Abstract
Naturally occurring admixture has now been documented in every major primate lineage, suggesting its key role in primate evolutionary history. Active primate hybrid zones can provide valuable insight into this process. Here, we investigate the history of admixture in one of the best-studied natural primate hybrid zones, between yellow baboons (Papio cynocephalus) and anubis baboons (Papio anubis) in the Amboseli ecosystem of Kenya. We generated a new genome assembly for yellow baboon and low coverage genome-wide resequencing data from yellow baboons, anubis baboons, and known hybrids (n=44). Using a novel composite likelihood method for estimating local ancestry from low coverage data, we found high levels of genetic diversity and genetic differentiation between the parent taxa, and excellent agreement between genome-scale ancestry estimates and a priori pedigree, life history, and morphology-based estimates (r2=0.899). However, even putatively unadmixed Amboseli yellow individuals carried a substantial proportion of anubis ancestry, presumably due to historical admixture. Further, the distribution of shared versus fixed differences between a putatively unadmixed Amboseli yellow baboon and an unadmixed anubis baboon, both sequenced at high coverage, are inconsistent with simple isolation-migration or equilibrium migration models. Our findings suggest a complex process of intermittent contact that has occurred multiple times in baboon evolutionary history, despite no obvious fitness costs to hybrids or major geographic or behavioral barriers. In combination with the extensive phenotypic data available for baboon hybrids, our results provide valuable context for understanding the history of admixture in primates, including in our own lineage.
Keywords: Amboseli baboons, admixture, hybridization, genome resequencing, local ancestry
Introduction
Naturally occurring admixture is of great interest in evolutionary biology as both a marker of the speciation process and a potential mechanism of evolutionary change (Anderson & Stebbins 1954; Arnold 1992, 1997; Barton 1989, 2001; Grant & Grant 1992; Lewontin & Birch 1966; Rieseberg 1997). Long thought to be relatively rare in animals, genetic evidence has combined with reports of hybrids in natural populations to suggest that, at least for some taxa, admixture may in fact be quite common (Arnold & Meyer 2006; Grant & Grant 1992). Indeed, naturally occurring admixture has now been documented in every major primate lineage, often through direct observations in the field (Arnold & Meyer 2006; Zinner et al. 2011). Meanwhile, interest in human evolutionary history has motivated development of a large suite of genomic tools for inferring admixture in the distant past (e.g., (Durand et al. 2011; Sankararaman et al. 2012; Wall et al. 2009). The emerging picture suggests that recently diverged primate taxa frequently mix when their populations come into contact, and that these events often produce viable and fertile offspring (including for species that diverged >3 million years ago: (Cortés-Ortiz et al. 2007; Jolly et al. 1997). Studies from active primate hybrid zones have demonstrated the importance of social interactions and population demographics in driving this process (Beehner & Bergman 2006; Bergman et al. 2008; Phillips-Conroy & Jolly 1986; Tung et al. 2012).
Such systems provide living models for understanding the phenotypic causes and consequences of admixture in recently diverged primates, including the roles played by social group composition and within-group social interactions. Thus far, however, population genomic analyses of admixture in primates have been uncoupled from the populations in which admixture events have actually been observed (Becquet et al. 2007; Prado-Martinez et al. 2013; Yan et al. 2011). Within these populations, genetic analyses have been limited to relatively small marker sets, restricting insights about the timing, rate, and impact of admixture to the very recent past. Thus, we have little sense of whether the evolutionary scenarios suggested by these data are consistent with the long-term history of admixture. For example, in hybrid zones between members of the baboon genus Papio, observations of naturally occurring hybridization have been variably interpreted as evidence of stable hybrid zones, recent range expansion, or temporally varying admixture rates over time (e.g., due to human activity or climate variation) (Alberts & Altmann 2001; Jolly 1993; Jolly et al. 2011; Phillips-Conroy & Jolly 1986; Tung et al. 2008). More powerful population genomic data sets can help resolve these alternatives by providing insight into when and how often the parental taxa have come into contact.
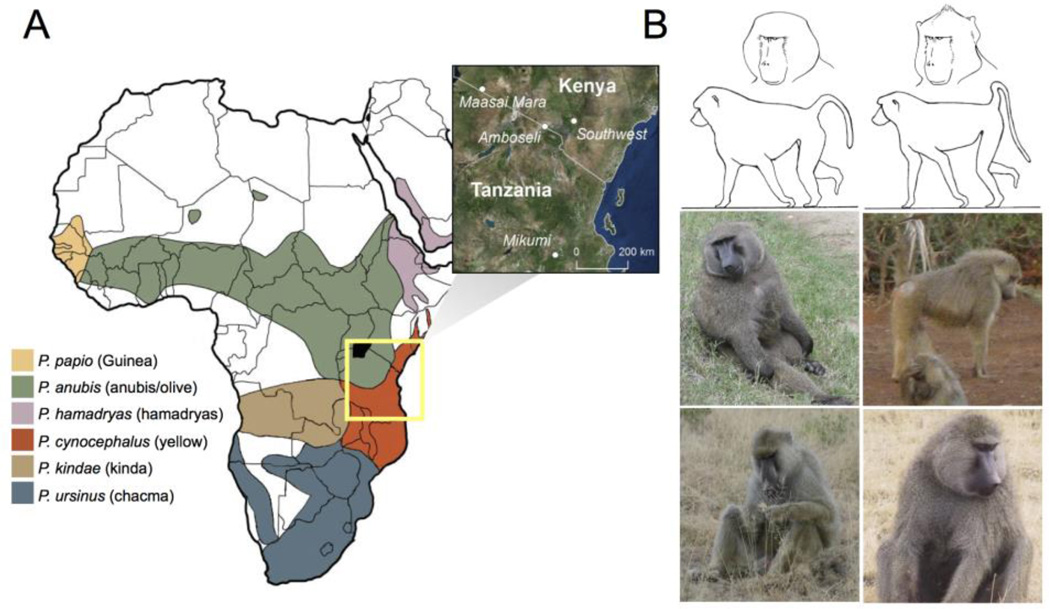
To do so here, we focused on a well-characterized baboon hybrid zone located in the Amboseli basin of southern Kenya. This population falls within a larger hybrid zone that is thought to stretch along the long boundary between yellow baboons (Papio cynocephalus) and anubis baboons (P. anubis) in East Africa. Typically for baboon hybrid zones, it occurs at the junction between otherwise geographically distinct ranges. Hybrids are both viable and fertile, and the parent taxa are readily distinguishable based on phenotypic characteristics (Figure 1) (Alberts & Altmann 2001; Charpentier et al. 2012). However, unlike most other primate hybrid zones, direct observations on the recent history of admixture are available. Specifically, continuous monitoring of the Amboseli population began in 1971, when observers reported a phenotypically uniform yellow baboon population (Alberts & Altmann 2012). Anubis immigrants began arriving in the Amboseli ecosystem in 1982, producing a population that remains majority yellow today, with approximately 1/3 of individuals showing evidence of admixture (Samuels & Altmann 1986; Tung et al. 2008). By combining this information with the extensive pedigree data from Amboseli (Alberts et al. 2006; Buchan et al. 2003), we were therefore able to focus our genetic sampling on animals with known recent ancestries (including both admixed and putatively unadmixed yellow baboons).
Figure 1.
(A) Distribution of the six commonly recognized baboon allotaxa in Africa and Arabia (following (Zinner et al. 2013)). The region surrounding the yellow-anubis hybrid zone is demarcated by the yellow square; inset shows approximate sampling locations for the samples included here (see also Supplementary Information). (B) Photographs of wild anubis (top left), yellow (top right), Amboseli yellow (lower left), and Amboseli hybrid (lower right) adult male baboons. Diagrams above the photographs show morphological characteristics that differ between anubis and yellow baboons (e.g., pelage shape, head shape). Seven such characteristics are used for morphological ancestry scoring in the Amboseli population (see Methods).
The history of admixture in Amboseli is of particular interest because of previous findings that raise questions about how the hybrid zone is maintained. In particular, both phenotypic assessment and earlier genetic analyses indicate that the yellow-anubis hybrid zone surrounding Amboseli is narrow, especially in comparison to the large geographic ranges of both parent taxa (Charpentier et al. 2012). In combination with the lack of clear geographic barriers to gene flow, the structure of the hybrid zone suggests a possible ecological selection gradient or tension zone. However, both species are found in a wide variety of ecological conditions, no fitness costs to hybridization have yet been documented, and some analyses in fact point to potential fitness benefits in the majority yellow groups in Amboseli (Charpentier et al. 2008; Tung et al. 2012); but see (Ackermann et al. 2006) for evidence of nonadditive effects on skeletal morphology in a captive hybrid colony) . Anubis ancestry is correlated with earlier maturation in both male and female baboons (a likely fitness advantage in growing populations, like Amboseli) (Charpentier et al. 2008), and, in males, is associated with an advantage in mate competition (Tung et al. 2012). Hence, fitness costs associated with mating behavior, which are thought to be important in maintaining another baboon hybrid zone in Ethiopia (between hamadryas baboons, P. hamadryas, and anubis baboons: (Bergman & Beehner 2003; Sugawara 1979) but see (Bergman et al. 2008) for an alternative interpretation), do not appear to restrict gene flow in Amboseli. The evolutionary processes that account for this combination—active gene flow, a thriving hybrid population, but an apparently geographically constrained hybrid zone—remain unclear. Understanding these processes will provide valuable insight into the role of admixture in primate evolution.
To achieve this goal, we used a population genomic strategy to investigate the history of admixture in Amboseli prior to recent observations. Specifically, we focused on patterns of genome-wide genetic divergence between yellow baboons, anubis baboons, and individuals sampled in Amboseli, and on the distribution of ancestry estimates (i.e., yellow vs. anubis ancestry) within Amboseli animals. We were particularly interested in whether these patterns are most consistent with: (i) recent secondary contact, suggesting that the narrow hybrid zone and the phenotypic characteristics of hybrids may be a consequence of recent anubis range expansion; (ii) equilibrium rates of gene flow, which would point to strong, as yet undetermined selection pressures acting on either side of the hybrid zone; or (iii) a more complex history of admixture, which would suggest that ecological or demographic factors drive varying rates of gene flow over time. Below, we first describe the resources we generated to pursue this analysis, including a new publicly available genome assembly for yellow baboon and a novel composite likelihood method for estimating local ancestry from low coverage sequencing data. We then report the results of applying this method to data generated from Amboseli individuals.
Materials and Methods
Genome assembly
Our primary goal was to evaluate the history of admixture in Amboseli by investigating the structure of local ancestry tracts and shared variation between species. Doing so required us to (i) assemble a reference genome for yellow baboon (the current anubis baboon genome assembly, Panu2.0, remains embargoed for population genomic analyses); and (ii) establish that yellow baboons and anubis baboons are sufficiently genetically differentiated to perform local ancestry estimation using low coverage sequencing data.
DNA was extracted from a yellow baboon (SWY) that was previously housed at the Southwest National Primate Research Center (individual 1×4811). Prior to sequencing, its ancestry was confirmed by microsatellite genotyping and comparison to previously characterized yellow, anubis, or hybrid populations (Figure S1; (Charpentier et al. 2012). We used this sample to produce Illumina sequencing libraries with 7 different insert sizes, ranging from 175 bp to 14 kb, using standard protocols (Table 1). All samples were sequenced on the Illumina HiSeq platform, either at the UCLA Neuroscience Genomics Core or at the UCSF Genomics Core. Most of the total coverage (38x) came from the short insert libraries (175 bp or 400 bp inserts), with the remaining 9x coming from long insert mate-pair libraries (Table 1).
Table 1.
Yellow baboon assembly coverage by each insert size
| Insert Size Used (bp) |
Raw Reads (108 pairs) |
Processed Reads (108 pairs) |
Proportion Unique |
Coverage |
|---|---|---|---|---|
| 0 (SE)a | 0 | 0.25 | 0.8 | |
| 175 | 3.4 | 3.2 | 0.91 | 17.8 |
| 400 | 3.5 | 3.4 | 0.93 | 19.0 |
| 3000 | 0.45 | 0.31 | 0.83 | 1.6 |
| 4300 | 1.4 | 0.74 | 0.85 | 3.8 |
| 5800 | 0.19 | 0.14 | 0.19 | 0.2 |
| 10000 | 1.2 | 0.79 | 0.72 | 3.4 |
| 14000 | 0.32 | 0.23 | 0.17 | 0.2 |
| Grand Total | 46.8 |
Represents reads in which only one end of the read pair was retained after trimming low quality bases; the “Processed Reads” and “Coverage” entries for this row therefore reflect the number and coverage contribution of single end reads, not read pairs.
To produce the assembly, we trimmed low quality bases (quality score < 17) from read ends using Trimmomatic (Bolger et al. 2014), and then used Corrector_HA from the SOAPec tool set (v. 2.01) (Luo et al. 2012b) to perform error correction (kmer size = 27, with kmer frequencies determined by KmerFreq_HA). Putative PCR duplicates were removed using FastUniq (Xu et al. 2012). We then assembled the draft genome using SOAPdenovo (v. 2.04) (Luo et al. 2012b), with a kmer size of 45. GapCLoser was used to fill in gaps created by the scaffolding process, and scaffolds smaller than 500 bp in length were removed. This resulted in a final assembly (Pcyn1.0) with an N50 contig size of 28.9 kbp, an N50 scaffold size of 887 kbp, and an unknown base (N) composition of 6.57% (Figure S2). In total, the scaffold length was 3.085 Gbp. Most of our analyses used a subset of this assembly, consisting of the 16,158 scaffolds that were ≥ 1 kbp in length. To check the coherency of the assembly, we used Cegma to search for 248 highly conserved genes. Of these genes, 95% were found in at least a partial form, while 85% were found in their entirety (Table S1).
Additional sequencing
We also generated 19.6x coverage sequence data from one Amboseli animal (HAP) and low coverage (mean 2.09x: Table S2) from 22 additional Amboseli individuals (all using 100 bp paired-end Illumina sequencing). Based on pedigree structure, life history, or morphological estimates, HAP and 10 of the low coverage samples were deemed to represent putatively unadmixed yellow individuals, and 9 individuals were deemed known hybrids. Specifically, the pedigree and life history data allowed us to estimate hybrid ancestry when an individual’s parents or grandparents were known anubis immigrants or hybrids, or when the individual’s ancestry could be traced back on both lineages before the advent of recent admixture (i.e., before 1982). For example, individual 60282’s mother was born in 1982, and was therefore likely to be an unadmixed Amboseli yellow baboon. Her father was born in 1988, and morphologically assessed as unadmixed yellow; further, her paternal grandmother was born in 1980, so also likely to be unadmixed. In combination with a morphological score for 60282 herself that was very close to “pure” unadmixed yellow, we therefore assigned 60282 an a priori estimate of 100% yellow ancestry. In contrast, 60326 was the son of a female assessed as unadmixed yellow and an immigrant male assessed as “pure” unadmixed anubis. 60326 was therefore assigned an a priori estimate of 50% yellow ancestry.
Pedigree data for Amboseli were obtained through a combination of direct observation (mother-offspring relationships are known as a result of tracking pregnancies that end in the appearance of a dependent infant) and microsatellite typing to assign paternity (Alberts et al. 2006; Buchan et al. 2003). Morphological scores were assigned in adulthood by experienced observers based on scoring of seven different characteristics (coat color, body shape, hair length, head shape, tail length and thickness, tail bend, and muzzle skin) (Alberts & Altmann 2001). Each characteristic was rated on a scale from 0 (pure yellow) to 2 (pure anubis), and the mean of each of the seven characteristics was assigned as the score for the individual as a whole. When multiple observers produced independent scores, we used the grand mean of scores as the final score. In the analyses reported here, we scaled morphological scores between 0 and 1 and inverted them so that higher numbers reflect increasing yellow ancestry instead of anubis ancestry. No morphological or pedigree information was available for three individuals in the sample.
To estimate site-specific genome-wide allele frequency differences (important for local ancestry estimation, below), we also performed low coverage sequencing (mean = 2.06x) from 13 unadmixed anubis baboons (6 from the Washington National Primate Research Center, WaNPRC, and 7 from the Maasai Mara National Reserve in Kenya) and 9 unadmixed yellow baboons from Mikumi National Park in Tanzania. WaNPRC baboons are most likely descendants of wild-caught individuals originally trapped by the Southwest Foundation for Research and Education (now Texas Biomedical Research Institute) near Darajani and Kibwezi, Kenya (see also Text S1). All 22 libraries were generated in the same manner as the Amboseli low coverage short read libraries.
Mapping and variant calling
All sequence reads, including the 23 Amboseli individuals (22 low coverage plus HAP), 22 unadmixed non-Amboseli individuals, the SWY individual, and reads downloaded for the anubis individual (SWA), were mapped to the set of Pcyn1.0 scaffolds >500 bp in length using the efficient short-read aligner bwa (bwa mem, with minimum seed length of 20) (Li & Durbin 2009). To identify genetic variants, we used the Genome Analysis Toolkit (GATK v. 3.3.0) to recalibrate base quality scores, identify potential indels, and realign reads around indels (DePristo et al. 2011; McKenna et al. 2010). We removed putative PCR duplicates using MarkDuplicates in Picard. Because there is no reference variant database available for baboons, we performed variant calling on read alignments without quality score recalibration using GATK’s UnifiedGenotyper (discarding indels) and kept the set of single nucleotide variants that passed the following hard filters: QD < 2.0; MQ < 35.0; FS > 60.0; HaplotypeScore >13.0; MQRankSum < −12.5; and ReadPosRankSum < −8.0 (following the strategy used in (Snyder-Mackler et al. 2015; Tung et al. 2014). All variants and genotypes were called in a joint analysis of all samples. For subsequent analyses, we filtered the set of 24.7 million raw variants further, as described below.
Estimating heterozygosity and Fst
For heterozygosity and Fst estimation, we limited our analysis to Pcyn1.0 scaffolds that were at least 1 kb in length (n = 16,158 scaffolds, with a total length of 3.075 Gb and 2.873 Gb of called bases). We then filtered the polymorphic sites to include only those with high-quality genotype calls in each of the three high-coverage samples. Specifically, we required variants to be biallelic SNPs with an overall variant level quality (QUAL) score > 50 and genotype quality (GQ) score ≥ 30 for each of the three samples. These criteria filtered out ~32.5% of putatively variable sites called by GATK. The number of bases for which we had uniquely mapped read data (i.e., non-zero coverage from uniquely mapped reads) ranged from 2.59 – 2.63 Gb across the three high-coverage samples. Thus, we estimated the total length of the genome for which we could accurately identify variable sites (i.e., the “accessible genome”) as 67.5% of the non-zero coverage for each sample. We used these values in the denominator when calculating per-base values of π. Fst values were calculated using the method of Hudson et al (Hudson et al. 1992).
Local ancestry model
For local ancestry estimation, we restricted our analysis to the first 200 kb of scaffolds that were at least 200 kb in length (n = 3,742 scaffolds; based on principal components projections, this more restricted data set loses little global information about ancestry: Figure S3). For each scaffold, we estimated yellow versus anubis baboon ancestry for each Amboseli individual using a modification of a previously described composite likelihood method (Wall et al. 2011b). We used this approach rather than existing haplotype-based methods (e.g., (Price et al. 2009) because the generalization to genotype likelihoods is straightforward. Specifically, for each SNP passing the filters described above, we estimated the allele frequencies in the ancestral populations using the genotype likelihood (PL) values generated by GATK as estimates of the probabilities of each possible genotype. We used the 13 low-coverage WaNPRC and Maasai Mara baboons to calculate allele frequencies for anubis baboon, and the 9 low-coverage Mikumi and the high-coverage SWY baboon to calculate allele frequencies for yellow baboon. We then filtered this set to retain only those SNPs for which the difference in estimated allele frequencies between anubis and yellow was at least 0.2.
Without genotype uncertainty, the likelihood of an ancestry assignment given the genotype data is simply based on the probability of observing the (single, reliable) genotype if the proposed ancestry assignment were correct, which is in turn based on the allele frequency estimates in the ancestral populations, p1 and p2. To incorporate genotype uncertainty, we extended the likelihood equation to incorporate the probability of observing each of the three possible genotypes (given the proposed ancestry assignment), weighted by the probability that the genotype itself was correct. Specifically, assuming that GATK’s PL values accurately reflect the true genotype probabilities, we calculated the likelihood of each potential ancestry assignment, Yi (where i corresponds to the number of alleles of yellow ancestry, and i = {0,1,2}) as:
where G is the genotype (a set of three possible values, since genotypes are known with uncertainty), and j refers to the number of alternate alleles in that genotype (hence, G0 refers to a homozygous reference genotype, G1 to a heterozygous genotype, and G2 to a homozygous alternate genotype). For example, the likelihood that an individual carries two yellow ancestry alleles at a variable site is given by:
To combine information across SNPs within each 200 kb scaffold or scaffold segment, we constructed a composite likelihood by multiplying the probabilities of observed genotypes across sites. While this approach implicitly assumes that the information across sites is independent (an unrealistic assumption), it should not affect the relative ranking of likelihoods for the alternative configurations. Hence, we assigned the ancestral configuration with the maximum composite likelihood as the true ancestry for each scaffold-individual combination, and tabulated the proportions of each ancestry assignment across the 3,742 scaffolds for each individual.
Simulations to assess the accuracy of local ancestry estimation
To test the applicability of our composite likelihood method to data of the type we generated, we ran simulations under a simple isolation-migration model using the same sample size and (approximate) coverage levels as in the actual data. We assumed a panmictic ancestral population that split 0.99 Ne generations ago into two equal-sized daughter populations that remained completely isolated until the present day. This divergence time was chosen since it produces an average Fst of 0.33, consistent with the measured Fst between the high coverage anubis and yellow baboons from SNPRC (SWY versus SWA). Each population was assigned a per-base pair value of θ = ρ = 0.0018, with θ again (roughly) based on the parameter estimates for the non-Amboseli high coverage individuals. We then estimated population-specific allele frequencies from 13 simulated anubis individuals, each with an average sequencing depth of 2x, and from 10 simulated yellow individuals (9 with an average coverage of 2x and 1 with high coverage [30x]). We assumed a Poisson-distributed number of reads covering each site, and used typical PL values from GATK for these coverage levels and allelic configurations (i.e., the typical PL values given specific numbers of reads supporting the reference or the alternate allele). We then performed local ancestry estimation as described above on additional low-coverage genomes sampled from the simulated daughter populations, and tabulated the accuracy of our method as a function of the size of the region we interrogated, over 1,000 replicate simulations. We also compared the results obtained from our genotype likelihood method with a simpler version which assumes that the most likely genotype at each SNP is the true genotype.
Additional local ancestry analyses
We assigned local ancestry estimates to 200 kb regions in the real data, which are predicted to yield highly accurate assignments based on our simulations. However, our simulation approach assumed that the true local ancestry does not shift within a given region, which is a reasonable assumption for relatively recent admixture, but may not hold for 200 kb tracts if admixture is much older. To qualitatively assess the physical scale over which local ancestry estimates change in the actual data, we therefore subdivided the 200 kb regions described above into two separate 100 kb regions and estimated the local ancestry for each of these sub-regions (using 100 kb regions is also likely to yield highly accurate ancestry assignments based on our simulations: Figure 2). We then tabulated the proportion of regions (for each individual) where these local ancestry estimates were discordant.
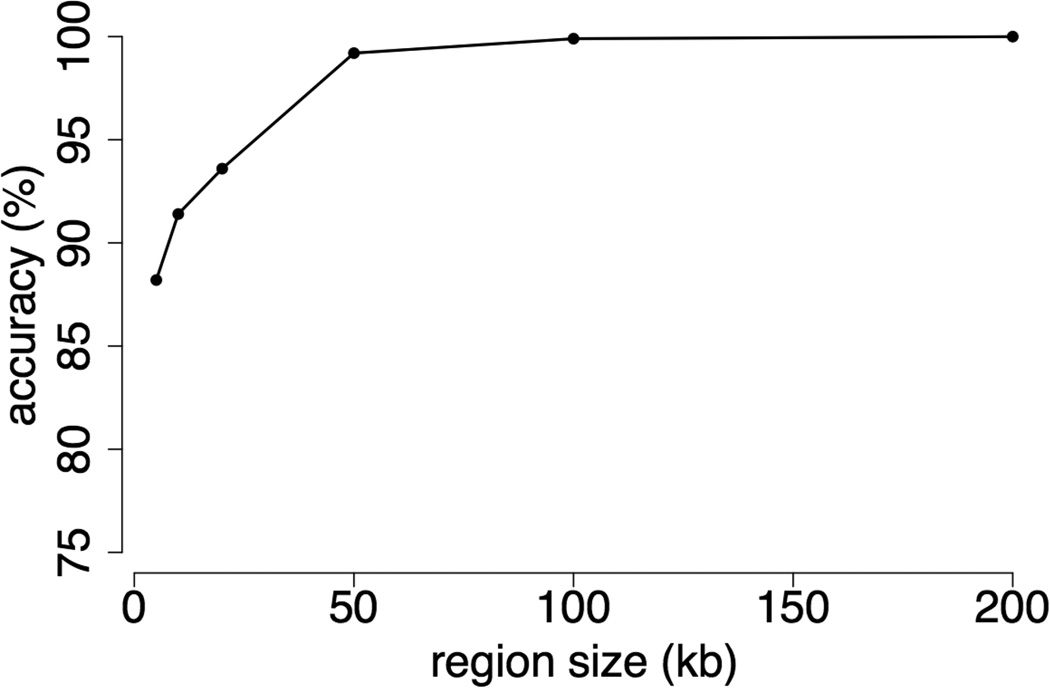
Figure 2.
Accuracy of the low-coverage, local ancestry composite likelihood method as a function of the size of the region under consideration, based on simulated data.
The length of local ancestry tracts is also informative about the history of admixture. Because of the fragmented nature of our yellow baboon assembly, our ability to make use of this information for the whole genome was limited. However, to provide some idea of this distribution for the largest scaffolds in the assembly (17 scaffolds >4 Mb in length), we also modified our local ancestry estimation procedure to estimate ancestry block boundaries from larger contiguous regions using a previously described heuristic method (Wall et al. 2011b). Briefly, we estimated local ancestry for 200 kb windows centered on every variable site. Then, for each SNP, we used majority-rule on all windows containing that SNP to make an ancestry call (i.e., 0, 1 or 2 alleles with yellow baboon ancestry) for that SNP. To reduce edge effects, we required the estimated boundaries to be at least 100 kb away from the edges of the scaffold.
Allele sharing across samples
To investigate shared polymorphisms and fixed differences between the Amboseli yellow high coverage individual (HAP) and the high coverage anubis individual (SWA), we used the same 200 kb regions as in the local ancestry analysis. We then tabulated the number of private polymorphisms (sites that were polymorphic in one individual but not the other; P1 and P2 for HAP and SWA, respectively), shared polymorphisms (sites that were polymorphic in both individuals, S), and fixed differences (sites that were monomorphic in both individuals, but for different bases, F) between HAP and SWA. Recent selective sweeps lead to a reduction in diversity around the selected site. To exclude regions that may have been recently swept, we required P1 > 20, P2 > 20, and (S+F) > 50 (i.e., regions that retained high diversity levels in both species). This filter eliminated 925 regions from our analysis, resulting 2,817 remaining regions.
We used an approximate summary likelihood approach for investigating the probability of observing the true distribution of S/(S+F) values under a specific evolutionary model, using the mean and variance as summary statistics. Qualitatively, summary likelihood replaces high-dimensional sequence data with one or more summary statistics, then uses maximum likelihood (on the summarized data) to estimate parameter values. Summary likelihood approaches have been used for some time in population genetics (e.g., (Fu & Li 1997; Weiss & von Haeseler 1998)) and can be thought of as the frequentist analogue of Approximate Bayesian Computation (ABC: (Beaumont et al. 2002; Tavaré et al. 1997)). Specifically, we compared our data to a simple isolation-migration model, in which a panmictic ancestral population splits at time T into two panmictic descendant populations, connected by symmetric migration rate M to the present day. We note that when T is arbitrarily large, this model is equivalent to an equilibrium island model. All three populations were assumed to be the same size. Because there is no fine-scale recombination map available for baboons, we used the recombination rate distribution described for humans. Specifically, we binned the HapMap YRI recombination rates into non-overlapping 200 kb windows, ordered them by increasing value, and split them into ten equal groups. We then calculated the average scaled recombination rate ρ (=4Ner) for each decile (Table S3). Our simulations drew on equal proportions of each of these ten recombination rate classes. To assess the robustness of our results to errors in these assumptions, we also tested a range of recombination rates with distribution proportional to the decile averages shown in Table S3. Using the simple isolation-migration model described above, our model has three freely varying parameters (T, M, k), where T is in units of 4N generations, M = 4Nm (where m is the migration rate per generation), and the scaled recombination rate for each decile is k multiplied by the numbers in Table S3. We simulated over a grid of values, with increments of 0.01 for T, 0.05 for M and 0.025 for k. For each parameter combination, we simulated 112,680 discrete 200 kb regions (i.e., 40 for each of the 2,817 actual regions). We used the ‘fixed S’ methodology (Hudson 1993) and tabulated S/(S+F) for each simulation. Then, we repeatedly sampled one simulation for each actual region and tabulated the mean and variance of the distribution of S/(S+F) values. The approximate likelihood of the data was estimated as the proportion of resamplings with mean and variance roughly equal to the observed values: 0.366 < mean < .372, and 0.099 < variance < 0.101. We performed 106 resamplings for each parameter combination. Finally, after estimating the likelihood surface, we constructed a profile likelihood for k, using standard asymptotic maximum likelihood assumptions to estimate confidence intervals and linear interpolation of log likelihoods for different values of k.
Results
Genome assembly and genome-wide genetic differentiation between yellow baboons and anubis baboons
We generated a reference genome from a high-coverage (47x) whole genome sequencing data set from a presumably unadmixed yellow baboon from Southwest National Primate Research Center (SWY; see Materials and Methods, Text S1, and Table 1). Specifically, we used a combination of short insert paired-end and long insert mate-pair reads to assemble a 33,203 scaffold (217,877 contig) yellow baboon assembly (Pcyn1.0) using SOAPdenovo v 2.04 (Luo et al. 2012a). The final assembly, restricted to scaffolds >500 bp in length, contained 3.09 Gbp, with an N50 scaffold size of 887 kb (for comparison, the N50 scaffold size for Panu2.0 is 529 kb). We used the subset of Pcyn1.0 scaffolds that were greater than 1 kb in length for all subsequent analyses, except where noted.
To evaluate levels of population differentiation between yellow and anubis baboons, we augmented the data set with two other higher coverage data sets: (i) short read data from an Amboseli yellow baboon whom pedigree and morphological assessment indicated was free of recent admixture (HAP: 19.6x mean coverage; Table S2); and (ii) short-read data from an olive baboon (SWA) available from NCBI’s Short Read Archive (21.4x mean coverage; Table S2). Genetic diversity levels (π) for each of these three individuals (based on ~15 million variants called on scaffolds that were at least 1 kb in length) were consistent with estimates based on much smaller data sets (Boissinot et al. 2014; Tung et al. 2009), with π equal to 0.206% in the anubis individual (SWA), 0.210% in the SNPRC yellow individual (SWY), and 0.251% in the Amboseli animal (HAP). We also found substantial genome-wide differentiation between yellow and anubis baboons in this set, with FST equal to 0.23 in the HAP vs. SWA comparison and 0.33 in the SWY vs. SWA comparison. Importantly, simulation results indicate that this level of differentiation should provide excellent power to identify local ancestry tracts using the approach developed here (Figure 2; see below).
Local ancestry estimation
Our previously described composite likelihood method (Wall et al. 2011a) performs local ancestry estimation by calculating the relative likelihood of each possible ancestry configuration (here, 0, 1, or 2 alleles of yellow baboon ancestry) across a genomic window that contains genetically differentiated sites. This method assumes that genotypes are called with certainty, such that ancestry configuration likelihoods depend only on the probability of sampling a genotype from each ancestral population (i.e., the ancestral allele frequencies). However, in low coverage data, genotypes are called with considerable uncertainty. To extend our method to accommodate these data, we therefore modified the likelihood equations to weight the genotype sampling probabilities by the probability of each possible genotype call (i.e., homozygous reference, heterozygous, or homozygous alternate), using the genotype likelihood information generated as part of the Genome Analysis Toolkit pipeline (see Materials and Methods for details) (DePristo et al. 2011; McKenna et al. 2010).
Results based on simulations indicate that, given a genome-wide FST = 0.33, a scaled mutation rate of θ = 1.8 / kb, and a mean 2x sequencing depth (i.e., properties similar to our real data sets, Table S2), this method assigns local ancestry almost perfectly for any tract length ≥100 kb (Figure 2). We therefore generated ~2x genome-wide coverage from an additional 13 anubis baboons, 9 non-Amboseli yellow baboons, and 22 Amboseli baboons (9 putatively admixed, 10 putatively unadmixed, and 3 unknown, based on pedigree data and morphological assessment, Table S4), and retained all reads that mapped to the first 200 kb of large scaffolds for subsequent analysis. After performing genotype calling and allele frequency estimation in GATK, we retained only those variants (~2.1 million, an average of 562 per 200 Kb region) that exhibited a difference in allele frequencies ≥0.2 between anubis and non-Amboseli yellow individuals. We then estimated yellow vs. anubis ancestry in each Amboseli animal (including the high coverage HAP genome) for 3,742 discrete 200 kb regions of the genome, using the composite likelihood approach.
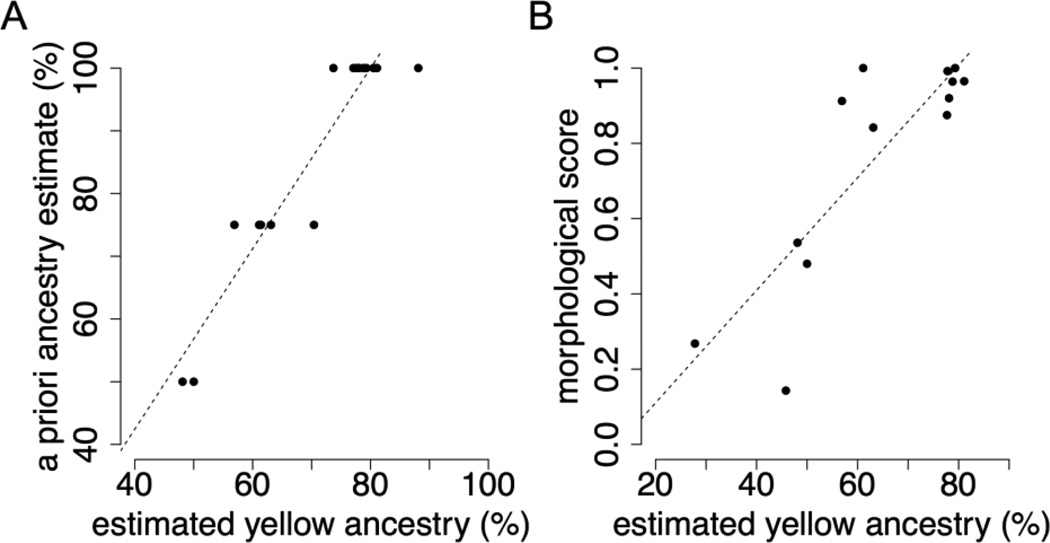
Global ancestry estimates based on the local ancestry results (i.e., the total proportion of yellow baboon ancestry in each individual) were strongly correlated with both morphological assessments of ancestry (based on scoring of 7 ancestry-informative features: r2 = 0.724, p = 2.27 × 10−4) (Alberts & Altmann 2001), and “a priori” estimates (based on combined pedigree, life history, and morphological data for the ancestors of sampled individuals: r2 = 0.899, p = 2.23 × 10−9) (Figure 3, Table S4). In other words, individuals that either had known anubis ancestors or were scored as morphologically more anubis-like also carried more 200 kb stretches of anubis or mixed anubis-yellow ancestry (Table S4). Surprisingly, however, our results suggest that even putatively unadmixed Amboseli individuals carry a substantial proportion of anubis ancestry, ranging from 12 – 26% (primarily in mixed ancestry states). Given that our simulations indicate nearly perfect performance of our method for large ancestry tracts (Figure 2), these results likely reflect a true biological signal, consistent with a history of admixture prior to the start of direct monitoring in 1971. Based on these results, we hypothesize that there has been occasional or intermittent gene flow between yellow and anubis baboons over thousands of generations, which would explain why the excess of anubis ancestry is found across all studied individuals.
Figure 3.
Estimated yellow baboon ancestry from the low coverage composite likelihood method agrees well with (A) a priori ancestry estimates (based on pedigree and life history data: r2 = 0.899, p = 2.23 × 10−9) and (B) morphological scores (r2 = 0.724, p = 2.27 × 10−4).
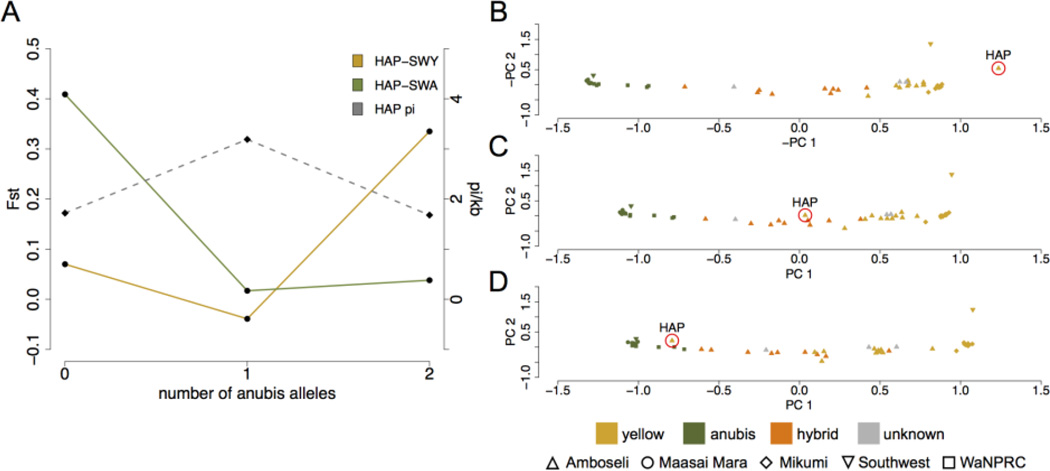
In support of this possibility, we estimated that HAP, a putatively unadmixed Amboseli animal, carries a genome with 19% anubis ancestry. Specifically, the composite likelihood method partitioned his genome into 67.6% pure yellow ancestry, 26.1% mixed ancestry, and 6.3% pure anubis ancestry. At regions estimated to be of anubis ancestry, FST between HAP and SWY is much higher than between HAP and SWA (0.335 versus 0.038), while at regions estimated to be of yellow ancestry, the pattern is reversed (0.07 versus 0.409). Further, in scaffolds estimated to be of mixed ancestry, heterozygosity is greatly elevated relative to scaffolds estimated to be of pure ancestry (Figure 4). Notably, within-species FST between the admixing anubis baboons and SWA is low (FST = 0.038 based on the segments of HAP’s genome inferred to be homozygous anubis) compared with the estimated FST between anubis and yellow baboons.
Figure 4.
The putatively yellow Amboseli baboon HAP exhibits a signature of historic admixture. (A) Fst levels between HAP and the SWY yellow baboon (yellow line) are low in regions inferred as homozygous yellow but high in regions of the genome inferred as homozygous anubis; the pattern is reversed for Fst comparisons between HAP and the SWA anubis baboon (green line). Genetic diversity in HAP is highest in regions of mixed ancestry (dashed gray line). (B–D) Principal components projections of genotype data on scaffolds where HAP is estimated to have homozygous yellow ancestry group him with other yellow baboons (B), but genotype data from scaffolds where HAP has heterozygous or homozygous anubis ancestry group him with hybrids (C) and anubis baboons (D), respectively. Colors depict a priori ancestry assignments based on population of origin or, for Amboseli, pedigree, life history, and morphological scores (see Methods and Table S2); shapes show population of origin. Note that –PC1 and –PC2 are plotted on the x and y-axes of (B) to maintain visual consistency with (C,D).
HAP’s father has an estimated birth year of 1974, long before the onset of recent admixture in 1982, as were both his maternal grandmother (b. 1969) and all possible candidate maternal grandfathers; further, of the four most likely maternal grandfathers, three were previously assessed as genetically similar to other Amboseli yellow baboons (Tung et al. 2008). Thus, while it is possible that HAP’s maternal grandfather was admixed if he immigrated to Amboseli from a different, previously admixed population, this scenario appears unlikely. Combined with the observation that HAP’s genome includes regions of homozygous anubis ancestry (i.e., inherited from both maternal and paternal lines), our data indicate that HAP is unlikely to be the product of recent admixture alone. Instead, the anubis component of his genome is likely to reflect, at least in part, a residual signature of admixture prior to the wave that began in 1982.
In principle, additional information on the timing of admixture can be obtained by examining the distribution of ancestry tract lengths – the impact of intragenic recombination means that older admixture will lead to smaller ancestry tracts (e.g., (Gravel 2012). Although the fragmented nature of our yellow baboon assembly limits our ability to evaluate the genome-wide distribution, we performed two additional analyses to qualitatively assess ancestry tract size within the Amboseli animals we sequenced. First, for the 3,742 200 Kb regions analyzed above, we separately estimated local ancestry for the first 100 Kb and the last 100 Kb. In the majority of cases (mean 77.6% ± 3.01%), ancestry estimates agreed between these sub-regions. However, for an appreciable percentage of regions (17 – 31 %: Table S5), assignments were discordant, suggesting that local ancestry tract lengths are, overall, generally quite short (e.g., < 1 Mb). Second, we modified our local ancestry estimation method to identify the boundaries of ancestry tracts for the 17 scaffolds that were >4 Mb in length. The mean ancestry tract length varied between 233 – 421 Kb across the 23 Amboseli individuals in our study (Table S5). While we cannot convert these lengths into genetic distances without a high-resolution genetic map, our results are broadly consistent with a demographic history of the Amboseli baboons that includes a substantial amount of admixture between yellow and anubis baboon ancestors hundreds of generations ago.
Patterns of genetic variation reject simple isolation-migration models for admixture in Amboseli
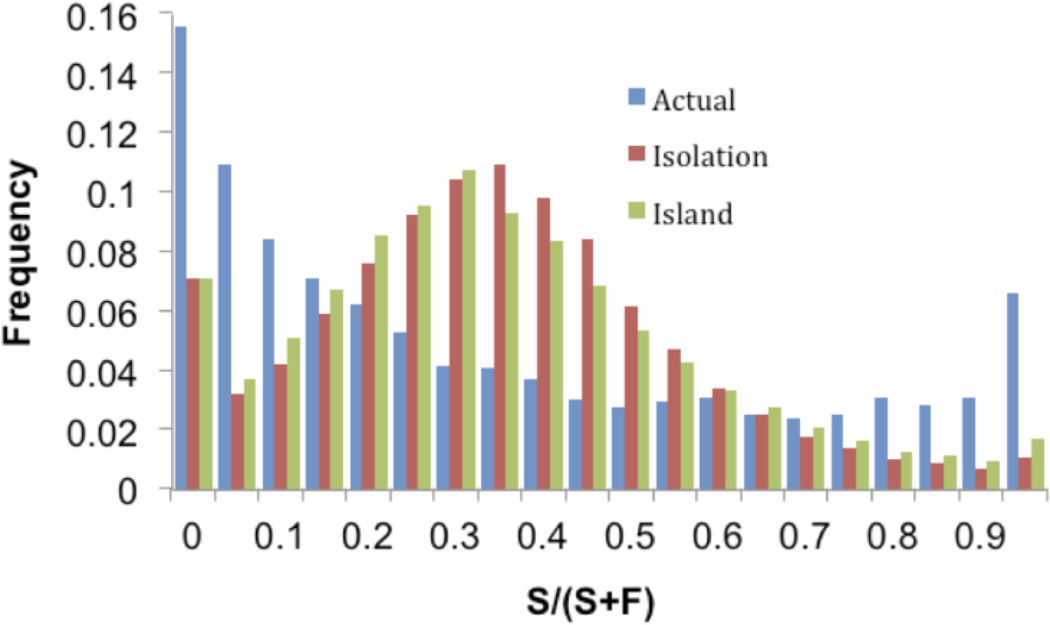
Finally, we used an orthogonal approach to examining the history of admixture in the Amboseli baboons, based on the numbers of shared polymorphisms (S) and fixed differences (F) between the higher coverage HAP and SWA (unadmixed anubis) individuals. Specifically, we looked at the distribution of S/(S+F) across 2,817 discrete 200 kb windows of the genome (a subset of those used to estimate local ancestry, after eliminating windows that may have been affected by recent selective sweeps: see Materials and Methods, Table S6). This statistic is related to FST: low values of S/(S+F) correspond to high values of FST and high levels of differentiation, while high values of S/(S+F) correspond to low values of FST and low levels of differentiation. However, unlike FST, this approach reduces the potentially confounding effects of variation in within-population heterozygosity.
We plotted the distribution of S/(S+F) values for the actual data, and compared it to distributions produced under a simple isolation-migration model and an equilibrium island model (Figure 5). While the means of the distributions are all roughly the same (0.369), the variances differ substantially. Specifically, the actual data have a much larger variance (0.100) than either of the simulated data sets (0.043 and 0.049 for the isolation-migration and island models respectively), with an excess of regions that exhibit higher and lower levels of genetic differentiation than expected under both simulated models. This difference suggests that our data arose from a demographic history inconsistent with these simple models. It also is unlikely to be accounted for by assumptions about the underlying recombination rate or the confounding effects of natural selection (see Discussion below). While it is beyond the scope of this paper to systematically examine more complicated models of demography, our simulations suggest that models of isolation followed by secondary contact can produce S/(S+F) distributions with mean and variance roughly the same as what we observed (see for example Figure S4). However, because a secondary contact model has four parameters and is used to estimate only two summary statistics, many parameter combinations can produce the same results. Thus, a different type of analysis (e.g., one with more summary statistics) would be necessary to estimate demographic parameters under more complex (and more realistic) models.
Figure 5.
Distribution of S/(S+F) values for the actual data (representing a comparison of HAP and SWA), an isolation-migration model (T = 0.52 Ne generations) and an equilibrium island model (M = 1.58). The actual data show an enrichment of low and high S/(S+F) values relative to the predictions of both alternative models.
Discussion
Together, our findings provide considerable new insight into the history of admixture and hybridization in this well-studied hybrid zone. Specifically, they extend the record of hybridization in the last three decades to include a longer history of admixture in the past—distant enough so that Amboseli animals phenotypically group with other yellow baboons, but with a residual impact that still contributes substantially to genetic variation in the population today. This result rejects a hypothesis based only on recent contact between these two species, and combines with our simulation results to suggest that the history of yellow-anubis hybridization in east Africa is more complex than the simple isolation-migration or equilibrium models we tested. Hence, the transition from a phenotypically yellow population to an admixed population observed in the 1980’s may be representative of a dynamic process that has occurred in this region multiple times before. This process would produce animals that we today recognize as Amboseli yellow baboons, but whose genomes are mosaics of regions inherited from anubis or yellow baboon ancestors. This leads to greater heterogeneity in patterns of population differentiation across the genome than the more uniform structure expected under models that propose consistent rates of admixture once admixture starts occurring. Indeed, our data support the hypothesis that the “ibean” morphotype of yellow baboons, of which the Amboseli baboons are often presented as the type example, originated from ancient admixture between anubis baboons and baboons of the “typical” yellow morphotype (found to the east and south of Amboseli, including in Mikumi) (Jolly 1993). Here, our ability to estimate the timing of these admixture events is currently limited by the lack of a high quality chromosomal assembly and sufficient data to perform haplotype phasing. As these resources come online, it should be possible to use complementary analyses (e.g., based on haplotype sharing, genome-wide ancestry block length, or sequential Markovian coalescent methods: (Gravel 2012; Schiffels & Durbin 2014) to reconstruct the history of admixture in this region in greater detail.
We note that our conclusions are based in part on a demographic interpretation for the mismatch between our actual data and the predictions of the island equilibrium and isolation-migration models. We favor this interpretation over two other possibilities—the confounding effects of natural selection, and substantial error in our recombination rate parameters—for the following reasons. First, pervasive natural selection could explain the excess of high divergence regions we observed if selective sweeps have been very common in one or both species; long-term balancing selection, on the other hand, could explain low divergence regions characterized by high levels of shared polymorphism. This explanation is unlikely because we explicitly removed from our analysis regions that were more consistent with recent selective sweeps; furthermore, long-lived balancing selection is thought to be quite rare (Leffler et al. 2013). Second, while it is theoretically possible that recombination rates in baboons are extremely different from recombination rates in humans (we used information from the YRI HapMap population here), they would have to be approximately five times smaller in baboons, on a per generation basis, to explain our results (the ratio of baboon to human recombination rates, k, would need to fall in a 95% CI between 0.17–0.23, Figure S5). This would contradict the standard belief that one crossover per chromosome arm per generation is needed for proper segregation of chromosomes during meiosis. Analogous simulations for SWY and SWA produced low recombination estimates as well (95% CI for k: 0.21 – 0.37), suggesting that historical gene flow between yellow and anubis baboons is not confined to the Amboseli population.
By excluding both a single episode of secondary contact and equilibrium rates of gene flow, our results help to refine the set of scenarios that could explain the collection of genomic and phenotypic observations emerging from the Amboseli hybrid zone. They suggest that either the hybrid zone has moved over time, producing changing rates of hybridization within Amboseli’s geographical bounds; that hybridization rates have temporally varied within a stable hybrid zone; or a combination of both. Both scenarios seem plausible, as researchers working in several primate hybrid zones have described changes in admixture rates even over the course of a few decades (Detwiler 2002; Phillips-Conroy & Jolly 1986). Possible explanations include anthropogenic activity, which can affect dispersal rates and the relative availability of conspecific versus heterospecific mates; climatic variation, which can affect selection pressure on hybrids if the parent species are ecologically differentiated; or differences in social and demographic conditions, which provide varying opportunities for heterospecific immigrants to succeed. Indeed, previous studies in both Amboseli and the Awash hybrid zone in Ethiopia suggest that the mating behavior (and, in Awash, the overall reproductive success) of hybrids and heterospecific baboons is highly dependent on the demographics of the groups to which they belong (Bergman et al. 2008; Tung et al. 2012). This raises the intriguing possibility (previously suggested for bird hybrid zones, but untested in group-living, socially complex primates) that varying ecological conditions may indirectly affect hybrid zone dynamics by influencing dominance and mating-related traits in an ancestry-dependent manner (Harr & Price 2014). A key outstanding question in this case is whether phenotypic differences between anubis and yellow baboons are indeed related to ecological specialization.
A strength of the Amboseli system is that such hypotheses can be tested through direct observations of living hybrids, thus providing insight into the phenotypic traits responsible for genomic signatures of admixture. In addition, the large FST values separating yellow and anubis baboons, the rich set of phenotypic data for this population, and the presence of multigenerational hybrids suggest the utility of this system for admixture mapping more generally. Baboons are one of the most important nonhuman primate models for human physiology, disease, and behavior (Jolly 2001; Rogers & Hixson 1997). They exhibit similar patterns of aging, obesity, cardiovascular disease, and vulnerability to socially induced stressors (Archie et al. 2014; Bronikowski et al. 2011; Comuzzie et al. 2003; Rogers & Hixson 1997). Further, they also exhibit traits of interest to human health, such as resistance to endotoxins that cause sepsis (Haudek et al. 2003; von Bülow et al. 1992; Zurovsky et al. 1987), that lie far outside the human phenotypic spectrum. Previous work has shown that, controlling for background kinship and population structure, the power to identify expression quantitative trait loci in Amboseli outstrips the power to map eQTL in a genetically diverse human population (Tung et al. 2014). Our finding that local ancestry tracts can be assigned with high confidence from low coverage data opens the door to additional strategies for understanding complex trait genetics in wild primates—a topic that we currently know almost nothing about.
Finally, we present several new tools and resources of more general interest to the research community. One of these is a new method for estimating local ancestry from low-coverage whole-genome sequence data. This new method uses genotype likelihoods in place of genotype calls, but our simulations find that local ancestry estimation assuming that the most likely genotype call is correct is almost as effective (Table SX). This suggests that local ancestry estimation may be reasonably accurate even in scenarios with <2X coverage or in species with less divergence than between yellow and anubis baboons.
Recent phylogenies for Papio indicate that yellow baboons and anubis baboons are among the most distantly related of the currently recognized baboon species, with an estimated divergence time ~1 – 2 million years ago (Boissinot et al. 2014; Zinner et al. 2013). Thus, the draft yellow baboon assembly we produced here should complement the fully assembled and annotated anubis baboon genome that is soon to be released. If existing estimates hold, yellow baboons may be among the most genetically diverse of the baboon species as well (Boissinot et al. 2014), meaning that the data sets we generated will help capture a substantial fraction of the genetic diversity within the genus. Notably, we identified a large set of putative high and low divergence regions separating anubis and Amboseli yellow baboons that could be indicative of a past history of selection (Table Sx). Combining this information with gene and functional element annotations could therefore shed considerable new light on targets of recent adaptation (or long-term balancing selection) in African savanna-dwelling primates—the environment in which our own ancestors also evolved (Jolly 2001).
Supplementary Material
Acknowledgments
We would like to thank the Kenya Wildlife Service, Institute of Primate Research, National Museums of Kenya, National Council for Science and Technology, members of the Amboseli-Longido pastoralist communities, Tortilis Camp, and Ker & Downey Safaris for their assistance in Kenya. We also thank Jeanne Altmann and Elizabeth Archie for their generous support and access to the Amboseli Baboon Research Project data set and samples; Raphael Mututua, Serah Sayialel, Kinyua Warutere, Mercy Akinyi, Tim Wango, and Vivian Oudu for invaluable assistance with the Amboseli baboon sample collection; Tauras Vilgalys and Laura Grieneisen for assistance with figures; the Coriell Institute and Integrated Primate Biomaterial and Information Resource for access to additional samples from the Masai Mara and Mikumi National Park; James Ha and the Washington National Primate Research Center for access to the WaNPRC samples; and two anonymous reviewers for constructive comments on an earlier version of this manuscript. This work was supported by National Science Foundation grants DEB-1405308 (to JT) and SMA-1306134 (to JT and NSM); NIH grants R24 OD017859 (to JDW and LC), P51 OD011133 (to LAC), and R01 GM115433 (to JDW); and NSF and NIH support for the long-term Amboseli field research, most recently NSF IOS-0919200, NSF DEB-0846286, NIH R01 AG034513, and NIH P01 AG031719.
Footnotes
Data accessibility
Sequence data for the SWA baboon is available from NCBI’s Short Read Archive (SRR927653-SRR927659). Resequencing data sets generated as part of this study are also deposited in NCBI SRA (BioProject PRJNA308870); the HAP data set is included in SRA accession SRP064514 (SRX1307910). The Pcyn1.0 assembly is available at https://abrp-genomics.biology.duke.edu/index.php?title=Other-downloads/Pcyn1.0. The software used to estimate local ancestry from low-coverage sequence data is available at http://www.github.com/jdwall02/LCLAE.
Author contributions
Conceptualization: J.D.W. and J.T.; Methodology: J.D.W., S.A.S., and J.T.; Investigation: J.D.W., S.A.S., N.S.M., K.N., and J.T.; Writing – Original Draft, J.D.W. and J.T.; Writing – Review & Editing, S.A.S., S.C.A., L.A.C., N.S.M., K.N., and L.C.; Funding Acquisition: J.D.W., S.C.A., L.A.C., L.C., and J.T.; Resources, S.C.A., L.A.C., L.C., and J.T.
References
- Ackermann RR, Rogers J, Cheverud JM. Identifying the morphological signatures of hybridization in primate and human evolution. Journal of Human Evolution. 2006;51:632–645. doi: 10.1016/j.jhevol.2006.07.009. [DOI] [PubMed] [Google Scholar]
- Alberts SC, Altmann J. Immigration and hybridization patterns of yellow and anubis baboons in and around Amboseli, Kenya. American Journal of Primatology. 2001;53:139–154. doi: 10.1002/ajp.1. [DOI] [PubMed] [Google Scholar]
- Alberts SC, Altmann J. The Amboseli Baboon Research Project: 40 Years of Continuity and Change. In: Kappeler PM, Watts DP, editors. Long-Term Field Studies of Primates. Berlin: Spring-Verlag; 2012. [Google Scholar]
- Alberts SC, Buchan JC, Altmann J. Sexual selection in wild baboons: from mating opportunities to paternity success. Animal Behaviour. 2006;72:1177–1196. [Google Scholar]
- Anderson E, Stebbins GL. Hybridization as an evolutionary stimulus. Evolution. 1954;8:378–388. [Google Scholar]
- Archie EA, Tung J, Clark M, Altmann J, Alberts SC. Social affiliation matters: both same-sex and opposite-sex relationships predict survival in wild female baboons. Proceedings of the Royal Society of London B: Biological Sciences. 2014;281:20141261. doi: 10.1098/rspb.2014.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold ML. Natural hybridization as an evolutionary process. Annual Review of Ecology and Systematics. 1992;23:237–261. [Google Scholar]
- Arnold ML. Natural hybridization and evolution. Oxford, UK: Oxford University Press; 1997. [Google Scholar]
- Arnold ML, Meyer A. Natural hybridization in primates: one evolutionary mechanism. Zoology. 2006;109:261–276. doi: 10.1016/j.zool.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Barton NH. Adaptation, speciation and hybrid zones. Nature. 1989;341:497–503. doi: 10.1038/341497a0. [DOI] [PubMed] [Google Scholar]
- Barton NH. The role of hybridization in evolution. Molecular Ecology. 2001;10:551–568. doi: 10.1046/j.1365-294x.2001.01216.x. [DOI] [PubMed] [Google Scholar]
- Beaumont MA, Zhang W, Balding DJ. Approximate Bayesian computation in population genetics. Genetics. 2002;162:2025–2035. doi: 10.1093/genetics/162.4.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becquet C, Patterson N, Stone AC, Przeworski M, Reich D. Genetic structure of chimpanzee populations. PLoS Genet. 2007;3:e66. doi: 10.1371/journal.pgen.0030066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beehner JC, Bergman TJ. Female behavioral strategies of hybrid baboons in the Awash National Park, Ethiopia. In: Swedell L, Leigh S, editors. Reproduction and Fitness in Baboons: Behavioral, Ecological, and Life History Perspectives. New York: Springer; 2006. pp. 53–79. [Google Scholar]
- Bergman TJ, Beehner JC. Hybrid zones and sexual selection: Insights from the Awash baboon hybrid zone (Papio hamadryas anubis¥ P. h. hamadryas) Sexual selection and reproductive competition in primates: new insights and directions. 2003;3:503–537. [Google Scholar]
- Bergman TJ, Phillips-Conroy JE, Jolly CJ. Behavioral variation and reproductive success of male baboons (Papio anubis× Papio hamadryas) in a hybrid social group. American Journal of Primatology. 2008;70:136–147. doi: 10.1002/ajp.20467. [DOI] [PubMed] [Google Scholar]
- Boissinot S, Alvarez L, Giraldo-Ramirez J, Tollis M. Neutral nuclear variation in baboons (genus Papio) provides insights into their evolutionary and demographic histories. American Journal of Physical Anthropology. 2014;155:621–634. doi: 10.1002/ajpa.22618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronikowski AM, Altmann J, Brockman DK, et al. Aging in the natural world: comparative data reveal similar mortality patterns across primates. Science. 2011;331:1325–1328. doi: 10.1126/science.1201571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JC, Alberts SC, Silk JB, Altmann J. True paternal care in a multi-male primate society. Nature. 2003;425:179–181. doi: 10.1038/nature01866. [DOI] [PubMed] [Google Scholar]
- Charpentier MJ, Fontaine MC, Cherel E, et al. Genetic structure in a dynamic baboon hybrid zone corroborates behavioural observations in a hybrid population. Molecular Ecology. 2012;21:715–731. doi: 10.1111/j.1365-294X.2011.05302.x. [DOI] [PubMed] [Google Scholar]
- Charpentier MJE, Tung J, Altmann J, Alberts SC. Age at maturity in wild baboons: genetic, environmental and demographic influences. Molecular Ecology. 2008;17:2026–2040. doi: 10.1111/j.1365-294X.2008.03724.x. [DOI] [PubMed] [Google Scholar]
- Comuzzie AG, Cole SA, Martin L, et al. The baboon as a nonhuman primate model for the study of the genetics of obesity. Obesity research. 2003;11:75–80. doi: 10.1038/oby.2003.12. [DOI] [PubMed] [Google Scholar]
- Cortés-Ortiz L, Duda TF, Canales-Espinosa D, et al. Hybridization in large-bodied New World primates. Genetics. 2007;176:2421–2425. doi: 10.1534/genetics.107.074278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature genetics. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detwiler KM. Hybridization between red-tailed monkeys (Cercopithecus ascanius) and blue monkeys (C. mitis) in East African forests. The guenons: Diversity and adaptation in African monkeys. 2002:79–97. [Google Scholar]
- Durand EY, Patterson N, Reich D, Slatkin M. Testing for ancient admixture between closely related populations. Molecular biology and evolution. 2011;28:2239–2252. doi: 10.1093/molbev/msr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y-X, Li W-H. Estimating the age of the common ancestor of a sample of DNA sequences. Molecular biology and evolution. 1997;14:195–199. doi: 10.1093/oxfordjournals.molbev.a025753. [DOI] [PubMed] [Google Scholar]
- Grant PR, Grant BR. Hybridization of bird species. Science. 1992;256:193–197. doi: 10.1126/science.256.5054.193. [DOI] [PubMed] [Google Scholar]
- Gravel S. Population genetics models of local ancestry. Genetics. 2012;191:607–619. doi: 10.1534/genetics.112.139808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harr B, Price T. Climate change: a hybrid zone moves north. Current Biology. 2014;24:R230–R232. doi: 10.1016/j.cub.2014.02.023. [DOI] [PubMed] [Google Scholar]
- Haudek SB, Natmessnig BE, Fürst W, et al. Lipopolysaccharide dose response in baboons. Shock. 2003;20:431–436. doi: 10.1097/01.shk.0000090843.66556.74. [DOI] [PubMed] [Google Scholar]
- Hudson RR, Slatkin M, Maddison WP. Estimation of levels of gene flow from DNA sequence data. Genetics. 1992;132:583–589. doi: 10.1093/genetics/132.2.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly CJ. Species, subspecies, and baboon systematics. In: Kimbel WH, Martin LB, editors. Species, species concepts, and primate evolution. New York: Plenum Press; 1993. pp. 67–107. [Google Scholar]
- Jolly CJ. A proper study for mankind: analogies from the papionin monkeys and their implications for human evolution. Yearbook of Physical Anthropology. 2001;116:177–204. doi: 10.1002/ajpa.10021. [DOI] [PubMed] [Google Scholar]
- Jolly CJ, Burrell AS, Phillips-Conroy JE, Bergey C, Rogers J. Kinda baboons (Papio kindae) and grayfoot chacma baboons (P. ursinus griseipes) hybridize in the Kafue river valley, Zambia. American Journal of Primatology. 2011;73:291–303. doi: 10.1002/ajp.20896. [DOI] [PubMed] [Google Scholar]
- Jolly CJ, Woolley-Barker T, Beyene S, Disotell TR, Phillips-Conroy JE. Intergeneric hybrid baboons. International Journal of Primatology. 1997;18:597–627. [Google Scholar]
- Leffler EM, Gao Z, Pfeifer S, et al. Multiple instances of ancient balancing selection shared between humans and chimpanzees. Science. 2013;339:1578–1582. doi: 10.1126/science.1234070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewontin RC, Birch LC. Hybridization as a source of variation for adaptation to new environments. Evolution. 1966;20:315–336. doi: 10.1111/j.1558-5646.1966.tb03369.x. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R, Liu B, Xie Y, et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience. 2012a;1:1–6. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R, Liu B, Xie Y, et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 2012b;1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome research. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips-Conroy JE, Jolly CJ. Changes in the Structure of the Baboon Hybrid Zone in the Awash-National-Park, Ethiopia. American Journal of Physical Anthropology. 1986;71:337–350. [Google Scholar]
- Prado-Martinez J, Sudmant PH, Kidd JM, et al. Great ape genetic diversity and population history. Nature. 2013;499:471–475. doi: 10.1038/nature12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Tandon A, Patterson N, et al. Sensitive detection of chromosomal segments of distinct ancestry in admixed populations. PLoS Genet. 2009;5:e1000519. doi: 10.1371/journal.pgen.1000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg LH. Hybrid origins of plant species. Annual Review of Ecology and Systematics. 1997;28:359–389. [Google Scholar]
- Rogers J, Hixson JE. Baboons as an animal model for genetic studies of common human disease. The American Journal of Human Genetics. 1997;61:489–493. doi: 10.1086/515527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels A, Altmann J. Immigration of a Papio-Anubis male into a group of Papio-Cynocephalus baboons and evidence for an Anubis-Cynocephalus hybrid zone in Amboseli, Kenya. International Journal of Primatology. 1986;7:131–138. [Google Scholar]
- Sankararaman S, Patterson N, Li H, Pääbo S, Reich D. The date of interbreeding between Neandertals and modern humans. 2012 doi: 10.1371/journal.pgen.1002947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffels S, Durbin R. Inferring human population size and separation history from multiple genome sequences. Nature genetics. 2014;46:919–925. doi: 10.1038/ng.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder-Mackler N, Majoros WH, Yuan ML, et al. Efficient genome-wide sequencing and low coverage pedigree analysis from non-invasively collected samples. bioRxiv. 2015 doi: 10.1534/genetics.116.187492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara K. Sociological study of a wild group of hybrid baboons between Papio anubis and P. hamadryas in the Awash Valley, Ethiopia. Primates. 1979;20:21–56. [Google Scholar]
- Tavaré S, Balding DJ, Griffiths RC, Donnelly P. Inferring coalescence times from DNA sequence data. Genetics. 1997;145:505–518. doi: 10.1093/genetics/145.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung J, Charpentier MJ, Mukherjee S, Altmann J, Alberts SC. Genetic effects on mating success and partner choice in a social mammal. American Naturalist. 2012;2012:113–129. doi: 10.1086/665993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung J, Charpentier MJE, Garfield DA, Altmann J, Alberts SC. Genetic evidence reveals temporal change in hybridization patterns in a wild baboon population. Molecular Ecology. 2008;17:1998–2011. doi: 10.1111/j.1365-294X.2008.03723.x. [DOI] [PubMed] [Google Scholar]
- Tung J, Primus A, Bouley AJ, et al. Evolution of a malaria resistance gene in wild primates. Nature. 2009;460:388–391. doi: 10.1038/nature08149. [DOI] [PubMed] [Google Scholar]
- Tung J, Zhou X, Alberts SC, Stephens M, Gilad Y. The Genetic Architecture of Gene Expression Levels in Wild Baboons. bioRxiv. 2014:008490. doi: 10.7554/eLife.04729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Bülow G, Puren A, Savage N. Interleukin-1 from baboon peripheral blood monocytes: altered response to endotoxin (lipopolysaccharide) and Staphylococcus aureus stimulation compared with human monocytes. European journal of cell biology. 1992;59:458–463. [PubMed] [Google Scholar]
- Wall JD, Jiang R, Gignoux C, et al. Genetic variation in Native Americans, inferred from Latino SNP and resequencing data. Mol Biol Evol. 2011a;28:2231–2237. doi: 10.1093/molbev/msr049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall JD, Jiang R, Gignoux C, et al. Genetic variation in Native Americans, inferred from Latino SNP and resequencing data. Molecular biology and evolution. 2011b;28:2231–2237. doi: 10.1093/molbev/msr049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall JD, Lohmueller KE, Plagnol V. Detecting ancient admixture and estimating demographic parameters in multiple human populations. Molecular biology and evolution. 2009;26:1823–1827. doi: 10.1093/molbev/msp096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss G, von Haeseler A. Inference of population history using a likelihood approach. Genetics. 1998;149:1539–1546. doi: 10.1093/genetics/149.3.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Luo X, Qian J, et al. FastUniq: a fast de novo duplicates removal tool for paired short reads. PLoS One. 2012;7:e52249. doi: 10.1371/journal.pone.0052249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan G, Zhang G, Fang X, et al. Genome sequencing and comparison of two nonhuman primate animal models, the cynomolgus and Chinese rhesus macaques. Nature biotechnology. 2011;29:1019–1023. doi: 10.1038/nbt.1992. [DOI] [PubMed] [Google Scholar]
- Zinner D, Arnold ML, Roos C. The strange blood: natural hybridization in primates. Evolutionary Anthropology: Issues, News, and Reviews. 2011;20:96–103. doi: 10.1002/evan.20301. [DOI] [PubMed] [Google Scholar]
- Zinner D, Wertheimer J, Liedigk R, Groeneveld LF, Roos C. Baboon phylogeny as inferred from complete mitochondrial genomes. American Journal of Physical Anthropology. 2013;150:133–140. doi: 10.1002/ajpa.22185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurovsky Y, Laburn H, Mitchell D, MacPhail A. Responses of baboons to traditionally pyrogenic agents. Canadian journal of physiology and pharmacology. 1987;65:1402–1407. doi: 10.1139/y87-220. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.