Abstract
The histone deacetylase (HDAC) family regulates several biological pathways through the deacetylation of lysine residues on histone and non-histone proteins. Mammals have 18 HDACs that are classified into four classes. Class I, II, and IV are zinc-dependent while class III is nicotinamide adenine dinucleotide (NAD+)-dependent lysine deacetylase or sirtuins. HDAC8, a class I HDAC family member, has been shown to have low deacetylation activity compared to other HDACs in vitro. Recent studies showed that several sirtuins, with low deacetylase activities can actually hydrolyze other acyl lysine modifications more efficiently. Inspired by this, we tested the activity of HDAC8 using a variety of different acyl lysine peptides. Screening a panel of peptides with different acyl lysine modifications, we found that HDAC8 can catalyze the removal of acyl groups with 2-16 carbons from lysine 9 of the histone H3 peptide (H3K9). Interestingly, the catalytic efficiencies (kcat/Km) of HDAC8 on octanoyl, dodecanoyl, and myristoyl lysine are several folds better than that on acetyl lysine. The increased catalytic efficiencies of HDAC8 on larger fatty acyl groups are due to the much lower Km values. T-cell leukemia Jurkat cells treated with a HDAC8 specific inhibitor, PCI-34051, exhibited an increase in global fatty acylation compared to control treatment. Thus, the defatty-acylation activity of HDAC8 is likely physiologically relevant. This is the first report of a zinc-dependent HDAC with defatty-acylation activity and identification of HDAC8 defatty-acylation targets will help to further understand the function of HDAC8 and protein lysine fatty acylation.
INTRODUCTION
Epigenetic modification of histones plays a crucial role in the regulation of gene expression. Histone acetylation and deacetylation on lysine residues is mediated by histone acetyltransferases (HATs) and histone deacetylases (HDACs), respectively.1 HDACs have been classified into four classes: class I, homologous to yeast Rpd3, consists of HDAC1, 2, 3 and 8; class II, homologous to yeast Hda1, is divided into two subclasses: IIa (HDAC4, 5, 7, 9) and IIb (HDAC6, 10) that contain one and two catalytic domains, respectively; class III HDACs are NAD+-dependent deacetylase, known as sirtuins; and class IV is comprised solely of HDAC11 which contains a deacetylase domain homologous to class I and class II.2
HDAC8, belonging to class I, is expressed in several human tissues (e.g. lung, heart, brain and kidney) and is localized in both the nucleus and the cytoplasm.3-5 HDAC8 deletion in mice causes perinatal lethality due to brain trauma, implying HDAC8 plays a role in skull development.6 The growth of certain cancer cell lines (A549, HeLa, and HCT116) is inhibited when HDAC8 is knocked down; this suggests that HDAC8 is important for cancer cell survival.7 HDAC8 expression level is positively correlated to poor prognosis and metastasis of neuroblastoma.8 Therefore, HDAC8 is considered as a promising therapeutic target for different cancers. Thus, understanding the function of HDAC8 may help the development of cancer therapeutics. However, so far, only two proteins have been validated as HDAC8 deacetylation targets, estrogen-related receptor alpha (ERRα)9 and structural maintenance of chromosomes protein 3 (SMC3).10
One of the key limiting factors towards understanding the function of HDAC8 is the weak deacetylase activity of HDAC8 in vitro. Previous kinetics studies of different HDACs (HDAC1, 2, 3, 6, 8 and 10)11 show that all of the HDACs have Km values between 20-40 μM, except for HDAC8 which has a significantly higher Km (>200 μM). Overall, HDAC8 has lowest kcat/Km value on acetyl lysine peptides (~60±7 M−1S−1) when compared to other HDACs tested.11 Recently, several class III HDACs, or sirtuins, that possess weak acetylation activity in vitro, have been shown to efficiently remove other acyl groups from lysine residues. For example, SIRT5 exhibits efficient activity to lysine desuccinylation, demalonylation and glutarylation12-14 and SIRT1, 2, 3 and SIRT6 are capable of hydrolyzing long chain fatty acyl groups from lysine residues.15-17 Therefore, we hypothesized that HDAC8 may have other enzymatic activities beyond deacetylation.
Here, we have synthesized a library of peptides with different acyl lysine modifications and examined the ability of HDAC8 to remove the acyl groups from the peptides. We have found that HDAC8 can catalyze the removal of long chain fatty-acyl groups from different peptide backbone sequences. The defatty-acylation activity of HDAC8 is several folds higher than its deacetylation activity. We have also obtained data suggesting that the defatty-acylase activity of HDAC8 is likely physiologically relevant.
RESULTS AND DISCUSSION
HDAC8 can remove long chain fatty acyl groups in vitro
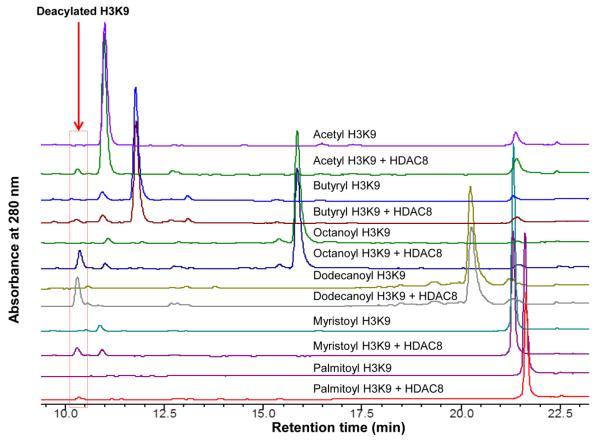
We overexpressed and purified recombinant HDAC8 from E. coli BL21 cells for studying the enzymatic activity of HDAC8. For peptide substrates (Table 1), we first focused on the H3K9 peptide sequence and varied the acyl groups. Using an HPLC-based assay, we found HDAC8 could remove both acetyl and several larger acyl groups (butyryl, octanoyl, dodecanoyl, myristoyl, and palmitoyl) from the lysine residue of H3K9 peptides (Table 1 and Fig. 1). However, HDAC8 could not remove the negatively charged succinyl or malonyl groups from H3K9 peptides. The bulkier biotinyl and lipoyl groups were also not good HDAC8 substrates (Table 1 and Supplementary Fig. S1).
Table 1.
The acyl peptide library for HDAC8 activity screening.
| Proteins | Peptide sequencesa, b | Acyl group | Hydrolyzed by HDAC8 |
|---|---|---|---|
| Histone H3K9 | KQTARKSTGGWW | acetyl | +++ |
| butyryl | + | ||
| octanoyl | +++ | ||
| dodecanoyl | +++ | ||
| myristoyl | +++ | ||
| palmitoyl | ++ | ||
| succinyl | − | ||
| malonyl | − | ||
| biotinyl | − | ||
| lipoyl | − | ||
| Histone H4K16 | KGGAKRHRKWW | acetyl | − |
| myristoyl | − | ||
| Histone H2BK12 | APAPKKGSKKWW | acetyl | ++ |
| myristoyl | ++ | ||
| Histone H3K18 | GGKAPRKQLATKAWW | acetyl | +++ |
| myristoyl | − | ||
| TNFα | EALPKKTGGPQWW | acetyl | − |
| myristoyl | − | ||
| Histone H2AK119 | VLLPKKTESHWW | acetyl | + |
| crotonyl | − | ||
| Histone H3K56 | YQKSTELLWW | acetyl | + |
| crotonyl | − | ||
| Histone H2BK5 | PEPSKSAPAPKWW | 2-hydroxyisobutyryl | − |
| Histone H3K122 | TIMPKDIQLAWW | 2-hydroxyisobutyryl | − |
| Histone H4K8 | RGKGGKGLGKGWW | 2-hydroxyisobutyryl | − |
| Histone H4K77 | TEHAKRKTVWW | 2-hydroxyisobutyryl | − |
| Gelsolin | GLGLSYLSSWW | N-terminal myristoyl | − |
| PAK2 | GAAKSLDKQKWW | − | |
| Gα protein | GGDASGEWW | − |
The modified residue is underlined
The Mass Spectrometry data were shown in Supplementary Figure S2
Figure 1.
HPLC traces of HDAC8 activity assay with different acyl peptides as substrates. HDAC8 can remove the acetyl group, and other long chain acyl groups with different efficiencies.
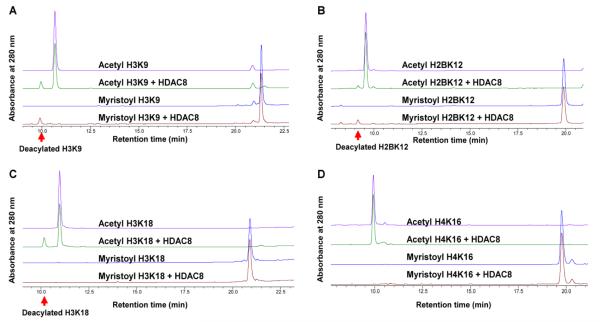
We then compared the deacetylation and demyristoylation activities on a few different peptide sequences. We were able to detect both deacetylation and demyristoylation activity on the H3K9 and H2BK12 peptides (Fig. 2A and 2B). However, only deacetylation activity was observed on H3K18 peptides (Fig. 2C), and neither activity was detected on H4K16 and TNFαK20 peptides (Fig. 2D and Supplementary Fig. S1). Therefore, HDAC8 enzymatic activity is dependent on the peptide sequence.
Figure 2.
HPLC traces showing HDAC8 selectivity on different histone peptides. (A-B) HDAC8 can catalyze deacetylation and demyristoylation on both H3K9 and H2BK12 peptides. (C) Only deacetylation activity was detected on H3K18 peptide. (D) No deacylation activity was detected on the H4K16 peptide.
Recently, lysine crotonylation and 2-hydroxyisobutyrylation have also been reported to occur on histone proteins.18, 19 Thus, we also tested HDAC8 on a few histone peptides bearing crotonyl (H2AK119, and H3K56) and 2-hydroxyisobutyryl groups (H2BK5, H3K122, H4K8, and H4K77) (Table 1). Except H3K56 crotonylation, all other sites have been reported to have the crotonyl or 2-hydroxyisobutyryl modifications. HDAC8 could remove acetyl groups, but not crotonyl groups from both the H2AK119 and H3K56 peptides (Table 1 and Supplementary Fig. S1). Similarly, HDAC8 could not remove 2-hydroxyisobutyryl groups from any of the peptides tested (Table 1 and Supplementary Fig. S1).
Protein N-terminal glycine myristoylation is a well-known PTM that is important for cell signaling. Given that HDAC8 can catalyze lysine demyristoylation, we also synthesized several peptides bearing N-terminal glycine myristoyl group, and tested whether HDAC8 could catalyze glycine demyristoylation. These peptides are from three proteins that are known to be modified by glycine myristoylation: gelsolin,20 p21-activated kinase 2 (PAK2),21 and Gα protein, transducin.22 However, none of the N-terminal glycine myristoylated peptides were HDAC8 substrates (Table 1 and Supplementary Fig. S1).
To further investigate the defatty-acylation activity of HDAC8, we carried out kinetics studies using H3K9 peptides with different acyl groups as the substrates. The kinetics parameters and the Michaelis-Menten plots of each acyl peptide substrate are shown in Table 2 and Supplementary Figure S3, respectively. The HDAC8 catalytic efficiencies (kcat/Km values) on the longer fatty acyl groups, octanoyl-, dodecanoyl-, and myristoyl-H3K9, are two to three times higher than that on the acetyl peptide. For deacetylation, we could not obtain the Km values as the initial reaction rate was linear with the acetyl peptide concentrations we used. In contrast, the Km value for the myristoyl H3K9 peptide was approximately 16 μM.
Table 2.
Kinetics parameters (± S.D.) of recombinant HDAC8 on different H3K9 acyl peptides.
| Substrate | kcat (S−1) | Km (uM) | kcat/Km (S−1.M−1) |
|---|---|---|---|
|
| |||
| Acetyl H3K9 | NDa | >600 | 58.0 ± 1.1 |
| NDa,b | >300b | 480.8b | |
|
| |||
| Butyryl H3K9 | NDa | >200 | 12.6 ± 0.3 |
|
| |||
| Octanoyl H3K9 | 0.0202 ± 0.0005 | 145.8 ± 10.3 | 139 ± 13 |
|
| |||
| Dodecanoyl H3K9 | 0.0123 ± 0.0013 | 69.5 ± 10.1 | 177 ± 7 |
|
| |||
| Myristoyl H3K9 | 0.0019 ± 0.0003 | 15.8 ± 5.1 | 120 ± 11 |
| 0.0755b | 60.5b | 1,246b | |
|
| |||
| Palmitoyl H3K9 | 0.00050 ± 0.00007 | 18.3 ± 7.4 | 28.8 ± 7 |
ND: Not determined since initial rate (V0) versus [Substrate] was linear. The kcat/Km value was obtained from the slope of the linear plot.
Recombinant HDAC8 from Sf9 insect cells
The catalytic efficiencies of HDAC8 on acetyl and myristoyl H3K9 peptides were still low compared to other HDACs. Our E. coli expression system for HDAC8 might have caused the low activity of HDAC8 due to a lack of certain post-translational modifications (PTMs).23 Therefore, we also used HDAC8 expressed from Sf9 insect cells for kinetics studies for both the acetyl- and myristoyl-H3K9 peptides. Indeed, we obtained higher kcat/Km values for both deacetylation and demyristoylation from HDAC8 expressed from Sf9 insect cells. Although the HDAC8 purified from insect cells gave much higher activity, the demyristoylation catalytic efficiency remained >2 times higher than that of deacetylation.
HDAC inhibitors, PCI-34051 and SAHA, can inhibit both the demyristoylation and deacetylation activities of HDAC8
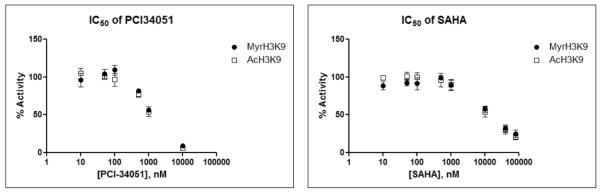
Several HDAC inhibitors have been developed which can inhibit the deacetylation activity of HDACs.24 We wanted to test whether they would inhibit the deacetylation and demyristoylation activities differently. We tested both a HDAC8 specific inhibitor, PCI-34051,25 and a class I and IIb-HDAC inhibitor, SAHA.26 As shown in Figure 3, both the demyristoylation and deacetylation activities of HDAC8 were inhibited by the two inhibitors in a dose response manner. PCI-34051 was able to inhibit both catalytic activities with ten times lower IC50 values compared to SAHA (Table 3). Therefore, PCI-34051 can efficiently inhibit both of demyristoylation and deacetylation activities of HDAC8. Notably, PCI-34051 inhibited the deacylase activity of HDAC8 on both the acetyl- and myristoyl-H3K9 peptides with comparable potency. We also obtained similar results using SAHA. Thus, it is possible that the inhibitors competitively bind to the peptide-binding pocket rather than the acetyl- or myristoyl-binding pocket of the enzyme.
Figure 3.
Inhibition curves of PCI-34051 and SAHA on the demyristoylation and deacetylation activities of HDAC8.
Table 3.
IC50 (± S.D.) of PCI-34051 and SAHA for HDAC8’s deacetylation and demyristoylation activities on H3K9 peptides.
| HDAC inhibitors |
IC50 ± SD (μM) | |
|---|---|---|
| Demyristoylation | Deacetylation | |
| PCI-34051 | 0.98 ± 0.04 | 1.15 ± 0.14 |
| SAHA | 9.92 ± 0.50 | 9.56 ± 0.58 |
Effect of the HDAC8 specific inhibitor (PCI-34051) on global fatty acylation level in mammalian cells
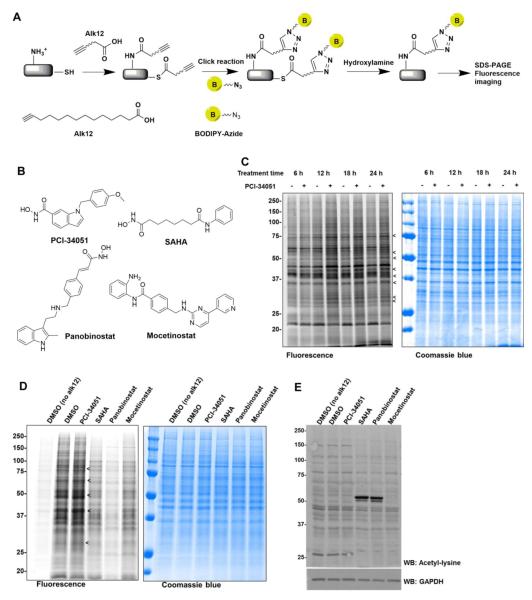
Since PCI-34051 can efficiently inhibit HDAC8 demyristoylation in vitro, we hypothesized that if the HDAC8 defatty-acylation activity is physiologically relevant, treating cells with PCI-34051 should increase protein lysine fatty-acylation level. To test this, we used a metabolic labeling method involving a bioorthogonal probe for protein myristoylation, an alkyne-tagged myristic acid analogue (Alk12).27 After treating different mammalian cell lines with PCI-34051, we cultured them in the presence of Alk12 which was metabolically incorporated into fatty-acylated proteins. The alkyne-labeled proteins were conjugated to a fluorescent tag (BODIPY-azide) via click chemistry. The proteins were then precipitated and incubated in 0.5 M hydroxylamine solution pH 8.0 at 95 °C for 7 min to remove fatty acyl modifications on cysteine residues.15 The fluorescently labeled proteins were then visualized after SDS-PAGE separation (Fig. 4A). We found that HeLa, HEK-293T, and MDA-MB 231 cells treated with PCI-34051 showed slightly increased fatty acylation compared to the control that was treated with DMSO (Supplementary Fig. S4). In contrast, the treatment of T lymphocyte Jurkat cells with PCI-34051 significantly increased the fatty acylation level (Fig. 4C). The increase in protein fatty acylation was time- and dose-dependent (Fig. 4C, Supplementary Fig. S5A and S5B). Moreover, similar results were obtained when we used Alk14, a palmitic acid analogue, as a metabolic labeling probe (Supplementary Fig. S5C).
Figure 4.
Global fatty acylation Jurkat cells treated with PCI-34051. (A) Method for the detection of global protein lysine fatty acylation in Jurkat cells. Cells were cultured with the myristic acid analogue (Alk12) to allow metabolic labeling to occur. BODIPY-azide (B-N3) was then conjugated to the alkyne group using click chemistry. The modifications on cysteine residues were removed by treating with 0.5 M hydroxylamine, pH 8.0, and the fluorescent signal was imaged after SDS-PAGE. (B) Chemical structures of different HDAC inhibitors. (C) Global fatty acylation of Jurkat cells treated with 50 μM PCI-34051 with different treatment times. The arrows point to several protein bands with elevated fatty acylation signals. (D) The increase in global fatty acylation was specific to PCI-34051. Jurkat cells treated with 25 μM of different HDAC inhibitors for 15 hrs. Only PCI-34051 treated cells showed higher fatty acylation when compared to the control and other HDAC inhibitor treated cells. (E) Western blot showing the global acetylation pattern of Jurkat cells treated with different HDAC inhibitors. SAHA and Parabinostat increased protein lysine acetylation while other inhibitors did not.
To further prove that the elevated fatty acylation level was specific to PCI-34051, we treated Jurkat cells with 25 μM of other three HDAC inhibitors, SAHA, Panobinostat (LBH589), and Mocetinostat (MGCD0103) (Fig. 4B). SAHA and Panobinostat are established inhibitors of class I and IIb-HDACs26, 28, 29 while Mocetinostat can only inhibit HDAC1, 2, 3 and 11.30 As we expected, only PCI-34051-treated Jurkat cells exhibited enhanced global protein lysine fatty acylation (Fig. 4D). We also detected the global lysine acetylation level by Western blot, and found that SAHA and Panobinostat predominantly increased the acetylation of protein bands around 55 kDa, while PCI-34051 and Mocetinostat did not noticeably increase protein lysine acetylation (Fig. 4E). Notably, the global fatty acylation labeling signal (measured by Alk12 labeling) from the cells treated with SAHA and Panobinostat was much lower than the control. The lower fatty acylation labeling signal could be due to the cytotoxicity of these inhibitors. To test this possibility, we also treated the cells with lower concentrations of these inhibitors, but still did not observe an increase in the fatty acylation signal measured by Alk12 labeling (Supplementary Fig. S5D). This result suggests that the HDAC8-specific inhibitor PCI-34051 is unique in the ability to increase protein fatty acylation. We showed here that HDAC8 can efficiently catalyze defatty-acylation, including deoctanoylation, dedodecanoylation, and demyristoylation. While some of the sirtuins, the NAD+-dependent HDACs, have been recognized as protein defatty-acylases,12,16,17 this is the first report of a zinc-dependent HDAC possessing this enzymatic activity. The myristoyl peptide has a much lower Km values compared to the acetyl peptide leading to a higher catalytic efficiency of demyristoylation over deacetylation.
Crystal structures of human HDAC8, in complex with different inhibitors, have been reported; they revealed a single α/β-domain with a core eight-stranded parallel β-sheet and 11 α-helices.7, 31, 32 Interestingly, the complexes between HDAC8 and the inhibitors displayed structural variation on the surface of HDAC8 around the binding pocket. This region has been suggested to be conformational flexible, and this property might allow HDAC8 to accommodate different substrates.31 This therefore may allow the binding of long chain fatty acyl peptides at the active site, enabling the defatty-acylation activity.
Protein fatty acylation has been known to play an essential role in cell signaling, membrane trafficking, protein-membrane interactions, and cellular localization.33 The majority of these fatty-acylated proteins have been reported to have N-glycine myristoylation or S-palmitoylation, while only a few proteins are reported to have lysine fatty acylation.15,34 Several NAD+-dependent deacetylase, SIRT1, SIRT2, SIRT3 and SIRT6, have been previously reported to possess lysine defatty-acylation activities.15-17 In this study, we also showed that zinc-dependent deacetylase, HDAC8, is another lysine defatty-acylase.
PCI-34051, a potent and selective HDAC8 inhibitor, can inhibit not only the deacetylation, but also the demyristoylation activity of HDAC8. PCI-34051 treatment in mammalian cells can enhance the global fatty acylation (measured by Alk12/Alk14 labeling) in a dose response manner, while other HDAC inhibitors did not. These results suggest that HDAC8 lysine defatty-acylation is likely physiologically relevant. The identification of an additional enzyme with lysine defatty-acylation activity suggests that lysine fatty acylation might be a more abundant modification than previously recognized, and it may play important roles in biology.
Thus far, only two direct HDAC8 targets, ERRα and SMC3, have been substantially validated. We identified the first zinc-dependent HDAC to possess defatty-acylase activity. The discovery of the defatty-acylase activity suggests that when searching for other HDAC8 targets we also need to consider the defatty-acylase activity of HDAC8. Our work will thus further facilitate the identification of additional substrate proteins for HDAC8 and help elucidate the physiological role of this enzyme.
METHODS
Material and reagents
Recombinant human HDAC8 from Sf9 cells was purchased from BPS Bioscience (cat#50008). HDAC inhibitors (PCI-34051, SAHA, Panobinostat, and Mocetinostat) were purchased from Selleckchem.com (cat#S2012, S1047, S1030, S1122, respectively). Solvents for peptide synthesis were purchased from Fisher unless otherwise indicated. Wang resin, Fmoc-protected amino acids, and derivatives were purchased from Chem-Impex.
Antibody
Anti-acetyl Lysine antibody (Rabbit polyclonal IgG) was purchased from Abcam (cat# 80178). The secondary antibody, rabbit anti-goat IgG-HRP, was purchased from SantaCruz (cat# 2768).
Cell Culture
HeLa, Human Embryonic Kidney 293T (HEK293T) and MDA-MB-231 cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% Fetal Bovine Serum (FBS) (Invitrogen). Jurkat cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 Media (Invitrogen) supplemented with 10% FBS. Cells were incubated in a humidified incubator at 37 °C with 5% CO2.
Cloning, expression and purification of HDAC8
Human HDAC8 cDNA was purchased from Open Biosystems (clone ID: 5761745). The full length cDNA was PCR-amplified by Platinum® Pfx DNA Polymerase (ThermoFisher) and subcloned into the pET28a vector using the EcoRI and XhoI restriction sites with the following primers: Sense: 5′-agtcagGAATTCATGGAGGAGCCGGAGGAACC-3′ Antisense: 5′-agtcagCTCGAGCTAGACCACATGCTTCAGATT-3′ The plasmid was transformed into E. coli BL21 Rosetta™ 2 competent cells (Novagen, Cat# 71402) for protein expression. The cells pellets were collected and washed with deionized water once before being dispersed in Buffer 1 (20 mM Tris-HCl pH 8.0, 500 mM NaCl, 10 mM MgCl2, 5 mM Imidazole, 5% glycerol, 10 μg/mL phenylmethylsulfonyl fluoride (PMSF)). The cells were lysed by passing through an EmulsiFlexTM-C3 cell disruptor (AVESTIN) three to four times, and the cell lysate was centrifuged to remove cell debris at 20,000 rpm at 4 °C for 45 min (Beckman Coulter). The supernatant containing HDAC8 was then loaded onto Ni-NTA agarose beads (QIAGEN, Cat#30210) and the beads were gently agitated at 4 °C for 2 hrs. The unbound supernatant was removed by gravity flow, and the beads were washed three times with Buffer 2 (20 mM Tris-HCl pH 8.0, 500 mM NaCl, 30 mM Imidazole). The protein was then eluted from the beads into eppendorf tubes with Buffer 3 (20 mM Tris-HCl pH 8.0, 500 mM NaCl, 10 mM KCl, and 50-300 mM Imidazole). Each collected fraction was run on SDS-PAGE to check for the presence of HDAC8 (molecular weight around 46 kDa). The fractions containing HDAC8 were combined and concentrated to 2-3 mL with an Amicon Ultra-4 10 kDa concentrator, and subsequently loaded onto a gel filtration column (Sephadex-75, GE healthcare), which was pre-equilibrated with 40 mM Tris-HCl pH 8.0, 110 mM NaCl, and 2.2 mM KCl. Each fraction, with UV absorption at 280 nm, was resolved by SDS-PAGE to detect HDAC8. The fractions containing HDAC8 with greater than 80% purity were pooled and concentrated using an Amicon Ultra-0.5 mL 10 kDa concentrator (EMD Millipore) and subsequently aliquoted into small volumes and stored at −80 °C.
In vitro HDAC8 activity and kinetics assay
The in vitro activity of HDAC8 was detected using an HPLC assay. Each enzymatic reaction consists of 25 μM of different acyl peptides in the reaction buffer (25 mM Tris-HCl pH 8, 137 mM NaCl, 2.7 mM KCl, 1 mM MgCl2) with a final volume of 60 μl. To begin a reaction, 1-5 μM of HDAC8 (depending on the activity) was added into the reaction and incubated at 37 °C for 1 hr. The reactions were quenched with 60 μl of quench buffer (100 mM hydrochloric acid, 160 mM acetic acid in 50% acetonitrile) and centrifuged at 17,000x g for 15 min at room temperature to remove HDAC8. The reaction was analyzed by reverse phase HPLC with a Kinetex 5U XB C18 column (100A, 150 mm × 4.60 mm, Phenomenex) monitoring at the wavelength 280 nm. The mobile phase A was water with 0.1% (v/v) trifluoroacetic acid (TFA), the mobile phase B was acetonitrile with 0.1% (v/v) TFA. The gradient used was 10 to 100% mobile phase B over 30 min with the flow rate of 0.5 mL/min. The product formation was verified by LC-MS (LCQ Fleet, Thermo Scientific).
The kinetics parameters of different acyl peptides were determined using the above reaction conditions and by varying the concentrations of each substrate as follows, 0-1,200 μM for acetyl H3K9; 0-400 μM for butyryl H3K9; 0-400 μM for octanoyl H3K9; 0-100 μM for dodecanoyl H3K9; 0-100 μM for myristoyl H3K9; 0-65 μM for palmitoyl H3K9. Each reaction was incubated with 1 μM HDAC8 at 37 °C for 1 hr. Each reaction was performed in duplicate to ensure reproducibility. After separation by HPLC, the product and remaining substrate peaks were quantified and converted to initial rates. The plots between substrate concentrations and the initial rates were fitted using GraphPad Prism® (± S.D., Standard Deviation).
In vitro inhibition assay of HDAC8 activities
Each reaction was performed with 1 μM of HDAC8 in the reaction buffer (25 mM Tris-HCl pH 8.0, 137 mM NaCl, 2.7 mM KCl, 1 mM MgCl2), which was pre-incubated with different concentrations of the inhibitors at room temperature for 15 min. Subsequently, 25 μM of the substrate peptide was added and the reaction was incubated at 37 °C for 60 min before HPLC analysis to determine the amount of product formed. The data (± S.D.) were fitted by using GraphPad Prism® to give the IC50 values shown in Table 3.
Acyl peptide synthesis
The different acyl peptide backbones were synthesized using standard solid phase peptide synthesis.35 Two tryptophan residues were added to the C-terminal of each peptide to enable facile detection of the peptide at 280 nm. A detailed method for acyl peptide synthesis can be found in the Supporting Information. The synthesized peptides were verified using LC-MS (LCQ Fleet, Thermo Scientific).
Metabolic labeling of mammalian cells treated with PCI-34051
The metabolic labeling method was modified from Yount et al 2011.27 A total of 1×106 Jurkat cells in each flask were grown overnight, and then incubated with 25 or 50 μM PCI-34051 or DMSO (control) for 15 hrs. The cells were then treated with 50 μM Alk12 (or Alk14) in the presence of the inhibitor for an additional 6 hrs. Cells were collected and washed with 1x phosphate saline buffer (PBS) three times. The cell pellets were lysed with 4% sodium dodecyl sulfate (SDS) buffer (50 mM triethanolamine pH 7.4, 150 mM NaCl, 4% (w/v) SDS) containing protease inhibitor cocktails (Sigma). A nuclease (Pierce™ Universal Nuclease for Cell Lysis) was added into the total lysate. The protein concentration was determined by the bicinchoninic acid (BCA) assay using the Pierce™ BCA Protein Assay Kit (ThermoFisher). A total of 50 μg of proteins from each sample was aliquoted, and adjusted to a final volume of 45 μl with 4% SDS buffer.
To perform the click reaction, 5.6 μl of the click chemistry reaction master mix was added to each sample (click reaction master mix per sample: 3 μl of 1 mM BODIPY Azide in DMF, 1 μl of 50 mM tris(2-carboxyethyl) phosphine hydrochloride (TCEP-HCl, Calbiochem) in water, 0.6 μl of 10 mM tris [(1-benzyl-1H-1,2,3-triazol-4-yl)methyl] amine (TBTA, Anal Tech) in DMF, 1 μl of 50 mM CuSO4 in water). After 1 hr at room temperature, 200 μl methanol, 75 μl chloroform and 150 μl water were added to each sample. After vortexing, the samples were centrifuged at 17,000 xg for 20 min at 4 °C. The supernatant was gently removed by pipetting, and to each pellet was added 1 mL of methanol. The samples were again vortexed and spun down at 17,000 xg for 10 min at 4°C. The methanol was removed, and the protein pellets were washed again with 1 mL methanol. After the second methanol wash, the protein pellets were air dried at room temperature for 10-15 min, and then resolubilized in 50 μl of 4% SDS buffer (50 mM triethanolamine pH 7.4, 150 mM NaCl, 4% (w/v) SDS). To each sample, 10 μl of 6x loading buffer (374 mM Tris-HCl pH 6.8, 12% SDS, 600 mM DTT, 60% v/v glycerol, 0.06% bromophenol blue) was added, and the samples were boiled at 95 °C for 5 min. To remove cysteine palmitoylation, 9 μl of each sample was mixed with 1 μl of 5 M Hydroxylamine pH 8.0, and heated at 95 °C for 7 min. The samples were then resolved on 12% SDS-PAGE gel. The gel was destained in 50% (v/v) acetic acid, 40% (v/v) methanol and 10% (v/v) water for at least 2 hrs at room temperature (or overnight at 4 °C), and destained in water for another 30 min. The fluorescent signal was visualized with a Typhoon 9400 Variable Mode Imager (GE Healthcare Life Sciences) using 488 nm excitation and 520 nm detection filters, and a PMT setting of 550 V. The signal was analyzed by Image Quant TL v2005.
Western Blot for protein lysine acetylation
Cells were collected and lysed with 4% SDS buffer containing Protease Inhibitor Cocktails and nuclease. Protein concentration was determined by BCA assay. Protein samples were separated by 12% SDS-PAGE and transferred to PVDF membrane (Bio-Rad) for 120 min. The membrane was blocked with 5% bovine serum albumin (BSA, Santa Cruz) and incubated with the primary antibody at 4 °C overnight. The membrane was incubated with the secondary antibody for another 1 hr at room temperature. The membrane was developed using ECL-Plus western blotting detection reagent (GE Healthcare). The signal was visualized using a Typhoon 9400 Variable Mode Imager (GE Healthcare) with 457 nm excitation and 526 nm detection filters, using a PMT of 600 V. The signal was analyzed by Image Quant TL v2005.
Supplementary Material
ACKNOWLEDGEMENT
This work is supported in part by National Institute of Health (GM098596, DK107868, and GM105933 to HL; and GM105933, DK107868, and GM115961 to YZ). PA is supported in part by a fellowship from the Royal Thai government.
Footnotes
AUTHOR CONTRIBUTIONS
PA and HL designed experiments and wrote the manuscript. NAS and BH synthesized Alk12, Alk14 and peptides. SPM, LD, and YZ synthesized peptides.
SUPPORTING INFORMATION. Figure S1-S5 and the method for synthesizing acyl peptides.
REFERENCES
- 1.Yang XJ, Seto E. HATs and HDACs: from structure, function and regulation to novel strategies for therapy and prevention. Oncogene. 2007;26:5310–5318. doi: 10.1038/sj.onc.1210599. [DOI] [PubMed] [Google Scholar]
- 2.Delcuve GP, Khan DH, Davie JR. Roles of histone deacetylases in epigenetic regulation: emerging paradigms from studies with inhibitors. Clin. Epigenetics. 2012;4:5. doi: 10.1186/1868-7083-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Waltregny D, Glenisson W, Tran SL, North BJ, Verdin E, Colige A, Castronovo V. Histone deacetylase HDAC8 associates with smooth muscle alpha-actin and is essential for smooth muscle cell contractility. FASEB J. 2005;19:966–968. doi: 10.1096/fj.04-2303fje. [DOI] [PubMed] [Google Scholar]
- 4.Yamauchi Y, Boukari H, Banerjee I, Sbalzarini IF, Horvath P, Helenius A. Histone deacetylase 8 is required for centrosome cohesion and influenza A virus entry. PLoS Pathog. 2011;7:e1002316. doi: 10.1371/journal.ppat.1002316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karolczak-Bayatti M, Sweeney M, Cheng J, Edey L, Robson SC, Ulrich SM, Treumann A, Taggart MJ, Europe-Finner GN. Acetylation of heat shock protein 20 (Hsp20) regulates human myometrial activity. J. Biol. Chem. 2011;286:34346–34355. doi: 10.1074/jbc.M111.278549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haberland M, Mokalled MH, Montgomery RL, Olson EN. Epigenetic control of skull morphogenesis by histone deacetylase 8. Genes Dev. 2009;23:1625–1630. doi: 10.1101/gad.1809209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vannini A, Volpari C, Filocamo G, Casavola EC, Brunetti M, Renzoni D, Chakravarty P, Paolini C, De Francesco R, Gallinari P, Steinkuhler C, Di Marco S. Crystal structure of a eukaryotic zinc-dependent histone deacetylase, human HDAC8, complexed with a hydroxamic acid inhibitor. Proc. Natl. Acad. Sci. USA. 2004;101:15064–15069. doi: 10.1073/pnas.0404603101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oehme I, Deubzer HE, Wegener D, Pickert D, Linke JP, Hero B, Kopp-Schneider A, Westermann F, Ulrich SM, von Deimling A, Fischer M, Witt O. Histone deacetylase 8 in neuroblastoma tumorigenesis. Clin. Cancer Res. 2009;15:91–99. doi: 10.1158/1078-0432.CCR-08-0684. [DOI] [PubMed] [Google Scholar]
- 9.Wilson BJ, Tremblay AM, Deblois G, Sylvain-Drolet G, Giguere V. An acetylation switch modulates the transcriptional activity of estrogen-related receptor alpha. Mol. Endocrinol. 2010;24:1349–1358. doi: 10.1210/me.2009-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deardorff MA, Bando M, Nakato R, Watrin E, Itoh T, Minamino M, Saitoh K, Komata M, Katou Y, Clark D, Cole KE, De Baere E, Decroos C, Di Donato N, Ernst S, Francey LJ, Gyftodimou Y, Hirashima K, Hullings M, Ishikawa Y, Jaulin C, Kaur M, Kiyono T, Lombardi PM, Magnaghi-Jaulin L, Mortier GR, Nozaki N, Petersen MB, Seimiya H, Siu VM, Suzuki Y, Takagaki K, Wilde JJ, Willems PJ, Prigent C, Gillessen-Kaesbach G, Christianson DW, Kaiser FJ, Jackson LG, Hirota T, Krantz ID, Shirahige K. HDAC8 mutations in Cornelia de Lange syndrome affect the cohesin acetylation cycle. Nature. 2012;489:313–317. doi: 10.1038/nature11316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schultz BE, Misialek S, Wu J, Tang J, Conn MT, Tahilramani R, Wong L. Kinetics and comparative reactivity of human class I and class IIb histone deacetylases. Biochemistry. 2004;43:11083–11091. doi: 10.1021/bi0494471. [DOI] [PubMed] [Google Scholar]
- 12.Du J, Zhou Y, Su X, Yu JJ, Khan S, Jiang H, Kim J, Woo J, Kim JH, Choi BH, He B, Chen W, Zhang S, Cerione RA, Auwerx J, Hao Q, Lin H. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science. 2011;334:806–809. doi: 10.1126/science.1207861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peng C, Lu Z, Xie Z, Cheng Z, Chen Y, Tan M, Luo H, Zhang Y, He W, Yang K, Zwaans BM, Tishkoff D, Ho L, Lombard D, He TC, Dai J, Verdin E, Ye Y, Zhao Y. The first identification of lysine malonylation substrates and its regulatory enzyme. Mol. Cell Proteomics. 2011;10:M111–012658. doi: 10.1074/mcp.M111.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tan M, Peng C, Anderson KA, Chhoy P, Xie Z, Dai L, Park J, Chen Y, Huang H, Zhang Y, Ro J, Wagner GR, Green MF, Madsen AS, Schmiesing J, Peterson BS, Xu G, Ilkayeva OR, Muehlbauer MJ, Braulke T, Muhlhausen C, Backos DS, Olsen CA, McGuire PJ, Pletcher SD, Lombard DB, Hirschey MD, Zhao Y. Lysine glutarylation is a protein posttranslational modification regulated by SIRT5. Cell Metab. 2014;19:605–617. doi: 10.1016/j.cmet.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang H, Khan S, Wang Y, Charron G, He B, Sebastian C, Du J, Kim R, Ge E, Mostoslavsky R, Hang HC, Hao Q, Lin H. SIRT6 regulates TNF-alpha secretion through hydrolysis of long-chain fatty acyl lysine. Nature. 2013;496:110–113. doi: 10.1038/nature12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feldman JL, Baeza J, Denu JM. Activation of the protein deacetylase SIRT6 by long-chain fatty acids and widespread deacylation by mammalian sirtuins. J. Biol. Chem. 2013;288:31350–31356. doi: 10.1074/jbc.C113.511261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Teng YB, Jing H, Aramsangtienchai P, He B, Khan S, Hu J, Lin H, Hao Q. Efficient demyristoylase activity of SIRT2 revealed by kinetic and structural studies. Sci. Rep. 2015;5:8529. doi: 10.1038/srep08529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tan M, Luo H, Lee S, Jin F, Yang JS, Montellier E, Buchou T, Cheng Z, Rousseaux S, Rajagopal N, Lu Z, Ye Z, Zhu Q, Wysocka J, Ye Y, Khochbin S, Ren B, Zhao Y. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146:1016–1028. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dai L, Peng C, Montellier E, Lu Z, Chen Y, Ishii H, Debernardi A, Buchou T, Rousseaux S, Jin F, Sabari BR, Deng Z, Allis CD, Ren B, Khochbin S, Zhao Y. Lysine 2-hydroxyisobutyrylation is a widely distributed active histone mark. Nat. Chem. Biol. 2014;10:365–370. doi: 10.1038/nchembio.1497. [DOI] [PubMed] [Google Scholar]
- 20.Sakurai N, Utsumi T. Posttranslational N-myristoylation is required for the anti-apoptotic activity of human tGelsolin, the C-terminal caspase cleavage product of human gelsolin. J. Biol. Chem. 2006;281:14288–14295. doi: 10.1074/jbc.M510338200. [DOI] [PubMed] [Google Scholar]
- 21.Vilas GL, Corvi MM, Plummer GJ, Seime AM, Lambkin GR, Berthiaume LG. Posttranslational myristoylation of caspase-activated p21-activated protein kinase 2 (PAK2) potentiates late apoptotic events. Proc. Natl. Acad. Sci. USA. 2006;103:6542–6547. doi: 10.1073/pnas.0600824103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sankaram MB. Membrane interaction of small N-myristoylated peptides: implications for membrane anchoring and protein-protein association. Biophys. J. 1994;67:105–112. doi: 10.1016/S0006-3495(94)80459-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baneyx F, Mujacic M. Recombinant protein folding and misfolding in Escherichia coli. Nat. Biotechnol. 2004;22:1399–1408. doi: 10.1038/nbt1029. [DOI] [PubMed] [Google Scholar]
- 24.West AC, Johnstone RW. New and emerging HDAC inhibitors for cancer treatment. J. Clin. Invest. 2014;124:30–39. doi: 10.1172/JCI69738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Balasubramanian S, Ramos J, Luo W, Sirisawad M, Verner E, Buggy JJ. A novel histone deacetylase 8 (HDAC8)-specific inhibitor PCI-34051 induces apoptosis in T-cell lymphomas. Leukemia. 2008;22:1026–1034. doi: 10.1038/leu.2008.9. [DOI] [PubMed] [Google Scholar]
- 26.Bradner JE, West N, Grachan ML, Greenberg EF, Haggarty SJ, Warnow T, Mazitschek R. Chemical phylogenetics of histone deacetylases. Nat. Chem. Biol. 2010;6:238–243. doi: 10.1038/nchembio.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yount JS, Zhang MM, Hang HC. Visualization and Identification of Fatty Acylated Proteins Using Chemical Reporters. Curr. Protoc. Chem. Biol. 2011;3:65–79. doi: 10.1002/9780470559277.ch100225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richon VM, Emiliani S, Verdin E, Webb Y, Breslow R, Rifkind RA, Marks PA. A class of hybrid polar inducers of transformed cell differentiation inhibits histone deacetylases. Proc. Natl. Acad. Sci. USA. 1998;95:3003–3007. doi: 10.1073/pnas.95.6.3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scuto A, Kirschbaum M, Kowolik C, Kretzner L, Juhasz A, Atadja P, Pullarkat V, Bhatia R, Forman S, Yen Y, Jove R. The novel histone deacetylase inhibitor, LBH589, induces expression of DNA damage response genes and apoptosis in Ph- acute lymphoblastic leukemia cells. Blood. 2008;111:5093–5100. doi: 10.1182/blood-2007-10-117762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fournel M, Bonfils C, Hou Y, Yan PT, Trachy-Bourget MC, Kalita A, Liu J, Lu AH, Zhou NZ, Robert MF, Gillespie J, Wang JJ, Ste-Croix H, Rahil J, Lefebvre S, Moradei O, Delorme D, Macleod AR, Besterman JM, Li Z. MGCD0103, a novel isotype-selective histone deacetylase inhibitor, has broad spectrum antitumor activity in vitro and in vivo. Mol. Cancer Ther. 2008;7:759–768. doi: 10.1158/1535-7163.MCT-07-2026. [DOI] [PubMed] [Google Scholar]
- 31.Somoza JR, Skene RJ, Katz BA, Mol C, Ho JD, Jennings AJ, Luong C, Arvai A, Buggy JJ, Chi E, Tang J, Sang BC, Verner E, Wynands R, Leahy EM, Dougan DR, Snell G, Navre M, Knuth MW, Swanson RV, McRee DE, Tari LW. Structural snapshots of human HDAC8 provide insights into the class I histone deacetylases. Structure. 2004;12:1325–1334. doi: 10.1016/j.str.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 32.Vannini A, Volpari C, Gallinari P, Jones P, Mattu M, Carfi A, De Francesco R, Steinkuhler C, Di Marco S. Substrate binding to histone deacetylases as shown by the crystal structure of the HDAC8-substrate complex. EMBO Rep. 2007;8:879–884. doi: 10.1038/sj.embor.7401047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Resh MD. Trafficking and signaling by fatty-acylated and prenylated proteins. Nat. Chem. Biol. 2006;2:584–590. doi: 10.1038/nchembio834. [DOI] [PubMed] [Google Scholar]
- 34.Stevenson FT, Bursten SL, Locksley RM, Lovett DH. Myristyl acylation of the tumor necrosis factor alpha precursor on specific lysine residues. J. Exp. Med. 1992;176:1053–1062. doi: 10.1084/jem.176.4.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu AY, Zhou Y, Khan S, Deitsch KW, Hao Q, Lin H. Plasmodium falciparum Sir2A preferentially hydrolyzes medium and long chain fatty acyl lysine. ACS Chem. Biol. 2012;7:155–159. doi: 10.1021/cb200230x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.