Abstract
Objective
Hepcidin is a peptide hormone produced by the liver that regulates systemic iron homeostasis. Hepcidin is also synthesized by tumors, where it contributes to tumor growth by increasing the tumoral retention of iron. Targeted reduction of hepcidin may therefore be useful in reducing tumor growth. H5F9-AM8 is an antibody in preclinical development for the anemia of chronic disease that reduces hepcidin synthesis by binding to RGMc, a co-receptor involved in the transcriptional induction of hepcidin by BMP6. We explored the ability of H5F9-AM8 to act as an anti-tumor agent.
Methods
Effects of anti-hemojuvelin antibody on hepcidin synthesis were assessed by qRTPCR in tissue culture and in tumor xenografts and livers of mice treated with H5F9-AM8 or saline. Tumor growth was assessed using caliper measurements. Serum iron was measured colorimetrically and tissue iron was measured using western blotting and inductively coupled mass spectrometry.
Results
In tissue culture, the anti-hemojuvelin antibody H5F9-AM8 significantly reduced BMP6-stimulated hepcidin synthesis in HepG2 and other cancer cells. In mice, H5F9-AM8 reduced hepcidin in the liver and increased serum iron, total liver iron, and liver ferritin. Although hepcidin in tumors was also significantly decreased, H5F9-AM8 did not reduce tumor iron content, ferritin, or tumor growth.
Conclusion
Anti-hemojuvelin antibody successfully reduces hepcidin in both tumors and livers but has different effects in these target organs: it reduces iron content and ferritin in the liver, but does not reduce iron content or ferritin in tumors, and does not inhibit tumor growth. These results suggest that despite their ability to induce hepcidin in tumors, the anti-tumor efficacy of systemic, non-targeted hepcidin antagonists may be limited by their ability to simultaneously elevate plasma iron. Tumor-specific hepcidin inhibitors may be required to overcome the limitations of drugs that target the synthesis of both systemic and tumor hepcidin.
Keywords: Hepatocellular carcinoma, Iron, Ferroportin, Bone morphogenetic protein, Ferritin, Hepcidin, Hemojuvelin, Repulsive guidance molecule
Introduction
Hepcidin is a circulating peptide hormone that serves as a master regulator of systemic iron homeostasis [1]. Hepcidin acts to regulate systemic iron by binding to ferroportin, a cell surface iron efflux pump present on the basolateral surface of enterocytes, hepatocytes and macrophages. Binding of hepcidin to ferroportin triggers ferroportin degradation, thus inhibiting the delivery of dietary iron into the circulation, as well as inhibiting the recycling of catabolized iron from macrophages and hepatocytes. The physiological consequence of hepcidin production is reduced delivery of iron to the circulation under conditions of iron excess.
Inappropriate synthesis of hepcidin contributes to a number of pathological conditions, notably the Anemia of Chronic Disease (ACD), iron-refractory iron deficiency anemia, and the anemia associated with chronic kidney disease [2,3]. The widespread prevalence of these conditions, particularly ACD, has led to the search for pharmacological inhibitors of hepcidin, a number of which are currently being tested in clinical trials [4].
Hepcidin antagonists may also be useful in inhibiting tumor growth. Although hepcidin synthesis occurs primarily in the liver, synthesis of hepcidin also occurs in the kidney, heart, adipose tissue, spinal cord, and other tissues [5]. In addition, tumors can synthesize hepcidin [6]. We and others have observed that the synthesis of hepcidin by tumors acts in an autocrine fashion to decrease ferroportin and thus restrict iron efflux out of tumor cells, promoting tumoral iron retention [7,8]. Since iron promotes tumor cell proliferation and metastasis [9], blocking hepcidin synthesis represents a potential strategy for limiting tumor growth, and might represent an additional use for commercially developed hepcidin antagonists.
A number of different strategies to inhibit hepcidin are being explored preclinically and clinically [4]. One strategy involves the direct blockade of hepcidin using anti-hepcidin antibodies or other direct inhibitors of hepcidin. Another strategy is to interfere with the productive interaction between hepcidin and ferroportin [10-12]. A third strategy is to block hepcidin synthesis, which is driven by Bone Morphogenetic Proteins (BMPs), particularly BMP6 [13]. BMP6 binds to a complex consisting of BMP type I and II receptors and a co-receptor, RGMc (hemojuvelin) [14]. Binding of BMP6 to its receptor triggers activation of SMAD 1/5/8. Activated SMAD 1/5/8 binds to SMAD4, translocates to the nucleus, and drives hepcidin transcription. IL6 can also augment hepcidin transcription through activation of STAT3 [15]. Agents that could potentially inhibit these pathways of hepcidin transcription therefore include BMP receptor antagonists, BMP co-receptor antagonists, and inhibitors of IL6 signaling [4].
H5F9-AM8 is a hepcidin inhibitor that shows promising preclinical activity [16]. H5F9-AM8 is a humanized monoclonal antibody directed at RGMc, the repulsive guidance co-receptor that mediates the initial steps of BMP-dependent transcriptional induction of hepcidin [16]. H59-AM8 also binds to a repulsive guidance molecule A (RGMa), which shares a 47% amino acid identity with RGMc [17]. As expected of an efficient hepcidin inhibitor, H5F9-AM8 effectively down regulates hepcidin and increases serum iron in rats and monkeys [16,18].
Although mechanisms that control hepcidin transcription in tumors have not been as comprehensively studied as those that control hepcidin synthesis in the liver, overlapping regulatory elements appear to be involved. For example, BMPs induce hepcidin synthesis in breast, prostate and liver cancer cells [7-19]. We therefore explored whether the hepcidin inhibitor H5F9-AM8, which targets the BMP co-receptor RGMc, could inhibit tumor growth in mice. We report that although H5F9-AM8 is effective at inhibiting hepcidin in tumor-bearing mice, H5F9-AM8 did not inhibit growth of liver tumor xenografts. We hypothesize that the increase in systemic iron induced by H5F9-AM8 limits the ability of H5F9-AM8 to effectively reduce tumoral iron, thus impeding the anti-tumor efficacy of H5F9-AM8. Our results suggest that agents that selectively target tumoral hepcidin synthesis will be required before hepcidin blockade can be used as an effective anti-tumor strategy.
Materials and Methods
Cell culture
The hepatocellular carcinoma cell lines HEP3B and HEPG2 were purchased from ATCC and cultured in EMEM medium (ATCC) supplemented with 10% FBS (Gemini Bio-Products). Cells were used after limited passage within 3 months of thawing. MDA-MB-231-luc-D3H2LN mammary gland adenocarcinoma cells (Caliper Life Sciences) were cultured in DMEM-F12 (Lonza) supplemented with 10% FBS. MCF7 cells (ATCC) were cultured in EMEM (ATCC) containing 10% FBS and 0.1 mg/mL insulin. DU145 prostate carcinoma cells were cultured in EMEM (ATCC) containing 10% benchmark FBS. All cells were maintained at 37°C in a humidified incubator with 5% CO2. Prior to use in these studies, DU145, MCF7, and MDA-MB-231 cell lines were authenticated using STR profiling (ATCC). For BMP treatment, cells were incubated in reduced serum conditions for 6 h before treatment with H5F9-AM8. BMP6 (R&D Systems) was added after 1 h of treatment with H5F9-AM8, and the cells were incubated an additional 24 h before RNA isolation.
H5F9-AM8
H5F9-AM8 is an affinity-matured antibody derived from ABT-207, a monoclonal antibody humanized from a rat hybridoma [16]. H5F9-AM8 binds to human, rat, monkey and mouse RGMc [16].
Animal experiments
All animal studies were conducted in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the Association for the Assessment and Accreditation of Laboratory Animal Care International (AAALAC). The experiment protocol was approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Connecticut Health Center. Outbred homozygous athymic nude female mice [Crl:NU(NCr)-Foxn1nu] were purchased from Charles River and divided into four groups with 10-13 mice/group. Groups consisted of non-tumor bearing mice treated with saline or H5F9-AM8, or tumor-bearing mice treated with saline or H5F9-AM8. To establish tumor xenografts, mice were subcutaneously inoculated in the flank with 2 million HepG2 cells suspended in a 1:1 mixture of matrigel (Membrane Matrix High Concentration (Corning)) and Phosphate Buffered Saline (PBS). Control animals were injected with PBS. One week post inoculation, treatment began. Tumor-bearing and non-tumor bearing mice were treated intraperitoneally with 10 mg/kg H5F9-AM8 or saline twice per week for a total of 3 weeks (n=10 minimum). Dose and schedule were based on previous results demonstrating systemic efficacy of H5F9-AM8 that plateaued at 0.2 mg/kg/wk in rats [16], a half-life of at least 4 days [16,18], and no obvious adverse effects at the highest dose used (20 mg/kg) [16]. There was a gene expression signature indicative of a mild oxidative stress response in rats treated with 20 mg/kg H5F9-AM8 [16], therefore we used a slightly reduced dose of 10 mg/kg administered twice a week. Observations and tumor measurements were performed twice weekly until sacrifice. Tumor volume was calculated using the following formula, π/6(X2)Y, where X=length and Y=width. Animals were sacrificed by CO2 inhalation and cervical dislocation. Livers and tumors were immediately harvested and flash frozen, and blood was collected for serum separation.
Iron assay
Blood was collected immediately after sacrifice by cardiac puncture and centrifuged in a BD Microtainer® tube with serum separator additive (Becton Dickinson). Serum was transferred to a new sterile tube and stored at −20°C overnight. Serum iron was determined by the Iron (Total) Assay (Sekisui Diagnostics) according to the manufacturer's recommendations. Human Based Serum Calibrator (Sekisui Diagnostics) was used to create a standard curve.
Western blots
Harvested tissues were flash frozen in liquid nitrogen and stored at −80°C until ready to process. Tissues were ground using a liquid nitrogen-cooled mortar and pestle and suspended in lysis buffer containing 10 mM Tris, pH 7.5, 150 mM NaCl, 0.1 mM EDTA (Gibco 15575-038), 0.5% Triton-X100 and freshly added Complete Proteinase Inhibitor (Thermo Scientific, 88265). Tissues in lysis buffer were homogenized using a Polytron homogenizer, and clarified by centriguation. Protein quantity was determined by BCA assay (ThermoFisher Scientific). All samples were reduced, separated by SDS-PAGE, and transferred to a nitrocellulose membrane (Bio-Rad). Antibodies against Ferritin H [20], Ferritin L (1:1000; Abcam), β-actin (1:20,000; Sigma), followed by secondary anti-mouse or anti-rabbit antibodies (1:2000-5000; Bio-Rad) were used.
Quantitative RTPCR
Total RNA was collected from cultured cells using High Pure RNA Isolation Kit (Roche Diagnostics Corporation). Total RNA was collected from mouse tissue using TRIzol Reagent (Ambion Life Technologies) followed by RNeasy clean-up with DNAse treatment (Qiagen) according to the manufacturer's instructions. Total RNA and purity was assessed using a nanospectrometer (Thermo Scientific NanoDrop Lite). cDNA was synthesized from 500 ng of RNA using a reverse transcription kit, TaqMan® Reverse Transcription Reagents (Thermo Fisher Scientific) and Oligo(dT) primer. Aliquots of cDNA were added to a reaction mixture containing 2X Universal SYBR® Green Supermix (Bio-Rad) and 400 nM primers. Target quantity was analyzed against a standard curve, using a ViiA 7 instrument and software (Applied Biosystems) and normalized to β-actin. The absence of DNA contamination was confirmed by carrying out amplification from cDNA without reverse transcriptase. The primers were designed with IDT PrimerQuest software (Integrated DNA Technologies, Inc.).
ICPMS
Mouse liver and tumor samples were frozen immediately after collection and shipped on dry ice for elemental analysis at the OHSU Elemental Analysis Core facility at Oregon Health and Science University. Briefly, tissues were digested with HNO3 and analysis was performed using inductively coupled plasma mass spectroscopy (ICPMS) on an Agilent 7700x equipped with an ASX 500 autosampler. Data were quantified using a 9-point calibration curve with external standards. Data were acquired in triplicates and averaged.
Statistics
Statistical comparisons were performed using tests for data that was normally distributed and Wilcoxian rank sum for non-parametric data. Outliers (defined as values ≥ 2 standard deviations from the mean) were excluded from analysis. P values ≤ 0.05 were accepted as significant.
Results
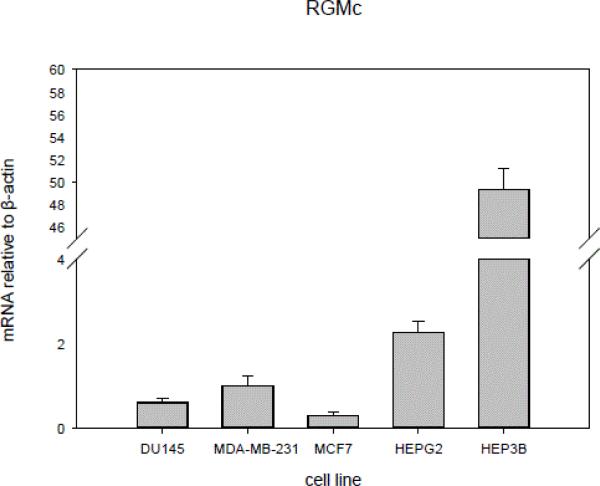
Hepcidin expression has been demonstrated in a variety of tumorigenic cell lines, including hepatocellular carcinoma, prostate cancer, and breast cancer. In order to test the anti-tumor efficacy of H5F9-AM8, we first screened representative human cancer cell lines for expression of RGMc, the target of H5F9-AM8. As shown in Figure 1, the hepatocellular carcinoma cell lines HEP3B and HepG2 exhibited a higher expression of RGMc than the prostate cell line DU145 or the breast cancer cell lines MCF7 and MB231. The remaining experiments were therefore conducted with HepG2 cells.
Figure 1.
HEPG2 and HEP3B cells express elevated levels of RGMc mRNA compared to other cell lines. mRNA levels of RGMc and β-actin were determined by qRTPCR in the indicated cell lines. Error bars represent standard deviations of triplicate determinations.
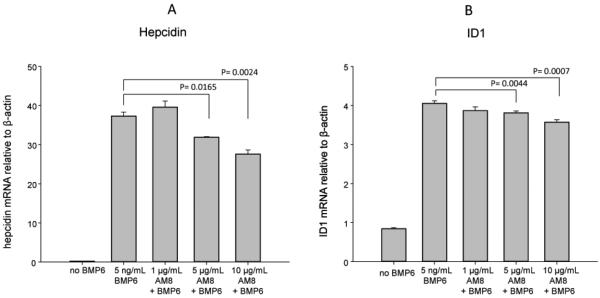
We confirmed that the BMP6 pathway, which is targeted by H5F9-AM8, regulates expression of hepcidin in HepG2 cells. Cells were treated with recombinant BMP6, and expression of ID1 and hepcidin, downstream targets of the BMP6 pathway, were measured by qRT-PCR. As shown in Figure 2, treatment with BMP6 induced both ID1 and hepcidin: induction of ID1 was approximately 4.7 fold, and hepcidin approximately 140 fold. Thus hepcidin is strongly induced by the BMP6 pathway, confirming previous reports [13]. We then tested whether H5F9-AM8 inhibited induction of hepcidin in vitro. HepG2 cells were pretreated with varying doses of H5F9-AM8 followed by treatment with BMP6 and measurement of hepcidin and ID1 transcripts by qRT-PCR (Figure 2). H5F9-AM8 exhibited a dose-dependent and statistically significant ability to inhibit BMP-mediated induction of both ID1 and hepcidin.
Figure 2.
H5F9-AM8 inhibits BMP6-mediated induction of hepcidin and ID1 in HEPG2 cells. Cells were treated as indicated and mRNA measured by qRTPCR. Means and standard deviations of triplicate determinations.
To test whether H5F9-AM8 could inhibit tumor growth in vivo, athymic nu/nu mice were inoculated subcutaneously with HepG2 cells. After one week, tumor-bearing mice were divided into two groups: one group was injected intraperitoneally with H5F9-AM8 twice a week for 3 weeks, and the control group was injected with saline. Non-tumor-bearing mice were similarly divided into an H5F9-AM8-treatment and saline control group (n=10 minimum for all groups). Tumor growth and parameters of iron metabolism were measured.
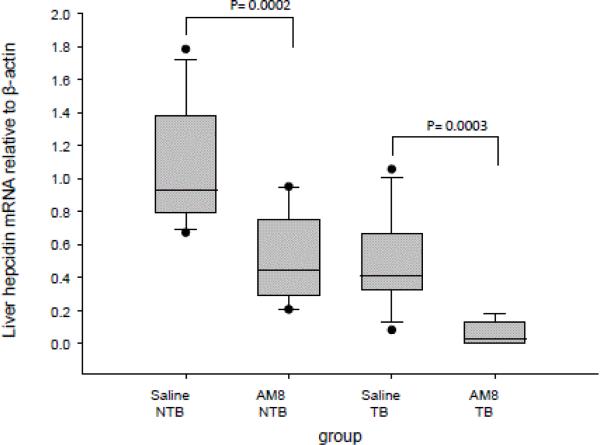
Since H5F9-AM8 had not been previously studied in immune-compromised mice, we tested whether H5F9-AM8 exhibited its anticipated effect of inhibiting hepcidin under our experimental conditions. We first measured hepcidin transcripts in mouse liver. As shown in Figure 3, H5F9-AM8 treatment significantly reduced hepcidin transcripts in the liver of both control and tumor-bearing mice. To test whether the reduction in hepcidin was functionally significant, we then measured serum iron. Since hepcidin blocks iron uptake and recycling, a reduction in hepcidin was expected to lead to an increase in serum iron.
Figure 3.
H5F9-AM8 reduces hepcidin in livers of non-tumor bearing and tumor-bearing mice. qRTPCR was performed to determine relative mRNA levels of hepcidin in liver tissue from mice treated with H5F9-AM8 or saline. Error bars represent the standard deviation of ≥ 8 mice (NTB saline, n=13; NTB AM8, n=12; TB saline, n=10; TB AM8 n=8). NTB, non-tumor bearing; TB, tumor-bearing.
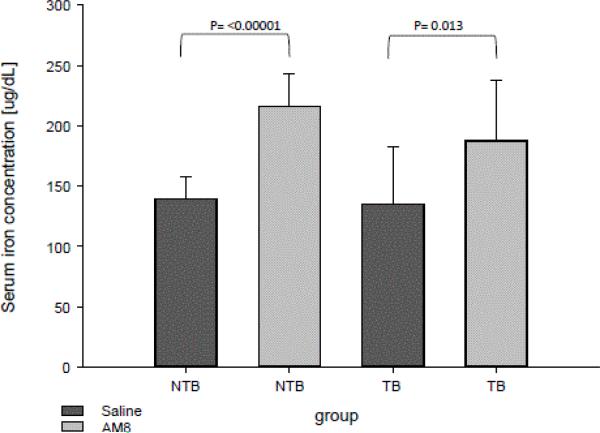
As shown in Figure 4, treatment of both control and tumor-bearing mice with H5F9-AM8 led to a significant increase in serum iron.
Figure 4.
H5F9-AM8 increases serum iron in non-tumor bearing and tumor-bearing mice. Error bars represent the standard deviation of ≥ 10 mice (NTB saline, n=13; NTB AM8, N=12; TB saline, n=10; TB AM8 n=10).
Measurement of total iron in livers of tumor-bearing mice using ICPMS also revealed a significant increase in iron content in the livers of H5F9-AM8-treated mice (Table 1). Levels of other trace metals were unchanged, except for magnesium, which appeared slightly decreased in H5F9-AM8-treated mice (Table 1).
Table 1.
H5F9-AM8 affects iron and magnesium in mouse liver.
| Metal Content of Livers from Control and Treated Mice | |||
|---|---|---|---|
| Control | AM8 | p Value | |
| Fe [μg/g] | 170.84 ± 40.9 | 438.57 ± 230.5 | 0.0020 |
| Cu [μg/g] | 4.23 ± 0.4 | 3.91 ± 0.4 | 0.0555 |
| Mg [μg/g] | 219.06 ± 19.0 | 196.04 ± 19.1 | 0.0147 |
| Mn [μg/g] | 1.05 ± 0.2 | 1.02 ± 0.2 | 0.7107 |
| Zn [μg/g] | 24.25 ± 2.4 | 22.93 ± 3.3 | 0.3167 |
Iron (Fe), copper (Cu), magnesium (Mg), manganese (Mn) and zinc (Zn) were measured in liver tissue using ICPMS. p Values represent 10 samples.
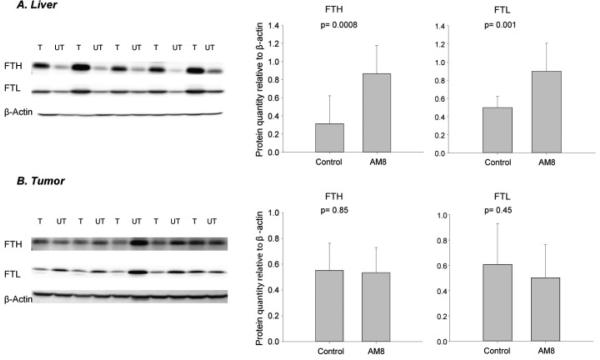
To confirm these results, we performed western blotting of liver tissue to assess levels of ferritin, a sensitive indicator of intracellular iron. Ferritin is an iron storage protein composed of two subunit types, ferritin H and ferritin L, both of which are translationally induced by iron [21]. Consistent with measurements of total iron, livers of H5F9-AM8-treated mice showed the expected increase in ferritin (Figure 5).
Figure 5.
H5F9-AM8 increases Ferritin H (FTH) and Ferritin L (FTL) in liver but not in tumors. A) Western blot of liver tissue from tumor bearing mice Treated (T) with H5F9-AM8 or Untreated (UT) with saline. Quantification is shown on right. B) Tumor tissue from mice T with H5F9-AM8 or control UT with saline. Error bars represent means and standard deviations of ≥ 9 mice/group.
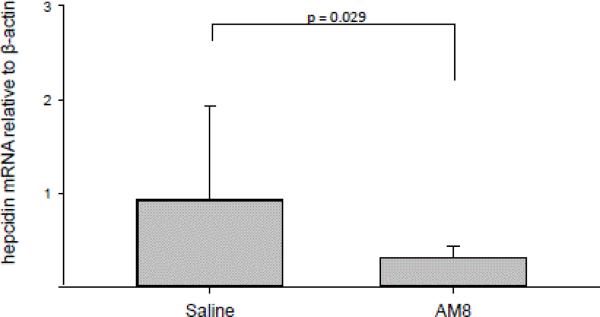
We then assessed effects of H5F9-AM8 on tumors. Levels of hepcidin mRNA in tumors were significantly decreased by H5F9-AM8 (Figure 6).
Figure 6.
H5F9-AM8 reduces levels of hepcidin mRNA in tumors. mRNA levels of hepcidin and β-actin were determined by qRTPCR. Error bars represent the standard deviation of 10 mice/group.
However, despite this decrease in hepcidin, tumor iron content was unchanged in treated versus control mice (Table 2).
Table 2.
H5F9 does not affect iron in mouse tumors.
| Metal Content of Tumors from Control and Treated Mice | |||
|---|---|---|---|
| Control | AM8 | p Value | |
| Fe [μg/g] | 266.34 ± 67.4 | 318.17 ± 79.5 | 0.1333 |
| Cu [μg/g] | 2.75 ± 1.1 | 3.09 ± 0.7 | 0.4250 |
| Mg [μg/g] | 128.09 ± 25.9 | 130.12 ± 19.4 | 0.8450 |
| Mn [μg/g] | 0.18 ± 0.1 | 0.17 ± 0.0 | 0.6999 |
| Zn [μg g] | 11.81 ± 2.2 | 12.19 ± 1.7 | 0.6729 |
Iron (Fe), copper (Cu), magnesium (Mg), manganese (Mn) and zinc (Zn) were measured in liver tissue using ICPMS. p Values represent 10 samples.
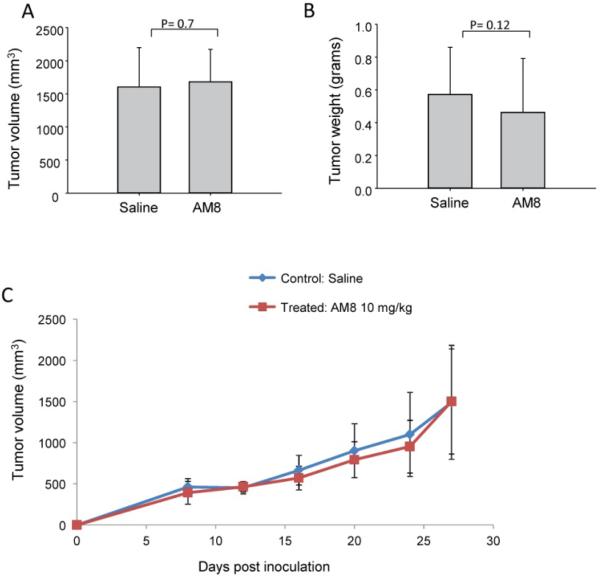
Further, and in contrast to the results seen in livers of the same mice, there was no change in the levels of either ferritin H or ferritin L in the tumors of mice that had been treated with H5F9-AM8 (Figure 5). Measurements of the rate of tumor growth, tumor size and tumor weight all revealed no change in H5F9-AM8-treated mice when compared to controls (Figure 7).
Figure 7.
H5F9-AM8 does not affect tumor volume, weight or rate of growth. A) Tumor volume was measured after 4 weeks of treatment with saline or H5F9-AM8 (n=10/group). B) Tumors were harvested after euthanasia and weighed (saline, n=10; AM8, n=9). C) Tumor volume was measured by digital caliper at the indicated timepoints (n=10/group). Error bars represent means and standard deviations.
Discussion
Mechanisms of iron acquisition and retention are enhanced in cancer cells [9], and the expression of genes that regulate iron metabolism are predictive of cancer patient outcome [22]. As a consequence, iron chelation is under active investigation as a potential anti-tumor strategy [23,24]. Here we explore whether inhibition of hepcidin, which impedes degradation of the iron efflux protein ferroportin and therefore depletes intracellular iron, could be used to inhibit tumor growth.
Our experiments used H5F9-AM8, an antibody directed at RGMc, a co-receptor that mediates hepcidin transcriptional induction by BMP6. Inhibition of hepcidin transcription offers an advantage over direct blockade of hepcidin, since the hepcidin peptide has a high turnover rate, necessitating the frequent administration of direct inhibitors [16,25]. We observed that H5F9-AM8 effectively inhibits systemic hepcidin synthesis in mice, leading to an increase in both circulating iron and storage iron in the liver (Figures 3-5) (Table 1). In addition to its effects on systemic hepcidin, we observed that H5F9-AM8 also reduced hepcidin synthesis in tumors (Figure 6).
However, despite these on-target effects, H5F9-AM8 was not effective in reducing tumor growth in mice. One potential explanation for this finding derives from the potential for systemic and local effects of hepcidin to act in opposition. Inhibition of hepcidin decreases cellular iron by stabilizing ferroportin, the only known cellular iron efflux pump [26]. Previous work from our laboratory has shown that hepcidin is up-regulated in cancer [6,7], and that blocking hepcidin with anti-hepcidin antibodies can reduce the proliferation of prostate cancer cells in vitro [7]. However, in vivo inhibition of hepcidin has systemic effects that may complicate this picture. Hepcidin blockade increases iron by accelerating the dietary uptake and recycling of iron, as evidenced by the increase in circulating iron and increased iron deposition in the liver observed in this study (Figure 4,5) (Table 1) and reported previously [16,27]. Thus, an increase in total available iron has the potential to contravene the decrease in tumor iron mediated by hepcidin inhibition, thus limiting the efficacy of anti-hepcidin reagents to inhibit tumor growth.
Different results were observed in a study that examined the ability of heparin to inhibit breast tumor growth [8]. In addition to its well-known anti-coagulant effects, heparin inhibits the activity of hepcidin [28]. When tumor-bearing mice were treated with heparin, hepatic hepcidin mRNA was reduced approximately 50%, and breast tumor growth approximately 20-50% [8]. Several factors may underlie the difference between our results and these findings. First, different tumor types were examined, and breast tumors may differ from hepatic tumors in their response to hepcidin blockade. Second, systemic iron was not explicitly examined in this study, and it is possible that heparin was less effective at increasing systemic iron than H5F9-AM8. Third, since heparin is much less specific than H5F9-AM8 in terms of its biological effects, its anti-tumor activity may be attributable to factors apart from or in addition to hepcidin blockade.
One strategy to overcome the limitations of drugs that target the synthesis of both systemic and tumor hepcidin is to develop specific inhibitors of tumor hepcidin. For example, synthesis of hepcidin in prostate cancer cells is dependent on a different pathway that utilizes BMP 4/7 in preference to BMP6. Hepcidin synthesis in these cells is sensitive to inhibition by SOSTDC1, a dual BMP and Wnt pathway antagonist that preferentially targets BMP4/7 [7]. Although the selectivity of SOSTDC1 has not been demonstrated, the existence of multiple pathways regulating hepcidin suggests that it may be possible to develop selective inhibitors of tumor hepcidin that can overcome the ability of systemic hepcidin inhibitors to provide an additional source of iron for tumor cells. Alternatively, selective delivery of anti-hepcidin reagents to tumors using tumor-targeting strategies may be considered [29].
Conclusion
Our experiments were designed to test the hypothesis that blockade of hepcidin synthesis in tumors would decrease degradation of ferroportin, increase iron efflux, and reduce tumor growth. To test this hypothesis we treated mice with H5F9-AM8, an antibody directed against hemojuvelin (RGMc), a co-receptor required for BMP-mediated induction of hepcidin transcription. This antibody successfully inhibited hepcidin synthesis in both the liver and tumor xenografts. However, the downstream consequences of reduced hepcidin differed in the liver and tumors: in the liver, inhibition of hepatic hepcidin increased iron, whereas in tumors, levels of iron were not significantly altered. Further, tumor growth was unaffected by H5F9-AM8. We speculate that tumor iron was not reduced by H5F9-AM8 because of the increase in circulating iron that resulted from inhibition of liver hepcidin. We suggest that tumor-selective inhibitors of hepcidin may be required to inhibit tumor growth. Approaches to the development of such reagents include targeted tumor delivery or blockade of tumor-specific pathways of hepcidin synthesis. In the appropriate tumor type, such targeted anti-hepcidin agents may function as successful anti-tumor drugs.
Acknowledgements and Funding
This work was supported by grants from the National Institutes of Health (R01CA171101, FMT), (R01CA188025, SVT) and a pilot project grant from AbbVie. ICPMS measurements were performed by Martina Ralle at the OHSU Elemental Analysis Core with partial support from NIH core grant S10RR025512.
References
- 1.Ganz T, Nemeth E. Hepcidin and iron homeostasis. Biochim Biophys Acta. 2012;1823:1434–1443. doi: 10.1016/j.bbamcr.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sun CC, Vaja V, Babitt JL, Lin HY. Targeting the hepcidinferroportin axis to develop new treatment strategies for anemia of chronic disease and anemia of inflammation. Am J Hematol. 2012;87:392–400. doi: 10.1002/ajh.23110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Babitt JL, Lin HY. Molecular mechanisms of hepcidin regulation: implications for the anemia of CKD. Am J Kidney Dis. 2010;55:726–741. doi: 10.1053/j.ajkd.2009.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blanchette NL, Manz DH, Torti FM, Torti SV. Modulation of hepcidin to treat iron deregulation: potential clinical applications. Expert Rev Hematol. 2016;9:169–186. doi: 10.1586/17474086.2016.1124757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins JF, Wessling-Resnick M, Knutson MD. Hepcidin regulation of iron transport. J Nutr. 2008;138:2284–2288. doi: 10.3945/jn.108.096347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pinnix ZK, Miller LD, Wang W, D'Agostino R, Jr, Kute T, et al. Ferroportin and iron regulation in breast cancer progression and prognosis. Sci Transl Med. 2010;2:43–56. doi: 10.1126/scisignal.3001127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tesfay L, Clausen KA, Kim JW, Hegde P, Wang X, et al. Hepcidin regulation in prostate and its disruption in prostate cancer. Cancer Res. 2015;75:2254–2263. doi: 10.1158/0008-5472.CAN-14-2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang S, Chen Y, Guo W, Yuan L, Zhang D, et al. Disordered hepcidin-ferroportin signaling promotes breast cancer growth. Cell Signal. 2014;26:2539–2550. doi: 10.1016/j.cellsig.2014.07.029. [DOI] [PubMed] [Google Scholar]
- 8.Torti SV, Torti FM. Iron and cancer: more ore to be mined. Nat Rev Cancer. 2013;13:342–355. doi: 10.1038/nrc3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mross K, Richly H, Fischer R, Scharr D, Büchert M, et al. First-inhuman phase I study of PRS-050 (Angiocal), an Anticalin targeting and antagonizing VEGF-A, in patients with advanced solid tumors. PLoS ONE. 2013;8:e83232. doi: 10.1371/journal.pone.0083232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwoebel F, van Eijk LT, Zboralski D, Sell S, Buchner K, et al. The effects of the anti-hepcidin Spiegelmer NOX-H94 on inflammation-induced anemia in cynomolgus monkeys. Blood. 2013;21:2311–2315. doi: 10.1182/blood-2012-09-456756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fung E, Sugianto P, Hsu J, Damoiseaux R, Ganz T, et al. High-throughput screening of small molecules identifies hepcidin antagonists. Mol Pharmacol. 2013;83:681–690. doi: 10.1124/mol.112.083428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andriopoulos B, Jr, Corradini E, Xia Y, Faasse SA, Chen S, et al. BMP6 is a key endogenous regulator of hepcidin expression and iron metabolism. Nat Genet. 2009;41:482–487. doi: 10.1038/ng.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Babitt JL, Huang FW, Wrighting DM, Xia Y, Sidis Y, et al. Bone morphogenetic protein signaling by hemojuvelin regulates hepcidin expression. Nat Genet. 2006;38:531–539. doi: 10.1038/ng1777. [DOI] [PubMed] [Google Scholar]
- 14.Nemeth E, Rivera S, Gabayan V, Keller C, Taudorf S, et al. IL-6 mediates hypoferremia of inflammation by inducing the synthesis of the iron regulatory hormone hepcidin. J Clin Invest. 2004;113:1271–1276. doi: 10.1172/JCI20945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Böser P, D Seemann, MJ Liguori, L Fan, L Huang, et al. Anti-repulsive Guidance Molecule C (RGMc) Antibodies Increases Serum Iron in Rats and Cynomolgus Monkeys by Hepcidin Downregulation. AAPS J. 2015;17:930–938. doi: 10.1208/s12248-015-9770-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Demicheva E, Cui YF, Bardwell P, Barghorn S, Kron M, et al. Targeting repulsive guidance molecule A to promote regeneration and neuroprotection in multiple sclerosis. Cell Rep. 2015;10:1887–1898. doi: 10.1016/j.celrep.2015.02.048. [DOI] [PubMed] [Google Scholar]
- 17.Kovac S, Böser P, Y Cui, Ferring-Appel D, D Casarrubea, et al. Anti-hemojuvelin antibody corrects anemia caused by inappropriately high hepcidin levels. Haematologica. 2016;101:173–176. doi: 10.3324/haematol.2015.140772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shanmugam NK, Cherayil BJ. Serum-induced up-regulation of hepcidin expression involves the bone morphogenetic protein signaling pathway. Biochem Biophys Res Commun. 2013;441:383–386. doi: 10.1016/j.bbrc.2013.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang W, Di X, Torti SV, Torti FM. Ferritin H induction by histone deacetylase inhibitors. Biochem Pharmacol. 2010;80:316–324. doi: 10.1016/j.bcp.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pantopoulos K, Porwal SK, Tartakoff A, Devireddy L. Mechanisms of mammalian iron homeostasis. Biochemistry. 2012;51:5705–5724. doi: 10.1021/bi300752r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller LD, Coffman LG, Chou JW, Black MA, Bergh J, et al. An iron regulatory gene signature predicts outcome in breast cancer. Cancer Res. 2011;71:6728–6737. doi: 10.1158/0008-5472.CAN-11-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lane DJ, Mills TM, Shafie NH, Merlot AM, Saleh Moussa R, et al. Expanding horizons in iron chelation and the treatment of cancer: role of iron in the regulation of ER stress and the epithelial-mesenchymal transition. Biochim Biophys Acta. 2014;1845:166–181. doi: 10.1016/j.bbcan.2014.01.005. [DOI] [PubMed] [Google Scholar]
- 23.Yu Y, Gutierrez E, Kovacevic Z, Saletta F, Obeidy P, et al. Iron chelators for the treatment of cancer. Curr Med Chem. 2012;19:2689–2702. doi: 10.2174/092986712800609706. [DOI] [PubMed] [Google Scholar]
- 24.Xiao JJ, Krzyzanski W, Wang YM, Li H, Rose M, et al. Pharmacokinetics of anti-hepcidin monoclonal antibody Ab 12B9m and hepcidin in cynomolgus monkeys. AAPS J. 2010;12:646–657. doi: 10.1208/s12248-010-9222-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nemeth E, Tuttle MS, Powelson J, Vaughn MB, Donovan A, et al. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science. 2004;306:2090–2093. doi: 10.1126/science.1104742. [DOI] [PubMed] [Google Scholar]
- 26.Poli M, Asperti M, Ruzzenenti P, Regoni M, Arosio P. Hepcidin antagonists for potential treatments of disorders with hepcidin excess. Front Pharmacol. 2014;5:86. doi: 10.3389/fphar.2014.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poli M, Girelli D, Campostrini N, Maccarinelli F, Finazzi D, et al. Heparin: a potent inhibitor of hepcidin expression in vitro and in vivo. Blood. 2011;117:997–1004. doi: 10.1182/blood-2010-06-289082. [DOI] [PubMed] [Google Scholar]
- 28.van Elk M, Murphy BP, Eufrásio-da-Silva T, O'Reilly DP, T Vermonden, et al. Nanomedicines for advanced cancer treatments: Transitioning towards responsive systems. Int J Pharm. 2016;515:132–164. doi: 10.1016/j.ijpharm.2016.10.013. [DOI] [PubMed] [Google Scholar]