphingolipids, regulate intracellular signaling cascades, and S-1-P is involved in pathophysiological processes such as apoptosis, the stress response, and inflammatory responses [51, 52], implicating that an altered SM content has been associated the changes of metabolic and signaling pathways associated with oncogenesis [50]. Findings in this study suggest a potential biological function of the perturbed phospholipid signature in EV shed from the gefitinib resistant NSCLC, which could be developed into biomarkers indicating drug resistance in clinical settings.
Known secondary messengers for cellular signal transduction, lipids and in particular phospholipids, associate with lipid rafts, which are specialized membrane domains enriched with cholesterol and glycosphingolipid [53]. Phospholipids may not only participate in lipid raft formation but also enhance oncogenesis [54]. As major regulators of phospholipid metabolism, several phospholipases such as phospholipdase A2, phospholipase C (PLC), and phospholipase, at the expression and/or activity levels, are positively correlated with clino-pathological features [55–57]. Phospholipid metabolism is altered in response to various chemotherapeutic agents such as specific inhibitors of PI3-Kinase-Akt, Ras-Erk/MAPK, phosphoinositide-specific phospholipase Cγ 1 (PI-PLCγ 1)-PKC signaling pathways, fatty acid synthase, and Heat Shock Protein 90 (Hsp90) signaling [58, 59]. Activation of choline kinase, which involves in biosynthesis of PC, has also been implicated in development of various cancers (e.g. breast cancer and ovarian cancer) [60–62]. Collectively, previous literature suggest that possibility that phospholipid metabolism in EV may represent phenotype of tumors [17–19, 63].
In conclusion, we have shown here that bioactive EV secreted from gefitinib resistant lung cancer cells contain lipid signature through the horizontal transfer from NSCLC. Here, we found interesting phospholipid changes that were directly related to gefitinib resistance, suggesting that EV components such as phospholipids may function as predictive biomarkers of gefitinib resistance. We are aware that phospholipids are challenging to pharmacological targeting, however, our understanding on the altered phosphorylation of key lipids will provide therapeutic opportunities in drug development aid to NSCLC treatment. Future studies evaluating the molecular evidence to identify patients at higher risk of relapse due to drug resistance in clinical trials are clearly warranted.
Supplementary Material
Acknowledgments
This work was supported by Proteogenomic Research Program, the Bio- and Medical Technology Development Program (Project No. 2012M3A9B6055305) through the National Research Foundation of Korea funded by the Korean Ministry of Education, Science and Technology (to K.P.K.), NIH grants 1R01DK100974-01 and Steven Spielberg Discovery Fund in Prostate Cancer Research Career Development Award (to J.K.).
Abbreviations
- DRL
differentially regulated lipids
- EGFR
epidermal growth factor receptor
- EV
extracellular vesicles
- LPC
lysophosphatidylcholines
- NSCLC
nonsmall-cell lung cancer
- PC
phosphatidylcholine
- PG
phosphatidylglycerols
- PI
phosphatidylinositols
- PLC
phospholipase C
- PI-PLCγ 1
phosphoinositide-specific phospholipase Cγ 1
- SM
sphingomyelines
- TKI
tyrosine kinase inhibitor
Footnotes
Additional supporting information may be found in the online version of this article at the publisher’s web-site
The authors have declared no conflict of interest.
References
- 1.Ulivi P, Zoli W, Capelli L, Chiadini E, et al. Target therapy in NSCLC patients: relevant clinical agents and tumour molecular characterisation. Mol Clin Oncol. 2013;1:575–581. doi: 10.3892/mco.2013.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rosell R, Bivona TG, Karachaliou N. Genetics and biomarkers in personalisation of lung cancer treatment. Lancet. 2013;382:720–731. doi: 10.1016/S0140-6736(13)61715-8. [DOI] [PubMed] [Google Scholar]
- 3.Petrelli F, Borgonovo K, Cabiddu M, Barni S. Efficacy of EGFR tyrosine kinase inhibitors in patients with EGFR-mutated non-small-cell lung cancer: a meta-analysis of 13 randomized trials. Clin Lung Cancer. 2012;13:107–114. doi: 10.1016/j.cllc.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 4.Pirker R. Novel drugs against non-small-cell lung cancer. Curr Opin Oncol. 2014;26:145–151. doi: 10.1097/CCO.0000000000000056. [DOI] [PubMed] [Google Scholar]
- 5.Bauml J, Mick R, Zhang Y, Watt CD, et al. Determinants of survival in advanced non–small-cell lung cancer in the era of targeted therapies. Clin Lung Cancer. 2013;14:581–591. doi: 10.1016/j.cllc.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi DS, Yang JS, Choi EJ, Jang SC, et al. The protein interaction network of extracellular vesicles derived from human colorectal cancer cells. J Proteome Res. 2012;11:1144–1151. doi: 10.1021/pr200842h. [DOI] [PubMed] [Google Scholar]
- 7.Choi DS, Lee JM, Park GW, Lim HW, et al. Proteomic analysis of microvesicles derived from human colorectal cancer cells. J Proteome Res. 2007;6:4646–4655. doi: 10.1021/pr070192y. [DOI] [PubMed] [Google Scholar]
- 8.Kim DK, Kang B, Kim OY, Choi DS, et al. EVpedia: an integrated database of high-throughput data for systemic analyses of extracellular vesicles. J Extracell Vesicles. 2013;2 doi: 10.3402/jev.v2i0.20384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi DS, Choi DY, Hong BS, Jang SC, et al. Quantitative proteomics of extracellular vesicles derived from human primary and metastatic colorectal cancer cells. J Extracell Vesicles. 2012;1 doi: 10.3402/jev.v1i0.18704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim J, Morley S, Le M, Bedoret D, et al. Enhanced shedding of extracellular vesicles from amoeboid prostate cancer cells: potential effects on the tumor microenvironment. Cancer Biol Ther. 2014;15:409–418. doi: 10.4161/cbt.27627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen WX, Zhong SL, Ji MH, Pan M, et al. MicroRNAs delivered by extracellular vesicles: an emerging resistance mechanism for breast cancer. Tumour Biol. 2014;35:2883–2892. doi: 10.1007/s13277-013-1417-4. [DOI] [PubMed] [Google Scholar]
- 12.Inal JM, Kosgodage U, Azam S, Stratton D, et al. Blood/plasma secretome and microvesicles. Biochim Biophys Acta. 2013;1834:2317–2325. doi: 10.1016/j.bbapap.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 13.Principe S, Hui AB, Bruce J, Sinha A, et al. Tumor-derived exosomes and microvesicles in head and neck cancer: implications for tumor biology and biomarker discovery. Proteomics. 2013;13:1608–1623. doi: 10.1002/pmic.201200533. [DOI] [PubMed] [Google Scholar]
- 14.Muralidharan-Chari V, Clancy JW, Sedgwick A, D’Souza-Schorey C. Microvesicles: mediators of extracellular communication during cancer progression. J Cell Sci. 2010;123:1603–1611. doi: 10.1242/jcs.064386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Voloshin T, Fremder E, Shaked Y. Small but mighty: microparticles as mediators of tumor progression. Cancer Microenviron. 2014:1–2. doi: 10.1007/s12307-014-0144-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi DY, You S, Jung JH, Lee JC, et al. Extracellular vesicles shed from gefitinib-resistant nonsmall cell lung cancer regulate the tumor microenvironment. Proteomics. 2014;14:1845–1856. doi: 10.1002/pmic.201400008. [DOI] [PubMed] [Google Scholar]
- 17.Kang S, Lee A, Park YS, Lee SC, et al. Alteration in lipid and protein profiles of ovarian cancer: similarity to breast cancer. Int J Gynecol Cancer. 2011;21:1566–1572. doi: 10.1097/IGC.0b013e318226c5f5. [DOI] [PubMed] [Google Scholar]
- 18.Kim IC, Lee JH, Bang G, Choi SH, et al. Lipid profiles for HER2-positive breast cancer. Anticancer Res. 2013;33:2467–2472. [PubMed] [Google Scholar]
- 19.Kim Y, Shanta SR, Zhou LH, Kim KP. Mass spectrometry based cellular phosphoinositides profiling and phospholipid analysis: a brief review. Exp Mol Med. 2010;42:1–11. doi: 10.3858/emm.2010.42.1.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim KP, Rafter JD, Bittova L, Han SK, et al. Mechanism of human group V phospholipase A2 (PLA2)-induced leukotriene biosynthesis in human neutrophils. A potential role of heparan sulfate binding in PLA2 internalization and degradation. J Biol Chem. 2001;276:11126–11134. doi: 10.1074/jbc.M004604200. [DOI] [PubMed] [Google Scholar]
- 21.Aboagye EO, Bhujwalla ZM. Malignant transformation alters membrane choline phospholipid metabolism of human mammary epithelial cells. Cancer Res. 1999;59:80–84. [PubMed] [Google Scholar]
- 22.Bogin L, Papa MZ, Polak-Charcon S, Degani H. TNF-induced modulations of phospholipid metabolism in human breast cancer cells. Biochim Biophys Acta. 1998;1392:217–232. doi: 10.1016/s0005-2760(98)00035-6. [DOI] [PubMed] [Google Scholar]
- 23.Ronen SM, Leach MO. Imaging biochemistry: applications to breast cancer. Breast Cancer Res. 2001;3:36–40. doi: 10.1186/bcr268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu L, Johnson C, Bakovic M. Stimulation of the human CTP: phosphoethanolamine cytidylyltransferase gene by early growth response protein 1. J Lipid Res. 2008;49:2197–2211. doi: 10.1194/jlr.M800259-JLR200. [DOI] [PubMed] [Google Scholar]
- 25.Smith EL, Schuchman EH. The unexpected role of acid sphingomyelinase in cell death and the pathophysiology of common diseases. FASEB J. 2008;22:3419–3431. doi: 10.1096/fj.08-108043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cummings BS. Phospholipase A2 as targets for anticancer drugs. Biochem Pharmacol. 2007;74:949–959. doi: 10.1016/j.bcp.2007.04.021. [DOI] [PubMed] [Google Scholar]
- 27.Inazu M. Choline transporter-like proteins CTLs/SLC44 family as a novel molecular target for cancer therapy. Biopharm Drug Dispos. 2014;35(8):431–449. doi: 10.1002/bdd.1892. [DOI] [PubMed] [Google Scholar]
- 28.Volinsky R, Kinnunen PK. Oxidized phosphatidylcholines in membrane-level cellular signaling: from biophysics to physiology and molecular pathology. FEBS J. 2013;280:2806–2816. doi: 10.1111/febs.12247. [DOI] [PubMed] [Google Scholar]
- 29.Monteiro-Cardoso VF, Silva AM, Oliveira MM, Peixoto F, Videira RA. Membrane lipid profile alterations are associated with the metabolic adaptation of the Caco-2 cells to aglycemic nutritional condition. J Bioenerg Biomembr. 2014;46:45–57. doi: 10.1007/s10863-013-9531-y. [DOI] [PubMed] [Google Scholar]
- 30.Jones DR, Divecha N. Linking lipids to chromatin. Curr Opin Genet Dev. 2004;14:196–202. doi: 10.1016/j.gde.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 31.Doria ML, Cotrim Z, Macedo B, Simoes C, et al. Lipidomic approach to identify patterns in phospholipid profiles and define class differences in mammary epithelial and breast cancer cells. Breast Cancer Res Treat. 2012;133:635–648. doi: 10.1007/s10549-011-1823-5. [DOI] [PubMed] [Google Scholar]
- 32.Shanta SR, Zhou LH, Park YS, Kim YH, et al. Binary matrix for MALDI imaging mass spectrometry of phospholipids in both ion modes. Anal Chem. 2011;83:1252–1259. doi: 10.1021/ac1029659. [DOI] [PubMed] [Google Scholar]
- 33.Separovic D, Joseph N, Breen P, Bielawski J, et al. Combining anticancer agents photodynamic therapy and LCL85 leads to distinct changes in the sphingolipid profile, autophagy, caspase-3 activation in the absence of cell death, and long-term sensitization. Biochem Biophys Res Commun. 2011;409:372–377. doi: 10.1016/j.bbrc.2011.04.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jones EE, Powers TW, Neely BA, Cazares LH, et al. MALDI imaging mass spectrometry profiling of proteins and lipids in clear cell renal cell carcinoma. Proteomics. 2014;14:924–935. doi: 10.1002/pmic.201300434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schone C, Hofler H, Walch A. MALDI imaging mass spectrometry in cancer research: combining proteomic profiling and histological evaluation. Clin Biochem. 2013;46:539–545. doi: 10.1016/j.clinbiochem.2013.01.018. [DOI] [PubMed] [Google Scholar]
- 36.Choi DS, Kim DK, Kim YK, Gho YS. Proteomics, transcriptomics and lipidomics of exosomes and ectosomes. Proteomics. 2013:10–11. doi: 10.1002/pmic.201200329. [DOI] [PubMed] [Google Scholar]
- 37.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 38.Edwards JL, Kennedy RT. Metabolomic analysis of eukaryotic tissue and prokaryotes using negative mode MALDI time-of-flight mass spectrometry. Anal Chem. 2005;77:2201–2209. doi: 10.1021/ac048323r. [DOI] [PubMed] [Google Scholar]
- 39.Gibb S, Strimmer K. MALDIquant: a versatile R package for the analysis of mass spectrometry data. Bioinformatics. 2012;28:2270–2271. doi: 10.1093/bioinformatics/bts447. [DOI] [PubMed] [Google Scholar]
- 40.Dieterle F, Ross A, Schlotterbeck G, Senn H. Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in 1H NMR metabonomics. Anal Chem. 2006;78:4281–4290. doi: 10.1021/ac051632c. [DOI] [PubMed] [Google Scholar]
- 41.Xia J, Wishart DS. Web-based inference of biological patterns, functions and pathways from metabolomic data using MetaboAnalyst. Nat Protoc. 2011;6:743–760. doi: 10.1038/nprot.2011.319. [DOI] [PubMed] [Google Scholar]
- 42.Lee GK, Lee HS, Park YS, Lee JH, et al. Lipid MALDI profile classifies non-small cell lung cancers according to the histologic type. Lung Cancer. 2012;76:197–203. doi: 10.1016/j.lungcan.2011.10.016. [DOI] [PubMed] [Google Scholar]
- 43.Dill AL, Ifa DR, Manicke NE, Costa AB, et al. Lipid profiles of canine invasive transitional cell carcinoma of the urinary bladder and adjacent normal tissue by desorption electrospray ionization imaging mass spectrometry. Anal Chem. 2009;81:8758–8764. doi: 10.1021/ac901028b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Son J, Lee G, Cha S. Direct analysis of triacylglycerols from crude lipid mixtures by gold nanoparticle-assisted laser desorption/ionization mass spectrometry. J Am Soc Mass Spectrom. 2014;25:891–894. doi: 10.1007/s13361-014-0844-9. [DOI] [PubMed] [Google Scholar]
- 45.Sudano MJ, Santos VG, Tata A, Ferreira CR, et al. Phosphatidylcholine and sphingomyelin profiles vary in Bos taurus indicus and Bos taurus taurus in vitro- and in vivo-produced blastocysts. Biol Reprod. 2012;87:130. doi: 10.1095/biolreprod.112.102897. [DOI] [PubMed] [Google Scholar]
- 46.Park YS, Yoo CW, Lee SC, Park SJ, et al. Lipid profiles for intrahepatic cholangiocarcinoma identified using matrix-assisted laser desorption/ionization mass spectrometry. Clin Chim Acta. 2011;412:1978–1982. doi: 10.1016/j.cca.2011.07.008. [DOI] [PubMed] [Google Scholar]
- 47.Thomas MC, Mitchell TW, Blanksby SJ. A comparison of the gas phase acidities of phospholipid headgroups: experimental and computational studies. J Am Soc Mass Spectrom. 2005;16:926–939. doi: 10.1016/j.jasms.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 48.Taniguchi M, Okazaki T. The role of sphingomyelin and sphingomyelin synthases in cell death, proliferation and migration-from cell and animal models to human disorders. Biochim Biophys Acta. 2014;1841:692–703. doi: 10.1016/j.bbalip.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 49.Barcelo-Coblijn G, Martin ML, de Almeida RF, Noguera-Salva MA, et al. Sphingomyelin and sphingomyelin synthase (SMS) in the malignant transformation of glioma cells and in 2-hydroxyoleic acid therapy. Proc Natl Acad Sci USA. 2011;108:19569–19574. doi: 10.1073/pnas.1115484108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Savic R, He X, Fiel I, Schuchman EH. Recombinant human acid sphingomyelinase as an adjuvant to sorafenib treatment of experimental liver cancer. PLoS One. 2013;8:e65620. doi: 10.1371/journal.pone.0065620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cheng JC, Bai A, Beckham TH, Marrison ST, et al. Radiation-induced acid ceramidase confers prostate cancer resistance and tumor relapse. J Clin Invest. 2013;123:4344–4358. doi: 10.1172/JCI64791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin HY, Delmas D, Vang O, Hsieh TC, et al. Mechanisms of ceramide-induced COX-2-dependent apoptosis in human ovarian cancer OVCAR-3 cells partially overlapped with resveratrol. J Cell Biochem. 2013;114:1940–1954. doi: 10.1002/jcb.24539. [DOI] [PubMed] [Google Scholar]
- 53.Freeman MR, Cinar B, Kim J, Mukhopadhyay NK, et al. Transit of hormonal and EGF receptor-dependent signals through cholesterol-rich membranes. Steroids. 2007;72:210–217. doi: 10.1016/j.steroids.2006.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Balla T. Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol Rev. 2013;93:1019–1137. doi: 10.1152/physrev.00028.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Song Y, Wilkins P, Hu W, Murthy KS, et al. Inhibition of calcium-independent phospholipase A2 suppresses proliferation and tumorigenicity of ovarian carcinoma cells. Biochem J. 2007;406:427–436. doi: 10.1042/BJ20070631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smrcka AV, Brown JH, Holz GG. Role of phospholipase Cepsilon in physiological phosphoinositide signaling networks. Cell Signal. 2012;24:1333–1343. doi: 10.1016/j.cellsig.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Samadi N, Bekele R, Capatos D, Venkatraman G, et al. Regulation of lysophosphatidate signaling by autotaxin and lipid phosphate phosphatases with respect to tumor progression, angiogenesis, metastasis and chemo-resistance. Biochimie. 2011;93:61–70. doi: 10.1016/j.biochi.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 58.Bruntz RC, Taylor HE, Lindsley CW, Brown HA. Phospholipase D2 mediates survival signaling through direct regulation of Akt in glioblastoma cells. J Biol Chem. 2014;289:600–616. doi: 10.1074/jbc.M113.532978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Musille PM, Kohn JA, Ortlund EA. Phospholipid– driven gene regulation. FEBS Lett. 2013;587:1238–1246. doi: 10.1016/j.febslet.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Paris L, Cecchetti S, Spadaro F, Abalsamo L, et al. Inhibition of phosphatidylcholine-specific phospholipase C downregulates HER2 overexpression on plasma membrane of breast cancer cells. Breast Cancer Res. 2010;12:R27. doi: 10.1186/bcr2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Iorio E, Mezzanzanica D, Alberti P, Spadaro F, et al. Alterations of choline phospholipid metabolism in ovarian tumor progression. Cancer Res. 2005;65:9369–9376. doi: 10.1158/0008-5472.CAN-05-1146. [DOI] [PubMed] [Google Scholar]
- 62.Gallego-Ortega D, Ramirez de Molina A, Ramos MA, Valdes-Mora F, et al. Differential role of human choline kinase alpha and beta enzymes in lipid metabolism: implications in cancer onset and treatment. PLoS One. 2009;4:e7819. doi: 10.1371/journal.pone.0007819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Llorente A, Skotland T, Sylvanne T, Kauhanen D, et al. Molecular lipidomics of exosomes released by PC-3 prostate cancer cells. Biochim Biophys Acta. 2013;1831:1302–1309. doi: 10.1016/j.bbalip.2013.04.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


