Abstract
The two‐spotted spider mite, Tetranychus urticae, and the carmine spider mite, Tetranychus cinnabarinus, are invasive and native species in China, respectively. Compared with T. cinnabarinus, T. urticae has expanded into most parts of China and has become the dominant species of spider mite since 1983, when it was first reported in China. However, the mechanism of the demographic conversion has not been illuminated. In this study, one T. urticae field population and one T. cinnabarinus field population were isolated from the same plant in the same field, and the toxicological characteristics were compared between these two species. Laboratory bioassays demonstrated that T. urticae was more tolerant to commonly used acaricides than T. cinnabarinus. The activities of detoxification enzymes were significantly greater in T. urticae, and the fold changes of enzymes activities in T. urticae were also greater following exposure to acaricides. Furthermore, more metabolism‐related genes were upregulated at a basal level, and more genes were induced in T. urticae following exposure to acaricides. The comparison of proteins and genes between both species led credence to the hypothesis that T. urticae was more resistant to acaricides, which was the reason explaining the expansion of invasive T. urticae against native T. cinnabarinus. Laboratory simulation experiments demonstrated that following the application of acaricides, the composition of a mixed T. urticae/T. cinnabarinus population would change from a T. cinnabarinus‐dominant to a T. urticae‐dominant population. This study not only reveals that T. urticae possesses stronger detoxification capacity than its sibling species T. cinnabarinus, which facilitated its persistent expansion in China, but also points to the need to accurately identify Tetranychus species and to develop species‐specific management strategies for these pests.
Keywords: acaricide, expansion, population structure, RNA‐seq, Tetranychus cinnabarinus, Tetranychus urticae
1. Introduction
The two‐spotted spider mite, Tetranychus urticae, and the carmine spider mite, Tetranychus cinnabarinus, are both economically important species of the genus Tetranychus (Chhillar, Gulati, & Bhatnagar, 2007; Grbić et al., 2011), which belongs to the class Arachnida, infraclass Acari, order Prostigmata, and family Tetranychidae. As sibling species, T. urticae and T. cinnabarinus can seriously damage fruit trees, vegetables, ornamentals, and weeds throughout the world (Bolland, Gutierrez, & Flechtmann, 1998; Jeppson, Keifer, & Baker, 1975). In addition to the economic damage of these two spider mites on agriculture, the taxonomic status of these mites as separate species remains controversial. Some acarologists, including those in China, believe that T. urticae and T. cinnabarinus are two separate species (Boudreaux, 1956; Brandenburg & Kennedy, 1981; Kuang & Cheng, 1990; Li, Chen, & Hong, 2009; Li, Lu, Feng, & He, 2009) because they have different morphological characteristics in their adult stages (e.g., body colors and setae on tibia) and they do not mate with each other naturally (Kuang & Cheng, 1990; Zhang & Jacobson, 2000). However, others consider that they belong to the same species with T. cinnabarinus being the red form of T. urticae (Auger, Migeon, Ueckermann, Tiedt, & Navarro, 2013; Dupont, 1979; Ehara, 1999; de Mendonça, Navia, Diniz, Auger, & Navajas, 2011).
In China, the two forms coexist, where the red form T. cinnabarinus is distributed throughout China and is considered to be native, while the green form T. urticae, which was first reported in 1983 in Beijing, is considered to be invasive (Dong, Guo, & Niu, 1987; Sun, Lian, Navajas, & Hong, 2012). Tetranychus urticae has recently expanded its distribution from its putative area of introduction in Beijing to many parts of the country, including Hebei, Liaoning, Jilin, Gansu, Anhui, and Yunnan provinces and elsewhere (Meng, Wang, Jiang, & Yi, 2001; Sun et al., 2012). Unexpectedly, the invading T. urticae has become the most important mite pest in apple orchards in the north of China (Cai, Cheng, & Sha, 2003). A recent study by Wang indicated that T. urticae and T. truncatus were the dominant species on vegetables in some areas in Beijing and Hebei, revising the traditional opinion that T. cinnabarinus was the major species on common vegetable plants (Wang, Zhang, Wu, Xie, & Xu, 2013).
Most toxicity studies have concluded that there are differential responses of the two mites to commonly used acaricides (including plant resource pesticides). For example, T. urticae has greater resistance to abamectin, spiromesifen, etoxazole, hexythiazox on strawberries than T. cinnabarinus in California, USA (Bi, Niu, Yu, & Toscano, 2016). T. urticae was also identified to be more tolerant than T. cinnabarinus to traditional acaricides, such as fenazaquin, pyridaben, propargite, azocyclotin, and hexythiazox (Gu, Zhang, Zhao, & Li, 2000). In comparison with T. cinnabarinus, T. urticae has a significantly decreased susceptibility to six acaricides, which has been shown to be true in both slide‐dip immersion and leaf‐dipping method testing, where remarkably, the tolerance of T. urticae to abamectin was 2,575‐fold higher than that of T. cinnabarinus (Zhao, Zhou, & Ren, 2006). Tetranychus urticae has also been shown to be more tolerant to natural products, where the LC50 values for crude extracts from Wikstroemia chamaedaphne using chloroform, petroleum ether, or ethyl acetate were higher for T. urticae than for T. cinnabarinus (You‐Nian et al., 2010).
The most common reasons for insect/mite resistance to insecticides are enhanced metabolic detoxification and target‐site insensitivity (Van Leeuwen, Vontas, Tsagkarakou, Dermauw, & Tirry, 2010). Metabolic resistance has been reported worldwide and usually involves detoxification enzymes such as cytochrome P450 monooxygenases (P450s or CYPs for genes), carboxy/cholinesterases (CarEs or CCEs), and glutathione S‐transferases (GSTs) (Van Leeuwen et al., 2010; Xu et al., 2014). Gene expression analysis is widely used to reveal regulatory mechanisms that control cellular processes in animal, plants, and microbes (Van Leeuwen, Dermauw, Grbic, Tirry, & Feyereisen, 2013), including the elucidation of the gene expression profiles of detoxification genes involved in the metabolic resistance of insecticides (Strode et al., 2008). In particular, recently developed massively parallel RNA‐Seq deep sequencing and digital gene expression (DGE) testing have substantially changed the way resistance‐relevant genes in insects are identified and characterized because these methods facilitate the investigation of the functional complexity of the transcriptome (Metzker, 2010). Moreover, RNA‐Seq offers a great depth of sequence coverage with reduced variability (Zhou et al., 2010).
Following the initial discovery of T. urticae, the range of this species has expanded throughout China and has become the dominant spider mite. The causes of the demographic change are not always clear and may be attributed to various biological mechanisms or anthropogenic factors, including differences in their respective reproductive success, and susceptibility to acaricides. Similar effects can be seen in other insect species, where it has been reported that the application of insecticides can lead to rapid shifts in the composition of leafminer complexes in laboratory and field‐based experiments (Gao, Reitz, Wei, Yu, & Lei, 2012). Although the greater resistance of T. urticae versus T. cinnabarinus to acaricides is generally recognized, it is unclear whether acaricides facilitate the expansion of T. urticae, resulting in T. urticae being the dominant species of spider mites.
In our study, we compare the relative fitness at different temperatures between T. urticae and T. cinnabarinus, as well as the susceptibility of the two mites collected from the same field crop to commonly used acaricides. Furthermore, the influence of acaricides on changes in the population composition of mixed populations of T. urticae and T. cinnabarinus was simulated under laboratory conditions. Finally, the biochemical characteristics and gene expression profiles of metabolic detoxification enzymes in both mite species were characterized and compared. The goal of our study is to elucidate the mechanism(s) that has resulted in the continuous expansion of T. urticae throughout China to become the dominant pest mite species.
2. Methods
2.1. Mites and acaricides
Tetranychus urticae population and Tetranychus cinnabarinus population were originally collected from the same commercial rose field in Kunming, Yunnan, China (25°23′N, 102°42′E) in May 2014, and named Tu‐YN, Tc‐YN, respectively (Figure 1). Yunnan is located within a subtropical zone with a yearly average temperature of 23°C and maximum temperature of 38.5°C. The two spider mite populations were regarded as having the same acaricide‐exposure background as they were collected from the same crop of roses, which had been sprayed mainly with abamectin and occasionally with other acaricides (e.g., propargite, pyridaben, or cyflumetofen). The Tc‐SS strain was used as a reference colony of T. cinnabarinus, which was originally established from more than 1,500 mites collected from a cowpea field in Beibei, Chongqing, China, in 1998 (Li, Chen et al., 2009; Li, Lu et al., 2009), and has since been continuously reared in the laboratory. All strains and stock cultures were reared on detached cowpea (Vigna unguiculata L.) leaves placed on water‐soaked cotton in petri dishes (9 cm diameter) in growth chambers.
Figure 1.

The two‐spotted spider mite, Tetranychus urticae, and the carmine spider mite, Tetranychus cinnabarinus are both economically significant species of the genus Tetranychus, belonging to the class Arachnida, infraclass Acari, order Prostigmata, and family Tetranychidae
The commonly used acaricides were obtained from their respective manufacturers as follows: abamectin, 95.0% technical material (TC) (Veyong Biotech Co., Ltd. Hebei, China); chlorfenapyr, 96.0% TC and cyflumetofen, 98.8% TC (FMC Plant Protective Co., Ltd. Suzhou, China); fenpropathrin, 97.0% TC, pyridaben, 95.0% TC and propargite, 92.2% TC (Zhengbang Bioch Co., Ltd. Jiangxi, China); tebufenpyrad, 95.5% TC (Mitsubishi Chemical Co., Ltd. Japan); and bifenazate, 95.5% TC (Chemtura Co., Ltd., USA).
2.2. Number of setae on tibia of leg I in female
Mites were slide‐mounted with Hoyer's solution and cured on a Flatting table (Leica M205A, Germany) at 50°C for 4–5 days. Mounted mites were observed with an Olympus BX51 (Japan) equipped with differential interference contrast. The tibia setation follows Kuang and Cheng (1990) with modifications of Norton (Norton, 1998) concerning coxal setae. Thirty adult females picked randomly were observed from the corresponding populations (Tc‐YN, Tu‐YN, and Tc‐SS, respectively), which had been reared under the same condition for 12 months.
2.3. Hybridization
Tetranychus spp. have haplodiploid sex determination, where unfertilized eggs develop into males, while zygotes will develop into females. Therefore, reproductive isolation can be detected based on the sex ratio of the offspring, as females can only be produced through a viable cross. Cross‐mating of T. urticae to T. cinnabarinus was conducted in six‐cm petri dish arenas containing a bean leaf disk (4 cm diameter) on water‐soaked cotton. One female deutonymph (Tc‐YN, Tu‐YN, or Tc‐SS) was transferred to each arena, and then three virgin males from another mite species were added. The males and female were removed 2 and 5 d after adult eclosion, respectively. The numbers of F1 females and males was recorded and if adult females were present among the F1 offspring, they were allowed to in‐cross and the numbers of F2 females and males were recorded through to the F3 generation.
2.4. Data collection of the expansion of Tetranychus urticae in China
Studies regarding T. urticae and T. cinnabarinus in China were collected by retrieving the term “Tetranychus urticae” and/or “Tetranychus cinnabarinus” in the CNKI database in Chinese and in the SCI database. Based on the historical reports from the literature, the geographical distributions of T. cinnabarinus and T. urticae were determined based on the reported locations and dates from which the mites were collected. The map of distribution was created using ArcGIS9.3 software (Environmental Systems Research institute, Redlands, CA, USA) (http://www.arcgis.com) and Adobe Illustrator CS5 (v 15.1.0.39) software (http://www.adobe.com/products/illustrator.html).
2.5. Biology of Tc‐YN and Tu‐YN under the different temperature conditions
Life‐history variables of Tc‐YN and Tu‐YN were measured using arenas, each consisting of a bean leaf disk (4 cm diameter) on water‐soaked cotton in a petri dish (6 cm diameter). The arenas were kept in climatic chambers at 16 ± 1°C/26 ± 1°C/33 ± 1°C, respectively, 50 ± 5% RH, and a 14:10 (L:D) regime. Five adult females were transferred to each arena for 4 hr to lay eggs, after which the females were removed and a single egg was left in each arena. A total of 50 arenas were prepared for each species per temperature. The arenas were examined every 24 hr, and the duration and mortality for each development stage were recorded.
Life‐tables were prepared from the estimated rates for age‐specific survival (l x) and age‐specific fecundity (m x), where l x and m x are the proportion of surviving females at age x and the number of oviposition events produced per female in the age interval x, respectively. The sum of the products (l x m x) then provides the net reproductive rate (R 0). The mean generation time (T, days) was calculated as: T = Σl x m x x/R 0. The innate capacity for increase (r m) was calculated as: r m = lnR 0/T (where e is the natural logarithm base), and the population doubling time (t, days) was computed using the formula: t = ln2/r m. Relative fitness (R f) was estimated as follows: R f = (R 0 of Tu‐YN)/(R 0 of Tc‐YN), an R f > 1 suggests that the Tu‐YN has a fitness advantage, whereas an R f < 1 suggests that Tu‐YN has a fitness defect compared with Tc‐YN (Groters, Tabashnik, & Finson, 1994; Li, Gao, Zheng, & Liang, 2000).
2.6. Toxicological tests and acaricide stimulation
Median lethal concentration (LC50) values for adult mites were measured using the modified residual coated vial (RCV) method recommended by Van Leeuwen, Stillatus, and Tirry (2004). Five acaricides (abamectin, tebufenpyrad, cyflumetofen, propargite, and bifenazate), which could kill the larva, were selected to carry out the larval bioassays, and these tests were based on the method described by Knight, Beers, Hoyt, and Riedl (1990). Thirty larvae were placed on a bean leaf disk (4 cm diameter) on water‐soaked cotton in a petri dish (6 cm diameter). Prepared solutions of acaricides (1 ml) were sprayed onto the leaf disks containing the mites using a Potter Spray Tower (Burkard Manufacturing, Rickmansworth, Herts, UK) at 68.9 kPa. The acaricide solutions were diluted in distilled water to at least seven concentrations that resulted in mite mortality ranging from 20 to 85%. Distilled water alone was used as a control, and all doses were independently replicated three times. Following acaricide application, mites were maintained on the leaf disks at 26 ± 1°C and a 14:10 (L:D) regime. Individual mites were assessed for mortality under a dissecting microscope 24–48 hr after treatment, when all larvae in the control treatment had developed into nymphs. LC50 values and their 95% confidence limits were calculated from probit regression using probit analysis software (POLO, leOra Software, Berkeley, CA, USA). If the 95% confidence limits of the LC50 values did not overlap between Tc‐YN and Tu‐YN, then the differential acaricide susceptibility was considered to be significant.
To test the response of mites to abamectin‐, fenpropathrin‐, and tebufenpyrad stimulation, newly emerged adults (2–3 days old) Tu‐YN and Tc‐YN were treated for 6 hr with an LC30 dosage as described by Shen, Shi, Xu, and He (2014). For this, adults were exposed to tubes coated with the acaricide and acetone served as the control. Surviving mites were collected for enzyme sources and RNA extraction. Each sample was independently replicated three times for enzyme activity assay and RNA sequencing.
2.7. Mixed population experiments
To generate mixed species populations, five male and five female adults (newly eclosed virgins) from each of the Tu‐YN and Tc‐YN populations were added to the same bean leaf disk (6 cm diameter) on water‐soaked cotton in a petri dish (9 cm diameter). Mites were held on the leaf disk at 26 ± 1°C and a 14:10 (L:D) photoperiod and allowed to mate and expand their populations for 15 days. After 15 days, the numbers of female adults belong to each species were recorded and the leaf disks were sprayed with either abamectin (4.5 mg/L, the recommended field dosage), cyflumetofen (80 mg/L, the recommended field dosage), or distilled water (as a control) using a Potter Spray Tower (Burkard Manufacturing, Rickmansworth, Herts, UK) at 68.9 kPa. The spray applications were repeated in intervals of 15 days for a total three times. The numbers of female adult mites of each species were measured immediately prior to each spray, and again for a final reading at 15 days following the last spray. All treatments were independently replicated three times.
2.8. P450 monooxygenase assay
P450 activity was measured according to Shang's method (Shang & Soderlund, 1984). A total of 200 female adult mites (Tu‐YN, Tc‐YN) were homogenized on ice in 1.5 ml phosphate‐buffered solution (PBS, 0.1 M, pH 7.8) and then centrifuged at 10,000 × g for 15 min at 4°C. The supernatant was then used for testing using nitroanisole (0.05 M in acetone) as the substrate, and NADPH was added to reaction. The reaction was allowed to proceed for 30 min at 37°C and then stopped with 1 M hydrochloric acid, extracted over chloroform and neutralized with 0.5 M NaOH. The optical densities of the reactions were then measured at 400 nm using a microplate reader (TECAN Co.). Wells without extracted enzyme were used as controls, and the amount of protein in the enzyme source was determined using the Bradford method, with bovine serum albumin as the standard. The specific activity was determined according to a nitrophenol standard curve and the protein concentration of the sample.
2.9. Glutathione S‐transferase activity assay
Glutathione S‐transferases activity was determined according to the method of Habig, Pabst, and Jakoby (1974) with modifications by Stumpf & Nauen (2002). A total of 200 female adult mites (Tu‐YN, Tc‐YN) were homogenized on ice in 1.5 ml PBS (0.04 M, pH 6.5), followed by centrifugation at 10,000 × g for 10 min at 4°C. CDNB (0.6 mM) and GSH (6 M) were used as substrates and added to the supernatant, which was incubated for 20 min at 37°C during which time, GSTs reacted with the reduced GSH. The optical density at 340 nm was immediately recorded at intervals of 30 s for 5 min using a microplate reader. The results were determined based on the protein concentration of sample, and the specific activity was converted from the OD value.
2.10. Carboxylesterase activity assay
The method reported by Van Asperen (Van Asperen, 1962) was adopted for testing CarE activity, in which 200 female adult mites (Tu‐YN, Tc‐YN) were homogenized on ice in 1.5 ml PBS (0.04 M, pH 7.0), then centrifuged at 10,000 × g for 10 min at 4°C. The supernatant was removed and placed on ice, and α‐naphthyl acetate (3 × 10−4 M) and 10−4 M physostigmine were used as the substrates. The reaction was incubated for 10 min at 37°C, then the color‐developing agent was added (mixed as follows: mass fraction 5% SDS: mass fraction 1% fast blue B salt = 5:2 (v/v)), and the OD value at 600 nm was recorded immediately. The specific activity of CarE was calculated based on the α‐naphthol standard curve and the protein concentration of sample.
2.11. RNA extraction
Total RNA was extracted from 2‐ and 3‐day‐old adult mites (Tu‐YN, Tc‐YN) using the RNeasy plus Micro Kit (Qiagen, Hilden, Germany). Genomic DNA was removed using a genomic DNA elimination column supplied with the kit. The quality of the RNA sample was verified by ensuring that the OD260/280 was within the range of 1.8–2.2 as measured by the NanoVue UV–vis spectrophotometer (GE Healthcare Bio‐Science, Uppsala, Sweden), and qualified samples also had a 28S to 18S rRNA ratio above 1.0 as measured by a 1% agarose gel electrophoresis and the Agilent 2100 Bioanalyzer (Palo Alto, CA, USA) with a minimum integrity value of 8.
2.12. cDNA library preparation and RNA‐Seq
Total RNA pools from each sample were then used for preparing cDNA libraries using the Ion Total RNA‐Seq kit V2 (Life Technologies Corporation, CA, USA). Double‐stranded cDNA was ligated to barcoded adapters and was sequenced using the Ion PI™ Chip (Ion torrent, Life technologies, CA, USA) at the Beijing Genomics Institute (Shenzhen, China). Libraries were run at a concentration of 4–5 pM. To ensure the accuracy of subsequent analysis, raw sequences were cleaned to remove adaptors and sequencing errors. Reads were removed if they contained the sequencing adaptor, more than 5% unknown nucleotides, or more than 20% of bases of low quality. This output was called clean reads, which was used for subsequent downstream analyses.
2.13. Gene mapping
The entire assembled genes were used to search for the best‐hit homologous proteins (BLASTX cutoff e‐value 1.0E‐5) in the T. urticae genome (Grbić et al., 2011). Ortholog prediction was performed by performing BLASTX and TBLASTN bidirectional comparisons between Tc‐YN and Tu‐YN (with a threshold cutoff e‐value ≤1.0E‐5) to identify the hits within the two species.
2.14. Differential gene expression analysis
RNA‐Seq data were mapped to the T. urticae genome (version 200909) (http://bioinformatics.psb.ugent.be/orcae/overview/Tetur) using the Torrent Mapping Alignment Program v3.4.1 (ThermoFisher Scientific, Grand Island, NY, USA) to obtain Reads Per Kb per Million reads (RPKM), and the differential gene expression analysis to identify differentially expressed genes (DEGs) between two different DGE libraries was conducted using the method described by Audic and Claverie (Audic & Claverie, 1997). The false discovery rate (FDR) method determines the p‐value threshold for multiple testing by controlling the FDR value. The criteria of FDR < 0.001 and the absolute value of log2 ratio >1 were used to judge the significance of gene expression differences. The full dataset has been submitted to NCBI‐GEO database under the experiment ID GSE75529. The DEG functional features were analyzed according to cluster of orthologous groups of proteins (COG) and KEGG orthology (E‐value, 10E‐5). The Blast2GO program (Conesa et al., 2005) was employed to obtain Gene Ontology (GO) annotations for the DEGs.
2.15. Cloning and sequence comparison of target of acaricide genes for Tu‐YN and Tc‐YN
The coding sequence (CDS) information of 14 genes (VGSC, GluCl‐01‐05, Rdl1‐3, SdhA‐E) (Table S10) targeted by acaricides was obtained from the NCBI database for Tc‐YN, and the T. urticae genome (Grbić et al., 2011) for Tu‐YN and used to design PCR primers (Table S10) to obtain full‐length cDNA of the 14 target genes in Tu‐YN and Tc‐YN. The sequences from NCBI and the T. urticae genome were considered as baseline acaricide‐susceptible sequences. Amplicons generated by PCR were then Sanger‐sequenced and aligned at the nucleotide and amino acid levels using ClustalW. Identified point mutations were then matched with reported mutations of target genes involved in acaricide resistance in mites (Ilias, Vontas, & Tsagkarakou, 2014; Kwon, Clark, & Lee, 2015). At least 10 samples from either Tc‐YN or Tu‐YN were sequenced for each gene.
2.16. Statistical analyses
The statistically significant differences of development duration, enzyme activity, and individual numbers in mixed population were calculated using independent‐sample t‐tests for all two‐sample comparisons with a p‐value <.05 using SPSS19.0 software, CSPSS, Chicago, IL, USA.
3. Results
3.1. Species identification
Following field collection, adult female mites with two spots and red bodies were identified as T. cinnabarinus and designated as Tc‐YN, while adult female mites with two spots and green bodies were identified as T. urticae and designated as Tu‐YN. From these two mite populations, 30 individuals were randomly selected from each for the counting of the setae on the tibia of leg I. In the Tc‐YN and Tc‐SS, the tibiae of the first pair legs of most adult females had 10 setae, with some individuals having 12 or 13 setae (Table 1). In the Tu‐YN population, however, only 10 setae were observed (Table 1, Figure S1), which is a feature of T. urticae population (Boudreaux, 1956; Kuang & Cheng, 1990). The morphological characters observed were similar to those reported by Kuang and Cheng (1990) and Zhang and Jacobson (2000) who reported that the number of setae on tibia I in adult females was a very useful and convenient method for separating T. cinnabarinus and T. urticae populations.
Table 1.
The number of setae on tibia of leg I in Tetranychus cinnabarinus and Tetranychus urticae
| Population (30 mite) | 10 setae | 12 setae | 13 setae | |||
|---|---|---|---|---|---|---|
| Mite | Percentage (%) | Mite | Percentage (%) | Mite | Percentage (%) | |
| Tc‐SS | 20 | 66.7 | 5 | 16.7 | 5 | 16.7 |
| Tc‐YN | 21 | 70.0 | 4 | 13.4 | 5 | 16.7 |
| Tu‐YN | 30 | 100.0 | – | – | – | – |
The results of cross‐breeding between T. urticae and T. cinnabarinus were presented in Table 2. Reciprocal cross‐mating between two T. cinnabarinus populations originating from the field (Tc‐YN)‐ and laboratory (Tc‐SS)‐produced normal progeny females (F1 to F3) showing no reproductive isolation between these two populations. Cross‐mating between the two T. cinnabarinus populations and Tu‐YN produced only males in the F1 generation, with the exception of one Tc‐SS × Tu‐YN’ mating, which generated F1 females with an unusual wax yellow color that could not generate F2 female offspring (Table 2, Figure S2). The results suggested that reproductive isolation between T. urticae and T. cinnabarinus was complete.
Table 2.
Cross‐breeding results between Tetranychus urticae and Tetranychus cinnabarinus from different populations (Numbers of females and males in F1, F2, and F3)
| Crosses (♀ × ♂) | Cross‐couples | Couples generating F1 ♀ | F1 ♀a | F1 ♂a | F1 ♀ (10mites) × brothers | F2 ♀ (10mites) × brothers | ||
|---|---|---|---|---|---|---|---|---|
| F2 ♀ | F2 ♂ | F3 ♀ | F3 ♂ | |||||
| Tc‐YN × Tu‐YN | 15 | 0 | 0 | 159 | – | – | – | – |
| Tu‐YN × Tc‐YN | 14 | 0 | 0 | 144 | – | – | – | – |
| Tc‐SS × Tc‐YN | 12 | 10 | 253 | 72 | 201 | 92 | 336 | 104 |
| Tc‐YN × Tc‐SS | 15 | 13 | 267 | 106 | 163 | 114 | 298 | 127 |
| Tc‐SS × Tu‐YN | 15 | 1 | 17b | 178 | 0 | 28 | – | – |
| Tu‐YN × Tc‐SS | 14 | 0 | 0 | 277 | – | – | – | – |
In T. cinnabarinus, the number of the female mite is greater than that of male, and the ratio range is generally (1–4):1 in an indoor population (Kuang & Cheng, 1990).
The body color of the 17 females was an unusual wax yellow, which was different from the typical adult female color of T. cinnabarinus and T. urticae (the typical adult female color for T. cinnabarinus was brownish red and that for T. urticae was light green) (Figure S2).
3.2. Distribution of Tetranychus urticae and Tetranychus cinnabarinus in China
From the map showing the distribution of T. urticae and T. cinnabarinus, only T. cinnabarinus was reported in China before 1983 (Figure 2a), T. urticae initially emerged in 1983 and expanded to nine districts of China gradually by the end of the last century (Figure 2b). Presently, T. urticae has expanded to 18 districts (Figure 2c). In recent years, many researchers observe a higher prevalence of T. urticae than T. cinnabarinus on multiple host plants, such as vegetables and ornamental plants, demonstrating that the invasive T. urticae has expanded successfully in China.
Figure 2.
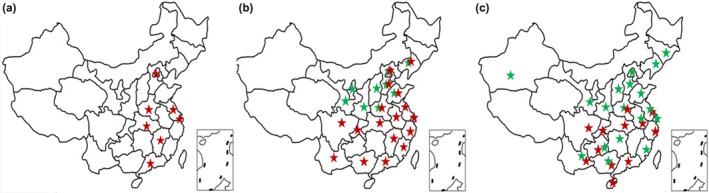
The distribution of Tetranychus cinnabarinus and Tetranychus urticae in China. (a) Sampling conducted during 1975–1982; (b) Sampling conducted during 1983–1999; (c) Sampling conducted during 2000–2014. Red and Green stars indicate T. cinnabarinus and T. urticae, respectively
3.3. Comparison of biological characteristics between Tc‐YN and Tu‐YN
Comparisons between Tc‐YN and Tu‐YN at 16°C indicated significant differences in the development duration (the duration of the period from egg to adult female), the net reproductive rate (R 0), the capacity for increase (r m), the population doubling time (t) and the average generation time (T). The main contrast between Tc‐YN and Tu‐YN was observed with respect to R 0, and the value for this for Tc‐YN was estimated to be 30.7% lower than for Tu‐YN. The relative fitness (R f) observed for Tu‐YN was 1.44, indicating the presence of a fitness advantage in Tu‐YN at lower temperatures compared with Tc‐YN (Table 3). At 26°C, no significant differences between Tc‐YN and Tu‐YN were recorded for all population parameters (Table 3). Under the high temperature condition (33°C), Tu‐YN exhibited slightly longer female time from egg to adult. No significant differences were observed for the parameter t and T. However, significant differences were recorded for the R 0 and r m parameters. For R 0, the value estimated for Tc‐YN was 79.3% higher than for Tu‐YN, while the R f observed for Tu‐YN was only 0.55, indicating the presence of a fitness cost in this species compared with Tc‐YN at 33°C (Table 3).
Table 3.
Development duration and population parameters of Tetranychus cinnabarinus and Tetranychus urticae at different temperatures
| Temperature (°C) | Mite | Development duration (mean ± SE, days) | Population parameter | Relative fitness R f | |||
|---|---|---|---|---|---|---|---|
| R 0 | r m | T | t | ||||
| 16 | Tc‐YN | 36.0 ± 1.2 | 35.56 | 0.08 | 46.76 | 9.08 | – |
| Tu‐YN | 31.5 ± 0.5a | 51.35 | 0.09 | 43.13 | 7.59 | 1.44 | |
| 26 | Tc‐YN | 11.1 ± 0.1 | 155.83 | 0.22 | 22.53 | 3.09 | – |
| Tu‐YN | 10.2 ± 0.1a | 140.47 | 0.24 | 20.29 | 2.84 | 0.90 | |
| 33 | Tc‐YN | 6.0 ± 0.1 | 104.73 | 0.39 | 11.95 | 1.78 | – |
| Tu‐YN | 6.4 ± 0.1a | 58.00 | 0.37 | 11.02 | 1.88 | 0.55 | |
R 0, Net reproduction rate; r m, Intrinsic rate of increase; T, Mean generation length (days); t, Population doubling time (days).
The data within a column under the same temperature condition are significantly different (p < .05).
3.4. Susceptibility of Tc‐YN and Tu‐YN to acaricides
The toxicities of abamectin and seven other acaricides to Tu‐YN and Tc‐YN were determined using the RCV method. The susceptibility of adult female mites to these acaricides is shown in Table 4. The LC50s of abamectin, fenpropathrin, tebufenpyrad, cyflumetofen, propargite, bifenazate, pyridaben, and chlorfenapyr for Tc‐YN were 2.49, 1072.97, 45.86, 23.16, 42.13, 62.08, 62.07, and 19.66 mg/L, respectively (Table 4). In comparison, the LC50 values for Tu‐YN were as similar or higher, being 8.50‐, 1.86‐, 0.93‐, 1.36‐, 4.75‐, 0.73‐, 2.85‐, and 2.87‐fold greater than that for Tc‐YN, respectively (Table 4). The toxicity of the selected acaricides against the larval mites is shown in Table 5. The LC50s of abamectin, tebufenpyrad, cyflumetofen, propargite, and bifenazate for the larvae of Tc‐YN were 0.044, 1.071, 0.329, 11.427, and 2.146 mg/L, respectively, while similar to the adult assays, the LC50s for the Tu‐YN larvae were higher, being 4.36‐, 2.56‐, 2.46‐, 3.04‐, and 2.30‐fold greater than Tc‐YN, respectively (Table 5). The results from the toxicity measurements revealed that Tu‐YN is more tolerant against the most of acaricides than Tc‐YN.
Table 4.
Susceptibility of Tetranychus cinnabarinus and Tetranychus urticae adults to selected acaricides
| Acaricides | Mite | Slope | χ2 a | LC50 (95% CL) (mg/L) | TRb |
|---|---|---|---|---|---|
| Abamectin | Tc‐YN | 1.22 ± 0.160 | 0.15c | 2.49 (1.91‐3.22) | 1.00 |
| Tu‐YN | 1.59 ± 0.211 | 1.03c | 21.19 (17.34‐25.86) | 8.50 | |
| Fenpropathrin | Tc‐YN | 1.91 ± 0.316 | 1.99c | 1072.97 (904.59‐1334.46) | 1.00 |
| Tu‐YN | 1.16 ± 0.23 | 0.45c | 1993.69 (1516.40‐3053.13) | 1.86 | |
| Tebufenpyrad | Tc‐YN | 1.28 ± 0.257 | 0.58c | 45.86 (34.03‐58.21) | 1.00 |
| Tu‐YN | 1.34 ± 0.257 | 0.76c | 42.84 (31.68‐53.75) | 0.93 | |
| Cyflumetofen | Tc‐YN | 2.79 ± 0.308 | 2.17c | 23.16 (20.40‐25.97) | 1.00 |
| Tu‐YN | 2.44 ± 0.289 | 1.97c | 31.50 (25.02‐39.59) | 1.36 | |
| Propargite | Tc‐YN | 3.93 ± 0.400 | 4.96c | 42.13 (34.41‐51.30) | 1.00 |
| Tu‐YN | 1.58 ± 0.239 | 2.46c | 199.92 (162.04‐254.79) | 4.75 | |
| Bifenazate | Tc‐YN | 2.04 ± 0.289 | 1.83c | 62.08 (52.99‐73.55) | 1.00 |
| Tu‐YN | 1.78 ± 0.261 | 3.63c | 45.43 (29.48‐62.41) | 0.73 | |
| Pyridaben | Tc‐YN | 2.39 ± 0.340 | 2.72c | 62.07 (54.01‐72.12) | 1.00 |
| Tu‐YN | 1.31 ± 0.290 | 0.35c | 176.85 (137.42‐249.79) | 2.85 | |
| Chlorfenapyr | Tc‐YN | 1.79 ± 0.239 | 1.62c | 19.66 (16.24‐24.37) | 1.00 |
| Tu‐YN | 1.74 ± 0.254 | 1.51c | 44.64 (36.80‐55.57) | 2.27 |
Pearson chi‐square, goodness‐of‐fit test.
TR: tolerance ratio = LC50 of Tu‐YN female adults/LC50 of Tc‐YN female adults.
The logarithm concentration‐probit regression line passed the chi‐square test. Chi‐squared distribution was used to test whether the regression between logarithm concentration and probit passes the goodness of fit, if passed (the calculated chi‐square values < expected χ2 (df)0.05, goodness‐of‐fit chi‐square is significant), meaning the current data represent the fact.
Table 5.
Susceptibility of Tetranychus cinnabarinus and Tetranychus urticae larvae to selected acaricides
| Acaricides | Mite | Slope ± SE | χ2 a | LC50 (95% CL) (mg/L) | TRb |
|---|---|---|---|---|---|
| Abamectin | Tc‐YN | 1.17 ± 0.15 | 1.96c | 0.04 (0.03‐0.06) | 1.00 |
| Tu‐YN | 1.24 ± 0.17 | 0.79c | 0.19 (0.15‐0.28) | 4.36 | |
| Tebufenpyrad | Tc‐YN | 0.84 ± 0.17 | 0.37c | 1.07 (0.69 ‐1.54) | 1.00 |
| Tu‐YN | 1.33 ± 0.18 | 0.63c | 2.74 (2.16 ‐3.58) | 2.56 | |
| Cyflumetofen | Tc‐YN | 1.31 ± 0.16 | 0.50c | 0.33 (0.25‐0.42) | 1.00 |
| Tu‐YN | 1.25 ± 0.17 | 1.32c | 0.81 (0.62‐1.05) | 2.46 | |
| Propargite | Tc‐YN | 3.36 ± 0.36 | 1.31c | 11.43 (10.25 ‐12.67) | 1.00 |
| Tu‐YN | 2.27 ± 0.35 | 0.17c | 34.82 (29.79 ‐42.71) | 3.04 | |
| Bifenazate | Tc‐YN | 1.45 ± 0.20 | 0.35c | 2.15 (1.72‐2.73) | 1.00 |
| Tu‐YN | 1.50 ± 0.21 | 1.28c | 4.94 (3.99‐6.45) | 2.30 |
Pearson chi‐square, goodness‐of‐fit test.
TR: tolerance ratio = LC50 of Tu‐YN larvae/LC50 of Tc‐YN larvae.
The logarithm concentration‐probit regression line passed the chi‐square test. Chi‐squared distribution was used to test whether the regression between logarithm concentration and probit pass the goodness of fit, if passed (the calculated chi‐square values < expected χ2 (df)0.05, goodness‐of‐fit chi‐square is significant), meaning the current data represent the fact.
3.5. The effect of acaricides on mixed species
Mixed populations of the two mite species were generated in the laboratory for simulating the effect of acaricides on the population composition in the field. In the control group (water treatment), the ratio of Tu‐YN to Tc‐YN was 1 (5:5) at the beginning of the mixed population (origination) and reduced to 0.89 after 15 days; however, it dramatically declined to 0.46 two months following origination (Table 6). The individuals of Tu‐YN in mixed populations were, respectively, 0.95‐ and 0.90‐fold of Tc‐YN in the abamectin and cyflumetofen treatment groups at the moment of pre‐exposure (15 days from origination); however, the species composition of the mixed population reversed after acaricide exposure; that is, the individuals of Tu‐YN were 4.44 and 1.85 times more abundant than Tc‐YN individuals in the mixed population following three applications of abamectin or cyflumetofen treatment, respectively (Table 6). The results of the simulation experiment revealed that in the absence of acaricide sprays, Tc‐YN would outcompete Tu‐YN; however, following the application of acaricides the population composition changed and Tu‐YN subsequently became the dominant mite species instead, even though all other experimental conditions remained unchanged.
Table 6.
The effect of abamectin and cyflumetofen applications on the population structure of Tetranychus cinnabarinus and Tetranychus urticae
| Samples | The number of female adult (Tc‐YN vs. Tu‐YN) | ||
|---|---|---|---|
| Control (water treatment) | Acaricide treatment | ||
| Abamectin treatment | Cyflumetofen treatment | ||
| Origination | 5 vs. 5a (1:1)b | 5 vs. 5 (1:1) | 5 vs. 5 (1:1) |
| Pre‐exposure (15 days from origination) | 118.0 vs. 105 (1:0.89) | 102 vs. 97.3 (1:0.95) | 105.3 vs. 95.0 (1:0.90) |
| Postexposure (3 times)(60 days from origination) | 525.0 vs. 235.0c (1:0.46) | 35.3 vs. 146.0c (1:4.44) | 66.6 vs. 123.0c (1:1.85) |
The number of female adults of each species in the mixed population (the number of Tc‐YN vs. the number of Tu‐YN). Numbers represent the mean of three repetitions.
The numbers in parentheses indicate the proportion of female adults from each species in the mixed population, normalized to Tc‐YN = 1.
Significantly different numbers of adult females for the two species in the mixed population.
3.6. The activities of detoxification enzymes (CarE, GST, and P450) in Tc‐YN and Tu‐YN
Compared with Tc‐YN, Tu‐YN has higher activities of detoxifying enzymes with the exception of GSTs. In Tu‐YN, the P450 activity toward nitroanisole and the CarE activities were 1.81‐ and 2.80‐fold higher than those of Tc‐YN, respectively, whereas GST activity was not significantly different between the two species (Figure 3).
Figure 3.
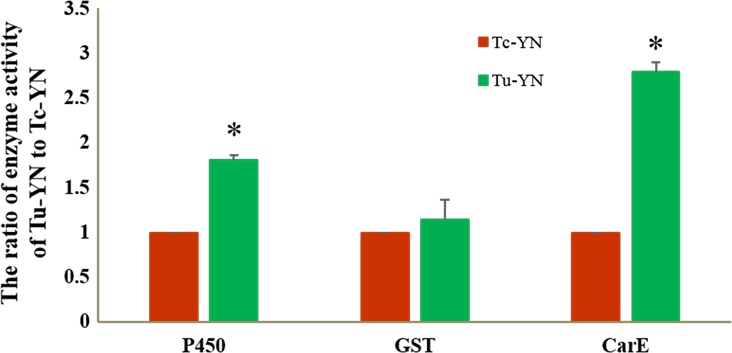
The ratio of enzyme activity (P450s, GSTs, CarE) of Tu‐YN to Tc‐YN. Error bars represent the standard error of the calculated mean ratios based on three biological replicates. Significant differences are indicated by an asterisk
3.7. Induction effects of acaricides on activities of detoxifying enzymes in Tu‐YN and Tc‐YN
After adult mites were treated with abamectin, fenpropathrin, and tebufenpyrad at the LC30 dosage, respectively, there were different induction effects on the activities of the main detoxification enzymes such as P450s, CarEs, and GSTs from the mites. Six hours following the abamectin treatment, the fold change of the activities of P450s, GSTs, and CarEs were 1.54‐, 1.53‐, and 1.23‐fold in Tu‐YN, and 1.13‐, 1.46‐, and 0.96‐fold in Tc‐YN, respectively (Figure 4a). In the fenpropathrin treatment group, the changes in increase in enzyme activity were 1.06‐, 1.99‐, and 1.22‐fold in Tc‐YN, and 1.32‐, 2.55‐, and 1.06‐fold in Tu‐YN, respectively (Figure 4b). Following tebufenpyrad exposure for 6 hr, the activity changes for P450s, GSTs, and CarEs in Tc‐YN were 1.04‐, 2.32‐, and 0.90‐fold, and 1.17‐, 1.61‐, and 1.07‐fold in Tu‐YN, respectively (Figure 4c). By and large, the responses of the three detoxification enzymes in Tu‐YN were stronger than those in Tc‐YN following the acaricide treatment; that is, the change in the activity of P450s in Tu‐YN was significantly greater than it was in Tc‐YN for all three treatment groups; furthermore, the increase in the activity of CarEs was higher for Tu‐YN than Tc‐YN for the abamectin‐treated group, and the increase in the activity of GSTs was greater in Tu‐YN than Tc‐YN for the fenpropathrin‐treated group, but lower for the tebufenpyrad‐treated group, and where the changes in enzyme activity showed no significant differences between Tc‐YN and Tu‐YN in other circumstances (Figure 4a–c).
Figure 4.
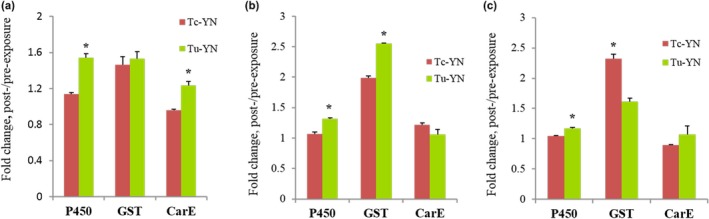
Induction of the specific activity of cytochrome P450s, glutathione S‐transferases (GSTs) and carboxylesterases (CarE) in Tc‐YN and Tu‐YN after treatment with various acaricides for 6 hr. (a) Abamectin treatment. (b) Fenpropathrin treatment. (c) Tebufenpyrad treatment. Asterisks on the error bars represent significant differences of the fold change between Tc‐YN and Tu‐YN within the same acaricide treatment
3.8. The results of RNA‐Seq for Tu‐YN and Tc‐YN
To measure the absolute mRNA expression levels of Tc‐YN and Tu‐YN and to identify transcripts that are differentially expressed under exposure to different acaricides, the gene expression profiles were analyzed using the DGE approach. The samples used for constructing the RNA‐Seq libraries (using the Ion Torrent Proton platform) consisted of three independent samples of both Tc‐YN and Tu‐YN, following exposure to either abamectin, fenpropathrin, or tebufenpyrad, as well as control nonexposed samples. The experiment was performed with three independently collected biological replicates, resulting in a total of 24 RNA‐Seq libraries for sequencing. After filtering low‐quality reads, the total numbers (mean) of clean reads of pre‐exposure, abamectin‐, fenpropathrin‐, and tebufenpyrad‐exposed mites were 13.7, 13.0, 13.1, and 13.8 million in Tc‐YN, respectively (Table S1). The total numbers (mean) of clean reads were 18.2, 16.5, 17.7, and 15.6 million in Tu‐YN, respectively (Table S1). For each sample, over 97% of total clean reads were successfully mapped onto the T. urticae genome without mismatch for further analysis (Table S1). Tetranychus cinnabarinus and Tetranychus urticae are two closely related species, which facilitates us taking the advantage of the sequenced T. urticae genome and using it as a reference.
For interlibrary comparison, read numbers were normalized to relative abundance as reads per kilobase transcriptome per million mapped reads (RPKM). The RPKM value (mean value of three biological replicates) of each gene was further used to compute the related coefficients between each sample. The expression correlations of genes showed a better accordance between the same species under different acaricide exposures, whereas the correlations were markedly lower between Tc‐YN and Tu‐YN, both with and without exposure to an acaricide (Figure S3). All raw and processed data have been deposited in the NCBI Gene Expression Omnibus under accession number GSE75529.
3.9. Functional annotation of differentially expressed genes (DEG) by GO, COG, and KEGG
Compared with Tc‐YN, a total of 974 DEGs were detected in Tu‐YN, with 538 upregulated and 436 downregulated genes (Figure 5). The DEGs were identified by BLASTx against the NCBI nonredundant protein database (Nr) and the T. urticae genome with a cutoff E‐value of 10−5. We also performed deep analysis based on DEGs, including Gene Ontology (GO) enrichment analysis, pathway enrichment analysis, and cluster analysis. Of the 974 DEGs between Tu‐YN and Tc‐YN, 258 DEGs were annotated into 950 GO terms; some of these DEGs participated in multiple GO terms. They were divided into three categories and 50 subcategories (Figure S4) including: biological process (646 GO terms, 22 subcategories), cellular component (155 GO terms, 15 subcategories), and molecular function (149 GO terms, 13 subcategories) (Figure S4).
Figure 5.
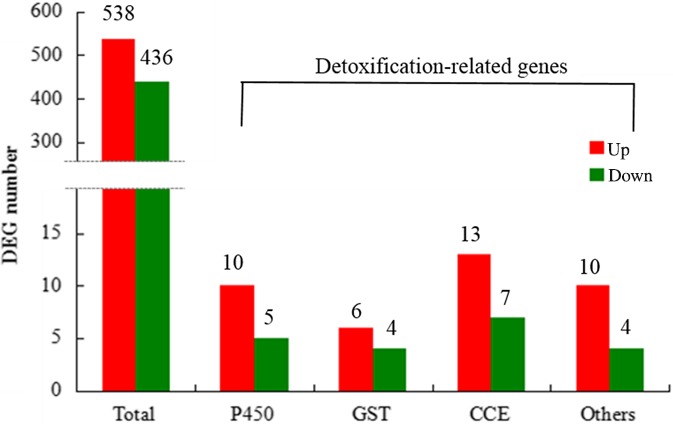
Summary of differently expressed genes (DEGs) in each pairwise comparison in Tu‐YN compared with Tc‐YN. The category “others” included SDR, Sialin, ID‐RCD, and ABC‐transporter genes
All DEGs were aligned to the clusters of orthologous groups (COG) database for functional prediction and classification. A total of 2,300 COG annotations were identified for 253 annotated DEGs, which were classified into 24 molecular families (Figure S5). Thus, some of these DEGs were associated with multiple COG annotations. Among these functional classes, the “general function prediction only” cluster constituted the largest group (131; 51.78%), followed by “carbohydrate transport and metabolism” (97; 38.34%), and “intracellular trafficking, secretion, and vesicular transport” (77; 30.44%).
To identify the metabolic pathways populated by these DEGs, all DEGs were mapped to the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. A total of 587 DEGs were annotated to 198 KEGG pathways (p‐value≤.05). The pathways with the most DEGs were “metabolic pathways” (116; 19.76%), followed by “lysosome” (47; 8.01%) and “retinol metabolism” (37; 6.3%). The pathway analysis showed that over 60% of DEGs are closely linked to the metabolism of xenobiotics and endogenous compounds (Table S2).
3.10. Focus on DEGs involved in insecticide detoxification
The DEGs relating to insecticide resistance generally encode detoxification enzymes. Generally, P450s, CCEs, and GSTs are the three primary enzymes involved in the detoxification of insecticides. Compared with Tc‐YN, the gene expression profiles of P450 (CYPs) genes revealed that 15 CYP genes showed significant transcription level variations (10 genes upregulated and five genes downregulated) in Tu‐YN (Figure 5). These differentially expressed CYP genes were distributed among all CYP clans (clan 2, 3, 4, and M). The majority of the upregulated CYP genes (6 of 10) were represented by clan 2, including tetur03 g09961 and tetur23 g00260, which both had an over 10‐fold increase in gene expression (Figure 9, Table S3). Three upregulated CYP genes belonged to clan 4, and another one was grouped to clan 3 (Table S3). comparing Tc‐YN with Tu‐YN, there were six upregulated and four downregulated GST genes, with three of the upregulated GST genes belonging to class mu and three of these genes belonging to class delta (Figure 5, Table S3). In this study, we found that 13 and 7 CCE genes were significantly up‐ and downregulated, respectively, in Tu‐YN vs. Tc‐YN (Figure 5, Table S3).
Figure 9.
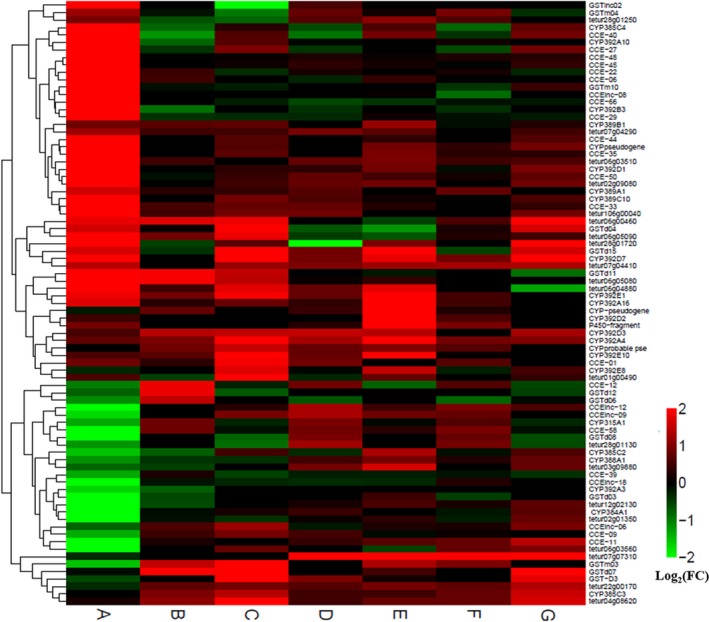
Clustering of metabolic enzyme genes differentially expressed across strains. Hierarchical clustering analysis‐based transcription levels was performed on 81 enzyme‐encoding genes showing significant differential transcription (Fisher's test p value <.001) in Tu‐YN compared with Tc‐YN for samples both exposed to acaricides and compared to their respective controls. Clustering was obtained using Euclidean distance calculated from all Log2 (fold changes) and complete linkage algorithm. Color scale from green to red indicates Log2 (fold changes) from ‐2 (fourfold under transcription) to 2 (fourfold over transcription). For each gene, either the gene ID from the T. urticae genome or the functional annotation of the gene is indicated. (a) Tu‐YN vs. Tc‐YN, (b) Tc‐YN exposed to abamectin vs. Tc‐YN, (c) Tu‐YN exposed to abamectin vs. Tu‐YN, (d) Tc‐YN exposed to fenpropathrin vs. Tc‐YN, (e) Tu‐YN exposed to fenpropathrin vs. Tu‐YN, (f) Tc‐YN exposed to tebufenpyrad vs. Tc‐YN, (g) Tu‐YN exposed to tebufenpyrad vs. Tu‐YN. tetur06 g04880 tetur106 g00040 tetur06 g05080 tetur06 g05090 tetur28 g01720 tetur02 g01350 tetur28 g01130 tetur12 g02130 tetur07 g07310 tetur22 g00170 represent RCD gene; tetur01 g00490 tetur06 g00460 tetur04 g08620 tetur28 g01250 represent ID‐RCD; tetur02 g09080 represent sialin; tetur07 g04290 tetur07 g04410 tetur03 g09880 tetur06 g03560 tetur06 g03510 represent ABC transporters. FC: Fold Change
There were several other biotransformation enzymes potentially participating in the detoxification process, such as short‐chain dehydrogenase/reductase (SDR), sialin, ABC transporters, and intradiol ring‐cleavage dioxygenase (ID‐RCDs). The genes that encode for these enzymes were classified as “others” in this study to separate them from the P450s, GSTs, and CCEs. In Tu‐YN vs. Tc‐YN, 10 “others” genes (five SDR genes, one sialin gene, three ABC‐transporter genes and one ID‐RCDs gene) were upregulated, and four (three SDR genes and one ABC‐transporter gene) were downregulated, respectively (Figures 5 and 9, Table S3). In total, there were 39 and 20 genes involved in biodegradation or transport (including P450s, GSTs, CCEs, and “others”) that were up‐ and downregulated, respectively, in Tu‐YN compared with Tc‐YN. This showed that the number of specifically upregulated detoxification and transport genes in Tu‐YN was nearly twice that of Tc‐YN (Figures 5 and 9).
3.11. Comparative analysis of the response to acaricides between Tc‐YN and Tu‐YN using RNA‐Seq
To obtain a global view of transcriptome responses following treatment with acaricides, we analyzed variations in the gene expression profile between acaricide‐treated and untreated mites (as a negative control group) using digital gene expression (DGE), which is a high‐throughput tag sequencing (Tag‐seq) method used to identify up‐ and downregulated genes between two datasets. To identify differentially expressed genes following exposure to acaricides, differences in gene expression were analyzed by pairwise comparisons between mites with and without acaricide exposure. We found that 35 and 133 genes were significantly up‐ and downregulated, respectively, in the control‐ vs. abamectin‐exposed in Tc‐YN, while in Tu‐YN 89 and 179 genes were up‐ and downregulated following abamectin exposure (Figure 6a). Among these differentially expressed genes, 73 genes were coregulated (22 upregulated and 51 downregulated) in both species following abamectin exposure. Among the differentially expressed genes in the two comparison groups, one P450 gene, three GSTs, and three “others” genes were upregulated in both Tc‐YN and Tu‐YN; however, seven P450s, six GSTs, two CCEs, and six “others” genes were upregulated in Tu‐YN, whereas only one P450, five GSTs, one CCE, and three “others” genes were upregulated in Tc‐YN. (Figures 6b and 9, Table S4).
Figure 6.
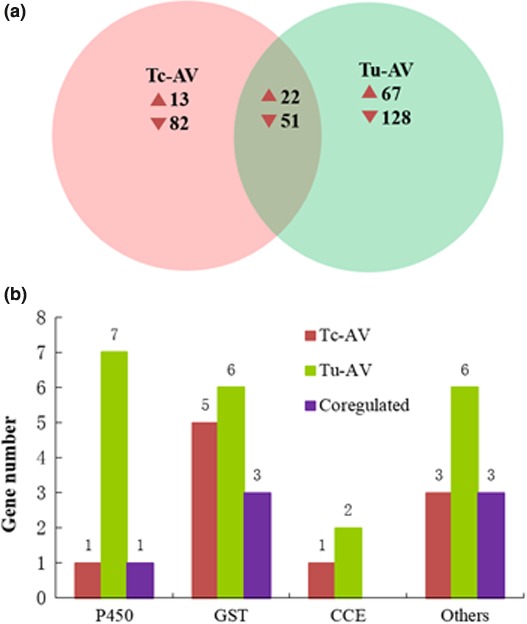
Summary of differentially expressed genes (DEGs) in each pairwise comparison following abamectin exposure. (a) Venn diagrams depicting overlap among DEGs of Tc‐YN and Tu‐YN exposed to abamectin, respectively. For each Venn diagram section, the numbers of transcripts differentially expressed in any strain with treatment as compared to control were indicated. (b) Distinctive and common differentially expressed (upregulated) genes of detoxification enzymes in the two previous comparisons. Tc‐AV: Tc‐YN were treated with abamectin; Tu‐AV: Tu‐YN were treated with abamectin. The category “others” included SDR, ID‐RCD, and ABC‐transporter genes
Following fenpropathrin exposure in Tc‐YN, 32 and 33 genes were significantly up‐ and downregulated in the treatment group compared with the unexposed group, while in the exposed Tu‐YN group there were 74 upregulated and 104 downregulated genes compared with the untreated Tu‐YN controls (Figure 7a). Among the differentially expressed genes in the two comparison groups, 11 P450s and five “others” genes were specifically upregulated in Tu‐YN (Figures 7b and 9), while only one P450 and one “others” gene were upregulated in Tc‐YN. Of the remaining detoxification enzyme categories, two GSTs genes and two CCE genes were specifically upregulated in Tu‐YN and Tc‐YN, respectively (Figure 7b, Table S5).
Figure 7.
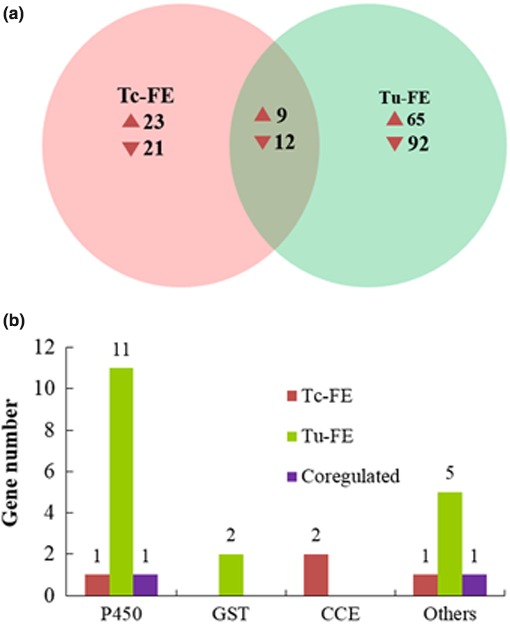
Summary of differentially expressed genes (DEGs) in each pairwise comparison following fenpropathrin exposure. (a) Venn diagram depicting the overlap among DEGs of Tc‐YN and Tu‐YN exposed to fenpropathrin. The numbers represent the differentially expressed transcripts for each species with treatment as compared to their respective control groups. (b) Distinctive and common differentially expressed (upregulated) genes for detoxification enzymes in the two previous comparisons. Tc‐FE: Tc‐YN were treated with fenpropathrin, and Tu‐FE, Tu‐YN were treated with fenpropathrin. The category “others” included SDR, ID‐RCD, and ABC‐transporter genes
Following tebufenpyrad exposure, there were 21 up‐ and 50 downregulated in Tc‐YN, whereas 59 genes were upregulated and 120 were downregulated in Tu‐YN (Figure 8a, Table S6). Of the upregulated genes in Tc‐YN, none were P450s, GSTs, or CCEs; however, three P450 and four GST genes were found to be upregulated in Tu‐YN (Figures 8b and 9). Furthermore, six “others” genes were specifically upregulated in tebufenpyrad‐treated Tu‐YN, while only one gene within the “others” category was upregulated in Tc‐YN (Figures 8b and 9).
Figure 8.
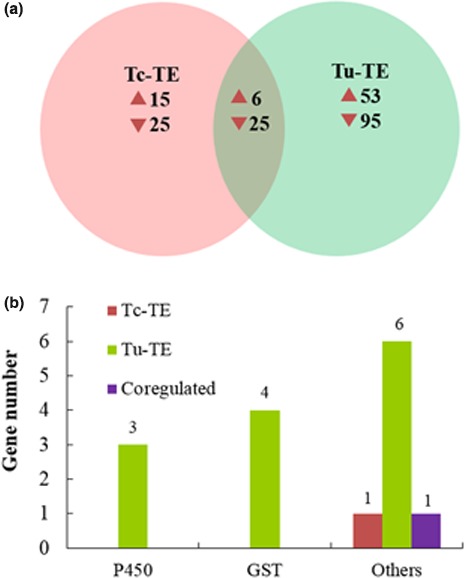
Summary of differentially expressed genes (DEGs) in each pairwise comparison following tebufenpyrad exposure. (a) Venn diagram depicting overlap among DEGs of Tc‐YN and Tu‐YN exposed to tebufenpyrad, respectively. The numbers represent the differentially expressed transcripts for each species with treatment as compared to their respective control groups. (b) Distinctive and common differentially expressed (upregulated) genes for detoxification enzymes in the two previous comparisons. Tc‐TE: Tc‐YN were treated with tebufenpyrad, Tu‐TE: Tu‐YN were treated with tebufenpyrad. The category “others” included SDR, ID‐RCD, and ABC‐transporter genes
Notably, a specific enrichment of genes belonging to certain metabolism pathways was observed in all DEGs, such as “metabolic pathways,” “metabolism of xenobiotics by cytochrome P450,” “drug metabolism—cytochrome P450,” “retinol metabolism,” “arachidonic acid metabolism,” “ascorbate and aldarate metabolism,” and “drug metabolism—other enzymes” (Tables S7–S9). The other notable phenomenon was that the DEGs that belonged to certain metabolism and sequestration pathways in Tu‐YN were much more abundant than those in Tc‐YN following acaricide exposure (Tables S7–S9).
3.12. Sequence alignment of target of acaricide genes in Tu‐YN and Tc‐YN
A total of 36 SNPs (nine nonsynonymous) were identified for the 14 acaricide‐target genes in Tu‐YN, while 32 SNPs (six nonsynonymous) were observed in Tc‐YN (Table 7). While multiple SNPs were identified for both mite species, none of the SNPs were located within the active site of the proteins encoded by these genes nor reported as a resistance‐related in previous studies (Ilias et al., 2014; Kwon et al., 2015). These results suggested that the identified SNPs are not correlated with acaricide resistance.
Table 7.
Identified SNPs of acaricide‐target gene sequences in Tu‐YN and Tc‐YN
| Target genea | Tu‐YN | Tc‐YN | ||||
|---|---|---|---|---|---|---|
| Nucleotideb | Amino acidc | Matchd | Nucleotide | Amino acid | Match | |
| VGSC | 11 | 1 | – | 10 | 1 | – |
| GluCl‐01 | 2 | 1 | – | 1 | 0 | – |
| GluCl‐02 | 1 | 1 | – | 1 | 1 | – |
| GluCl‐03 | 1 | 1 | – | 0 | 0 | – |
| GluCl‐04 | 2 | 0 | – | 3 | 1 | – |
| GluCl‐05 | 0 | 0 | – | 1 | 0 | – |
| Rdl1 | 3 | 2 | – | 2 | 1 | – |
| Rdl2 | 4 | 0 | – | 3 | 1 | – |
| Rdl3 | 1 | 0 | – | 1 | 0 | – |
| SdhA | 6 | 2 | – | 5 | 1 | – |
| SdhB | 1 | 0 | – | 2 | 0 | – |
| SdhC | 4 | 1 | – | 2 | 0 | – |
| SdhD | 0 | 0 | – | 1 | 0 | – |
| SdhE | 0 | 0 | – | 0 | 0 | – |
| Total | 36 | 9 | – | 32 | 6 | – |
VGSC: voltage‐gated sodium channel gene; GluCl‐01~GluCl‐05: 5 glutamate‐gated chloride channel genes; Rdl1–Rdl3: 3 GABA receptor genes; SdhA–SdhE: 5 succinate dehydrogenase complex genes.
Alignment between tested cDNA sequence and baseline at the level of nucleotide.
Alignment between tested cDNA sequence and baseline at the level of amino acid.
Identified point mutation from amino acid sequence was matched with the reported mutations of target genes involved in acaricide resistance in mites; “–” indicates not match.
4. Discussion
The differentiation between T. urticae and T. cinnabarinus is often difficult and controversial; however, in China, this is simplified in practice. That is, the green (female adult) with two black brown spots is named the two‐spotted spider mite and viewed as an invasive species, while the red (female adult) with two black brown spots is named the carmine spider mite and viewed as a native species (Kuang & Cheng, 1990; Sun et al., 2012). For more accuracy, the number of setae on tibia I and cross with T. cinnabarinus maintaining in laboratory were studied for identifying the species of collected field population in this work. According to Kuang and Cheng (Kuang & Cheng, 1990), the number of setae on tibia I for T. urticae population is held constant at 10, while in T. cinnabarinus the majority of individuals have 10 with a smaller number having 12 or 13. In this study, the proportion of individual with 10 setae on tibia I was 100% in the T. urticae field population, while it was only 70 and 66.7% in the T. cinnabarinus field and laboratory populations, respectively. In addition, the cross between the T. cinnabarinus and laboratory population could continuously produce F1, F2, and F3 females, whereas the cross between the T. urticae and laboratory populations could not produce normal female offspring. This demonstrated that reproduction isolation existed between T. urticae and the laboratory T. cinnabarinus population. Taken together, these results confirmed that the collected green field population was T. urticae and the red field population was T. cinnabarinus.
So far, studies investigating the complete distribution of T. urticae and T. cinnabarinus in China are absent because of the historical nature of these events, although the majority of researchers realize that it is increasingly more difficult to collect T. cinnabarinus in the field while it is becoming easier to collect T. urticae. An alternative method to determine the geographical range of T. urticae and T. cinnabarinus was adopted in our study in order to characterize the expansion of T. urticae from its first report in China. This method was to compile all of the literature ranging from 1975 to 2014 that reported on T. urticae and/or T. cinnabarinus. These reports were collected, and the sample collection place (province) was confirmed and mapped. From the distribution map of the two mites, it can be shown that T. urticae expanded from the first reported location, Beijing, to several of the Northern provinces from 1983 to 1999. T. urticae further expanded persistently and rapidly to the majority of China from 2000 to 2014, opposite to the range of T. cinnabarinus, which contracted its distribution from the end of the 20th century up until the present (Figure 2). This alternative approach to mapping the distribution of T. urticae and T. cinnabarinus in China cannot describe the true distribution changes in the two mite species in China as different sampling methods and research objectives could bias the results. However, the method is adequate to confirm that T. urticae has expanded successfully, as it has been reported from more collection locations than T. cinnibarinus, despite having originally invaded only in Beijing. We suggest that T. cinnabarinus is contracting its distribution based on two facts. First, the practice of in‐field observations from ourselves and peers found that the collection of T. urticae is presently much easier than that for T. cinnabarinus; second, a 5‐year‐long survey has demonstrated that T. urticae has supplanted T. cinnabarins as the dominant species of mites on vegetables in Beijing and Hebei provinces (Wang et al., 2013). All of the above‐provided indirect evidence that the range of the invasive T. urticae is expanding and outcompeting the native T. cinnabarinus in China.
In ecological theory, when one species show a competitive advantage against others in the same environmental conditions, the advantage could be attributable to the better ability of the more advantageous species to fit to the environment (Reitz & Trumble, 2002; Roush & Daly, 1990), also known as a fitness advantage. The environmental factors for arthropods includes temperature, food, pesticides, where these factors affect the competition and distribution of arthropods differently under different conditions. To date, no one has reported on the factors nor the mechanisms responsible for the expansion of T. urticae in China, prior to our study. However, we can find clues pointing to probable factors that facilitated the expansion of T. urticae based on existing studies, which imply that the application of acaricides may serve as the facilitator of the expansion of T. urticae, resulting in the displacement/replacement of T. cinnabarinus. Among these probable factors are host plant preference, temperature, and acaricide tolerance. First of all, the host plants could not be excluded as important factors. Tetranychus urticae and Tetranychus cinnabarinus are both polyphagous mites, capable of feeding and cofeeding on the same host (Bolland et al., 1998; Saito, 1979); however, the competitive capacity was found to vary between T. urticae and T. cinnabarinus when they fed on different host plants (Li & Cheng, 2011; Liu & Sun, 1998; Tomczyk, Kropczynska, & Elst, 1995). Theoretically, the distribution of host plants may be an extremely important factor in which one species has the overall advantage; nevertheless, we could not judge the role of host plant type on the distribution of these two species due to the lack of relevant data from investigation to date. In addition, temperature may be excluded as an impact factor as T. urticae is typically distributed in high latitude regions, which are characterized by lower temperatures, while T. cinnabarinus prefers regions of lower latitude (Dupont, 1979; Goka & Takafuji, 1991; Takafuji, So, & Tsuno, 1991). The higher fecundity of T. urticae at lower temperatures (16°C) and lower fecundity at higher temperatures (33°C) compared with T. cinnabarinus were also confirmed in our study (Table 3). We can imagine from their nature that T. cinnabarinus would benefit from global warming and enlarge its distribution wider than T. urticae; however, this is not true (Figure 2c). Over the past three decades, T. urticae gradually spread into the south of China; that is, T. urticae spread from high latitude regions of lower temperatures to low latitude regions of higher temperatures in China. Therefore, the factor of temperature cannot be considered crucial for the competitive capacity of T. urticae expansion to become the dominant spider mite in China.
Existing studies conformably revealed that T. urticae is less sensitive to most acaricides than T. cinnabarinus and that the difference in sensitivity to abamectin, an acaricide with extensive use in China, is especially significant (Bi et al., 2016; Gu et al., 2000; Zhao et al., 2006). Similar bioassay results were obtained in our study (Tables 4 and 5). These highly consistent results encourage us to make a bold inference that it is the higher tolerance against acaricides, in particular against abamectin, which is the major factor that has resulted in the expansion of T. urticae over T. cinnabarinus in China. That is, the application of acaricides, especially the extensive use of abamectin, facilitates the expansion of the geographical range of T. urticae in China. This inference is further strengthened with the fact that mite control relies mainly on the application of acaricides, with abamectin having been applied for nearly three decades as an essential component of insecticide and acaricide usage in China (Sun & Meng, 2009). The history of abamectin usage in China occurred in three stages: the first stage was from 1993 to 1998, when abamectin was sparingly applied only for economically important crops, as abamectin production was limited by fermentation technology; in the second stage, 1998–2005, abamectin was applied more broadly, benefiting from a lower cost of production, which resulted from improvements in fermentation and acaracide technologies; the third stage began in 2005, when a ban on the application of highly toxic pesticides was granted by the government, and abamectin use increased further and was more broadly applied to field crops and other plants of economic importance, as it was considered to be an environmentally friendly pesticide for the control of mites, insects, and nematodes. Most important of all, our study provides strong evidence that the application of acaricides biases the species composition of a mixed mite population toward T. urticae and that the positive effect of abamectin exposure on T. urticae population bias was the most significant factor (Table 6), albeit field conditions are more complex than those designed for in‐laboratory simulation experiments.
While the logical reasoning that the expansion of T. urticae in China may be attributable to the historical usage of acaricides, this is the first study to provide an in‐depth analysis to elucidate the relationship between acaricides and their effect on the population dynamics of T. urticae and T. cinnabarinus. Differential susceptibility to insecticides has been linked with changes in the demographics of other pest complexes. For example, the displacement of the B biotype of Bemisia tabaci (Gennadius) by the Q biotype in many regions where they have both invaded, has been attributed to the greater insecticide resistance of the Q biotype (Chu, Wan, Zhang, & Brown, 2010; Guo et al., 2014). This difference allows the Q biotype to overcome the competitive advantage that the B biotype has in the absence of insecticide pressures (Elbaz et al., 2012; Pascual & Callejas, 2004). Similarly, Gao, Lei, Abe, and Reitz (2011), Gao et al. (2012) suggested that differential susceptibility to commonly used insecticides could account for the replacement of Liriomyza sativae by Liriomyza trifolii on Hainan Island of southern China (Gao et al., 2011, 2012). However, the underpinning mechanisms behind the differential susceptibility to insecticides for these Bemisia spp. and Liriomyza spp. were not studied in depth or elucidated. It is reasonable for us to compare the activities of P450s, GSTs, and CCEs between T. urticae and T. cinnabarinus as they are the main enzymes functioning in the detoxification and metabolizing of exogenous chemicals, such as a variety of insecticides (Bass & Field, 2011; Li, Schuler, & Berenbaum, 2007; Qiu, 2005). Our results showed that the activity of GSTs did not differ significantly between T. urticae and T. cinnabarinus; however, the activities of P450s and CarEs were significantly higher (1.81‐ and 2.80‐fold, respectively) in T. urticae than in T. cinnabarinus. Furthermore, the change in enzyme activity showed a significant difference between T. cinnabarinus and T. urticae in six of nine cases of in‐laboratory acaricide‐stimulated experiments, in which T. urticae responded more strongly in five cases while T. cinnabarinus responded more positively in only one case (Figure 4). These results from the enzyme activity studies revealed from a biochemical perspective that T. urticae possesses a stronger capacity for the detoxification of acaricides than T. cinnabarinus.
At a molecular level, the transcriptome contains the complete repertoire of mRNAs transcribed by a living cell, that is, the sum of genetic information transcribed from the genomic DNA (Xu et al., 2014). Comparative expression profiling between T. cinnabarinus and T. urticae provides a priori information that can reveal the underlying mechanisms mediating the stronger acaricide tolerance of T. urticae over T. cinnabarinus, although the direct functions of DEGs requires further study. DGE analysis showed that there were 102 upregulated DEGs in T. urticae vs. T. cinnabarinus, with 39 of the upregulated genes encoding detoxification enzymes, while only 20 genes were as such in T. cinnabarinus (Figure 5). Following exposure to abamectin, fenpropathrin and tebufenpyrad, respectively, the amount of DEGs (up‐ and downregulated) in T. urticae were 1.59‐, 2.74‐, and 2.52‐fold greater than in T. cinnabarinus, with the upregulated DEGs in T. urticae being 2.54‐, 2.31‐, and 2.81‐fold greater than T. cinnabarinus. Among the upregulated genes in T. urticae, the detoxification enzyme groups of P450s, GSTs, and CCEs had 1.65‐, 3.33‐, and 7.00‐fold greater expression than in T. cinnabarinus, respectively (Figures 5, 6, 7). While not all DEGs from the acaricide‐stimulation RNA‐Seq datasets had available functional annotation, a specific enrichment of genes belonging to certain metabolism and sequestration pathways was identified for both T. cinnabarinus and T. urticae, where the abundance of upregulated detoxification or transportation genes was greater in T. urticae than T. cinnabarinus (Tables S7–S9). All of the above results strongly suggested that compared with T. cinnabarinus, T. urticae possesses a greater potential and ability to mitigate the stress from acaricides by possessing more upregulated genes (including metabolism‐related genes) at a basal gene expression level, and more genes that respond by intensively increasing their expression levels following acaricide exposure. This is the underlying molecular reason that T. urticae possess a stronger tolerance to acaricides than T. cinnabarinus. An important thing need to point out is the detoxifying genes switching them on more in the presence of three acaricides compared with no acaricide exposure, which may be explained that higher expression of detoxifying genes would come at a cost and it may only be beneficial to express these genes more when a pesticide is present.
In addition to our investigation of well‐known detoxification gene families involved in acaricide resistance, our study on target‐site modification revealed that the number of identified SNP differences within the acaricide‐target genes from the two field populations was not different too much. Furthermore, none of the identified SNPs were consistent with the reported mutations of target genes known to confer target‐site resistance against pesticides (Ilias et al., 2014; Kwon et al., 2015). The sequence alignment of target genes in T. urticae and in T. cinnabarinus implied that target‐site modification probably played a very minor role in conferring the higher acaricide tolerance in T. urticae over T. cinnabarinus.
In summary, this paper provides evidence that the activities and expression levels of detoxification enzymes were generally greater in T. urticae than in T. cinnabarinus. This enhanced metabolic detoxification may be an important reason why T. urticae is more resistant than T. cinnabarinus to acaricides. Furthermore, this greater resistance to acaricides may explain the phenomenon that acaricides facilitate the continuous expansion of T. urticae as the dominant spider mite in many locations in China, excluding other aspects as an essential factor. The population competition experiments further supported this hypothesis that the competitive displacement of T. cinnabarinus by T. urticae is mediated by human activities. Not only do our results reveal that T. urticae possesses stronger detoxification capacity than its sibling species T. cinnabarinus, which facilitated its persistent expansion in China, but they also point to the need to accurately identify Tetranychus species and to develop species‐specific management strategies for these pests.
Data Archival Location
All raw and processed data are available from the NCBI Gene Expression Omnibus (GEO) under project no. PRJNA304476 with accession number GSE75529.
Conflict of Interest
None declared.
Supporting information
Acknowledgments
This research was funded in part by the National Nature Sciences Foundation (31672085, 31470115), the Innovation Fund for Graduate Students of Chongqing (CYB16072), and the Fundamental Research Funds for the Central Universities (XDJK2016A005). We thank Dr. Bradley from University of California for constructive suggestions for polishing this work and Dr. Li Shi from Southwest University for preparation of figures.
Lu W, Wang M, Xu Z, et al. Adaptation of acaricide stress facilitates Tetranychus urticae expanding against Tetranychus cinnabarinus in China. Ecol Evol. 2017;7:1233–1249. doi: 10.1002/ece3.2724.
References
- Audic, S. , & Claverie, J. M. (1997). The significance of digital gene expression profiles. Genome Research, 7, 986–995. [DOI] [PubMed] [Google Scholar]
- Auger, P. , Migeon, A. , Ueckermann, E. A. , Tiedt, L. , & Navarro, M. N. (2013). Evidence for synonymy between Tetranychus urticae and Tetranychus cinnabarinus (Acari, Prostigmata, Tetranychidae): Review and new data. Acarologia, 53, 383–415. [Google Scholar]
- Bass, C. , & Field, L. M. (2011). Gene amplification and insecticide resistance. Pest Management Science, 67, 886–890. [DOI] [PubMed] [Google Scholar]
- Bi, J. L. , Niu, Z. M. , Yu, L. , & Toscano, N. C. (2016). Resistance status of the carmine spider mite, Tetranychus cinnabarinus and the twospotted spider mite, Tetranychus urticae to selected acaricides on strawberries. Insect Sci., 23, 88–93. [DOI] [PubMed] [Google Scholar]
- Bolland H. R., Gutierrez J., & Flechtmann C. H. (Eds.) (1998). World catalogue of the spider mite family (Acari: Tetranychidae). Leiden, the Netherlands: Brill Academic Publishing. [Google Scholar]
- Boudreaux, H. B. (1956). Revision of the two‐spotted spider mite (Acarina, Tetranychidae) complex, Tetranychus telarius (Linnaeus). Annals of the Entomological Society of America, 49, 43–48. [Google Scholar]
- Brandenburg, R. , & Kennedy, G. (1981). Differences in dorsal integumentary lobe densities between Tetranychus urticae Koch and Tetranychus cinnabarinus (Boisduval)(Acarina: Tetranychidae) from northeastern North Carolina. International Journal of Acarology, 7, 231–234. [Google Scholar]
- Cai, S. , Cheng, L. , & Sha, L. (2003). Changes of chlorophyll content in host plants infested with Two‐Spotted spider mite (Tetranychus urticae Koch). Chinese Journal Tropical Crop, 24, 54–57. [Google Scholar]
- Chhillar B., Gulati R., & Bhatnagar P. (Eds.) (2007). Agricultural acarology. New Delhi, India: Daya Books. [Google Scholar]
- Chu, D. , Wan, F. H. , Zhang, Y. J. , & Brown, J. K. (2010). Change in the biotype composition of Bemisia tabaci in Shandong Province of China from 2005 to 2008. Environmental Entomology, 39, 1028–1036. [DOI] [PubMed] [Google Scholar]
- Conesa, A. , Götz, S. , García‐Gómez, J. M. , Terol, J. , Talón, M. , & Robels, M. (2005). Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics, 21, 3674–3676. [DOI] [PubMed] [Google Scholar]
- Dong, H. , Guo, Y. , & Niu, L. (1987). Species identification of three common spider mites through cross breeding in China. Acta Phytophy Sin, 14, 157–161. [Google Scholar]
- Dupont, L. M. (1979). On gene flow between Tetranychus urticae Koch, 1836 and Tetranychus cinnabarinus (Boisduval) Boudreaux, 1956 (Acari: Tetranychidae): Sononomy between the two species. Entomologia Experimentalis et Applicata, 25, 297–303. [Google Scholar]
- Ehara, S. (1999). Revision of the spider mite family Tetranychidae of Japan (Acari, Prostigmata). Species Diversity: An International Journal for Taxonomy, Systematics, Speciation, Biogeography, and Life History Research of Animals, 4, 63–141. [Google Scholar]
- Elbaz, M. , Halon, E. , Malka, O. , Malitsky, S. , Blum, E. , Aharoni, A. , & Morin, S. (2012). Asymmetric adaptation to indolic and aliphatic glucosinolates in the B and Q sibling species of Bemisia tabaci (Hemiptera: Aleyrodidae). Molecular Ecology, 21, 4533–4546. [DOI] [PubMed] [Google Scholar]
- Gao, Y. , Lei, Z. , Abe, Y. , & Reitz, S. R. (2011). Species displacements are common to two invasive species of leafminer fly in China, Japan, and the United States. Journal of Economic Entomology, 104, 1771–1773. [DOI] [PubMed] [Google Scholar]
- Gao, Y. , Reitz, S. R. , Wei, Q. , Yu, W. , & Lei, Z. (2012). Insecticide‐mediated apparent displacement between two invasive species of leafminer fly. PLoS One, 7, e36622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goka, K. , & Takafuji, A. (1991). Genetical studies on the diapause of the two‐spotted spider mite Tetranychus urticae Koch (2). Applied Entomology and Zoology, 26, 77–84. [Google Scholar]
- Grbić, M. , Van Leeuwen, T. , Clark, R. M. , Rombauts, S. , Rouze, P. , Grbić, V. …, de Peer, Y. V. (2011). The genome of Tetranychus urticae reveals herbivorous pest adaptations. Nature, 479, 487–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groters, F. R. , Tabashnik, B. E. , & Finson, N. (1994). Fitness costs of resistance to Bacillus thuringiensis in the diamondback moth (Plutella xylostella). Evolution, 48, 197–201. [DOI] [PubMed] [Google Scholar]
- Gu, Y. , Zhang, Y. , Zhao, C. , & Li, S. (2000). Toxicity comparison on a few acaricides to three species spider mites. Journal of Laiyang Agricultural College, 17, 203–206. [Google Scholar]
- Guo, L. , Xie, W. , Wang, S. , Wu, Q. , Li, R. , & Wang, N. (2014). Detoxification enzymes of Bemisia tabaci B and Q: Biochemical characteristics and gene expression profiles. Pest Management Science, 70, 1588–1594. [DOI] [PubMed] [Google Scholar]
- Habig, W. H. , Pabst, M. J. , & Jakoby, W. B. (1974). Glutathione S‐transferases the first enzymatic step in mercapturic acid formation. Journal of Biological Chemistry, 249, 7130–7139. [PubMed] [Google Scholar]
- Ilias, A. , Vontas, J. , & Tsagkarakou, A. (2014). Global distribution and origin of target site insecticide resistance mutations in Tetranychus urticae . Insect Molecular Biology, 48, 17–28. [DOI] [PubMed] [Google Scholar]
- Jeppson, L. R. , Keifer, H. H. , & Baker, E.W. (Eds). (1975). Mites injurious to economic plants. Berkeley, CA: University of California Press. [Google Scholar]
- Knight, A. , Beers, E. , Hoyt, S. , & Riedl, H. (1990). Acaricide bioassays with spider mites (Acari: Tetranychidae) on pome fruits: Evaluation of methods and selection of discriminating concentrations for resistance monitoring. Journal of Economic Entomology, 83, 1752–1760. [Google Scholar]
- Kuang, H. , & Cheng, L. (1990). Studies on differentiation between two sibling species Tetranychus cinnabarinus and T. urticae . Acta Entomologica Sinica, 33, 109–115. [Google Scholar]
- Kwon, D. H. , Clark, J. M. , & Lee, S. H. (2015). Toxicodynamic mechanisms and monitoring of acaricide resistance in the two‐spotted spider mite. Pesticide Biochemistry and Physiology, 121, 97–101. [DOI] [PubMed] [Google Scholar]
- Li, T. , Chen, X. L. , & Hong, X. Y. (2009). Population genetic structure of Tetranychus urticae and its sibling species Tetranychus cinnabaribus (Acari: Tetranychidae) in China as inferred from microsatellite data. Annals of the Entomological Society of America, 102, 674–683. [Google Scholar]
- Li, Y. , & Cheng, L. (2011). Interspecies competition between Tetraychus urticae Koch and T. cinnabarinus Boisduval fed with cotton. Journal of Tropical Organisms, 2, 214–218. [Google Scholar]
- Li, T. W. , Gao, X. W. , Zheng, B. Z. , & Liang, P. (2000). Study on genetics of avermectins resistance and population fitness in Plutella xylostella . Acta Entomologica Sinica, 43, 255–263. [Google Scholar]
- Li, M. , Lu, W. C. , Feng, H. Z. , & He, L. (2009). Molecular characterization and expression of three heat shock protein70 genes from the carmine spider mite, Tetranychus cinnabarinus (Boisduval). Insect Molecular Biology, 18, 183–194. [DOI] [PubMed] [Google Scholar]
- Li, X. , Schuler, M. , & Berenbaum, M. (2007). Molecular mechanisms of metabolic resistance to synthetic and natural xenobiotics. Annual Review of Entomology, 52, 231–253. [DOI] [PubMed] [Google Scholar]
- Liu, T. , & Sun, J. J. (1998). Interspecies competition between Tetranychus urticae Koch and T. cinnabarinus (Boisduval). Plant Prot, 24, 6–10. [Google Scholar]
- de Mendonça, R. S. , Navia, D. , Diniz, I. R. , Auger, P. , & Navajas, M. (2011). A critical review on some closely related species of Tetranychus sensu stricto (Acari: Tetranychidae) in the public DNA sequences databases. Experimental and Applied Acarology, 55, 1–23. [DOI] [PubMed] [Google Scholar]
- Meng, H. S. , Wang, K. Y. , Jiang, X. Y. , & Yi, M. Q. (2001). Occurrence characteristics of Tetranychus urticae and its control methods. Entomological Knowledge, 38, 52–54. [Google Scholar]
- Metzker, M. L. (2010). Sequencing technologies‐the next generation. Nature Reviews Genetics, 11, 31–46. [DOI] [PubMed] [Google Scholar]
- Norton, R. A. (1998). Morphological evidence for the evolutionary origin of Astigmata (Acari: Acariformes). Experimental and Applied Acarology, 22, 559–594. [Google Scholar]
- Pascual, S. , & Callejas, C. (2004). Intra‐and interspecific competition between biotypes B and Q of Bemisia tabaci (Hemiptera: Aleyrodidae) from Spain. Bulletin of Entomological Research, 94, 369–375. [DOI] [PubMed] [Google Scholar]
- Qiu, X. (2005). Insecticide resistance: Genetics, genomics and implications for pest control. Acta Entomologica Sinica, 48, 960–967. [Google Scholar]
- Reitz, S. R. , & Trumble, J. T. (2002). Competitive displacement among insects and arachnids. Annual Review of Entomology, 47, 435–465. [DOI] [PubMed] [Google Scholar]
- Roush, R. , & Daly, J. (1990). The role of population genetics in resistance research and management In Roush R. & Daly J. (Eds.), Pesticide resistance in arthropods (pp. 97–152). New York, USA: Springer. [Google Scholar]
- Saito, Y. (1979). Comparative studies on life histories of three species of spider mites (Acarina: Tetranychidae). Applied Entomology and Zoology, 14, 83–94. [Google Scholar]
- Shang, C. C. , & Soderlund, D. M. (1984). Monooxygenase activity of tobacco budworm (Heliothis virescens F.) larvae: Tissue distribution and optimal assay conditions for the gut activity. Comparative Biochemistry and Physiology Part B: Comparative Biochemistry, 79, 407–411. [Google Scholar]
- Shen, G. M. , Shi, L. , Xu, Z. F. , & He, L. (2014). Inducible Expression of Mu‐Class Glutathione S‐Transferases Is Associated with Fenpropathrin Resistance in Tetranychus cinnabarinus . International Journal of Molecular Sciences, 15, 22626–22641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strode, C. , Wondji, C. S. , David, J. P. , Hawkes, N. J. , Lumjuan, N. , Nelson, D. R. , Ranson, H. , et al. (2008). Genomic analysis of detoxification genes in the mosquito Aedes aegypti . Insect Biochemistry and Molecular Biology, 38, 113–123. [DOI] [PubMed] [Google Scholar]
- Stumpf, N. , & Nauen, R. (2002). Biochemical markers linked to abamectin resistance in Tetranychus urticae (Acari: Tetranychidae). Pesticide Biochemistry and Physiology, 72, 111–121. [Google Scholar]
- Sun, J. T. , Lian, C. , Navajas, M. , & Hong, X. Y. (2012). Microsatellites reveal a strong subdivision of genetic structure in Chinese populations of the mite Tetranychus urticae Koch (Acari: Tetranychidae). BMC Genetics, 13, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, J. , & Meng, S. (2009). Status and development trend of avermectin in Chinese market. Chinese World Pesticides, 31, 18–22. [Google Scholar]
- Takafuji, A. , So, P. M. , & Tsuno, N. (1991). Inter‐and intra‐population variations in diapause attribute of the two‐spotted spider mite, Tetranychus urticae Koch, in Japan. Researches on Population Ecology, 33, 331–344. [Google Scholar]
- Tomczyk, A. , Kropczynska, D. , & Elst, I. (1995). Studies on competitive displacement of Tetranychus urticae by Tetranychus cinnabarinus on cucumber and gerbera In Tomczyk A. & Kropczynska D. (Eds.), The Acari. Physiological and ecological aspects of Acari‐host relationships (pp. 305–314). Warsaw, Poland: Dabor. [Google Scholar]
- Van Asperen, K. (1962). A study of housefly esterases by means of a sensitive colorimetric method. Journal of Insect Physiology, 8, 401–416. [Google Scholar]
- Van Leeuwen, T. , Dermauw, W. , Grbic, M. , Tirry, L. , & Feyereisen, R. (2013). Spider mite control and resistance management: Does a genome help? Pest Management Science, 69, 156–159. [DOI] [PubMed] [Google Scholar]
- Van Leeuwen, T. , Stillatus, V. , & Tirry, L. (2004). Genetic analysis and cross‐resistance spectrum of a laboratory‐selected chlorfenapyr resistant strain of two‐spotted spider mite (Acari: Tetranychidae). Experimental and Applied Acarology, 32, 249–261. [DOI] [PubMed] [Google Scholar]
- Van Leeuwen, T. , Vontas, J. , Tsagkarakou, A. , Dermauw, W. , & Tirry, L. (2010). Acaricide resistance mechanisms in the two‐spotted spider mite Tetranychus urticae and other important Acari: A review. Insect Biochemistry and Molecular Biology, 40, 563–572. [DOI] [PubMed] [Google Scholar]
- Wang, S. L. , Zhang, Y. J. , Wu, Q. J. , Xie, W. , & Xu, B. Y. (2013). Dominant species identification of spider mites on vegetables in some areas in Beijing and Hebei. Journal of Environmental Entomology, 36, 481–486. In Chinese with English abstract. [Google Scholar]
- Xu, Z. , Zhu, W. , Liu, Y. , Liu, X. , Chen, Q. , Peng, M. , & He, L. (2014). Analysis of insecticide resistance‐related genes of the Carmine spider mite Tetranychus cinnabarinus based on a de novo assembled transcriptome. PLoS One, 9, e94779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You‐Nian, W. , Chun‐Ya, B. , Yong‐Sheng, J. , Jian‐jun, R. , Hui‐li, G. , & Lei, Z. (2010). Acaricidal activities of Wikstroemia chamaedaphne extracts against Tetranychus urticae and Tetranychus cinnabarinus (Acari: Tetranychidae) In Bioinformatics and Biomedical Engineering (iCBBE). 2010 4th International Conference, Chengdu, China: pp. 1–5. [Google Scholar]
- Zhang, Z. Q. , & Jacobson, R. J. (2000). Using adult female morphological characters for differentiating Tetrancyhus urticae complex (Acari: Tetranychidae) from greenhouse tomato crops in UK. Systematic and Applied Acarology, 5, 69–76. [Google Scholar]
- Zhao, Y. , Zhou, Y. , & Ren, J. (2006). Comparing susceptibility of Tetranychus urticae and Tetranychus cinnabarinus to common miticides. Agrochemicals, 45, 418–419. [Google Scholar]
- Zhou, X. , Ren, L. , Meng, Q. , Li, Y. , Yu, Y. , & Yu, J. (2010). The next‐generation sequencing technology and application. Protein & Cell, 1, 520–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


