CONSPECTUS
Pathogens are recognized by the innate immune system in part via their unique and complex RNA signatures. A key sensor in human innate immunity is the RNA-activated protein kinase PKR, which has two double-stranded RNA (dsRNA) binding motifs (dsRBMs) at its N-terminus. Early studies described PKR as being activated potently by long stretches of perfect dsRNA, a signature typical of viruses. More recently, we and others have found that PKR is also activated by RNAs having structural defects such as bulges and internal loops. This article describes advances in our understanding of the ability of PKR to detect diverse foreign RNAs and how that recognition plays significant roles in discriminating self from non-self. The experiments discussed employ a wide range of techniques including activation assays, native polyacrylamide gel electrophoresis (PAGE), protein footprinting, and small angle X-ray scattering (SAXS). We discuss how misfolding and dimerization of RNA lead to activation of PKR. We also present recent findings on the activation of PKR by varied bacterial functional RNAs including ribozymes and riboswitches, which are among the few structured RNAs known to interact with PKR in a site-specific manner. Molecular models for how these structured RNAs activate PKR are provided. Studies by SAXS revealed that PKR straightens bent RNAs. Most external and internal RNA cellular modifications introduced in vitro and found naturally, such as the m7G cap and m6A group, abrogate activation of PKR, but other modifications, such as 5’-ppp and 2’-fluoro groups, are immunostimulatory and potential anticancer agents. Genome-wide studies of RNA folding in vitro and in vivo have provided fresh insights into general differences in RNA structure amongst bacteria, viruses, and human. These studies suggest that in vivo, cellular human RNAs are less folded than once thought, unwound by helicases, destabilized by m6A modifications, and often bound up with proteins—conditions known to abrogate activation of PKR. It thus appears that non-self RNAs are detected as unmodified, naked RNAs with appreciable secondary and tertiary structure. Observation that PKR is activated by structured but otherwise diverse RNAs is consistent both with the broad-spectrum nature of innate immunity and with the non-specific recognition of RNA by the dsRBM family. These findings provide a possible explanation for the apparent absence of protein-free structured human RNAs, such as ribozymes and riboswitches.
INTRODUCTION
One of the most important tasks any organism faces is defending itself against attack by pathogens. Innate immunity is an ancient form of self-defense and provides an organism with broad-spectrum protection against pathogens.1 Discriminating self from non-self requires detection of molecules possessing inherent diversity in their molecular configuration and conformation. The innate immune system is comprised of pattern recognition receptors (PRR’s) that recognize bacteria, viruses, fungi, and protozoa.2 These receptors recognize specific pathogen-associated molecular patterns (PAMPs) that induce signaling proteins, inflammatory cytokines, and type-I interferons (IFN-I), which ultimately limit viral replication in cells.3 PAMPs can embrace any pathogen-associated molecular motif including glycans, lipopolysaccharides, specific proteins, and nucleic acids.
Ribonucleic acid (RNA) is comprised of four similar nitrogenous heterocycles, A, G, C, and U, and a sugar-phosphate backbone. It thus has limited covalent molecular diversity, and moreover the intrinsic covalent makeup of RNA is shared among all forms of life. Where then does RNA’s diversity come from? Because it is single stranded, RNA can fold back on itself and form complex secondary and tertiary structures. This structural diversity is so rich that RNA can be an enzyme (ribozyme) or a specific and high affinity small molecule sensor (aptamer and riboswitch).4,5 Furthermore, RNAs can be modified both externally and internally to enhance their covalent molecular diversity.6 As such, two ways to distinguish self from non-self are through differences in the folds and covalent modifications of cellular and pathogenic RNA.
A common way in which RNA is detected in the cell is via RNA-binding proteins. RNA binding motifs run the gamut from sequence-specific motifs that bind single-stranded RNA to non-sequence-specific motifs that bind double-stranded RNA. Single-stranded sequence-specific RNA binding proteins are common in the human genome and include such motifs as the KH and RRM. There are thought to be more than 420 RBPs in the human genome and ~270 of them are sequence-specific.7 These proteins serve roles in gene regulation, splicing, capping, modification and export/localization. In contrast, proteins involved in innate immunity need to recognize a vast swath of RNAs and so are often comprised of non-sequence specific motifs.8 Key among these is the double-stranded RNA (dsRNA)-binding motif (dsRBM) that recognizes RNA in the minor groove, which has the RNA-specific 2’-hydroxyls but is generally devoid of sequence-specific information.9 The dsRBM is present in a number of dsRNA-binding proteins including Drosha, DGCR8, Dicer, and TRBP, which are involved in miRNA maturation, and ADAR, which is involved in RNA editing.9
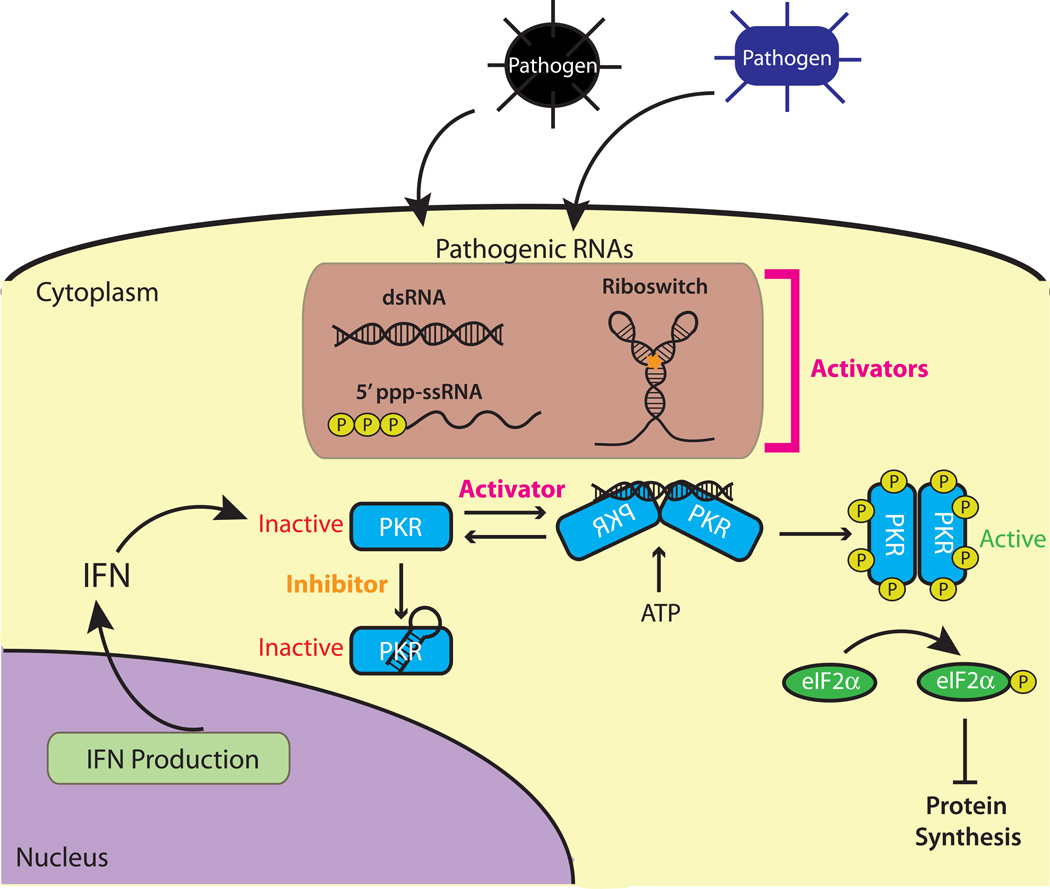
The RNA-activated protein kinase PKR is an innate immune sensor and has two dsRBMs at its N-terminus and a set of kinase domains at its C-terminus. In the presence of long dsRNA, typically of viral origin, PKR dimerizes via binding of RNA by its dsRBMs and autophosphorylates to become activated.10 Once phosphorylated, PKR can phosphorylate eIF2α, which inhibits the initiation of translation and can lead to apoptosis.10 An overview of the roles of PKR in the cell is provided in Figure 1. Remarkably, PKR is evolving more rapidly than other proteins in the human genome, apparently because the pathogens PKR recognizes are themselves evolving rapidly.11
Figure 1.
Roles of PKR in the cell. PKR is latent in healthy cells and upon pathogen infection interferons are produced, which upregulate PKR. These pathogens release their RNAs, which promote PKR dimerization and autophosphorylation. PKR autophosphorylates eukaryotic initiation factor 2α (eIF2α), which leads to inhibition of protein synthesis and apoptosis. PKR is inactivated by smaller RNAs, potentially from self- or non-self.
In the absence of pathogens, PKR exists in a latent non-phosphorylated state (Figure 1). The latent state can be enforced by binding either short structured RNAs or inhibitory proteins such as TRBP.12 Upon infection by pathogens, interferon is produced, which increases PKR production. PKR is activated by binding RNA of a minimal length of ~33 bp required for dimerization,13–15 which leads to an innate immune response.12 Although PKR is primarily found in the cytoplasm where it regulates translation, about 20% of PKR is nuclear where it has been shown to regulate RNA splicing, mRNA stability, and ribosome biogenesis and affect diseases including acute myeloid leukemia AML.16–18
The focus of this review is the molecular discrimination of self and non-self at the level of RNA recognition, with an emphasis on our recent studies in which RNA structure, misfolding, and modification participate in this process. We also highlight recent studies on the interaction of PKR with bacterial RNAs. It is our hope that this will lay a foundation to help understand the varied functions of PKR as well as other dsRBM-related proteins.
PKR IS ACTIVATED BY RNA MISFOLDING AND DIMERIZATION
Regulation of PKR by RNA secondary structure is not limited to perfectly double-stranded RNA. Many complex functional RNAs with structural imperfections activate PKR, including HIV-TAR (human immunodeficiency virus-trans activation response element), the HDV (hepatitis delta virus) ribozyme, and tRNA, each of which contains multiple helical defects (Figure 2).19–23 Studies from our lab with these RNAs revealed that activation of PKR is dependent upon misfolding via RNA dimerization. In each case, monomeric RNA does not activate PKR, either because it is globular, as in the HDV ribozyme and tRNA, or because it has too few base pairs, as in HIV-TAR. Dimerization of nucleic acids is common and is often, but not always, accompanied by loss in RNA function, where it serves as an example of RNA misfolding.24,25
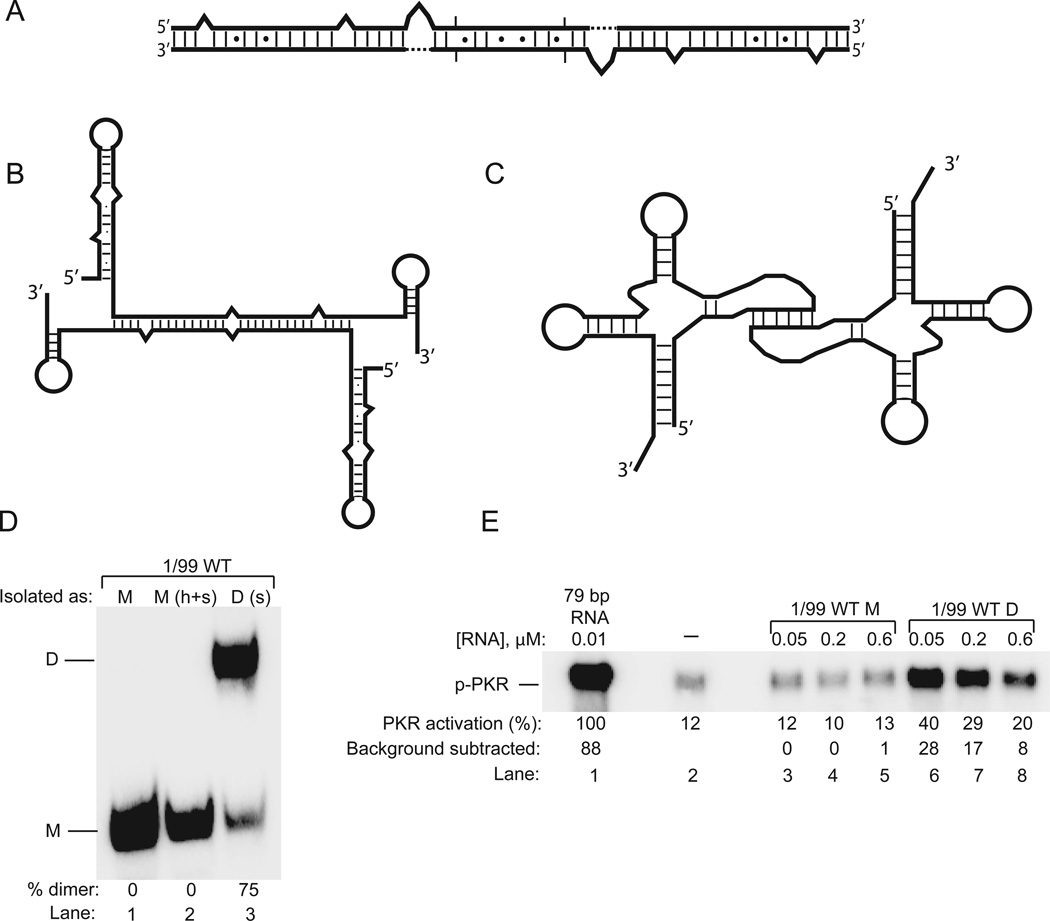
Figure 2.
Activation of PKR by RNA dimers. (A) HIV-I TAR RNA dimer, which is ~23 bp as a monomer and ~46 bp as a dimer. (Adapted with permission from ref. 19. Copyright 2009 Elsevier.) (B) HDV ribozyme dimer, which has 14 bp in the intramolecular hairpin regions and 26 bp in the intermolecular double-stranded region. (C) tRNALeu dimer, which has 20 bp total as a monomer and 42 bp as a dimer. (Adapted with permission from ref. 21. Copyright 2013 Creative Commons.) (D) Native PAGE gel showing the migration of the HDV ribozyme monomer and dimer. ‘h’ is heat treatment; ‘s’ is high salt treatment. (E) PKR activity assay for HDV ribozyme monomeric and dimeric forms in comparison to 79 bp dsRNA. Shown is an SDS-PAGE gel and ‘p-PKR’ is phosphorylated PKR. (Panels B, D, and E are adapted with permission from ref. 20. Copyright 2012 Elsevier.)
The HIV-1 TAR RNA resides in the 5’-end of the HIV-1 RNA genome and is known to dimerize.26 Reports about the effect of TAR RNA on PKR activation were conflicting, with some studies claiming it is an inhibitor of PKR27 and others that it is an activator.28 We were able to isolate TAR RNA in non-interconverting monomeric and dimeric forms.19 This was a key technical advancement and was accomplished using native polyacrylamide gel electrophoresis (PAGE), with confirmation of the molecularity of the isolated RNA by analytical ultracentrifugation (AUC). The monomeric form of TAR RNA inhibited PKR autophosphorylation by competing for PKR that had dimerized on longer RNAs. In contrast, the dimeric form of TAR RNA potently activated PKR at low to moderate RNA dimer concentrations, and, like all RNA activators, inhibited PKR activation at high concentrations, presumably by titrating PKR dimers out to monomers. Dimerization of TAR doubles the number of base pairs per RNA from ~23 to ~46 (Figure 2A). Gain of activation upon RNA dimerization could be understood in light of the classical activation of PKR requiring at least 33 bp and reconciles the disagreement on the regulatory status of TAR-RNA by showing that PKR is both inhibited and activated depending on the molecularity of TAR.
Hepatitis delta virus (HDV) is a 1.7 kb single-stranded RNA that replicates by a double rolling circle mechanism.29,30 Both the genomic and antigenomic forms of HDV contain ribozymes, which linearize the genome during replication. Earlier studies showed that a ribozyme-containing segment of the genomic viral RNA, which folds back on itself to form a ribozyme-inactivating rod, activated PKR.22,23 We isolated monomeric and dimerized forms of ~100 nt ribozyme-containing constructs by native PAGE and found that monomeric RNA, which folded into the native structure of the ribozyme according to structure mapping, did not activate PKR whereas the dimer, which folded into an extended species (Figure 2B), potently activated PKR.20 Sample native gel and activation data are provided in Figure 2D and E. The dimer prevents native tertiary structure and approximately doubles the number of contiguous base pairs, approaching the activating length of 33 bp, which can be augmented by peripheral helices (Figure 2B).
In a third effort to test the generality of RNA dimers to activate PKR, we examined the A14G pathogenic mutant of mitochondrial tRNA-leucine mt-tRNALeu, which dimerizes via its D stem-loop (Figure 2C).31 We isolated monomeric and dimeric forms of an unmodified version of this tRNA by native PAGE and again found that dimeric RNA is a potent activator of PKR, inducing phosphorylation levels similar to those of long perfectly double-stranded RNA. Monomeric tRNA, on the other hand, did not activate PKR even at high concentrations.21 The dimer disrupts native tertiary structure and approximately doubles the number of base pairs, from 20 to 42 bp (Figure 2C).32 As described below, covalent RNA modifications occur much less frequently in mitochondrial tRNAs than cytoplasmic tRNAs. Since tRNAs are prone to misfold, modifications in cytoplasmic tRNAs, which are known to favor native tertiary structure, may function, at least in part, to suppress tRNA dimerization thereby preventing an unprovoked innate immune response.21
Each of the structures for these three dimer RNAs contains helical imperfections such as bulges and internal loops (Figure 2A–C). We conducted a systematic analysis of the effect of bulges and internal loops on the activation of PKR.33 Overall, bulges decreased activation of PKR by ~2- to 10-fold, although effects were generally rescuable by higher concentrations of RNA or longer times of incubation. In the background of dsRNA, larger bulges disfavored PKR activation more strongly, and centrally located bulges were most detrimental. For multiple bulges, those that were cis to each other (i.e. on the same strand) activated PKR more potently than bulges that were trans, while higher ionic strength increased discrimination against RNA structural defects. Finally, we note that helical defects are tolerated by being extruded, with PKR straightening any bulge-induced bending of the RNA. This notion is supported by early native PAGE results and more recent SAXS studies from our lab.34,35
There is a strong link between protein misfolding and human disease, for example in transmissible spongiform encephalopathies (TSEs) and Alzheimer’s disease.36 Links between RNA misfolding and disease are less well established. Nonetheless, tRNA mutations in the mitochondria are associated with devastating human disease,37 and many viruses are known to dimerize their genomes during replication.38
COVALENT MODIFICATIONS OF RNA REGULATE PKR ACTIVATION
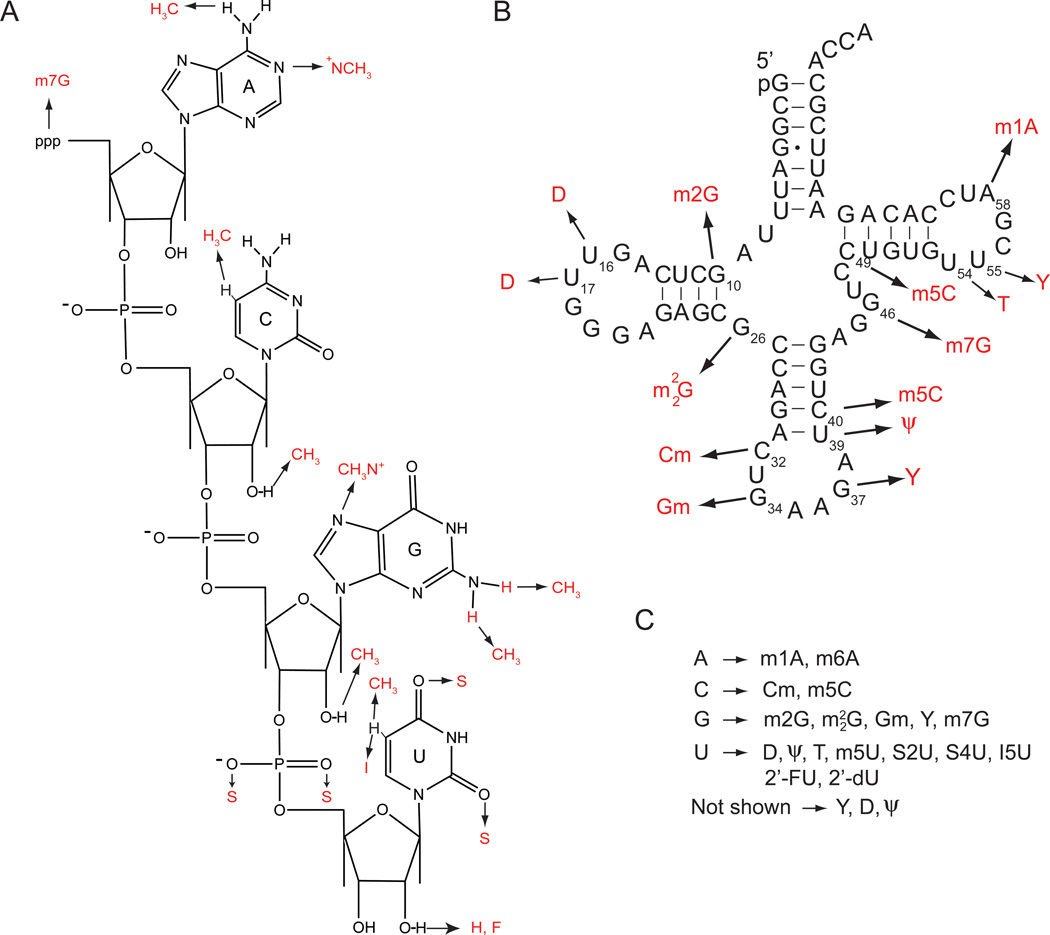
Covalent modifications are widespread in RNA. They are most abundant and diverse in tRNA, but also occur in rRNA, and more recently have been found throughout mRNA.39 Naturally occurring modifications occur at the 5’-end of the RNA and internally at the ribose sugar and nucleobases (Figure 3A). Modifications to RNA can also be introduced synthetically and have been used to prepare immunostimulatory RNAs (isRNA) for turning on the innate immune system to treat cancer and chronic infections.40,41
Figure 3.
Naturally occurring chemical modifications that regulate activation of PKR. (A) Internal and external modifications discussed herein. (B) Naturally occurring RNA modifications in yeast tRNAPhe tested en masse. (C) Modifications of the four nucleobases in panel A. Also listed are several additional modifications only in panel B: Y (wyosine), D (dihydrouridine), and Ψ (pseudouridine).
Effects of a large number of covalent RNA modifications on PKR activation have been tested (Figure 3).40,42–44 Several studies investigated the activation of PKR by external modifications. We reported that short unstructured RNAs could activate PKR just as strongly as long dsRNA14 provided a 5’-triphosphate (ppp), which is found in many pathogenic RNAs, was present.42 A m7G cap, which is found in most human cellular RNAs, did not activate PKR. A recent investigation by Toroney et. al. focused on overlap of the binding sites for 5’-triphosphate, dsRNA, and ATP. We found absence of selectivity for the 5’-most nucleobase, consistent with broad-spectrum protection, but strong selectivity for ATP, indicating that the 5’-ppp and ATP binding sites do not overlap.44 Lastly, evidence was provided that ppp-ssRNAs interact with multiple domains of PKR according to activation, binding, and crosslinking studies.
Several studies from our lab sought to understand whether internal modifications affect activation of PKR by RNA. We found that activation is abrogated when ssRNA and dsRNA are fully substituted with diverse modifications, including 2’-deoxy sugar substitutions and s2U, s4U, ψ, m5U, I5U, and m6A nucleobase substitutions.40 We also tested modified tRNAs for their ability to activate PKR, including naturally occurring cellular tRNAs, which are heavily modified, and mitochondrial tRNA (mt-tRNA), which are modestly modified.21 We found that T7-transcribed yeast tRNA (unmodified) and total mt-tRNA activated PKR potently in vitro, whereas total tRNAs from E. coli, yeast, and wheat did not. Likewise, PKR was activated by unmodified tRNAPhe but not by naturally modified yeast tRNAPhe both in vitro and in vivo.21 Modifications in yeast tRNAPhe include m1A, ψ, T, m5C, m7G, Y, 2’-methoxyC, 2’-methoxyG, di-m2G, D, and m2G. In collaboration with Kariko and Weissman, we obtained similar results with pseudouridine (ψ), where ψ incorporation into mRNA enhanced translation by diminishing PKR activation.45 Overall these findings support suppression of PKR and the innate immune system by cellular RNA modifications.
Two recent studies provide additional support of a role for covalent modification of cellular RNAs in suppression of PKR function. Yuen and co-workers found that suppression of m6A epigenetic marks on mRNA via mutations in NIPBL, a cohesin loader, led to activation of PKR,46 while George and co-workers reported that suppression of ADAR1, which performs most A-to-I deaminations in RNA, led to enhanced phosphorylation of eIF2α.47 An emerging view is that RNA modifications are marks that differentiate cellular self-RNAs from pathogenic non-self RNAs, thus keeping PKR in a latent state. This is reminiscent of the restriction-modification immune systems of bacteria.48
While most covalent modifications of RNA abrogate activation of PKR, certain modifications maintained or enhanced activation of PKR. As mentioned, a 5’-triphosphate leads to enhanced levels of PKR activation.43 In addition, we showed that phosphorothioates and 2’-fluoro modifications of uridine lead to activation of PKR.40 A 2’-fluoro group can serve as a hydrogen bond acceptor in the minor groove of dsRNA where the dsRBM of PKR binds, providing a molecular rationale for this observation. We previously proposed that such modified RNAs, which have enhanced stability in vivo, might serve as immunostimulatory RNAs (isRNA) for treating cancer.40 Notably, recent work from Lee, Sullenger, and co-workers illustrates that RNAs containing both 2’-fluoro pyrimidines and a 5’-triphosphate (so-called “2’F5’ppp RNA”) increase cell death in human cancer cells and that innate immune-sensing PRRs, including PKR, are strongly upregulated in this process.41
BACTERIAL RNA ACTIVATES INNATE IMMUNITY VIA PKR
To date, PKR has been principally recognized as being activated by viral RNAs. However, recent reports indicate that PKR is phosphorylated in cells, either human cardiac myocytes or a fibrosarcoma cell line, that are infected with RNA from bacteria, which can lead to apoptosis.49,50 For certain bacteria, molecules are secreted into the cytoplasm of the host during the early stage of infection, and it has been proposed that these molecules could include nucleic acids, providing the potential for PKR to detect bacterial RNAs through this mechanism.51,52 In addition, RIG-I, another PRR, is activated by RNA secreted from Listeria, suggesting a general role for the innate immune system in protection from bacterial pathogens.53 Lastly, certain bacteria undergo a lytic stage in which their RNA contents are released into the infected cell, providing another means of detection.54
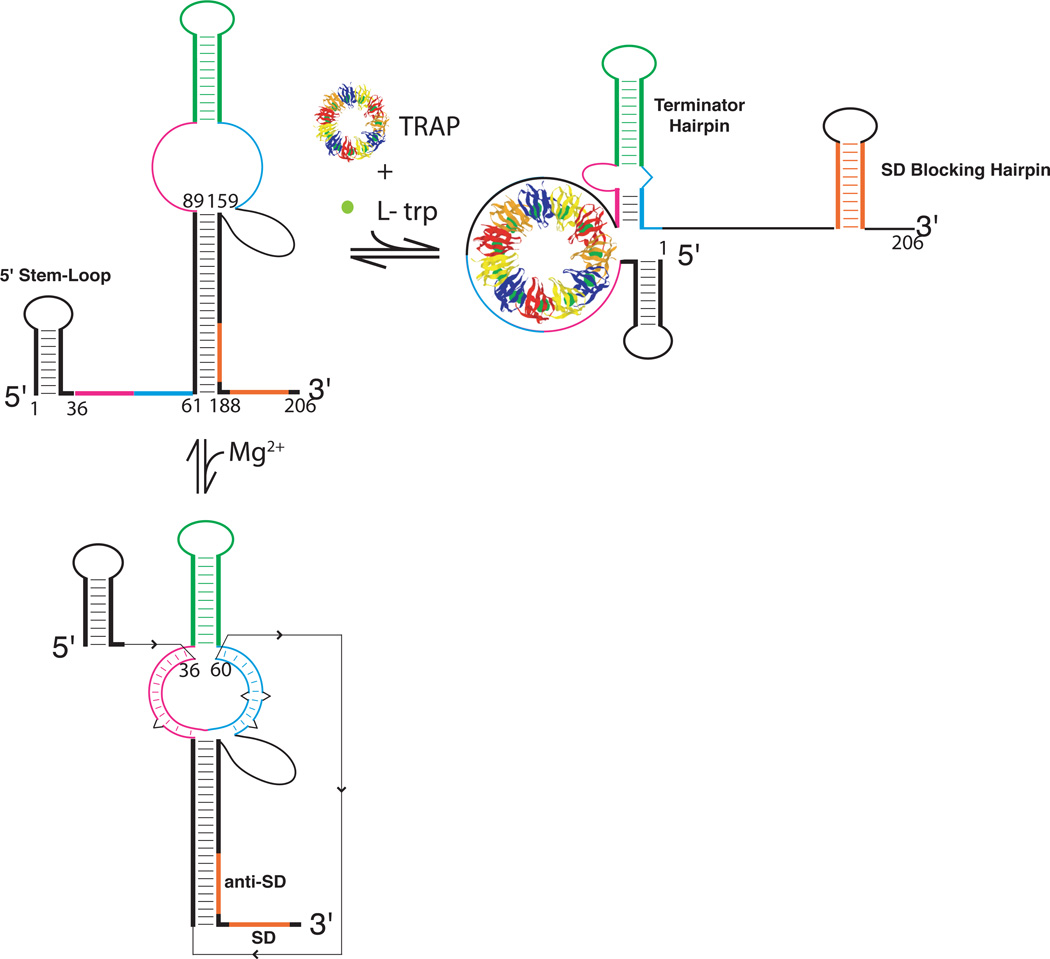
In order to better understand how PKR is activated by bacterial RNA, effects of the trp 5’-UTR translation control RNA from B. subtilis were investigated.55 This RNA contains many elements typical of a structural, regulatory bacterial RNA including stem-loops, competing terminator and antiterminator hairpins, long-range tertiary structure, and extensive dsRNA and ssRNA regions (Figure 4). In addition, this RNA regulates L-trp levels in Bacillus subtilis through structural rearrangement mediated by the RNA-binding protein, TRAP, which itself binds L-trp. These structural rearrangements lead to formation of a Shine–Dalgarno blocking hairpin, providing additional elements typical of bacterial RNA.
Figure 4.
Multiple folds of the B. subtilis trp 5’UTR RNA lead to activation of PKR. A 206 nt transcript with multiple features including a 5’-stem loop, long dsRNA region, and a terminator hairpin. This RNA binds the protein TRAP, which is activated by excess L-trp, and leads to the translation control structure shown at the right, where the Shine-Dalgarno sequence is blocked. In addition, the naked RNA has a magnesium-dependent tertiary (pseudoknot) feature shown in blue and pink in the lower panel. (Adapted with permission from ref. 55. Copyright 2015 Elsevier.)
Studies on this system provided the first evidence that multiple structural features of this prototypical bacterial leader potently activate PKR.55 The 5’-stem loop, terminator hairpin, and Shine–Dalgarno blocking hairpin all activated PKR and displayed flanking tail and 5’-triphosphate dependence. Disruption of a long-range tertiary interaction had little effect on PKR activity. Moreover, potent activation of PKR by the trp 5’-UTR and by total E. coli RNA was maintained upon lowering magnesium concentrations to human physiological levels.55
To more broadly understand PKR’s interaction with bacterial RNA, we recently investigated its interaction with three functional bacterial RNAs with extensive secondary and tertiary structures and diverse regulatory mechanisms: the V. cholerae cyclic-di GMP riboswitch, the B. anthracis glmS riboswitch-ribozyme, and the C. bolteae twister ribozyme, where the riboswitches change structure upon ligand binding.56 We found that all of these bacterial RNAs activate PKR under both human and bacterial physiological magnesium conditions with remarkably high potency considering their complex structures. Maximal PKR activity ranged from 80–140% that of long dsRNA, requiring just 5–10-fold higher RNA levels.
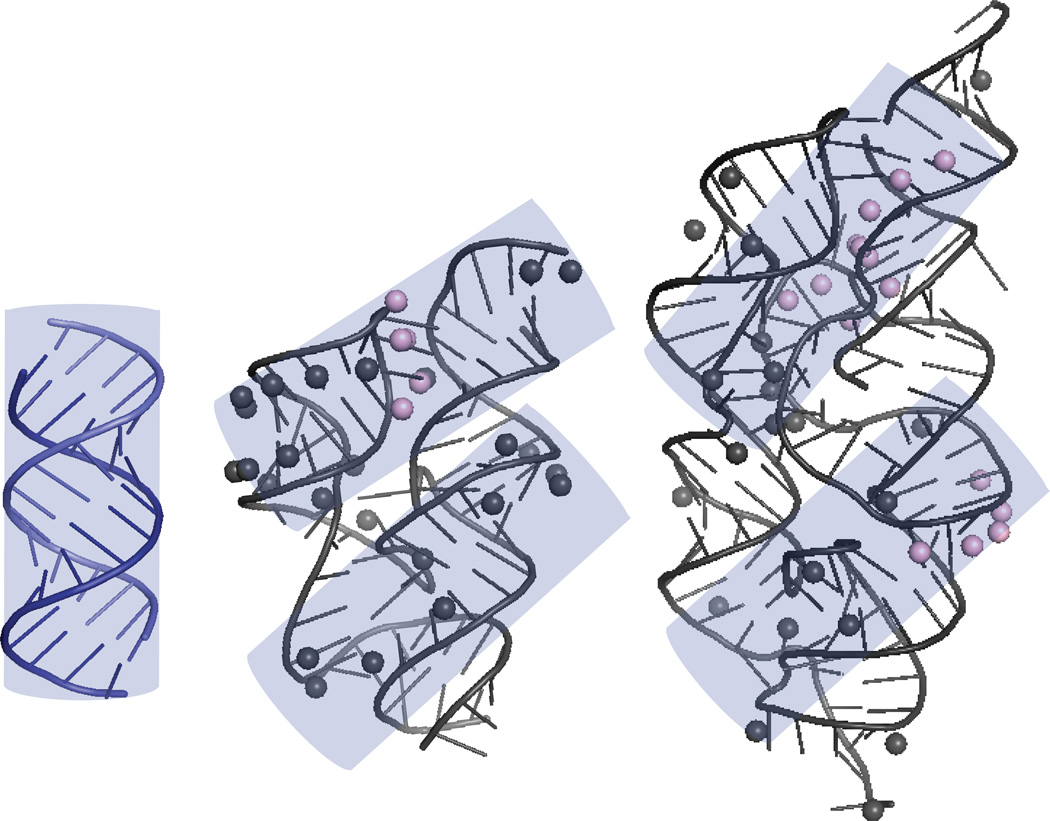
To understand structurally how these RNAs interact with PKR, we conducted ribonuclease structure mapping and PKR footprinting studies. Results suggested that PKR dimerizes on the peripheral helices of the natively folded cyclic-di GMP riboswitch and glmS riboswitch-ribozyme (Figure 5). We modeled the length of dsRNA required for each monomer of PKR to bind (~16 bp)15 onto the structure mapping/PKR footprinting data of the cyclic di-GMP riboswitch and glmS riboswitch-ribozyme. Remarkably, two footprints were found in A-form-like helical segments on each of the RNAs (Figure 5). We also attempted to map PKR onto the Vc2 cdiGMP riboswitch G83C mutant, which inhibits tertiary interactions and leaves the RNA in an extended undocked conformation. No apparent footprinting was found, most likely due to PKR not being locked onto a single binding site. This suggests that PKR does indeed dimerize on the tertiary structures of intact functional bacterial RNAs, leading to its activation.56 These are among the first reports of RNAs that activate PKR via their tertiary structure.57
Figure 5.
Footprinting of PKR onto riboswitches. (Left) Model of 16 bp dsRNA, the minimal length needed to bind a PKR monomer. (Middle and Right) A cylinder representing the 16 bp dsRNA is placed twice onto bacterial RNAs that activate PKR. Vc2 cdiGMP riboswitch (middle) and glmS riboswitch-ribozyme (right) are modeled according to structure mapping and PKR footprinting data. Pink balls are 2’OHs that are always protected and black balls are 2’OHs that are protected by PKR. Two footprints are possible for each RNA. Another potential binding site on the glmS riboswitch-ribozyme is found by rotating the bottom cylinder ~45° counterclockwise about the axis coming out of the page.56
CONCLUSIONS
The innate immune system is the first line of defense against invading pathogens. It is both ancient in origin and general in recognizing pathogens. RNA is also ancient according to the ‘RNA World Hypothesis’ where self-replicating RNA molecules are thought to be the precursors to all life on Earth. It is thus possible that RNA and the innate immune system co-evolved. Some of the biggest questions regarding the mechanism of innate immunity are ones of molecular recognition: How is self differentiated from non-self in terms of species-specific molecular patterns?
Genome-wide studies of RNA folding have emerged in the last few years, and fresh insights into general differences in RNA structure among bacteria, viruses, and human are being generated.58–61 These studies have revealed that in vivo, eukaryotic RNAs are less structured than once imagined owing in part to the presence of helicases and m6A epigenetic modifications, each of which unfolds RNA. Since activation of PKR is generally promoted by RNA structure, this view provides a molecular model for why these cellular modifications suppress PKR activation. Pseudouridine also suppresses PKR activation,21,45 but in fact ψ stabilizes RNA structure, including all mismatches.62,63 Given the extra hydrogen bonding group in ψ, these modifications may drive RNA into non-A-form geometries, which disfavor canonical positioning of a dsRBM. In addition, covalent RNA modifications that favor compact native tertiary structure, especially for tRNA, may disfavor competing RNA dimerization, thereby preventing an autoimmune response. An important future direction will be understanding the molecular bases for how RNA modifications affect PKR activation.
A model emerges in which non-self RNAs are detected as those naked, unmodified RNAs with appreciable secondary and tertiary structure. Observation that PKR is activated by a diverse set of structured, unmodified RNAs—including bacterial leaders, riboswitches, and ribozymes— is consistent with the broad-spectrum response nature of innate immunity and with the nonspecific recognition of RNA by the dsRBM family. Exogenous mRNAs and aptamers can be modified covalently to either avoid or stimulate the immune systems, as desired.40,41,45 We suggest that the apparent absence of protein-free ribozymes and riboswitches in the human genome may be because such RNAs trigger innate immunity and so are under negative selection.
Acknowledgments
We would like to thank our many collaborators on this work including Craig Cameron, Jim Cole, Katalin Kariko, Linda Spremulli, and Drew Weissman, as well as members of the Bevilacqua lab who have contributed to PKR research especially Ananya Anmangandla, Laurie Heinicke, Subba Rao Nallagatla, and Rebecca Toroney. We thank Sally Assmann for critically reading the manuscript. A portion of this research was supported by National Institutes of Health Grant R01GM110237.
Biographies
Chelsea M. Hull was born in 1987 and raised in Kingston, NY. She received her B.S. in chemistry from SUNY New Paltz in 2009 and her Ph.D. in chemistry from the Pennsylvania State University in 2016 with Philip Bevilacqua, where her principal research was investigating the mechanism by which PKR recognizes different levels and types of RNA structure.
Philip C. Bevilacqua was born in 1965 and raised in North Collins, NY. He received his B.S. degree in chemistry and physics from John Carroll University in 1987. He carried out graduate studies as a Sproull fellow with Douglas Turner at the University of Rochester, studying RNA folding by transient kinetics and graduating with a Ph.D. in chemistry in 1993. He conducted postdoctoral studies as a Jane Coffin Childs Fellow with Thomas Cech at the University of Colorado at Boulder until 1997 when he joined the Pennsylvania State University, where he is currently Professor of Chemistry and Professor of Biochemistry & Molecular Biology. Research in his laboratory is centered on understanding functions of RNA in nature at the molecular level.
Footnotes
The authors declare no competing financial interest.
REFERENCES
- 1.Gura T. Innate immunity. Ancient system gets new respect. Science. 2001;291:2068–2071. doi: 10.1126/science.291.5511.2068. [DOI] [PubMed] [Google Scholar]
- 2.Creagh EM, O’Neill LA. TLRs, NLRs and RLRs: a trinity of pathogen sensors that cooperate in innate immunity. Trends Immunol. 2006;27:352–357. doi: 10.1016/j.it.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 3.Kawasaki T, Kawai T, Akira S. Recognition of nucleic acids by pattern-recognition receptors and its relevance in autoimmunity. Immunol. Rev. 2011;243:61–73. doi: 10.1111/j.1600-065X.2011.01048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Serganov A, Nudler E. A decade of riboswitches. Cell. 2013;152:17–24. doi: 10.1016/j.cell.2012.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilson TJ, Liu Y, Lilley DMJ. Ribozymes and the mechanisms that underlie RNA catalysis. Front. Chem. Sci. Engin. 2016:1–8. [Google Scholar]
- 6.Yi C, Pan T. Cellular dynamics of RNA modification. Acc. Chem. Res. 2011;44:1380–1388. doi: 10.1021/ar200057m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ray D, Kazan H, Cook KB, Weirauch MT, Najafabadi HS, Li X, Gueroussov S, Albu M, Zheng H, Yang A, Na H, Irimia M, Matzat LH, Dale RK, Smith SA, Yarosh CA, Kelly SM, Nabet B, Mecenas D, Li W, Laishram RS, Qiao M, Lipshitz HD, Piano F, Corbett AH, Carstens RP, Frey BJ, Anderson RA, Lynch KW, Penalva LO, Lei EP, Fraser AG, Blencowe BJ, Morris QD, Hughes TR. A compendium of RNA-binding motifs for decoding gene regulation. Nature. 2013;499:172–177. doi: 10.1038/nature12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rebsamen M, Kandasamy RK, Superti-Furga G. Protein interaction networks in innate immunity. Trends Immunol. 2013;34:610–619. doi: 10.1016/j.it.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 9.Tian B, Bevilacqua PC, Diegelman-Parente A, Mathews MB. The double-stranded-RNA-binding motif: interference and much more. Nat. Rev. Mol. Cell Biol. 2004;5:1013–1023. doi: 10.1038/nrm1528. [DOI] [PubMed] [Google Scholar]
- 10.Zhang F, Romano PR, Nagamura-Inoue T, Tian B, Dever TE, Mathews MB, Ozato K, Hinnebusch AG. Binding of double-stranded RNA to protein kinase PKR is required for dimerization and promotes critical autophosphorylation events in the activation loop. J. Biol. Chem. 2001;276:24946–24958. doi: 10.1074/jbc.M102108200. [DOI] [PubMed] [Google Scholar]
- 11.Elde NC, Child SJ, Geballe AP, Malik HS. Protein kinase R reveals an evolutionary model for defeating viral mimicry. Nature. 2009;457:485–489. doi: 10.1038/nature07529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singh M, Castillo D, Patel CV, Patel RC. Stress-induced phosphorylation of PACT reduces its interaction with TRBP and leads to PKR activation. Biochemistry. 2011;50:4550–4560. doi: 10.1021/bi200104h. [DOI] [PubMed] [Google Scholar]
- 13.Manche LGSR, Schmedt C, Mathews MB. Interactions between double-stranded RNA regulators and the protein kinase DAI. Mol. Cell. Biol. 1992;12:5238–5248. doi: 10.1128/mcb.12.11.5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng X, Bevilacqua PC. Activation of the protein kinase PKR by short double-stranded RNAs with single-stranded tails. RNA. 2004;10:1934–1945. doi: 10.1261/rna.7150804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lemaire PA, Anderson E, Lary J, Cole JL. Mechanism of PKR Activation by dsRNA. J. Mol. Biol. 2008;381:351–360. doi: 10.1016/j.jmb.2008.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blalock WL, Piazzi M, Bavelloni A, Raffini M, Faenza I, D’Angelo A, Cocco L. Identification of the PKR nuclear interactome reveals roles in ribosome biogenesis, mRNA processing and cell division. J. Cell. Phys. 2014;229:1047–1060. doi: 10.1002/jcp.24529. [DOI] [PubMed] [Google Scholar]
- 17.Oshima M, Iwama A. Nuclear, not cytoplasmic, PKR maneuvers in AML. Blood. 2015;126:1523–1524. doi: 10.1182/blood-2015-08-661421. [DOI] [PubMed] [Google Scholar]
- 18.Cheng X, Byrne M, Brown KD, Konopleva MY, Kornblau SM, Bennett RL, May WS. PKR inhibits the DNA damage response, and is associated with poor survival in AML and accelerated leukemia in NHD13 mice. Blood. 2015;126:1585–1594. doi: 10.1182/blood-2015-03-635227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heinicke LA, Wong CJ, Lary J, Nallagatla SR, Diegelman-Parente A, Zheng X, Cole JL, Bevilacqua PC. RNA dimerization promotes PKR dimerization and activation. J. Mol. Biol. 2009;390:319–338. doi: 10.1016/j.jmb.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heinicke LA, Bevilacqua PC. Activation of PKR by RNA misfolding: HDV ribozyme dimers activate PKR. RNA. 2012;18:2157–2165. doi: 10.1261/rna.034744.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nallagatla SR, Jones CN, Ghosh SK, Sharma SD, Cameron CE, Spremulli LL, Bevilacqua PC. Native tertiary structure and nucleoside modifications suppress tRNA’s intrinsic ability to activate the innate immune sensor PKR. PLoS One. 2013;8:e57905. doi: 10.1371/journal.pone.0057905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robertson HD, Manche L, Mathews MB. Paradoxical interactions between human delta hepatitis agent RNA and the cellular protein kinase PKR. J Virol. 1996;70:5611–5617. doi: 10.1128/jvi.70.8.5611-5617.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Circle DA, Neel OD, Robertson HD, Clarke PA, Mathews MB. Surprising specificity of PKR binding to delta agent genomic RNA. RNA. 1997;3:438–448. [PMC free article] [PubMed] [Google Scholar]
- 24.Holbrook SR, Cheong C, Tinoco I, Jr, Kim SH. Crystal structure of an RNA double helix incorporating a track of non-Watson-Crick base pairs. Nature. 1991;353:579–581. doi: 10.1038/353579a0. [DOI] [PubMed] [Google Scholar]
- 25.Nowakowski J, Shim PJ, Prasad GS, Stout CD, Joyce GF. Crystal structure of an 82-nucleotide RNA-DNA complex formed by the 10–23 DNA enzyme. Nat. Struct. Biol. 1999;6:151–156. doi: 10.1038/5839. [DOI] [PubMed] [Google Scholar]
- 26.Andersen ES, Contera SA, Knudsen B, Damgaard CK, Besenbacher F, Kjems J. Role of the trans-activation response element in dimerization of HIV-1 RNA. J. Biol. Chem. 2004;279:22243–22249. doi: 10.1074/jbc.M314326200. [DOI] [PubMed] [Google Scholar]
- 27.Gunnery S, Rice AP, Robertson HD, Mathews MB. Tat-responsive region RNA of human immunodeficiency virus 1 can prevent activation of the double-stranded-RNA-activated protein kinase. Proc. Natl. Acad. Sci. U S A. 1990;87:8687–8691. doi: 10.1073/pnas.87.22.8687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maitra RK, McMillan NA, Desai S, McSwiggen J, Hovanessian AG, Sen G, Williams BR, Silverman RH. HIV-1 TAR RNA has an intrinsic ability to activate interferon-inducible enzymes. Virology. 1994;204:823–827. doi: 10.1006/viro.1994.1601. [DOI] [PubMed] [Google Scholar]
- 29.Lai MM. The molecular biology of hepatitis delta virus. Annu. Rev. Biochem. 1995;64:259–286. doi: 10.1146/annurev.bi.64.070195.001355. [DOI] [PubMed] [Google Scholar]
- 30.Alvarado-Mora MV, Locarnini S, Rizzetto M, Pinho JR. An update on HDV: virology, pathogenesis and treatment. Antivir. Ther. 2013;18:541–548. doi: 10.3851/IMP2598. [DOI] [PubMed] [Google Scholar]
- 31.Wittenhagen LM, Kelley SO. Dimerization of a pathogenic human mitochondrial tRNA. Nat. Struct. Biol. 2002;9:586–590. doi: 10.1038/nsb820. [DOI] [PubMed] [Google Scholar]
- 32.Roy MD, Wittenhagen LM, Kelley SO. Structural probing of a pathogenic tRNA dimer. RNA. 2005;11:254–260. doi: 10.1261/rna.7143305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heinicke LA, Nallagatla SR, Hull CM, Bevilacqua PC. RNA helical imperfections regulate activation of the protein kinase PKR: effects of bulge position, size, and geometry. RNA. 2011;17:957–966. doi: 10.1261/rna.2636911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng X, Bevilacqua PC. Straightening of bulged RNA by the double-stranded RNA-binding domain from the protein kinase PKR. Proc. Natl. Acad. Sci. U S A. 2000;97:14162–14167. doi: 10.1073/pnas.011355798. 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patel S, Blose JM, Sokoloski JE, Pollack L, Bevilacqua PC. Specificity of the double-stranded RNA-binding domain from the RNA-activated protein kinase PKR for double-stranded RNA: insights from thermodynamics and small-angle X-ray scattering. Biochemistry. 2012;51:9312–9322. doi: 10.1021/bi300935p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu. rev. of biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 37.Lott MT, Leipzig JN, Derbeneva O, Xie HM, Chalkia D, Sarmady M, Procaccio V, Wallace DC. mtDNA variation and analysis using MITOMAP and MITOMASTER. Curr. Protoc. Bioinformatics. 2013;1:1–26. doi: 10.1002/0471250953.bi0123s44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson SF, Telesnitsky A. Retroviral RNA dimerization and packaging: the what, how, when, where, and why. PLoS Pathog. 2010;6:1001–1007. doi: 10.1371/journal.ppat.1001007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Agris P, Crain PF, Rozenski J, Fabris D, Vendrix FAP. [accessed May 16, 2016];RNA Modifications Database. http://mods.rna.albany.edu.
- 40.Nallagatla SR, Bevilacqua PC. Nucleoside modifications modulate activation of the protein kinase PKR in an RNA structure-specific manner. Rna. 2008;14:1201–1213. doi: 10.1261/rna.1007408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee Y, Urban JH, Xu L, Sullenger BA, Lee J. 2’Fluoro modification differentially modulates the ability of RNAs to activate pattern recognition receptors. Nucleic Acid Ther. 2016 doi: 10.1089/nat.2015.0575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Puthenveetil S, Whitby L, Ren J, Kelnar K, Krebs JF, Beal PA. Controlling activation of the RNA-dependent protein kinase by siRNAs using site-specific chemical modification. Nucleic Acids Res. 2006;34:4900–4911. doi: 10.1093/nar/gkl464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nallagatla SR, Hwang J, Toroney R, Zheng X, Cameron CE, Bevilacqua PC. 5’-triphosphate-dependent activation of PKR by RNAs with short stem-loops. Science. 2007;318:1455–1458. doi: 10.1126/science.1147347. [DOI] [PubMed] [Google Scholar]
- 44.Toroney R, Hull CM, Sokoloski JE, Bevilacqua PC. Mechanistic characterization of the 5’-triphosphate-dependent activation of PKR: lack of 5’-end nucleobase specificity, evidence for a distinct triphosphate binding site, and a critical role for the dsRBD. RNA. 2012;18:1862–1874. doi: 10.1261/rna.034520.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anderson BR, Muramatsu H, Nallagatla SR, Bevilacqua PC, Sansing LH, Weissman D, Kariko K. Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Res. 2010;38:5884–5892. doi: 10.1093/nar/gkq347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yuen KC, Xu B, Krantz ID, Gerton JL. NIPBL controls RNA biogenesis to prevent activation of the stress kinase PKR. Cell Rep. 2016;14:93–102. doi: 10.1016/j.celrep.2015.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.George CX, Ramaswami G, Li JB, Samuel CE. Editing of cellular self-RNAs by adenosine deaminase ADAR1 suppresses innate immune stress responses. J. Biol. Chem. 2016;291:6158–6168. doi: 10.1074/jbc.M115.709014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vasu K, Nagaraja V. Diverse functions of restriction-modification systems in addition to cellular defense. Microbiol Mol. Biol. Rev. 2013;77:53–72. doi: 10.1128/MMBR.00044-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bleiblo F, Michael P, Brabant D, Ramana CV, Tai T, Saleh M, Parrillo JE, Kumar A, Kumar A. Bacterial RNA induces myocyte cellular dysfunction through the activation of PKR. J. Thor. Dis. 2012;4:114–125. doi: 10.3978/j.issn.2072-1439.2012.01.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bleiblo F, Michael P, Brabant D, Ramana CV, Tai T, Saleh M, Parrillo JE, Kumar A, Kumar A. JAK kinases are required for the bacterial RNA and poly I:C induced tyrosine phosphorylation of PKR. Int. J. Clin. Exp. Med. 2013;6:16–25. [PMC free article] [PubMed] [Google Scholar]
- 51.Sander LE, Davis MJ, Boekschoten MV, Amsen D, Dascher CC, Ryffel B, Swanson JA, Muller M, Blander JM. Detection of prokaryotic mRNA signifies microbial viability and promotes immunity. Nature. 2011;474:385–389. doi: 10.1038/nature10072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eigenbrod T, Dalpke AH. Bacterial RNA: An underestimated stimulus for innate immune responses. J. Immunol. 2015;195:411–418. doi: 10.4049/jimmunol.1500530. [DOI] [PubMed] [Google Scholar]
- 53.Abdullah Z, Schlee M, Roth S, Mraheil MA, Barchet W, Bottcher J, Hain T, Geiger S, Hayakawa Y, Fritz JH, Civril F, Hopfner KP, Kurts C, Ruland J, Hartmann G, Chakraborty T, Knolle PA. RIG-I detects infection with live Listeria by sensing secreted bacterial nucleic acids. EMBO J. 2012;31:4153–4164. doi: 10.1038/emboj.2012.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Park J, Zhang Y, Chen C, Dudley EG, Harvill ET. Diversity of secretion systems associated with virulence characteristics of the classical bordetellae. Microbiol. 2015;161:2328–2340. doi: 10.1099/mic.0.000197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hull CM, Bevilacqua PC. Mechanistic analysis of activation of the innate immune sensor PKR by bacterial RNA. J. Mol. Biol. 2015;427:3501–3515. doi: 10.1016/j.jmb.2015.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hull CM, Anmandangla A, Bevilacqua PC. Bacterial riboswitches and ribozymes potently activate the human innate immune sensor PKR. ACS Chem. Biol. 2016;11:1118–1127. doi: 10.1021/acschembio.6b00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ben-Asouli Y, Banai Y, Pel-Or Y, Shir A, Kaempfer R. Human interferon-gamma mRNA autoregulates its translation through a pseudoknot that activates the interferon-inducible protein kinase PKR. Cell. 2002;108:221–232. doi: 10.1016/s0092-8674(02)00616-5. [DOI] [PubMed] [Google Scholar]
- 58.Kwok CK, Tang Y, Assmann SM, Bevilacqua PC. The RNA structurome: transcriptome-wide structure probing with next-generation sequencing. Trends Biochem. Sci. 2015 doi: 10.1016/j.tibs.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 59.Rouskin S, Zubradt M, Washietl S, Kellis M, Weissman JS. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature. 2014;505:701–705. doi: 10.1038/nature12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Spitale RC, Flynn RA, Zhang QC, Crisalli P, Lee B, Jung JW, Kuchelmeister HY, Batista PJ, Torre EA, Kool ET, Chang HY. Structural imprints in vivo decode RNA regulatory mechanisms. Nature. 2015;519:486–490. doi: 10.1038/nature14263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roost C, Lynch SR, Batista PJ, Qu K, Chang HY, Kool ET. Structure and thermodynamics of N6-methyladenosine in RNA: a spring-loaded base modification. J. Am. Chem. Soc. 2015;137:2107–2115. doi: 10.1021/ja513080v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hudson GA, Bloomingdale RJ, Znosko BM. Thermodynamic contribution and nearest-neighbor parameters of pseudouridine-adenosine base pairs in oligoribonucleotides. RNA. 2013;19:1474–1482. doi: 10.1261/rna.039610.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kierzek E, Malgowska M, Lisowiec J, Turner DH, Gdaniec Z, Kierzek R. The contribution of pseudouridine to stabilities and structure of RNAs. Nucleic Acids Res. 2014;42:3492–3501. doi: 10.1093/nar/gkt1330. [DOI] [PMC free article] [PubMed] [Google Scholar]