Abstract
Metabolomics continues to make rapid progress through the development of new and better methods and their applications to gain insight into the metabolism of a wide range of different biological systems from a systems biology perspective. Customization of NMR databases and search tools allows the faster and more accurate identification of known metabolites, whereas the identification of unknowns, without a need for extensive purification, requires new strategies to integrate NMR with mass spectrometry, cheminformatics, and computational methods. For some applications, the use of covalent and non-covalent attachments in the form of labeled tags or nanoparticles can significantly reduce the complexity of these tasks.
Graphical Abstract
INTRODUCTION
Metabolomics, also known as metabonomics, has emerged as a key discipline to address a wide range of scientific questions in biological, biomedical, environmental, agricultural, and nutritional research. This is largely because of technological advances in high-resolution analytical instrumentation, especially nuclear magnetic resonance (NMR) and mass spectrometers (MS), which allow the separate detection of hundreds of signals belonging to different metabolites in complex metabolite mixtures.[1] Detected signals can be directly analyzed by chemometric and statistical analysis tools for the determination of patterns that are characteristic for samples from different subgroups, such as wild type vs. mutant, young vs. old, or healthy vs. diseased.[2] Such pattern-based approaches allow the rapid diagnostic application of metabolomics without the need to identify the chemical content of a sample. In order to relate observed changes to the underlying biochemical pathways, it is crucial to be able to accurately and comprehensively identify the metabolites present in the samples. This step often starts with the querying of experimental NMR or MS signals against metabolomics databases, which contain spectral information of many of the known metabolites. Over recent years, many metabolite repositories and querying platforms have undergone substantial expansions,[3–7] which led to greatly improved querying results both in terms of increased true positive rates and decreased false positive rates. Known metabolites that can be produced through alternative pathways can be tracked by the use of 13C-or 15N-enriched precursors, such as 13C6-glucose or 13C5,15N2-glutamine by using stable isotope-resolved metabolomics (SIRM) analysis. The precise labeling patterns of the products provides important insights into the regulatory mechanisms of these pathways in healthy vs. cancer cells.[8]
Still, many of the signals found in NMR and MS spectra of most metabolomics samples belong to molecules that are not present in metabolomics databases.[9] Identification of these “unknown” molecules is notably hard and is widely recognized as a central bottleneck toward further progress in the metabolomics field. NMR spectroscopy has a long track record in the structure elucidation of unknown biological molecules and has been commonly used for structure determination of isolated compounds in the context of natural product research.[10] However, the separation process is time consuming, labor-intensive and some metabolites can be modified or lose their activity during extraction.[11] In the context of metabolomics, a primary goal is to perform the efficient and accurate identification of both known and unknown metabolites in their complex mixture environment with little or no prior purification.
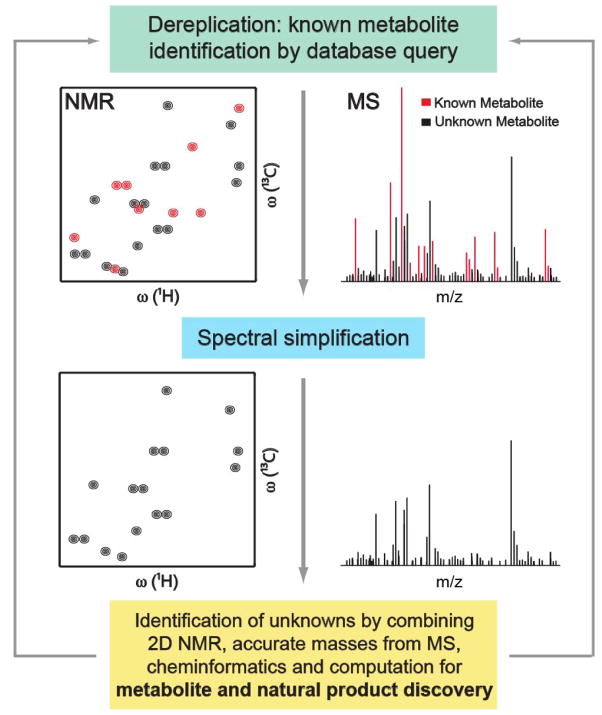
Metabolomics databases permit the rapid identification of a maximal number of metabolites. The remaining unexplained NMR and MS peaks belong to unknowns. This two-step strategy reduces the number of NMR and MS signals that have to be analyzed during structure elucidation in the presence of a complex mixture by dereplication, i.e. the discard of known compounds.[12]. Newly discovered metabolites are then added to metabolomics databases providing easy access to an ever larger number of known metabolites to study their role under various biological conditions (Figure 1). Over the past couple of years important advances have been made in this area. This review summarizes some of these advances and points out remaining challenges.
Figure 1.
Integrated metabolomics workflow for the identification of known and unknown metabolites in complex mixtures. Combined use of metabolomics databases with experimental NMR and MS spectra (e.g., NMR/MS Translator [27]) allows the rapid identification of a maximal number of known metabolites present in the mixture, while unidentified signals are used as fingerprints of unknowns. Next, structures of unknown metabolites can be elucidated or vastly narrowed down by the combined use of multidimensional NMR, MS, cheminformatics, and computation (e.g., SUMMIT MS/NMR [35]).
Identification of known metabolites by multidimensional NMR
The metabolite identification procedure in metabolomics usually starts with querying experimental signals against metabolomics databases to assign as many metabolites as possible. One-dimensional (1D) 1H NMR spectroscopy is the most popular NMR experiment and one of the most widely used metabolomics experiments overall, allowing high-throughput applications by measuring hundreds or even thousands of samples, since it takes only of the order of a few minutes per sample. However, identification of metabolites in complex mixtures solely based on 1D 1H NMR can be challenging because of severe peak overlap in crowded regions of the spectra. Alternatively, more reliable metabolite identification can be achieved by going from 1D to two-dimensional (2D) NMR experiments, whereby spectral resolution is enhanced at the expense of prolonged experiment times.[13–15] In fact, a growing number of metabolomics studies relies almost exclusively on 2D NMR experiments for the accurate and comprehensive identification of metabolites.[16–20]
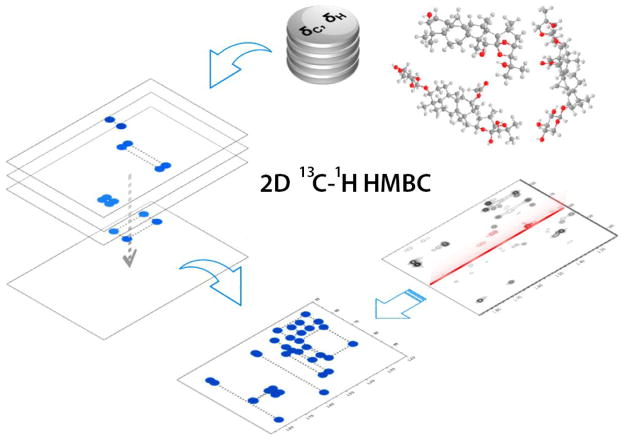
A recent development in NMR-based metabolite identification has been the introduction of customized metabolomics databases designed for the querying of particular types of 2D NMR experiments, which has resulted in improved compound identification accuracy. The SpinCouple metabolomics database has been introduced recently for the analysis of 2D J-resolved (Jres) 1H NMR spectra.[21] SpinCouple as well as the Birmingham Metabolite Library[22] allow querying of 2D Jres 1H NMR spectra of complex metabolite mixtures against 2D Jres 1H NMR spectra of metabolite standards. A 2D 13C-1H heteronuclear multiple bond correlation (HMBC) database was introduced for the identification of triterpenes from Actaea species. This database was generated by analyzing four commonly observed HMBC correlations in triterpene structures, which together produce a rectangular pattern with four HMBC cross-peaks unique to each triterpene (Figure 2). By using these patterns as “barcodes”, the authors successfully identified different catalogued triterpenes in A. racemosa extract.[23]
Figure 2.
2D 13C-1H HMBC barcoding approach. The 2D NMR data of reference compounds were analyzed to generate a database of rectangular patterns (barcodes on the left) with four HMBC cross-peaks unique to each triterpene. Next, triterpene components in A. racemosa extract were identified by in silico matching of experimental NMR signals with the reference barcodes (bottom). Adapted with permission from [23]. Copyright 2014 American Chemical Society.
The 13C-TOCCATA customized database is optimized for the querying of 13C-13C TOCSY spectra of uniformly 13C labeled metabolomics samples,[24] whereas the 1H(13C)-TOCCATA customized database permits the querying of 1H-1H TOCSY and 13C-1H HSQC-TOCSY spectra of complex metabolite mixtures at natural 13C abundance.[25] A novelty in the TOCCATA databases is that they sort the spectral information of each metabolite into individual spin systems and slowly interchanging isomers. Since any selected cross-section of the 2D TOCSY spectrum reflects the 1D spectrum of a single spin system or an individual isomer rather than the entire 1D spectrum of a given compound, it increases the accuracy (reliability) and confidence of metabolite identification. For a closely related set of metabolites, the COLMAR 13C-1H HSQC metabolomics database has been introduced for querying 13C-1H HSQC spectra. COLMAR 13C-1H HSQC database sorts HSQC spectra of metabolites into their individual slowly interchanging isomeric states: for example, α- and β-glucose are queried like two separate molecules. This improves the query result, since it is insensitive to the relative populations of different isomers which sometimes can be quite uneven, i.e. the unambiguous detection of one isomer per compound is all that is needed for query if the other isomer(s) is below the detection limit.[26] As these databases continue to grow, it will be important to integrate them so that multiple experiments collected on the same sample by NMR, as well as other methods, can be simultaneously queried, which will further improve the accuracy and confidence of the top hits of these queries.
Identification of known metabolites by hybrid NMR/MS
Depending on the type of sample at hand, querying of experimental 2D NMR spectra against metabolomics databases often achieves unambiguous identification of a majority of the catalogued metabolites. However, this strategy fails when two or more metabolites have very similar chemical shifts, which is often because they share a highly similar structure as is the case, e.g., for creatine vs. creatine-phosphate or ADP vs. ATP. On the other hand, these metabolites often have different mass-to-charge (m/z) ratios and, hence, it should be possible to differentiate them when including information from mass spectrometry (MS) data. An automated hybrid NMR/MS approach, the “NMR/MS Translator”, has recently been introduced for this task. The NMR/MS Translator first generates metabolite candidates from experimental 1D and/or 2D NMR spectra by NMR database query, which is followed by the prediction of the masses (m/z) of all likely ions and adducts of metabolite candidates with their characteristic isotope distributions. The expected m/z ratios are then compared with the experimental MS1 spectrum for the direct assignment of those signals of the mass spectrum that correspond to the metabolites generated from the NMR spectra. In this way, the MS and NMR spectra are simultaneously assigned in a manner that can be fully automated. By co-analysis of chemical shift and accurate mass data, 88 metabolites could be identified that were consistent with both NMR and MS data, thereby increasing the confidence of these metabolites over the use of either method alone.[27]
Identification of unknown metabolites by multidimensional NMR
Identification of unknown metabolites is a critically important objective as it provides information about altered or new biochemical pathways, along with their associated enzymes, or the previously unknown influx of exogenous metabolites. Their identification in complex mixtures requires deconvolution of the NMR spectrum into subsets of signals where each subset can be attributed to an individual metabolite. This can be done by physical separation of a mixture, for example by high-performance liquid chromatography (HPLC), followed by the acquisition of the NMR and MS spectra of individual fractions. This approach has proven useful in natural product discovery pipelines, e.g. using commercial LC-MS-SPE-NMR instruments,[28, 29] but the utility of this approach is limited in the context of high-throughput applications. Alternatively, the NMR spectrum of a mixture can be deconvoluted into individual components by acquiring high-resolution 2D NMR spectra. Experiments providing connectivity information such as 2D 1H-1H TOCSY, 2D 13C-1H HSQC-TOCSY, and 2D 13C-1H HMBC allow deconvolution of the signals of each mixture component without the need for physical separation.[30]
2D 13C-13C CT-TOCSY spectroscopy has been used for the deconvolution and characterization of the backbone topologies of unknown molecules in metabolomics samples toward the elucidation of metabolite structures in complex mixtures. In a fully 13C-labeled E. coli cell lysate, it was possible to identify 112 individual carbon backbone topologies.[31] A similar strategy has been successfully applied to fully 13C-labeled C. elegans cell lysate by using the 2D 13C-13C INADEQUATE experiment, which correlates pairs of directly J-coupled 13C spins in the metabolites.[32] Determination of the carbon backbone topology of each spin system of a molecule represents the first step for structure elucidation. Next, one needs to connect multiple backbone topologies of each molecule by using long-range correlation experiments; moreover, one has to identify the (non-carbon) substituents (functional groups) that are attached to the carbon skeleton.[33] This has been achieved recently by combining 3D (H)CCH-TOCSY, 3D (H)CCH-COSY, 2D 13C-1H HSQC, 2D 13C-13C COSY, and 2D 13C-1H D-HMBC experiments. The authors used this set of experiments to elucidate complete structures of metabolites in a 13C-enriched R. japonicum extract. In addition, they used quantum chemical calculation to verify the identified metabolites by predicting chemical shifts of the structures for comparison with experiment.[34] Some of the structures of uncatalogued metabolites identified by this approach are depicted in Figure 3. These structure elucidation approaches have been demonstrated in 13C-labeled organism for which they work particularly well. Since many types of metabolomics samples cannot be easily 13C-labeled, alternative approaches that work also for natural abundance 13C samples, such as human urine and serum, are clearly needed.
Figure 3.
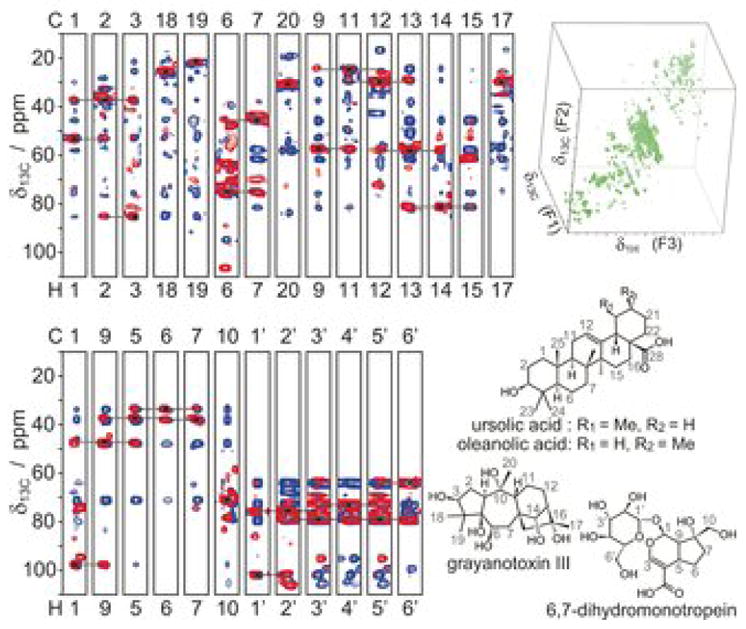
Chemical structures of uncatalogued metabolites identified in a 13C-enriched R. japonicum plant cell lysate by using 3D NMR spectroscopy and density functional theory-based chemical shift prediction. Adapted with permission from [34].
Identification of unknown metabolites by hybrid MS/NMR
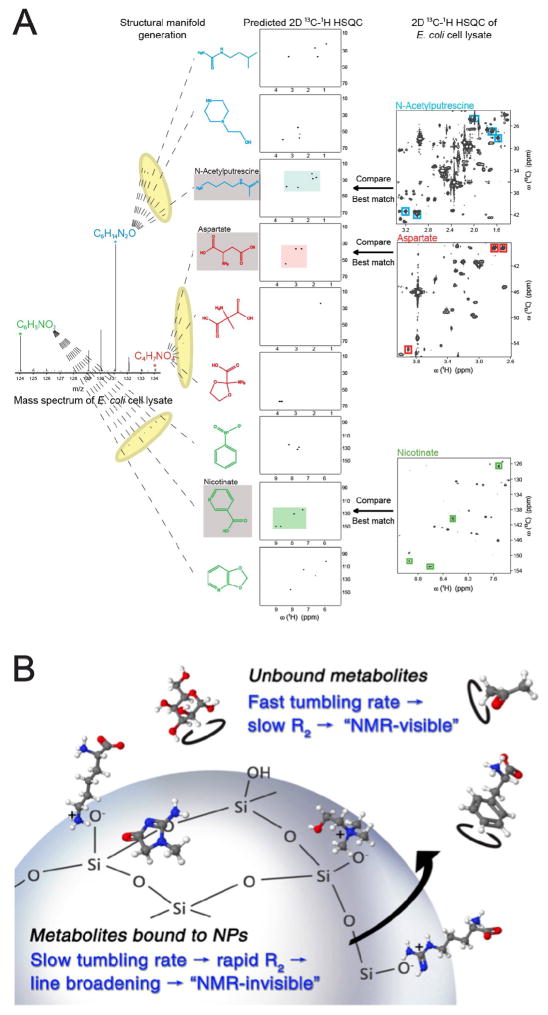
A hybrid MS/NMR metabolite identification strategy, SUMMIT MS/NMR, has been proposed, which works for both natural abundance 13C samples as well as 13C-labeled samples. The approach first extracts accurate masses of all detected metabolites from high-resolution mass spectra and generates all structures consistent with the derived chemical formulas (‘structural manifold’). The comparison of the predicted NMR spectra of all candidate structures with the experimental NMR spectra of the same sample permits identification of the structures present in the complex mixture of interest (Figure 4A). SUMMIT MS/NMR was applied to E. coli cell extract, where it correctly identified a wide range of different types of metabolites.[35] The results suggest that SUMMIT MS/NMR might be suitable for high-throughput applications for the discovery of new metabolites in biological and biomedical mixtures, without the need for experimental MS and NMR metabolite databases or extensive metabolite purification for the elucidation of the structures of unknown metabolites.
Figure 4.
(a) Schematic representation of the SUMMIT MS/NMR approach for the rapid and accurate identification of unknown metabolites in complex mixtures. Adapted with permission from [35]. Copyright 2015 American Chemical Society. (b) Nanoparticle-assisted NMR metabolomics approach for the selective suppression of the NMR signals of metabolites based on their charge and hydrophobicity. When metabolites interact (even only transiently) with the surface of charged silica nanoparticles they undergo rapid transverse relaxation, which results in substantial line broadening or complete peak disappearance in the NMR spectrum.[40]
Physical and chemical methods for spectral simplification
For highly complex mixtures, such as urine, the selection or suppression of certain classes of compounds with common chemical or physical properties can be of advantage. [36] Two 15N-labeled agents, namely the cholamine tag and the aminooxy probe, have been introduced that selectively and covalently attach to carboxyl and carbonyl group containing metabolites, respectively.[37, 38] This makes signals of these metabolites directly visible in 2D 15N-1H HSQC NMR spectra and mass spectra because of the introduction of a permanent charge, thereby linking NMR and MS signals of the modified metabolites. Other functional groups, such as amines and hydroxyls, have also been covalently tagged with 13C-labeled or 31P-containing tags for their unique detection.[39] Another recent approach uses attractive non-covalent interactions between certain types of metabolites and anionic or cationic silica nanoparticles (SNPs). The addition of the SNPs to the NMR sample leads to the disappearance of NMR signals of those metabolites that interact with the SNPs.[40] Even transiently bound metabolites experience strong line broadening of the metabolite resonances due to the very slow rotational diffusion of the much larger SNPs (Figure 4B). This approach selectively suppresses the NMR signals of metabolites that have opposite charge than the SNPs as well as certain metabolites that contain hydrophobic groups, such as multiple methyl groups. The disappearance of NMR peaks in the 1D or 2D spectra facilitates the analysis of the remaining peaks, while at the same time it permits the attribution of specific physical-chemical properties to metabolites affected by the SNP presence, especially electric charge, which is otherwise hard to measure by NMR at constant pH. The addition of SNPs to serum also helps eliminate protein from the NMR sample to produce clean spectra for quantitative analysis.[41] It should be possible to specifically target certain metabolite(s) of interest by coating nanoparticles with ligands that bind to them specifically. In fact, such a chemosensing strategy has been demonstrated recently for relatively simple synthetic mixtures.[42] This research direction has strong potential by exploiting synergies between synthetic chemistry, nanoscience, and metabolomics.
OUTLOOK
NMR-based metabolomics is making rapid progress on multiple fronts. The expansion of NMR databases with their content customized for different types of NMR experiments remains an important, although time-consuming task. Identification of unknowns clearly remains one of the biggest challenges of present day metabolomics. The combined use of MS and NMR methods to the same sample,[43, 44] along with advanced statistical,[45–48] cheminformatics[27],[35] and spectral prediction methods[49,50], promises to considerably shorten the arduous, but rewarding path to the structure determination of unknowns. All these scientific and technological advances provide rich and urgently needed information about the metabolic make-up of life. In the biomedical field, they already play increasingly important roles in disease diagnostics and will be gradually employed to guide intervention.
Highlights.
Power of 2D NMR permits the study of both known and unknown metabolites.
Known metabolites are reliably identified using customized metabolomics databases.
Combining NMR, MS, computation with cheminformatics offers roadmap toward unknowns.
Use of additives such as reactive agents or charged nanoparticles can be beneficial.
Acknowledgments
We thank Mouzhe Xie for preparing Figure 4B. This work was supported by the National Institutes of Health (grant R01 GM 066041 and SECIM grant U24 DK097209-01A1).
Footnotes
CONFLICTS OF INTEREST
There are no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bingol K, Brüschweiler R. Two elephants in the room: new hybrid nuclear magnetic resonance and mass spectrometry approaches for metabolomics. Curr Opin Clin Nutr Metab Care. 2015;18:471–477. doi: 10.1097/MCO.0000000000000206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Veselkov KA, McKenzie JS, Nicholson JK. Multivariate Data Analysis Methods for NMR-based Metabolic Phenotyping in Pharmaceutical and Clinical Research. eMagRes. 2015;4:323–334. [Google Scholar]
- 3.Ulrich EL, Akutsu H, Doreleijers JF, Harano Y, Ioannidis YE, Lin J, Livny M, Mading S, Maziuk D, Miller Z, et al. BioMagResBank. Nucleic Acids Res. 2008;36:D402–408. doi: 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cui Q, Lewis IA, Hegeman AD, Anderson ME, Li J, Schulte CF, Westler WM, Eghbalnia HR, Sussman MR, Markley JL. Metabolite identification via the Madison Metabolomics Consortium Database. Nat Biotechnol. 2008;26:162–164. doi: 10.1038/nbt0208-162. [DOI] [PubMed] [Google Scholar]
- 5.Chikayama E, Sekiyama Y, Okamoto M, Nakanishi Y, Tsuboi Y, Akiyama K, Saito K, Shinozaki K, Kikuchi J. Statistical indices for simultaneous large-scale metabolite detections for a single NMR spectrum. Anal Chem. 2010;82:1653–1658. doi: 10.1021/ac9022023. [DOI] [PubMed] [Google Scholar]
- 6.Tautenhahn R, Cho K, Uritboonthai W, Zhu ZJ, Patti GJ, Siuzdak G. An accelerated workflow for untargeted metabolomics using the METLIN database. Nat Biotechnol. 2012;30:826–828. doi: 10.1038/nbt.2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, Liu Y, Djoumbou Y, Mandal R, Aziat F, Dong E, et al. HMDB 3. 0--The Human Metabolome Database in 2013. Nucleic Acids Res. 2013;41:D801–807. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8*.Fan TWM, Lane AN. Applications of NMR spectroscopy to systems biochemistry. Prog Nucl Magn Reson Spectrosc. 2016;92–93:18–53. doi: 10.1016/j.pnmrs.2016.01.005. A comprehensive review covering NMR methods to profile metabolites, their isotopomer distributions and biochemical pathways by stable isotope-resolved metabolomics (SIRM) analysis along with applications to different biological systems. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bingol K, Bruschweiler-Li L, Li DW, Zhang B, Xie M, Brüschweiler R. Emerging new strategies for successful metabolite identification in metabolomics. Bioanalysis. 2016;8:557–573. doi: 10.4155/bio-2015-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robinette SL, Brüschweiler R, Schroeder FC, Edison AS. NMR in metabolomics and natural products research: two sides of the same coin. Acc Chem Res. 2012;45:288–297. doi: 10.1021/ar2001606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Srinivasan J, Kaplan F, Ajredini R, Zachariah C, Alborn HT, Teal PE, Malik RU, Edison AS, Sternberg PW, Schroeder FC. A blend of small molecules regulates both mating and development in Caenorhabditis elegans. Nature. 2008;454:1115–1118. doi: 10.1038/nature07168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaudencio SP, Pereira F. Dereplication: racing to speed up the natural products discovery process. Nat Prod Rep. 2015;32:779–810. doi: 10.1039/c4np00134f. [DOI] [PubMed] [Google Scholar]
- 13.Bingol K, Zhang F, Bruschweiler-Li L, Brüschweiler R. Quantitative analysis of metabolic mixtures by two-dimensional 13C constant-time TOCSY NMR spectroscopy. Anal Chem. 2013;85:6414–6420. doi: 10.1021/ac400913m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bingol K, Brüschweiler R. Multidimensional approaches to NMR-based metabolomics. Anal Chem. 2014;86:47–57. doi: 10.1021/ac403520j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dubey A, Rangarajan A, Pal D, Atreya HS. Pattern recognition-based approach for identifying metabolites in nuclear magnetic resonance-based metabolomics. Anal Chem. 2015;87:7148–7155. doi: 10.1021/acs.analchem.5b00990. [DOI] [PubMed] [Google Scholar]
- 16.Castillo DA, Kolesnikova MD, Matsuda SP. An effective strategy for exploring unknown metabolic pathways by genome mining. J Am Chem Soc. 2013;135:5885–5894. doi: 10.1021/ja401535g. [DOI] [PubMed] [Google Scholar]
- 17.Guennec AL, Giraudeau P, Caldarelli S. Evaluation of fast 2D NMR for metabolomics. Anal Chem. 2014;86:5946–5954. doi: 10.1021/ac500966e. [DOI] [PubMed] [Google Scholar]
- 18.Pudakalakatti SM, Dubey A, Jaipuria G, Shubhashree U, Adiga SK, Moskau D, Atreya HS. A fast NMR method for resonance assignments: application to metabolomics. J Biomol NMR. 2014;58:165–173. doi: 10.1007/s10858-014-9814-6. [DOI] [PubMed] [Google Scholar]
- 19.Wen H, An YJ, Xu WJ, Kang KW, Park S. Real-time monitoring of cancer cell metabolism and effects of an anticancer agent using 2D in-cell NMR spectroscopy. Angew Chem Int Ed. 2015;54:5374–5377. doi: 10.1002/anie.201410380. [DOI] [PubMed] [Google Scholar]
- 20.Jin X, Kang S, Tanaka S, Park S. Monitoring the Glutathione Redox Reaction in Living Human Cells by Combining Metabolic Labeling with Heteronuclear NMR. Angew Chem Int Ed. 2016;55:1–5. doi: 10.1002/anie.201601026. [DOI] [PubMed] [Google Scholar]
- 21*.Kikuchi J, Tsuboi Y, Komatsu K, Gomi M, Chikayama E, Date Y. SpinCouple: development of a web tool for analyzing metabolite mixtures via two-dimensional J-resolved NMR database. Anal Chem. 2016;88:659–665. doi: 10.1021/acs.analchem.5b02311. This sizeable database allows querying of 2D Jres 1H NMR spectra of complex metabolite mixtures. 2D Jres 1H NMR is one of the oldest homonuclear 1H experiments, which at natural 13C abundance has higher sensitivity than a 2D 13C-1H HSQC. Since its resolution is much lower, it is particularly useful for the analysis of low concentration, low complexity mixtures. [DOI] [PubMed] [Google Scholar]
- 22.Ludwig C, Easton JM, Lodi A, Tiziani S, Manzoor SE, Southam AD, Byrne JJ, Bishop LM, He S, Arvanitis TN, et al. Birmingham Metabolite Library: a publicly accessible database of 1-D H-1 and 2-D H-1 J-resolved NMR spectra of authentic metabolite standards (BML-NMR) Metabolomics. 2012;8:8–18. [Google Scholar]
- 23*.Qiu F, McAlpine JB, Lankin DC, Burton I, Karakach T, Chen SN, Pauli GF. 2D NMR barcoding and differential analysis of complex mixtures for chemical identification: the Actaea triterpenes. Anal Chem. 2014;86:3964–3972. doi: 10.1021/ac500188j. This study introduces a 2D 13C-1H HMBC-based database with querying for the identification of known triterpenes in plant extracts. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bingol K, Zhang F, Bruschweiler-Li L, Brüschweiler R. TOCCATA: a customized carbon total correlation spectroscopy NMR metabolomics database. Anal Chem. 2012;84:9395–9401. doi: 10.1021/ac302197e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bingol K, Bruschweiler-Li L, Li DW, Brüschweiler R. Customized metabolomics database for the analysis of NMR 1H-1H TOCSY and 13C-1H HSQC-TOCSY spectra of complex mixtures. Anal Chem. 2014;86:5494–5501. doi: 10.1021/ac500979g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26*.Bingol K, Li DW, Bruschweiler-Li L, Cabrera OA, Megraw T, Zhang F, Brüschweiler R. Unified and isomer-specific NMR metabolomics database for the accurate analysis of 13C-1H HSQC spectra. ACS Chem Biol. 2015;10:452–459. doi: 10.1021/cb5006382. This database combines data from BMRB and HMDB and annotates metabolites isomer specifically, thereby improving the querying result since it is insensitive to the relative populations of different isomers. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27*.Bingol K, Brüschweiler R. NMR/MS Translator for the enhanced simultaneous analysis of metabolomics mixtures by NMR spectroscopy and mass spectrometry: application to human urine. J Proteome Res. 2015;14:2642–2648. doi: 10.1021/acs.jproteome.5b00184. This study introduces a hybrid NMR/MS approach that offers the rapid and accurate identification of known metabolites detected in both NMR and MS spectra of the same sample. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van der Hooft JJJ, de Vos RCH, Ridder L, Vervoort J, Bino RJ. Structural elucidation of low abundant metabolites in complex sample matrices. Metabolomics. 2013;9:1009–1018. [Google Scholar]
- 29.Sumner LW, Lei Z, Nikolau BJ, Saito K. Modern plant metabolomics: advanced natural product gene discoveries, improved technologies, and future prospects. Nat Prod Rep. 2015;32:212–229. doi: 10.1039/c4np00072b. [DOI] [PubMed] [Google Scholar]
- 30.Bingol K, Brüschweiler R. Deconvolution of chemical mixtures with high complexity by NMR consensus trace clustering. Anal Chem. 2011;83:7412–7417. doi: 10.1021/ac201464y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bingol K, Zhang F, Bruschweiler-Li L, Brüschweiler R. Carbon backbone topology of the metabolome of a cell. J Am Chem Soc. 2012;134:9006–9011. doi: 10.1021/ja3033058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clendinen CS, Pasquel C, Ajredini R, Edison AS. 13C NMR metabolomics: INADEQUATE network analysis. Anal Chem. 2015;87:5698–5706. doi: 10.1021/acs.analchem.5b00867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ito K, Tsutsumi Y, Date Y, Kikuchi J. Fragment assembly approach based on graph/network theory with quantum chemistry verifications for assigning multidimensional NMR signals in metabolite mixtures. ACS Chem Biol. 2016;11:1030–1038. doi: 10.1021/acschembio.5b00894. [DOI] [PubMed] [Google Scholar]
- 34*.Komatsu T, Ohishi R, Shino A, Kikuchi J. Structure and metabolic-flow analysis of molecular complexity in a 13C-labeled tree by 2D and 3D NMR. Angew Chem Int Ed. 2016;55:6000–6003. doi: 10.1002/anie.201600334. This study combines 13C-labeling, multidimensional NMR and density functional theory-based chemical shift prediction for the structure elucidation of unknown metabolites in complex mixtures. [DOI] [PubMed] [Google Scholar]
- 35*.Bingol K, Bruschweiler-Li L, Yu C, Somogyi A, Zhang F, Brüschweiler R. Metabolomics beyond spectroscopic databases: a combined MS/NMR strategy for the rapid identification of new metabolites in complex mixtures. Anal Chem. 2015;87:3864–3870. doi: 10.1021/ac504633z. This study introduces a hybrid NMR/MS strategy toward complete structure elucidation of unknown metabolites in complex mixtures without the need for extensive purification. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Correa J, Pinto LF, Riguera R, Fernandez-Megia E. Predicting PSR filters by transverse relaxation enhancements. Anal Chem. 2015;87:760–767. doi: 10.1021/ac5037186. [DOI] [PubMed] [Google Scholar]
- 37.Tayyari F, Gowda GA, Gu H, Raftery D. 15N-cholamine--a smart isotope tag for combining NMR- and MS-based metabolite profiling. Anal Chem. 2013;85:8715–8721. doi: 10.1021/ac401712a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lane AN, Arumugam S, Lorkiewicz PK, Higashi RM, Laulhe S, Nantz MH, Moseley HN, Fan TW. Chemoselective detection and discrimination of carbonyl-containing compounds in metabolite mixtures by 1H-detected 15N nuclear magnetic resonance. Magn Reson Chem. 2015;53:337–343. doi: 10.1002/mrc.4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39*.Gowda GAN, Raftery D. Can NMR solve some significant challenges in metabolomics? J Magn Reson. 2015;260:144–160. doi: 10.1016/j.jmr.2015.07.014. A comprehensive review covering the latest NMR and NMR/MS methods for quantitation and identification of metabolites. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40*.Zhang B, Xie M, Bruschweiler-Li L, Bingol K, Brüschweiler R. Use of charged nanoparticles in NMR-based metabolomics for spectral simplification and improved metabolite identification. Anal Chem. 2015;87:7211–7217. doi: 10.1021/acs.analchem.5b01142. This study introduces electrically charged silica nanoparticles as chemical agents to differentiate mixture components in NMR spectra based on their electric charge and hydrophobicity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang B, Xie M, Bruschweiler-Li L, Brüschweiler R. Nanoparticle-assisted removal of protein in human serum for metabolomics studies. Anal Chem. 2016;88:1003–1007. doi: 10.1021/acs.analchem.5b03889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salvia MV, Ramadori F, Springhetti S, Diez-Castellnou M, Perrone B, Rastrelli F, Mancin F. Nanoparticle-assisted NMR detection of organic anions: from chemosensing to chromatography. J Am Chem Soc. 2015;137:886–892. doi: 10.1021/ja511205e. [DOI] [PubMed] [Google Scholar]
- 43.Bouatra S, Aziat F, Mandal R, Guo AC, Wilson MR, Knox C, Bjorndahl TC, Krishnamurthy R, Saleem F, Liu P, et al. The human urine metabolome. PLoS One. 2013;8:e73076. doi: 10.1371/journal.pone.0073076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44*.Dame ZT, Aziat F, Mandal R, Krishnamurthy R, Bouatra S, Borzouie S, Guo AC, Sajed T, Deng L, Lin H, et al. The human saliva metabolome. Metabolomics. 2015;11:1864–1883. One of a growing number of comprehensive analyses of human biofluids by using NMR, MS and other analytical techniques along with dedicated metabolomics databases. [Google Scholar]
- 45.Crockford DJ, Holmes E, Lindon JC, Plumb RS, Zirah S, Bruce SJ, Rainville P, Stumpf CL, Nicholson JK. Statistical heterospectroscopy, an approach to the integrated analysis of NMR and UPLC-MS data sets: application in metabonomic toxicology studies. Anal Chem. 2006;78:363–371. doi: 10.1021/ac051444m. [DOI] [PubMed] [Google Scholar]
- 46.Pan Z, Gu H, Talaty N, Chen H, Shanaiah N, Hainline BE, Cooks RG, Raftery D. Principal component analysis of urine metabolites detected by NMR and DESI-MS in patients with inborn errors of metabolism. Anal Bioanal Chem. 2007;387:539–549. doi: 10.1007/s00216-006-0546-7. [DOI] [PubMed] [Google Scholar]
- 47.Marshall DD, Lei S, Worley B, Huang Y, Garcia-Garcia A, Franco R, Dodds ED, Powers R. Combining DI-ESI-MS and NMR datasets for metabolic profiling. Metabolomics. 2015;11:391–402. doi: 10.1007/s11306-014-0704-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48*.Hao J, Liebeke M, Sommer U, Viant MR, Bundy JG, Ebbels TM. Statistical correlations between NMR spectroscopy and direct Infusion FT-ICR mass spectrometry aid annotation of unknowns in metabolomics. Anal Chem. 2016;88:2583–2589. doi: 10.1021/acs.analchem.5b02889. Systematic analysis of the correlation of 1D 1H NMR and direct infusion MS signals of 26 metabolites over 200 metabolomics samples. This study showed that in a majority of cases the highest correlated NMR and MS signals belong to the same metabolite. Therefore, statistical correlation can be utilized to inter-relate NMR and MS signals of unknowns in complex mixtures. When combined with cheminformatics and computation, this can provide the structures of unknowns. [DOI] [PubMed] [Google Scholar]
- 49.Willoughby PH, Jansma MJ, Hoye TR. A guide to small-molecule structure assignment through computation of 1H and 13C NMR chemical shifts. Nat Protoc. 2014;9:643–660. doi: 10.1038/nprot.2014.042. [DOI] [PubMed] [Google Scholar]
- 50.Chikayama E, Shimbo Y, Komatsu K, Kikuchi J. The effect of molecular conformation on the accuracy of theoretical 1H and 13C chemical shifts calculated by ab initio methods for metabolic mixture analysis. J Phys Chem B. 2016;120:3479–3487. doi: 10.1021/acs.jpcb.5b12748. [DOI] [PubMed] [Google Scholar]