Fign regulates cultured astrocyte migration by severing tyrosinated microtubules (MTs). Inhibition of cellular migration induced by Fign knockdown can be rescued with concomitant knockdown of kinesin-12. A working model is given for the MT reconfiguration underlying cellular migration elicited by the cooperation of two distinct MT-related proteins.
Abstract
Microtubule (MT) organization is essential for many cellular events, including mitosis, migration, and cell polarity. Fidgetin (Fign), an ATP-dependent, MT-severing protein, contributes to the regulation of MT configuration by cutting and trimming MT polymers. Functions of Fign have been indicated in neurite outgrowth, mitosis, meiosis, and cellular migration. Here we focus on migration of astrocytes. We find that Fign plays an essential role in cultured astrocyte migration by preferentially targeting MTs (or regions of MTs) that are rich in tyrosinated tubulin, a marker for especially dynamic MTs or especially dynamic regions of MTs. Inhibition of cellular migration induced by Fign knockdown can be rescued with concomitant knockdown of kinesin-12, a motor protein best known for its role in mitosis. We propose a novel working model for MT reconfiguration underlying cellular migration elicited by the functional cooperation of two distinct MT-related proteins.
INTRODUCTION
Cellular migration is an important behavior for wound healing, tissue repair, and regeneration. During cellular migration, the cytoskeleton, which is principally composed of microtubules (MTs) and actin filaments, undergoes reorganization. Cortical actin polymers are early reactors to extracellular cues, contributing to the formation of filopodia and lamellipodia. MT behaviors, including sliding, polymerization, and depolymerization, contribute to the polarity of cellular migration (Watanabe et al., 2005). These behaviors are regulated by various MT-related proteins, including molecular motor proteins, MT-severing proteins, and MT-end-binding proteins, as well as extracellular and intracellular signaling factors (van der Vaart et al., 2009; Sharp and Ross, 2012). The tubulin subunits that comprise the MT are subjected to various posttranslational modifications (PTMs; Sharp and Ross, 2012; Roll-Mecak, 2015). PTMs are often indicators of the stability properties of the polymers. Detyrosination and acetylation usually demarcate stable MTs, whereas tyrosination and deacetylation correlate with labile MTs (Sirajuddin et al., 2014; Roll-Mecak, 2015). Stable and labile polymers, rich in modified and unmodified subunits, respectively, can also manifest as regions on the same MT, as reported for MTs in the axon (Baas and Ahmad, 2013).
Fidgetin (Fign) is a relatively conserved AAA enzyme with MT-severing capability in vitro (Mukherjee et al., 2012). Previous studies found that MT-severing proteins take part in various cellular events, such as mitosis/meiosis, morphogenesis, wound healing, and migration (Sharp and Ross, 2012). However, the functional roles of Fign in those cellular events are poorly understood. The seminal study on Drosophila Fign indicated that during mitosis, Fign severs MTs so as to create MT flux back to the centrosome (Zhang et al., 2007). A subsequent study from the same group, performed on human cells, suggested that Fign suppresses MT growth from the centrosome and stimulates poleward tubulin flux (Mukherjee et al., 2012). Two Fign-like genes, Fign-like 1 and 2, are expressed, in vertebrates (Cox et al., 2000). Of interest, Fign-like-2 was identified as a key regulator of cell migration in mammalian cells (Charafeddine et al., 2015). No functional studies for Fign in neuronal cells were available until a recent report on cultured rat embryonic cortical neurons found that, unlike katanin-P60, which targets acetylated MTs for its severing activity (Sudo and Baas, 2010), Fign targets unacetylated MTs and thereby regulates neuronal process number, axonal growth, and axonal branches (Leo et al., 2015). One of the most provocative discoveries from this study is that vertebrate Fign and Drosophila Fign target opposing PTM statuses, with the former targeting unacetylated MTs and the latter targeting acetylated MTs (Leo et al., 2015).
It remains unclear whether and how vertebrate Fign participates in cell migration—for example, in clinically important events such as spinal cord injury. In this study, we used cultured rat astrocytes to examine this question and probe for the potential underlying mechanism.
RESULTS
Depletion of Fign inhibits the migration of cultured astrocytes derived from rat spinal cord
In a first set of studies on cultured astrocytes, we investigated whether depletion of Fign alters cellular migration. The astrocyte is the major cell type in the spinal cord that proliferates and migrates. We chose them for these studies not only because of their functional importance to clinical situations such as spinal cord injury but also because they are very flat and have broad lamellar regions that are excellent for imaging analyses. First, we investigated the effect of Fign depletion on cultured astrocytes derived from postnatal day 1 (P1) rat spinal cords. Efficient knockdown of Fign from cultured astrocytes with small interfering RNA (siRNA) was confirmed by quantitative real-time PCR (qRT-PCR) 24 h after transfection (Figure 1A) and Western blotting 72 h after transfection (Figure 1B). The siRNA significantly reduced Fign expression, with a 73% decrease at the mRNA level and a 52% decrease at the protein level.
FIGURE 1:
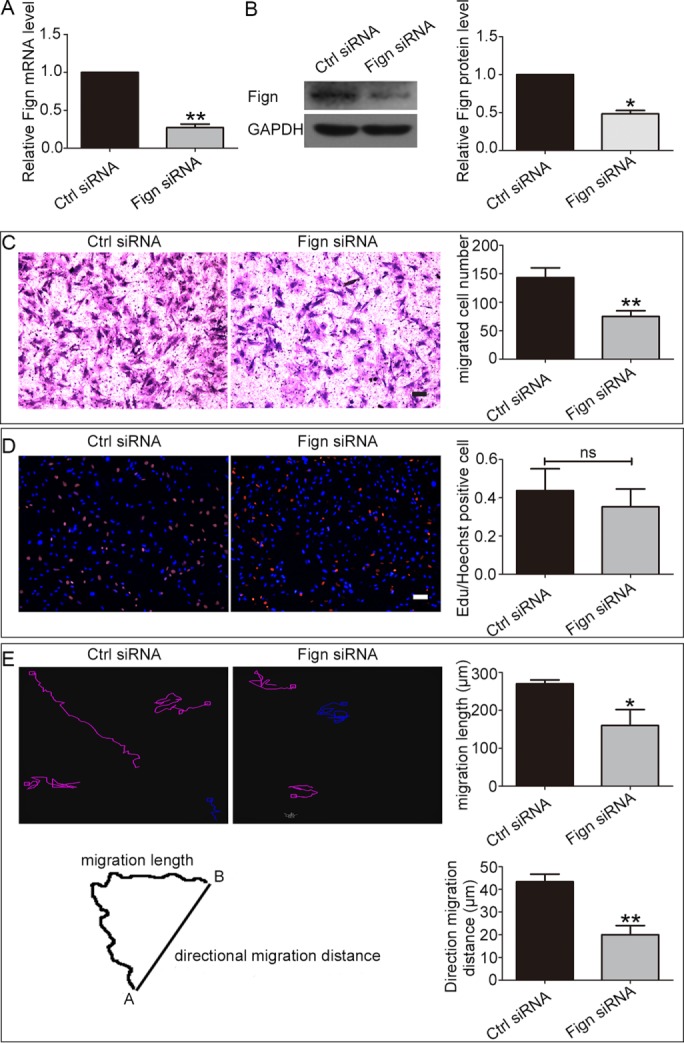
Depletion of Fign inhibits migration of rat cultured astrocytes. (A) The efficiency of Fign siRNA treatment for 24 h in cultured astrocytes was detected by qRT-PCR. The data are shown as mean ± SE. The relative mRNA level of the control (Ctrl) siRNA–treated astrocytes is normalized as 1, and Fign siRNA = 0.27 ± 0.04; n = 3, **p < 0.01. (B) The efficiency of Fign siRNA treated for 72 h in cultured astrocytes was detected by Western blotting. Left, representative Western blotting; right, quantification. Data are shown as mean ± SE. The relative protein level of Ctrl siRNA–treated astrocytes is normalized as 1, and Fign siRNA = 0.48 ± 0.06; n = 4, *p < 0.05. (C) Representative Transwell images of crystal violet staining (red) of transferred cultured astrocytes after siRNA treatment for 72 h with Ctrl siRNA and Fign siRNA. Bar, 100 μm. Right, statistical analysis showing migrated cell number; data are shown as mean ± SE. Ctrl siRNA = 143.2 ± 17.29, Fign siRNA = 75.1 ± 9.93, n = 5, **p < 0.01. (D) Left, representative images of EdU staining of astrocyte proliferation after siRNA treatment for 72 h; right, statistical analysis shows no significant difference between Ctrl siRNA and Fign siRNA treatment. Red (EdU staining), proliferating cells; blue (Hoechst staining), total cells. Bar, 100 μm. Data are shown as mean ± SE. Ctrl siRNA = 0.43 ± 0.11, Fign siRNA = 0.35 ± 0.09, n = 5; ns, no significance; p > 0.05. (E) Left, top, astrocytes transfected with Ctrl siRNA or Fign siRNA were treated as described in Materials and Methods and then imaged at 10-min intervals. Left, bottom, tracks of four randomly picked cells for each group, showing migration length (total track distance) and directional migration distance (from start to end point) of a cell. Right, top, migration length of Ctrl siRNA– and Fign siRNA–treated cells. The data are shown as mean ± SE, n = 28 cells for each condition from three independent experiments, Ctrl siRNA = 270 ± 10.0 μm, Fign siRNA = 160.0 ± 42.03 μm, *p < 0.05. Right, bottom, directional migration distance of Ctrl and Fign siRNA–treated cells. The data are presented as mean ± SE, n = 26 cells for each condition from three independent experiments, Ctrl siRNA = 43 ± 3.3 μm, Fign siRNA = 20.00 ± 4.0 μm, **p < 0.01.
To probe cell migration, we performed Transwell assays as in our previous study (Feng et al., 2016). In contrast to the increase in Drosophila S2 cell migration observed by knocking down Fign-like 2 (Charafeddine et al., 2015), we found that Fign depletion reduced astrocyte migration, with a 47.5% decrease compared with the control (migrated cells were stained with crystal violet in red; Figure 1C). Considering that cell proliferation is the other potential variable in this assay, we applied the 5-ethynyl-2’-deoxyuridine (EdU) incorporation DNA assay to cultured astrocytes depleted of Fign with siRNA as in our previous study (Feng et al., 2016). The EdU assay showed no significant difference in cell proliferation between the control group and Fign siRNA group (Figure 1D; cells were stained with EdU in red and Hoechst in blue). In addition to Transwell assays, live-cell imaging was used to confirm the effect of Fign depletion on astrocyte migration. Live-cell imaging of individual cells offers a powerful tool to record the detailed trajectory of their movements (Zhang et al., 2013). Quantitative analyses from recording the trajectory of individual astrocytes indicated that Fign siRNA–treated cells displayed both slower and less directional movement than the controls (Figure 1E; data given as mean ± SE; migration length, Ctrl siRNA = 270 ± 10.0 μm, Fign siRNA = 160 ± 42.0 μm, p < 0.05; directional migration distance, Ctrl siRNA = 43 ± 3.3 μm, Fign siRNA = 20 ± 4.1 μm, p < 0.01). Taken together, the results show that depletion of Fign significantly compromises the movement of spinal cord–derived astrocytes.
Fign targets tyrosinated tubulin in the cell cortical region of astrocytes
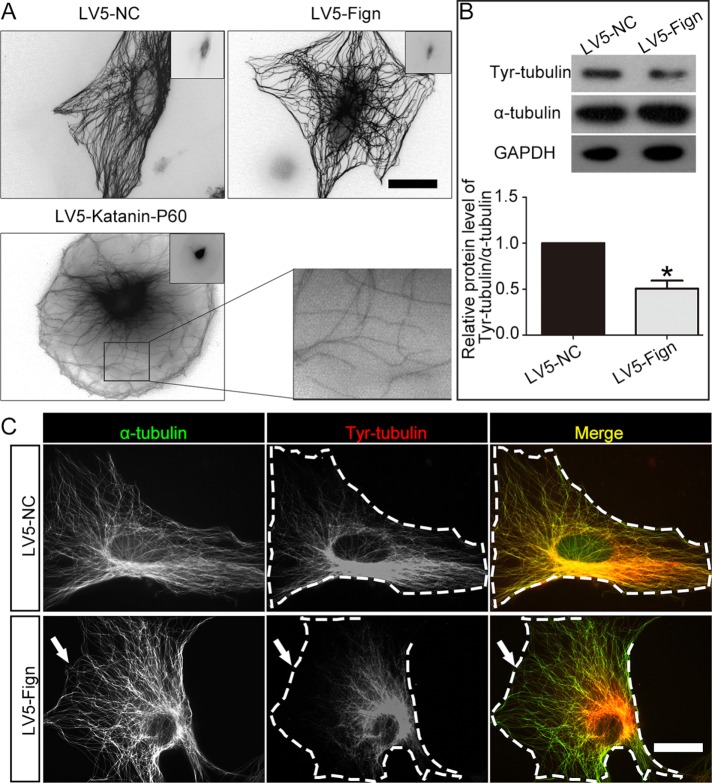
Previous studies suggested that different MT-related proteins target distinct MT PTMs (Ikegami and Setou, 2010). Recent evidence on cultured rat cortical neurons suggests that Fign cuts in regions of the polymer rich in unacetylated tubulin (Leo et al., 2015). However, its severing preference in other cell types has not been explored. To begin to examine this issue in cultured astrocytes, we generated and introduced lentiviral particles that encode Fign (LV5-Fign) into astrocyte cultures for 3 d. Unlike the case with overexpression of katanin or spastin (Karabay et al., 2004; Yu et al., 2008)—two other MT-severing proteins—no discernible MT severing was found after Fign overexpression in astrocytes (Figure 2A, top); we used katanin-P60 overexpression as a positive control (Figure 2A, bottom). Although no visible MT fragments from Fign overexpression were identified by immunocytochemistry using anti–α-tubulin antibody, we also used Western blotting. Various PTM-MT populations were evaluated using antibodies against acetylated tubulin (acetyl-tubulin), detyrosinated tubulin (detyr-tubulin), and tyrosinated tubulin (tyr-tubulin) after Fign was overexpressed. As shown in Figure 2B, overexpression of Fign reduced total MT mass by reducing tyr-MTs significantly without affecting detyr-MTs and acetyl-MTs (unpublished data). By immunostaining, as shown in Figure 2C, we found that Fign overexpression dramatically removed tyr-MTs close to the cortical region of astrocytes, whereas detyr-MTs or acetyl-MTs remained intact (unpublished data). Consistent with the previous report (Leo et al., 2015), Fign appears to specifically cut labile MTs, which are mainly composed of tyr-MTs. Moreover, overexpression of Fign in cultured astrocytes in particular severed tyr-MTs close to the peripheral region of the cells (Figure 2C, arrows).
FIGURE 2:
Overexpression of Fign reduces tyr-MTs in astrocytes. (A) Representative images of severing MT. LV-5-Fign lentivirus was introduced into astrocytes for 3 d and immunostained by anti–α-tubulin, and LV-5-katanin lentivirus was used as a functional control of severing MTs. No MT severing was observed, and no short MT fragments were found in LV5-Fign–treated astrocytes, but severing MTs were found in the group of LV5-katanin-P60–overexpressed cells (left, bottom; bar, 20 μm). Right, bottom, magnification of severed MTs. (B) Top, representative Western blotting result showing that LV5-Fign–overexpression cultures have a lower ratio of tyr-tubulin to α-tubulin than LV5-NC cultures. Bottom, statistical data showing a decrease of tyr-tubulin protein after Fign overexpression. The data are shown as mean ± SE. The relative tyr-tubulin protein level (α-tubulin as the control) of LV-NC–overexpressed astrocytes was normalized as 1. LV5-Fign = 0.48 ± 0.04, n = 3, *p < 0.05. (C) Effect of LV5-Fign overexpression on modified tubulins. Astrocytes treated with LV5-NC (negative control; NC) or LV5-Fign lentivirus for 3 d were fixed and immunostained with tyr-tubulin (red) and α-tubulin (green). No short MTs were detected in Fign-overexpression groups for LV5-Fign (bottom) but decreased significantly in the cell edge with tyr-tubulin (white arrows) compared with the LV5-NC–treated astrocytes (top). Bar, 20 μm.
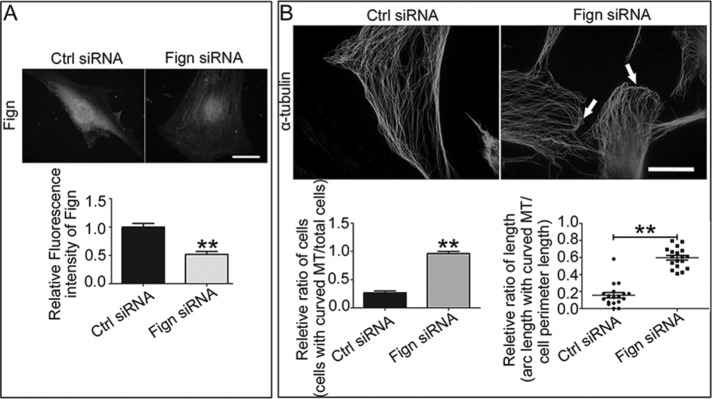
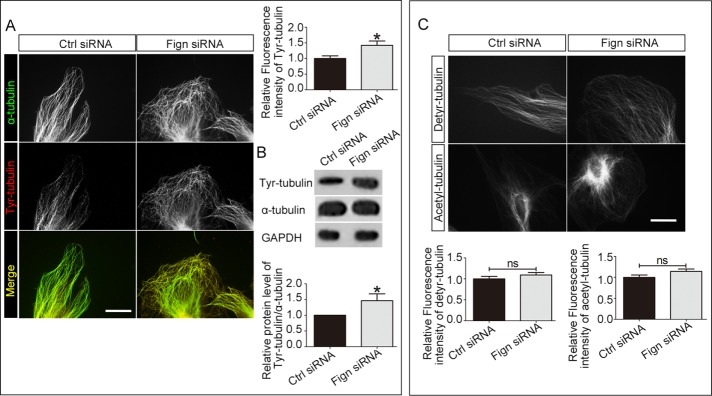
Next we investigated the change of MTs in cultured astrocytes when Fign was depleted. Successful Fign knockdown in astrocytes by siRNA was validated (Figure 3A). We treated the cells with Fign siRNA for 72 h before we fixed the cells. We found no significant change in actin cytoskeleton in Fign-knockdown astrocytes (unpublished data). However, an unusual MT organization in the peripheral region was identified in which the MTs were bent and curved (Figure 3B, arrows, Fign siRNA). This specific MT rearrangement is substantially different from what is normally seen in control cells, in which the MT arrays keep their radial distribution when they approach the edge and stay perpendicular to the cell membrane (Figure 3B, Ctrl siRNA). Referring to a previous study (Charafeddine et al., 2015), we quantified the ratio of cells with MT curving to total cells and the ratio of arc length with curved MT to cell perimeter length. We found that Fign siRNA treatment resulted in a 256% increase in the cell number with turnover MTs and a 275% increase of the arc length with curved MT. In line with our overexpression studies (Figure 2C), depletion of Fign significantly increased the amount of tyr-MTs, which is more prominent in the peripheral region of the cell (Figure 4A). Statistical data showed a 47% increase of the fluorescence intensity for Tyr-MT mass in Fign siRNA–treated cells. This significant 42% increase of tyr-MTs was also confirmed by Western blotting (Figure 4B). As a parallel control, statistical analysis of immunostaining results showed no significant change for detyrosinated or acetylated MT mass when Fign was depleted (Figure 4C). In conclusion, Fign targets severing tyr-MTs in cultured astrocytes from rat spinal cord, with a particular effect on MT reorganization at the peripheral region of cultured astrocytes, which might affect their migration.
FIGURE 3:
Depletion of Fign leads to abnormal MT curving parallel to the plasma membrane in the cortical region of cultured astrocytes. (A) Fign protein was dramatically decreased after Fign knockdown in single cells. Top, representative images of immunostaining by Fign antibody in cultured astrocytes treated with Ctrl or Fign siRNA for 3 d (bar, 20 μm). Bottom, quantified data shown as mean ± SE. The relative fluorescence intensity of Ctrl siRNA–treated astrocytes was normalized as 1. Fign siRNA = 0.52 ± 0.05, n = 30, **p < 0.01. (B) Top, representative images of immunostaining by α-tubulin antibody in astrocytes treated with Ctrl siRNA or Fign siRNA. In contrast to no significant change of the tubulin skeleton in the Ctrl group, knockdown of Fign resulted in MT curving in the cell edge (white arrows indicate curved MT at the cortex region; bar, 20 μm. Left, bottom, ratio of cells with MT curving to total cells after Ctrl or Fign siRNA treatment; the data are shown as mean ± SE. Ctrl siRNA = 0.27 ± 0.03, Fign siRNA = 0.96 ± 0.04, n = 18, **p < 0.01. Right, bottom, ratio of arc length for curved MT to cell perimeter length after Ctrl or Fign siRNA treatment; the data are shown as mean ± SE. Ctrl siRNA = 0.16 ± 0.03, Fign siRNA = 0.60 ± 0.03, n = 18, **p < 0.01.
FIGURE 4:
Depletion of Fign increases tyr-MTs in the cortical region of cultured astrocytes. (A) Left, representative images of α-tubulin (green)– and tyr-tubulin (red)–expressing astrocytes treated with Ctrl siRNA and Fign siRNA. Compared to Ctrl group, depletion of Fign resulted in MTs curved in cell cortex regions, and these MTs are mainly composed of tyr-tubulin. Bar, 20 μm. Right, top, quantification of the relative fluorescence intensity of tyr-tubulin protein. Compared to the Ctrl siRNA–treated cells, Fign siRNA increased tyr-tubulin mass. The data are shown as mean ± SE. The relative fluorescence intensity of Ctrl siRNA–treated astrocytes is normalized as 1. Fign siRNA = 1.42 ± 0.14, n = 18, *p < 0.05. (B) Representative Western blotting result. Top, Fign siRNA treatment increased the ratio of tyr-tubulin compared with Ctrl siRNA treatment for 3 d. Bottom, tyr-tubulin protein increased significantly after Fign knockdown. The data are presented as mean ± SE. The relative tyr-tubulin protein level (α-tubulin as the control) of Ctrl siRNA–treated astrocytes was normalized as 1. Fign siRNA = 1.46 ± 0.22, n = 4, *p < 0.05. (C) Top, astrocytes treated with Ctrl or Fign siRNA for 3 d, fixed, and immunostained for detyrosinated or acetylated tubulin. To further illustrate that Fign depletion reduced tyr-tubulin mass, detyrosinated and acetylated tubulins were chosen as the controls. Bar, 20 μm. Bottom, quantification, with relative fluorescence intensity of Ctrl siRNA–treated astrocytes normalized as 1. There were no significantly different alterations for detyrosinated tubulin (bottom, left; Fign siRNA = 1.09 ± 0.07) or acetylated tubulin (bottom, right; Fign siRNA = 1.14 ± 0.06). n = 30; ns, not significant; p > 0.05.
Depletion of kinesin-12 rescues the migratory defect of astrocytes induced by Fign knockdown
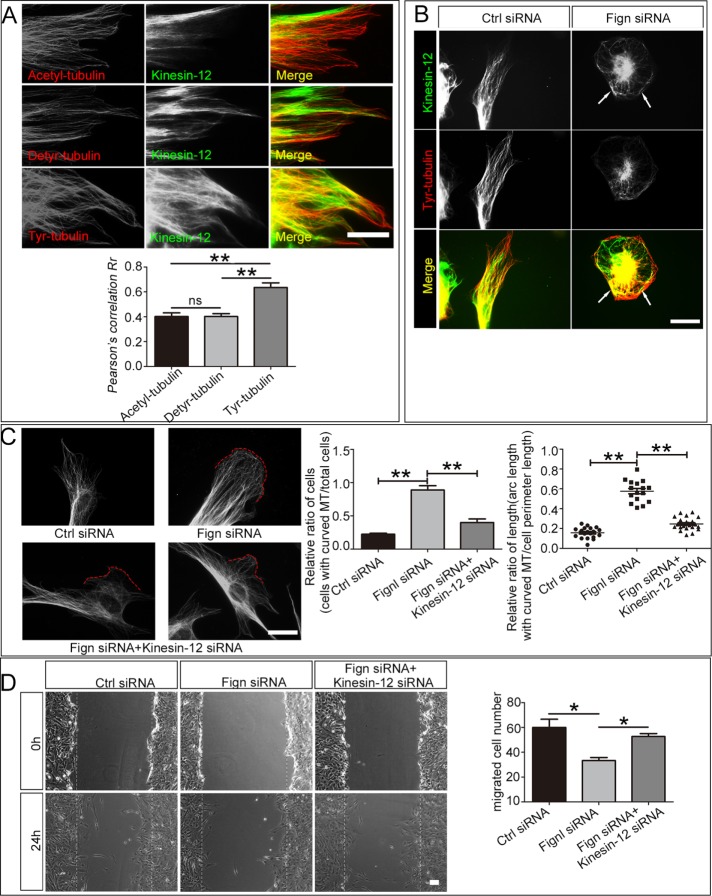
Kinesin-12 is a MT-based motor protein that contributes to mitosis by modulating spindle bipolarity (Sturgill and Ohi, 2013; Drechsler and McAinsh, 2016). We recently found that kinesin-12 knockdown promotes the migration of astrocytes by interacting with myosin-IIB, one of the actin-dependent motor proteins in the cell’s cortical region (Feng et al., 2016). We wondered whether kinesin-12 participates in MT reorganization together with Fign in the cell cortex. Immunostaining shows that kinesin-12 colocalized with tyr-MTs more significantly than with the other PTM-MTs, especially at the cell cortical region (Figure 5A). In particular, kinesin-12 colocalizes with curved tyr-tubulin–modified parts induced by Fign siRNA treatment (Figure 5B, arrows). It is conceivable that kinesin-12 serves as the sliding motor protein for the tyr-MTs at the cell peripheral region, whereas Fign serves as the severing protein for them. To test whether kinesin-12’s effect on Fign depletion induces a migratory defect, we applied gene-specific siRNAs to knock down kinesin-12 and Fign simultaneously. Kinesin-12 depletion rescued the MT-curving effect caused by Fign depletion (Figure 5C). We quantified the ratio of cell number with curved MTs (angle >302) against the cell membrane among all cells by referring to the study of Ruiz-Saenz et al. (2013). The ratio (shown in Figure 5C) decreases in the double-knockdown group compared with the Fign-knockdown group (Figure 5C, middle, graph). We also calculated the ratio of curved MT length to total MT length and found that it also decreased (statistical data in Figure 5C, right). These results indicated that kinesin-12 plays a pivotal role in serving as the sliding MT motor that causes the elongated tyr-MTs to curve up at the cell peripheral region when Fign is depleted. Next we examined the functional effects of the double knockdown on cell migration. The results of a wound-healing assay showed that kinesin-12 knockdown rescued the migratory defect of Fign-depleted astrocytes (Figure 5D). We compared the numbers of cells that enter the cell-free region after the scratch. Statistical data showed a 44% decrease in cell numbers in the Fign siRNA–treated group compared with control and only a 12% decrease in the group treated with Fign siRNA plus kinesin-12 siRNA, which indicated a partial rescue in the double-knockdown group.
FIGURE 5:
Kinesin-12 depletion reduced abnormal MT curving and rescued cell migration. (A) Colocalization analysis of kinesin-12 protein with different modified tubulins. Astrocytes were cultured on glass coverslips. After 24 h, the cells were fixed and immunostained by acetylated, detyrosinated, or tyrosinated tubulin together with kinesin-12 antibody. Top, representative images show immunostaining results of kinesin-12 antibody with acetyl-tubulin, detyr-tubulin, or Tyr-tubulin antibodies. Bar, 40 μm. To ascertain the degree of colocalization, statistical analysis of Pearson r correlations was used to quantify kinesin-12 and the different modifications of tubulin using ImageJ. Kinesin-12 colocalized with MTs rich in tyr-tubulin (0.64 ± 0.04) more than detyr-tubulin (0.40 ± 0.02) and acetylated tubulin (0.40 ± 0.03); n = 30; ns, not significant; **p < 0.01. (B) Representative images of kinesin-12 location in astrocytes after Fign depletion for 3 d. Immunofluorescence results show a consistent pattern of kinesin-12 followed by changeable tubulin, especially in the cell edge, where MT curved or bundled after Fign siRNA treatment (white arrows). (C) The effect of kinesin-12 on MT curving. Left, representative images of immunostaining by α-tubulin antibody in astrocytes treated with Ctrl siRNA, Fign siRNA, or Fign siRNA plus kinesin-12 siRNA. Double knockdown with Fign and kinesin-12 can partly rescue MT curving. In Ctrl group, MTs are perpendicular to the cell edge, whereas in the Fign siRNA group, MTs are parallel to the cell edge. Double depletion of Fign and kinesin-12 can recue MT curving (red line represents curving part). Bar, 20 μm. Middle, ratio of cells with MT curving to total cells after Ctrl or Fign siRNA or Fign siRNA plus kinesin-12 siRNA treatment. The data are shown as mean ± SE. Ctrl siRNA = 0.22 ± 0.01 (n = 20); Fign siRNA = 0.88 ± 0.06 (n = 15); Fign siRNA plus kinesin-12 siRNA = 0.39 ± 0.05 (n = 20), **p < 0.01. Right, ratio of arc length with curved MT to cell perimeter length after Ctrl, Fign siRNA, or Fign siRNA plus kinesin-12 siRNA treatment. The data are shown as mean ± SE. Ctrl siRNA = 0.16 ± 0.01; Fign siRNA = 0.58 ± 0.04; Fign siRNA plus kinesin-12 siRNA = 0.25 ± 0.01; n = 30, **p < 0.01. (D) Left, representative images of wound healing. Kinesin-12 depletion rescues cell migration deficiency caused by Fign protein knockdown (bar, 100 μm). Right, cell number entering the wound field after Ctrl, Fign siRNA, Fign siRNA plus kinesin-12 siRNA treatment. The data are shown as mean ± SE. Ctrl siRNA = 60.0 ± 6.67, Fign siRNA = 33.3 ± 2.40, Fign siRNA plus kinesin-12 siRNA = 52.7 ± 2.41; n = 3, *p < 0.05.
DISCUSSION
In the axon, MT-severing proteins regulate the length of MTs by cutting them in either their stable or labile domain, depending on the particular severing protein. In other cell types, whether stable and labile MT polymers exist as distinct domains on the same MT is unclear, but it makes sense that the longer the MT, the more stable it would be toward its minus end. Here we demonstrate further evidence that vertebrate Fign preferentially severs labile MT polymers, which could be MTs that are entirely or mostly labile or labile domains of MTs that also have a substantial stable domain. Fign-like 2, a closely related protein, was previously shown to participate in fibroblast movement but with knockdown producing the opposite phenotype to the one documented here for Fign knockdown from astrocytes. Unlike the case with Fign-like 2, whose depletion enhances fibroblast movement, the knockdown of Fign from cultured astrocytes inhibits their migration. Despite the close relationship between these two MT-severing proteins, their sequences apparently vary enough to result in this opposite effect on cell motility. Another possibility is cell type specificity in the response. We observed no defect of cell proliferation in the Fign-depleted astrocytes even though previous studies indicated that MT severing by Fign close to the centrosome is important to maintaining the integrity of the mitotic spindle in anaphase A (Mukherjee et al., 2012). Further investigation to examine Fign’s role, if any, in mitosis of astrocytes is needed, as is more information on potential differences between the properties of Fign and Fign-like 2 in different cell types.
As the major cell type in the CNS, astrocytes play an important role in the response of the CNS to injury in addition to serving a number of structural and functional roles. Astrocytes migrate into lesions of the injured spinal cord and form a physical barrier that significantly impedes axonal regeneration (Lin et al., 2014). Inhibition of astroglial scar formation could thereby create a relatively more favorable environment for nerve regeneration, allowing axons to partially overcome normally inhibitory cues (Li et al., 2016). Consistent with a recent report on cortical neurons (Leo et al., 2015), we observed no visible MT fragments resulting from severing when Fign was overexpressed in astrocytes (Figure 2). However, Leo et al. (2015) found that depletion of Fign gave rise to longer axons in cultured rat fetal cortical neurons, as well as a lower ratio of acetylated to total tubulin, indicating functional consequences of Fign levels. In the present study, we found that Fign targets tyrosinated MTs (or tyrosinated regions of MTs) for severing, especially those that are close to the plasma membrane. Tyrosinated tubulin is tubulin that has not (yet) been subjected to posttranslational detyrosination, an enzyme-driven process that generally occurs on MTs after they are stabilized. Thus MTs (or regions of MTs) that are especially rich in tyrosinated tubulin are typically labile (i.e., highly dynamic). The idea is that Fign produces a kind of trimming effect of the labile ends of the MTs, and hence Fign activity does not manifest as short MT fragments because the fragments depolymerize instantaneously upon the severing event. In neurons, the effects of Fign knockdown could be helpful clinically for augmenting axonal growth after injury, whereas in astrocytes, the effects could assist nerve regeneration in a different way: by reducing glial scar formation.
Our studies do not reveal whether it is the tyrosination state of the labile MTs that is recognized by Fign or whether Fign recognizes some other aspect of the labile MTs. In other words, Fign targets the tyrosinated MTs (or domains of MTs) that extend into the periphery of the astrocyte, but it may not be Fign targets that MT polymer because it is tyrosinated. Leo et al. (2015) presented some evidence that it may be the lack of acetylation of labile MTs that is recognized by Fign. Our Western blots show an increase in tyrosinated tubulin but not unacetylated tubulin when Fign is depleted from astrocytes, but this could reflect lower levels of acetylation of MTs in astrocytes relative to neurons. More work needs to be done on this.
The depletion of Fign from astrocytes in our study not only increased the tyr-MT level but also led to enhanced bending and curving of the MT polymers when they reached the cell periphery, where they normally remain relatively perpendicular to the plasma membrane (Figures 3 and 4). Presumably, the excessive growth of the MTs close to the leading edge underlies the migration defect of the astrocytes. This phenotype is reminiscent of the Drosophila katanin-knockdown phenotype, in which many bent MTs were seen in the peripheral regions (Zhang et al., 2011).
There is abundant evidence that dynamic activities of actin polymers, as well as the interaction between MTs and actin polymers, significantly affect cell movement. Actin often interacts with the plus end of the MTs, possibly via certain MT-end-binding proteins or even some motor proteins, to regulate cell migration (Bartolini et al., 2012; Morris et al., 2014; Cammarata et al., 2016). Cell migratory defects from Fign depletion are reminiscent of similar results from studies on depletion of small GTPases, such as Rac, at the leading edge of migratory cells. It has been reported that inhibition of Rac activity alters the reconfiguration of both MT actin filaments that are required for protrusion formation and focal adhesion readjustment (Waterman-Storer et al., 1999). Rapid polymerization of F-actin drives lamellipodia/lamella protrusions at the cell leading edge. Activated Rac 1, a member of the Rho family of small GTPases, accumulates in lamellipodia and stimulates local actin polymerization to promote cell migration (Nishimura et al., 2012). Activation of Rac 1 induces the formation of lamellipodia pioneer MTs (Wittmann et al., 2003). The results taken together show that, in migratory astrocytes, prolonged tyr-MTs induced by Fign deficiency, especially those close to the leading edge, curve parallel to the plasma membrane via certain forces. Miscommunication of the MTs and actin polymers arising from those abnormal curled or curved MTs at the leading edge substantially inhibits cell migration.
Kinesin-12, a kinesin best known for its role in mitosis, has been identified as a binding partner for myosin-IIB in astrocytes. Kinesin-12 mainly colocalizes with myosin-IIB at the lamellar region of cultured astrocytes, where it may play an important role as a functional mediator between MTs and actin filaments (Feng et al., 2016). Kinesin-12 is also considered a MT-sliding motor, contributing to axon growth and branch formation (Liu et al., 2010). Recently researchers reported that kinesin-12 operates on parallel MTs (Drechsler and McAinsh, 2016). Therefore we considered that kinesin-12 may provide the potential force responsible for the increase parallel curved MTs at the leading edge by virtue of its function and localization. Our initial immunocytochemical data revealed that kinesin-12 colocalizes more with tyr-MTs than any posttranslationally modified MTs. This colocalization seemed to be strengthened when Fign was depleted, presumably due to the increased tyr-MT level in the region.
An interesting question is why the elongated MTs at the cell periphery induced by Fign depletion undergo enhanced buckling and bending. Previous studies indicated that MTs in living cells can be bent by constant large-scale compressive forces; however, the bending wavelength can occasionally be reduced possibly via lateral reinforcement from surrounding elastic cytoskeletal elements (Wang et al., 2001; Gupton et al., 2002; Schaefer et al., 2002; Brangwynne et al., 2006). It is conceivable that the elongated MTs at the cell periphery lose sufficient support previously obtained from the surrounding environment. However, evidence from filament-gliding assays suggested that molecular motor proteins on MTs may contribute to the forces that bend the polymers, to the extent that they may even cause them to form loops (Liu et al., 2011). Our results demonstrated that depletion of kinesin-12 significantly rescued the cell migration defect possibly by reducing the curved and buckled MT polymer close to the peripheral region. This indicated that kinesin-12 might be an essential player in generating forces on the Fign-depletion–induced elongated MTs and causing the MTs to bend and curve. It is likely that kinesin-12 carries out this function by being an intermediator between MTs and actin filaments, given that it interacts with both polymers. Here we speculate that Fign regulates labile MT lengths in the cell cortical region in cultured migratory astrocytes (Figure 6). When Fign is present, most of the labile MTs stay perpendicular to the plasma membrane, and cells are able to move by responding to extracellular cues, whereas when Fign is absent, those labile MTs extend excessively and curve parallel to the plasma membrane by the sliding force of kinesin-12, thus compromising normal cellular movements (Figure 5D). Investigation into the concomitant changes of actin filaments and actin-related proteins followed by Fign depletion may contribute more insight.
FIGURE 6:
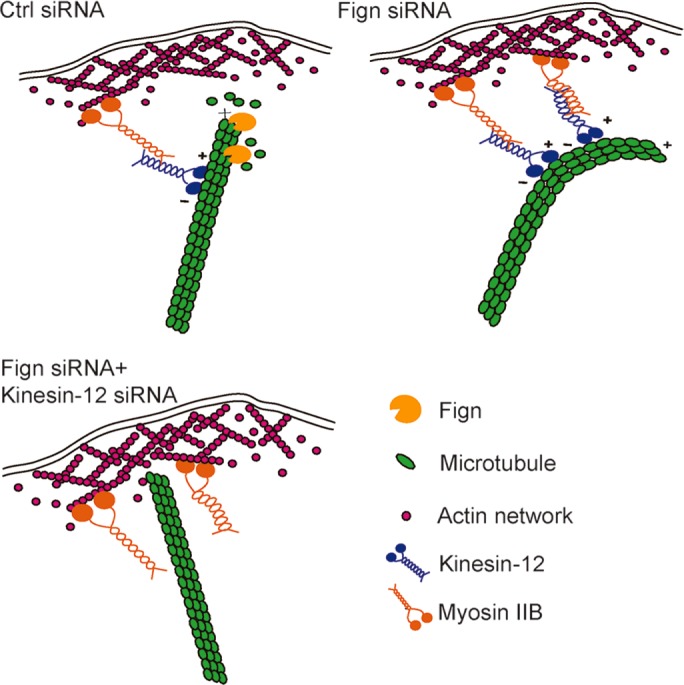
Model of how Fign regulates labile MT length in the cell cortical region in cultured migratory astrocytes. Fign plays a role in cultured astrocyte migration by targeting MTs rich in tyrosinated tubulin. When Fign is present, labile MTs rich in tyrosinated tubulin in the cell cortical region can be pruned properly and stay perpendicular to the plasma membrane, and the cells are able to move in response to extracellular cues. When Fign is depleted, those labile MTs become curved parallel to the plasma membrane by the sliding force of kinesin-12, so that cell movement is significantly decreased. If kinesin-12 protein is depleted together with Fign, the sliding forces derived from kinesin-12 and myosin-IIB interaction are unburdened. Cell migration can be partly rescued.
MATERIALS AND METHODS
Cell culture and reagents
Newborn rat pups (P1) were obtained from the Laboratory Animal Center of Nantong University (Nantong, China). All animal surgeries were conducted in accord with Institutional Animal Care guidelines, as well as with National Institutes of Health (Bethesda, MD) guidelines. Primary astrocytes from rat spinal cord were prepared as previously described (de Vellis and Cole, 2012), cultured in DMEM (Invitrogen) supplemented with 10% fetal bovine serum (FBS; Invitrogen), 0.5 mM glutamine (Invitrogen), and 1% penicillin–streptomycin (Invitrogen), and incubated in a humidified atmosphere of 95% air and 5% CO2 at 37°C. Briefly, spinal cords from P1 rat pups were isolated aseptically, and meninges were removed. The tissues were dissected out, digested, and then gently dropped through a sterile 75-μm Nitex mesh. The cell suspension was plated into tissue culture dishes. When cells became confluent, the cultures were shaken at 150 rpm for 16 h to purify the cultures. The purification of the subcultured astrocytes was confirmed by Glial fibrillary acidic protein (GFAP; Signalway Antibody) immunocytochemical staining, and astrocyte cultures were considered appropriate for use when they were 95% positive for GFAP.
Gene overexpression and siRNA treatment
The Fign and katanin-P60 full-length CDS constructs were obtained by RT-PCR from E18 rat spinal cords using gene-specific primers (listed in Table 1) designed from rat Fign (NM_001106484.1) and katanin-P60 (NM_001004217.4) genes. After being confirmed by DNA sequencing, the constructs of Fign and katanin-P60 was used to generate overexpression virus. Fign virus (LV5-Fign) and katanin-P60 virus (LV5-katanin-P60) were designed and produced by Genepharma (Suzhou, China) and applied into cultured astrocytes with the ratio of 1:10 (virus to medium). For the 24-well plates, 3 × 104 cells were plated and cultured for 18 h before addition of 20 µl of virus with 200 µl of culture medium (including 1/1000 Polybrene). After 24 h of incubation, we changed with fresh medium for the cell cultures. Three days after LV5-Fign or LV5-katanin p60 overexpression, astrocytes were replated on 0.1 mg/ml poly-l-lysine (PLL; Gibco)–coated slides to evaluate MTs with immunostaining (Liu et al., 2010).
TABLE 1:
Sequences of primers for constructs, siRNA, and qRT-PCR used in this study.
| Name | Sequence (5′-3′) |
|---|---|
| Fign full-length sense | ccgctcgagatgatcagtagcaccagtgtttatg (XhoI) |
| Fign full-length antisense | tccccgcggctgactgcaaccaaacattttg (SacII) |
| Katanin-P60 full-length sense | ccgctcgagctatgagcatgagtcttctaatgatt (XhoI) |
| Katanin-P60 full-length antisense | tccccgcggtctagcatgatccaaactcaact (SacII) |
| Ctrl siRNA sense | uucuccgaacgugucacguTT |
| Ctrl siRNA antisense | acgugacacguucggagaaTT |
| Fign siRNA sense | ggaucguauuugcauucuaTT |
| Fign siRNA antisense | uagaaugcaaauacgauccTT |
| Kinesin-12 siRNA sense | cagccauaauugcaaauguTT |
| Kinesin-12 siRNA antisense | acauuugcaauuauggcugTT |
| Fign qRT-PCR sense | tttgtgccaccagtaaacca |
| Fign qRT-PCR antisense | acgagcagtgcaaactcctt |
| GAPDH qRT-PCR sense | ccatcactgccactcagaagact |
| GAPDH qRT-PCR antisense | acattgggggtaggaacacg |
Letters in underlined italics indicate induced restriction enzyme–recognized sites.
All siRNAs used in this study were designed and synthesized by Biomics Biotechnologies (Nantong, China). The sequences of the universal control (Ctrl) siRNA, gene-specific Fign siRNA, and Fign qRT-PCR primers are listed in Table 1. The cultured astrocytes were transfected using Lipofectamine2000 (Invitrogen), according to the manufacturer’s instructions. After transfection for 24 h, the cells were replated on 35-mm culture dishes that had been coated with 0.1 mg/ml PLL, after which the cultures were expanded for the indicated number of days. The efficiency of Fign siRNA transfection was >80% as indicated by control siRNA–tagged fluorescein fluorescence, and the knockdown levels in the entire population were confirmed by qRT-PCR and Western blotting. After 2–3 d in culture, to permit protein depletion, cells were analyzed for morphological and functional effects.
Analyses of cell migration and proliferation
The methods of wound healing, Transwell, and live-cell imaging were used to evaluate astrocyte migration after siRNA treatment as described (Yu et al., 2012; Feng et al., 2016).
For the wound-healing assay, the astrocytes were dissociated, counted, resuspended in the fresh, prewarmed culture medium, and plated at a density of 30% in 35-mm plates. After cell transfection for 48 h, astrocytes were replated into a 24-well dish at a density of 70–90% and then wounded using 100-μl tips. After rinsing with phosphate-buffered saline (PBS) and changing with 2% FBS medium, cell migration behavior was recorded and imaged at 0 and 24 h. The number of astrocytes moving into the scratch-wound zone was used to evaluate the rate of migration.
For the Transwell experiment, astrocyte migration was examined using 6.5-mm Transwell chambers with 8-μm pores (Costar, Cambridge, MA). A 200-μl amount of 2% FBS-medium containing 3 × 104 astrocytes was transferred into the top chambers of each Transwell and was added together with 600 μl of complete medium into the lower cell-free chambers. After the cells were allowed to migrate for the indicated time, the nonmigrated cells on the upper surface of each membrane were cleaned with a cotton swab. Cells adhering to the bottom surface of each membrane were stained with 0.1% crystal violet, imaged, and counted using a DMR inverted microscope. Assays were performed three times, using triplicate wells once.
For live-cell imaging, astrocytes were cultured in a glass-bottomed culture dish (P35G-0-10-C; MatTek). During imaging, cells were maintained in complete medium at 37°C with a CO2-supported chamber. Images of living cells were taken with a DeltaVision microscopy system at 1 frame per 10 min. Measurements and statistical analyses were performed using ImageJ and GraphPad Prism. Statistical significance was determined by Student’s t test.
As described in our previous study (Feng et al., 2016), we evaluated astrocyte proliferation using a Cell-Light EdU DNA Cell Proliferation Kit (Ribobio, Guangzhou, China), following the manufacturer’s protocol. Briefly, the astrocytes were resuspended in fresh, prewarmed culture medium and plated at a density of 1 × 105 cells/ml in 96-well plates. At the indicated time after cell transfection, 50 µM EdU reagent was applied to the cells. After incubation for an additional 24 h, the cells were fixed with 4% formaldehyde in PBS for 30 min. The cells were then assayed, and astrocyte proliferation (ratio of EdU-positive cells to all cells) was analyzed by using images of randomly selected fields obtained on a DMR fluorescence microscope (Leica Microsystems, Bensheim, Germany).
Western blotting
For Western blotting, protein samples that had been quantified by the bicinchoninic acid method were separated using SDS–PAGE. After transfer to a polyvinylidene fluoride membrane (Millipore), the membrane was blocked with 5% nonfat dried milk in Tris-buffered saline (TBS; pH 7.4) and incubated overnight with the primary antibodies anti-α-tubulin (1:2000; ab18251, rabbit, polyclonal; Abcam, Cambridge, United Kingdom), tyrosinated tubulin (YL1/2) rat monoclonal antibody (1:2000; ab6160, rat, monoclonal; Abcam), anti–detyrosinated tubulin (AA12; 1:2000; ab24622, mouse, monoclonal, Abcam), anti–acetylated tubulin (6-11B-1; 1:2000; ab14610, mouse, monoclonal; Abcam), and anti-Fign antibody (1:400; sc-68343, rabbit, polyclonal; Santa Cruz Biotechnology) at 4°C. After washing with TBST (TBS with 0.1% Tween 20), IRDye-800–conjugated, affinity-purified goat anti-mouse immunoglobulin G (IgG; 1:3000; Rockland, Philadelphia, PA), goat anti-rabbit IgG (1:3000; Rockland), or goat anti-rat IgG (1:3000; Rockland) antibodies were used at room temperature for 2 h. The blot was covered with x-ray film and imaged using a scanner, and the data were analyzed with PDQuest 7.2.0 software (Bio-Rad). For normalization and relative quantitative analysis of target proteins expression, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or α-tubulin was used as the control protein.
Immunostaining
For investigating MT dynamics, cultures were briefly washed with prewarmed PBS three times and then simultaneously fixed and extracted with 4% paraformaldehyde, 0.2% glutaraldehyde, 1× PHEM (60 mM PIPES, 25 mM HEPES, 10 mM EGTA, and 2 mM MgSO4) buffer, and 0.1% Triton X-100 for 20 min. Cultures were then washed with PBS three times and blocked with 10% normal goat serum with 10 mg/ml bovine serum albumin for 1 h at 37°C (Feng et al., 2016). After blocking, cells transfected with Fign siRNA or overexpression virus were incubated with anti–α-tubulin (1:1000), anti–tyrosinated tubulin (1:1000), and anti-Fign (1:200) antibody overnight (4°C for 16 h). After 30 min of rewarming on the next day, cultures were rinsed with PBS three times and then incubated in the corresponding secondary antibodies (Cy3-conjugated goat anti-mouse IgG at 1:800, Cy3-conjugated goat anti-rabbit IgG at 1:800, 488-conjugated goat anti-mouse IgG at 1:600, 488-conjugated goat anti-rabbit IgG at 1:600, and Cy3-conjugated goat anti-rat IgG at 1:800; Jackson ImmunoResearch) for 2 h at room temperature. Then cells were washed with PBS three times and mounted in a medium that reduces photobleaching. Fluorescence images were obtained on an Olympus microscope using the identical camera, microscope, and imaging criteria, including gain, brightness and contrast, and exposure time.
The colocalization analysis of kinesin-12 protein with different modified tubulins followed Feng et al. (2016). Briefly, cells were fixed and stained for tyrosinated, detyrosinated, or acetylated tubulin. Images of both channels were obtained at 100× using a Leica microscope, transported to 16-bit TIFF, and analyzed in ImageJ for colocalization using colocalization analysis within flat regions of the cells. Background was subtracted from the region of interest using a factor of three within ImageJ before colocalization analysis. Pearson’s r correlation mean value is reported.
Statistical analyses
Comparisons between different experimental groups were analyzed by using repeated-measures analysis of variance (ANOVA) with GraphPad Prism 5.0 software (GraphPad Software, La Jolla, CA). All data were tested for normal distribution. If the data were normally distributed and had similar variances, then one-way ANOVA was performed to compare means among all measured variables. Data are expressed as mean ± SE of three to five separate experiments for each assay. p < 0.05 is considered statistically significant.
Acknowledgments
We thank Peter W. Baas of Drexel University (Philadelphia, PA) for helping to revise the manuscript. This study was supported by grants from the National Natural Science Foundation of China (31171007, 31371078) and the Priority Academic Program Development of Jiangsu Provincial Department of Education, Scientific Innovation Project (KYZZ_0354 in 2014) for Postgraduate Students in Jiangsu Province to Z.H.
Abbreviations used:
- AAA
ATPase associated with various cellular activities
- Acetyl-tubulin
acetylated tubulin
- Ctrl siRNA
control small interfering RNA
- Detyr-tubulin
detyrosinated tubulin
- E18
embryonic day 18
- Fign
fidgetin
- LV5-Fign
lentivirus 5-fidgetin
- LV5-NC
lentivirus 5-negative control
- PTM
posttranslational modifications
- Tyr-MT
tyrosinated microtubule
- Tyr-tubulin
tyrosinated tubulin.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-09-0628) on December 14, 2016.
REFERENCES
- Baas PW, Ahmad FJ. Beyond Taxol: microtubule-based treatment of disease and injury of the nervous system. Brain. 2013;136:2937–2951. doi: 10.1093/brain/awt153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolini F, Ramalingam N, Gundersen GG. Actin-capping protein promotes microtubule stability by antagonizing the actin activity of mDia1. Mol Biol Cell. 2012;23:4032–4040. doi: 10.1091/mbc.E12-05-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne CP, MacKintosh FC, Kumar S, Geisse NA, Talbot J, Mahadevan L, Parker KK, Ingber DE, Weitz DA. Microtubules can bear enhanced compressive loads in living cells because of lateral reinforcement. J Cell Biol. 2006;173:733–741. doi: 10.1083/jcb.200601060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammarata GM, Bearce EA, Lowery LA. Cytoskeletal social networking in the growth cone: how +TIPs mediate microtubule-actin cross-linking to drive axon outgrowth and guidance. Cytoskeleton (Hoboken) 2016;73:461–476. doi: 10.1002/cm.21272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charafeddine RA, Makdisi J, Schairer D, O’Rourke BP, Diaz-Valencia JD, Chouake J, Kutner A, Krausz A, Adler B, Nacharaju P, et al. Fidgetin-like 2: a microtubule-based regulator of wound healing. J Invest Dermatol. 2015;135:2309–2318. doi: 10.1038/jid.2015.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox GA, Mahaffey CL, Nystuen A, Letts VA, Frankel WN. The mouse fidgetin gene defines a new role for AAA family proteins in mammalian development. Nat Genet. 2000;26:198–202. doi: 10.1038/79923. [DOI] [PubMed] [Google Scholar]
- de Vellis J, Cole R. Preparation of mixed glial cultures from postnatal rat brain. Methods Mol Biol. 2012;814:49–59. doi: 10.1007/978-1-61779-452-0_4. [DOI] [PubMed] [Google Scholar]
- Drechsler H, McAinsh AD. Kinesin-12 motors cooperate to suppress microtubule catastrophes and drive the formation of parallel microtubule bundles. Proc Natl Acad Sci USA. 2016;113:E1635–E1644. doi: 10.1073/pnas.1516370113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Hu Z, Chen H, Hua J, Wu R, Dong Z, Qiang L, Liu Y, Baas PW, Liu M. Depletion of kinesin-12, a myosin-IIB-interacting protein, promotes migration of cortical astrocytes. J Cell Sci. 2016;129:2438–2447. doi: 10.1242/jcs.181867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupton SL, Salmon WC, Waterman-Storer CM. Converging populations of F-actin promote breakage of associated microtubules to spatially regulate microtubule turnover in migrating cells. Curr Biol. 2002;12:1891–1899. doi: 10.1016/s0960-9822(02)01276-9. [DOI] [PubMed] [Google Scholar]
- Ikegami K, Setou M. Unique posttranslational modifications in specialized microtubule architecture. Cell Struct Funct. 2010;35:15–22. doi: 10.1247/csf.09027. [DOI] [PubMed] [Google Scholar]
- Karabay A, Yu W, Solowska JM, Baird DH, Baas PW. Axonal growth is sensitive to the levels of katanin, a protein that severs microtubules. J Neurosci. 2004;24:5778–5788. doi: 10.1523/JNEUROSCI.1382-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leo L, Yu W, D’Rozario M, Waddell EA, Marenda DR, Baird MA, Davidson MW, Zhou B, Wu B, Baker L, et al. Vertebrate fidgetin restrains axonal growth by severing labile domains of microtubules. Cell Rep. 2015;12:1723–1730. doi: 10.1016/j.celrep.2015.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Cao Y, Shen F, Wang Y, Bai L, Guo W, Bi Y, Lv G, Fan Z. Mdivi-1 inhibits astrocyte activation and astroglial scar formation and enhances axonal regeneration after spinal cord injury in rats. Front Cell Neurosci. 2016;10:241. doi: 10.3389/fncel.2016.00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin B, Xu Y, Zhang B, He Y, Yan Y, He MC. MEK inhibition reduces glial scar formation and promotes the recovery of sensorimotor function in rats following spinal cord injury. Exp Ther Med. 2014;7:66–72. doi: 10.3892/etm.2013.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Tuzel E, Ross JL. Loop formation of microtubules during gliding at high density. J Phys Condensed Matter. 2011;23:374104. doi: 10.1088/0953-8984/23/37/374104. [DOI] [PubMed] [Google Scholar]
- Liu M, Nadar VC, Kozielski F, Kozlowska M, Yu W, Baas PW. Kinesin-12, a mitotic microtubule-associated motor protein, impacts axonal growth, navigation, and branching. J Neurosci. 2010;30:14896–14906. doi: 10.1523/JNEUROSCI.3739-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris EJ, Nader GP, Ramalingam N, Bartolini F, Gundersen GG. Kif4 interacts with EB1 and stabilizes microtubules downstream of Rho-mDia in migrating fibroblasts. PLoS One. 2014;9:e91568. doi: 10.1371/journal.pone.0091568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, Diaz Valencia JD, Stewman S, Metz J, Monnier S, Rath U, Asenjo AB, Charafeddine RA, Sosa HJ, Ross JL, et al. Human Fidgetin is a microtubule severing the enzyme and minus-end depolymerase that regulates mitosis. Cell Cycle. 2012;11:2359–2366. doi: 10.4161/cc.20849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura Y, Applegate K, Davidson MW, Danuser G, Waterman CM. Automated screening of microtubule growth dynamics identifies MARK2 as a regulator of leading edge microtubules downstream of Rac1 in migrating cells. PLoS One. 2012;7:e41413. doi: 10.1371/journal.pone.0041413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roll-Mecak A. Intrinsically disordered tubulin tails: complex tuners of microtubule functions? Semin Cell Dev Biol. 2015;37:11–19. doi: 10.1016/j.semcdb.2014.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Saenz A, van Haren J, Sayas CL, Rangel L, Demmers J, Millan J, Alonso MA, Galjart N, Correas I. Protein 4.1R binds to CLASP2 and regulates dynamics, organization and attachment of microtubules to the cell cortex. J Cell Sci. 2013;126:4589–4601. doi: 10.1242/jcs.120840. [DOI] [PubMed] [Google Scholar]
- Schaefer AW, Kabir N, Forscher P. Filopodia and actin arcs guide the assembly and transport of two populations of microtubules with unique dynamic parameters in neuronal growth cones. J Cell Biol. 2002;158:139–152. doi: 10.1083/jcb.200203038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp DJ, Ross JL. Microtubule-severing enzymes at the cutting edge. J Cell Sci. 2012;125:2561–2569. doi: 10.1242/jcs.101139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirajuddin M, Rice LM, Vale RD. Regulation of microtubule motors by tubulin isotypes and post-translational modifications. Nat Cell Biol. 2014;16:335–344. doi: 10.1038/ncb2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgill EG, Ohi R. Kinesin-12 differentially affects spindle assembly depending on its microtubule substrate. Curr Biol. 2013;23:1280–1290. doi: 10.1016/j.cub.2013.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudo H, Baas PW. Acetylation of microtubules influences their sensitivity to severing by katanin in neurons and fibroblasts. J Neurosci. 2010;30:7215–7226. doi: 10.1523/JNEUROSCI.0048-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vaart B, Akhmanova A, Straube A. Regulation of microtubule dynamic instability. Biochem Soc Trans. 2009;37:1007–1013. doi: 10.1042/BST0371007. [DOI] [PubMed] [Google Scholar]
- Wang N, Naruse K, Stamenovic D, Fredberg JJ, Mijailovich SM, Tolic-Norrelykke IM, Polte T, Mannix R, Ingber DE. Mechanical behavior in living cells consistent with the tensegrity model. Proc Natl Acad Sci USA. 2001;98:7765–7770. doi: 10.1073/pnas.141199598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Noritake J, Kaibuchi K. Regulation of microtubules in cell migration. Trends Cell Biol. 2005;15:76–83. doi: 10.1016/j.tcb.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Waterman-Storer CM, Worthylake RA, Liu BP, Burridge K, Salmon ED. Microtubule growth activates Rac1 to promote lamellipodial protrusion in fibroblasts. Nat Cell Biol. 1999;1:45–50. doi: 10.1038/9018. [DOI] [PubMed] [Google Scholar]
- Wittmann T, Bokoch GM, Waterman-Storer CM. Regulation of leading edge microtubule and actin dynamics downstream of Rac1. J Cell Biol. 2003;161:845–851. doi: 10.1083/jcb.200303082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Qian T, Wang Y, Zhou S, Ding G, Ding F, Gu X. miR-182 inhibits Schwann cell proliferation and migration by targeting FGF9 and NTM, respectively at an early stage following sciatic nerve injury. Nucleic Acids Res. 2012;40:10356–10365. doi: 10.1093/nar/gks750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Qiang L, Solowska JM, Karabay A, Korulu S, Baas PW. The microtubule-severing proteins spastin and katanin participate differently in the formation of axonal branches. Mol Biol Cell. 2008;19:1485–1498. doi: 10.1091/mbc.E07-09-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Grode KD, Stewman SF, Diaz-Valencia JD, Liebling E, Rath U, Riera T, Currie JD, Buster DW, Asenjo AB, et al. Drosophila katanin is a microtubule depolymerase that regulates cortical-microtubule plus-end interactions and cell migration. Nat Cell Biol. 2011;13:361–370. doi: 10.1038/ncb2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Rogers GC, Buster DW, Sharp DJ. Three microtubule severing enzymes contribute to the “Pacman-flux” machinery that moves chromosomes. J Cell Biol. 2007;177:231–242. doi: 10.1083/jcb.200612011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Shao H, Zhu T, Xia P, Wang Z, Liu L, Yan M, Hill DL, Fang G, Chen Z, et al. DDA3 associates with microtubule plus ends and orchestrates microtubule dynamics and directional cell migration. Sci Rep. 2013;3:1681. doi: 10.1038/srep01681. [DOI] [PMC free article] [PubMed] [Google Scholar]