Abstract
Objectives
The genus Aeromonas is known to causes diseases such as food poisoning, sepsis, and wound infection. However, the mode of Aeromonas transmission from environment to humans is not clearly understood. To evaluate the health risks of Aeromonas spp. in environmental freshwater, the number, proportion and putative virulence factors of Aeromonas species were investigated in Okinawa Prefecture, Japan.
Methods
Environmental freshwater samples were collected from three dams, two springs and three private wells. Aeromonas strains were identified by the biochemical method and the viable count was calculated. The production of extracellular enzymes and the virulence genes were investigated for possessing putative virulence factors using representative isolates.
Results
At least seven species of already-known Aeromonas isolates as well as unidentified Aeromonas spp. with/without arginin dehydrolase (ADH) exist in water at these sites. Aeromonas spp. was found to exist at over 1000 CFU/100 ml in one spring and two wells. A. veronii biovar sobria and A. jandaei were the predominant species in dams, and A. hydrophila and/or A. eucrenophila were predominant in wells. Almost all the sampled Aeromonas species produced protease, gelatinase, lipase, esterase and DNase, but A. caviae, A. caviae-like bacteria, and A. eucrenophila had low hemolytic activity. Most sampled A. hydrophila strains possessed both aerolysin gene (aer) and hemolysin gene (hlyA), but A. caviae and A. eucrenophila strains did not possess either gene.
Conclusions
Since these results indicated that several Aeromonas species having potential pathogenicity exist in environmental water in Okinawa, surveys are recommended as a public health measure.
Keywords: Aeromonas species, Environmental water, Distribution, Proportion, Putative virulence factors
Introduction
As recently as 27 years ago, only five species of the genus Aeromonas were recognized [1] but the classification is now subdivided, and at least 32 genospecies have been named [2]. Some species of the genus Aeromonas known to cause food poisoning, sepsis, wound infection and other diseases [3] have been found in human environments such as fresh water, coastal marine water, and soil [4]. Serious Aeromonas infections can occur in exposed patients with underlying diseases such as hepatitis, diabetes and/or malignant tumors [3, 5]. In addition, there have been reports of Aeromonas infections through trauma [6, 7] and near-drowning events [8] caused by recreational activities such as boating, fishing and diving.
Aeromonas spp. has also been isolated after natural disasters. High numbers (approximately 107 CFU/ml) of Aeromonas spp. were detected in floodwater samples during Hurricane Katrina in New Orleans in August 2005 [9]. Aeromonas species were found in 145 (22.6 %) of 641 isolates from patients with skin and soft-tissue infections during the tsunami in southern Thailand in December 2004 [10].
Several Aeromonas species secrete a number of extracellular proteins that are known virulence factors, including amylase, chitinase, elastase, aerolysin, nuclease, gelatinase, lecithinase, lipase and protease, found in four species [11]. Sechi et al. [12] also characterized slime, hemolysin, gelatinase, and protease, and adhesion to epithelial cells, and the virulence genes such as the cytolytic enterotoxin (AHCYTOEN), type IV pilus (Tap), and bundle forming pilus (BfpA and BfoG) in clinical and environmental isolates of three Aeromonas species.
The mode of transmission of pathogenic Aeromonas from environment to humans is still not clearly understood. Since the detailed distribution and proportions of Aeromonas species in aquatic environments are also not known precisely, a survey was required to classify the species living in the actual environment. Knowledge of the concentrations and virulence factors of Aeromonas spp. is important also for management of Aeromonas infections but there have been few relevant investigations in aquatic environments.
To evaluate public health risks of Aeromonas spp. in the environment, we investigated the number and proportion of Aeromonas species in the aquatic environment to learn the extent to which Aeromonas species exist in dams, springs and private wells used in daily life in Okinawa Prefecture, Japan. Further, we investigated whether the Aeromonas isolates possess any putative virulence factors.
Materials and methods
Isolation of bacteria and measurement of viable count
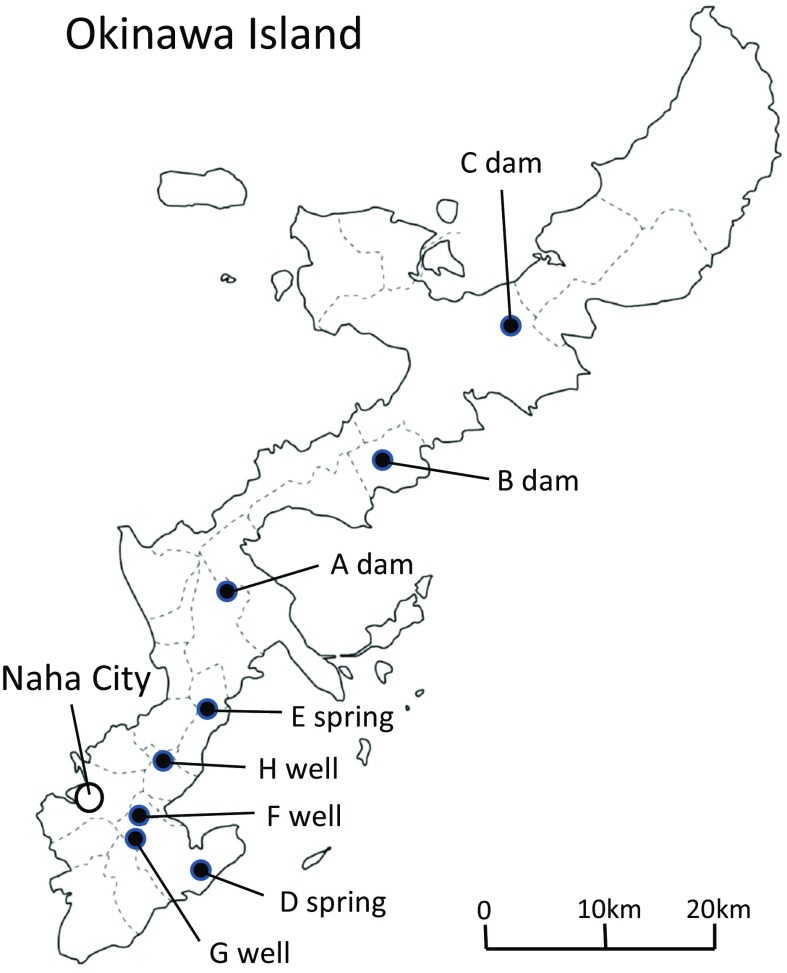
Okinawa Prefecture (population about 1,431,000) is located in the south-west part of Japan, in the subtropical zone. Environmental freshwater samples were collected from April to December 2009 at three dams, two springs and three private wells on Okinawa Island, the main island in Okinawa Prefecture (Fig. 1). The reasons for choosing the dams, springs, and wells as sampling points are as follows: (1) the water behind dams is used as drinking water after filtration and disinfection, (2) since the springs are also tourist resorts, visitors may consume the water, (3) water from the wells is used in personal daily life. Three samples each of 500 ml environmental freshwater were collected in sterilized bottles from a single point in Okinawa Island, respectively. The water temperature was measured at each sampling point. The measurement of viable count of Aeromonas spp. was performed as follows. One hundred ml and 10 ml of the water were centrifuged for 30 min at 1700g and resulting pellets were mixed with 1 ml of saline to produce 100 and 10 times concentrated samples, respectively. One hundred microliter samples of 100, 10 times, and the original concentration were each inoculated in triplicate using a spreader on Ryan’s Aeromonas Medium (RAM: Oxoid Ltd., Hampshire, England) plates and Xylose Deoxycholate Citrate Agar (XDCA: self-compounded) plates. The agar plates were incubated overnight at 35 °C. Suspect Aeromonas colonies from segments (whole, 1/2, 1/4, or 1/8) of the RAM and XDCA plates containing up to 300 colonies per plate were transferred to nutrient agar plates and incubated at 35 °C overnight. These colonies were screened by oxidase test, catalase test and gram staining, and suspect colonies were characterized by resistance to vibriostatic agent O/129 disc tests (150 μg/ml; Oxoid Ltd, Hampshire, England) using triple sugar iron agar (TSI: Eiken Chemical Co., Ltd., Tokyo, Japan) and sulfide indole motility medium (SIM: Eiken Chemical Co., Ltd., Tokyo, Japan); Voges-Proskauer and methyl red tests; growth in 0 % NaCl; tests for esculin hydrolysis, citrate, urocanic acid, and DL-lactate utilization, presence of lysine decarboxylase, ornithine decarboxylase, and arginine dihydrolase; production of acid and gas from glucose, and acid production from lactose, sucrose, arabinose, myo-inositol, mannitol, salicin, cellobiose, rhamnose, and mannose. The strains were identified according to the biochemical scheme of Abbott et al. [13] and Bergey’s manual [14]. The viable count of Aeromonas spp. was calculated using the identified positive colonies from the direct agar plates. The total viable count in the environmental water was calculated by plating on trypticase soy agar (TSA: BD, Sparks MD, USA) regulated to 3 % agar concentration, using samples of 100, 10 times and original concentration. The most probable number (MPN) of fecal coliform group was calculated using EC broth (Eiken Chemical Co., Ltd., Tokyo, Japan) with 10 ml and 1 ml of the original water, and 1 ml each of samples diluted to 10 and 100 times.
Fig. 1.
Freshwater sampling points. Environmental freshwater samples were collected from three dams (a, b and c), two springs (d and e) and three private wells (f, g and h) of Okinawa Island in Okinawa Prefecture. Naha city is the prefectural capital of Okinawa Prefecture
Production of extracellular enzymes
The tests for β-hemolysin and protease were carried out with modifications by Khan et al. [15] using 5 % sheep blood cells and 3 % skim milk, respectively. The test for gelatinase was performed by the method of Frazier [16] using agar plates including gelatin. The test for lipase was performed by the method of Davis and Ewing [17] using agar plates including corn oil. The test for esterase was performed by the method of Sierra [18] using agar plates including Tween 80. The test for DNase was carried out using commercially available deoxyribonucleic acid (DNA) agar (Eiken Chemical Co., Ltd., Tokyo, Japan).
PCR assay
To extract DNA, a colony on a nutrient agar plate was suspended in 150 microliters of Tris–EDTA (TE) buffer and boiled for 10 min. The suspension was centrifuged at 17,000g for 10 min with refrigeration and the supernatant was used as template DNA for PCR. Primer set Aero-F (5′-gag cga gaa ggt gac cac caa gaa c-3′) and Aero-R (5′-ttc cag tcc cac cac ttc act tca c-3′) designed by Nam and Joh [19] were used to amplify a 417 bp aerolysin gene (aer) fragment. Another primer set H1 (5′-ggc cgg tgg ccc gaa gat acg gg-3′) and H2 (5′-ggc ggc gcc gga cga gac ggg-3′) designed by Wong et al. [20] were used to amplify a 597 bp hemolysin gene (hlyA) fragment. A modified protocol for the amplification of the aer and hlyA genes from Aeromonas isolates was used. To amplify the aer gene, 2.5 μl of the template DNA was mixed with 2.5 μl of 10× Taq reaction mixture, 2 μl each of the 2.5 mM dNTPs, 0.125 μl each of the 50 μM primers, 0.2 μl of 5 U/μl Taq polymerase and 17.55 μl of distilled water (Total 25 μl) in a 200 μl microtube. Each reagent used for the PCR mixture was from a commercially available PCR reagent set (TaKaRa Ex Taq: Takara Bio Inc., Shiga, Japan) except the primers, which were synthesized (Hokkaido System Science Co., Ltd., Hokkaido, Japan). Each microtube was heated as a pre-denaturation step at 94 °C for 5 min, and as a DNA amplification step for 35 cycles at 94, 60 and 72 °C for 40, 20 and 30 s, respectively, as a final extension step at 72 °C for 5 min in a thermal cycler (Program Temp Control System PC-320: ASTEC, Fukuoka, Japan). PCR products were separated by electrophoresis in 1.5 % agarose gels and stained with 1 μg/ml ethidium bromide. The products were then observed under UV light, and a single band at 417 bp was determined to be positive for aer gene. To amplify the hlyA gene, a PCR mixture was prepared in the same way as the mixture for the aer gene except using half the primer volume. The amplification was performed as follows: Pre-denaturation at 94 °C for 2 min, DNA amplification for 30 cycles at 94, 68 and 72 °C for 40, 5 and 5 s, and final extension at 72 °C for 3 min. The verification of PCR products of 597 bp was performed by the above method.
Results
We investigated the viable count of Aeromonas spp. and other bacteria to understand the number of these bacteria in environmental freshwater being utilized in daily life (Table 1). Aeromonas spp. was found at over 1000 CFU/100 ml in environmental water at three points, E spring, F well and H well. Although the proportion of Aeromonas spp. in the total viable count was below 2.3 % in the other sampling points, it was 21.8 % in the E spring. The fecal coliform group was highest in the wells.
Table 1.
Viable count of Aeromonas spp. and other bacteria in water samples
| A dam | B dam | C dam | D spring | E spring | F well | G well | H well | |
|---|---|---|---|---|---|---|---|---|
| Month of water sampling | Aug | Aug | Dec | Apr | Apr | Sep | Sep | Dec |
| Water temperature (°C) | 32.0 | 31.4 | 18.0 | 22.5 | 20.5 | 26.5 | 27.0 | 22.8 |
| Total viable count (CFU/100 ml) | 7000 | 6000 | 9500 | 2000 | 11,000 | 61,000 | 6000 | 140,000 |
| Fecal coliform group (MPN/100 ml) | 4 | <3.0 | <3.0 | <3.0 | <3.0 | 7 | 4 | 43 |
| Viable count of Aeromonas spp. (CFU/100 ml) | 88 | 40 | 220 | 15 | 2400 | 1200 | 5 | 2200 |
| Viable count of Aeromonas spp./Total viable count (%) | 1.3 | 0.7 | 2.3 | 0.8 | 21.8 | 2.0 | 0.1 | 1.6 |
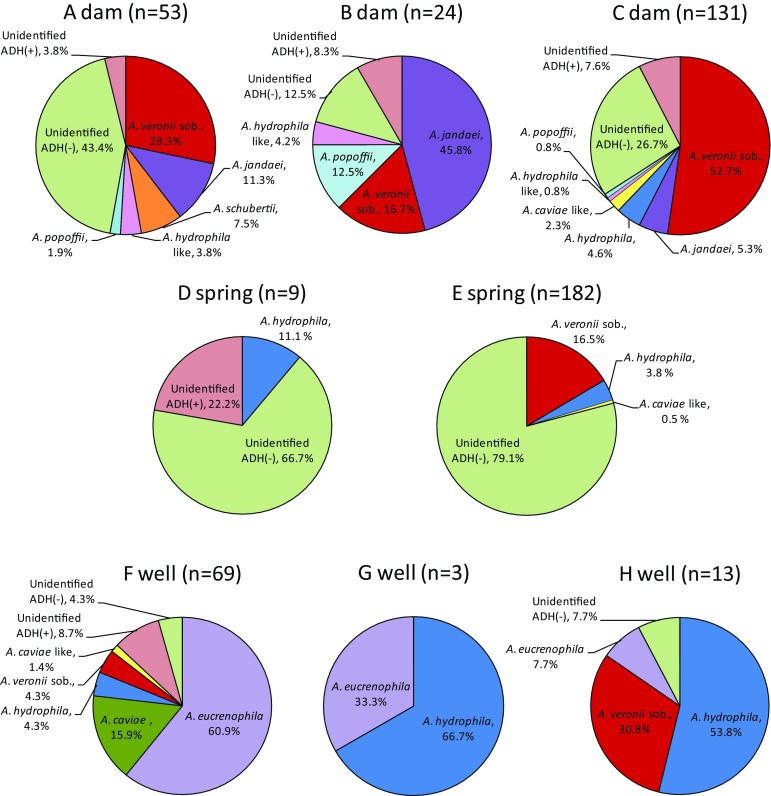
Regarding the prevalence of Aeromonas species in each sampling point, the unidentified Aeromonas spp. of arginin dehydrolase (ADH) negative (−) was the predominant species in springs and A. veronii bv. sobria and A. jandaei were predominant in dams. In wells A. hydrophila and/or A. eucrenophila were the predominant species, with A. eucrenophila accounting for 60.9 % of all Aeromonas spp. in F well, a much higher frequency than was isolated at other points (Fig. 2). Although unidentified Aeromonas spp. of ADH(−) was the most frequent isolate from A dam, they were classified into seven types by biochemical patterns (data not shown), indicating the presence of seven distinct unidentified strains of Aeromonas spp. Likewise, in C dam the unidentified Aeromonas spp. of ADH(−) included 11 types of biochemical patterns, and the unidentified Aeromonas spp. of ADH(+) included six types (data not shown). One of the three types of biochemical character pattern of unidentified Aeromonas spp. ADH(−) was 33.3 % at the D spring, and one of three types of unidentified Aeromonas spp. ADH(−) was 78.0 % at the E spring (data not shown). The total result of isolated Aeromonas species found by direct plating is shown in Table 2. We isolated 565 Aeromonas strains from water samples from dams, springs and wells. The already-known isolates included at least seven species of A. hydrophila, A. caviae, A. eucrenophila, A. veronii bv. sobria, A. jandaei, A. schubertii and A. popoffii. Unidentified Aeromonas spp. of ADH(+) and ADH(−) were found in 26 and 223 of total isolates respectively. A. hydrophila and A. veronii bv. sobria were both isolated from six of the eight points. Some species were more frequent in dams than in springs and wells. A. jandaei, A. popoffii and A. hydrophila-like bacteria were isolated only from the three dams, and A. eucrenophila was isolated only from the three wells. Although F well and H well had very high viable counts, the recovered strains of Aeromonas spp. were low (Table 1). The reason may be that because of the high viable count, concentrations of 10 times and original were used for measurement of Aeromonas species in both agar plates. Therefore, there were few isolated colonies, but the counts would likely be higher if these samples had been concentrated to 100 times like other sites.
Fig. 2.
Proportion of isolated Aeromonas species from direct plating. The proportion of Aeromonas species in each sampling point was investigated using direct plating, with colonies on 3 RAM and 3 XDCA plates using water samples of 100 times concentration except for 10 times sample of F well and original sample of H well. A. veronii sob. is A. veronii biovar sobria. A. hydrophila-like and A. caviae-like bacteria are difficult isolates that distinguishes A. hydrophila from A. bestiarum and A. caviae from A. media, respectively. Unidentified ADH(+) is the unidentified Aeromonas spp. of arginin dehydrolase (ADH) positive. Unidentified ADH(−) is the unidentified Aeromonas spp. of arginin dehydrolase (ADH) negative
Table 2.
Isolated Aeromonas species from water samples of dams, springs and wellsa
| Isolated Aeromonas | A dam | B dam | C dam | D spring | E spring | F well | G well | H well | Total |
|---|---|---|---|---|---|---|---|---|---|
| A. hydrophila | 6 | 2 | 11 | 12 | 2 | 7 | 40 | ||
| A. hydrophila likeb | 3 | 1 | 1 | 5 | |||||
| A. caviae | 19 | 19 | |||||||
| A. caviae likec | 3 | 6 | 1 | 10 | |||||
| A. eucrenophila | 58 | 1 | 1 | 60 | |||||
| A. veronii biovar sobria | 18 | 4 | 83 | 32 | 3 | 4 | 144 | ||
| A. jandaei | 6 | 13 | 7 | 26 | |||||
| A. schubertii | 4 | 4 | |||||||
| A. popoffii | 4 | 3 | 1 | 8 | |||||
| Unidentified ADH(+)d | 3 | 2 | 10 | 2 | 9 | 26 | |||
| Unidentified ADH(−)e | 28 | 3 | 34 | 7 | 147 | 3 | 1 | 223 | |
| Total | 66 | 26 | 145 | 11 | 196 | 105 | 3 | 13 | 565 |
a Aeromonas colonies were harvested from plates at the sample concentration used for calculation of viable count, and from other plates which were lower concentrations than the plates used for calculation
bThere are difficult isolates that distinguish A. hydrophila from A. bestiarum
cThere are difficult isolates that distinguish A. caviae from A. media
dUnidentified ADH(+) is Aeromonas spp. of arginin dehydrolase (ADH) positive
eUnidentified ADH(−) is Aeromonas spp. of arginin dehydrolase (ADH) negative
Representative Aeromonas spp. isolates were investigated to determine if they possessed any kind of putative virulence factors (Table 3). Nearly all Aeromonas species produced protease, gelatinase, lipase, esterase and DNase, however A. caviae, A. caviae-like bacteria and A. eucrenophila belonging to A. caviae complex had low hemolytic activity. Since most Aeromonas hemolysins described to date are related to one of two toxins of the aerolysin [21] and AHH1 hemolysin [22], we investigated aer and hlyA genes by the PCR method (Table 4). All A. hydrophila strains possessed aer and/or hlyA genes, with 83.3 % having both aer and hlyA genes. No A. caviae and A. eucrenophila strains possessed both genes, but 50 % of A. caviae-like isolates had the aer gene only, and the hlyA gene was not present in any. Most (80 %) of the A. veronii bv. sobria isolates possessed only the aer gene, with most of the rest having both aer and hlyA genes. Isolates of both A. jandaei and A. popoffii were 100 % positive for β-hemolysin, but 77.8 and 62.5 % had the aer (−)/hlyA(−) pattern, respectively. Most of the unidentified Aeromonas spp. of ADH(+) and ADH(−) possessed aer and/or hlyA genes.
Table 3.
Detection of putative virulence factors from Aeromonas spp.
| Isolated Aeromonas | Number of strains (%) | β-Hemolysis (sheep) | Protease (skim milk) | Gelatinase (gelatin) | Lipase (corn oil) | Esterase (Tween 80) | DNase |
|---|---|---|---|---|---|---|---|
| A. hydrophila | 24 (100) | 24 (100) | 24 (100) | 24 (100) | 24 (100) | 24 (100) | 24 (100) |
| A. hydrophila likea | 5 (100) | 3 (60.0) | 5 (100) | 5 (100) | 5 (100) | 5 (100) | 5 (100) |
| A. caviae | 7 (100) | 2 (28.6) | 7 (100) | 7 (100) | 7 (100) | 7 (100) | 7 (100) |
| A. caviae likeb | 10 (100) | 1 (10.0) | 10 (100) | 10 (100) | 10 (100) | 10 (100) | 10 (100) |
| A. eucrenophila | 6 (100) | 1 (16.7) | 6 (100) | 6 (100) | 6 (100) | 6 (100) | 6 (100) |
| A. veronii biovar sobria | 30 (100) | 29 (96.7) | 30 (100) | 30 (100) | 30 (100) | 30 (100) | 30 (100) |
| A. jandaei | 9 (100) | 9 (100) | 9 (100) | 9 (100) | 9 (100) | 9 (100) | 9 (100) |
| A. popoffii | 8 (100) | 8 (100) | 8 (100) | 8 (100) | 8 (100) | 8 (100) | 8 (100) |
| Unidentified ADH(+)c | 19 (100) | 14 (73.7) | 19 (100) | 19 (100) | 19 (100) | 19 (100) | 19 (100) |
| Unidentified ADH(−)d | 34 (100) | 28 (82.4) | 33 (97.1) | 34 (100) | 33 (97.1) | 33 (97.1) | 34 (100) |
| Total | 152 (100) | 119 (78.3) | 151 (99.3) | 152 (100) | 151 (99.3) | 151 (99.3) | 152 (100) |
aThere are difficult isolates that distinguish A. hydrophila from A. bestiarum
bThere are difficult isolates that distinguish A. caviae from A. media
cArginin dehydrolase (ADH) positive
dArginin dehydrolase (ADH) negative
Table 4.
Detection of aer and hlyA genes from Aeromonas spp.
| Isolated Aeromonas | No. of strains (%) | aer(+)/hlyA(+) | aer(+)/hlyA(−) | aer(−)/hlyA(+) | aer(−)/hlyA(−) |
|---|---|---|---|---|---|
| A. hydrophila | 24 (100) | 20 (83.3) | 1 (4.2) | 3 (12.5) | 0 (0) |
| A. hydrophila likea | 5 (100) | 0 (0) | 2 (40.0) | 0 (0) | 3 (60.0) |
| A. caviae | 7 (100) | 0 (0) | 0 (0) | 0 (0) | 7 (100) |
| A. caviae likeb | 10 (100) | 0 (0) | 5 (50.0) | 0 (0) | 5 (50.0) |
| A. eucrenophila | 6 (100) | 0 (0) | 0 (0) | 0 (0) | 6 (100) |
| A. veronii biovar sobria | 30 (100) | 5 (16.7) | 24 (80.0) | 0 (0) | 1 (3.3) |
| A. jandaei | 9 (100) | 0 (0) | 2 (22.2) | 0 (0) | 7 (77.8) |
| A. popoffii | 8 (100) | 0 (0) | 3 (37.5) | 0 (0) | 5 (62.5) |
| Unidentified ADH(+)c | 19 (100) | 7 (36.8) | 6 (31.6) | 0 (0) | 6 (31.6) |
| Unidentified ADH(−)d | 34 (100) | 3 (8.8) | 25 (73.5) | 1 (2.9) | 5 (14.7) |
| Total | 152 (100) | 35 (23.1) | 68 (44.7) | 4 (2.6) | 45 (29.6) |
aThere are difficult isolates that distinguish A. hydrophila from A. bestiarum
bThere are difficult isolates that distinguish A. caviae from A. media
cArginin dehydrolase (ADH) positive
dArginin dehydrolase (ADH) negative
Discussion
The genus Aeromonas has been isolated by Holmes et al. from aquatic environments such as rivers, lakes, ponds, seawater (estuaries), drinking water, groundwater, wastewater, and sewage in various stages of treatment [23]. In our study, Aeromonas spp. were found to exist at over 1000 CFU/100 ml in one spring and two wells, far higher than the <1 CFU/ml in ground water that has been reported by Holmes et al. [23]. Aeromonas in the E spring water accounted for 21.8 % of the total viable count. This water is used to make tea consumed at a traditional community event. Although Aeromonas numbers in dams were similar to what Holmes et al. reported, and also in lakes (1–102 CFU/ml) [23], various Aeromonas species were isolated from these sites. The water from dams in Okinawa Prefecture is filtered and disinfected for distribution as public water supplies. The D and E springs receive many sightseers since they were designated among “Selected 100 Exquisite and Well-Conserved Water Sites’’ by the Japanese Ministry of Environment. Visitors often consume the raw water because it appears to be clear and pure. The water from wells is used mainly for watering garden plants, washing cars, and washing harvested vegetables, so household members can be exposed to Aeromonas spp. in these ways. In brief, raw water without chlorination from these sampling points, except for dams, is frequently consumed or contacted by people.
Several studies have reported Aeromonas isolation from aquatic environments [4, 24, 25] but these contained little data on the proportion of various component species, and no other studies done in Japan with such data were found. Investigation of Aeromonas species in the environment revealed the proportion of the bacteria in environmental water, as shown in Fig. 2 of this study. Although our sampling points were geographically distant, the isolation of A. hydrophila and A. veronii bv. sobria from six of eight points, respectively (Fig. 1 and Table 2), suggests that both these species exist widely in aquatic environments in Okinawa. A. jandaei, A. popoffii and A. hydrophila-like bacteria were isolated from three dams, but these species were not isolated from springs and wells. These species may exist in environments where water has become eutrophic due to photosynthesis of phytoplankton, fertilizer runoff or other reasons, or because the water of dams is muddier than springs and wells. On the other hand, A. eucrenophila was isolated from three wells but not from dams and springs. Since well water appears clear, A. eucrenophila may prefer an environment where there is little organic matter and/or it not illuminated by sunlight. This is supported by A. eucrenophila being found in unpolluted surface and groundwater such as oligosaprobic streams, and in clean well water [14]. There was no apparent influence of water temperature in the sampled range, as seen in Table 1.
Reports indicate that ADH-positive species of Aeromonas are prevalent in strains isolated from clinical samples, and ADH-negative species are more prevalent in environmental strains [13], but the unidentified Aeromonas spp. of ADH(+) and ADH(−) strains in our study showed almost identical levels of protease, gelatinase, lipase, esterase, and DNase. The positive rate for β-hemolysin was somewhat higher in the unidentified ADH(−) Aeromonas spp. than the ADH(+) strain. We investigated aer and hlyA genes in isolated species to test the existence of two hemolytic toxins because these genes may contribute to the virulence of A. hydrophila [26]. Although the primer set of Pollard et al. could detect the aer gene from only A. hydrophila [27], we could detect it from various species using the primer set of Nam and Joh [19]. The positive rate found for the hlyA gene in A. caviae, A. caviae-like bacteria, and A. eucrenophila was 0 %, compared to 35 % found by of Heuzenroeder et al. [26] in A. caviae. The reason may lie in the difference of the sample source, because most positive samples of Heuzenroeder et al. were from fish rather than from water. On the other hand, the results for β-hemolysin and hlyA gene of the A. jandaei and A. popoffii do not agree with each other in Tables 3 and 4, suggesting that there may be a hemolytic gene besides aer and hlyA genes in the both species.
At least seven species of already-known Aeromonas isolates and the unidentified Aeromonas spp. with/without ADH exist in water of dams, springs and private wells that are in regular use in Okinawa. Six of the seven (excepting A. eucrenophila) already-known species have been reported in clinical Aeromonas isolates [28]. A. hydrophila and A. veronii bv. sobria were widely isolated at most of the sampling points. A. hydrophila, A. caviae and A. veronii bv. sobria are among the species mainly responsible for causing disease in humans [3, 29]. Infection caused by aeromonads in Okinawa is, therefore, also a possibility because species are widespread in freshwater environments.
Recently Khajanchi et al. found suggestive evidence of successful colonization and infection by particular strains of certain Aeromonas species after transmission to humans from water [30]. Voss et al. found that 13 of 28 Aeromonas wound and soft tissue infections during a 4-year period from 1985 to 1989 were associated with a water-related injury, and 43 % of the total could be directly related to water from a lake or river [7]. In Okinawa, 38 cases of extraintestinal Aeromonas infections were reported in a 5-year period from April 2008 to March 2013 in Urasoe General Hospital in Urasoe city (population about 114,000) adjacent to the prefectural capital Naha city [31]. Further, an afebrile 15-year-old girl who had been healthy until the onset of the illness died from A. veronii bv. sobria infection. The infection route was unknown [32]. Since these reports suggest the likelihood of transmission of Aeromonas species to humans from environmental freshwater, we believe that they support the significance of our study.
Through this study, we have quantified the prevalence, proportion, and the presence of various putative virulence factors of Aeromonas species in aquatic environments in Okinawa Prefecture. We clarified the trend of distribution of Aeromonas species in the environment. Our findings may help direct public health efforts to reduce Aeromonas infection transmitted from water to humans by pinpointing such risk factors as leisure activities in shallow waters at dams, children playing in springs, and using water from private wells for gardening or washing vegetables or cars. Aeromonas species are also potential pathogens when traumatic injury is exposed to environmental water, for immunocompetent as well as immunocompromised hosts. A survey of environmental water for Aeromonas spp. is, therefore, suggested.
Acknowledgments
We thank Dr F. Takamine formerly of the Laboratory of Microbiology, School of Health Sciences, Faculty of Medicine, University of the Ryukyus for her technical advice. We also thank Ms I. Oshiro of the Laboratory of Microbiology, School of Health Sciences, Faculty of Medicine, University of the Ryukyus for her technical help.
Compliance with ethical standards
Conflict of interest
The authors have no conflicts of interest.
References
- 1.Janda JM, Duffey PS. Mesophilic aeromonads in human disease: current taxonomy, laboratory identification, and infectious disease spectrum. Rev Infect Dis. 1988;10:980–997. doi: 10.1093/clinids/10.5.980. [DOI] [PubMed] [Google Scholar]
- 2.http://www.bacterio.cict.fr/. Accessed 25 Jan 2016.
- 3.Janda JM, Abbott SL. Evolving concepts regarding the genus Aeromonas: an expanding panorama of species, disease presentations, and unanswered questions. Clin Infect Dis. 1998;27:332–344. doi: 10.1086/514652. [DOI] [PubMed] [Google Scholar]
- 4.Nishikawa Y, Kishi T. Isolation and characterization of motile Aeromonas from human, food and environmental species. Epidemiol Infect. 1988;101:213–223. doi: 10.1017/S0950268800054121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ko WC, Chuang YC. Aeromonas bacteremia: review of 59 episodes. Clin Infect Dis. 1995;20:1298–1304. doi: 10.1093/clinids/20.5.1298. [DOI] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention (CDC) Aeromonas wound infections associated with outdoor activities—California. Morb Morta Wkly Rep. 1990;39:334–335. [PubMed] [Google Scholar]
- 7.Voss LM, Rhodes KH, Johnson KA. Musculoskeletal and soft tissue Aeromonas infection: an environmental disease. Mayo Clin Proc. 1992;67:422–427. doi: 10.1016/S0025-6196(12)60387-5. [DOI] [PubMed] [Google Scholar]
- 8.Bossi-Küpfer M, Genini A, Peduzzi R, Demarta A. Tracheobronchitis caused by Aeromonasveronii biovar sobria after near-drowning. J Med Microbiol. 2007;56:1563–1564. doi: 10.1099/jmm.0.47202-0. [DOI] [PubMed] [Google Scholar]
- 9.Presley SM, Rainwater TR, Austin GP, Platt SG, Zak JC, Cobb GP, et al. Assessment of pathogens and toxicants in New Orleans, LA following hurricane Katrina. Environ Sci Technol. 2006;40:468–474. doi: 10.1021/es052219p. [DOI] [PubMed] [Google Scholar]
- 10.Hiransuthikul N, Tanisiriwat W, Lertutsahakul K, Vibhagool A, Boonma P. Skin and soft-tissue infections among tsunami survivors in southern Thailand. Clin Infect Dis. 2005;41:e93–e96. doi: 10.1086/497372. [DOI] [PubMed] [Google Scholar]
- 11.Pemberton JM, Kidd SP, Schmidt R. Secreted enzymes of Aeromonas. FEMS Microbiol Lett. 1997;152:1–10. doi: 10.1111/j.1574-6968.1997.tb10401.x. [DOI] [PubMed] [Google Scholar]
- 12.Sechi LA, Deriu A, Falchi MP, Fadda G, Zanetti S. Distribution of virulence genes in Aeromonas spp. isolated from Sardinian waters and from patients with diarrhoea. J Appl Microbiol. 2002;92:221–227. doi: 10.1046/j.1365-2672.2002.01522.x. [DOI] [PubMed] [Google Scholar]
- 13.Abbott SL, Cheung WKW, Janda JM. The genus Aeromonas: biochemical characteristics, atypical reaction, and phenotypic identification schemes. J Clin Microbiol. 2003;41:2348–2357. doi: 10.1128/JCM.41.6.2348-2357.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin-Carnahan A, Joseph SW. Order XII Family 1. Aeromonadaceae. In: Brenner DJ, Krieg NR, Staley JT, Garrity GM, editors. Bergey’s manual of systematic bacteriology, The Proteobacteria, Part B the Gammaproteobacteria. 2nd ed. New York: Springer; 2005. p. 556–80.
- 15.Khan R, Takahashi E, Nakura H, Ansaruzzaman M, Banik S, Ramamurthy T, et al. Toxin production by Aeromonassobria in natural environments: river water vs. seawater. Acta Med Okayama. 2008;62:363–371. doi: 10.18926/AMO/30947. [DOI] [PubMed] [Google Scholar]
- 16.Frazier WC. A method for detection of changes in gelatin due to bacteria. J Infect Dis. 1926;39:302–309. doi: 10.1093/infdis/39.4.302. [DOI] [Google Scholar]
- 17.Davis BR, Ewing WH. Lipolytic, pectolytic, and alginolytic activities of Enterobacteriaceae. J Bacteriol. 1964;88:16–19. doi: 10.1128/jb.88.1.16-19.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sierra G. A simple method for the detection of lipolytic activity of micro-organisms and some observations on the influence of the contact between cells and fatty substrates. Antonie Van Leeuwenhoek. 1957;23:15–22. doi: 10.1007/BF02545855. [DOI] [PubMed] [Google Scholar]
- 19.Nam IY, Joh K. Rapid detection of virulence factors of Aeromonas isolated from a trout farm by hexaplex-PCR. J Microbiol. 2007;45:297–304. [PubMed] [Google Scholar]
- 20.Wong CYF, Heuzenroeder MW, Flower RLP. Contribution of extracellular factors to the virulence of Aeromonas hydrophila in the suckling mouse model. Microbiol. 1998;144:291–298. doi: 10.1099/00221287-144-2-291. [DOI] [PubMed] [Google Scholar]
- 21.Howard SP, Garland WJ, Green MJ, Buckley JT. Nucleotide sequence of the gene for the hole-forming toxin aerolysin of Aeromonas hydrophila. J Bacteriol. 1987;169:2869–2871. doi: 10.1128/jb.169.6.2869-2871.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirono I, Aoki T. Nucleotide sequence and expression of an extracellular hemolysin gene of Aeromonas hydrophila. Microb Pathog. 1991;11:189–197. doi: 10.1016/0882-4010(91)90049-G. [DOI] [PubMed] [Google Scholar]
- 23.Holmes P, Niccolls LM, Sartory DP. The ecology of mesophilic Aeromonas in the aquatic environment. In: Austin B, Altwegg M, Gosling PJ, Joseph S, editors. The genus Aeromonas. West Sussex: Wiley; 1996. pp. 127–150. [Google Scholar]
- 24.Chacon MR, Figueras MJ, Castro-Escarpulli G, Soler L, Guarro J. Distribution of virulence genes in clinical and environmental isolates of Aeromonas spp. Antonie Van Leeuwenhoek. 2003;84:269–278. doi: 10.1023/A:1026042125243. [DOI] [PubMed] [Google Scholar]
- 25.Igbinosa IH, Okoh AI. Detection and distribution of putative virulence associated genes in Aeromonas species from freshwater and wastewater treatment plant. J Basic Microbiol. 2013;53:895–901. doi: 10.1002/jobm.201200351. [DOI] [PubMed] [Google Scholar]
- 26.Heuzenroeder MW, Wong CYF, Flower RLP. Distribution of two hemolytic toxin genes in clinical and environmental isolates of Aeromonas spp.: correlation with virulence in a suckling mouse model. FEMS Microbiol Lett. 1999;174:131–136. doi: 10.1111/j.1574-6968.1999.tb13559.x. [DOI] [PubMed] [Google Scholar]
- 27.Pollard DR, Johnson WM, Lior H, Tyler SD, Rozee KR. Detection of the Aerolysin gene in Aeromonas hydrophila by the polymerase chain reaction. J Clin Microbiol. 1990;28:2477–2481. doi: 10.1128/jcm.28.11.2477-2481.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Janda JM, Abbott SL. The genus Aeromonas: taxonomy, pathogenicity, and infection. Clin Microbiol Rev. 2010;23:35–73. doi: 10.1128/CMR.00039-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu CJ, Wu JJ, Yan JJ, Lee HC, Lee NY, Chang CM, et al. Clinical significance and distribution of putative virulence markers of 116 consecutive clinical Aeromonas isolates in southern Taiwan. J Infect. 2007;54:151–158. doi: 10.1016/j.jinf.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 30.Khajanchi BK, Fadl AA, Borchardt MA, Berg RL, Horneman AJ, Stemper ME, et al. Distribution of virulence factors and molecular fingerprinting of Aeromonas species isolates from water and clinical samples: suggestive evidence of water-to-human transmission. Appl Environ Microbiol. 2010;76:2313–2325. doi: 10.1128/AEM.02535-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shimoji N, Murayama M, Furuno Y, Uechi A, Tamaki I, Tedokon M, et al. Basic analysis in the case of Aeromonas bacteria detected in areas other than the intestinal tract. Japanese J Med Technol. 2015;64:295–301. [Google Scholar]
- 32.Shiina Y, Ii K, Iwanaga M. An Aeromonas veronii biovar sobria infection with disseminated intravascular gas production. J Infect Chemother. 2004;10:37–41. doi: 10.1007/s10156-003-0274-2. [DOI] [PubMed] [Google Scholar]