Abstract
Sleep and circadian rhythms studies have recently benefited from metabolomics analyses, uncovering new connections between chronobiology and metabolism. From untargeted mass spectrometry to quantitative nuclear magnetic resonance spectroscopy, a diversity of analytical approaches has been applied for biomarker discovery in the field. In this review we consider advances in the application of metabolomics technologies which have uncovered significant effects of sleep and circadian cycles on several metabolites, namely phosphatidylcholine species, medium-chain carnitines, and aromatic amino acids. Study design and data processing measures essential for detecting rhythmicity in metabolomics data are also discussed. Future developments in these technologies are anticipated vis-à-vis validating early findings, given metabolomics has only recently entered the ring with other systems biology assessments in chronometabolism studies.
Graphical Abstract
Introduction
Daily changes in organismal biology are highly conserved across species throughout evolution. Early scientific work performed on circadian rhythms originated from observations of daily leaf movements in heliotrope plants in 1729[1], opening doors to discoveries of circadian clocks and rest/activity cycles in higher organisms. Exploration of temporal changes in metabolism extends back to the earliest days of metabolic discovery, for example the reports from Forsgren that the ratios of bile and glycogen synthesis in rabbits exhibited diurnal variability[2]. The core genetic machinery which produces daily oscillations of transcripts and proteins has since been discovered and consists of transcription-translation feedback loops, driven through a Bmal-Clock protein complex in mammals[3]. Analysis of a small number of biomarker metabolites, such as melatonin and cortisol[4], are now commonly used to characterize diurnal patterns in both sleep and circadian research.
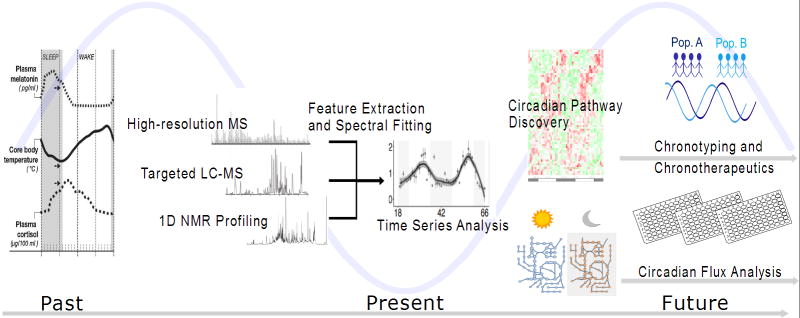
Traditionally, sleep and circadian rhythms have been studied by segregated research communities. Sleep studies typically focus on alterations in physiology which correlate with sleep status[5,6], while circadian designs employ higher time resolution sampling to assess oscillatory patterns in physiology[7], considering amplitude, phase, and period metrics. Thus, the sleep field is anchored from a clinical and epidemiological perspective, and the circadian field is driven by molecular discovery of basic clock mechanisms. Metabolism offers a tantalizing connection between these fields as the application of metabolomics technology cuts more broadly and deeply into physiology. An excellent review of key biological findings and interpretations from metabolomics’ studies on circadian rhythms has recently been published[8]. Here we discuss how diverse analytical platforms have advanced multiplexed chrono-metabolic biomarker studies. We also discuss future steps and technological developments required for chronobiology metabolomics to truly find the light in translational biology (Figure 1).
Figure 1.
Timeline of metabolomics in chronometabolic studies, both current approaches and future directions.
Source: Melatonin and cortisol data reprinted with permission from reference [4].
Mass spectrometry as an exploratory tool in chronometabolism
Metabolic variation in certain classes of low abundance metabolites such as glucocorticoids[9], catecholamines[10], and bulk lipids[11] has been long-established in both circadian and sleep contexts. It follows then that mass spectrometry (MS) forms an important component in the high-throughput detection of relevant metabolites. Collectively, advancements in complementary MS and nuclear magnetic resonance (NMR) spectroscopy approaches have facilitated recent chronobiology discoveries as will be discussed with a focus on human studies in the next two sections.
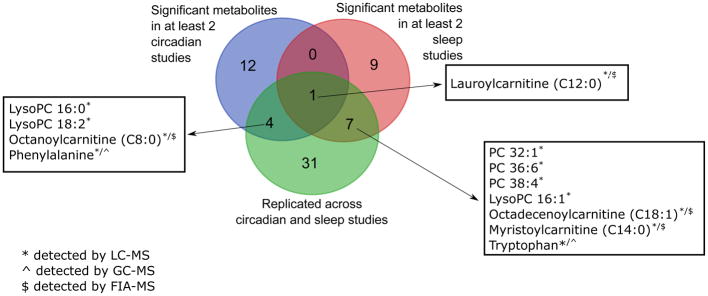
The diversity in both front-end separation and MS detection tools, combined with experimental design heterogeneity, has yielded a rich set of putative metabolite markers for further investigation. Figure 2 and Supplemental Table 1 summarize the results from nine key human chronobiology metabolomics studies[12–20] which have identified carnitines, aromatic amino acids, phosphatidylcholines and lysophosphatidylcholines as substantially enriched overlapping metabolite classes that oscillate across circadian time and are perturbed in sleep disruption studies (Figure 2). This evidence would suggest a significant interaction of circadian rhythms and sleep with metabolism through fatty acid metabolism, possibly for both energetic homeostasis via acylcarnitine oxidation and signaling roles that phoshatidylcholines may exert in and across tissues. The aromatic amino acids are both precursors of important neurotransmitters such as serotonin and catecholamines, of relevance to known diurnal changes in cognition and blood pressure respectively.
Figure 2.
Number of significant metabolites replicated across at least two circadian (blue) or sleep (red) studies and overlapping in at least one circadian and sleep study (green). Any overlaps across the three groups implies these metabolites are found in at least three of the nine considered papers.
These studies have primarily used liquid chromatography (LC)-MS metabolomics, although other approaches such as gas chromatography (GC)-MS or flow-injection (FIA)-MS[17] have been also been employed. The GC-MS studies have been part of a targeted commercial platform which combines two forms of LC-MS/MS (LC solvents appropriated for separate positive or negative mode ionizations) with GC-MS[14,16] to identify smaller and more polar metabolites including amino acids, sugars, and steroids. Other human metabolomics studies have instead focused on reverse-phase LC-MS and/or lipidomics, generating unique lists of lipid and other largely nonpolar features which change in sleep and circadian contexts[12,13,15,19]. These lipid profiles have identified subsets of unique circadian phenotypes within a healthy human population[13], as well as facilitated an approach for identifying characteristics of the internal body clock starting with roughly 4000 untargeted features[15]. Another approach combined untargeted quadrupole time-of-flight (qToF)-MS with hydrophilic interaction chromatography (HILIC) as well as reverse-phase lipidomics and high resolution GC-MS to yield a set of unique biomarkers of sleep debt [18], notably including larger lipid species only detected by lipidomics platforms. An exciting extension of circadian metabolomics has recently been performed in real-time breath analysis[21], which has direct clinical implications. While a high enrichment of metabolite features displayed rhythmicity (>36% of features), these masses were not identified as known metabolites. However, as mass resolution, sensitivity, and spectral databases continue to improve, these high-resolution MS analyses can identify metabolites with increased throughput and potential novelty[22].
Recently, metabolomics has yielded more causal links of the circadian clock to targeted metabolite outputs in mice[23,24], and also uncovered unique signaling roles of a specific lipid (PC 18:0/18:1) in regulating diurnal variability in metabolism across tissues[25]. Thus the field currently consists of a collection of untargeted analyses, where larger swaths of metabolites are detected without bias, and targeted analyses, which contain fewer but more confidently identified metabolites. We want to stress that while some of these methods are used in a complementary manner, even two or three collective analyses can only represent a fraction of the entire metabolome, and there exists considerable room for growth in exploratory analyses of the chronobiology metabolome. A total of 328 unique metabolites are significant in the aforementioned human sleep and circadian metabolomics studies (Supplemental Table 1). Encouragingly, 13.4% of these metabolites are found in at least one circadian and one sleep study (Figure 2, Supplemental Table 1). Still, the majority of metabolites found to date are not replicated, highlighting the extent of validation required in both study designs and analytical approaches. Somewhat provocatively, our own work has shown that repeated sleep study protocol and metabolomics analysis yields reproducibility of only about 4.5%, though it is unclear if this inconsistency derives from real biology or technical variability[18]. More quantitative and targeted approaches, including quadrupole-based MS techniques, which benefit from recent enhancements in scanning electronics, and NMR, will further advance the field towards actionable biomarkers in diagnostic and therapeutic settings.
NMR metabolomics as a clinical tool for chronobiology
Robustness, minimal sample processing, straightforward quantification and availability of structural insight are some of the key features of high-resolution NMR spectroscopy relevant to metabolomics. In human biomarker studies, NMR has been used to probe time-of-day variation in saliva metabolic profiles[26] and under different sleep conditions in urine [20]. NMR can be used quantitatively as a rapid and robust primary filter for analyzing circadian oscillations in polar metabolite rich systems. In this way, Giskeødegård et al. have demonstrated that the circadian urine metabotypes of individuals post sleep deprivation are distinct from their well-rested control state (Supplemental Table 1)[20]. In addition, the study also reports distinct excretory metabotypes pre- and post-sleep deprivation. Worth noting is that NMR identified structural isomers in these results, including sugars, which is difficult by LC-MS.
Non-invasive magnetic resonance spectroscopy has been used to demonstrate the relation of human sleep architecture with bioenergetics[27] and alterations in glia[28]. High-resolution NMR can be used to investigate intact cells and tissues and has been used to investigate temporal metabolism of intact red blood cells (RBCs) and hepatocytes, albeit without the time resolution or span needed for a true circadian study[29,30]. Anucleate RBCs are of special interest in this regard as they have functional non-transcriptional molecular clocks based on peroxiredoxin cycling[31]. Furthermore, profiling of tissue samples by High Resolution Magic Angle Spinning (HRMAS)-NMR may shed further light on chronobiology of intact tissues[32].
The major limiting factor in adoption of NMR metabolomics is the lack of sensitivity compared to MS. Using conventional probes, low micromolar concentration of polar metabolites in complex biological mixtures can be reliably quantified using sophisticated profiling techniques[33]. Fortunately, there are recent developments in NMR methods such as micro-coil NMR and hyperpolarization that may improve the detection limit and decrease the acquisition time significantly[34]. A number of important compounds found in human sleep and circadian biomarker studies can be already detected by NMR such as aromatic and branched chain amino acids, histidine, and creatine (Figure 2, Supplemental Table 1), amongst which phenylalanine and tryptophan have already been identified across multiple studies.
The true quantitative nature of NMR remains under-exploited, mainly due to the complex spectral overlap of biological mixtures and peak shift across samples[35]. Multidimensional NMR may be used to deal with the overlap issue, but comes at the expense of the acquisition time and sensitivity. Technically, efforts are ongoing to increase the detection limit and decrease the acquisition time of multidimensional NMR (reviewed in this issue by Giraudeau and co-authors), which has potential for significant contributions to the field of circadian metabolomics. One-dimensional NMR remains the most common method to leverage the quantitative capacity of NMR[33], and can be combined with quantitative approaches to assess metabolite concentrations[36,37,33]. Recently, we have used high temporal resolution (2 hour sampling) NMR-based targeted profiling of culture media and cells from human osteosarcoma (U2 OS) cell lines to demonstrate that circadian rhythms of glucose and glutamine metabolism are strongly affected by Myc oncogene expression[38]. We have also used quantitative targeted profiling of the secretome of U2 OS cells to show the presence of linear metabolic patterns latent in a typical circadian metabolic experiment, and suggest that detrending of such data prior to circadian analysis is potentially advantageous[39].
Experimental Design and Data Processing Considerations for Time-Dependent Data
The basic aim of circadian analysis is to detect biological variance over a defined time series, therefore separating analytical and biological variation is critical. Concerns of signal-to-noise have been noted in microarray analysis, where 18.3% of transcripts significantly oscillate with a median 1.51 fold change[40]. Encouragingly, the same set of liver samples yields a median 1.98 fold change in over 50% of detected metabolites (unpublished). Regardless of the approach taken, in order to assess true positives in oscillating metabolites, sufficient power must be incorporated into the experiment. For example, sampling every one or two hours greatly increases the probability of detecting a truly oscillating metabolite compared to every four or six hours. Recent work suggests that sampling every 2 hours over 48 hours provides a reasonable compromise between time resolution and resource allocation[41]. The scalability concerns of large sample numbers inherent in proper circadian study designs present key data processing considerations, including the use of proper quality control samples to mitigate unwanted variance due to instrumental drift and removal of spurious metabolic features, which has been reviewed in excellent detail by Dunn et al. for liquid chromatography mass spectrometry (LC-MS) metabolomics[42].
Multiple statistical approaches have been developed for extracting temporal rhythms and compared[43], each with their own strengths and weaknesses. One particularly strong algorithm, JTK_CYCLE[44], is the most popular method in the studies discussed in this review and remains a cutting-edge choice for analysis of rhythmicity. Fortuitously, analyzing waveform data takes advantage of biological phenomena to reduce sampling requirements. Given that these algorithms search for repeating patterns in time-series data, replicates in the number of days offers advantages over replicate time points within a single day [41]. Thus these concerns for experimental scale can be partly mitigated by study design, recent statistical methods, and improved metabolomics instrumentation.
Future Directions
The comparative analysis of metabolites from sleep and circadian studies presented in Figure 2 raise some important questions to be addressed. Given that medium-chain carnitines, lysophosphatidylcholines, and phosphatidylcholines are clearly enriched amongst metabolite classes, the mechanism driving these changes needs to be defined[45]. For example, lauroylcarnitine is significant in five of the nine studies in Figure 2, but it is unclear what role this molecule may play in chronobiology. These chemical classes are readily observed in LC-MS analyses and may have emerged due to analytical and platform bias of more reverse-phase LC-MS and electrospray ionization analyses used thus far in chronometabolism, however given the unique roles lipids play in biology for energy production and signaling, these metabolites likely truly sit at the nexus of sleep, circadian rhythms, and metabolism. Deeper and more diverse metabolomics analyses are still needed to elucidate the significance of other metabolite classes, including more specific detection methods for sugars, steroids, metabolites conjugated through biotransformations, and other nonpolar aromatic compounds not detected in most circadian and sleep studies to date. We believe some of these other compound classes are also important players in chronobiology, and will be added to the overlapping list of metabolites in Figure 2 as the field expands. Moving forward, circadian biologists striving for mechanistic elucidation of these overlapping metabolites can benefit from robust LC-MS analyses previously used, while further exploration of chrono-metabolism should focus on diversity in analytical detection methods.
Chronobiology has seen a wave of systems biology data in recent years, at each level of the central dogma[19,46,47]. While tools exist to integrate metabolite and transcription data for pathway mapping[48] including circadian-specific datasets[49], they have little use yet broadly in the field. Rich databases for microarray and RNASeq circadian studies have been developed[50], which opens the door for developing analogous metabolomics databases as the number of these studies continues to grow with ever-improving metabolite identification and high-resolution MS data.
The vast majority of high-throughput chronobiology studies to date have been performed using static metabolite profiling. While advantageous for biomarker detection, the field still lacks mechanistic understanding of the steady-state metabolite changes observed in these studies. Flux analysis has yielded valuable insights into defining the activity of reactions and pathways which contain the metabolites of interest, and exciting new advancements in isotope tracing analysis should clarify the true phenotypic output of the clock. Improved precision and sensitivity in LC-MS metabolomics along with untargeted isotope-based metabolomics platforms[51,52] that are further discussed within this issue by Fabien Lestisse has great potential for unbiased screening of clock kinetics. NMR has long been used for metabolic flux analysis and through such analyses, our group has shown that GABA-transaminase independently regulates metabolic and sleep homeostasis in Drosophila neurons[53]. The recent advent of multidimensional NMR technologies coupled to ultrafast acquisition such as nonuniform sampling (NUS) promises to further enhance NMR-based metabolic flux analysis[54]. Targeted MS labeling studies have also been performed in the context of sleep and neurotransmitter modulation in neurons[55,56], and has just recently been applied to interrogate in vitro the role of the clock in carcinogenesis[57], which we expect to garner greater use in the near future.
Ultimately, these tools may unearth drug targets which modulate circadian clocks and augment metabolic disease[58]. Furthermore, the new technologies in remote monitoring and wearable technology, combined with sequencing and metabolite data in patients can decipher inter-individual variability in clock outputs and metabolic status, laying the foundation for the dream of precision medicine.
Conclusions
Metabolomics has greatly elucidated the chronobiology-metabolism connection, with the advent of broader and deeper metabolome analyses and rhythmicity detection algorithms. Now equipped with a set of promising metabolite hits, further validation is required to understand mechanistically the interplay of circadian clocks, sleep, and metabolism before leveraging this information for therapeutic purposes. Exciting advancements in MS metabolite profiling and new NMR acquisition techniques will clarify metabolic kinetics through flux analysis and provide a deeper systems-level understanding of metabolic networks.
Supplementary Material
Highlights.
Metabolism is a nexus of sleep and circadian processes as observed by metabolomics
An overlapping set of metabolites is emerging from sleep and circadian studies
These studies have unique data processing, design and statistical considerations
Validation and increased coverage are required for robust chrono-metabolism markers
Future mechanistic studies will be needed to complement biomarkers
Acknowledgments
We would like to thank John Hogenesch, Amita Sehgal, and Garret FitzGerald for their comments on the manuscript. We would also like to thank Michael Hughes and Nicholas Lahens for their assistance in determining transcript and metabolite fold changes, and Saikumari Krishnaiah for valuable discussions. The project described was supported by the National Center for Research Resources, Grant UL1RR024134, and is now at the National Center for Advancing Translational Sciences, Grant UL1TR000003. The content is solely the responsibility of the official views of the NIH. Supported in part by the Institute for Translational Medicine and Therapeutics’ (ITMAT) Transdisciplinary Program in Translational Medicine and Therapeutics. S.D.R was supported through a Pharmacology T32 Training Grant (T32 GM008076).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.de Marian J-J. Observation botanique. Hist l’Academie R des Sci. 1729 [Google Scholar]
- 2.Fosgren E. On the Relationship between the Formation of Bile and Glycogen in the Liver of Rabbit. Skand Arch Physiol. 1928;53:137–151. [Google Scholar]
- 3.Bass J. Circadian topology of metabolism. Nature. 2012;491:348–356. doi: 10.1038/nature11704. [DOI] [PubMed] [Google Scholar]
- 4.Glickman G. Circadian rhythms and sleep in children with autism. Neurosci Biobehav Rev. 2010;34:755–68. doi: 10.1016/j.neubiorev.2009.11.017. [DOI] [PubMed] [Google Scholar]
- 5.Bonnet MH, Arand DL. Clinical effects of sleep fragmentation versus sleep deprivation. Sleep Med Rev. 2003;7:297–310. doi: 10.1053/smrv.2001.0245. [DOI] [PubMed] [Google Scholar]
- 6.da Silva RAPC. Sleep disturbances and mild cognitive impairment: A review. Sleep Sci. 2015;8:36–41. doi: 10.1016/j.slsci.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saini C, Brown SA, Dibner C. Human peripheral clocks: applications for studying circadian phenotypes in physiology and pathophysiology. Front Neurol. 2015;6:95. doi: 10.3389/fneur.2015.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8**.Brown SA. Circadian Metabolism: From Mechanisms to Metabolomics and Medicine. Trends Endocrinol Metab. 2016;27:415–426. doi: 10.1016/j.tem.2016.03.015. This recent review dives deep into the latest findings from circadian metabolism studies and will provide the reader with a thorough background on the biological mechanisms of clocks. This review also emphasizes the role of metabolomics in driving these recent discoveries, and future translational applications within the field. [DOI] [PubMed] [Google Scholar]
- 9.Weitzman ED, Fukushima D, Nogeire C, Roffwarg H, Gallagher TF, Hellman L. Twenty-four hour pattern of the episodic secretion of cortisol in normal subjects. J Clin Endocrinol Metab. 1971;33:14–22. doi: 10.1210/jcem-33-1-14. [DOI] [PubMed] [Google Scholar]
- 10.Lewy AJ, Sack RL. The dim light melatonin onset as a marker for circadian phase position. Chronobiol Int. 1989;6:93–102. doi: 10.3109/07420528909059144. [DOI] [PubMed] [Google Scholar]
- 11.Hollister L, Wright A. Diurnal variation of serum lipids. J Atheroscler Res. 1965;5:445–50. doi: 10.1016/s0368-1319(65)80016-3. [DOI] [PubMed] [Google Scholar]
- 12.Ang JE, Revell V, Mann A, Mäntele S, Otway DT, Johnston JD, Thumser AE, Skene DJ, Raynaud F. Identification of human plasma metabolites exhibiting time-of-day variation using an untargeted liquid chromatography-mass spectrometry metabolomic approach. Chronobiol Int. 2012;29:868–81. doi: 10.3109/07420528.2012.699122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13*.Chua EC-P, Shui G, Lee IT-G, Lau P, Tan L-C, Yeo S-C, Lam BD, Bulchand S, Summers SA, Puvanendran K, et al. Extensive diversity in circadian regulation of plasma lipids and evidence for different circadian metabolic phenotypes in humans. Proc Natl Acad Sci U S A. 2013;110:14468–73. doi: 10.1073/pnas.1222647110. The most comprehensive lipidomics LC-MS analyses of plasma from healthy individuals across circadian time to date. The authors demonstrate that individuals can be stratified into unique circadian metabolic phenotypes based on specific subsets of lipids. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dallmann R, Viola AU, Tarokh L, Cajochen C, Brown SA. The human circadian metabolome. Proc Natl Acad Sci U S A. 2012;109:2625–9. doi: 10.1073/pnas.1114410109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kasukawa T, Sugimoto M, Hida A, Minami Y, Mori M, Honma S, Honma K, Mishima K, Soga T, Ueda HR. Human blood metabolite timetable indicates internal body time. Proc Natl Acad Sci U S A. 2012;109:15036–41. doi: 10.1073/pnas.1207768109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bell LN, Kilkus JM, Booth JN, Bromley LE, Imperial JG, Penev PD. Effects of sleep restriction on the human plasma metabolome. Physiol Behav. 2013;122:25–31. doi: 10.1016/j.physbeh.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Den Berg R, Mook-Kanamori DO, Donga E, Van Dijk M, Van Dijk JG, Lammers GJ, Van Kralingen KW, Prehn C, Adamski J, Romijn JA, et al. A single night of sleep curtailment increases plasma acylcarnitines: Novel insights in the relationship between sleep and insulin resistance. Arch Biochem Biophys. 2016;589:145–151. doi: 10.1016/j.abb.2015.09.017. [DOI] [PubMed] [Google Scholar]
- 18**.Weljie AM, Meerlo P, Goel N, Sengupta A, Kayser MS, Abel T, Birnbaum MJ, Dinges DF, Sehgal A. Oxalic acid and diacylglycerol 36:3 are cross-species markers of sleep debt. Proc Natl Acad Sci U S A. 2015;112:2569–74. doi: 10.1073/pnas.1417432112. LC-MS and GC-MS were both utilized for an extensive metabolomics analysis, yielding the first quantitative human markers of chronic sleep restriction. Two markers were conserved across humans and rats. An additional strength of the study was complete replication of experimental and analytical procedures in the rat study, demonstrating the importance of validation in metabolomics analyses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19**.Davies SK, Ang JE, Revell VL, Holmes B, Mann A, Robertson FP, Cui N, Middleton B, Ackermann K, Kayser M, et al. Effect of sleep deprivation on the human metabolome. Proc Natl Acad Sci U S A. 2014;111:10761–6. doi: 10.1073/pnas.1402663111. This study applied LC-MS in plasma of healthy individuals with adequate sampling resolution for circadian analysis also yielded the first metabolite markers of total sleep deprivation. This powerful study design facilitated the detection of both metabolites that are significantly rhythmic and dampened during forced wakefulness. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20**.Giskeødegård GF, Davies SK, Revell VL, Keun H, Skene DJ. Diurnal rhythms in the human urine metabolome during sleep and total sleep deprivation. Sci Rep. 2015;5:14843. doi: 10.1038/srep14843. Using the same design as Davies et al., 2014, NMR spectroscopy uncovered metabolites perturbed in urine of healthy human volunteers after forced wakefulness. Of note, some of these metabolites were also found to be altered in other sleep deprivation and circadian studies using mass spectrometry, generating a list of cross-platform hits. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martinez-Lozano Sinues P, Tarokh L, Li X, Kohler M, Brown SA, Zenobi R, Dallmann R. Circadian variation of the human metabolome captured by real-time breath analysis. PLoS One. 2014;9:e114422. doi: 10.1371/journal.pone.0114422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yore MM, Syed I, Moraes-Vieira PM, Zhang T, Herman MA, Homan EA, Patel RT, Lee J, Chen S, Peroni OD, et al. Discovery of a class of endogenous mammalian lipids with anti-diabetic and anti-inflammatory effects. Cell. 2014;159:318–32. doi: 10.1016/j.cell.2014.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adamovich Y, Rousso-Noori L, Zwighaft Z, Neufeld-Cohen A, Golik M, Kraut-Cohen J, Wang M, Han X, Asher G. Circadian clocks and feeding time regulate the oscillations and levels of hepatic triglycerides. Cell Metab. 2014;19:319–30. doi: 10.1016/j.cmet.2013.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24*.Zwighaft Z, Aviram R, Shalev M, Rousso-Noori L, Kraut-Cohen J, Golik M, Brandis A, Reinke H, Aharoni A, Kahana C, et al. Circadian Clock Control by Polyamine Levels through a Mechanism that Declines with Age. Cell Metab. 2015;22:874–885. doi: 10.1016/j.cmet.2015.09.011. Given enzymes involved in polyamine metabolism oscillate across circadian time, the authors performed targeted MS analysis in mice to identify polyamine levels display rhythmicity and themselves modulate rhythms through interactions with core clock proteins. Additionally, these rhythms decline in aging mice, highlighting one of the first studies to uncover a potential therapeutic application of metabolite analysis in circadian rhythms. [DOI] [PubMed] [Google Scholar]
- 25.Liu S, Brown JD, Stanya KJ, Homan E, Leidl M, Inouye K, Bhargava P, Gangl MR, Dai L, Hatano B, et al. A diurnal serum lipid integrates hepatic lipogenesis and peripheral fatty acid use. Nature. 2013;502:550–4. doi: 10.1038/nature12710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dame ZT, Aziat F, Mandal R, Krishnamurthy R, Bouatra S, Borzouie S, Guo AC, Sajed T, Deng L, Lin H, et al. The human saliva metabolome. Metabolomics. 2015;11:1864–1883. [Google Scholar]
- 27.Plante DT, Trksak GH, Jensen JE, Penetar DM, Ravichandran C, Riedner BA, Tartarini WL, Dorsey CM, Renshaw PF, Lukas SE, et al. Gray matter-specific changes in brain bioenergetics after acute sleep deprivation: a 31P magnetic resonance spectroscopy study at 4 Tesla. Sleep. 2014;37:1919–27. doi: 10.5665/sleep.4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cross NE, Lagopoulos J, Duffy SL, Cockayne NL, Hickie IB, Lewis SJG, Naismith SL. Sleep quality in healthy older people: relationship with 1H magnetic resonance spectroscopy markers of glial and neuronal integrity. Behav Neurosci. 2013;127:803–10. doi: 10.1037/a0034154. [DOI] [PubMed] [Google Scholar]
- 29.Dabos KJ, Parkinson JA, Hewage C, Nelson LJ, Sadler IH, Hayes PC, Plevris JN. 1H NMR spectroscopy as a tool to evaluate key metabolic functions of primary porcine hepatocytes after cryopreservation. NMR Biomed. 2002;15:241–50. doi: 10.1002/nbm.765. [DOI] [PubMed] [Google Scholar]
- 30.Mehta M, Sonawat HM, Sharma S. Malaria parasite-infected erythrocytes inhibit glucose utilization in uninfected red cells. FEBS Lett. 2005;579:6151–8. doi: 10.1016/j.febslet.2005.09.088. [DOI] [PubMed] [Google Scholar]
- 31.O’Neill JS, Reddy AB. Circadian clocks in human red blood cells. Nature. 2011;469:498–503. doi: 10.1038/nature09702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giskeødegård GF, Cao MD, Bathen TF. High-Resolution Magic-Angle-Spinning NMR Spectroscopy of Intact Tissue. 2015:37–50. doi: 10.1007/978-1-4939-2377-9_4. [DOI] [PubMed] [Google Scholar]
- 33.Weljie AM, Newton J, Mercier P, Carlson E, Slupsky CM. Targeted Profiling: Quantitative Analysis of 1 H NMR Metabolomics Data. Anal Chem. 2006;78:4430–4442. doi: 10.1021/ac060209g. [DOI] [PubMed] [Google Scholar]
- 34.Nagana Gowda GA, Raftery D. Can NMR solve some significant challenges in metabolomics? 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alonso A, Marsal S, Julià A. Analytical methods in untargeted metabolomics: state of the art in 2015. Front Bioeng Biotechnol. 2015;3:23. doi: 10.3389/fbioe.2015.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Puchades-Carrasco L, Palomino-Schätzlein M, Pérez-Rambla C, Pineda-Lucena A. Bioinformatics tools for the analysis of NMR metabolomics studies focused on the identification of clinically relevant biomarkers. Brief Bioinform. 2016;17:541–52. doi: 10.1093/bib/bbv077. [DOI] [PubMed] [Google Scholar]
- 37.Hao J, Liebeke M, Astle W, De Iorio M, Bundy JG, Ebbels TMD. Bayesian deconvolution and quantification of metabolites in complex 1D NMR spectra using BATMAN. Nat Protoc. 2014;9:1416–1427. doi: 10.1038/nprot.2014.090. [DOI] [PubMed] [Google Scholar]
- 38*.Altman BJ, Hsieh AL, Sengupta A, Krishnanaiah SY, Stine ZE, Walton ZE, Gouw AM, Venkataraman A, Li B, Goraksha-Hicks P, et al. MYC Disrupts the Circadian Clock and Metabolism in Cancer Cells. Cell Metab. 2015;22:1009–19. doi: 10.1016/j.cmet.2015.09.003. High time resolution NMR metabolomics was applied to an in vitro model of circadian rhythms to demonstrate the overlaps in the transcriptional regulatory roles of the oncogene MYC and the core genetic clock. The clock is thus implicated in mitigating the biosynthetic advantage of MYC-driven cancers, and likely to be thus an unobserved aberration in many human tumors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sengupta A, Krishnaiah S, Rhoades S, Growe J, Slaff B, Venkataraman A, Olarerin-George A, Van Dang C, Hogenesch J, Weljie A. Deciphering the Duality of Clock and Growth Metabolism in a Cell Autonomous System Using NMR Profiling of the Secretome. Metabolites. 2016;6:23. doi: 10.3390/metabo6030023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hughes ME, DiTacchio L, Hayes KR, Vollmers C, Pulivarthy S, Baggs JE, Panda S, Hogenesch JB. Harmonics of Circadian Gene Transcription in Mammals. PLoS Genet. 2009;5:e1000442. doi: 10.1371/journal.pgen.1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li J, Grant GR, Hogenesch JB, Hughes ME. Considerations for RNA-seq Analysis of Circadian Rhythms. Circadian Rhythm Biol Clocks. 2015;551:349–367. doi: 10.1016/bs.mie.2014.10.020. [DOI] [PubMed] [Google Scholar]
- 42.Dunn WB, Broadhurst D, Begley P, Zelena E, Francis-McIntyre S, Anderson N, Brown M, Knowles JD, Halsall A, Haselden JN, et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat Protoc. 2011;6:1060–83. doi: 10.1038/nprot.2011.335. [DOI] [PubMed] [Google Scholar]
- 43.Zhang Z, Wu G, Zhu J, Yu J, Zhou L, Huang JZ. Evaluation of Five Methods for Genome-Wide Circadian Gene Identification. J Biol Rhythms. 2014;29:231–242. doi: 10.1177/0748730414537788. [DOI] [PubMed] [Google Scholar]
- 44.Hughes ME, Hogenesch JB, Kornacker K. JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J Biol Rhythms. 2010;25:372–80. doi: 10.1177/0748730410379711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45*.Arble DM, Bass J, Behn CD, Butler MP, Challet E, Czeisler C, Depner CM, Elmquist J, Franken P, Grandner MA, et al. Impact of Sleep and Circadian Disruption on Energy Balance and Diabetes. A Summary of Workshop Discussions. Sleep. 2015;38:1849–60. doi: 10.5665/sleep.5226. A workshop was recently held at the National Institute for Diabetes and Digestive and Kidney Diseases with a focus on sleep and circadian rhythms in energy balance and metabolic disorders. Recent findings, opportunities, and objectives for translational science are highlighted. Emphasis is placed on proper human study designs to better understand the chronometabolic connection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mauvoisin D, Wang J, Jouffe C, Martin E, Atger F, Waridel P, Quadroni M, Gachon F, Naef F. Circadian clock-dependent and -independent rhythmic proteomes implement distinct diurnal functions in mouse liver. Proc Natl Acad Sci U S A. 2014;111:167–72. doi: 10.1073/pnas.1314066111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang R, Lahens NF, Ballance HI, Hughes ME, Hogenesch JB. A circadian gene expression atlas in mammals: Implications for biology and medicine. Proc Natl Acad Sci. 2014 doi: 10.1073/pnas.1408886111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kamburov A, Cavill R, Ebbels TMD, Herwig R, Keun HC. Integrated pathway-level analysis of transcriptomics and metabolomics data with IMPaLA. Bioinformatics. 2011;27:2917–2918. doi: 10.1093/bioinformatics/btr499. [DOI] [PubMed] [Google Scholar]
- 49.Patel VR, Eckel-Mahan K, Sassone-Corsi P, Baldi P. CircadiOmics: integrating circadian genomics, transcriptomics, proteomics and metabolomics. Nat Methods. 2012;9:772–3. doi: 10.1038/nmeth.2111. [DOI] [PubMed] [Google Scholar]
- 50.Pizarro A, Hayer K, Lahens NF, Hogenesch JB. CircaDB: a database of mammalian circadian gene expression profiles. Nucleic Acids Res. 2013;41:D1009–13. doi: 10.1093/nar/gks1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang X, Chen Y-J, Cho K, Nikolskiy I, Crawford PA, Patti GJ. X13CMS: global tracking of isotopic labels in untargeted metabolomics. Anal Chem. 2014;86:1632–9. doi: 10.1021/ac403384n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kluger B, Bueschl C, Neumann N, Stückler R, Doppler M, Chassy AW, Waterhouse AL, Rechthaler J, Kampleitner N, Thallinger GG, et al. Untargeted profiling of tracer-derived metabolites using stable isotopic labeling and fast polarity-switching LC-ESI-HRMS. Anal Chem. 2014;86:11533–7. doi: 10.1021/ac503290j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maguire SE, Rhoades S, Chen WF, Sengupta A, Yue Z, Lim JC, Mitchell CH, Weljie AM, Sehgal A. Independent effects of γ-aminobutyric acid transaminase (GABAT) on metabolic and sleep homeostasis. J Biol Chem. 2015;290:20407–20416. doi: 10.1074/jbc.M114.602276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reardon PN, Marean-Reardon CL, Bukovec MA, Coggins BE, Isern NG. 3D TOCSY-HSQC NMR for Metabolic Flux Analysis Using Non-Uniform Sampling. Anal Chem. 2016;88:2825–2831. doi: 10.1021/acs.analchem.5b04535. [DOI] [PubMed] [Google Scholar]
- 55.Qu H, Konradsen JR, Van Hengel M, Wolt S, Sonnewald U. Effect of glutamine and GABA on [U-13C]glutamate metabolism in cerebellar astrocytes and granule neurons. J Neurosci Res. 2001;66:885–90. doi: 10.1002/jnr.10055. [DOI] [PubMed] [Google Scholar]
- 56.McKenna MC, Sonnewald U. GABA alters the metabolic fate of [U-13C]glutamate in cultured cortical astrocytes. J Neurosci Res. 2005;79:81–7. doi: 10.1002/jnr.20309. [DOI] [PubMed] [Google Scholar]
- 57**.Papagiannakopoulos T, Bauer MR, Davidson SM, Bartlebaugh J, Vander Heiden MG, Correspondence TJ, Heimann M, Subbaraj L, Bhutkar A, Jacks T. Circadian Rhythm Disruption Promotes Lung Tumorigenesis. Cell Metab. 2016;24:324–331. doi: 10.1016/j.cmet.2016.07.001. These authors are the first to employ an isotope-labeling approach in in vitro models of circadian rhythms to more clearly understand the impact of core clock genes on metabolism. We expect isotope-assisted metabolomics and flux analysis to be increasingly vital to future studies of sleep and circadian rhythms. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.He B, Nohara K, Park N, Park YS, Guillory B, Zhao Z, Garcia JM, Koike N, Lee CC, Takahashi JS, et al. The Small Molecule Nobiletin Targets the Molecular Oscillator to Enhance Circadian Rhythms and Protect against Metabolic Syndrome. Cell Metab. 2016;23:610–621. doi: 10.1016/j.cmet.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.