Abstract
Burkholderia pseudomallei (Bp) and Burkholderia mallei (Bm), the agents of melioidosis and glanders, respectively, are Tier 1 biothreats. They infect humans and animals, causing disease ranging from acute and fatal to protracted and chronic. Chronic infections are especially challenging to treat, and the identification of in vitro phenotypic markers which signal progression from acute to persistent infection would be extremely valuable. First, a phenotyping strategy was developed employing colony morphotyping, chemical sensitivity testing, macrophage infection, and lipopolysaccharide fingerprint analyses to distinguish Burkholderia strains. Then mouse spleen isolates collected 3–180 days after infection were characterized phenotypically. Isolates from long-term infections often exhibited increased colony morphology differences and altered patterns of antimicrobial sensitivity and macrophage infection. Some of the Bp and Bm persistent infection isolates clearly displayed enhanced virulence in mice. Future studies will evaluate the potential role and significance of these phenotypic markers in signaling the establishment of a chronic infection.
Electronic supplementary material
The online version of this article (doi:10.1007/s00203-016-1303-8) contains supplementary material, which is available to authorized users.
Keywords: Burkholderia, Glanders, Melioidosis, Mouse, Spleen isolates
Introduction
Burkholderia pseudomallei (Bp) and Burkholderia mallei (Bm), the etiologic agents of melioidosis and glanders, respectively, are classified as Tier 1 bacterial select agents. Bm is an obligate animal pathogen which causes a debilitating and often fatal zoonotic disease of equines. It has been eradicated in most countries, but is still found in parts of Africa, the Middle East, Eastern Europe, Asia, and South America. In contrast, Bp is a saprophytic, free-living organism which causes endemic infections in tropical regions such as Southeast Asia and Northern Australia. Both agents can infect humans and animals by several routes. Infections occur upon exposure to contaminated water, soil, or secretions, and through skin abrasions, inhalation, or ingestion. The diseases are manifested by protean, often nonspecific generalized symptoms such as fever and malaise, ulcerating lesions of the skin and mucus membranes, pneumonia, granulomatous abscesses in multiple organs, and septicemia. Without effective treatment, the course of these diseases may range from acute and rapidly fatal to a very protracted and frequently chronic form, as described below (Dance 2014; Gregory and Waag 2008; Welkos et al. 2015; Wiersinga et al. 2012).
These pathogens are global concerns for several reasons including the wide environmental range of Bp, the challenges posed in diagnosing the diseases and identifying the agents, treatment complications due to inherent and acquired antibiotic resistance, and their potential for adversarial use (Currie et al. 2000, 2010; Dance 2014; Dance et al. 1991; Fritz and Waag 2012; Naha et al. 2012, 2014; Vidyalakshmi et al. 2008; Wiersinga et al. 2012). In addition, these pathogens are potential biothreat agents because of their high aerosol infectivity and ability to cause severe disease with often nonspecific symptoms (Fritz and Waag 2012; Wiersinga et al. 2012).
These highly pathogenic Burkholderia species are capable of eliciting a wide range of infection states, i.e., acute, chronic, recurrent and latent (Currie et al. 2000; Nigg 1963; Tarlow and Lloyd 1971). Acute infection can manifest as a severe fulminant disease with overwhelming septicemia and pneumonia. Chronic forms are characterized as persistent infections which recrudesce clinically at varying intervals. They are characterized by signs and symptoms similar to, but milder than, those of the acute disease. These chronic infections are commonly associated with immunocompromising conditions, e.g., diabetes, and can persist for years. Reoccurring illness is also observed and can potentially be due to reinfection or relapse of a persistent infection. Latent infections are asymptomatic and can remain subclinical or alternately progress to acute melioidosis up to decades after the initial exposure. All of these forms, especially the more persistent ones, can be very challenging to diagnose and treat effectively (Dance 2014; Fritz and Waag 2012; Wiersinga et al. 2012).
To prevent the conversion of an active primary infection into a chronic or subclinical form, it is necessary to identify bacterial and host phenotypic changes which might mark infection transition to a long-term persistent state. The purpose of this study was to develop a scheme to identify in vitro bacterial phenotypes potentially associated with persistent infection and determine its capacity to distinguish isolates which persist in chronically or subclinically infected mice. These objectives are based on the hypothesis that the transition of an infection from an acute to persistent, long-term form involves adaptive changes in bacteria facilitating their resistance to antibacterial host responses. This hypothesis is supported by numerous studies on Bp and other Gram-negative bacteria and their ability to adapt to, or persist in, harsh environments and stressful host conditions. Previous studies documented resistance to extended periods of anaerobiosis, pH extremes, phagolysosomal contents such as antimicrobial peptides, antibiotics, and other stressors (Butt et al. 2014; Chen et al. 2014; Fauvart et al. 2011; Goodyear et al. 2012; Hamad et al. 2011; Hayden et al. 2012; Keren et al. 2004; Kint et al. 2012; Price et al. 2013; Pumpuang et al. 2011; Romero et al. 2006) and attempted to identify reliable in vitro markers for long-term colonization or chronic infection (Chantratita et al. 2007, 2012; Chen et al. 2014; Hayden et al. 2012; Tandhavanant et al. 2010; Velapatino et al. 2012). Phenotypic switching, as manifested typically by colony morphology variation, is a well-known phenomenon in the highly pathogenic Burkholderia. In 1924, Stanton and Fletcher first reported the presence of two colony morphotypes, a rough and mucoid form, isolated from a patient infected with Bp (previously Bacillus whitmori) (Stanton et al. 1924). Nicholls and Cantab (1930) isolated and characterized similar variants of B. whitmori from abscess material from an infected cow. The morphotypes were reversible, with the rough being more abundant and stable than the mucoid type. The colony types were associated with in vitro phenotypic differences such as production of alkaline conditions in broth, and evidence suggesting differences in virulence was described. Numerous more recent studies have supported the hypothesis that different colony morphotypes potentially reflect adaptive changes that enhance fitness in a particular environment (Austin et al. 2015; Chantratita et al. 2007; Rogul and Carr 1972; Tandhavanant et al. 2010; Velapatino et al. 2012; Wikraiphat et al. 2015; Shea, A., unpublished).
In this study, a panel of phenotyping tests for differentiating splenic mouse isolates of Bp and Bm was developed in efforts to identify markers for distinguishing isolates responsible for persistent infection. The tests included colony morphology differentiation, chemical and peptide sensitivity, macrophage infection and cytotoxicity, and LPS characterization. The goal was to identify markers which could then be further investigated to determine how Burkholderia transitions from an acute to chronic infection.
Materials and methods
Bacterial strains, media, and chemicals
The strains of Bp and Bm and their sources are described in Supplementary Table 1. Bm strains Turkey 1 and 10229 and Bp strains 316c and 4845 were obtained from the USAMRIID Therapeutics Core (TC) group. The Bm strain used as the prototype was Bm FMH, a relatively recent human clinical isolate derived from strain Bm ATCC 23344/China 7. This strain, Bp prototype strains K96243 and 1026b, and the remaining seven Bp strains described in the table were obtained from the USAMRIID Unified Culture Collection (UCC). All the Burkholderia strains were human clinical isolates with the exception of Bp 4845 and possibly Bm Turkey 1. Two of the three Bm strains were virulent in the hamster model of glanders (D. DeShazer, unpublished data). Additional stocks of Bm strain ATCC 23344/China 7 and Bp strain K96243 were obtained from fellow investigators [gifts from D. Waag (DW) and D. DeShazer (DD)] and used where indicated in the text. Plated media were available commercially (Thermo Fisher-Remel, Lenexa, KS) as described below, or prepared in-house. The differential/nonselective media included sheep blood agar (SBA) plates and glycerol tryptone agar (GTA). The four differential/selective media used were: OFPBL (oxidation–fermentation base polymyxin B–bacitracin–lactose) agar plates; PC/BCA (Pseudomonas/Burkholderia cepacia agar) plates with polymyxin B, ticarcillin, and dye to detect alkaline pyruvate metabolism; BCSA (Burkholderia cepacia selective agar) plates with polymyxin B, gentamicin, vancomycin, sucrose, and lactose with dye to detect acid production (for Bp); and Ashdown’s agar (AA) plates containing dyes and gentamicin (for Bp) or no antibiotic (for Bm). All were available commercially (Thermo Fisher-Remel) except GTA and AA plates which were manually prepared (Ashdown 1979; Fritz et al. 2000). Liquid growth media were glycerol tryptone broth (GTB) (Fritz et al. 2000) and cation-adjusted Mueller–Hinton broth (MHB) (BBL™, BD Diagnostics Franklin Lake, NJ). Chemicals were obtained from Sigma-Aldrich (St. Louis, MO), and antimicrobial peptides were acquired from the following sources: Sigma/Fluka, Bachem (Torrance, CA), Biopeptek (Malvern, PA), Synthetic Biomolecules (San Diego, CA), and Peptides International (Louisville, KY).
Chemical sensitivity and enzyme production
Three platforms are used for sensitivity testing and enzyme detection: (1) GEN III MicroPlates™ (Biolog, Inc., Hayward, CA) which include 23 antimicrobial chemicals for screening sensitivity/growth inhibition; (2) agar plate assays for assessing sensitivity to hydrogen peroxide (representative of reactive oxygen species) and for detecting protease production using skim milk agar plates, as described (Sokol et al. 1979); and (3) microtiter tests to evaluate sensitivity to selected chemicals and antimicrobial peptides (AMPs) in which growth or inhibition was determined by measuring optical density (OD630).
The GEN III plates were inoculated as described by the manufacturer. After incubation for 48 h (Bp) or 72 h (Bm), growth was detected by measuring OD630. The data were scored in accordance with the Biolog instructions: Wells with growth similar to the plate positive control well were scored resistant (R), well readings less than half the positive control were sensitive (S), and readings which were borderline (±0.1 OD630 units below or above the R or S cutoffs) or variable were scored as R/S.
Chemicals selected for testing in the microtiter assays included NaCl (1, 4, and 8 %), nalidixic acid (5 and 50 µg/mL), surfactant Niaproof 4 (0.027 and 0.10 %), reactive oxygen species (ROS) inducer paraquat dichloride (2.5 µM), and reactive N2 intermediate (RNI) sodium nitrite (2 mM). The Bm and Bp strains were differentially responsive to the selected concentrations in preliminary tests; both were sensitive to 2 mM sodium nitrite as shown previously (Tandhavanant et al. 2010); and Bp strains were generally sensitive to 2.5 µM paraquat similarly to Escherichia coli (Cho et al. 2012). Twelve antimicrobial peptides (AMPs) were screened: cecropin A and P1, mastoparan 7, LL-37, magainin, histatin 5, melittin, HNP-1, hBD-2, BMAP-18, bactenecin, and CA-MA (Fox et al. 2012; Kanthawong et al. 2009; Madhongsa et al. 2013; Tandhavanant et al. 2010). Four AMPs (cecropin A, mastoparan 7, magainin, and melittin) were downselected for testing with all strains since they exhibited greatest antibacterial activity for the Burkholderia. Some strains were tested with an additional four because of differential sensitivity (LL-37, BMAP-18, bactenecin, and CA-MA). Due to the generally high level of resistance, a single concentration of each AMP was tested (200 µM for cecropin A and magainin and 100 µM for the rest due to reduced solubility in liquid media MHB and GTB) (Kanthawong et al. 2009; Tandhavanant et al. 2010). E. coli strains ATCC 25922 and Bp K96243 were used in the assays to verify activity. After addition of the antimicrobials to the trays, the wells were inoculated with strains adjusted to a concentration of approximately 1 × 106 CFU/mL in MHB (Bp) or GTB (Bm). The trays were incubated and read as described for the Biolog trays, and the absorbance results were recorded as resistant (R, OD630 >75 % positive growth control); sensitive (S, OD630 <50 % positive control); borderline (R/S, OD630 >50 % and <75 % positive control); or very sensitive (S+, OD630 ≤2× the uninoculated negative control wells containing medium alone).
A separate assay for sensitivity to a combination of the enzymes lactoferrin and lysozyme (20 and 200 µg/mL, respectively), or each alone, was also conducted as described previously (Ellison and Giehl 1991; Tandhavanant et al. 2010). Microtiter plates containing PBS alone or PBS with lactoferrin, lysozyme, or lactoferrin plus lysozyme were inoculated with bacterial suspensions, and the plates were incubated for 48 h at 37 °C. The effect of the enzymes on growth was detected by measuring optical density (OD630) and by comparison of the densities of the treated wells to that of the untreated well (PBS alone).
Characterization of Burkholderia survival and cytotoxicity in macrophages
Phagocytosis assays were developed to measure the ability of Bp and Bm to infect macrophages and to induce cell damage. The ability of the strains to replicate inside macrophages was also determined by plating for viable counts at various time points post-phagocytosis. The procedure was similar to those described previously (Arjcharoen et al. 2007; Burtnick et al. 2011; Kespichayawattana et al. 2000; Mulye et al. 2014; Tandhavanant et al. 2010; Welkos et al. 2015). The J774.A1 murine-derived macrophage-like cell line was cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) with glucose, glutamine, and 10 % fetal bovine serum in 24-well plates (with or without coverslips). The cells were incubated for 2 days at 37 °C with 5 % CO2, at which time growth to 90–95 % confluence was achieved. On the day before the experiment, glycerol tryptone broth (GTB) was inoculated with growth from fresh SBA plate cultures of the bacteria and the flasks were incubated overnight at 37 °C with shaking at 200 rpm. Optical density measurements of the cultures were taken, suspensions adjusted to OD620 of 1.0 (~1×109/mL) were prepared, and the macrophages were inoculated with Burkholderia at a target multiplicity of infection (MOI) of 5–10 bacteria per macrophage for Bm and <5 or 20 bacteria per macrophage for Bp. The infected macrophages were then incubated at 37 °C for 1 h to allow bacterial phagocytosis. The cells were washed three times to remove unphagocytosed bacteria, and fresh medium was added which contained 250–500 µg/mL of kanamycin to inactivate any residual extracellular bacteria. The cells were incubated for an additional 2 h, washed to remove the kanamycin, and then either lysed to determine the extent of phagocytosis or refed with medium containing 20 µg/mL of kanamycin. The latter cultures were then incubated for a total of 7–8 h (Bp) or 20–22 h (Bm) to determine the level of replication and cytotoxicity. The incubation times were determined based on differences in growth and infection rate. As will be described below, the shorter incubation times for Bp facilitated detection of variations in the responses of different strains. Bp strains generally grew and killed the cells more rapidly than Bm causing excessive cell loss when incubated for the time used in the Bm assays. The lysed macrophage samples were diluted and plated on SBA plates to determine quantitative viable counts of the intracellular bacteria. To account for differences in strain MOIs, the viable CFU counts were also normalized to the inoculum and expressed as percentages. In separate wells, the extent of cell cytotoxicity was measured by trypan blue (TB) dye uptake and/or by staining with propidium iodide (PI). Live cells exclude TB and PI and are unstained under phase (TB) or fluorescence (PI) microscopy, whereas dead cells are permeable and have blue (TB) or bright red (PI)-stained nuclei. Cells on coverslips were alternately stained with Diff-Quik™ histologic stain to assess macrophage condition (normal versus necrotic, apoptotic, or multi-nucleated appearance of cells and nuclei), extent of formation of multi-nucleated giant cells (MNGCs), and the relative level of residual bacterial infection. It should be noted that the nuclei in advanced-stage MNGCs are often necrotic, poorly visible, and no longer stainable by PI or TB; however, they were detectable by Diff-Quik™ stain.
Based on studies with different strains, the following phenotypes were developed for the initial screening of strains and in vivo isolates of Burkholderia: (1) relative bacterial survival, as determined by viable plate counts of lysed cells harvested at 6–8 h (Bp) or 20–22 h (Bm) post-phagocytosis in comparison to the concentration of bacteria present in the inoculum; (2) cytotoxicity, as assessed by live–dead staining with TB and PI to estimate the proportion of dead cells and determine the percentage of cells lost compared to the uninfected monolayer in a low power field (100× final magnification), and by staining to detect morphologic changes (Diff-Quik™); and (3) MNGC, as determined by the relative proportion of MNGCs compared to normal cells and by the nuclear morphologies (% nuclei contained in MNGCs of the total nuclei present in a higher power field [600× final magnification]). For the microscopy data, cells in a minimum of six separate fields, two for each of three Diff-Quik-stained coverslips were counted to obtain the MNGC parameters and to compare the extent of cell loss; three fields in duplicate TB- or PI-stained wells were examined to determine the proportion of dead cells and estimate of percentage cell loss. Strains selected for further analysis as a result of the screening were evaluated in assays which measured bacterial adherence and extent of uptake, as well as the time course of intracellular viability. Infected macrophage samples were lysed and diluted/plated immediately after the 1-h phagocytosis period, at the 3-h time point (after 2-h incubation in antibiotic), and after incubation of infected cells for a total of 8 h (Bp) or 20–22 h (Bm).
LPS characterization
Differences in the gel banding profiles of LPS from different strains or spleen isolates of Bm and Bp were examined using silver-stained 10–20 % tricine gels and western blots probed with various Bm- and Bp-specific monoclonal and polyclonal antibodies, by methods described previously (Welkos et al. 2015). Silver staining was conducted using the method described by Tsai and Frasch (1982).
Animal challenges
BALB/c mice (female, 7–10 weeks of age at time of challenge) were obtained from the National Cancer Institute, NCI/Charles River (Frederick, MD), and used in groups of 10 for challenge by aerosol or intraperitoneal (IP) routes as part of LD50 determinations and prospective serial collection experiments. IP challenges were conducted with various doses of Bp or Bm grown in GTB, as described previously (Welkos et al. 2015). The bacteria were quantified via OD620 estimations and delivered IP in 200 µL of GTB. The delivered doses were then verified by plate counts on SBA. The animals were monitored for clinical signs and symptoms for 60 days, except where indicated below. Early endpoint euthanasia was uniformly employed to limit pain and distress (Welkos et al. 2015). Aerosol challenges were done by whole-body exposure to aerosols generated by nebulization of GTB-diluted overnight cultures in a modified Henderson apparatus, as described previously (Jeddeloh et al. 2003; Waag and DeShazer 2004).
Bacterial isolates were obtained from mice surviving challenge by the aerosol or IP routes; spleens collected from mice euthanized at various times after challenge were homogenized and cultured on SBA plates for recovery of viable Burkholderia, as described previously (Amemiya et al. 2015).
Research was conducted under an IACUC-approved protocol in compliance with the Animal Welfare Act and other federal statutes and regulations relating to animals and experiments involving animals and adheres to the principles stated in the Guide for the Care and Use of Laboratory Animals, National Research Council, 2011. The facility where this research was conducted is fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International.
Statistical analysis
Differences in the viable counts obtained from infected macrophages were determined by t test or, when more than two strains were compared, by ANOVA and Tukey multi-comparison post-tests. The mean OD630 values determined from replicate GEN III microtiter plate experiments were evaluated for sensitivity compared to the growth control by ANOVA and multi-comparison post-tests. These statistical analyses were done with GraphPad Prism versions 5.2 and 6.0. The day 21 and 60 survival data of mice challenged by IP route with Burkholderia were analyzed for each strain, and the data were evaluated statistically to compare parent strain and spleen derivative virulence potencies. Bayesian probit analysis was used to calculate median lethal dose (LD50) and 95 % credible interval estimates and to construct dose response curves. The statistical analyses were performed using Stan 2.1.0 and R.3.1.1 software (SD Team 2013, 2014; Welkos et al. 2015).
Results
Characterization of a phenotyping panel for Burkholderia strains
The development and evaluation of assays to be used for phenotyping animal isolates were conducted using strains that were designated as “prototypes.” For Bm, the chosen strain was Bm ATCC 23344/China 7 and a derivative isolated from a relatively recent case of human glanders, FMH (Srinivasan et al. 2001 and Supplementary Table 1). Strains K96243 and 1026b were designated as the Bp prototypes. These strains have all been described extensively in the literature, have been sequenced, and are considered to be representative of typical virulent strains of Burkholderia. As a result, they are frequently employed in animal models. First, the phenotypes of the prototypes were compared to that of other strains of the same species. Bm strains ATCC 23344/China 7, FMH, Turkey 1, and 10229 were screened in the initial phenotyping stages. Besides the Bp prototype strains, Bp strains 316c, 4845, Bp22, and MSHR5855 were also examined initially.
Colony morphotyping
To characterize colony morphology of Bp and Bm, six selective and/or differential media were used: SBA, GTA, AA, OFPBL, BCA, and BCSA. In addition, skim milk agar was included to assay for protease production. Because Bm strains are susceptible to gentamicin, a modified version of Ashdown’s medium lacking the antibiotic was used for Bm strains. The use of six media allowed strains to be differentiated morphologically in at least seven characteristics, i.e., colony size, color, moistness, opacity, circumference shape, surface texture, and changes in agar color (the extent of hemolysis activity on SBA plates or change in indicator color due to bacterial metabolic activity, e.g., acid production). Colony phenotyping for Bp strains is illustrated in Fig. 1. Whereas a variety of different randomly occurring phenotypes were readily distinguished among the Bp strains, the strains of Bm exhibited few morphological differences (data not shown). The variable production of different colony morphotypes often observed for Bp strains was especially apparent using SBA, GTA, and AA. The variant colonies demonstrated on the SBA plate (Fig. 1a) are those of Bp strain MSHR668; strain K96243 was used to illustrate growth by Bp on OFPBL, AA, BCA and BCSA selective media, respectively (Fig. 1b–e). Despite the random morphotypic heterogeneity often observed in Bp strains, the flat, rough colonies which appear in the majority on the AA plate (Fig. 1c) illustrated one of the morphotypes commonly observed on AA plates among the Bp strains studied here. Specifically, this morphotype was a relatively flat, rough, often dry, and striated colony type, which was observed for many strains, albeit in varying relative abundance. Similar morphotypes were among those described previously by Chantratita et al. (2007) in a set of seven morphotypes (I–VII). The colonies we observed in the Bp parent strains varied in color and central colony texture but resembled variants of Morphotype I (rough colony center) or IV (striated, with smooth umbonated center), and had a purple color ranging from light to dark, as described for Morphotypes I or IV and Morphotype II, respectively. The colonies exhibited differences between plating on AA in color, moisture, and surface texture, as illustrated by the 406e parent strain in Supplementary Table 5A and B. However, this flat, rough colony type and variants thereof may represent the major colony phenotype isolated from patients with melioidosis and the colony from which switching to other colony morphotypes occurred in vivo, as reported by Chantratita et al. (2007). In addition to AA plates, this morphotype was characterized by relatively flat, nonmucoid colonies on SBA plates, as described previously (Wikraiphat et al. 2015), and by acid production on OPFBL plates. A form similar to this flat, rough morphotype was produced in varying proportions by all of the six strains described below in the mouse isolate studies (K96243, 1026b, 1106a, 406e, MSHR305, and HBPUB10134a) and by Bp22, MSHR5855, 316c, and 4845.
Fig. 1.
Colony morphology of Bp strains on selective/differential media. a The growth of Bp strain MSHR668 on SBA plates illustrates the colony morphology variation that is commonly observed for Bp. On this plate, three colony types are shown: the majority of the colonies are small, round, and smooth (#1); the next most numerous are larger, shinier, and less opaque, and have a translucent edge (#2); and there are two “water splat” colonies that are large, flat, and translucent/clear, and have an irregular edge (#3). The growth of Bp strain K96243 illustrates morphologies produced by Bp on several selective media (b–e). b OFPBL plate. c AA plate showing a common colony morphology of Bp strains. d BCA plate. Strains metabolizing the peptones and pyruvate produce alkaline conditions changing the agar indicator color from orange to pink. e BCSA plate, used for Bp only. This medium distinguishes strains which metabolize the peptones (alkaline color change) or catabolize the carbohydrates (acidic color change from deep pink to yellow, as shown for K96243)
Chemical sensitivity testing and enzyme production
Chemical sensitivity tests in the Biolog GEN III panel were evaluated as a candidate phenotyping tool. Several strains of Bm and Bp were used to determine optimal incubation times, assess the range of responses, and establish the scoring. The responses of prototype strains Bm ATCC 23344/China 7 and Bp K96243 were compared, and the results indicated that Bp is potentially more resistant to potential environmental, intra-phagocyte, and antimicrobial stresses than Bm (Supplementary Table 2). Bm and Bp were similarly resistant to some antibiotics (rifampin, lincomycin, and vancomycin), and Bp was more resistant than Bm to other antibiotics (troleandomycin, nalidixic acid, and minocycline). Both species exhibited a similar sensitivity to NaCl concentrations (variable growth in 1 % but not 4 or 8 %) and were fully resistant to mild pH stress (pH 6). Bm was moderately resistant, while Bp was fully resistant to low pH stress (pH 5). Bp K96243 was more resistant to potassium tellurite and to the surfactant Niaproof 4 than Bm ATCC 23344/China 7.
Further studies were done with additional Burkholderia strains. Bm strain ATCC 23344/China 7 did not show any differences associated with the source of the strain (different culture collections of the original or human isolate FMH), and there were few variations in sensitivity between Bm strain 23344 and the other two strains tested (Bm Turkey 1 and 10229) (data not shown). In assays with 10 strains of Bp (K96243, 1026b, 316c, 4845, MSHR668, 1106a, 406e, HBPUB10134a, MSHR305, and Bp22), several phenotypes exhibited strain-specific differences in sensitivity, to include the antibiotics nalidixic acid, minocycline, and aztreonam, as well as Niaproof 4 and potassium tellurite (Table 1). Inter-experimental variability in sensitivities to aztreonam, potassium tellurite, and Niaproof 4 was noted for some of the Bp strains and to a lesser extent for Bm strains. As described below, potentially confounding effects of this variability were alleviated in all phenotypic comparisons of in vivo isolates with the challenge strain by the inclusion of the parent strain in all in vitro assays.
Table 1.
Comparison of chemical sensitivities of different B. pseudomallei strains: Biolog phenotyping panel
| Chemical | K96243/DW | K96243/DD | 1026b | 316c | 4845 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | |
| Pos. ctrl. | 1.644 | 0.141 | 1.558 | 0.019 | 1.706 | 0.064 | 1.719 | 0.140 | 1.567 | 0.256 |
| 1 % NaCl | 1.279 | 0.153 | 1.492 | 0.015 | 1.540 | 0.138 | 1.673 | 0.002 | 1.380 | 0.016 |
| Sodium lactate | 1.841 | 0.167 | 1.737 | 0.027 | 1.820 | 0.082 | 1.826 | 0.025 | 1.669 | 0.191 |
| Troleandomycin | 1.356 | 0.309 | 1.280 | 0.068 | 1.406 | 0.296 | 1.332 | 0.511 | 1.253 | 0.443 |
| Lincomycin | 1.636 | 0.145 | 1.735 | 0.027 | 1.824 | 0.071 | 1.912 | 0.018 | 1.783 | 0.058 |
| Vancomycin | 1.730 | 0.181 | 1.692 | 0.083 | 1.826 | 0.077 | 1.795 | 0.134 | 1.729 | 0.063 |
| Nalidixic acid | 1.204 | 0.233 | −0.097 | 0.009 | −0.059 | 0.053 | 1.679 | 0.146 | 1.249 | 0.188 |
| Aztreonam | 0.097 | 0.040 | 0.647 | 0.312 | 0.237 | 0.106 | 0.318 | 0.170 | 0.253 | 0.332 |
| pH 6 | 1.734 | 0.109 | 1.658 | 0.074 | 1.774 | 0.069 | 1.807 | 0.102 | 1.671 | 0.162 |
| 4 % NaCl | −0.063 | 0.066 | −0.121 | 0.010 | 0.026 | 0.089 | 0.052 | 0.218 | −0.052 | 0.148 |
| Fusidic acid | 0.024 | 0.063 | −0.117 | 0.000 | −0.088 | 0.038 | 0.224 | 0.212 | −0.060 | 0.121 |
| Rifampin | 1.667 | 0.187 | 1.617 | 0.121 | 1.809 | 0.042 | 1.847 | 0.188 | 1.725 | 0.173 |
| Guanidine HCl | 0.155 | 0.056 | 0.187 | 0.007 | 0.242 | 0.104 | 0.337 | 0.106 | 0.294 | 0.049 |
| Tetrazolium violet | 2.908 | 0.022 | 2.919 | 0.025 | 2.892 | 0.021 | 2.955 | 0.027 | 2.859 | 0.008 |
| LiCl | −0.022 | 0.151 | −0.096 | 0.004 | 0.006 | 0.123 | 0.062 | 0.120 | −0.038 | 0.142 |
| Sodium butyrate | 0.074 | 0.119 | −0.098 | 0.002 | 0.050 | 0.096 | 0.197 | 0.145 | −0.021 | 0.060 |
| pH 5 | 1.828 | 0.152 | 1.769 | 0.066 | 1.837 | 0.062 | 1.692 | 0.254 | 1.797 | 0.126 |
| 8 % NaCl | −0.141 | 0.015 | −0.009 | 0.141 | −0.113 | 0.024 | −0.116 | 0.011 | −0.149 | 0.007 |
| d-Serine | −0.072 | 0.028 | −0.102 | 0.002 | 0.236 | 0.155 | 0.221 | 0.025 | 0.063 | 0.057 |
| Minocycline | 1.267 | 0.252 | −0.098 | 0.012 | −0.074 | 0.020 | 1.695 | 0.117 | 1.377 | 0.107 |
| Niaproof 4 | 0.481 | 0.073 | 0.589 | 0.005 | 0.153 | 0.293 | 0.949 | 0.325 | 0.516 | 0.550 |
| Tetrazolium blue | 3.023 | 0.049 | 2.983 | 0.104 | 2.980 | 0.081 | 3.056 | 0.069 | 2.996 | 0.003 |
| Potassium tellurite | 0.877 | 0.971 | 1.605 | 0.010 | 1.002 | 0.970 | 1.291 | 0.787 | 1.645 | 0.046 |
| Sodium bromate | −0.121 | 0.017 | −0.102 | 0.011 | −0.101 | 0.016 | −0.106 | 0.015 | −0.136 | 0.008 |
The criteria for sensitivity are similar to those defined by Biolog, Inc.
Bold = growth densities (OD630) less than half that of positive control well are sensitive
Unchanged = growth densities similar to positive control are resistant
Bold italics = density compared to positive control is borderline or variable (different experiments or strain sources)
In addition to the Biolog panel, Bm and Bp prototype strains were assayed independently for sensitivity to selected chemicals. Initial findings with Bm FMH and Bp K96243 indicated that both were sensitive to 2 mM sodium nitrite (RNI), as shown previously (Tandhavanant et al. 2010). Also, Bm was more sensitive than Bp to H2O2, and both were similar in sensitivities to NaCl and Niaproof 4; Bp K96243 was more sensitive to these two chemicals in the individual assays than in the Biolog plates (Table 1, parent strains in Table 2 and Supplementary Tables 2 and 3, and data not shown). The concentrations of chemicals used in the Biolog microplate assays are proprietary and results can only be qualitatively compared to assays which use defined concentrations (B. Bochner, personal commun.). Responses of Bm and Bp were variable to 5 µg/mL nalidixic acid, but Bm strains were generally sensitive and Bp strains were generally resistant (parent strains in Table 2, Supplementary Tables 3 and 4, and data not shown). All strains were sensitive to 50 µg/mL nalidixic acid. Thus, the chemical sensitivity data indicated that Bp appeared to be generally more resistant to potential environmental and host antimicrobial stresses than Bm. Due to evidence of strain variability, the subsequent analyses of phenotypic changes in strains recovered during infection required that responses of the original parent strain be tested in each assay. The direct comparison of parent and in vivo isolates responses allowed relative differences to be detected.
Table 2.
In vitro phenotype screening of in vivo isolates of B. mallei: representative results
| Straina | Biologb | Individual chemical sensitivitiesc | Antimicrobial peptidesc | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. resistant/total (23) | Change | H2O2: 0.02, 0.1 % | NaCl: 1, 4 % | Nalidixic acid: 5, 50 µg/mL | Niaproof 4: 0.027, 0.10 % | RNI: 2 mM | Cecropin P1 | Cecropin A | LL-37 | Mastoparan 7 | Magainin | Melittin | Histatin 5 | Proteolytic activity (mm) | |
| Spleen isolates: Bm FMH aerosols | |||||||||||||||
| Bm FMH parent | 9d | none | 4.5, 4.5 | S, S+ | S, S+ | S+, S+ | S | R | R/S | R | R/S | R/S | S | R | 1.5 |
| 4, 5 | S, S+ | S/R, S+ | S+, S+ | S | S | nd | S | R | S | S | nd | nd | |||
| 26-2 | 10 | R-nal. acid | NCe | S, S+ | R, S | S+, S+ | S | nde | R− | nd | S+ | R | R | nd | 2.5–3 |
| 26-3 | 10 | R-nal. acid | 4.0, 7 | S, S+ | R, S | S, S+ | S | nd | R− | nd | S+ | R | R | nd | 2.5 |
| Bm FMH parent | 9d | 4, 5 | S, S+ | R/S, S+ | S+, S+ | S | nd | S | R | S | S | nd | nd | ||
| 45-7 | 9, diffs | R-nal.A; S-linco | NC | NC | R/S+ | S, S+ | NC | nd | NC | NC | S + | NC | NC | nd | nd |
| 45-9 | 9, diffs | R/S-nal.A; S-Nacl | NC | NC | NC | NC | NC | NC | nd | NC | R | NC | NC | nd | nd |
| Bm FMH parent | 9d | – | 4.5, 6 | S+, S+ | S, S+ | S+, S+ | S+ | nd | S | nd | S+ | S | S | nd | nd |
| 60-1 | NC | None | 4.5, 6 | S+, S+ | S, S+ | S+, S+ | S+ | nd | R | nd | S | S | R | nd | “ |
| 60-3 | NC | None | 5, 5(haze) | S+, S+ | R, S | S+, S+ | S | nd | S | nd | S | R | R | nd | “ |
| 60-3 | NC | None | 4.5, 5(haze) | S, S+ | R, S | S+, S+ | S | nd | R | nd | S | R | R | nd | “ |
aIsolates are from spleens of mice infected with Bm FMH. With the exception of the infecting strain (parent), the strains are identified by the day post-inoculation on which spleens were collected from the infected mice and by the isolate number. Responses of the isolates which varied from the parent are bolded (more R) or in bold italics (more S)
bSensitivities were determined with the GEN III panel of 23 chemicals (Biolog). The Biolog criteria were used for resistance (R, >50 % positive control OD630), sensitive (S, <50 % pos. control), and borderline or variable (R/S). “Diffs” indicates that the strain was resistant to the same total no. chemicals as the parent but differed in specific chemical sensitivities
cSensitivities to specific chemicals and antimicrobial peptides are determined by OD630 readings: values >75 % positive control (resistant, R), >50 % and <75 % pos. ctrl (borderline, R/S), <50 % (sensitive, S), and <×2 negative control (highly sensitive, S+). For the H2O2 plates, the values (mm) are the zones of cleared growth proximal to the H2O2 spot
dIn Biolog experiments, Bm strain FMH was resistant to nine conditions: 1 % NaCl, Na lactate, pH 5 and 6, tetrazolium blue and purple, vancomycin, rifampin, and lincomycin. It was variable but usually sensitive to nalidixic acid in the Biolog and individual assays (low, 5 µg/mL; and high, 50 µg/mL). Bm FMH was generally sensitive to 13 conditions: NaCl (4 % and 8 %), fusidic acid, d-serine, guanidine HCl, Niaproof 4, LiCl, potassium tellurite, aztreonam, Na butyrate, nalidixic acid, Na bromate, and troleandomycin
eNC = same as parent (no change); nd = not done
The Burkholderia have been reported to be notably resistant to most AMPs at concentrations ≤200 µM. Although Bp has been shown to be sensitive in vitro to the peptide LL-37 (Kanthawong et al. 2009; Tandhavanant et al. 2010), we observed resistant to borderline resistant responses to LL-37 in repeated tests with Bm FMH and most Bp strains. These findings are exemplified for the parent strain data in Table 2 and Supplementary Tables 3 and 4. Sensitivities to five more AMPs were initially evaluated with the prototype strains. Bm and Bp were resistant to histatin 5, Bp was resistant more often than Bm to cecropin A and melittin, and both were often relatively sensitive to mastoparan 7 (data not shown). In skim milk agar assays for protease production, prototype strain Bp K96243 produced larger zones of clearing than Bm 23344/China 7, possibly due to the secretion of more protease by Bp and/or its faster rate of growth (data not shown). Since the Burkholderia demonstrated some variability in AMP sensitivity between experiments, the impact of this on later analyses of the sensitivities of infected mice isolates was mitigated by direct comparisons to the activity of the parent strain in each assay.
Macrophage phenotypes of the Bm and Bp strains
The J774.A1 cell line was used to develop several macrophage phenotypes for comparing different strains of Bm and Bp. Several parameters were identified including relative bacterial survival over time within the macrophage and several infection-induced cytotoxic effects involving cell killing and MNGC production. In screening assays, the number of viable Bm recovered was much less than the number in the inoculum. In contrast, Bp prototype strain K96243 proliferated and exhibited a large increase in CFU compared to the input concentration, i.e., 600-fold after 20 h (Fig. 2), but also after incubation for much shorter times, as described below. As shown in Fig. 2, for the Bm strains, there was a large decline in viable counts recovered 20 h after infection (126-, 112-, and 267-fold, respectively, for FMH parent and isolates 1-1-3 and 1-2-4 from infected spleens). At t20, for all three Bm strains, <1 % of the input bacterial inoculum could be recovered (0.8, 0.9, and 0.4 %, respectively). In contrast, the Bp strain K96243 exhibited a 5.8-fold (581 %) increase in number of CFU/well at t20 compared to the initial concentration added at t0. Despite the lower MOI of the K96243 inoculum (p < 0.8 × 10−5), the number of viable bacteria recovered at 20 h was significantly greater than the viable organisms recovered of the Bm strains (p < 0.001 each, by ANOVA and Tukey post-test paired comparisons). This pattern of survival and proliferation in macrophages of K96243 was also observed for other Bp strains from diverse sources (Supplementary Table 1) (Welkos et al. 2015; data not shown). Furthermore, both Bm FMH and Bp K96243 induced MNGC formation, yet Bp was significantly more cytotoxic and the infection resulted in ≥90 % killing and loss of adherent cells from the monolayer. However, Bm cytotoxicity was observed to differ significantly in association with the MOI and was generally less for MOIs <5 and more for MOIs >5 in our model system. In contrast, the cytotoxicity associated with strains of Bp was less affected by the MOI (discussed further below). The more extensive cytotoxicity demonstrated for K96243 was also observed for other Bp strains from diverse sources (Supplementary Table 1) such as 1026b, Bp22, and MSHR5848 (Welkos et al. 2015; data not shown).
Fig. 2.
Comparison of macrophage survival of Bp and Bm strains. The J774.A1 macrophage cell line was infected with Bp strain K96243 at a MOI of 4.3; or with Bm strain FMH or its spleen isolates Bm 1-1-3 or Bm 1-2-4 at MOIs of 6.8, 7.95, and 7.1, respectively. The paired bars correspond to the mean viable counts from triplicate wells determined at t0 (macrophage inocula) and after 20-h incubation of infected macrophages (t20). Spleen isolates Bm 1-1-3 and Bm 1-2-4 were collected from IP-infected mice on days 3 and 16, respectively
Analysis of in vivo isolates of Burkholderia
The phenotypic characterization strategy was employed to examine in vivo isolates of Bm and Bp for changes which might be associated with progression of an acute infection to a more persistent form. Isolates from spleens collected from 3 to 180 days post-challenge from mice involved in aerosol or IP LD50 studies were analyzed. Spleen isolates obtained from infections by either route were generally observed to be similar in their phenotypic comparisons to the corresponding parent challenge strain. Because of the in vitro variability of the pathogenic Burkholderia spp., the original challenge strain was included in all in vitro assays for comparison with the spleen isolates. To categorize the in vivo isolates, we empirically defined three phases of infection: an acute phase (≤21 days post-challenge), a midterm phase (21–59 days), and a long-term phase (≥60 days).
Comparison of isolates and parent strains by colony morphology phenotyping
Bacteria isolated from mice euthanized during the acute phase of infection with either Bm or Bp did not differ significantly from the parent challenge strain in their colony morphotypes (data not shown). To confirm this observation, mice were infected IP with 5 LD50 doses of either strain 1106a or HBPUB10134a and survivors were serial sampled on days 3, 7, or 14 post-infection for spleen colony isolations. These two strains represent mice that were, respectively, the least and most virulent by the intraperitoneal route (Welkos et al. 2015). For both strains, the spleen isolates exhibited no significant differences from the challenge strain, as typically observed for early stage isolates. Similar findings were observed for isolates obtained on days two through six after aerosol challenge with HBPUB10134a (data not shown).
For spleen isolates from aerosol challenges with the Bm FMH strain, there was an apparent increase in the overall number of morphotypic differences between spleen isolates and parent with longer times post-challenge. For example, most Bm FMH isolates collected ≤day 45 varied little from the parent challenge strain, while isolates collected 119 days after challenge were larger on SBAP, OFPBL, and BCA plates (data not shown). Similarly, for some Bp strains there was an increase in the overall number of small differences in colony morphology with time post-challenge for isolates. For example, isolates of Bp strain 1106a collected at times later than day 49 exhibited numerous alterations compared to the parent in colony size (several media) and morphology (e.g., opacity and moistness). For three Bp strains (1106a, MSHR668, and 406e), there was a reduced incidence of multiple colony variants on SBA, GTA, and/or AA displayed by isolates; i.e., whereas the parent exhibited two distinct variants differing in several characteristics, only one of the variant types was observed in all the isolates. This is demonstrated for strain 406e in Supplementary Table 5A and B. For some of the strains, such as MSHR305 and HBPUB10134a, there was a paucity of mice surviving at later time points, and among the survivors, few if any had residually infected spleens. However, isolates obtained 33 and 35 days post-infection with MSHR305 were larger on both SBAP and AA (day 33) or on SBAP (day 35), and half of those tested had enhanced metabolic activity on BCA plates, as indicated by color change in this medium illustrated in Fig. 1d (data not shown). Thus, it can be concluded that Bp isolates routinely exhibited differences in colony morphology from the parent strain, yet an overall common pattern of change in isolate colony morphotype was not observed among the Bm/Bp strains examined. Therefore, we considered the presence of any distinct difference in colony morphotype compared to parent colony morphology to be important as one of the bases of selecting isolates for subsequent phenotypic analyses. Colony morphology differences together with GEN III plate differences (described below) were used as criteria for selecting strains for subsequent, more complex phenotype assays.
Comparison of isolates and parent strains in antimicrobial sensitivities
Spleen isolates were initially screened for sensitivity to 23 antimicrobial chemicals using GEN III plates, and selected isolates were then tested in the additional antimicrobial sensitivity assays. All assays included the corresponding parental challenge strain for intra-experimental comparisons. Isolates obtained from spleens collected between 3 and 16 days after challenge did not differ significantly in sensitivity from the parent Bm FMH or Bp strain. Such isolates included, for example, those from spleens collected 3 and 16 days after IP challenge with Bm FMH (Fig. 2), strains from samples obtained 2–14 days after IP injection of Bp strains 1106a or HBPUB10134a, and spleens collected 2–6 days after aerosol and/or IP exposure to HBPUB10134a or 1026b, as described above.
Results with spleen isolates acquired from survivors on or after day 26 after aerosol exposure to Bm FMH are illustrated in Table 2. These isolates often differed in sensitivity, typically being more resistant than the parent to several antimicrobials, especially nalidixic acid and the AMPs cecropin A, magainin, and melittin. Isolates from the spleens of mice infected with the seven Bp strains were obtained from 17 experiments (12 aerosol challenges of 7 strains and 5 IP challenges of 5 strains). The results of chemical sensitivity screening of the isolates are summarized in Table 3 and Supplementary Table 6. The latter details the sensitivity differences between strains for the chemicals in Table 3. The data for strain HBPUB10134a are not included in these tables since isolates from the acute stage of infection were only available (≤day 14). The changes in chemical sensitivities of the in vivo isolates which are described in Table 3 and Supplementary Table 6 can be summarized as follows. Differences in sensitivities of spleen isolates compared to the Bp parent strain were observed most frequently for two antibiotics, nalidixic acid and minocycline, for which isolates from five or three of six strains tested, respectively, differed in sensitivity from the parent strain. Isolates with enhanced resistance to nalidixic acid were recovered for all five of the strains. Isolates with altered sensitivities compared to the Bp parent strain were detected most commonly for four of the chemicals tested (Niaproof 4, nitrite, NaCl, and potassium tellurite) and the AMPs mastoparan 7, BMAP-18, and magainin. As shown in Table 3, overall, these Bp isolates most often exhibited an increased resistance to the antimicrobial substances compared to the parent. For example, isolates with greater resistance to Niaproof 4 were recovered for five of the six Bp strains evaluated and isolates with greater resistance to NaCl were detected for all three Bp strains from which bacteria with altered NaCl sensitivities were obtained. These findings can be further illustrated with the isolate sensitivity results of strains K96243 and 1026b. Supplementary Table 3 describes sensitivity patterns of K96243 isolates obtained on day 70 after aerosol exposure. Despite parental strain variability, these isolates were generally enhanced in their resistance to potassium tellurite, Niaproof 4, and several AMPs (mastoparan 7 and potentially LL-37, melittin, and BMAP-18) but more sensitive to fusidic acid in the GEN III assays. The K96243 isolates were sensitive to mastoparan 7 at day 28 and resistant at days 49 and 70 (Supplementary Table 3 and data not shown), suggesting a time-associated response. The responses of the day 30 and day 60 isolates from challenge experiments with Bp strain 1026b are shown in Supplementary Table 4. The data suggested an increase in the resistances of 1026b isolates with time post-challenge as seen with nalidixic acid and various AMPs such as bactenecin and BMAP-18. These findings were intriguing though additional serial kill studies will be required to confirm a time-associated pattern of change among other Bp strains. It will be necessary to evaluate many isolates at each of the time points post-challenge from mice. There was no apparent time-associated pattern in antimicrobial resistances of the in vivo isolates of strain Bm FMH (Table 2).
Table 3.
Summary of differences between parent B. pseudomallei and in vivo isolates in chemical sensitivities
| Chemical | No. strains with: variant isolates/total no. strainsa | Increased in | |
|---|---|---|---|
| Sensitivity | Resistance | ||
| Niaproof 4 | 6/6 | 3b,c | 5b,c |
| Nalidixic acid | 5/6 | 1 | 5b |
| Mastoparan 7 | 4/6 | 1 | 4b |
| BMAP-18 | 3/4e | 0 | 3 |
| Minocycline | 3/6 | 2 | 2b |
| Nitrite | 3/6 | 2 | 1 |
| NaCl | 3/6 | 0 | 3 |
| Potassium tellurite | 3/6 | 1b | 3b |
| Magainin | 2/6 | 0 | 2 |
| Melittin | 2/6 | 1 | 1 |
| d-Serine | 2/6 | 1 | 1 |
| Others | 1/6d,e | – | – |
aSpleen isolates from aerosol or IP challenge experiments of seven Bp strains were obtained (17 total experiments). The variant responses of the isolates consisted of significant increases in either sensitivity (S) or resistance (R) to the antimicrobial chemical compared to the challenge strain, as described in the methods. The data do not include isolates exhibiting no differences from the parent. For strain HBPUB10134a, no isolates were obtained from survivor spleens cultured later than 14 days after challenge, so the data are not included in the table
bThe sensitivity of isolates collected at early and later time points differed (nalidixic acid, potassium tellurite, Niaproof 4, and mastoparan 7) or individual isolates from a given challenge experiment collection responded differently, being either more sensitive or more resistant than the parent (minocycline, Niaproof 4, potassium tellurite, and mastoparan 7) as described in the text
cDifferent responses by the same isolate were observed in Biolog and antimicrobial sensitivity microtiter assays, but chemical concentrations in the Biolog system are proprietary and unavailable
dFor one Bp strain each, the sensitivity to the chemical of an isolate(s) varied from that of the parent as follows: aztreonam (R), guanidine HCl (R), LL-37 (R), CA-MA (R), and bactenecin (variable)
ePeptides BMAP-18, CA-MA, bactenecin, and LL-37 were tested in four of the six strains only
Comparison of isolates and parent strains in macrophage phenotypes
To prospectively evaluate macrophage virulence during the acute phase of infection, mice were intraperitoneally challenged with Bp 1106a and serial killed on days 3, 7, and 14 post-infection, as described above. The 1106a spleen isolates exhibited a progressive change in macrophage virulence. The day 3 isolate was similar in extent of phagocytosis, survival, and cytotoxicity to the parent, and the day 7 isolate was phagocytosed to a greater extent but otherwise similar in macrophage parameters (data not shown). However, the day 14 isolate of 1106a (Bp 14-1) was consistently more cytotoxic for, and better able to multiply within, macrophages than the parent (data not shown). In addition, Bp 14-1 was phagocytosed fivefold better, and the 8-h recovery of viable cells was more than twice that of the 1106a parent (Supplementary Figure 1). The progressive change from the parent of in vivo isolates in regard to macrophage virulence may be strain associated. For example, early isolates of Bp HBPUB10134a, a strain with more than a three-log greater IP virulence than 1106a [4], did not differ significantly from the parent in macrophage survival and cytotoxicity (data not shown).
The macrophage phenotypes of spleen isolates from mice with longer term infections were then characterized. Bm FMH isolates collected at days 26 and 45 after aerosol challenge differed only minimally in cytotoxicity and survival compared to the parent. However, day 60 isolates were distinctly reduced in survival and/or several cytotoxicity parameters, as illustrated in Table 4 and data not shown. The cytotoxicity induced by the parent strain was dose related in four of five experiments; a relatively low MOI produced significant bacterial survival and cell cytotoxicity in one experiment for unknown reasons.
Table 4.
In vitro macrophage-associated phenotypes of isolates from B. mallei-infected animals
| Straina | Expt. | MOI | Bacterial survival (%)b | Cytotoxicityc | Cell morphologyd | ||
|---|---|---|---|---|---|---|---|
| % cell loss (TB, DQ) | % dead (PI or TB stain) | Relative proportion MNGC | Major nuclear morphology | ||||
| Bm FMH parent | 9 | 7.7 | 0.90 | >50 | ~50 | ≥10 % | <50/>50 % normal/cytotoxic |
| 11 | 2.9 | 5 × 10−4 | <1 | 1–2 | Rare, <1 % | >90/<10 % normal/cytotoxic | |
| 12 | 4.3 | ~1 × 10−4 | <1 | <1 | None | >90/<10 % normal/cytotoxic | |
| 36 | 9.6 | 0.20 | ~40 | ~65 | 16.5 | 37/63 % normal/cytotoxic | |
| 63 | 2.3 | 8.80 | 90 | 87 | 20.6 | 36/64 % normal/cytotoxic | |
| Isolate 60-5 | 9 | 7.9 | 0.70 | NC | 12 | Rare, <1 % | >50/<50 % normal/cytotoxic |
| 11 | 2.3 | 5 × 10−4 | NC | <1 | None | NC | |
| 12 | 3.9 | <1 × 10 −5 | NC | <1 | NC | NC | |
| 36 | 8.8 | 0.25 | ~30 | ~65 | 7.10 % | 47/53 % normal/cytotoxic | |
| Isolate 60-3 | 63 | 2.2 | 2.70 | 50 | 4 | 16.8 % | 38/62 % normal/cytotoxic |
NC no change/same response as parent strain
aBALB/c mice were exposed by aerosol to parent strain Bm FMH in an LD50 experiment. Colony isolates were collected from spleens of surviving mice 60 days after exposure. Results are those of two isolates, 60-3 and 60-5, which had differed more from parent in other phenotypes (colony morphology and chemical susceptibility) compared to four other isolates. All were from two mice in the lowest aerosol dose group (32 CFU). Data are representative of seven experiments. Bolded values are those notably different from parent values
bThe bacterial survival is expressed as the change in viable counts after 20–23 h compared to the input inoculum (% of input no. CFU)
cCytotoxicity was determined after 20–23-h incubation and is shown as the % cells lost [by trypan blue and Diff-Quik™ (DQ) stains]. The % dead cells was determined by PI and trypan blue staining. The values do not include cells/MNGCs in an advanced state of necrosis with degraded, unstainable DNA
dThe values describe approximate proportion of MNGCs among total cells/field and the major relative appearance of nuclei [typical large and intact vs. cytotoxic (necrotic/apoptotic or within an MNGC)]
In contrast to the results with Bm, the survival and cytotoxicity of the isolates in comparison to the Bp parent appeared to be strain dependent. Of the strains of Bp from which isolates from long-term infections could be recovered, a trend toward enhanced macrophage virulence was observed consistently for strains 1106a and MSHR668 and typically, but less consistently, for K96243. The responses of the 1026b isolates were variable, and there were usually no differences observed for isolates of parent strain 406e (Fig. 3). Figure 3a shows the results of a spleen isolate recovered on day 70 after aerosol exposure with strain K96243. Isolate Bp 70-1 was phagocytosed to a greater extent compared to K96243 for all three time points. Greater cell death and detachment was also associated with Bp 70-1 at 8-h (Table 5). The greater cell loss associated with Bp 70-1 at 8-h suggests that the 8 h viable counts might have underestimated the strain’s replication. To account for differences in strain MOIs, the counts were normalized to the inoculum (no. CFU added at t0) and expressed as percentages. These normalized values, presented in Supplementary Table 7, confirmed the differences shown here. Isolates recovered 49 days post-exposure also exhibited better survival than the parent K96243 strain (data not shown).
Fig. 3.
Comparison of macrophage survival of Bp parent strain and spleen isolate pairs. J774.A1 cells were incubated with the parent strain or isolate for 1 h. For a–d, f, the data shown are the mean viable counts (triplicate wells) recovered after 1-h uptake, after incubation of the infected cells in the presence of kanamycin for 2 h to remove unphagocytosed bacteria (3 h total), and after incubation for a total of 8 h. a Bp strain K96243 and a mouse spleen isolate collected 70 days after exposure (Bp 70-1) with MOIs of 13.3 and 11.0, respectively. The viable counts determined at the three times points were statistically different (1 and 3 h: p < 0.0001; 8 h: p = 0.0016). b Bp strain MSHR668 and mouse spleen isolates. The MOIs were 38.5 (MSHR668) and 32.3 (Bp 41-1). Each set of bars are the data for the challenge strain (MSHR668) and for a spleen isolate from a mouse surviving aerosol challenge and euthanized on day 41 (Bp 41-1); p = 0.004 for 8 h. Inset macrophages were infected with strain MSHR668 or isolates 41-1 or 41-4 at MOIs of 12.2, 11.2, and 13.1, respectively. The infected cells were incubated and treated as described above, and the viable counts obtained after a total of 7 h are shown. They are compared to the initial no. of CFU added to the wells (t0 inoculum); p < 0.001 for 7 h for 41-1. c Bp strain 1106a and a mouse spleen isolate collected 62 days after aerosol exposure (Bp 62-3), at MOIs of 12.3 and 14.1, respectively. The viable counts of Bp 62-3 determined at the three times points were significantly greater than those of 1106a (1 and 3 h: p < 0.0001; 8 h: p = 0.013). D. Bp strain 1026b and a spleen isolate from a mouse surviving aerosol challenge and euthanized on day 60 (Bp 60-3). The MOIs were 9.5 and 11.4, respectively. The viable counts of the parent strain were slightly greater than those of the isolate after 1-h incubation (p = 0.08, not significant) and after 3-h incubation (p = 0.008). Greater quantities of isolate than parent were recovered after 8 h although the absolute differences were not statistically significant. e Macrophages were infected with strain MSHR305 or a spleen isolate collected 32 days after exposure (Bp 33-2), at MOIs of 15.4 and 17.5, respectively. The infected cells were incubated for 1 h and treated with kanamycin for 2 h, and the viable counts obtained after a total of 7 h are shown. The quantities of bacteria of the two strains recovered from the cells at 7 h were not significantly different (1.7 and 2.5 %, respectively). f Bp strain 406e and mouse spleen isolate Bp 60-4 at MOIs of 17.1 and 15.5, respectively. The viable counts obtained at all three time points for the parent and isolate were not significantly different (p > 0.05). The observed differences for these experiments were confirmed using comparisons of the normalized viable CFU counts
Table 5.
Phenotypes of macrophages infected with B. pseudomallei parent and spleen isolates—cytotoxicity comparisons
| Strain | Panela | MOI | % cell lossb | % cells dead (TB)b | No. MNGC or necrotic cells (% of total)c | No. MNGC nuclei (% of total)c |
|---|---|---|---|---|---|---|
| K96243 | a | 13.3 | 40–50 | 17 | 4.4 | 43.6 |
| Bp 70-1 | 11.0 | 60–65 | 32.4 | 5.8 | 58.8 | |
| MSHR668 | b inset | |||||
| expt. #1 | 12.2 | 35–40 | 4.5 | 7.6 | 32.0 | |
| Bp 41-1 | 11.2 | 60–70 | 20.0 | 13.0 | 54.0 | |
| Bp 41-4 | 13.1 | 35–40 | 12.5 | 14.0 | 58.0 | |
| MSHR668 | b | |||||
| expt. #2 | 38.5 | 20 | nd | 20.7 | 67.8 | |
| Bp 41-1 | 32.3 | 18 | nd | 18.0 | 73.2 | |
| 1106a | c | 12.3 | 40 | 27.9 | 21.9 | 70.8 |
| Bp 62-3 | 14.1 | 60–65 | 73.2 | 41.8 | 85.4 | |
| 1026b | d | 9.5 | 25–30 | 24 | 8.8 | 35.3 |
| Bp 60-3 | 11.4 | 25–30 | 25 | 9.6 | 47.4 | |
| MSHR305 | e | 15.4 | 30 | 2.5 | 3.5 | 10.0 |
| Bp 33-2 | 17.5 | 35 | 2.5 | 3.5 | 10.0 | |
| 406e | f | 17.1 | 40 | 10.3 | 5.6 | 21.8 |
| Bp 60-4 | 15.5 | 45 | 22.3 | 5.0 | 25.8 |
nd not done
aFigure 3, panels a–f
bCytotoxicity is shown as the % cells lost from the macrophage layer by trypan blue (TB) staining and the % dead cells among those remaining by TB and/or PI staining. However, necrotic cells and advanced-stage MNGCs with degraded nuclei are unstained by TB, and the values underestimate the actual proportion of dead cells
cThe values describe the approximate relative proportion of MNGCs (%) among the total cells in six fields, two from each of three replicate coverslips (×600); and the major nuclear phenotype, considering all the nucli in the fields (typical large nuclei vs. nuclei present in MNGC with intact stained nuclei or in necrotic fused cell masses with lightly stained nuclei), expressed as percentages. Counts were done using Diff-Quik-stained cells. The more extensive cell death and detachment induced by some strains may minimize the extent of these values
In experiments with strain MSHR668 and two day 41 isolates, the isolates again replicated better than the challenge strain (Fig. 3b; Supplementary Table 7); 7–8 h counts of the isolates were significantly greater than those of the parent. The isolates were more cytotoxic at lower MOIs, as shown by the greater proportion of TB-stained dead cells and the greater number and larger size of MNGCs (Table 5). The cytotoxic effects on macrophages often associated with Bp infection are illustrated by MSHR668 isolate Bp 41-1-infected cells in Fig. 4. The detachment of J774.A1 cells and the formation and appearance of MNGCs are shown in panels A–B and C–D, respectively. Isolates recovered 60 days after IP challenge exhibited similar macrophage phenotypes (data not shown). The results with strain 1106a and an isolate collected 62 days after challenge again exemplified the increase in isolate macrophage infectivity and cytotoxicity, as shown in Fig. 3c. Isolate Bp 62-3 was phagocytosed to a greater extent. Both strains doubled during the 5-h incubation after removal of antibiotic, but the greater loss of macrophages during this time in the Bp 62-3-infected culture likely resulted in an underestimation of the 8 h viable counts. The isolate was more cytotoxic for the macrophages, inducing more cell death and detachment from the wells (Table 5).
Fig. 4.
Changes in the morphology and adherence of macrophages infected with Bp. Cells were infected with spleen isolate Bp 41-1 of strain MSHR668 and stained with Diff-Quik™ after 7-h incubation. The cultures are shown under low power (×100, a, b) and higher power (×600, c, d). a Uninfected cells of normal semi-confluent J774.A1 cells. b Cells infected with isolate Bp 41-1 showing detachment/loss of cells and presence of numerous multi-nucleated giant cells (MNGCs, indicated by arrows) and clusters of fused necrotic cells. c, d Cells infected with isolate Bp 41-1 and illustrating the appearance of MNGCs with numerous intact nuclei (c) and MNGCs in a more advanced state of decay with more necrotic, poorly stained nuclei (d)
As shown in Fig. 3d and Table 5, a day 60 isolate from strain 1026b was variably more active in macrophages than the parent strain, albeit marginally. Although Bp 60-3 was phagocytosed to a slightly lesser extent than 1026b (1-h and 3-h viable counts), the isolate replicated more than twice as fast as the 1026b parent post-phagocytosis, as determined by the greater normalized 8-h counts (742 vs. 1839 % of the inoculum for parent and isolate, respectively; Supplementary Table 7). Isolate Bp 60-3 was also slightly more cytotoxic, as shown by the MNGC parameters (Table 5 and data not shown). The macrophage phenotypes of strain MSHR305 and an isolate collected 33 days after aerosol challenge (Bp 33-2) suggest the isolate was similar to the parent in its capacity to replicate in the macrophages and induce a cytotoxic response (Fig. 3e; Table 5). Additional day 33 isolates exhibited variable macrophage phenotypes (data not shown), perhaps suggesting that persistent infection had not been established. Finally, the spleen isolates evaluated from mice challenged with Bp strain 406e did not induce macrophage phenotypes that were consistently different from the challenge strain, as determined with spleen isolates from days 34 and 60 after infection (Fig. 3f; Supplementary Table 7; Table 5; and data not shown.
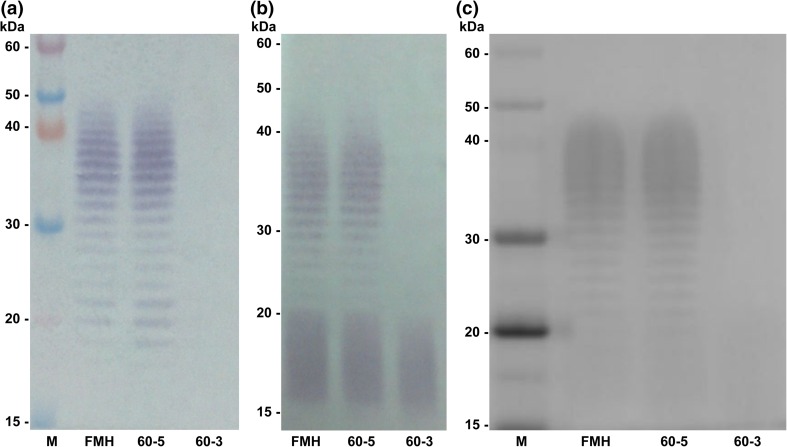
LPS phenotyping
LPS phenotyping was explored as an additional tool for distinguishing different strains of Bp and Bm as well as their in vivo isolates. Distinct strain-specific differences in the LPS banding profiles of numerous other strains of each species were distinguished previously (Welkos et al. 2015). In this study, LPS electrophoretic banding profiles of 20 Bp and 13 Bm isolates were compared to those of their respective parent strains using species-specific monoclonal antibodies for LPS (11G3-1 for Bp and 8G3-1B11 for Bm). One spleen isolate of Bm FMH obtained on day 60 post-infection displayed a change in LPS phenotype, from smooth to rough, during the course of infection (Fig. 5). Figure 5a shows a western blot with parent strain FMH, a day 60 isolate that did not change its LPS phenotype (60-5), and the 60-3 isolate that appeared to change from smooth to rough. The transition from a smooth to rough LPS phenotype results from the loss of O polysaccharide (OPS). This result was further supported by a western blot using polyclonal antibody ABE 335 where OPS was absent and lipid A was visible, confirming the presence of LPS (Fig. 5b). To rule out the possibility that the OPS simply changed its structure, thus preventing antibody recognition, silver staining was performed (Fig. 5c). No OPS could be detected for 60-3 by silver straining, thereby confirming the rough phenotype.
Fig. 5.
LPS phenotyping comparing the Bm FMH parent strain with two 60-day mouse spleen isolates. Western blots using monoclonal antibody 8G3-1B11 (a) and polyclonal antibody ABE 335 (b) show the loss of O polysaccharide in isolate 60-3. This LPS change was confirmed by silver staining (c)
Comparison of mouse virulence of Burkholderia strains and their spleen isolates
An initial study to compare the virulence and morbidity of strains of Bp and Bm with their in vivo isolates was conducted in an established Burkholderia mouse model (Welkos et al. 2015). In the current study, BALB/c mice were challenged by the IP route with dilutions of a strain of Bp/Bm or with a selected spleen isolate. The LD50 values and relative virulence potencies of parent versus isolate were evaluated statistically using the survival data accrued by days 21 and 60 (Table 6). Isolates collected from midterm survivors did not always show statistically significant differences from their respective parent strains. However, the day 60 LD50 of K96243 isolate Bp 49-4 was tenfold less, suggesting an enhanced virulence of the isolate compared to the parent strain. As for the long-term survivors, Bp 60-2 was significantly more virulent than its parent strain MSHR668, i.e., its day 21 LD50 was 46-fold less than that of the parent strain (Table 6; Fig. 6a). The greater virulence of the isolate was also demonstrated by its greater potency at all lethal doses up to 90 % mortality (probability ≤0.9). In Fig. 6b, the lightly shaded area indicates the range of lethal doses of Bp 60-2 that were associated with significantly greater virulence compared to MSHR668. Larger doses of MSHR668 were required to produce the same lethality achieved with lower doses of isolate. Finally, even though the day 21 LD50 values of the parent Bm FMH strain and isolate Bm 60-5 were similar, the day 60 LD50 for isolate Bm 60-5, albeit not statistically significant, was almost fourfold less than that of parent strain (Table 6).
Table 6.
Statistical comparison of relative virulence for mice of Burkholderia strains and their in vivo isolates
| Strain | Day 21a | Day 60a | ||||
|---|---|---|---|---|---|---|
| LD50 | 95 % credible interval | LD50 | 95 % credible interval | |||
| Lower | Upper | Lower | Upper | |||
| Bp MSHR668 | 3.9 × 102 | 1.7 × 102 | 8.6 × 102 | 0.3 | 0 | 1.6 × 101 |
| Bp 41-1 | 4.7 × 102 | 1.2 × 102 | 2.2 × 103 | 5.1 | 0.1 | 3.8 × 101 |
| Bp 60-2 | 8.4 b | 0.7 | 4.3 × 101 | 3.4 | 0.5 | 1.2 × 101 |
| Bp K96243 | 2.3 × 104 | 1.2 × 104 | 4.7 × 104 | 1.9 × 103 | 4.7 × 102 | 6.1 × 103 |
| Bp 49-4 | 4.3 × 104 | 2.3 × 104 | 7.7 × 104 | 1.9 × 10 2c | 0.4 | 2.2 × 103 |
| Bp 1106a | 3.3 × 104 | 1.7 × 104 | 6.4 × 104 | 1.2 × 104 | 3.3 × 103 | 4.0 × 104 |
| Bp 62-3 | 4.7 × 104 | 1.9 × 104 | 1.2 × 105 | 1.2 × 104 | 7.0 × 103 | 2.0 × 104 |
| Bm FMH | 3.3 × 107 | 1.4 × 107 | 7.6 × 107 | 1.1 × 107 | 2.1 × 106 | 7.2 × 107 |
| Bm 60-5 | 2.1 × 107 | 1.2 × 107 | 3.7 × 107 | 2.9 × 10 6d | 7.7 × 105 | 1.2 × 107 |
aBALB/c mice were challenged by the IP route with a strain of Bp or Bm, or a spleen isolate of the strain, and the mice were monitored for morbidity and mortality for 60 days. The 21- and 60-day survival data were analyzed for each strain, and the data were evaluated statistically to compare parent strain and spleen derivative relative virulence potencies and LD50 values (no. CFU) by using Bayesian probit analysis, as described in the methods. The bolded LD50 values are those which were significantly different from the parent strain LD50 value
bThe day 21 LD50 value of Bp 60-2 was less than that of parent strain MSHR668 (probability ≥95 %) and the isolate exhibited greater potency at all lethal doses up to 90 % (probability of mortality ≤0.9)
cThe day 21 LD50 values of K96243 and its isolate Bp 49-4 were not significantly different. The day 60 LD50 of the isolate was about tenfold less than that of the parent, although not significantly different
dAlthough the day 60 LD50 for isolate Bm 60-5 was almost fourfold less than that of the parent strain, the values were not significantly different
Fig. 6.
Comparison of relative virulence for mice of Bp strain MSHR668 and spleen isolate Bp 60-2 collected on day 60 after infection. a Mice were challenged by the IP route with different doses of the bacteria, and the daily morbidity/mortality data through day 60 were used to determine the strain LD50 values. The isolate LD50 was significantly less than that of the parent (Table 6). b Statistical pairwise comparisons of relative potency of MSHR668 and its isolate at each lethality level along the dose–survival curve
Discussion
A number of in vitro assays were used to identify potential phenotypic markers for chronic Bp and Bm infection. Although a relatively small number of strains were fully characterized in vitro and over an extended period in vivo, the strains described represented a diversity of Bp from different patient sources and geographical locations to include the two foci that have been of predominant research concern: Thailand and Northern Australia. Characterization of bacteria isolated during the later stages of a long-term infection with Bp or Bm revealed frequent differences in all of the characterized phenotypes of the isolates compared to the original parent strains. The results of antimicrobial sensitivity assays commonly showed an increase in resistance in both Bm and Bp isolates. Whereas there was evidence for a positive association between development of increased resistance to particular antibiotics, chemicals, and AMPs in Bp isolates recovered at early compared to late infection times, there was no such trend clearly established with Bm isolates. In macrophage assays, infectivity/cytotoxicities of Bp isolates from mice with extended infections were greater than that of the parent. In contrast, Bm isolates were reduced in macrophage replication and cytotoxicity. In LPS phenotypic analysis, a potential loss of OPS expression in Bm was discovered during the course of infection. Finally, preliminary studies in a mouse model suggested that enhanced pathogenicity was associated with phenotypic variation of isolates from persistent infections.
Many host cells which provide a niche for persistent Burkholderia infection also possess activities and antibacterial products (e.g., AMPs and reactive oxygen and nitrogen species) important in innate immunity. Thus, intracellular pathogens would need to overcome such defenses. In our antimicrobial sensitivity assays, differences in the sensitivities to certain antibiotics, chemicals, and AMPs between isolates and parent Bp and Bm strains were often observed. Antimicrobial peptides are small cationic molecules which are part of the innate host defenses of many organisms. These AMPs are not specific for a particular target and their bactericidal activity usually involves membrane disruption (Andreu and Rivas 1998; Fox et al. 2012; Higashijima et al. 1988; Kreil 1973; Madhongsa et al. 2013; Skerlavaj et al. 1996; Steiner et al. 1981; Vila-Farres et al. 2012; Zasloff 2002, 1987). They are potential therapeutic agents for antibiotic-resistant organisms such as Bp and Bm (Kanthawong et al. 2009; Kreil 1973; Madhongsa et al. 2013). However, Burkholderia spp. have been reported to be relatively resistant to AMPs, although the sensitivity of Bp appears to be strain-specific (Fox et al. 2012; Kanthawong et al. 2009; Tandhavanant et al. 2010). For instance, notable sensitivity of Bp to the human cathelicidin peptide LL-37, or to hybrid peptides containing this AMP, has been observed (Fox et al. 2012; Kanthawong et al. 2009; Tandhavanant et al. 2010). Although the Bp and Bm strains used in this study were overall relatively resistant to AMPs, they were often more sensitive to some AMPs than were their spleen isolates (i.e., For mastoparan 7, BMAP-18, and magainin). Therefore, altered sensitivities of isolates to certain AMPs, such as mastoparan 7 and BMAP-18, could potentially be used as candidate markers for screening isolates from persistent Burkholderia infections. Nonetheless, and as expected, increased isolate resistance to specific AMPs was strain dependent, as demonstrated for 1026b and K96243.
Intracellular Bp has also been shown to evade or suppress production of reactive oxygen and nitrogen species, as well as other harsh chemicals (Chantratita et al. 2007; Dowling et al. 2010; Loprasert et al. 2003; Mulye et al. 2014; Tandhavanant et al. 2010; Utaisincharoen et al. 2001, 2004). In our study, Bp spleen isolates from three strains harbored sensitivities to sodium nitrite that differed from the parent. But whereas isolates of one strain were more resistant than the parent, those from two strains exhibited increased sensitivity. However, for most chemicals, the in vivo isolates displayed phenotypes of enhanced resistance, as exemplified by the responses to elevated NaCl concentrations, potassium tellurite, and variably to the surfactant Niaproof 4. Detergents such as Niaproof 4 often have opsonophagocytic activity and may play a role in innate immunity (Wright 1997, 2003). A direct association between time post-challenge and the number or extent of sensitivity differences exhibited by isolates was not always apparent but was suggested for not only several AMPs, but also the antibiotic nalidixic acid and potentially for Niaproof 4 and potassium tellurite, as described above. The isolate response data en toto suggested that extended maintenance of Bp in vivo may be facilitated by development of an increased resistance to antibacterial environments such as the macrophage phagolysosome.
As illustrated by our findings, the identification of infection-associated in vitro phenotypes is influenced by the phenotypic variability of the Burkholderia, and especially Bp, which is often associated with reversible epigenetic alterations (Chantratita et al. 2007, 2012; Velapatino et al. 2012; Vipond et al. 2013). The ability of Bp to produce different colony morphotypes is well known (Nicholls and Cantab 1930; Rogul and Carr 1972; Stanton et al. 1924) and potentially reflects adaptive changes that enhance fitness in a particular environment. Chantratita et al. (2007) observed that different Bp colony morphologies could be recovered from individual sites in patients with melioidosis. They identified and characterized seven colony morphologies. Type I could switch to the other six types in apparent response to environmental stresses such as iron limitation (Chantratita et al. 2007). Morphotypes I, II, and III were associated with differential ability to survive and persist in cell culture and mice and with resistance to peroxide and AMPs (Chantratita et al. 2007; Tandhavanant et al. 2010). Morphotypes associated with adaptive fitness differed significantly in the expression profiles of up- and down-regulated proteins (Chantratita et al. 2012). However, the finding that specific colony morphotypes are predictive of enhanced resistance to certain stresses may be strain related. Analyses of additional Bp strains revealed heterogeneity in responses between and within strains (Chantratita et al. 2007; Tandhavanant et al. 2010), and further analyses may be needed to confirm associations between in vitro phenotype differences and in vivo survival and persistence. More recently, Wikraiphat et al. (2015) described mucoid (M) and nonmucoid (NM) variants from the same sample which exhibited antigenically different OPS. The association between colony type switching and conformational differences in OPS was determined in analysis of several M/NM pairs from clinical samples under different in vitro growth conditions. Although the mechanism regulating colony/OPS variation and differential growth fitness is not entirely clear, the association between colony type and OPS modification might be the basis of the colony variation described previously (Nicholls and Cantab 1930; Srinivasan et al. 2001). As reported by Austin et al. (2015), Bp strain K96243 exhibited three colony variants which differed not only in colony morphology but also in other in vitro phenotypes. The isogenic colony types (one white and two yellow forms) were relatively stable but reversible, and they grew differentially in vitro under low oxygen or acidic environments. Only one type, albeit highly attenuated, could persist in the harsh conditions of the murine stomach and colonize the mucosa. A transcriptional regulator controlling colony variation was identified, though it is unknown whether this gene or a global master regulator controls this as well as other examples of reversible phenotypic switching in Bp. Lastly, Velapatino et al. (2012) analyzed proteins from Bp isolates recovered from a patient during the acute and relapse stages of infection. Colony Morphotype I (Chantratita et al. 2007; Wikraiphat et al. 2015) was recovered from the primary infection and an additional morphotype (III) was recovered during the relapse phase. Many proteins were differentially regulated for the relapse isolate compared to acute stage isolates, e.g., enhanced production of factors which detoxify the damaging RNI nitric oxide and proteins required for anaerobic growth. Prospective studies with Bp strains harboring mutations in relevant differentially expressed genes are needed to link expression with in vivo persistence. Strains of Burkholderia species other than Bp are also known to exhibit phenotypic variability. Romero et al. (2006) showed that in vitro passaging of Bm strains originally obtained from animal or human sources produces a diverse population of clones with alterations in mRNA levels of many genes. The changes in phenotypic expression were associated with insertions or deletions in DNA repeat regions which occurred rapidly. Such events may contribute to the instability of the Bm genome and enhance bacterial fitness. They might also partially explain the variability observed in this and other studies between and within Bp and Bm strains in colony morphology and in response to antimicrobial compounds (Austin et al. 2015; Chantratita et al. 2007; Tandhavanant et al. 2010; Velapatino et al. 2012; Wikraiphat et al. 2015). Members of the Burkholderia cepacia complex (BCC) exhibit colony morphology switching associated with their ability to colonize the lungs and cause severe infection in patients with cystic fibrosis (Bernier et al. 2008; Vial et al. 2010). For instance, phase switching by B. ambifaria resulted in alterations in colony morphology, biofilm formation, plant root colonization, and virulence. The data suggested that B. ambifaria adapts to the very different environments of patient lungs and the rhizosphere by reversible phase variation.
Phagocytic components of the cellular immune system are known to have a dual role in infection. They can serve both as critical components of host immune protection against Burkholderia infection and as facilitators of bacterial persistence in vivo (Galyov et al. 2010; Gregory and Waag 2008; Mulye et al. 2014; Valvano et al. 2005). For instance, depleting mice of macrophages has been shown to increase their susceptibility to melioidosis (Breitbach et al. 2006), and inducible nitrous acid synthase was essential for clearance of Bm from macrophages (Brett et al. 2008). These results suggest a role for macrophages in host protection. However, Bp and Bm produce virulence factors (secreted proteins of the T3SS and T6SS-1, BimA, and others) which allow the bacteria to escape the phagolysosome, multiply in the cytoplasm, and spread by direct cell-to-cell passage (Brett et al. 1994; Chuaygud et al. 2008; Dowling et al. 2010; Galyov et al. 2010; Gregory and Waag 2008; Kespichayawattana et al. 2000; Stevens et al. 2005a, b; Utaisincharoen et al. 2004; Valvano et al. 2005; Whitlock et al. 2008, 2009). Thus, the expression of numerous virulence factors which promote bacterial survival and inhibit cellular antibacterial mechanisms may contribute to the persistence of these agents in a relatively sheltered intracellular host niche (Butt et al. 2014).
Several studies have highlighted genetic and phenotypic characteristics potentially associated with persistence of Bp and Bm (Hayden et al. 2012; Price et al. 2013; Velapatino et al. 2012). Hayden et al. compared genome sequences of Bp strain pairs from patients consisting of the primary infecting strain and an isolate from relapsed infection occurring long after a successful response to antibiotic treatment (Hayden et al. 2012). They observed numerous genetic changes (indels, SNPs, and structural variations) for all the primary-relapse pairs. The only consistent alteration was in a homolog of the TetR family of global transcriptional regulators. At least one of the relapse isolates exhibited much higher MICs to several antibiotics relative to the primary strain. Their studies supported the hypothesis that this homolog and similar genes play a role in Bp adaptive survival in vivo. In studies with Bm, Romero et al. reported extensive genomic variability and rapid alterations in RNA expression in isolates obtained after passage in animal and human subjects (Romero et al. 2006). Price et al. also found significant genomic changes in Bp strains isolated almost 12 years apart from a chronically infected human survivor (Price et al. 2013). These changes appeared to mark an adaptive evolution that occurred over time and included inactivation of major virulence factor and regulatory genes required for environmental survival. This apparent reductive evolution appeared to mimic the process thought to have led to the genomic losses and conversion of Bm from a free-living microbe such as Bp into an obligate host pathogen.
In this study, although isolates of Bm FMH obtained early after infection displayed variable macrophage cytotoxicity and survival, most collected on day 60 were clearly reduced in both toxicity and intracellular growth. The basis for the attenuated phenotype is not known, but it conceivably reflects a shift in the infection toward a more chronic or subclinical intracellular form less prone to stimulate host immune responses. In contrast, of the five Bp strains for which colonies from mid- to long-term infections were available, isolates from three (MSHR668, 1106a, and less consistently, K96243) were more virulent for macrophages than the parent strain in their ability to multiply within or produce cytotoxic responses. However, the other Bp strains were more variable so the significance of the relationship between macrophage phenotypes and mouse virulence requires further study. It must be noted that macrophage cultures cannot replace or fully model pathogenesis, and animal models are needed for preliminary confirmation of significant findings obtained from cell culture. Furthermore, results using macrophage models may differ depending on the source of macrophage used and other factors. Whereas the results shown here and those of various other investigators employed murine-derived macrophage-like J774.A1 cells (Chantratita et al. 2007, 2012; Haussler et al. 2003; Kespichayawattana et al. 2000; Stevens et al. 2003; Tandhavanant et al. 2010; Wand et al. 2011; Welkos et al. 2015), others used cell lines such as murine RAW264.7 cells or human monocyte-like U937 cells (Arjcharoen et al. 2007; Brett et al. 2008; Burtnick et al. 2010, 2011; Chantratita et al. 2012; Pegoraro et al. 2014; Stevens et al. 2002; Tandhavanant et al. 2010; Utaisincharoen et al. 2000, 2004). Overall, we observed relatively reduced survival of Bm in J774.A1 macrophages compared to that of Bp, as reported previously (Brett et al. 2008). However, Bm generally appeared to be capable of greater J774.A1 macrophage survival and extensive cytotoxic activity at higher MOIs, yet these attributes were negatively impacted at higher MOIs in studies using a RAW264.7 model (Brett et al. 2008). The source of the macrophages and differences in model parameters likely explain some of the differences in results. Macrophage models are important tools in pathogenesis research with the caveat that they likely provide a model which is restricted to specific defined aspects of infection.
LPS differences can also be used as phenotypic markers, and previous studies suggest that Burkholderia LPS changes could be associated with chronic infection (Evans et al. 1999; Price et al. 2013). In this study, we provide evidence that Bm has the ability to lose OPS during the course of infection. This is intriguing because OPS modification and loss has been established as a hallmark of chronic Pseudomonas aeruginosa infection (Smith et al. 2006). In addition, rough Burkholderia cepacia strains lacking OPS have been associated with chronic infection in the lungs of cystic fibrosis patients (Evans et al. 1999). There are many possible reasons why losing OPS could benefit Burkholderia during infection. P. aeruginosa and Salmonella typhimurium mutants lacking OPS have been shown to increase outer membrane vesicle (OMV) formation (Salkinoja-Salonen and Nurmiaho 1978; Smit et al. 1975). OMVs are important virulence factor delivery mechanisms used by a wide variety of bacterial pathogens and can help the bacteria disseminate throughout the body (Kuehn and Kesty 2005). OMVs are also secreted by S. typhimurium when grown intracellularly (Garcia-del Portillo et al. 1997). In addition, once inside the macrophage, the bacteria may no longer have a need for OPS. Bp and Bm OPS has been shown to play a role in serum resistance (Brett et al. 2007; DeShazer et al. 1998). However, a mutation that disrupts OPS synthesis may not have a negative effect on bacterial survival once inside the macrophage because the bacteria are protected from serum. The loss of OPS could also play a role in evading the host immune response during relapse of an infection. OPS is highly immunogenic, and antibodies are produced by the host to recognize OPS from specific bacterial species, including Bp and Bm (Ho et al. 1997; Trevino et al. 2006). The loss of OPS could be highly advantageous during relapse of infection when the bacteria exit the macrophage, as the immune system would have a delayed response in recognizing the bacteria. Due to these possible implications, it is clear that the potential role of OPS in Burkholderia chronic infection warrants further investigation. Future studies include sequencing the OPS biosynthetic gene cluster of Bm FMH 60-3 to identify the genetic basis for the loss of OPS. In addition, the LPS of isolates from more well-established chronic Bp and Bm mouse models will be examined to look for additional instances of LPS changes during the course of infection. Rough isolates/strains will also be passaged through mice to determine the effect of OPS loss on virulence.
In initial attempts to detect a relationship between altered phenotypes and pathogenicity, the mouse virulence of Bp/Bm strains and their respective isolates was compared. The challenge data for three strains and their in vivo isolates suggested that bacteria isolated from persistently infected animals can differ significantly from the original strain in virulence, as indicated by LD50 and lethal dose potency comparisons. This was clearly demonstrated with one isolate (MSHR668 isolate 60-2) and potentially for the isolates of Bp strain K96243 and Bm strain FMH. Taken together, the data suggest that some isolates from mice persistently infected with certain Burkholderia strains might be enhanced in virulence compared to the parent and that this enhanced pathogenicity was associated with phenotypic variation. It was interesting that isolates of two of the three Bp strains (MSHR668 and K96243), with a tendency for enhanced survival and cytotoxicity for macrophages, were also more virulent for mice. The inverse was observed for the day 60 Bm isolate 60-5 which was more virulent for mice but, as shown also for Bm isolate 60-3, proliferated less and/or was less cytotoxic for macrophages. Perhaps the different results of Bp and Bm in vivo isolates in macrophages are associated with the lesser ability of Bm to destroy the host cell (and lose an in vivo niche) compared to Bp in the J774.A1 macrophage model. Further investigations are obviously needed. However, it may be possible to take advantage of several of the measurable aspects of the bacterial-macrophage interaction to show that a significant change (increase/reduction) in a parameter of infection, such as a change in the proportion of MNGCs, represents a potential marker to signal a conversion of a relatively acute to more persistent stage in the pathogenesis of infection.
It is not unexpected that the virulence of the in vivo isolates often appeared to be enhanced compared to that of the parent strain. The laboratory culture of pathogenic bacterial strains has been shown to result in the accumulation of genetic changes and thus genetically mixed cultures which can impact virulence. Genomic plasticity and instability has been extensively characterized not only for Bp and Bm but also for Yersinia pestis (Buchrieser et al. 1998; Galyov et al. 2010; Perry and Fetherston 1997; Romero et al. 2006; Sahl et al. 2015; Tumapa et al. 2008). The passage of nonuniform cultures of some pathogens such as Y. pestis and Legionella pneumophila in vivo selects for the most fit clones and has been used to increase genetic uniformity and restore virulence (Bozue et al. 2011; Cirillo et al. 1999; Welkos et al. 1997). This has also been shown or suggested more recently for Bp and Bm (Chen et al. 2014; Inglis et al. 2000; Waag and DeShazer 2004). However, the Burkholderia appear to exploit their capacity for genomic variability by generating diverse populations which accumulate in vivo in response to differing host environments (Price et al. 2010, 2013; Romero et al. 2006). Thus, the impact of in vivo passage on the Burkholderia could be complex and strain or host dependent.
Overall, this study has identified potentially useful and often specific phenotypic markers of long-term Burkholderia infection in several different phenotypic classes (morphotype, antimicrobial sensitivities, macrophage infection, LPS, and murine virulence). Additional studies are needed to confirm the consistency and range of these phenotypes in chronic infection models and examine the potential role for them in establishment of a chronic infection. However, in vivo residence was clearly associated with changes in resistance and responses to many potential host antibacterial defenses. The wide diversity of the antibacterial chemicals and other stressors impacted by these altered Bp responses suggests that the host environmental factors and/or bacterial switch mechanisms driving a conversion to resistance are multifactorial.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Comparison of macrophage survival of Bp strain 1106a and mouse spleen isolate Bp 14-1. J774.A1 cells were inoculated with MOIs of 16.6 and 11.0, respectively, and incubated for 1 h. The data shown are the mean viable counts (triplicate wells) recovered after the 1-h uptake, incubation of the infected cells in the presence of kanamycin for 2 h (3 h), and after incubation for a total of 8 h. The isolate had been obtained from the spleen of a mouse surviving IP challenge with 1106 s and euthanized on day 14. The viable counts recovered from the isolate-infected cells were greater than those of the parent strain at all three time points (p ≤ 0.0013) (TIFF 133 kb)
Acknowledgments
We thank Drs. Joel Bozue and David DeShazer for their valuable comments and review of this manuscript. The expert technical assistance of Titilayo Omotade and Jennifer Dankmeyer is appreciated. We also thank Sylvia Trevino for kindly providing antibodies and animal samples. Opinions, interpretations, conclusions, and recommendations are those of the authors and are not necessarily endorsed by the US Army. Funding was provided by Joint Science and Technology Office-Chemical and Biological Defense/Defense Threat Reduction Agency CCAR# CB3846 PPE-1 Burkholderia.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Amemiya K, Dankmeyer JL, Fetterer DP, Worsham PL, Welkos SL, Cote CK. Comparison of the early host immune response to two widely diverse virulent strains of Burkholderia pseudomallei that cause acute or chronic infections in BALB/c mice. Microb Pathog. 2015;86:53–63. doi: 10.1016/j.micpath.2015.07.004. [DOI] [PubMed] [Google Scholar]
- Andreu D, Rivas L. Animal antimicrobial peptides: an overview. Biopolymers. 1998;47:415–433. doi: 10.1002/(SICI)1097-0282(1998)47:6<415::AID-BIP2>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Arjcharoen S, Wikraiphat C, Pudla M, Limposuwan K, Woods DE, Sirisinha S, Utaisincharoen P. Fate of a Burkholderia pseudomallei lipopolysaccharide mutant in the mouse macrophage cell line RAW 264.7: possible role for the O-antigenic polysaccharide moiety of lipopolysaccharide in internalization and intracellular survival. Infect Immun. 2007;75:4298–4304. doi: 10.1128/IAI.00285-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashdown LR. An improved screening technique for isolation of Pseudomonas pseudomallei from clinical specimens. Pathology. 1979;11:293–297. doi: 10.3109/00313027909061954. [DOI] [PubMed] [Google Scholar]
- Austin CR, Goodyear AW, Bartek IL, Stewart A, Sutherland MD, Silva EB, Zweifel A, Vitko NP, Tuanyok A, Highnam G, Mittelman D, Keim P, Schweizer HP, Vazquez-Torres A, Dow SW, Voskuil MI. A Burkholderia pseudomallei colony variant necessary for gastric colonization. MBio. 2015 doi: 10.1128/mBio.02462-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernier SP, Nguyen DT, Sokol PA. A LysR-type transcriptional regulator in Burkholderia cenocepacia influences colony morphology and virulence. Infect Immun. 2008;76:38–47. doi: 10.1128/IAI.00874-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozue J, Mou S, Moody KL, Cote CK, Trevino S, Fritz D, Worsham P. The role of the phoPQ operon in the pathogenesis of the fully virulent CO92 strain of Yersinia pestis and the IP32953 strain of Yersinia pseudotuberculosis. Microb Pathog. 2011;50:314–321. doi: 10.1016/j.micpath.2011.02.005. [DOI] [PubMed] [Google Scholar]
- Breitbach K, Klocke S, Tschernig T, van Rooijen N, Baumann U, Steinmetz I. Role of inducible nitric oxide synthase and NADPH oxidase in early control of Burkholderia pseudomallei infection in mice. Infect Immun. 2006;74:6300–6309. doi: 10.1128/IAI.00966-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brett PJ, Mah DC, Woods DE. Isolation and characterization of Pseudomonas pseudomallei flagellin proteins. Infect Immun. 1994;62:1914–1919. doi: 10.1128/iai.62.5.1914-1919.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brett PJ, Burtnick MN, Snyder DS, Shannon JG, Azadi P, Gherardini FC. Burkholderia mallei expresses a unique lipopolysaccharide mixture that is a potent activator of human Toll-like receptor 4 complexes. Mol Microbiol. 2007;63:379–390. doi: 10.1111/j.1365-2958.2006.05519.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brett PJ, Burtnick MN, Su H, Nair V, Gherardini FC. iNOS activity is critical for the clearance of Burkholderia mallei from infected RAW 264.7 murine macrophages. Cell Microbiol. 2008;10:487–498. doi: 10.1111/j.1462-5822.2007.01063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchrieser C, Brosch R, Bach S, Guiyoule A, Carniel E. The high-pathogenicity island of Yersinia pseudotuberculosis can be inserted into any of the three chromosomal asn tRNA genes. Mol Microbiol. 1998;30:965–978. doi: 10.1046/j.1365-2958.1998.01124.x. [DOI] [PubMed] [Google Scholar]
- Burtnick MN, DeShazer D, Nair V, Gherardini FC, Brett PJ. Burkholderia mallei cluster 1 type VI secretion mutants exhibit growth and actin polymerization defects in RAW 264.7 murine macrophages. Infect Immun. 2010;78:88–99. doi: 10.1128/IAI.00985-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burtnick MN, Brett PJ, Harding SV, Ngugi SA, Ribot WJ, Chantratita N, Scorpio A, Milne TS, Dean RE, Fritz DL, Peacock SJ, Prior JL, Atkins TP, Deshazer D. The cluster 1 type VI secretion system is a major virulence determinant in Burkholderia pseudomallei. Infect Immun. 2011;79:1512–1525. doi: 10.1128/IAI.01218-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt A, Higman VA, Williams C, Crump MP, Hemsley CM, Harmer N, Titball RW. The HicA toxin from Burkholderia pseudomallei has a role in persister cell formation. Biochem J. 2014;459:333–344. doi: 10.1042/BJ20140073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chantratita N, Wuthiekanun V, Boonbumrung K, Tiyawisutsri R, Vesaratchavest M, Limmathurotsakul D, Chierakul W, Wongratanacheewin S, Pukritiyakamee S, White NJ, Day NP, Peacock SJ. Biological relevance of colony morphology and phenotypic switching by Burkholderia pseudomallei. J Bacteriol. 2007;189:807–817. doi: 10.1128/JB.01258-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chantratita N, Tandhavanant S, Wikraiphat C, Trunck LA, Rholl DA, Thanwisai A, Saiprom N, Limmathurotsakul D, Korbsrisate S, Day NP, Schweizer HP, Peacock SJ. Proteomic analysis of colony morphology variants of Burkholderia pseudomallei defines a role for the arginine deiminase system in bacterial survival. J Proteomics. 2012;75:1031–1042. doi: 10.1016/j.jprot.2011.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YS, Shieh WJ, Goldsmith CS, Metcalfe MG, Greer PW, Zaki SR, Chang HH, Chan H, Chen YL. Alteration of the phenotypic and pathogenic patterns of Burkholderia pseudomallei that persist in a soil environment. Am J Trop Med Hyg. 2014;90:469–479. doi: 10.4269/ajtmh.13-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho MH, Shin YW, Chun JH, Hong KJ, Na BK, Rhie GE, Seong BL, Yoo CK. Expression and characterization of an iron-containing superoxide dismutase from Burkholderia pseudomallei. J Microbiol. 2012;50:1029–1033. doi: 10.1007/s12275-012-2267-2. [DOI] [PubMed] [Google Scholar]
- Chuaygud T, Tungpradabkul S, Sirisinha S, Chua KL, Utaisincharoen P. A role of Burkholderia pseudomallei flagella as a virulent factor. Trans R Soc Trop Med Hyg. 2008;102(Suppl 1):S140–S144. doi: 10.1016/S0035-9203(08)70031-2. [DOI] [PubMed] [Google Scholar]
- Cirillo JD, Cirillo SL, Yan L, Bermudez LE, Falkow S, Tompkins LS. Intracellular growth in Acanthamoeba castellanii affects monocyte entry mechanisms and enhances virulence of Legionella pneumophila. Infect Immun. 1999;67:4427–4434. doi: 10.1128/iai.67.9.4427-4434.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie BJ, Fisher DA, Anstey NM, Jacups SP. Melioidosis: acute and chronic disease, relapse and re-activation. Trans R Soc Trop Med Hyg. 2000;94:301–304. doi: 10.1016/S0035-9203(00)90333-X. [DOI] [PubMed] [Google Scholar]
- Currie BJ, Ward L, Cheng AC. The epidemiology and clinical spectrum of melioidosis: 540 cases from the 20 year Darwin prospective study. PLoS Negl Trop Dis. 2010;4:e900. doi: 10.1371/journal.pntd.0000900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dance D. Treatment and prophylaxis of melioidosis. Int J Antimicrob Agents. 2014;43:310–318. doi: 10.1016/j.ijantimicag.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dance DA, Wuthiekanun V, Chaowagul W, Suputtamongkol Y, White NJ. Development of resistance to ceftazidime and co-amoxiclav in Pseudomonas pseudomallei. J Antimicrob Chemother. 1991;28:321–324. doi: 10.1093/jac/28.2.321. [DOI] [PubMed] [Google Scholar]
- DeShazer D, Brett PJ, Woods DE. The type II O-antigenic polysaccharide moiety of Burkholderia pseudomallei lipopolysaccharide is required for serum resistance and virulence. Mol Microbiol. 1998;30:1081–1100. doi: 10.1046/j.1365-2958.1998.01139.x. [DOI] [PubMed] [Google Scholar]
- Dowling AJ, Wilkinson PA, Holden MT, Quail MA, Bentley SD, Reger J, Waterfield NR, Titball RW, Ffrench-Constant RH. Genome-wide analysis reveals loci encoding anti-macrophage factors in the human pathogen Burkholderia pseudomallei K96243. PLoS ONE. 2010;5:e15693. doi: 10.1371/journal.pone.0015693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison RT, 3rd, Giehl TJ. Killing of gram-negative bacteria by lactoferrin and lysozyme. J Clin Invest. 1991;88:1080–1091. doi: 10.1172/JCI115407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans E, Poxton IR, Govan JR. Lipopolysaccharide chemotypes of Burkholderia cepacia. J Med Microbiol. 1999;48:825–832. doi: 10.1099/00222615-48-9-825. [DOI] [PubMed] [Google Scholar]
- Fauvart M, De Groote VN, Michiels J. Role of persister cells in chronic infections: clinical relevance and perspectives on anti-persister therapies. J Med Microbiol. 2011;60:699–709. doi: 10.1099/jmm.0.030932-0. [DOI] [PubMed] [Google Scholar]
- Fox MA, Thwaite JE, Ulaeto DO, Atkins TP, Atkins HS. Design and characterization of novel hybrid antimicrobial peptides based on cecropin A, LL-37 and magainin II. Peptides. 2012;33:197–205. doi: 10.1016/j.peptides.2012.01.013. [DOI] [PubMed] [Google Scholar]
- Fritz DL, Waag DM. Glanders. In: Swearengen JR, editor. Biodefense research methodology and animal models. Boca Raton: CRC Press; 2012. pp. 99–111. [Google Scholar]
- Fritz DL, Vogel P, Brown DR, Deshazer D, Waag DM. Mouse model of sublethal and lethal intraperitoneal glanders (Burkholderia mallei) Vet Pathol. 2000;37:626–636. doi: 10.1354/vp.37-6-626. [DOI] [PubMed] [Google Scholar]
- Galyov EE, Brett PJ, DeShazer D. Molecular insights into Burkholderia pseudomallei and Burkholderia mallei pathogenesis. Annu Rev Microbiol. 2010;64:495–517. doi: 10.1146/annurev.micro.112408.134030. [DOI] [PubMed] [Google Scholar]
- Garcia-del Portillo F, Stein MA, Finlay BB. Release of lipopolysaccharide from intracellular compartments containing Salmonella typhimurium to vesicles of the host epithelial cell. Infect Immun. 1997;65:24–34. doi: 10.1128/iai.65.1.24-34.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodyear A, Bielefeldt-Ohmann H, Schweizer H, Dow S. Persistent gastric colonization with Burkholderia pseudomallei and dissemination from the gastrointestinal tract following mucosal inoculation of mice. PLoS ONE. 2012;7:e37324. doi: 10.1371/journal.pone.0037324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory BC, Waag DM. Glanders. In: Bozue J, Cote C, Glass P, editors. Textbook of military medicine: medical aspect of biological warfare. Washington: Borden Institute; 2008. pp. 121–146. [Google Scholar]
- Hamad MA, Austin CR, Stewart AL, Higgins M, Vazquez-Torres A, Voskuil MI. Adaptation and antibiotic tolerance of anaerobic Burkholderia pseudomallei. Antimicrob Agents Chemother. 2011;55:3313–3323. doi: 10.1128/AAC.00953-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haussler S, Rohde M, von Neuhoff N, Nimtz M, Steinmetz I. Structural and functional cellular changes induced by Burkholderia pseudomallei rhamnolipid. Infect Immun. 2003;71:2970–2975. doi: 10.1128/IAI.71.5.2970-2975.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden HS, Lim R, Brittnacher MJ, Sims EH, Ramage ER, Fong C, Wu Z, Crist E, Chang J, Zhou Y, Radey M, Rohmer L, Haugen E, Gillett W, Wuthiekanun V, Peacock SJ, Kaul R, Miller SI, Manoil C, Jacobs MA. Evolution of Burkholderia pseudomallei in recurrent melioidosis. PLoS ONE. 2012;7:e36507. doi: 10.1371/journal.pone.0036507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashijima T, Uzu S, Nakajima T, Ross EM. Mastoparan, a peptide toxin from wasp venom, mimics receptors by activating GTP-binding regulatory proteins (G proteins) J Biol Chem. 1988;263:6491–6494. [PubMed] [Google Scholar]
- Ho M, Schollaardt T, Smith MD, Perry MB, Brett PJ, Chaowagul W, Bryan LE. Specificity and functional activity of anti-Burkholderia pseudomallei polysaccharide antibodies. Infect Immun. 1997;65:3648–3653. doi: 10.1128/iai.65.9.3648-3653.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglis TJ, Rigby P, Robertson TA, Dutton NS, Henderson M, Chang BJ. Interaction between Burkholderia pseudomallei and Acanthamoeba species results in coiling phagocytosis, endamebic bacterial survival, and escape. Infect Immun. 2000;68:1681–1686. doi: 10.1128/IAI.68.3.1681-1686.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeddeloh JA, Fritz DL, Waag DM, Hartings JM, Andrews GP. Biodefense-driven murine model of pneumonic melioidosis. Infect Immun. 2003;71:584–587. doi: 10.1128/IAI.71.1.584-587.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanthawong S, Nazmi K, Wongratanacheewin S, Bolscher JG, Wuthiekanun V, Taweechaisupapong S. In vitro susceptibility of Burkholderia pseudomallei to antimicrobial peptides. Int J Antimicrob Agents. 2009;34:309–314. doi: 10.1016/j.ijantimicag.2009.05.012. [DOI] [PubMed] [Google Scholar]
- Keren I, Kaldalu N, Spoering A, Wang Y, Lewis K. Persister cells and tolerance to antimicrobials. FEMS Microbiol Lett. 2004;230:13–18. doi: 10.1016/S0378-1097(03)00856-5. [DOI] [PubMed] [Google Scholar]
- Kespichayawattana W, Rattanachetkul S, Wanun T, Utaisincharoen P, Sirisinha S. Burkholderia pseudomallei induces cell fusion and actin-associated membrane protrusion: a possible mechanism for cell-to-cell spreading. Infect Immun. 2000;68:5377–5384. doi: 10.1128/IAI.68.9.5377-5384.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kint CI, Verstraeten N, Fauvart M, Michiels J. New-found fundamentals of bacterial persistence. Trends Microbiol. 2012;20:577–585. doi: 10.1016/j.tim.2012.08.009. [DOI] [PubMed] [Google Scholar]
- Kreil G. Biosynthesis of melittin, a toxic peptide from bee venom. Amino-acid sequence of the precursor. Eur J Biochem. 1973;33:558–566. doi: 10.1111/j.1432-1033.1973.tb02716.x. [DOI] [PubMed] [Google Scholar]
- Kuehn MJ, Kesty NC. Bacterial outer membrane vesicles and the host-pathogen interaction. Genes Dev. 2005;19:2645–2655. doi: 10.1101/gad.1299905. [DOI] [PubMed] [Google Scholar]
- Loprasert S, Sallabhan R, Whangsuk W, Mongkolsuk S. Compensatory increase in ahpC gene expression and its role in protecting Burkholderia pseudomallei against reactive nitrogen intermediates. Arch Microbiol. 2003;180:498–502. doi: 10.1007/s00203-003-0621-9. [DOI] [PubMed] [Google Scholar]
- Madhongsa K, Pasan S, Phophetleb O, Nasompag S, Thammasirirak S, Daduang S, Taweechaisupapong S, Lomize AL, Patramanon R. Antimicrobial action of the cyclic peptide bactenecin on Burkholderia pseudomallei correlates with efficient membrane permeabilization. PLoS Negl Trop Dis. 2013;7:e2267. doi: 10.1371/journal.pntd.0002267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulye M, Bechill MP, Grose W, Ferreira VP, Lafontaine ER, Wooten RM. Delineating the importance of serum opsonins and the bacterial capsule in affecting the uptake and killing of Burkholderia pseudomallei by murine neutrophils and macrophages. PLoS Negl Trop Dis. 2014;8:e2988. doi: 10.1371/journal.pntd.0002988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naha K, Dasari S, Kusugodlu R, Prabhu M. Cranial melioidosis with extradural extension after a fall in the bathroom. Australas Med J. 2012;5:455–458. doi: 10.4066/AMJ.2012.1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naha K, Shastry BA, Saravu K. Colonization or spontaneous resolution: expanding the role for Burkholderia pseudomallei. Asian Pac J Trop Med. 2014;7:250–252. doi: 10.1016/S1995-7645(14)60031-6. [DOI] [PubMed] [Google Scholar]
- Nicholls L, Cantab BC. Melioidosis with special reference to the dissociation of Bacillus whittori. Br J Exp Pathol. 1930;XI:393–399. [Google Scholar]
- Nigg C. Serologic studies on subclinical melioidosis. J Immunol. 1963;91:18–28. [PubMed] [Google Scholar]
- Pegoraro G, Eaton BP, Ulrich RL, Lane DJ, Ojeda JF, Bavari S, DeShazer D, Panchal RG. A high-content imaging assay for the quantification of the Burkholderia pseudomallei induced multinucleated giant cell (MNGC) phenotype in murine macrophages. BMC Microbiol. 2014;14:98. doi: 10.1186/1471-2180-14-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry RD, Fetherston JD. Yersinia pestis—etiologic agent of plague. Clin Microbiol Rev. 1997;10:35–66. doi: 10.1128/cmr.10.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price EP, Hornstra HM, Limmathurotsakul D, Max TL, Sarovich DS, Vogler AJ, Dale JL, Ginther JL, Leadem B, Colman RE, Foster JT, Tuanyok A, Wagner DM, Peacock SJ, Pearson T, Keim P. Within-host evolution of Burkholderia pseudomallei in four cases of acute melioidosis. PLoS Pathog. 2010;6:e1000725. doi: 10.1371/journal.ppat.1000725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price EP, Sarovich DS, Mayo M, Tuanyok A, Drees KP, Kaestli M, Beckstrom-Sternberg SM, Babic-Sternberg JS, Kidd TJ, Bell SC, Keim P, Pearson T, Currie BJ. Within-host evolution of Burkholderia pseudomallei over a twelve-year chronic carriage infection. MBio. 2013 doi: 10.1128/mBio.00388-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pumpuang A, Chantratita N, Wikraiphat C, Saiprom N, Day NP, Peacock SJ, Wuthiekanun V. Survival of Burkholderia pseudomallei in distilled water for 16 years. Trans R Soc Trop Med Hyg. 2011;105:598–600. doi: 10.1016/j.trstmh.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogul M, Carr SR. Variable ammonia production among smooth and rough strains of Pseudomonas pseudomallei: resemblance to bacteriocin production. J Bacteriol. 1972;112:372–380. doi: 10.1128/jb.112.1.372-380.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero CM, DeShazer D, Feldblyum T, Ravel J, Woods D, Kim HS, Yu Y, Ronning CM, Nierman WC. Genome sequence alterations detected upon passage of Burkholderia mallei ATCC 23344 in culture and in mammalian hosts. BMC Genomics. 2006;7:228. doi: 10.1186/1471-2164-7-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahl JW, Allender CJ, Colman RE, Califf KJ, Schupp JM, Currie BJ, Van Zandt KE, Gelhaus HC, Keim P, Tuanyok A. Genomic characterization of Burkholderia pseudomallei isolates selected for medical countermeasures testing: comparative genomics associated with differential virulence. PLoS ONE. 2015;10:e0121052. doi: 10.1371/journal.pone.0121052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salkinoja-Salonen M, Nurmiaho EL. The effect of lipopolysaccharide composition on the ultrastructure of Pseudomonas aeruginosa. J Gen Microbiol. 1978;105:23–28. doi: 10.1099/00221287-105-1-119. [DOI] [PubMed] [Google Scholar]
- SD Team (2013) Atna: a C++ library for probability and sampling
- SD Team (2014) RStan: the R interface to Stan
- Skerlavaj B, Gennaro R, Bagella L, Merluzzi L, Risso A, Zanetti M. Biological characterization of two novel cathelicidin-derived peptides and identification of structural requirements for their antimicrobial and cell lytic activities. J Biol Chem. 1996;271:28375–28381. doi: 10.1074/jbc.271.45.28375. [DOI] [PubMed] [Google Scholar]
- Smit J, Kamio Y, Nikaido H. Outer membrane of Salmonella typhimurium: chemical analysis and freeze-fracture studies with lipopolysaccharide mutants. J Bacteriol. 1975;124:942–958. doi: 10.1128/jb.124.2.942-958.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EE, Buckley DG, Wu Z, Saenphimmachak C, Hoffman LR, D’Argenio DA, Miller SI, Ramsey BW, Speert DP, Moskowitz SM, Burns JL, Kaul R, Olson MV. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc Natl Acad Sci USA. 2006;103:8487–8492. doi: 10.1073/pnas.0602138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol PA, Ohman DE, Iglewski BH. A more sensitive plate assay for detection of protease production by Pseudomonas aeruginosa. J Clin Microbiol. 1979;9:538–540. doi: 10.1128/jcm.9.4.538-540.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan A, Kraus CN, DeShazer D, Becker PM, Dick JD, Spacek L, Bartlett JG, Byrne WR, Thomas DL. Glanders in a military research microbiologist. N Engl J Med. 2001;345:256–258. doi: 10.1056/NEJM200107263450404. [DOI] [PubMed] [Google Scholar]
- Stanton AT, Fletcher W, Kanagarayer K. Two cases of melioidosis. J Hyg (Lond) 1924;23(268–276):267. doi: 10.1017/s0022172400034197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner H, Hultmark D, Engstrom A, Bennich H, Boman HG. Sequence and specificity of two antibacterial proteins involved in insect immunity. Nature. 1981;292:246–248. doi: 10.1038/292246a0. [DOI] [PubMed] [Google Scholar]
- Stevens MP, Wood MW, Taylor LA, Monaghan P, Hawes P, Jones PW, Wallis TS, Galyov EE. An Inv/Mxi-Spa-like type III protein secretion system in Burkholderia pseudomallei modulates intracellular behaviour of the pathogen. Mol Microbiol. 2002;46:649–659. doi: 10.1046/j.1365-2958.2002.03190.x. [DOI] [PubMed] [Google Scholar]
- Stevens MP, Friebel A, Taylor LA, Wood MW, Brown PJ, Hardt WD, Galyov EE. A Burkholderia pseudomallei type III secreted protein, BopE, facilitates bacterial invasion of epithelial cells and exhibits guanine nucleotide exchange factor activity. J Bacteriol. 2003;185:4992–4996. doi: 10.1128/JB.185.16.4992-4996.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens JM, Ulrich RL, Taylor LA, Wood MW, Deshazer D, Stevens MP, Galyov EE. Actin-binding proteins from Burkholderia mallei and Burkholderia thailandensis can functionally compensate for the actin-based motility defect of a Burkholderia pseudomallei bimA mutant. J Bacteriol. 2005;187:7857–7862. doi: 10.1128/JB.187.22.7857-7862.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens MP, Stevens JM, Jeng RL, Taylor LA, Wood MW, Hawes P, Monaghan P, Welch MD, Galyov EE. Identification of a bacterial factor required for actin-based motility of Burkholderia pseudomallei. Mol Microbiol. 2005;56:40–53. doi: 10.1111/j.1365-2958.2004.04528.x. [DOI] [PubMed] [Google Scholar]
- Tandhavanant S, Thanwisai A, Limmathurotsakul D, Korbsrisate S, Day NP, Peacock SJ, Chantratita N. Effect of colony morphology variation of Burkholderia pseudomallei on intracellular survival and resistance to antimicrobial environments in human macrophages in vitro. BMC Microbiol. 2010;10:303. doi: 10.1186/1471-2180-10-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarlow MJ, Lloyd J. Melioidosis and chronic granulomatous disease. Proc R Soc Med. 1971;64:19–20. doi: 10.1177/003591577106400111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevino SR, Permenter AR, England MJ, Parthasarathy N, Gibbs PH, Waag DM, Chanh TC. Monoclonal antibodies passively protect BALB/c mice against Burkholderia mallei aerosol challenge. Infect Immun. 2006;74:1958–1961. doi: 10.1128/IAI.74.3.1958-1961.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CM, Frasch CE. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal Biochem. 1982;119:115–119. doi: 10.1016/0003-2697(82)90673-X. [DOI] [PubMed] [Google Scholar]
- Tumapa S, Holden MT, Vesaratchavest M, Wuthiekanun V, Limmathurotsakul D, Chierakul W, Feil EJ, Currie BJ, Day NP, Nierman WC, Peacock SJ. Burkholderia pseudomallei genome plasticity associated with genomic island variation. BMC Genomics. 2008;9:190. doi: 10.1186/1471-2164-9-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utaisincharoen P, Tangthawornchaikul N, Kespichayawattana W, Anuntagool N, Chaisuriya P, Sirisinha S. Kinetic studies of the production of nitric oxide (NO) and tumour necrosis factor-alpha (TNF-alpha) in macrophages stimulated with Burkholderia pseudomallei endotoxin. Clin Exp Immunol. 2000;122:324–329. doi: 10.1046/j.1365-2249.2000.01386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utaisincharoen P, Tangthawornchaikul N, Kespichayawattana W, Chaisuriya P, Sirisinha S. Burkholderia pseudomallei interferes with inducible nitric oxide synthase (iNOS) production: a possible mechanism of evading macrophage killing. Microbiol Immunol. 2001;45:307–313. doi: 10.1111/j.1348-0421.2001.tb02623.x. [DOI] [PubMed] [Google Scholar]
- Utaisincharoen P, Anuntagool N, Arjcharoen S, Limposuwan K, Chaisuriya P, Sirisinha S. Induction of iNOS expression and antimicrobial activity by interferon (IFN)-beta is distinct from IFN-gamma in Burkholderia pseudomallei-infected mouse macrophages. Clin Exp Immunol. 2004;136:277–283. doi: 10.1111/j.1365-2249.2004.02445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valvano MA, Keith KE, Cardona ST. Survival and persistence of opportunistic Burkholderia species in host cells. Curr Opin Microbiol. 2005;8:99–105. doi: 10.1016/j.mib.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Velapatino B, Limmathurotsakul D, Peacock SJ, Speert DP. Identification of differentially expressed proteins from Burkholderia pseudomallei isolated during primary and relapsing melioidosis. Microbes Infect. 2012;14:335–340. doi: 10.1016/j.micinf.2011.11.011. [DOI] [PubMed] [Google Scholar]
- Vial L, Groleau MC, Lamarche MG, Filion G, Castonguay-Vanier J, Dekimpe V, Daigle F, Charette SJ, Deziel E. Phase variation has a role in Burkholderia ambifaria niche adaptation. ISME J. 2010;4:49–60. doi: 10.1038/ismej.2009.95. [DOI] [PubMed] [Google Scholar]
- Vidyalakshmi K, Chakrapani M, Shrikala B, Damodar S, Lipika S, Vishal S. Tuberculosis mimicked by melioidosis. Int J Tuberc Lung Dis. 2008;12:1209–1215. [PubMed] [Google Scholar]
- Vila-Farres X, Garcia de la Maria C, Lopez-Rojas R, Pachon J, Giralt E, Vila J. In vitro activity of several antimicrobial peptides against colistin-susceptible and colistin-resistant Acinetobacter baumannii. Clin Microbiol Infect. 2012;18:383–387. doi: 10.1111/j.1469-0691.2011.03581.x. [DOI] [PubMed] [Google Scholar]
- Vipond J, Kane J, Hatch G, McCorrison J, Nierman WC, Losada L. Sequence determination of Burkholderia pseudomallei strain NCTC 13392 colony morphology variants. Genome Announc. 2013 doi: 10.1128/genomeA.00925-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waag DM, DeShazer D. Glanders: new insights into an old disease. In: Lebeda FJ, Lindler E, Korch GW, Totowa NJ, editors. Infectious diseases: biological weapons defense: infectious diseases and counterterrorism. New York: Humana Press; 2004. pp. 209–238. [Google Scholar]
- Wand ME, Muller CM, Titball RW, Michell SL. Macrophage and Galleria mellonella infection models reflect the virulence of naturally occurring isolates of B. pseudomallei, B. thailandensis and B. oklahomensis. BMC Microbiol. 2011;11:11. doi: 10.1186/1471-2180-11-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welkos SL, Friedlander AM, Davis KJ. Studies on the role of plasminogen activator in systemic infection by virulent Yersinia pestis strain C092. Microb Pathog. 1997;23:211–223. doi: 10.1006/mpat.1997.0154. [DOI] [PubMed] [Google Scholar]
- Welkos SL, Klimko CP, Kern SJ, Bearss JJ, Bozue JA, Bernhards RC, Trevino SR, Waag DM, Amemiya K, Worsham PL, Cote CK. Characterization of Burkholderia pseudomallei strains using a murine intraperitoneal infection model and in vitro macrophage assays. PLoS ONE. 2015;10:e0124667. doi: 10.1371/journal.pone.0124667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock GC, Estes DM, Young GM, Young B, Torres AG. Construction of a reporter system to study Burkholderia mallei type III secretion and identification of the BopA effector protein function in intracellular survival. Trans R Soc Trop Med Hyg. 2008;102(Suppl 1):S127–S133. doi: 10.1016/S0035-9203(08)70029-4. [DOI] [PubMed] [Google Scholar]
- Whitlock GC, Valbuena GA, Popov VL, Judy BM, Estes DM, Torres AG. Burkholderia mallei cellular interactions in a respiratory cell model. J Med Microbiol. 2009;58:554–562. doi: 10.1099/jmm.0.007724-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiersinga WJ, Currie BJ, Peacock SJ. Melioidosis. N Engl J Med. 2012;367:1035–1044. doi: 10.1056/NEJMra1204699. [DOI] [PubMed] [Google Scholar]
- Wikraiphat C, Saiprom N, Tandhavanant S, Heiss C, Azadi P, Wongsuvan G, Tuanyok A, Holden MT, Burtnick MN, Brett PJ, Peacock SJ, Chantratita N. Colony morphology variation of Burkholderia pseudomallei is associated with antigenic variation and O-polysaccharide modification. Infect Immun. 2015;83:2127–2138. doi: 10.1128/IAI.02785-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright JR. Immunomodulatory functions of surfactant. Physiol Rev. 1997;77:931–962. doi: 10.1152/physrev.1997.77.4.931. [DOI] [PubMed] [Google Scholar]
- Wright JR. Pulmonary surfactant: a front line of lung host defense. J Clin Invest. 2003;111:1453–1455. doi: 10.1172/JCI200318650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zasloff M. Magainins, a class of antimicrobial peptides from Xenopus skin: isolation, characterization of two active forms, and partial cDNA sequence of a precursor. Proc Natl Acad Sci USA. 1987;84:5449–5453. doi: 10.1073/pnas.84.15.5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of macrophage survival of Bp strain 1106a and mouse spleen isolate Bp 14-1. J774.A1 cells were inoculated with MOIs of 16.6 and 11.0, respectively, and incubated for 1 h. The data shown are the mean viable counts (triplicate wells) recovered after the 1-h uptake, incubation of the infected cells in the presence of kanamycin for 2 h (3 h), and after incubation for a total of 8 h. The isolate had been obtained from the spleen of a mouse surviving IP challenge with 1106 s and euthanized on day 14. The viable counts recovered from the isolate-infected cells were greater than those of the parent strain at all three time points (p ≤ 0.0013) (TIFF 133 kb)