Abstract
Post-translational modifications (PTMs) regulate numerous proteins and are important for many biological processes. Lysine 4-oxononanoylation (4-ONylation) is a newly discovered histone PTM that prevents nucleosome assembly under oxidative stress. Whether there are cellular enzymes that remove 4-ONyl from histones remains unknown, which hampers the further investigation of the cellular function of this PTM. Here, we report that mammalian SIRT2 can remove 4-ONyl from histones and other proteins in live cells. A crystal structure of SIRT2 in complex with a 4-ONyl peptide reveals a lone pair-π interaction between Phe119 and the ketone oxygen of the 4-ONyl group. This is the first time that a mechanism to reverse 4-ONyl lysine modification is reported and will help to understand the role of SIRT2 in oxidative stress responses and the function of 4-ONylation.
Protein post-translational modifications (PTMs) play crucial roles in regulating a wide range of biological processes, such as gene transcription, DNA replication, and chromosome segregation.1 Histones can be modified by several reversible PTMs, such as acetylation, methylation, phosphorylation, and ubiquitination. Recently, a new acyl modification, 4-oxononanoyl (4-ONyl) lysine, by 4-oxo-2-nonenal (4-ONE), is reported to occur on histone H3 (Figure 1A).2 The 4-ketoamide adduct has polar ketone oxygen that is different from other short chain fatty acyl groups. 4-ONE can be generated by the oxidation of cellular lipids upon stress and react with histone lysines.2 This 4-ketoamide adduct at H3K27 and H3K23 can be stimulated by lipopolysaccharides, disrupt chromatin dynamics and alter histone turnover.2 However, no eraser has been known to remove this 4-ONyl lysine modification on histones.
Figure 1.
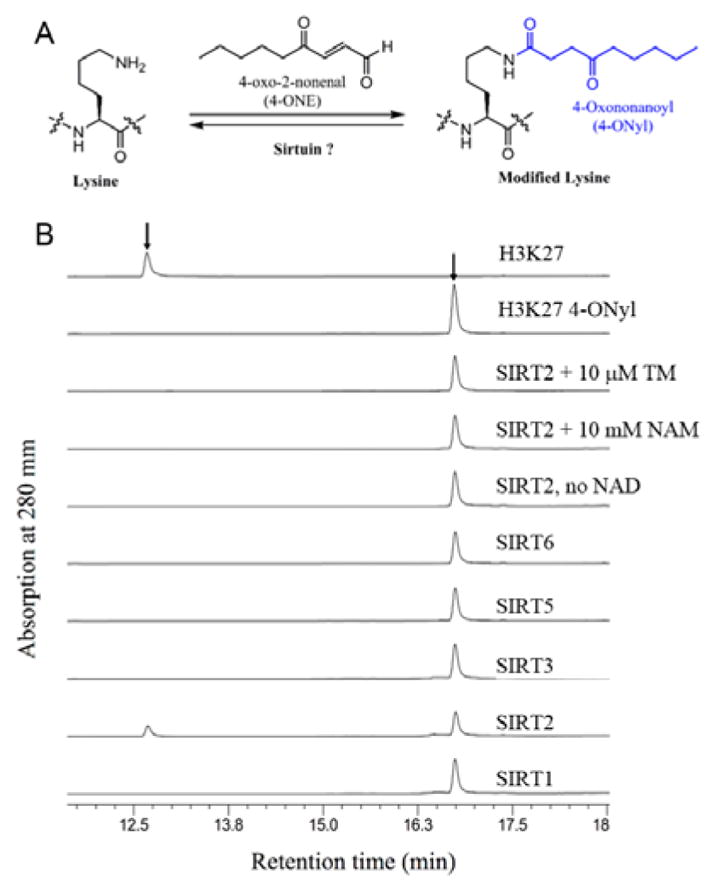
SIRT2 can hydrolyze 4-ONyl modification on lysine. (A) 4-ONyl modification on protein lysine residues. (B) HPLC traces showing 4-ONyl H3K27 can be hydrolyzed by SIRT2, but not other sirtuins.
Sirtuins, the class III histone deacetylases (HDACs), are enzymes with nicotinamide adenine dinucleotide (NAD)-dependent deacetylation activities.3–5 Mammalian sirtuins (SIRT1-7) display widespread subcellular localizations. SIRT1, SIRT6, and SIRT7 are predominantly nuclear; SIRT2 is mainly in cytoplasm while SIRT3-5 are mainly in mitochondria.6 Interestingly, SIRT2 was also found to deacetylate H3K18 upon bacterial infection or deacetylate H4K16 in the G2/M transition to ensure chromatin condensation.7,8 Emerging evidence demonstrate sirtuins as deacylases that remove a variety of acyl groups on lysine. SIRT1-3 can depropionylate and debutyrylate.9,10 SIRT3 is reported to have decrotonylation activity and SIRT4 is reported to act as a lipoamidase regulating pyruvate dehydrogenase complex activity.11,12 SIRT5 can hydrolyze succinyl, malonyl and glutaryl lysines.13,14 SIRT6 can efficiently remove long chain fatty acyl groups on TNFα.15 Interestingly, biochemical studies also defined SIRT2 as demyristoylase.16,17 These findings broaden the substrates scope of sirtuins as deacylases.
Since the 4-ONyl modification is similar to fatty acyl groups and sirtuins can act as defatty-acylase,18 we wonder whether sirtuins are capable of hydrolyzing 4-ONyl lysine and regulate the change of the 4-ONylation level on endogenous histones. To investigate the hypothesis, we screened the activities of several sirtuins on H3k27 4-ONyl peptide. The results demonstrated that with SIRT2, but not other sirtuins, can hydrolyze H3k27 4-ONyl (Figure 1B). The activity of SIRT2 on H3k27 4-ONyl was NAD-dependent (Figure 1B). Nicotinamide (NAM, a pansirtuin inhibitor) or the TM molecule19 (a SIRT2-specific inhibitor), inhibited the hydrolysis of H3k27 4-ONyl (Figure 1B). We also measured the kinetics of SIRT2 on H3K27 4-ONyl and acetyl peptides. The catalytic efficiency of SIRT2 on H3K27 4-ONyl was comparable to that of deacetylation (Table 1). Similarly, SIRT2 could catalyze the hydrolysis of H3K23 4-ONyl and acetyl with similar efficiencies (Table 1). SIRT2 also shows the similar Kd values for 4-ONyl and acetyl H3K27 and H3K23 peptides (Figure S3).
Table 1.
The kinetic parameters on 4-ONyl lysine peptides.
| SIRT2 | acyl lysine peptide | kcat (s−1) | Km (μM) | kcat/Km (s−1 M−1) |
|---|---|---|---|---|
| WT | H3K27 4-ONyla | 0.0248 ± 0.0013 | 48 ± 9 | 522 |
| H3K27 acetyla | 0.0538 ± 0.0024 | 38 ± 7 | 1400 | |
| H3K23 4-ONylb | 0.0121 ± 0.0010 | 12 ± 6 | 1010 | |
| H3K23 acetylb | 0.0644 ± 0.0026 | 21 ± 4 | 3130 | |
| F119A | H3K27 4-ONyla | 0.0133 ± 0.0019 | 104 ± 22 | 127 |
| H3K27 (Acetyl)a | 0.0089 ± 0.0003 | 55 ± 9 | 162 | |
| H3K23 4-ONylb | 0.0134 ± 0.0019 | 124 ± 5 | 108 | |
| H3K23 (Acetyl)b | 0.0249 ± 0.0011 | 55 ± 8 | 452 |
Peptide sequence: TKAARK*SAPATWW.
Peptide sequence: KQLATK*AARKSWW.
To find out more about how SIRT2 recognizes the 4-ONyl group, we obtained the crystals of SIRT2 with both H3K27 4-ONyl and H3K23 4-ONyl peptides. A crystal of SIRT2 in complexes with the H3K23 (4-ONyl) and carba-NAD diffracted to 2.0 Å resolution after optimization. Crystallographic data collection statistics are shown in supplemental Table S1. The asymmetric unit consisted of 2 molecules, each containing one SIRT2-H3K23 4-ONyl in complex with carba-NAD (Figure 2A). The two globular domains of SIRT2 composed of an NAD-binding Rossmann fold and a zinc-binding motif are similar to other sirtuins (Figure 2A).13,15,20 The H3K23 4-ONyl bound to SIRT2 in a way similar to that of H3K9 myristoyl binding to SIRT2 (PDB 4R8M) (Figure S2).16 The 4-ONyl group was accommodated in a pocket of SIRT2 formed by hydrophobic residues Phe96, Phe119, Phe131, Thr134, Tyr139, Phe190 and Ile232 (Figure 2B). The catalytic His187 interacted with the oxygen of amide bond (Figure 2B). Interestingly, the benzene ring of Phe119 interacted with the ketone oxygen of 4-ONyl group. The distance d between the carbonyl oxygen atom and the phenyl ring-centroid was 3.4 Å and the dihedral angle ω between the planes defined by the O=CRR′ moiety and the aromatic ring was 60.5°, indicating a lone pair-π interaction with a standard distance and angle constraints (Figure 2C,D).21,22 Mutation of this Phe to Ala (F119A) significantly decreased the activity of SIRT2 on 4-ONyl and acetyl peptides (Table 1).
Figure 2.
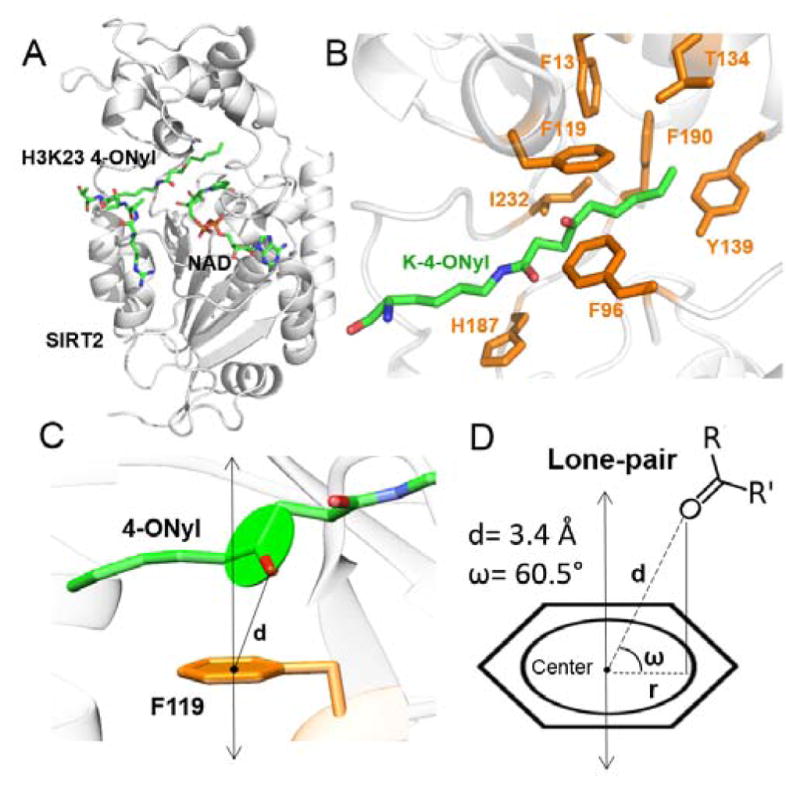
The structural basis for recognition of 4-ONyl lysine by SIRT2. (A) Overall structure of SIRT2 (gray) in complexes with H3K23 4-ONyl peptide and carba-NAD (green). (B) The binding pocket formed by hydrophobic residues (orange). (C) A side view of the lone pair-π interaction between Phe119 and the 4-ONyl group. (D) Scheme showing the distance (d) and dihedral angle ω between the ketone oxygen and the phenyl ring of Phe119.
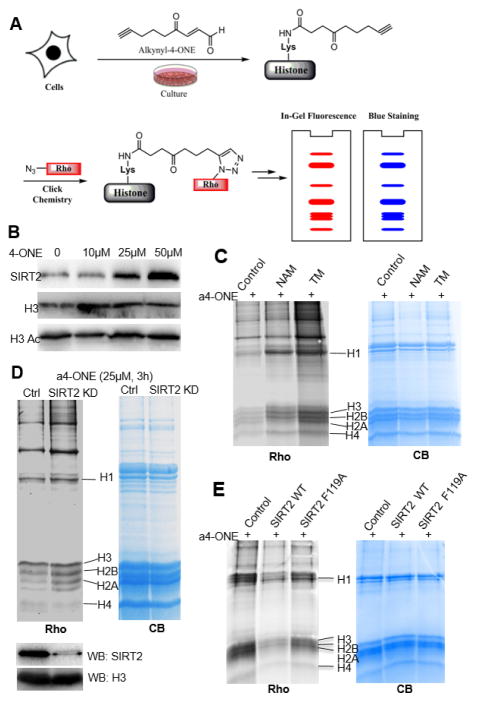
Next, we investigated whether SIRT2 could remove 4-ONyl group from histones in cells. Mouse embryonic fibroblast (MEF) cells were used for all the cellular experiments. SIRT2 was detected on extracted histones (Figure 3B), indicating that SIRT2 interacts with histones. Interestingly, treating cells with the increasing concentration of 4-ONE, the end product of lipid peroxidation,2,23 led to increased SIRT2 on histones (Figure 3B). To monitor the changes of protein 4-ONylation levels in cells, a chemical reporter, alkynyl-4-ONE (a4-ONE), was used to label the Lys-4-ONylated proteins.2 Histones were then extracted and subjected to an azide-alkyne click chemistry to conjugate the alkynyl-4-ONE labeled proteins with a rhodamine dye (Figure 3A and Figure S4). The alkynyl-4-ONE could label histones in a dose dependent manner (Figure S1). In contrast, histone acetylation level showed no obvious changes upon the increased 4-ONE treatment (Figure 3B). Treating cells with nicotinamide and TM molecule inhibitors led to the increase of alkynyl-4-ONE labeling on histones (Figure 3C). The potent and SIRT2-specific inhibitor, TM, led to much stronger labeling, suggesting that SIRT2 regulates lysine 4-ONylation in cells. To further confirm this, we carried out the labeling experiments in control and SIRT2 knockdown (KD) MEF cells. As shown in Figure 3D, the labeling in SIRT2 KD cells was stronger than that in control cells, suggesting the reduction of SIRT2 resulted in the accumulation of the 4-ONylation on histones and non-histone proteins. To make sure that SIRT2 can directly remove the 4-ONylation on these proteins, we added purified recombinant SIRT2 into histones extracted from SIRT2 KD cells that were treated with a4-ONE. SIRT2 treatment led to dramatic reduction of the alkynyl-4-ONE labeling on histones, supporting SIRT2 directly remove the 4-ONylation on these proteins (Figure 3E). The SIRT2 F119A mutant with diminished activity on (4-ONyl) peptide also had much weaker activity on the extracted histones (Figure 3E). Taken together, we conclude that SIRT2 can interact with histones and hydrolyze 4-ONyl lysine in living cells.
Figure 3.
SIRT2 regulates alkynyl-4-ONyl level on histones. (A) A scheme of showing the use of a chemical reporter, alkynyl-4-ONE, to detect histone 4-ONylation. (B) Western blot (WB) showing increased SIRT2 and the similar acetyl level on H3 (H3 Ac) in extracted histones upon 4-ONE treatment (10, 25 and 50 μM for 3 h). H3 was used as a loading control. (C) Modification of histones by alkynyl-4-ONyl in the absence and presence of SIRT2 inhibitors. (D) Analysis of alkynyl-4-ONE labeling on histones from control and SIRT2 knockdown cells. SIRT2 knockdown was confirmed by Western blot and H3 was used as loading control. MEF cells were labeled with alkynyl-4-ONE for 3 h. Histones were extracted and click chemistry was carried out to attach rhodamine. The modification level was readout by rhodamine fluorescence (Rho) after SDS-PAGE. (E) The ability of SIRT2 WT and F119A mutant to remove alkynyl-4-ONyl from proteins isolated from SIRT2 knockdown cells. The Coomassie blue (CB) stained gel was used to show equal loading in different lanes.
To make sure the endogenous 4-ONyl modification on histones (modification without adding exogenous 4-ONE) could also be regulated by SIRT2, we examined the endogenous 4-ONyl modification in SIRT2 WT and knockout (KO) MEF cells by LC-MS/MS. Lipopolysaccharide was used to treat cells and then histones were extracted for LC-MS/MS analysis. We consistently detected the 4-ONyl modification on histone H3K23 as reported (Figure S5A and S5B).2 The quantification result showed that the 4-ONyl H3K23 level in SIRT2 KO MEFs was 1.64-fold higher than that in WT MEFs (Figure 4). The LC-MS/MS analysis demonstrates SIRT2 is able to regulate endogenous 4-ONylation on histones.
Figure 4.
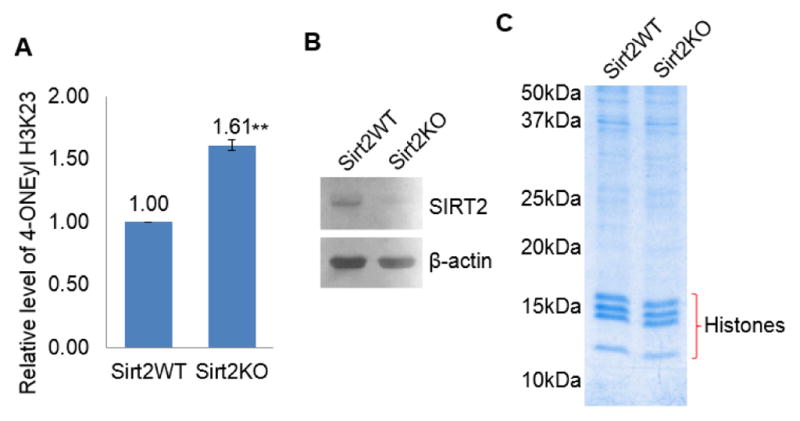
SIRT2 regulates endogenous 4-ONyl level on histones in mouse embryonic fibroblasts (MEFs). (A) MS quantification of 4-ONyl H3K23 levels in SIRT2 WT and KO MEFs based. The levels of 4-ONyl H3K23 is normalized by unmodified H3 Y41-R49 peptide. The results were from two independent biological replicates. (B) Western blot analysis showing the levels of SIRT2 in WT and KO MEFs. (C) Coomassie blue-stained gel showing histones extracted from SIRT2 WT and KO MEFs that were used for the MS quantification.
Upon stress, the 4-ketoamide adduct on histones disrupts the interaction between the histone H3 and DNA, therefore preventing the nucleosome assembly2. Thus, the ability of SIRT2 to remove the 4-ONyl groups on histones serves to mitigate the negative impact of protein 4-ONylation caused by oxidative stress. Our studies demonstrate that SIRT2 is able to hydrolyze 4-ONyl lysine on endogenous histones in living cells and reveal a lone pair-π interaction for the substrates recognition. This is first time a mechanism to reverse protein lysine 4-ONylation is reported. It broadens the activity scope of sirtuins and more importantly provides a new research direction to understand the role of SIRT2 in oxidative stress.
Supplementary Material
Acknowledgments
This work was supported in part by HKU-SPF foundation (Code. 201409176152 to Y.W.); National Natural Science Foundation of China (No. 21302027 and 21662010 to B.H.); Innovation Team of Natural Science Foundation of Department of Education of Guizhou Province (No. QJHRCTDZ [2015] 57 to B.H.), and the National Institute of Health (DK107868 to H.L.). The crystallographic data were collected on BL17U of Shanghai Synchrotron Radiation Facility. Atomic coordinates and structure factors have been deposited with the Protein Data Bank under accession codes 5G4C (SIRT2/H3K23-4-ONyl/carba-NAD)
Footnotes
Author Contributions 175
J.J., B.H., and Y.W. contributed equally to this work.
Notes
The authors declare no competing financial interests.
This Supporting Information is available free of charge on the ACS Publication website at DOI: 10.1021/jacs.6b04977.
References
- 1.Kouzarides T. Cell. 2007;128:693. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Galligan JJ, Rose KL, Beavers WN, Hill S, Tallman KA, Tansey WP, Marnett LJ. J Amer Chem Soc. 2014;136:11864. doi: 10.1021/ja503604t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frye RA. Biochem Biophys Res Commun. 2000;273:793. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]
- 4.Sauve AA, Wolberger C, Schramm VL, Boeke JD. Annu Rev Biochem. 2006;75:435. doi: 10.1146/annurev.biochem.74.082803.133500. [DOI] [PubMed] [Google Scholar]
- 5.Michan S, Sinclair D. Biochem J. 2007;404:1. doi: 10.1042/BJ20070140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. Mol Biol Cell. 2005;16:4623. doi: 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eskandarian HA, Impens F, Nahori MA, Soubigou G, Coppee JY, Cossart P, Hamon MA. Science. 2013;341:1238858. doi: 10.1126/science.1238858. [DOI] [PubMed] [Google Scholar]
- 8.Vaquero A, Scher MB, Lee DH, Sutton A, Cheng HL, Alt FW, Serrano L, Sternglanz R, Reinberg D. Genes Dev. 2006;20:1256. doi: 10.1101/gad.1412706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garrity J, Gardner JG, Hawse W, Wolberger C, Escalante-Semerena JC. J Biol Chem. 2007;282:30239. doi: 10.1074/jbc.M704409200. [DOI] [PubMed] [Google Scholar]
- 10.Smith BC, Denu JM. J Biol Chem. 2007;282:37256. doi: 10.1074/jbc.M707878200. [DOI] [PubMed] [Google Scholar]
- 11.Bao X, Wang Y, Li X, Li XM, Liu Z, Yang T, Wong CF, Zhang J, Hao Q, Li XD. eLife. 2014:3. doi: 10.7554/eLife.02999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mathias RA, Greco TM, Oberstein A, Budayeva HG, Chakrabarti R, Rowland EA, Kang Y, Shenk T, Cristea IM. Cell. 2014;159:1615. doi: 10.1016/j.cell.2014.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Du J, Zhou Y, Su X, Yu JJ, Khan S, Jiang H, Kim J, Woo J, Kim JH, Choi BH, He B, Chen W, Zhang S, Cerione RA, Auwerx J, Hao Q, Lin H. Science. 2011;334:806. doi: 10.1126/science.1207861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tan M, Peng C, Anderson KA, Chhoy P, Xie Z, Dai L, Park J, Chen Y, Huang H, Zhang Y, Ro J, Wagner GR, Green MF, Madsen AS, Schmiesing J, Peterson BS, Xu G, Ilkayeva OR, Muehlbauer MJ, Braulke T, Muhlhausen C, Backos DS, Olsen CA, McGuire PJ, Pletcher SD, Lombard DB, Hirschey MD, Zhao Y. Cell Metabolism. 2014;19:605. doi: 10.1016/j.cmet.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang H, Khan S, Wang Y, Charron G, He B, Sebastian C, Du J, Kim R, Ge E, Mostoslavsky R, Hang HC, Hao Q, Lin H. Nature. 2013;496:110. doi: 10.1038/nature12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teng YB, Jing H, Aramsangtienchai P, He B, Khan S, Hu J, Lin H, Hao Q. Sci Rep. 2015;5:8529. doi: 10.1038/srep08529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feldman JL, Dittenhafer-Reed KE, Kudo N, Thelen JN, Ito A, Yoshida M, Denu JM. Biochemistry. 2015;54:3037. doi: 10.1021/acs.biochem.5b00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feldman JL, Baeza J, Denu JM. J Biol Chem. 2013;288:31350. doi: 10.1074/jbc.C113.511261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jing H, Hu J, He B, Negron Abril YL, Stupinski J, Weiser K, Carbonaro M, Chiang YL, Southard T, Giannakakou P, Weiss RS, Lin H. Cancer Cell. 2016;29:607. doi: 10.1016/j.ccell.2016.03.011. [DOI] [PubMed] [Google Scholar]
- 20.Yuan H, Marmorstein R. J Biol Chem. 2012;287:42428. doi: 10.1074/jbc.R112.372300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Egli M, Sarkhel S. Accounts Chem Res. 2007;40:197. doi: 10.1021/ar068174u. [DOI] [PubMed] [Google Scholar]
- 22.Jain A, Purohit CS, Verma S, Sankararamakrishnan R. J Phys Chem B. 2007;111:8680. doi: 10.1021/jp072742l. [DOI] [PubMed] [Google Scholar]
- 23.Shibata T, Iio K, Kawai Y, Shibata N, Kawaguchi M, Toi S, Kobayashi M, Kobayashi M, Yamamoto K, Uchida K. J Biol Chem. 2006;281:1196. doi: 10.1074/jbc.M509065200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.