Dear Editor,
Using well-characterised, but old and carefully frozen genital tract research samples may be a cost-effective way of performing metagenomic studies, but risks loss of low abundance (but relevant) bacterial species DNA. Moi et al.1 used 16S rRNA and UreDNA sequencing to detect ureaplasmas in frozen urine samples collected from 362 men with NGU in 2010–2011. They found that urethral inflammatory responses to ureaplasmas were less severe than to Chlamydia trachomatis and Mycoplasma genitalium.
In a pilot study in 2015, we used 16S rRNA sequencing to evaluate bacterial communities in 20 stored self-taken vaginal samples collected in 2004–2008 for the Prevention of Pelvic Infection (POPI) chlamydia screening trial.2 The mean age of the women whose samples were tested was 18 years (range: 16 to 26), and three were positive for C. trachomatis (including one co-infected with Neisseria gonorrhoeae) and one for M. genitalium. Despite up to 11 years of storage in Gen-Probe Aptima transport medium at −80°C, DNA integrity was adequate in all the 20 samples analysed including four postal samples.
The full-length bacterial 16S rRNA gene was amplified using New England Biolab Q5 high-fidelity polymerase kit. Equimolar library mix was prepared and sequenced using PacBio SMRT sequencing technology. Downstream bioinformatics analysis was performed using Mothur (version 1.34.4).
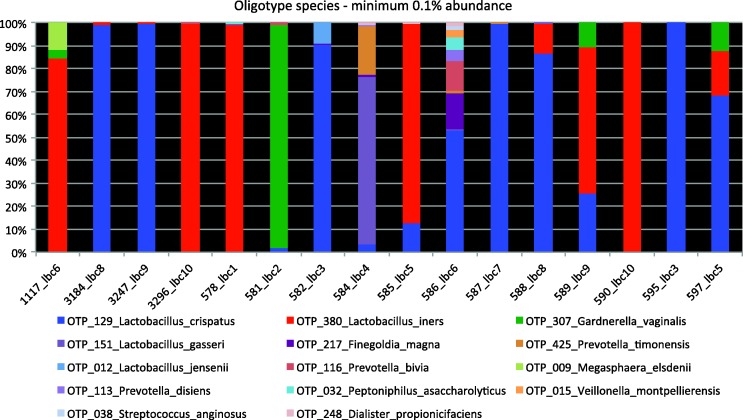
We obtained 73,590 raw sequences from 22 samples, including one negative and one positive control, of which 41,166 were of high quality. Four samples with low sequence number (<500 sequences) were excluded. In the remaining 16 samples, 65 unique oligotype species were identified, of which 14 had a minimum abundance of 0.1% (Figure 1). Burkholderia species were identified as contaminants and removed.
Figure 1.
Species identified from 16 self-taken vaginal swabs after up to 11 years of storage.
Five oligotype species had a minimum abundance of 1%: Lactobacillus crispatus (52.8%), Lactobacillus iners (33.7%), Gardnerella vaginalis (3.5%), Lactobacillus gasseri (2.9%), and Finegoldia magna (1.1%). Six samples were dominated by L. iners (red bar), and seven samples by L. crispatus (blue bar). Sample 581_lbc2, previously assessed as bacterial vaginosis (BV) intermediate, was dominated by G. vaginalis. Patient 1117, the one positive for C. trachomatis and N. gonorrhoeae, had the highest number of Megasphaera elsdenii, which has been reported in BV. Patient 586_Ibc6 described her ethnicity as black other and had the largest bacterial diversity among all patients analysed.
We note that C. trachomatis and N. gonorrhoeae were not detected in the samples. This was not unexpected given that we sequenced from vulvo-vaginal swabs, and the source of whole organism for these pathogens in the female reproductive tract is the cervix. Although nucleic acid amplification tests using specific primers for C. trachomatis and N. gonorrhoeae are highly accurate in vulvo-vaginal swabs, the relative low abundance of their genomic DNA in these samples would mean that the generic 16S rRNA primers were likely to preferentially bind to higher load organisms.
Like Moi et al.,1 we found that carefully characterised samples can be used after many years of storage for the investigation of research questions using 16S rRNA sequencing or PCR.3–5
Declaration of conflicting interests
The authors declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: Professor Oakeshott is a member of the NIHR South London Collaboration for Leadership in Applied Health Research and Care, and the UKCRC-funded esti2 consortium which is led by Dr ST Sadiq.
Funding
The authors received no financial support for the research, authorship, and/or publication of this article.
Ethical approval
Bromley research ethics committee reference: 07/Q0705/16.
References
- 1.Moi H, Reinton N, Randjelovic I, et al. Urethral inflammatory response to ureaplasma is significantly lower than to Mycoplasma genitalium and Chlamydia trachomatis. Int J STD AIDS. 2016. Epub ahead of print 24 August. DOI: 10.1177/0956462416666482. [DOI] [PubMed] [Google Scholar]
- 2.Oakeshott P, Kerry S, Aghaizu A, et al. Randomised controlled trial of screening for Chlamydia trachomatis to prevent pelvic inflammatory disease: the POPI (prevention of pelvic infection) trial. Br Med J 2010; 340: 1642–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oakeshott P, Aghaizu A, Hay P, et al. Is Mycoplasma genitalium in women the “new chlamydia”? Community-based prospective cohort study. Clin Infect Dis 2010; 51: 1160–1166. [DOI] [PubMed] [Google Scholar]
- 4.Oakeshott P, Aghaizu A, Reid F, et al. Frequency and risk factors for prevalent, incident, and persistent genital carcinogenic human papillomavirus infection in sexually active women: community based cohort study. BMJ 2012; 344: e4168–4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reid F, Oakeshott P, Kerry SR, et al. Chlamydia related bacteria (Chlamydiales) in early pregnancy: community-based cohort study. Clin Microbiol Infect 2016. Epub ahead of print 20 October. DOI: 10.1016/j.cmi.2016.10.011. [DOI] [PMC free article] [PubMed]