Abstract
Gene fusions and their encoded products (fusion RNAs and proteins) are viewed as one of the hallmarks of cancer. Traditionally, they were thought to be generated solely by chromosomal rearrangements. However, recent discoveries of trans-splicing and cis-splicing events between neighboring genes, suggest that there are other mechanisms to generate chimeric fusion RNAs without corresponding changes in DNA. In addition, chimeric RNAs have been detected in normal physiology, complicating the use of fusions in cancer detection and therapy. On the other hand, “intergenically spliced” fusion RNAs represent a new repertoire of biomarkers and therapeutic targets. Here, we review current knowledge on chimeric RNAs and implications for cancer detection and treatment, and discuss outstanding questions for the advancement of the field.
Keywords: Chimeric RNA, trans-splicing, cis-SAGe, fusion, intergenic splicing
Gene Fusions and Chimeric RNAs
Gene fusions are a common phenomenon in cancer [1-3]. Many fusions resulting from chromosomal rearrangements are driver mutations in tumors and are currently used as biomarkers or drug targets [1, 3-7]. For example, BCR-ABL, which results from a translocation between chromosomes 9 and 22 [2], is the target for Gleevec in chronic myeloid leukemia [8]. Other examples include EML4-ALK, a target for crizotinib in lung cancer [9], and PAX3-FOXO1, a biomarker for alveolar rhabdomyosarcoma [4].
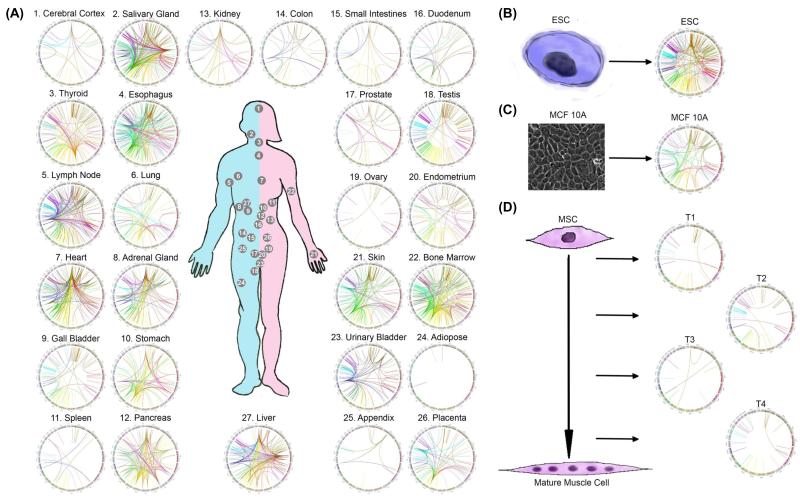
Until recently, the assumption has been that all gene fusions and fusion products (RNA and protein) were exclusive to cancer. This dogma is being challenged as more groups are demonstrating the presence of fusion RNAs and proteins in non-pathological situations [10-13]. In addition, the fusion RNAs are produced in the absence of changes at the DNA level, supporting claims of alternative mechanisms for generating fusion transcripts [10, 12, 14-16]. For instance, our recent analysis of RNA-Seq libraries covering 30 different non-neoplastic human tissues and cells detected thousands of fusion transcripts, with 291 fusions being expressed in more than one tissue/cell sample [17] (Figure 1). Even though not all fusions were validated, the large number of fusion transcripts existing in non-cancer tissues/cells exceeded our original estimation [17]. The large number of fusion RNAs in normal physiology can complicate the use of fusion RNAs as biomarkers and/or drug targets, and raise alarm for the common practice of calling any fusions identified in cancer samples as “cancer-fusion”. In the next sections, we discuss alternative mechanisms to generate chimeric fusion RNAs, give an overview of current methods and challenges to identify fusion transcripts, and hint at potential connections between these new mechanisms and chromosomal rearrangements. Finally, we discuss the use of fusion RNAs in the context of cancer detection and therapy.
Figure 1. Chimeric RNAs in Normal Tissues and Cells.
RNA-Sequencing of (A) libraries covering 27 non-neoplastic human tissues; (B) human embryonic stem cells(ESC); (C) MCF10A breast epithelial cells; and (D) mesenchymal stem cell (MSC) collected at four differentiation steps (T1-T4) was analyzed by SoapFuse algorithms2. Fusion events are shown by Circos plots. The fused transcripts are illustrated here as a line that connects two parental genes.
Production of Chimeric RNAs
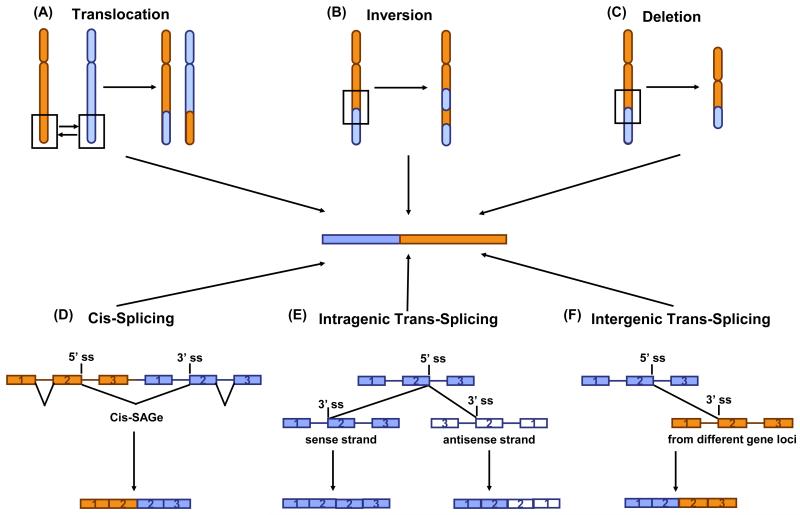
Chimeric transcripts are traditionally thought to be generated by chromosomal rearrangements (Figure 2A). Chromosomal translocations, for example, are a well-documented source of gene fusions. One of the best known examples of fusion genes, BCR-ABL, is produced from the reciprocal chromosomal translocation between chromosomes 9 and 22 [2]. Another way to generate fusion RNAs is by chromosomal inversions (Figure 2B). Fusions resulting from chromosomal inversions were found in various types of tumors. For example, EML4-ALK, which results from (2)(p21p23) inversion, is associated with a small subset of non-small cell lung cancer [18, 19]. Other fusion genes resulting from inversions, such as MLL-CALM (11)(q14q23), CBFB-MYH11 (16)(p13q22), and NUP98-DDX10 (11)(p15q22) occur in acute myeloid leukemia [20-22]. Interstitial deletion is another source for producing fusions (Figure 2C). One example is TMPRSS2-ERG, which is frequently found in prostate cancer [23, 24]. Other cases include MLL-FOXR1 and PAFAH1B2-FOXR1, found exclusively in neuroblastoma [25].
Figure 2. Key Figure. Production of Chimeric RNAs from Chromosomal Rearrangements and Intergenic Splicing.
(A) Translocation. Rearrangement of parts from two non-homologous chromosomes can join two separated genes. (B) Inversion. Segment of a chromosome spanning two genes is reversed end to end, leading to the formation of fusion genes. (C) Deletion. Deleting interstitial part between two separate genes can result in gene fusion. (D) Cis-splicing between adjacent genes. The transcription machinery reads through two neighboring genes, transcribing in the same direction. After splicing, the exons from difference genes are spliced together to produce a chimeric RNA. (E) Intragenic trans-splicing. Two pre-mRNA transcripts from the same genomic locus participate in the process. The two pre-mRNA transcripts can be transcribed from the same strand or different strands. Intragenic trans-splicing can result in exon duplication of sense-antisense fusion. (F) Intergenic trans-splicing. Two pre-mRNA transcripts from two different genomic loci are spliced together.
Besides chromosomal rearrangements, RNA processing events, such as cis- and trans-splicing, also contribute to formation of chimeric RNAs. Cis-splicing between adjacent genes (cis-SAGe) is a RNA processing event that occurs within a single pre-mRNA, where the transcription machinery reads through the intergenic regions of two neighboring genes. It is essentially an alternative splicing between exons of neighboring genes [26] (Figure 2D). Although only a handful examples of cis-spliced RNA chimeras were experimentally confirmed in mammalian cells [27], in-silico analysis and paired-end RNA-sequencing have successfully identified many chimeric RNAs composed of two neighboring genes, which could originate from transcriptional read-through [27-33]. In fact, it is postulated that 4%-5% of tandem gene pairs in the human genome can participate in this process and form chimeric RNAs [28, 34]. We found that cis-SAGe fusions tend to occur between neighboring genes that are located within 30kb distance, and that these fusions often involve the second-to-the-last exon of the 5′ gene, and the second exon of the 3′ gene (2′-2′ rule) [35]. The 2′-2′ rule comes as no surprise, since the second-to-the-last exon of the 5′ gene has the closest splicing donor site to the nearest splicing acceptor site (second exon of 3′ gene). With a small number of randomly selected neighboring gene pairs located within 30kb, we were able to detect fusion transcripts in 20% of the candidates with primers designed following the 2′-2′ rule [35]. Considering that around 4500 head-to-tail gene pairs in the genome are within 30kb distance to each other, cis-SAGe may indeed be a wide spread phenomenon.
Trans-splicing is another non-canonical splicing process from which chimeric RNAs can be generated. Unlike cis-splicing, trans-splicing joins exons from two different primary RNA transcripts together. There are two types of trans-splicing based on the source of primary RNA transcripts: intragenic trans-splicing and intergenic trans-splicing. Intragenic trans-splicing takes place between two copies of primary RNAs transcribed from the same genome locus, and can lead to exon-duplication and sense-antisense fusion [13, 36, 37] (Figure 2E). Intergenic trans-splicing takes place between two pre-mRNAs transcribed from different gene loci [10, 12, 38] (Figure 2F). Trans-splicing is a common process in lower species, such as unicellular organisms, nematodes, or trypanosomes, with up to 70% of the genes in these organisms participating in the process [39-42]. The mechanism of trans-splicing in these organisms, specifically known as spliced-leader (SL) trans-splicing, involves transferring a short leader sequence of 15-50 nucleotides from a specialized non-coding RNA molecule to the 5′ end of the mRNAs. SL trans-splicing does not increase the transcriptome complexity with additional functional transcripts, since the 5′ end is non-coding; instead, it is used to promote the stability and translation of mRNA [43].
Although SL trans-splicing is not present in humans and other higher eukaryotes, the occurrence of non-SL trans-splicing is evident. In humans, examples of intragenic trans-splicing, such as estrogen receptor alpha, have been reported [36, 37]. Four intragenically trans-spliced fusion RNAs were detected in human embryonic stem cells, one of which contributes to maintenance of pluripotency [13]. More notably, there is also evidence of intergenic trans-splicing events in higher organisms [10, 38, 44]. Among them, chimeras of JAZF1-SUZ12 [10, 38] and PAX3-FOXO1 [12] fusions were found in normal cells as well as in tumors. The difference is that these fusion transcripts are product of trans-splicing in normal cells, whereas they are product of chromosomal translocations in tumors [10, 12, 38].
Intergenically Spliced Chimeric RNAs: Complicating the Use of Fusion RNAs in Cancer Detection and Therapy
Many gene fusions resulting from chromosomal rearrangement are used as markers in the clinic for diagnosing and monitoring cancer. For instance, PAX3-FOXO1 resulting from the t(2;13)(q35;q14) translocation [4] is detected in 55% of pediatric patients with alveolar rhabdomyosarcoma (ARMS) [45], and it is used as a diagnostic aid in many pathology laboratories worldwide [46].
Fusion products can also be ideal therapeutic targets. Prominent examples include BCR-ABL in chronic myelogenous leukemia [2] with the development of Gleevec as a paradigm for targeted therapy [8], and the rapid targeting of ALK gene fusion products with crizotinib after its discovery in lung cancer [47].
The detection of gene fusions has relied on Southern blot analysis, fluorescence in situ hybridization (FISH), and on PCR based assays. Southern Blot and FISH have multiple disadvantages, including low sensitivity, being labor intensive, and having a slow turnaround. In contrast, PCR based assays are sensitive, and quick and relatively easier to use. However, for most gene fusions, the chromosomal breakpoints span a large region of DNA, making direct PCR of DNA containing the recombination site not practical. However, because the breakpoints tend to fall within a few introns, the mature chimeric fusion RNA is uniform, despite the heterogeneity of the recombination sites within the DNA. In these situations, RT-PCR based assays are used for clinical diagnosis and monitoring of residual disease, under the assumption that detecting a fusion transcript at the RNA level is equivalent to detecting the gene fusion at the DNA level, and thus an indication of cancer.
The discovery of intergenically spliced chimeric RNAs in normal cells has, however, posed challenges. For instance, trans-spliced JAZF1-SUZ12 fusion RNA was detected in normal endometrial stromal cells. Interestingly, the mature fusion RNA is identical to the one resulting from the t(7;17) chromosomal translocation in endometrial stromal sarcoma [10]. In another example, PAX3-FOXO1 fusion RNA, believed to be a signature of t(2;13) ARMS, was detected transiently during muscle differentiation, but no evidence of t(2;13) was detected in the normal cells [12]. USP9Y-TTTY15, which was identified as a frequent fusion in prostate tumors in the Chinese population [48], was later found to be a cis-splicing between neighboring genes that occurs in normal tissues [49]. SLC45A3-ELK4 e4e2 form can also be detected in non-malignant prostate tissues [49, 50]. The assumption that all fusion(RNA)s are cancer-fusions resulted in an explosion in the deposition of fusion genes in the Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer in the Cancer Genome Anatomy Project [51]1. The number of entries raised more than 4-fold from April 2015 (2,276 entries) to June 2016 (10,256 entries). This is mostly due to the wide application of RNA-Sequencing to cancer samples. However, the vast majority of studies did not include sufficient number of normal, non-cancer control samples. Rushing to translate these fusions into cancer biomarkers will result in a disastrous amount of false positives. We found at least 13 fusions listed in the database of April 2015 that are present in multiple adult tissues in non-cancer donors [17].
We believe that the vast majority of the newly identified fusion RNAs should be carefully validated before they can be used as biomarkers in clinical settings. At the same time, we believe that the fusions validated in a large number of clinical cases, and by additional evidence at DNA level, are ideal tools for molecular diagnosis when used properly. Two issues need to be considered. 1. Even though some chimeric RNAs could be detected in the tissues of healthy individuals [52-57], they tend to be at a much lower level than the chimeric RNAs detected in cancer. This could be due to lower expression of intergenically spliced fusion RNAs compared with overexpression of the fusion RNAs generated by chromosomal translocation. It could also be due to the small percentage of normal cells that produce the intergenically spliced fusion, compared with a relatively homogenous cell population in a tumor mass. In either case, a threshold can be used as a cutoff to eliminate false positives in cancer diagnosis, as in the situation of BCR-ABL. 2. Knowing the details of expression of a specific fusion may provide additional guidance. For instance, although PAX3-FOXO1 can be found during skeletal muscle differentiation, it is only detected transiently [12]. This is in contrast to a constant expression of the fusion due to t(2;13) in ARMS. In this case, in a situation of differential diagnosis involving areas with no active muscle growth, a positive detection indicates ARMS.
When identical fusions exist in normal physiology, using these chimeric RNAs as biomarkers for monitoring residual disease is problematic. Additional evidence for the fusion occurring at the DNA level can be informative. A targeted enrichment followed by DNA sequencing facilitates the detection of the fusion. However, caution should still be taken as even chromosomal rearrangements may not be unique to cancer.
Therapeutically, caution should be taken when developing drugs targeting fusion products that also occur in normal physiology. For instance, many small inhibitors of PAX3-FOXO1 chimeric proteins can suppress malignant phenotypes of ARMS and may be attractive therapeutic strategies in this subset of patients [58, 59]. However, since the fusion occurs during skeletal muscle differentiation, potential side effects on muscle development in a pediatric population should be considered. Again, understanding the details of expression of fusion genes will facilitate a rational treatment approach to avoid side effects.
Intergenically Spliced Chimeric RNAs: a New Repertoire for Biomarkers and Therapeutic Targets
The human genome has about 20,000 genes, similar to the number in c. elegans and drosophila. Clearly, gene number is not enough to explain the multi-level differences between humans and lower organisms. From an evolutionary perspective, intergenic splicing is a potential strategy to expand the functional genome together with other non-canonical splicings [60]. At the same time, intergenic splicing is subject to mis-regulation in disease settings. It is estimated that one-third of disease-causing mutations disrupt splicing [61-63]. The abnormal splicing can be caused by mutations at or near the splicing site of the gene itself, or by mutations in other genes that are involved in splicing, such as splicing factors. For example, SF3B1 is one of the most frequently mutated genes in chronic lymphocytic leukemia (CLL), as well as in many other cancer types [64-66]; and 45-85% of myelodysplasias harbor mutations in splicing machinery, including U2AF35, ZRSR2, SRSF2 and SF3B1 [67]. Presumably, mutations that affect regular splicing will also affect intergenic splicing. In addition, whole genome sequencing studies of many cancers, including lung, breast and prostate carcinoma, show fewer numbers of coding gene mutations than expected on average in any tumor sample [65, 68-70], leaving room for additional mechanisms involved in tumor biology. It is thus speculated that certain intergenically spliced chimeric RNA may be produced in cancer due to mis-regulation of the process, and they may be normal markers and/or drug targets [71]. Supporting this idea, a recurrent chimeric RNA, YPEL5-PPP1CB, was found in 28% of CLL. Interestingly, whole-genome sequencing and Southern blotting demonstrated no evidence for a genomic fusion between YPEL5 and PPP1CB [16], thus making it a putative trans-spliced fusion in CLL. SLC45A3-ELK4 e1e2 form is an example of cis-SAGe fusion in the absence of corresponding fusions at DNA level. Its level correlates with prostate cancer development [15, 71]. In addition, Wen et al. identified seven fusions in acute myeloid leukemia with normal karyotyping [72]. These examples and others support that abnormality of intergenically spliced chimeric RNAs may play an underappreciated role in cancer biology. In addition to mutations in the 20,000 genes in our genome, abnormally produced chimeric RNAs represent a new repertoire for biomarker and therapeutic target discovery.
Methods and Challenges in the Detection of Chimeric RNAs
Although traditional laboratory methods, such as FISH, Southern blot, and karyotyping were used effectively to discover gene fusions in hematological malignancies and soft-tissue sarcomas, they require knowledge of the fusions being looked for. Recent progress in microarray analyses and deep-sequencing efforts have been used to discover previously unrecognized gene fusions and fusion RNAs in common carcinomas, including TMPRSS2-ERG in prostate cancer [6], and EML4-ALK in lung cancer [18]. Although Whole Genome Sequencing (WGS) detected several important gene fusion events [50–52], this technique will not detect intergenically spliced chimeric RNAs. In contrast, RNA-Seq will detect both traditional fusions and fusion RNAs that only occur at the RNA level. RNA-Seq also allows the detection of multiple alternative splice variants resulting from fusions. These and additional factors, such as low cost and quick turnaround time, make RNA-Seq very popular in detecting fusion transcripts. However, RNA-Sequencing has also limitations: i) it cannot detect fusion events involving non-transcribed events; ii) tissue-specificity and the broad dynamic range of expression in the human transcriptome are two factors that complicate RNA-Seq data analysis [73]; iii) during library preparation, template switching can occur, generating artificial products of fusion molecules [74, 75]; and iv) detecting fusion events not occurring at the DNA level is a double-edged sword. Some of the fusion detection tools, such as Comrad, CRAC, and nFuse, require WGS and RNA-Seq reads to focus on fusion events that occur at DNA level.
Even though there are about 33 computational tools available, there are various challenges associated with these tools. Their performance varies depending on the sequencing reads length, and reads quality. A large amount of time and computational memory is often required. In addition to missing true fusion events, these tools can also produce false positives. More importantly, in the recent comparative studies of Liu et al. [76], and our own [77], small overlaps were found when different software tools were used to detect fusion transcripts. More sensitive, accurate and efficient tools are urgently needed.
Trans-Splicing and Chromosomal Rearrangement
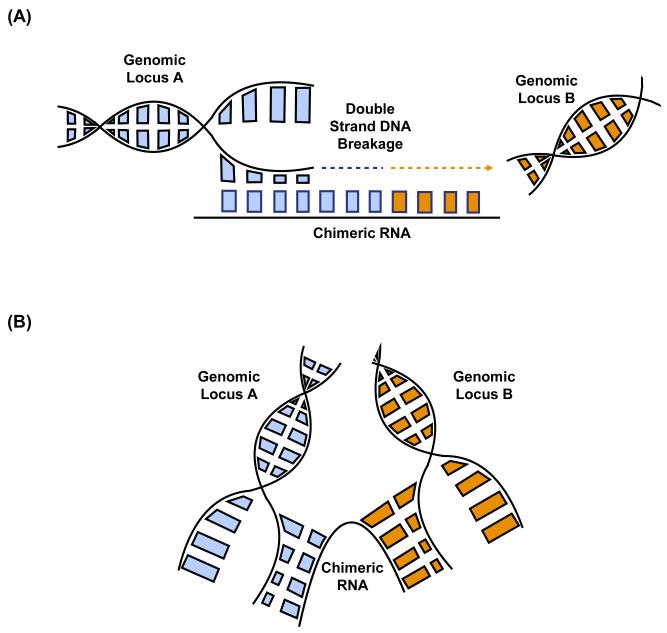
The occurrence of RNA trans-splicing in normal cells and identical fusion transcripts produced by chromosomal rearrangement in tumors raised the possibility of RNA-mediated DNA rearrangement, speculated by us [10] and others [78]. This is a process that occurs in other species, such as ciliates and yeast [79, 80]. There are two ways by which chimeric RNAs can facilitate genomic translocation. First, the fusion RNAs can act as templates for the repair of double strand DNA break. When the broken ends of DNA are synthesized across the chimeric RNA, the newly synthesized DNA will share sequence homology with the chimera (Figure 3A). Interestingly, multiple papers have reported the induction of small RNA molecules around the double-stranded DNA breakage site in both plants and animals, suggesting a role of RNA in DNA repair [81-83]. Furthermore, Keskin et al. demonstrated that endogenous RNA mediates DNA recombination and repair in yeast [84]. Another way is when chimeric RNAs serve as scaffolds that bring two genomic loci into proximity (Figure 3B). This RNA-DNA interaction may facilitate strand breaks and genomic translocation.
Figure 3. Mechanisms of RNA-mediated DNA Rearrangement.
(A) Chimeric RNAs act as repair templates for double strand DNA breakage. Chimeric RNAs pair with one strand of DNA at the double strand breakage site. The repair machinery uses chimeric RNA as the template, and the newly synthesized DNA will share sequence similarity with the chimeric RNA. (B) Chimeric RNAs act as scaffolds to bring two genomic loci into proximity, which might promote the breakage and fusion between the two gene loci. ss-splicing site.
The occurrence of a same chimeric RNA in both normal physiology and tumor malignancies might also be explained by the “true-true, unrelated” model [26]. In this model, there is no causal relationship between trans-spliced fusion RNA and chromosomal rearrangement. Instead, both DNA rearrangements and trans-spliced RNA chimeras are formed under the influence of the same factor(s), which we postulate to be the physical proximity of the two participating gene loci in three dimensions, the openness of chromatin to facilitate active transcription, or active transcription, among other possible mechanisms. More research is clearly needed to elucidate the relationship between trans-spliced chimeric RNAs and chromosomal translocation.
Concluding Remarks
Chimeric fusion RNAs are hybrid RNAs where fragments from two previously separated genomic loci are juxtaposed. These fusion RNAs and their encoded fusion proteins have been widely used in the clinical setting as biomarkers and drug targets in many types of cancer. Chimeric RNAs can be generated from chromosomal rearrangements, including translocations, deletions and inversions. Recent research demonstrated that they could also be generated from trans-splicing or cis-splicing between adjacent genes. The implications of these intergenically spliced chimeric RNAs are multifaceted. On one hand, their presence in normal tissues and cells complicates the use of fusion RNAs in cancer diagnosis and treatment. On the other hand, they represent a new repertoire for the discovery of new biomarkers and drug targets. Traditional methods for fusion detection include Southern blotting, FISH and PCR-based techniques. Current development of next-generation sequencing, especially paired-end RNA sequencing, provides an effective high throughput way to discover many other fusion RNAs. However, due to sequencing platforms and software tools, these methods can generate false negatives and false positives. The field of intergenically spliced chimeric RNAs is still in its infancy. We don’t have yet a precise knowledge of the mechanisms of RNA trans-splicing or cis-SAGe (see outstanding questions). Are there particular factors involved in these non-canonical splicing events or do they use the same machinery of regular splicing? How are they regulated? Why does a fusion form between certain parental gene pairs, at certain positions, and in certain tissues or conditions? Future studies should uncover more fusion RNAs, elucidate the mechanisms of origin, and the connection of these fusions to DNA rearrangements. This information will guide better applications in the diagnosis and treatment of cancer.
Outstanding Questions.
-
What are the mechanisms of non-SL trans-splicing and cis-SAGe?
Currently, the mechanisms of intergenic splicing in vertebrates remain largely unknown. Which are the factors involved in the process? How is the precise splicing location between partner genes selected in specific tissues?
-
Can we develop better software tools or methods to detect intergenic splicing events?
Traditional detecting methods, such as Southern blotting and FISH, have low throughput and low sensitivity. PCR-based techniques are sensitive; yet, there are artifacts associated with template switch. Paired-end RNA-sequencing offers a high throughput way for the detection of chimeras; however, there are problems associated with current software tools and algorithms. Therefore, further optimization of current detection methods is needed to improve sensitivity and accuracy.
-
How is trans-splicing connected to chromosomal rearrangement?
JAZF1-SUZ12 and PAX3-FOXO1 are two examples where the same RNA fusions are found in cancer and normal cells. The fusion transcripts in neoplastic conditions are derived from chromosomal translocation, whereas the fusions in normal cells are the results of trans-splicing. It may be an indication of RNA-mediated DNA rearrangement where chimeric RNAs facilitates the translocation at the DNA level by acting either as repair templates for double strand DNA breaks or as scaffolds that bring two genomic loci into proximity. Alternatively, two processes are “true-true, unrelated”?
Trends Box.
Chimeric fusion RNAs can be generated by intergenic splicing, including trans-splicing and cis-splicing between adjacent genes.
Fusion RNAs (and proteins) are traditionally viewed as specific to cancer, but they are also present in normal physiology.
Intergenically spliced fusion RNAs can complicate the application of fusion RNAs in cancer detection and treatment.
Intergenically spliced fusion RNAs can be mis-regulated in cancer. These fusion RNAs represent a new repertoire for the discovery of new biomarkers and therapeutic targets.
Acknowledgements
We would like to acknowledge support from the National Cancer Institute, grant [CA190713], HL is a Research Scholar Grant [126405-RSG-14-065-01-RMC] from the American Cancer Society, and a St. Baldrick’s V Scholar.
Glossary
- Chimeric fusion RNAs
hybrid RNAs in which fragments from two previously separated genomic loci are juxtaposed.
- Chromosomal rearrangement
a type of chromosome structural change involving several different classes of events, such as translocations, inversions, deletions, and duplications.
- Trans-splicing
a type of intergenically splicing where exons from two different primary RNA transcripts are spliced together.
- Cis-splicing between Adjacent Genes (cis-SAGe)
a type of intergenically splicing where exons from two neighboring genes from one primary RNA transcript are spliced together.
- RNA-Sequencing
next generation sequencing technique to study whole transcriptomes.
Resources
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- 1.Rabbitts TH. Chromosomal translocations in human cancer. Nature. 1994;372:143–149. doi: 10.1038/372143a0. [DOI] [PubMed] [Google Scholar]
- 2.Rowley JD. Letter: A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243:290–293. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- 3.Heim S, Mitelman F. Molecular screening for new fusion genes in cancer. Nature genetics. 2008;40:685–686. doi: 10.1038/ng0608-685. [DOI] [PubMed] [Google Scholar]
- 4.Downing JR, et al. Multiplex RT-PCR assay for the differential diagnosis of alveolar rhabdomyosarcoma and Ewing's sarcoma. The American journal of pathology. 1995;146:626–634. [PMC free article] [PubMed] [Google Scholar]
- 5.Rowley JD. The role of chromosome translocations in leukemogenesis. Seminars in hematology. 1999;36:59–72. [PubMed] [Google Scholar]
- 6.Tomlins SA, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 7.Kowarz E, et al. Premature transcript termination, trans-splicing and DNA repair: a vicious path to cancer. American journal of blood research. 2011;1:1–12. [PMC free article] [PubMed] [Google Scholar]
- 8.Druker BJ, et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med. 2001;344:1031–1037. doi: 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- 9.Shaw AT, et al. Effect of crizotinib on overall survival in patients with advanced non-small-cell lung cancer harbouring ALK gene rearrangement: a retrospective analysis. Lancet Oncol. 2011;12:1004–1012. doi: 10.1016/S1470-2045(11)70232-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li H, et al. A neoplastic gene fusion mimics trans-splicing of RNAs in normal human cells. Science. 2008;321:1357–1361. doi: 10.1126/science.1156725. [DOI] [PubMed] [Google Scholar]
- 11.Chase A, et al. TFG, a target of chromosome translocations in lymphoma and soft tissue tumors, fuses to GPR128 in healthy individuals. Haematologica. 2010;95:20–26. doi: 10.3324/haematol.2009.011536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuan H, et al. A chimeric RNA characteristic of rhabdomyosarcoma in normal myogenesis process. Cancer Discov. 2013;3:1394–1403. doi: 10.1158/2159-8290.CD-13-0186. [DOI] [PubMed] [Google Scholar]
- 13.Wu CS, et al. Integrative transcriptome sequencing identifies trans-splicing events with important roles in human embryonic stem cell pluripotency. Genome research. 2014;24:25–36. doi: 10.1101/gr.159483.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Relaix F, et al. The transcriptional activator PAX3-FKHR rescues the defects of Pax3 mutant mice but induces a myogenic gain-of-function phenotype with ligand-independent activation of Met signaling in vivo. Genes & development. 2003;17:2950–2965. doi: 10.1101/gad.281203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y, et al. Chimeric transcript generated by cis-splicing of adjacent genes regulates prostate cancer cell proliferation. Cancer discovery. 2012;2:598–607. doi: 10.1158/2159-8290.CD-12-0042. [DOI] [PubMed] [Google Scholar]
- 16.Velusamy T, et al. Recurrent reciprocal RNA chimera involving YPEL5 and PPP1CB in chronic lymphocytic leukemia. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:3035–3040. doi: 10.1073/pnas.1214326110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Babiceanu M, et al. Recurrent chimeric fusion RNAs in non-cancer tissues and cells. Nucleic acids research. 2016;44:2859–2872. doi: 10.1093/nar/gkw032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soda M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 19.Li Y, et al. Clinical significance of EML4-ALK fusion gene and association with EGFR and KRAS gene mutations in 208 Chinese patients with non-small cell lung cancer. PloS one. 2013;8:e52093. doi: 10.1371/journal.pone.0052093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wechsler DS, et al. A novel chromosomal inversion at 11q23 in infant acute myeloid leukemia fuses MLL to CALM, a gene that encodes a clathrin assembly protein. Genes, chromosomes & cancer. 2003;36:26–36. doi: 10.1002/gcc.10136. [DOI] [PubMed] [Google Scholar]
- 21.Morerio C, et al. Inversion (11)(p15q22) with NUP98-DDX10 fusion gene in pediatric acute myeloid leukemia. Cancer genetics and cytogenetics. 2006;171:122–125. doi: 10.1016/j.cancergencyto.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 22.Haferlach C, et al. AML with CBFB-MYH11 rearrangement demonstrate RAS pathway alterations in 92% of all cases including a high frequency of NF1 deletions. Leukemia. 2010;24:1065–1069. doi: 10.1038/leu.2010.22. [DOI] [PubMed] [Google Scholar]
- 23.Hermans KG, et al. TMPRSS2:ERG fusion by translocation or interstitial deletion is highly relevant in androgen-dependent prostate cancer, but is bypassed in late-stage androgen receptor-negative prostate cancer. Cancer research. 2006;66:10658–10663. doi: 10.1158/0008-5472.CAN-06-1871. [DOI] [PubMed] [Google Scholar]
- 24.Perner D, et al. Association between sun-exposure, smoking behaviour and plasma antioxidant levels with the different manifestation of skin ageing signs between Japanese and German women--a pilot study. Journal of dermatological science. 2011;62:138–140. doi: 10.1016/j.jdermsci.2011.02.010. [DOI] [PubMed] [Google Scholar]
- 25.Santo EE, et al. Oncogenic activation of FOXR1 by 11q23 intrachromosomal deletion-fusions in neuroblastoma. Oncogene. 2012;31:1571–1581. doi: 10.1038/onc.2011.344. [DOI] [PubMed] [Google Scholar]
- 26.Jividen K, Li H. Chimeric RNAs generated by intergenic splicing in normal and cancer cells. Genes Chromosomes Cancer. 2014;53:963–971. doi: 10.1002/gcc.22207. [DOI] [PubMed] [Google Scholar]
- 27.Wang K, et al. RBM6-RBM5 transcription-induced chimeras are differentially expressed in tumours. BMC Genomics. 2007;8:348. doi: 10.1186/1471-2164-8-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parra G, et al. Tandem chimerism as a means to increase protein complexity in the human genome. Genome Res. 2006;16:37–44. doi: 10.1101/gr.4145906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Denoeud F, et al. Prominent use of distal 5' transcription start sites and discovery of a large number of additional exons in ENCODE regions. Genome research. 2007;17:746–759. doi: 10.1101/gr.5660607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siepel A, et al. Targeted discovery of novel human exons by comparative genomics. Genome research. 2007;17:1763–1773. doi: 10.1101/gr.7128207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kannan K, et al. Recurrent chimeric RNAs enriched in human prostate cancer identified by deep sequencing. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:9172–9177. doi: 10.1073/pnas.1100489108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nacu S, et al. Deep RNA sequencing analysis of readthrough gene fusions in human prostate adenocarcinoma and reference samples. BMC medical genomics. 2011;4:11. doi: 10.1186/1755-8794-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hernandez-Torres F, et al. Intron retention and transcript chimerism conserved across mammals: Ly6g5b and Csnk2b-Ly6g5b as examples. BMC genomics. 2013;14:199. doi: 10.1186/1471-2164-14-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Akiva P, et al. Transcription-mediated gene fusion in the human genome. Genome research. 2006;16:30–36. doi: 10.1101/gr.4137606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qin F, et al. Discovery of CTCF-Sensitive Cis-Spliced Fusion RNAs between Adjacent Genes in Human Prostate Cells. PLoS Genet. 2015;11:e1005001. doi: 10.1371/journal.pgen.1005001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murphy LC, et al. Investigation of the origin of variant, truncated estrogen receptor-like mRNAs identified in some human breast cancer biopsy samples. Breast cancer research and treatment. 1993;26:149–161. doi: 10.1007/BF00689688. [DOI] [PubMed] [Google Scholar]
- 37.Pink JJ, et al. Cloning and characterization of a 77-kDa oestrogen receptor isolated from a human breast cancer cell line. British journal of cancer. 1997;75:17–27. doi: 10.1038/bjc.1997.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gingeras TR. Implications of chimaeric non-co-linear transcripts. Nature. 2009;461:206–211. doi: 10.1038/nature08452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sutton RE, Boothroyd JC. Evidence for trans splicing in trypanosomes. Cell. 1986;47:527–535. doi: 10.1016/0092-8674(86)90617-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krause M, Hirsh D. A trans-spliced leader sequence on actin mRNA in C. elegans. Cell. 1987;49:753–761. doi: 10.1016/0092-8674(87)90613-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nilsen TW. Evolutionary origin of SL-addition trans-splicing: still an enigma. Trends in genetics : TIG. 2001;17:678–680. doi: 10.1016/s0168-9525(01)02499-4. [DOI] [PubMed] [Google Scholar]
- 42.Lasda EL, Blumenthal T. Trans-splicing. Wiley interdisciplinary reviews. RNA. 2011;2:417–434. doi: 10.1002/wrna.71. [DOI] [PubMed] [Google Scholar]
- 43.Hastings KE. SL trans-splicing: easy come or easy go? Trends in genetics : TIG. 2005;21:240–247. doi: 10.1016/j.tig.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 44.Finta C, Zaphiropoulos PG. Intergenic mRNA molecules resulting from trans-splicing. J Biol Chem. 2002;277:5882–5890. doi: 10.1074/jbc.M109175200. [DOI] [PubMed] [Google Scholar]
- 45.Sorensen PH, et al. PAX3-FKHR and PAX7-FKHR gene fusions are prognostic indicators in alveolar rhabdomyosarcoma: a report from the children's oncology group. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2002;20:2672–2679. doi: 10.1200/JCO.2002.03.137. [DOI] [PubMed] [Google Scholar]
- 46.Missiaglia E, et al. PAX3/FOXO1 fusion gene status is the key prognostic molecular marker in rhabdomyosarcoma and significantly improves current risk stratification. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2012;30:1670–1677. doi: 10.1200/JCO.2011.38.5591. [DOI] [PubMed] [Google Scholar]
- 47.Shaw AT, et al. Effect of crizotinib on overall survival in patients with advanced non-small-cell lung cancer harbouring ALK gene rearrangement: a retrospective analysis. Lancet Oncol. 12:1004–1012. doi: 10.1016/S1470-2045(11)70232-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ren S, et al. RNA-seq analysis of prostate cancer in the Chinese population identifies recurrent gene fusions, cancer-associated long noncoding RNAs and aberrant alternative splicings. Cell research. 2012;22:806–821. doi: 10.1038/cr.2012.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ren G, et al. Transcription-mediated chimeric RNAs in prostate cancer: time to revisit old hypothesis? OMICS. 2014;18:615–624. doi: 10.1089/omi.2014.0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nacu S, et al. Deep RNA sequencing analysis of readthrough gene fusions in human prostate adenocarcinoma and reference samples. BMC Med Genomics. 4:11. doi: 10.1186/1755-8794-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mitelman F, J.B.a.M.F.E. Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer. 2015 [Google Scholar]
- 52.Mori H, et al. Chromosome translocations and covert leukemic clones are generated during normal fetal development. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:8242–8247. doi: 10.1073/pnas.112218799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Biernaux C, et al. Detection of major bcr-abl gene expression at a very low level in blood cells of some healthy individuals. Blood. 1995;86:3118–3122. [PubMed] [Google Scholar]
- 54.Bose S, et al. The presence of typical and atypical BCR-ABL fusion genes in leukocytes of normal individuals: biologic significance and implications for the assessment of minimal residual disease. Blood. 1998;92:3362–3367. [PubMed] [Google Scholar]
- 55.Eguchi-Ishimae M, et al. Breakage and fusion of the TEL (ETV6) gene in immature B lymphocytes induced by apoptogenic signals. Blood. 2001;97:737–743. doi: 10.1182/blood.v97.3.737. [DOI] [PubMed] [Google Scholar]
- 56.Quina AS, et al. PML-RARA fusion transcripts in irradiated and normal hematopoietic cells. Genes, chromosomes & cancer. 2000;29:266–275. doi: 10.1002/1098-2264(2000)9999:9999<::aid-gcc1030>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 57.Uckun FM, et al. Clinical significance of MLL-AF4 fusion transcript expression in the absence of a cytogenetically detectable t(4;11)(q21;q23) chromosomal translocation. Blood. 1998;92:810–821. [PubMed] [Google Scholar]
- 58.Jothi M, et al. Small molecule inhibition of PAX3-FOXO1 through AKT activation suppresses malignant phenotypes of alveolar rhabdomyosarcoma. Molecular cancer therapeutics. 2013;12:2663–2674. doi: 10.1158/1535-7163.MCT-13-0277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Loupe JM, et al. Inhibiting phosphorylation of the oncogenic PAX3-FOXO1 reduces alveolar rhabdomyosarcoma phenotypes identifying novel therapy options. Oncogenesis. 2015;4:e145. doi: 10.1038/oncsis.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sibley CR, et al. Lessons from non-canonical splicing. Nat Rev Genet. 2016 doi: 10.1038/nrg.2016.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lopez-Bigas N, et al. Are splicing mutations the most frequent cause of hereditary disease? FEBS Lett. 2005;579:1900–1903. doi: 10.1016/j.febslet.2005.02.047. [DOI] [PubMed] [Google Scholar]
- 62.Singh RK, Cooper TA. Pre-mRNA splicing in disease and therapeutics. Trends Mol Med. 2012;18:472–482. doi: 10.1016/j.molmed.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Daguenet E, et al. The pathogenicity of splicing defects: mechanistic insights into pre-mRNA processing inform novel therapeutic approaches. EMBO Rep. 2015;16:1640–1655. doi: 10.15252/embr.201541116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Quesada V, et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat Genet. 2012;44:47–52. doi: 10.1038/ng.1032. [DOI] [PubMed] [Google Scholar]
- 65.Cancer Genome Atlas, N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Biankin AV, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491:399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yoshida K, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- 68.Berger MF, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–220. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cancer Genome Atlas Research, N. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stephens PJ, et al. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature. 2009;462:1005–1010. doi: 10.1038/nature08645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rickman DS, et al. SLC45A3-ELK4 is a novel and frequent erythroblast transformation-specific fusion transcript in prostate cancer. Cancer Res. 2009;69:2734–2738. doi: 10.1158/0008-5472.CAN-08-4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wen H, et al. New fusion transcripts identified in normal karyotype acute myeloid leukemia. PloS one. 2012;7:e51203. doi: 10.1371/journal.pone.0051203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Taylor BS, Ladanyi M. Clinical cancer genomics: how soon is now? J Pathol. 2011;223:318–326. doi: 10.1002/path.2794. [DOI] [PubMed] [Google Scholar]
- 74.Houseley J, Tollervey D. Apparent non-canonical trans-splicing is generated by reverse transcriptase in vitro. PLoS One. 2010;5:e12271. doi: 10.1371/journal.pone.0012271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McManus CJ, et al. Regulatory divergence in Drosophila revealed by mRNA-seq. Genome research. 2010;20:816–825. doi: 10.1101/gr.102491.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu C, et al. FusionQ: a novel approach for gene fusion detection and quantification from paired-end RNA-Seq. BMC bioinformatics. 2013;14:193. doi: 10.1186/1471-2105-14-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kumar S, et al. Comparative assessment of methods for the fusion transcripts detection from RNA-Seq data. Sci Rep. 2016;6:21597. doi: 10.1038/srep21597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zaphiropoulos PG. Trans-splicing in Higher Eukaryotes: Implications for Cancer Development? Front Genet. 2012;2:92. doi: 10.3389/fgene.2011.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nowacki M, et al. RNA-mediated epigenetic programming of a genome-rearrangement pathway. Nature. 2008;451:153–158. doi: 10.1038/nature06452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Storici F, et al. RNA-templated DNA repair. Nature. 2007;447:338–341. doi: 10.1038/nature05720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Francia S, et al. Site-specific DICER and DROSHA RNA products control the DNA-damage response. Nature. 2012;488:231–235. doi: 10.1038/nature11179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Michalik KM, et al. A small RNA response at DNA ends in Drosophila. Nucleic acids research. 2012;40:9596–9603. doi: 10.1093/nar/gks711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wei W, et al. A role for small RNAs in DNA double-strand break repair. Cell. 2012;149:101–112. doi: 10.1016/j.cell.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 84.Keskin H, et al. Transcript-RNA-templated DNA recombination and repair. Nature. 2014;515:436–439. doi: 10.1038/nature13682. [DOI] [PMC free article] [PubMed] [Google Scholar]