Abstract
Reactive oxygen species (ROS), which are both a natural byproduct of oxidative metabolism and an undesirable byproduct of many environmental stressors, can damage all classes of cellular macromolecules and promote diseases from cancer to neurodegeneration. The actions of ROS are mitigated by the transcription factor NRF2, which regulates expression of antioxidant genes via its interaction with cis-regulatory antioxidant response elements (AREs). However, despite the seemingly straightforward relationship between the opposing forces of ROS and NRF2, regulatory precision in the NRF2 network is essential. Genetic variants that alter NRF2 stability or alter ARE sequences have been linked to a range of diseases. NRF2 hyperactivating mutations are associated with tumorigenesis. On the subtler end of the spectrum, single nucleotide variants (SNVs) that alter individual ARE sequences have been linked to neurodegenerative disorders including progressive supranuclear palsy and Parkinson’s disease, as well as other diseases. Although the human health implications of NRF2 dysregulation have been recognized for some time, a systems level view of this regulatory network is beginning to highlight key NRF2-targeted AREs consistently associated with disease.
Keywords: NRF2, NFE2L2, ARE, Oxidative stress, Polymporphism, Mutation, GWAS, Cancer, MAPT, Parkinson disease
1. NRF2-ARE-mediated gene regulation
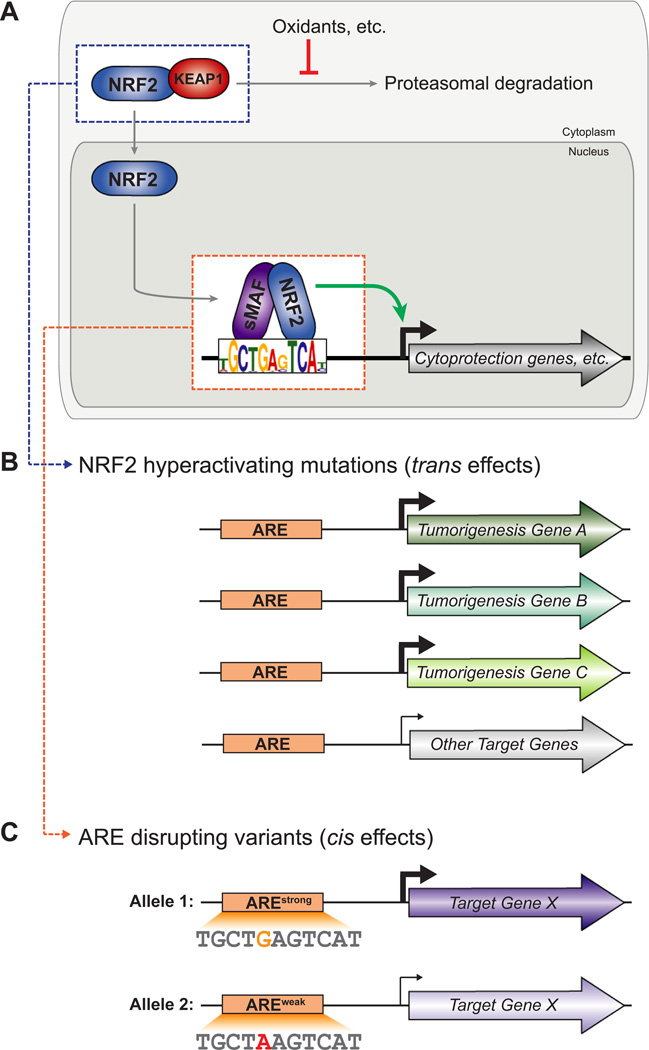
Many cellular challenges – chemical, metabolic, and physical – disrupt redox homeostasis and generate excess reactive oxygen species (ROS). ROS have the potential to damage macromolecules including proteins, lipids, and DNA. The latter effect is especially harmful because it can cause DNA mutations with long-term consequences, but significant oxidative stress can also lead to apoptotic or necrotic cell death. Thus oxidative damage is a significant contributor to chronic diseases, from cancer to neurodegenerative disease [1–4]. In response to oxidative stress, cells activate a panel of cytoprotective genes, including antioxidant and detoxifying enzymes, that counteract ROS and ROS-induced damage to the cell. NRF2, a Cap-n-Collar (CNC) basic leucine zipper (bZIP) transcription factor encoded by the gene NFE2L2, is a master regulator of the transcriptional response to oxidative stress [5]. NRF2 is structurally and functionally conserved from insects to humans, and it must dimerize with one of the three small MAF (sMAF) bZIP proteins (MAFF, MAFG, MAFK) to bind ARE sequences and regulate gene expression [6]. NRF2 is widely expressed, but when ROS levels are low, nuclear NRF2 is kept low by direct interaction with the inhibitory protein KEAP1, which sequesters NRF2 in the cytoplasm and targets it for proteasomal degradation [7–10]. However, ROS modify KEAP1 and impair its ability to target NRF2 for degradation. So as ROS levels increase (oxidative stress), KEAP1’s ability to inhibit NRF2 decreases and nuclear NRF2 increases; the increase in nuclear NRF2 drives upregulation of many cytoprotective genes (Figure 1A) [10,11].
Figure 1. Regulatory effects of NRF2 network variation.
(A) General schematic of NRF2 regulatory pathway. See text for details. (B) Mutations that disrupt NRF2-KEAP1 interactions lead to NRF2 hyperactivation and are associated with tumorigenesis. Select NRF2 target genes are consistently upregulated in tumors with hyperactivated NRF2. (C) Variants that disrupt individual ARE sequences create alleles with stronger ARE activity and weaker ARE activity. Such variants can alter NRF2 target gene expression and, in some cases, disease risk.
Although gene sets necessary for responding to and recovering from oxidative stress are key NRF2 targets, these are not the only genes regulated by NRF2 [12–14]. The NRF2 regulatory network also contains multiple autoregulatory loops, including a critical negative feedback loop: NRF2 transcriptionally activates its own repressor, KEAP1 [14,15]. The NRF2-KEAP1 negative feedback loop is deeply conserved, present in organisms ranging from Drosophila to human [14,16]. This ancient negative feedback loop highlights the importance of keeping theNRF2 pathway in check, and suggests precise regulation of NRF2’s nuclear concentration is paramount. The rules governing transcription factor interactions with DNA within the nucleus are complex, but are largely a function of transcription factor concentration and its binding affinity for a given target DNA sequence [17,18]. Presumably, the autoregulatory loops modulating NRF2’s nuclear concentration ensure that NRF2-ARE binding and cis-regulatory output at all NRF2 target genes is finely tuned for a wide range of stress conditions.
NRF2, in combination with one of the sMAF proteins, regulates gene expression by binding ARE sequences in its target genes’ enhancer or promoter regions. The ARE is also referred to as the electrophile response element (EpRE) or the CNC-sMAF-binding element (CsMBE) [19,20]. Although the term ARE is more restrictive than the latter two terms – NRF2-ARE transactivation is responsive to more than just oxidative stress – it is the most commonly used term for the NRF2-sMAF-binding element. The original ARE consensus sequence was defined as GCnnnSTCAY (where S = G or C, and Y = C or T) [21,22]. Consistent with the consensus motif, genome-wide NRF2 binding sites identified using ChIP-seq (chromatin immunoprecipitation followed by high throughput sequencing) are strongly enriched for the sequence TGCTGAGTCAY [12,13,23]. Thus in vivo NRF2 DNA binding is largely driven by a higher information content version of the original ARE consensus. The sequence identified by ChIP-seq is functionally relevant, because both NRF2-sMAF DNA binding (in vitro and in vivo) and NRF2-mediated regulatory output are correlated with a target ARE’s similarity to TGCTGAGTCAY [14,23,24].
Considering the finely tuned nature of NRF2-mediated gene expression, it is likely that genetic variants disrupting either controls on NRF2 nuclear concentration (Figure 1B) or NRF2-bound ARE sequences (Figure 1C) would have significant implications for NRF2 target gene expression. Indeed, variants of both types have now been characterized. These variants can have a marked effect on NRF2-mediated gene expression, and are associated with a range of pathologies.
2. Trans-regulatory effects of NRF2 or KEAP1 mutation
NRF2 activity is cytoprotective. Loss of NRF2 is associated with genomic instability and tumorigenesis, whereas activation of NRF2 is chemopreventive and promotes longevity [25–27]. Yet NRF2 activation beyond a certain threshold can be detrimental: somatic mutations that disrupt NRF2-KEAP1 interaction promote cancer progression [28–32]. Mutations altering either the NRF2 binding domain of KEAP1 or the KEAP1 binding domain of NRF2 were first observed in lung cancer [28,29,33]. Mechanistically, disruption of protein domains at the NRF2-KEAP1 interface prevents efficient targeting of NRF2 for proteasomal degradation, which in turn leads to NRF2 accumulation and constitutive activation of the pathway. Lung tissue is particularly prone to NRF2 hyperactivating mutations, but such mutations are also found in head and neck squamous cell carcinomas, endometrial cancer, and many other solid tumor types [32,34–37]. In addition to somatic mutation, DNA hypermethylation at the KEAP1 locus is also associated with KEAP1 repression, increased NRF2 activity, and tumorigenesis [38–42].
The exact mechanisms underlying NRF2’s oncogenic properties remain unclear, but likely involve aberrant induction of NRF2 target genes (Figure 1B). Importantly, data from The Cancer Genome Atlas (TCGA) indicate that mutations altering the KEAP1 binding domain of NRF2 are associated with similar patterns of gene upregulation across diverse tumor types (Table 1) [43,44]. Because these gene expression changes are a result of genetic variation at an unlinked locus (the NFE2L2 gene), they represent the trans-effects of mutations at NFE2L2. Many of the upregulated genes are near genomic regions bound by NRF2 based on human ChIP-seq data, so these presumably represent direct NRF2 targets (Table 1).
Table 1. Genes consistently upregulated in tumors with hyperactivated NRF2.
Bold genes are direct NRF2 targets based on human ChIP-seq data [13,14]. Non-bold genes are direct NRF2 targets based on additional experimental evidence [48]. “Additional genes” are those with little evidence for direct regulation by NRF2. Tumor expression data are from [43].
| Gene symbol | Gene name | Tumor type |
|---|---|---|
| Upregulated in 4/4 NRF2 hyperactivated tumor types: | ||
| ABCB6 | ATP binding cassette subfamily B member 6 | B, H, L, U |
| ALDH3A1 | Aldehyde dehydrogenase 3 family member A1 | B, H, L, U |
| FECH | Ferrochelatase | B, H, L, U |
| GCLM | Glutamate-cysteine ligase, modifier subunit | B, H, L, U |
| ME1 | Malic enzyme 1 | B, H, L, U |
| NQO1 | NAD(P)H quinone dehydrogenase 1 | B, H, L, U |
| PGD | Phosphogluconate dehydrogenase | B, H, L, U |
| SRXN1 | Sulfiredoxin 1 | B, H, L, U |
| TALDO1 | Transaldolase 1 | B, H, L, U |
| TKT | Transketolase | B, H, L, U |
| TXNRD1 | Thioredoxin reductase 1 | B, H, L, U |
| OSGIN1 | Oxidative stress induced growth inhibitor 1 | B, H, L, U |
| AKR1C1 | Aldo-keto reductase family 1 member C1 | B, H, L, U |
| EPHX1 | Epoxide hydrolase 1 | B, H, L, U |
| G6PD | Glucose-6-phosphate dehydrogenase | B, H, L, U |
| Additional genes: AKR1B10, AKR1C2, AKR1C3, CABYR, CES1, CYP4F3, JAKMIP3, PANX2, TRIM16L | ||
| Upregulated in 3/4 NRF2 hyperactivated tumor types: | ||
| GCLC | Glutamate-cysteine ligase catalytic subunit | B, H, U |
| GSR | Glutathione reductase | B, H, L |
| PRDX1 | Peroxiredoxin 1 | B, H, L |
| SLC7A11 | Solute carrier family 7 member 11 | B, H, L |
| TXN | Thioredoxin | B, H, L |
| PTGR1 | Prostaglandin reductase 1 | B, H, L |
| Additional genes: AGPAT9, CBR3, CES4, CLDN8, CYP4F11, FTHL3, KIAA0319, MAP2, MDGA1, NAMPT, RBM19, RIT1, SAMD5, SPP1, TDP2, TSPAN7, WNT5A | ||
| Upregulated in 2/4 NRF2 hyperactivated tumor types: | ||
| ABHD4 | Abhydrolase domain containing 4 | H, L |
| ADAM17 | ADAM metallopeptidase domain 17 | B, L |
| COA6 | Cytochrome c oxidase assembly factor 6 | B, L |
| FTH1 | Ferritin heavy chain 1 | H, U |
| KEAP1 | Kelch like ECH associated protein 1 | B, L |
| MAF-G | MAF bZIP transcription factor G | B, L |
| PIR | Pirin | B, U |
| SLC12A8 | Solute carrier family 12 member 8 | H, L |
| SLC3A2 | Solute carrier family 3 member 2 | B, L |
| TLK1 | Tousled like kinase 1 | H, L |
| TMTC3 | Transmembrane and tetratricopeptide repeat containing 3 | B, L |
| ZNF746 | Zinc finger protein 746 | B, L |
| ABCC1 | ATP binding cassette subfamily C member 1 | B, H |
| ABCC3 | ATP binding cassette subfamily C member 3 | H, L |
| ADH7 | Alcohol dehydrogenase 7 | B, L |
| CBR1 | Carbonyl reductase 1 | B, H |
| GPX2 | Glutathione peroxidase 2 | H, L |
| IDH1 | Isocitrate dehydrogenase 1 | H, L |
| PRDX6 | Peroxiredoxin 6 | H, L |
| Additional genes: ABCA4, ADAM23, AKR1B15, ANXA10, ASF1A, ASPH, C14orf149, CREG1, DNAJB4, EPS8, EPT1, ETFB, FAM190A, FBXO30, GLI2, GSTM3, GSTM4, HHIPL2, LOC729082, MAP1B, MEGF9, NECAB2, NSUN3, PHEX, PHKB, RAP1GAP, RNF217, SLC47A2, SLC9A3R1, SMOC2, SOST, TNPO1, TPD52L1, TRIM16, TSKU, UGT1A7 | ||
Tumor type abbreviations: B = Bladder Urothelial Carcinoma; H = Head–Neck Squamous Cell Carcinoma; L = Lung Squamous Cell Carcinoma; U = Uterine Corpus Endometrial Carcinoma.
Oncogenic NRF2 provides a selective advantage to cells across diverse tissue environments, and the TCGA data demonstrate that it also casts a transcriptional shadow that is relatively consistent across these environments. Thus it is likely that subsets of the consistently upregulated genes are responsible for the metabolic, proliferative, and chemoresistance advantages afforded to cells with constitutive NRF2 activity. Multiple genes in Table 1 represent high priority candidates. For example, NRF2 targets necessary for generating the antioxidants glutathione and thioredoxin are consistently overexpressed in NRF2 gain-of-function tumors, and both antioxidants are essential for cancer initiation and progression [45]. Synthesis and regeneration of glutathione is largely controlled by the catalytic and modifier subunits of the glutamate-cysteine ligase enzyme (encoded by GCLC and GCLM, respectively), the cysteine transporter subunit encoded by SLC7A11, and glutathione reductase (encoded by GSR). Thioredoxin activity is induced by expression of TXN, which codes for this antioxidant, and regenerated by thioredoxin reductase (encoded by TXNRD1). But antioxidant genes are not the only group of NRF2 targets with potential cancer relevance. Upregulation of enzymes in the pentose phosphate pathway, including 6-phosphogluconate dehydrogenase (PGD), glucose-6-phosphate dehydrogenase (G6PD), transaldolase (TALDO1), and transketolase (TKT), drives metabolic reprogramming and NRF2-dependent proliferation in lung adenocarcinoma cells (A549) and other cell lines [46,47]. Additionally, a number of the NRF2 targets that are repeatedly induced in cancer likely promote chemoresistance by increasing drug metabolism (ALDH3A1, NQO1, AKR1C1, EPHX1) or transport (ABCB6) [48]. Combined, the genes that are consistently responsive to NRF2 activity in diverse tissue contexts can explain much of NRF2’s oncogenic potential.
The molecular profile of oncogenic NRF2 hyperactivation, at least with regard to gene expression signatures, is largely independent of a tumor’s tissue of origin. The upregulated genes in NRF2-driven cancer are responsible for many of NRF2’s core cytoprotective functions in non-pathological situations, so it makes sense that they can be activated in many cell types. Nevertheless, what mechanistically differentiates this significant subset of NRF2-targeted AREs from its other targets remains unclear. At the cancer-induced AREs, NRF2 binding and transactivation is ostensibly not inhibited by other tissue-specific regulatory networks (transcriptional repressors, chromatin environment, etc.), and this lack of constraint can have dire consequences.
3. Cis-regulatory effects of ARE variation
Genetic variants that disrupt individual NRF2-targeted ARE sequences are expected to have more specific effects than variants that alter overall levels of nuclear NRF2. Whereas disruption of overall NRF2 levels impacts many genes in the network (Table 1), a cis-regulatory ARE variant will primarily alter expression of the gene regulated by the ARE (Figure 1C). Although the effects of cis-regulatory variation are more precise, they are not without biological consequence. Most disease-associated variants identified by genome-wide association studies (GWAS) fall in non-protein coding DNA. Most of the non-coding disease-associated variants are innocuous – they simply reach significance because they are co-inherited with a functional variant – but those with functional relevance often disrupt transcription factor binding sites [49–51]. This is indeed the case for NRF2: polymorphic ARE motifs have recently been linked to allele-specific enhancer activity, gene expression, and disease risk [24,52].
A position weight matrix scanning approach identified over two million ARE sequences in the human genome, but NRF2 and sMAF ChIP-seq data suggest that less than 2% are functional AREs [52]. This is consistent with numbers observed for other human transcription factors [18,53]. Approximately 7.5% (2689 out of 35,659) of the AREs consistently bound by NRF2 and/or sMAFs contain a potential ARE-altering single nucleotide variant (SNV), and 14 of the variable AREs are in linkage disequilibrium (i.e., co-inherited) with disease-associated variants identified by GWAS [52]. Disease-associated, variant AREs represent instances where cis-regulatory variation in the NRF2 network might have a phenotypic impact (disease risk); a subset of these ARE-altering SNVs are outlined in Table 2. A connection to cancer is still evident: two polymorphic AREs are associated with testicular germ cell tumors. However variant AREs are also linked to disease beyond cancer. Interestingly, the list includes hits for neurodegenerative disorders including progressive supranuclear palsy (PSP), Parkinson disease (PD), and corticobasal degeneration, as well as gastrointestinal disorders including celiac disease and colitis. These disease associations are based on common germline SNVs (minor allele frequency greater than 1%), but rare inherited variants that disrupt AREs could also be important [24].
Table 2. Disease-associated, ARE-disrupting single nucleotide variants.
Summary of significant SNVs identified in [52]. SNVs represented are those falling within 2 base pairs of an ARE containing the GCnnnnTCA core sequence, and with a position weight matrix (PWM) match score >10. For the ARE Sequences column, SNVs are underlined and highlighted in red/blue – the variant that generates a stronger PWM match is highlighted red, and the weaker PWM match is highlighted blue. Allele frequency data are from the 1000 Genomes Project.
| SNP ID | ARE sequence | Allele frequency | Nearest gene | Disease association(s) |
|---|---|---|---|---|
| rs242561 | 0.90 | MAPT | Progressive Supranuclear Palsy; Parkinson’s Disease; Corticobasal |
|
| 0.10 | Degeneration; Interstitial Lung Disease | |||
| rs241032 | 0.57 | CRHR1-IT1 | Parkinson’s Disease | |
| 0.43 | ||||
| rs6426833 | 0.59 | RNF186 | Ulcerative Colitis | |
| 0.41 | ||||
| rs17035378 | 0.52 | PLEK | Celiac Disease | |
| 0.48 | ||||
| rs369184 | 0.85 | TEX14 | Testicular Germ Cell Tumor | |
| 0.15 | ||||
| rs4818832 | 0.64 | YBEY | Testicular Germ Cell Tumor | |
| 0.36 | ||||
| Additional high priority polymorphic AREs: rs6426519, rs9603754, rs9884209, rs12638492, rs13067040, rs16857611, rs62033400, rs62094906 | ||||
One ARE-altering SNV, rs242561, falls within an NRF2-bound ARE at the MAPT locus [52]. The major allele of rs242561 creates a mismatch ARE (CGCTGAGTCAC – variant sequence is underlined) and the minor allele creates a perfect ARE (TGCTGAGTCAC). Thus, most people carry one or two copies of a mismatch NRF2-targeted ARE in a MAPT enhancer region. A smaller subset of the population carries a perfect-match ARE at this MAPT enhancer. MAPT encodes the protein tau, which plays a central role in multiple neurodegenerative diseases, and rs242561 is in linkage disequilibrium with variants associated PSP, PD, and corticobasal degeneration [54–56]. Importantly, the minor allele of rs242561 (perfect-match ARE) is associated with decreased risk of all three aforementioned neurodegenerative disorders, acts as a hypermorphic ARE in reporter assays, and is associated with increased expression of a protective isoform of MAPT [52,57–59]. Although it is possible that additional non-coding variants affect MAPT expression and neurodegenerative disease risk, these data suggest that the ARE impacted by rs242561 plays a significant functional role at this locus.
The above example suggests that inherited variation in specific NRF2-targeted ARE sequences can influence gene expression and, ultimately, disease risk. However, somatic variation in ARE sequences might also have an impact on disease. Cancer genome sequencing data from TCGA indicate that somatic variants disrupting ARE-like sequences are under positive selection in cancer cells [60]. This intriguing finding places the ARE among a small subset of transcription factor binding motifs commonly mutated in cancer. Thus, ARE mutations and aberrant expression of select NRF2 target genes are also likely to play an important functional role in cancer.
When considering the effects of ARE variation, one must consider that our current models of NRF2 DNA binding and regulatory output are incomplete. Identification of ARE sequences are dependent on model used (position weight matrix or other models). Some models focus on the core ARE sequence described above, while others include significant stretches of flanking sequence – both may be functionally relevant. In addition, there are many variant AREs associated with gene expression changes that have not yet been linked to a disease phenotype (SL and MS, not shown). It is likely that many of these changes are not strong enough to have a phenotypic impact; however some may be revealed as important in future disease association studies.
It is also important to recognize that ARE activity is context-dependent. That is, ARE activity generally increases under various stress conditions. Therefore the cis-regulatory effects of some ARE variants might only be evident under conditions where AREs are active. Oxidative stress, from both exogenous and endogenous sources, is an early and ongoing contributor to many diseases, so it makes sense that some variant AREs will appear significant in standard disease association studies. This explanation holds for rs242561, as oxidative stress plays a significant role in the pathology of neurodegenerative diseases such as PD and PSP [61–64]. However, certain ARE variants may only display significant disease associations in the presence of other genetic variants or environmental stressors that disrupt redox homeostasis. GWAS are become increasingly expansive, allowing for exploration of gene-gene and gene-environment interactions, so it is possible that additional disease-associated ARE variants will be identified in future studies.
4. A role for additional ARE binding proteins?
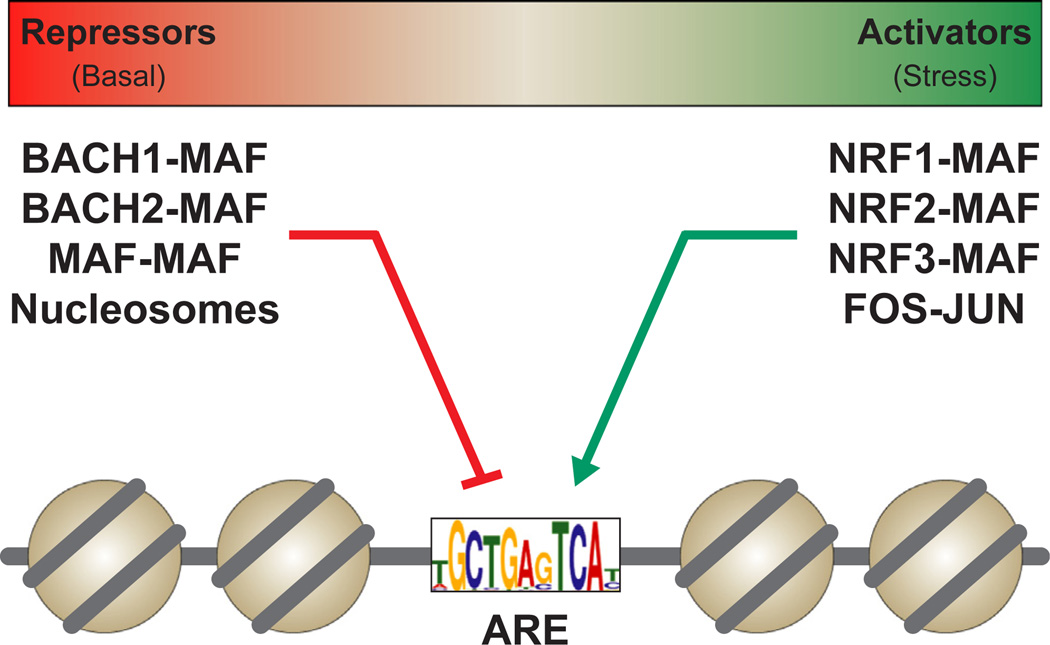
Precise regulation of NRF2-ARE binding is clearly important. Nuclear NRF2 concentration and ARE quality (i.e., similarity to sequence TGCTGAGTCAY) are significant contributors to this protein-DNA interaction, however additional variables must be considered. NRF2 is part of a family of CNC transcription factors, all with very similar DNA binding properties. Mammalian genomes contain six CNC proteins: four transcriptional activators (NFE2, NRF1, NRF2, NRF3), and two repressors (BACH1, BACH2) [5]. With the exception of NFE2, which regulates developmental transitions in the hematopoietic system [65–67], all CNC factors have been implicated as regulators of stress responsive genes [5]. And like NRF2, the other CNC proteins all dimerize with sMAF proteins, and all bind ARE sequences to regulate gene expression [6].
Compared to NRF2, less is known about the other four stress responsive CNC factors. All are expressed in a variety of cell types except for BACH2, which is most prevalent in the brain and B cells [5]. Current models of CNC mediated gene expression posit that regulatory output at an ARE is the result of competition between activator and repressor CNC factors, with activator CNC factors dominating in stress conditions [68–72]. However, this competition model is based largely on the opposing actions of NRF2 and BACH1 at AREs associated with two canonical antioxidant genes (NQO1 and HMOX1), and may not apply equally to all CNC target genes [68,71,73]. Further complicating matters, non-CNC proteins can also modulate ARE activity. The ARE, like all cis-regulatory sequences, can be repressed by nucleosomes, which hinder transcription factor access to a binding motif. ARE-like motifs can also be directly repressed by MAF homodimers [74] (see for a [75] comprehensive review). Additionally, stress responsive AP-1 protein complexes, which consist of heterodimers of proteins from the FOS, JUN, and ATF families, bind a target sequence very similar to the ARE [76].
A complete understanding of NRF2-mediated gene regulation must take additional ARE-binding factors into account (Figure 2). Models that integrate cooperation and competition for ARE binding among the activating and repressive factors will certainly further our understanding of the normal, homeostatic functions of NRF2. More comprehensive models might also explain why only a subset of AREs is consistently misregulated in cancer, or why some ARE-disrupting SNVs affect disease risk while others do not. The genomics era has yielded tremendous insights into NRF2 biology. Approaches that view NRF2 in the context of additional ARE-binding factors, and integrate with disease related functional genomics data (e.g., TCGA and GWAS data), will provide a comprehensive view of the regulatory mechanisms at play in this network in both physiological and pathological contexts.
Figure 2. Potential ARE-binding transcription factor complexes.
Current models suggest ARE regulatory output is driven by competition between activator and repressor CNC-MAF proteins. The impact of additional ARE binding proteins and nucleosomes, and whether this model extends equally to a wide range of functional AREs, remains unclear.
Acknowledgments
We apologize to our colleagues whose work is not cited herein due to space limitations. This work was supported by funding from the National Institutes of Health (R35GM119553 to MS), the Whiteside Institute for Clinical Research (MS), and the University of Minnesota Foundation (MS).
Reference
Papers of particular interest, published within the period of review, have been highlighted as:
* of special interest
** of outstanding interest
- 1.Waris G, Ahsan H. Reactive oxygen species: role in the development of cancer and various chronic conditions. J Carcinog. 2006;5:14. doi: 10.1186/1477-3163-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barja G. Free radicals and aging. Trends Neurosci. 2004;27:595–600. doi: 10.1016/j.tins.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 3.Jimenez-Del-Rio M, Velez-Pardo C. The bad, the good, and the ugly about oxidative stress. Oxid Med Cell Longev. 2012;2012:163913. doi: 10.1155/2012/163913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Halliwell B. Biochemistry of oxidative stress. Biochem Soc Trans. 2007;35:1147–1150. doi: 10.1042/BST0351147. [DOI] [PubMed] [Google Scholar]
- 5.Sykiotis GP, Bohmann D. Stress-activated cap‘n’collar transcription factors in aging and human disease. Sci Signal. 2010;3:re3. doi: 10.1126/scisignal.3112re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kannan MB, Solovieva V, Blank V. The small MAF transcription factors MAFF, MAFG and MAFK: current knowledge and perspectives. Biochim Biophys Acta. 2012;1823:1841–1846. doi: 10.1016/j.bbamcr.2012.06.012. [DOI] [PubMed] [Google Scholar]
- 7.Zipper LM, Mulcahy RT. The Keap1 BTB/POZ dimerization function is required to sequester Nrf2 in cytoplasm. J Biol Chem. 2002;277:36544–36552. doi: 10.1074/jbc.M206530200. [DOI] [PubMed] [Google Scholar]
- 8.Wakabayashi N, Itoh K, Wakabayashi J, Motohashi H, Noda S, Takahashi S, Imakado S, Kotsuji T, Otsuka F, Roop DR, et al. Keap1-null mutation leads to postnatal lethality due to constitutive Nrf2 activation. Nat Genet. 2003;35:238–245. doi: 10.1038/ng1248. [DOI] [PubMed] [Google Scholar]
- 9.McMahon M, Itoh K, Yamamoto M, Hayes JD. Keap1-dependent proteasomal degradation of transcription factor Nrf2 contributes to the negative regulation of antioxidant response element-driven gene expression. J Biol Chem. 2003;278:21592–21600. doi: 10.1074/jbc.M300931200. [DOI] [PubMed] [Google Scholar]
- 10.Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, Yamamoto M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dinkova-Kostova AT, Holtzclaw WD, Cole RN, Itoh K, Wakabayashi N, Katoh Y, Yamamoto M, Talalay P. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc Natl Acad Sci U S A. 2002;99:11908–11913. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malhotra D, Portales-Casamar E, Singh A, Srivastava S, Arenillas D, Happel C, Shyr C, Wakabayashi N, Kensler TW, Wasserman WW, et al. Global mapping of binding sites for Nrf2 identifies novel targets in cell survival response through ChIP-Seq profiling and network analysis. Nucleic Acids Res. 2010;38:5718–5734. doi: 10.1093/nar/gkq212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chorley BN, Campbell MR, Wang X, Karaca M, Sambandan D, Bangura F, Xue P, Pi J, Kleeberger SR, Bell DA. Identification of novel NRF2-regulated genes by ChIP-Seq: influence on retinoid X receptor alpha. Nucleic Acids Res. 2012;40:7416–7429. doi: 10.1093/nar/gks409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lacher SE, Lee JS, Wang X, Campbell MR, Bell DA, Slattery M. Beyond antioxidant genes in the ancient Nrf2 regulatory network. Free Radic Biol Med. 2015;88:452–465. doi: 10.1016/j.freeradbiomed.2015.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee OH, Jain AK, Papusha V, Jaiswal AK. An auto-regulatory loop between stress sensors INrf2 and Nrf2 controls their cellular abundance. J Biol Chem. 2007;282:36412–36420. doi: 10.1074/jbc.M706517200. [DOI] [PubMed] [Google Scholar]
- 16.Sykiotis GP, Bohmann D. Keap1/Nrf2 signaling regulates oxidative stress tolerance and lifespan in Drosophila. Dev Cell. 2008;14:76–85. doi: 10.1016/j.devcel.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Biggin MD. Animal transcription networks as highly connected, quantitative continua. Dev Cell. 2011;21:611–626. doi: 10.1016/j.devcel.2011.09.008. [DOI] [PubMed] [Google Scholar]
- 18.Slattery M, Zhou T, Yang L, Dantas Machado AC, Gordan R, Rohs R. Absence of a simple code: how transcription factors read the genome. Trends Biochem Sci. 2014;39:381–399. doi: 10.1016/j.tibs.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Friling RS, Bensimon A, Tichauer Y, Daniel V. Xenobiotic-inducible expression of murine glutathione S-transferase Ya subunit gene is controlled by an electrophile-responsive element. Proc Natl Acad Sci U S A. 1990;87:6258–6262. doi: 10.1073/pnas.87.16.6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Otsuki A, Suzuki M, Katsuoka F, Tsuchida K, Suda H, Morita M, Shimizu R, Yamamoto M. Unique cistrome defined as CsMBE is strictly required for Nrf2-sMaf heterodimer function in cytoprotection. Free Radic Biol Med. 2016;91:45–57. doi: 10.1016/j.freeradbiomed.2015.12.005. [DOI] [PubMed] [Google Scholar]
- 21.Rushmore TH, Morton MR, Pickett CB. The antioxidant responsive element. Activation by oxidative stress and identification of the DNA consensus sequence required for functional activity. J Biol Chem. 1991;266:11632–11639. [PubMed] [Google Scholar]
- 22.Wasserman WW, Fahl WE. Functional antioxidant responsive elements. Proc Natl Acad Sci U S A. 1997;94:5361–5366. doi: 10.1073/pnas.94.10.5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirotsu Y, Katsuoka F, Funayama R, Nagashima T, Nishida Y, Nakayama K, Engel JD, Yamamoto M. Nrf2-MafG heterodimers contribute globally to antioxidant and metabolic networks. Nucleic Acids Res. 2012;40:10228–10239. doi: 10.1093/nar/gks827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuosmanen SM, Viitala S, Laitinen T, Perakyla M, Polonen P, Kansanen E, Leinonen H, Raju S, Wienecke-Baldacchino A, Narvanen A, et al. The effects of sequence variation on genome-wide NRF2 binding–new target genes and regulatory SNPs. Nucleic Acids Res. 2016;44:1760–1775. doi: 10.1093/nar/gkw052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang H, Davies KJ, Forman HJ. Oxidative stress response and Nrf2 signaling in aging. Free Radic Biol Med. 2015 doi: 10.1016/j.freeradbiomed.2015.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oh DH, Rigas D, Cho A, Chan JY. Deficiency in the nuclear-related factor erythroid 2 transcription factor (Nrf1) leads to genetic instability. FEBS J. 2012;279:4121–4130. doi: 10.1111/febs.12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gan L, Johnson JA. Oxidative damage and the Nrf2-ARE pathway in neurodegenerative diseases. Biochim Biophys Acta. 2014;1842:1208–1218. doi: 10.1016/j.bbadis.2013.12.011. [DOI] [PubMed] [Google Scholar]
- 28.Singh A, Misra V, Thimmulappa RK, Lee H, Ames S, Hoque MO, Herman JG, Baylin SB, Sidransky D, Gabrielson E, et al. Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS Med. 2006;3:e420. doi: 10.1371/journal.pmed.0030420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shibata T, Ohta T, Tong KI, Kokubu A, Odogawa R, Tsuta K, Asamura H, Yamamoto M, Hirohashi S. Cancer related mutations in NRF2 impair its recognition by Keap1-Cul3 E3 ligase and promote malignancy. Proc Natl Acad Sci U S A. 2008;105:13568–13573. doi: 10.1073/pnas.0806268105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Konstantinopoulos PA, Spentzos D, Fountzilas E, Francoeur N, Sanisetty S, Grammatikos AP, Hecht JL, Cannistra SA. Keap1 mutations and Nrf2 pathway activation in epithelial ovarian cancer. Cancer Res. 2011;71:5081–5089. doi: 10.1158/0008-5472.CAN-10-4668. [DOI] [PubMed] [Google Scholar]
- 31.Kim YR, Oh JE, Kim MS, Kang MR, Park SW, Han JY, Eom HS, Yoo NJ, Lee SH. Oncogenic NRF2 mutations in squamous cell carcinomas of oesophagus and skin. J Pathol. 2010;220:446–451. doi: 10.1002/path.2653. [DOI] [PubMed] [Google Scholar]
- 32.Cancer Genome Atlas Research N. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Padmanabhan B, Tong KI, Ohta T, Nakamura Y, Scharlock M, Ohtsuji M, Kang MI, Kobayashi A, Yokoyama S, Yamamoto M. Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol Cell. 2006;21:689–700. doi: 10.1016/j.molcel.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 34.Ganan-Gomez I, Wei Y, Yang H, Boyano-Adanez MC, Garcia-Manero G. Oncogenic functions of the transcription factor Nrf2. Free Radic Biol Med. 2013;65:750–764. doi: 10.1016/j.freeradbiomed.2013.06.041. [DOI] [PubMed] [Google Scholar]
- 35.Menegon S, Columbano A, Giordano S. The dual roles of NRF2 in cancer. Trends Mol Med. 2016;22:578–593. doi: 10.1016/j.molmed.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 36.Cancer Genome Atlas N. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gibson WJ, Hoivik EA, Halle MK, Taylor-Weiner A, Cherniack AD, Berg A, Holst F, Zack TI, Werner HM, Staby KM, et al. The genomic landscape and evolution of endometrial carcinoma progression and abdominopelvic metastasis. Nat Genet. 2016;48:848–855. doi: 10.1038/ng.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang D, Ma Y, Yang X, Xu X, Zhao Y, Zhu Z, Wang X, Deng H, Li C, Gao F, et al. Hypermethylation of the Keap1 gene inactivates its function, promotes Nrf2 nuclear accumulation, and is involved in arsenite-induced human keratinocyte transformation. Free Radic Biol Med. 2015;89:209–219. doi: 10.1016/j.freeradbiomed.2015.07.153. [DOI] [PubMed] [Google Scholar]
- 39.Martinez VD, Vucic EA, Pikor LA, Thu KL, Hubaux R, Lam WL. Frequent concerted genetic mechanisms disrupt multiple components of the NRF2 inhibitor KEAP1/CUL3/RBX1 E3-ubiquitin ligase complex in thyroid cancer. Mol Cancer. 2013;12:124. doi: 10.1186/1476-4598-12-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martinez VD, Vucic EA, Thu KL, Pikor LA, Lam S, Lam WL. Disruption of KEAP1/CUL3/RBX1 E3-ubiquitin ligase complex components by multiple genetic mechanisms: association with poor prognosis in head and neck cancer. Head Neck. 2015;37:727–734. doi: 10.1002/hed.23663. [DOI] [PubMed] [Google Scholar]
- 41.Zhang P, Singh A, Yegnasubramanian S, Esopi D, Kombairaju P, Bodas M, Wu H, Bova SG, Biswal S. Loss of Kelch-like ECH-associated protein 1 function in prostate cancer cells causes chemoresistance and radioresistance and promotes tumor growth. Mol Cancer Ther. 2010;9:336–346. doi: 10.1158/1535-7163.MCT-09-0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang R, An J, Ji F, Jiao H, Sun H, Zhou D. Hypermethylation of the Keap1 gene in human lung cancer cell lines and lung cancer tissues. Biochem Biophys Res Commun. 2008;373:151–154. doi: 10.1016/j.bbrc.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 43.Araya CL, Cenik C, Reuter JA, Kiss G, Pande VS, Snyder MP, Greenleaf WJ. Identification of significantly mutated regions across cancer types highlights a rich landscape of functional molecular alterations. Nat Genet. 2016;48:117–125. doi: 10.1038/ng.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ding J, McConechy MK, Horlings HM, Ha G, Chun Chan F, Funnell T, Mullaly SC, Reimand J, Bashashati A, Bader GD, et al. Systematic analysis of somatic mutations impacting gene expression in 12 tumour types. Nat Commun. 2015;6:8554. doi: 10.1038/ncomms9554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harris IS, Treloar AE, Inoue S, Sasaki M, Gorrini C, Lee KC, Yung KY, Brenner D, Knobbe-Thomsen CB, Cox MA, et al. Glutathione and Thioredoxin antioxidant pathways synergize to drive cancer initiation and progression. Cancer Cell. 2015 doi: 10.1016/j.ccell.2014.11.019. [DOI] [PubMed] [Google Scholar]
- 46.Mitsuishi Y, Taguchi K, Kawatani Y, Shibata T, Nukiwa T, Aburatani H, Yamamoto M, Motohashi H. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell. 2012;22:66–79. doi: 10.1016/j.ccr.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 47.Kowalik MA, Guzzo G, Morandi A, Perra A, Menegon S, Masgras I, Trevisan E, Angioni MM, Fornari F, Quagliata L, et al. Metabolic reprogramming identifies the most aggressive lesions at early phases of hepatic carcinogenesis. Oncotarget. 2016 doi: 10.18632/oncotarget.8632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hayes JD, Dinkova-Kostova AT. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem Sci. 2014;39:199–218. doi: 10.1016/j.tibs.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 49.Ward LD, Kellis M. Interpreting noncoding genetic variation in complex traits and human disease. Nat Biotechnol. 2012;30:1095–1106. doi: 10.1038/nbt.2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Edwards SL, Beesley J, French JD, Dunning AM. Beyond GWASs: illuminating the dark road from association to function. Am J Hum Genet. 2013;93:779–797. doi: 10.1016/j.ajhg.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Albert FW, Kruglyak L. The role of regulatory variation in complex traits and disease. Nat Rev Genet. 2015;16:197–212. doi: 10.1038/nrg3891. [DOI] [PubMed] [Google Scholar]
- 52.Wang X, Campbell MR, Lacher SE, Cho HY, Wan M, Crowl CL, Chorley BN, Bond GL, Kleeberger SR, Slattery M, et al. A polymorphic antioxidant response element links NRF2/sMAF binding to enhanced MAPT expression and reduced risk of Parkinsonian disorders. Cell Rep. 2016 doi: 10.1016/j.celrep.2016.03.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wunderlich Z, Mirny LA. Different gene regulation strategies revealed by analysis of binding motifs. Trends Genet. 2009;25:434–440. doi: 10.1016/j.tig.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nalls MA, Pankratz N, Lill CM, Do CB, Hernandez DG, Saad M, DeStefano AL, Kara E, Bras J, Sharma M, et al. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat Genet. 2014;46:989–993. doi: 10.1038/ng.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kouri N, Ross OA, Dombroski B, Younkin CS, Serie DJ, Soto-Ortolaza A, Baker M, Finch NC, Yoon H, Kim J, et al. Genome-wide association study of corticobasal degeneration identifies risk variants shared with progressive supranuclear palsy. Nat Commun. 2015;6:7247. doi: 10.1038/ncomms8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hoglinger GU, Melhem NM, Dickson DW, Sleiman PM, Wang LS, Klei L, Rademakers R, de Silva R, Litvan I, Riley DE, et al. Identification of common variants influencing risk of the tauopathy progressive supranuclear palsy. Nat Genet. 2011;43:699–705. doi: 10.1038/ng.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhong Q, Congdon EE, Nagaraja HN, Kuret J. Tau isoform composition influences rate and extent of filament formation. J Biol Chem. 2012;287:20711–20719. doi: 10.1074/jbc.M112.364067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Trabzuni D, Wray S, Vandrovcova J, Ramasamy A, Walker R, Smith C, Luk C, Gibbs JR, Dillman A, Hernandez DG, et al. MAPT expression and splicing is differentially regulated by brain region: relation to genotype and implication for tauopathies. Hum Mol Genet. 2012;21:4094–4103. doi: 10.1093/hmg/dds238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ramasamy A, Trabzuni D, Gibbs JR, Dillman A, Hernandez DG, Arepalli S, Walker R, Smith C, Ilori GP, Shabalin AA, et al. Resolving the polymorphism-in-probe problem is critical for correct interpretation of expression QTL studies. Nucleic Acids Res. 2013;41:e88. doi: 10.1093/nar/gkt069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Melton C, Reuter JA, Spacek DV, Snyder M. Recurrent somatic mutations in regulatory regions of human cancer genomes. Nat Genet. 2015;47:710–716. doi: 10.1038/ng.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Martinez A, Portero-Otin M, Pamplona R, Ferrer I. Protein targets of oxidative damage in human neurodegenerative diseases with abnormal protein aggregates. Brain Pathol. 2010;20:281–297. doi: 10.1111/j.1750-3639.2009.00326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Johnson DA, Johnson JA. Nrf2 – a therapeutic target for the treatment of neurodegenerative diseases. Free Radic Biol Med. 2015;88:253–267. doi: 10.1016/j.freeradbiomed.2015.07.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen PC, Vargas MR, Pani AK, Smeyne RJ, Johnson DA, Kan YW, Johnson JA. Nrf2-mediated neuroprotection in the MPTP mouse model of Parkinson’s disease: critical role for the astrocyte. Proc Natl Acad Sci U S A. 2009;106:2933–2938. doi: 10.1073/pnas.0813361106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Beraud D, Hathaway HA, Trecki J, Chasovskikh S, Johnson DA, Johnson JA, Federoff HJ, Shimoji M, Mhyre TR, Maguire-Zeiss KA. Microglial activation and antioxidant responses induced by the Parkinson’s disease protein alpha-synuclein. J Neuroimmune Pharmacol. 2013;8:94–117. doi: 10.1007/s11481-012-9401-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gasiorek JJ, Blank V. Regulation and function of the NFE2 transcription factor in hematopoietic and non-hematopoietic cells. Cell Mol Life Sci. 2015;72:2323–2335. doi: 10.1007/s00018-015-1866-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fujita R, Takayama-Tsujimoto M, Satoh H, Gutierrez L, Aburatani H, Fujii S, Sarai A, Bresnick EH, Yamamoto M, Motohashi H. NF-E2 p45 is important for establishing normal function of platelets. Mol Cell Biol. 2013;33:2659–2670. doi: 10.1128/MCB.01274-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Forsberg EC, Downs KM, Bresnick EH. Direct interaction of NFE2 with hypersensitive site 2 of the beta-globin locus control region in living cells. Blood. 2000;96:334–339. [PubMed] [Google Scholar]
- 68.Reichard JF, Motz GT, Puga A. Heme oxygenase-1 induction by NRF2 requires inactivation of the transcriptional repressor BACH1. Nucleic Acids Res. 2007;35:7074–7086. doi: 10.1093/nar/gkm638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Niture SK, Khatri R, Jaiswal AK. Regulation of Nrf2-an update. Free Radic Biol Med. 2014;66:36–44. doi: 10.1016/j.freeradbiomed.2013.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Igarashi K, Sun J. The heme-Bach1 pathway in the regulation of oxidative stress response and erythroid differentiation. Antioxid Redox Signal. 2006;8:107–118. doi: 10.1089/ars.2006.8.107. [DOI] [PubMed] [Google Scholar]
- 71.Dhakshinamoorthy S, Jain AK, Bloom DA, Jaiswal AK. Bach1 competes with Nrf2 leading to negative regulation of the antioxidant response element (ARE)-mediated NAD(P)H: quinone oxidoreductase 1 gene expression and induction in response to antioxidants. J Biol Chem. 2005;280:16891–16900. doi: 10.1074/jbc.M500166200. [DOI] [PubMed] [Google Scholar]
- 72.Boutten A, Goven D, Artaud-Macari E, Boczkowski J, Bonay M. NRF2 targeting: a promising therapeutic strategy in chronic obstructive pulmonary disease. Trends Mol Med. 2011;17:363–371. doi: 10.1016/j.molmed.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 73.MacLeod AK, McMahon M, Plummer SM, Higgins LG, Penning TM, Igarashi K, Hayes JD. Characterization of the cancer chemopreventive NRF2-dependent gene battery in human keratinocytes: demonstration that the KEAP1-NRF2 pathway, and not the BACH1-NRF2 pathway, controls cytoprotection against electrophiles as well as redox-cycling compounds. Carcinogenesis. 2009;30:1571–1580. doi: 10.1093/carcin/bgp176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dhakshinamoorthy S, Jaiswal AK. Small maf (MafG and MafK) proteins negatively regulate antioxidant response elementmediated expression and antioxidant induction of the NAD(P) H: Quinone oxidoreductase1 gene. J Biol Chem. 2000;275:40134–40141. doi: 10.1074/jbc.M003531200. [DOI] [PubMed] [Google Scholar]
- 75.Katsuoka F, Yamamoto M. Small Maf proteins (MafF, MafG, MafK): History, structure and function. Gene. 2016;586:197–205. doi: 10.1016/j.gene.2016.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Eferl R, Wagner EF. AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer. 2003;3:859–868. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]