Abstract
Purpose
The aim of this study was to explore the association of the DNA-methyltransferase (DNMT)-3A and DNMT3B promoter polymorphisms with the risk of human spontaneous abortion after assisted reproduction techniques (ARTs) and natural conception.
Methods
We collected tissues from women who underwent abortion procedures: (a) chorionic villus samples (CVS) and muscle samples (MS) from spontaneous abortions conceived by ART and natural cycle (study group), n = 152; and (b) CVS and MS from normal early pregnancy and second trimester (control group), n = 155. The single-nucleotide polymorphism (SNP) −448A > G in the DNMT3A promoter region and −149C/T polymorphism of DNMT3B were determined by polymerase chain reaction–restriction fragment length polymorphism (PCR-RFLP) and confirmed by sequencing.
Results
The allele frequency of −448A among pregnancy loss group and control group was 34.2 % vs. 16.5 %, respectively. Compared with GG carriers, the DNMT3A −448AA homozygotes had an about 16-fold increased risk of spontaneous abortion [odds ratio (OR) = 16.130, 95 % confidence interval (CI), 3.665–70.984], and AG heterozygotes had an OR of 2.027 (95 % CI, 1.247–3.293). However, the distribution of −448A > G in individuals derived from ART pregnancies was not statistically significantly compared with those derived from spontaneous pregnancies (P = 0.661). For DNMT3B, we observed genotype frequencies of 100 % (TT) in the study group and the control group.
Conclusions
The DNMT3A −448A > G polymorphism may be a novel functional SNP and contribute to its genetic susceptibility to spontaneous abortion in Chinese women, and ART may not affect the distribution of −448A > G in pregnancy loss and normal pregnancy. The observed TT genotype of DMNT3B suggests that this is the predominant genotype of this population. The findings provide new insights into the etiology of human spontaneous abortion.
Electronic supplementary material
The online version of this article (doi:10.1007/s10815-016-0837-7) contains supplementary material, which is available to authorized users.
Keywords: DNA methylation, Spontaneous abortion, DNA-methyltransferase, Single-nucleotide polymorphisms, Assisted reproductive technology
Introduction
Spontaneous abortion (SA) is a frustrating clinical problem, of which the exact etiology has not been well identified yet. An estimated 25 % of women attempting pregnancy have at least one spontaneous abortion, and about 10 % of all clinical pregnancies are lost in the first and early second trimesters [1]. Among those various possible etiological factors associated with SA, aberrant gene imprinting has been considered to be one of the possible causes [2]. Genomic imprinting is an epigenetic process, and one of the most studied epigenetic modifications is DNA methylation of cytosine residues within CpG dinucleotides [3]. In mammals, there are at least two developmental periods, in germ cells and in preimplantation embryos, in which methylation patterns are reprogrammed genome-wide, generating cells with a broad developmental potential [4–6]. Both animal and human studies have demonstrated that assisted reproduction technique (ART) procedures may disturb the normal imprinting process and cause methylation defects in individual genes, which might affect subsequent embryonic development and contribute to SA [7–9]. Previously, Zheng presented abnormal methylation patterns of PEG1, H19, LIT1, and SNRPN in aborted human chorionic villus. However, an increased occurrence of aberrant methylation in chorionic villus from ART pregnancies was not observed [10, 11].
DNA methylation, one of the most commonly studied epigenetic phenomena, is essential for normal embryonic development [12], and it plays important roles in the regulation of gene expression, X chromosome inactivation, genomic imprinting, chromatin modification, and silencing of endogenous retrovirus [13]. The methylation requires adding a methyl group to the 5′ position of the cytosine ring in the CpG dinucleotides to form 5-methylcytosine, which is established and maintained by DNA-methyltransferases (DNMTs). The DNMTs family includes DNMT1, DNMT2, DNMT3A, DNMT3B, and DNMT3L [14, 15]. DNMT3A and DNMT3B are both de novo methyltransferases and play an important role in the embryogenesis and the generation of aberrant methylation in carcinogenesis [16]. Inactivation of both Dnmt3a and Dnmt3b caused early embryonic lethality, and most homozygous mutant mice of Dnmt3a became runted and died at about 4 weeks of age. In contrast, no viable mice knockout of the Dnmt3b genes were recovered at birth [17]. In addition, [Dnmt3a (−/−), Dnmt3b (+/−)] mice failed to establish maternal methylation imprints, resulting in miscarriages. Both Dnmt3a and Dnmt3l are required for spermatogenesis; together, Dnmt3l may cooperate with Dnmt3 family methyltransferases to carry out de novo methylation of maternally imprinted genes in oocytes [18, 19].
To date, a number of studies have suggested that single-nucleotide polymorphisms (SNPs) within the promoter region of DNMT3A and DNMT3B genes could lead to modifications of gene expression levels and methylation patterns by changing promoter activities, thereby increasing the risk to various cancers [20–22]. Among those studies about polymorphisms of DNMT3s, DNMT3A −448A/G (GenBank accession no. NT_022184.14:g.4381840) and DNMT3B −149G/T (C46359T, GenBank accession no. AL035071) are two of the most widely studied SNPs. However, the roles of DNMT3A and DNMT3B polymorphisms in the etiology of SA have never been specifically investigated before. In order to explore the association of DNMT3A and DNMT3B promoter polymorphisms with the risk of spontaneous abortion loss after assisted reproduction techniques and natural conception, we compared DNMT3A and DNMT3B promoter polymorphism expression in chorionic villus samples (CVS) and muscle samples (MS) between spontaneous abortions and non-spontaneous abortions.
Materials and methods
Participants and samples
Informed consent was obtained from all individual participants included in the study. CVS were collected from women who underwent abortion procedures in the Department of Gynecology and Obstetrics in Nanfang Hospital of Southern Medical University from May 2008 to July 2011. MS were obtained from aborted fetuses and stillbirths during diagnostic examination at the Department of Pathology. The gestational age of each fetus was determined by the last menstrual period. Exclusion criteria included chromosomal abnormalities, endocrine diseases, infections and anatomical abnormalities of the genital tract, immunological diseases, trauma, signs of other concurrent medical complications, and any chemical agent intake before sample collection. Samples with aneuploidy were also excluded.
The samples were divided into two groups according to source: (a) 135 CVS and 17 MS from SA/stillbirth conceived by ART and spontaneous way (study group, 62 ART pregnancies and 90 natural conceptions), n = 152. In cases of SA, an intrauterine sac without fetal heartbeat was observed. (b) 142 CVS and 13 MS from normal early pregnancy (gestation age < 12 weeks) and second trimester (control group, 73 ART pregnancies and 82 natural conceptions), n = 155. Fetal heartbeat was observed in these pregnancies. These abortions were performed at the patient’s request for personal or social reasons. The duration of pregnancy ranged from 6 to 26 weeks, and the maternal age ranged from 18 to 45 years.
CVS were collected immediately after abortion or multi-fetal reduction and washed with physiological saline. Biopsies from the musculus quadriceps were dissected within 24–48 h after abortion. Then all the samples were stored at −80 °C until analyses.
DNA extraction and genotyping of SNPs
Genomic DNA was extracted after sample collection by proteinase K digestion, using a Genomic DNA Purification Kit (Promega, Madison, WI, USA). The transition of A > G of DNMT3A SNP creates a TaaI (Fermentas, Thermo Scientific, USA) restriction site, and for DNMT3B the variant T allele had an AvrII (Takara, Dalian, China) restriction site. Polymerase chain reaction–restriction fragment length polymorphism (PCR-RFLP) was used to detect this A-G transition in the promoter of DNMT3A at −448A > G and the C-T transition in the promoter of DNMT3B at −149C/T [20, 21, 23]. The following primers were used to amplify the 358 bp and 380 bp PCR products: 5′-ACACACCGCCCTCACCCCTT-3′ (forward) and 5′-TCCAGCAATCCCTGCCCACA-3′ (reverse) for DNMT3A, and 5′-TGCTGTGACAGGCAGAGCAG-3′ (forward) and 5′-GGTAGCCGGGAACTCCACGG-3′ (reverse) for DNMT3B. A total of 25 μL of PCR products was obtained, which comprised 100–200 ng of sample DNA, 2× Premix rTaq (Takara, Dalian, China), and 0.5 μM each primer. PCR cycle conditions consisted of an initial melting step of 95 °C for 2 min, followed by 40 cycles of 94 °C for 30 s, 65 °C for 45 s, 72 °C for 45 s, and a final extension step of 72 °C for 5 min. The amplified products were visualized by electrophoresis in 2 % agarose gels. The PCR products were digested with TaaI (for 20 min at 65 °C) for DNMT3A and with AvrII (for 20 min at 37 °C) for DNMT3B. Then the digested products were separated on a 4.0 % agarose gel and the RFLP bands visualized under ultraviolet light with ethidium bromide staining (additional data are given in Online Resource). For quality control, genotyping analysis was performed blind, with respect to case/control status, and repeated twice for all subjects. The results of genotyping were 100 % concordant. In order to confirm the genotyping results, randomly selected PCR-amplified DNA samples (n = 10, respectively, for each group) were examined by DNA sequencing, and the results were also 100 % concordant.
Statistical analyses
Patients and controls were compared using Student’s t test for continuous variables and chi square (χ 2) test for categorical variables. Quantitative data were calculated as mean ± SD. The Hardy-Weinberg equilibrium was tested with a goodness-of-fit χ 2 test to compare the observed genotype frequencies and the expected genotype frequencies among subjects. In addition, the SNPs of DNMT3A −448A/G and DNMT3B −149G/T were divided into three classes, namely, wild-type homozygotes, variant heterozygotes, and variant homozygotes. A comparison of the genotypes and allele distributions among the study groups was performed by means of two-sided contingency tables using χ 2 test or Fisher’s exact test. The odds ratio (OR) and 95 % confidence interval (CI) were calculated using the Statistical Package for the Social Sciences 16.0 statistical analyses program. All statistical tests were two-tailed and P <0.05 was considered statistically significant.
Results
Subject characteristics
A total of 307 subjects, 152 patients with SA/stillbirth (mean age, 31.4 ± 4.0 years; gestation weeks, 10.1 ± 4.4; range, 6–26 weeks) and 155 women undergoing selective pregnancy termination for non-medical reasons (mean age, 30.6 ± 4.2 years; gestation weeks, 9.4 ± 3.7; range, 6–24 weeks), were included in this study. No significant differences were observed regarding maternal age or gestation age between the study group and the control group (P = 0.120 and P = 0.148, respectively). The clinical characteristics of all subjects are summarized in Table 1. There was no significant difference in the gestational weeks distribution, but 23.0 % of the study group were ≥35 years, significantly higher than that of the control group (14.2 %) (P = 0.047).
Table 1.
Frequency distributions of clinical characteristics in spontaneous abortion patients (the study group) and the control group
| Characteristics | Study group (n = 152) | Control group (n = 155) | P valuea | ||
|---|---|---|---|---|---|
| Number | (%) | Number | (%) | ||
| Gestation | 0.409 | ||||
| <12 weeks | 135 | (88.8) | 142 | (91.6) | |
| ≥12 weeks | 17 | (11.2) | 13 | (8.4) | |
| Maternal age | 0.047 | ||||
| <35 years | 117 | (77.0) | 133 | (85.8) | |
| ≥35 years | 35 | (23.0) | 22 | (14.2) | |
aTwo-sided χ 2 test
Association of SNPs with risk of pregnancy loss
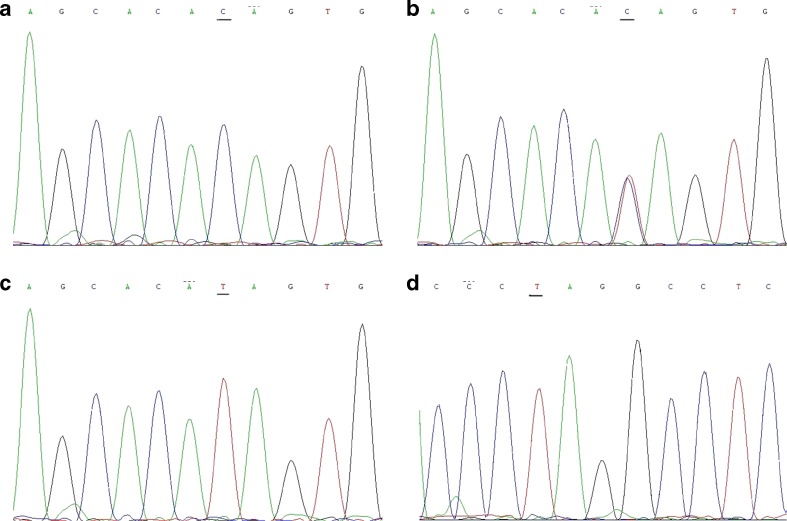
The −448A > G polymorphism in the promoter of DNMT3A gene and the −149C/T polymorphism of DNMT3B were first investigated in the CVS and MS from Chinese women with SA or normal pregnancy by PCR-RFLP. All subjects were successfully genotyped for the DNMT3A polymorphism and the SNP of DNMT3B. The DNMT3A genotypes AA, AG, and GG were detected in spontaneous abortions and the controls. For DNMT3B, we observed genotype frequencies of 100 % (TT) in study populations and control groups. The genotyping by PCR-RFLP analysis was completely confirmed by DNA sequencing analysis (Fig. 1).
Fig. 1.
Sequencing results for the PCR products from different genotypes of DNMT3A (a–c) and DNMT3B (d). The single-nucleotide polymorphism sites are underlined. The results were completely matched to the corresponding results derived from PCR-restriction fragment length polymorphism genotyping. a GG wild type (reverse strand), b AG variants (reverse strand), c AA variants (reverse strand), d TT variants (original sequence)
The DNMT3A A allele frequencies and genotype distributions in the two groups are summarized in Table 2, and the pregnancy loss risks related to the −448A > G genotype are also shown. We observed genotype frequencies of 45.4 % (GG), 40.8 % (AG), and 13.8 % (AA) in the SA and 68.4 % (GG), 30.3 % (AG), and 1.3 % (AA) in the control subjects, respectively, and the differences between the two groups were statistically significant (P < 0.001). The variant A allele frequency was 34.2 % for the pregnancy loss group and 14.5 % for the control subjects, and this difference was statistically significant (P < 0.001). The DNMT3A −448A > G polymorphisms were distributed in the Hardy-Weinberg equilibrium. The OR and their 95 % CI were calculated using the more common homozygous variant genotype as the reference group (−448 GG genotypes). Compared to the reference group, AA homozygotes had a >16-fold increased risk of pregnancy loss (OR, 16.130; 95 % CI, 3.128–14.034, P < 0.001) and AG heterozygotes an OR of 2.027 (95 % CI, 1.247–3.293, P = 0.004).
Table 2.
DNMT3A genotype and allele frequencies and their association with spontaneous abortion (SA)
| Genotypes | SA group (n = 152) |
Control group (n = 155) |
Odds ratio (95 % CI) |
P valuea | ||
|---|---|---|---|---|---|---|
| Number | (%) | Number | (%) | |||
| DNMT3A −448A > G | ||||||
| GG | 69 | (45.4) | 106 | (68.4) | 1.0 | |
| AG | 62 | (40.8) | 47 | (30.3) | 2.027 (1.247–3.293) | 0.004 |
| AA | 21 | (13.8) | 2 | (1.3) | 16.130 (3.665–70.984) | 0.000 |
| A allele | 34.2 % | 16.5 % | ||||
GG wild-type homozygotes, AG heterozygotes, AA variant homozygotes
aTwo-sided χ 2 test
When the analyses of DNMT3A were stratified by the maternal age and gestation, the AA and AG genotypes were associated with an increased risk of pregnancy loss (for AA, OR 16.897, 95 % CI = 3.810–74.925, P < 0.001; for AG, OR 2.293, 95 % CI = 1.371–3.834, P = 0.001) at <12 weeks, and the AA and AG genotypes were also associated with an increased risk of pregnancy loss (for AA, OR 28.868, 95 % CI = 3.734–223.156, P < 0.001; for AG, OR 1.900, 95 % CI = 1.111–3.251, P = 0.019) at <35 years. However, the distributions of −448A > G were not statistically significant among the gestational weeks ≥12 or the maternal age ≥35 years (Table 3).
Table 3.
Stratification analysis of DNMT3A genotype frequencies, ORs, and 95 % CIs
| Genotype | Spontaneous abortions (%) | Controls (%) | OR (95 % CI) | P value |
|---|---|---|---|---|
| Gestation | ||||
| <12 weeks | ||||
| GG | 58 (38.2) | 98 (63.2) | 1.0 | |
| AG | 57 (37.5) | 42 (27.1) | 2.293 (1.371–3.834) | 0.001 |
| AA | 20 (13.2) | 2 (1.3) | 16.897 (3.810–74.925) | 0.000 |
| ≥12 weeks | ||||
| GG | 11 (7.2) | 8 (5.2) | 1.0 | |
| AG | 5 (3.3) | 5 (3.2) | 0.727 (0.156–3.386) | 0.684 |
| AA | 1 (0.7) | 0 (0) | – | 0.240 |
| Age | ||||
| <35 years | ||||
| GG | 53 (34.9) | 90 (58.1) | 1.0 | |
| AG | 47 (30.9) | 42 (27.1) | 1.900 (1.111–3.251) | 0.019 |
| AA | 17 (11.2) | 1 (0.6) | 28.868 (3.734–223.156) | 0.000 |
| ≥35 years | ||||
| GG | 16 (10.5) | 16 (10.3) | 1.0 | |
| AG | 15 (9.9) | 5 (3.2) | 3.000 (0.880–10.229) | 0.074 |
| AA | 4 (2.6) | 1 (0.6) | 4.000 (0.402–39.827) | 0.211 |
Association of SNPs with conception modes
The frequency of −448A > G between the ART-derived pregnancies and the spontaneous pregnancies was analyzed. All subjects analyzed in this part were <12 weeks and those from the second trimester were excluded, considering that all samples ≥12 weeks are from spontaneous pregnancies. As shown in Table 4, the distribution of −448A > G in individuals derived from ART pregnancies was not statistically significantly compared with those derived from spontaneous pregnancies (P = 0.661). When the analyses of DNMT3A were stratified by pregnancy outcomes (SA or induced abortion), there was no difference in the distribution of −448A > G neither.
Table 4.
Distribution of −448A > G DNMT3A genotypes in ART-derived pregnancies and natural conceptions
| ART pregnancies (n = 135) | Natural conceptions (n = 142) | P value | |||||
|---|---|---|---|---|---|---|---|
| GG (%) | AG (%) | AA (%) | GG (%) | AG (%) | AA (%) | ||
| Total (n = 277) | 79 (58.5) | 47 (34.8) | 9 (6.7) | 77 (54.2) | 52 (36.6) | 13 (9.2) | 0.661 |
| Spontaneous abortion (n = 135) | 27 (43.5) | 27 (43.5) | 8 (12.9) | 31 (42.5) | 30 (41.1) | 12 (16.4) | 0.844 |
| Induced abortion (n = 142) | 52 (71.2) | 20 (27.4) | 1 (1.4) | 46 (66.7 %) | 22 (31.9) | 1 (1.4) | 0.839 |
Discussion
In this study, we first found that the A variant genotype of DNMT3A was associated with a significantly increased risk of pregnancy loss. Although the mechanism of this association is unknown, one possible explanation is that this G-A transition, which increases the DNMT3A promoter activity, may up-regulate gene expression that involves an aberrant de novo methylation of CpG islands in some development genes. Furthermore, we found that ART did not affect the distribution of DNMT3A promoter polymorphism −448A > G in spontaneous abortion and normal pregnancy, indicating that ART may not influence the methylation pattern of imprinting genes, which might affect subsequent embryonic development.
During the process of genomic imprinting, specific loci acquire differential DNA methylation in a parent-of-origin manner. It is believed that this differential methylation is then used by cells to maintain the mono-allelic expression patterns that characterize the imprinted gene [3]. Imprinted genes are thought to be particularly important in development, growth, behavior, and stem cells [24]. Abnormal methylation patterns of imprinted genes have been demonstrated to be associated with fetal growth abnormalities [25].
DNMT3A and DNMT3B, the important methyltransferases, can catalyze DNA methylation and serve an essential function in embryonic development and the generation of aberrant methylation in tumorigenesis. In rodents, Dnmt3b and Dnmt3a were highly expressed in embryonic stem cells. However, after cell differentiation, the expressions of these genes decreased significantly in somatic cells and maintained in the low level [14]. Inactivation of both Dnmt3a and Dnmt3b disrupted de novo methylation in mouse embryonic stem cells and genome-wide de novo methylation occurring during early development, but it had no discernible effect on the maintenance of preexisting methylation patterns [17]. Kaneda et al. reported that offspring from Dnmt3a conditional mutant females die in utero and lack methylation and allele-specific expression at all maternally imprinted loci examined. Dnmt3a conditional mutant males showed impaired spermatogenesis and lacked methylation at two of three paternally imprinted loci examined in spermatogonia [26]. In germline stem cells, the loss of Dnmt3a/Dnmt3b resulted in hypomethylation in SineB1 and spermatogenic defects [27]. Dnmt3a/Dnmt3b-deficient hematopoietic stem cells lost self-renewal activity and were incapable of long-term reconstitution in irradiated animals [28]. Thus, we hypothesized that aberrant expression of DNMT3A and DNMT3B might lead to the defects of genomic imprinting, thereby affecting normal gametogenesis and fetal development.
With the completion of the human genome project, the relationship between individual genetic variation and disease has been paid more and more attention. SNP, one of the most common human genetic variations, is associated with many phenotypic differences in the human body and the susceptibility to drugs or diseases. It is helpful to identify candidate genes and key regulatory factors of certain diseases by combining the SNPs of some specific localization points in the genome with the features of related diseases, such as pregnancy loss. It is suggested that IL-17F polymorphism (rs763780), two receptor gene polymorphisms [KDR (Q472H) and PROKR2 (V331M)], and the T657C polymorphism of the SYCP3 gene might be associated with a high risk of idiopathic recurrent pregnancy loss in humans [29–31]. To date, public databases have proposed several candidate SNPs in the DNMT3A and 3B genes. Among these SNPs, A/G in the 448 bp and G/T in the 149 bp upstream of the transcription start site of the promoter region, respectively, have been widely explored. Fan et al. used the luciferase assay to prove that the promoter activity of the −448A allele of DNMT3A was significantly higher than that of the −448G allele, which also increased the risk of gastric cancer [20]. In addition, the C-T transition polymorphism in the DNMT3B promoter was found to significantly increase the promoter activity of DNMT3B gene, thereby increasing the risk of lung cancer [21]. As far as we know, no paper was available on the role of DNMT3A and 3B polymorphisms in the risk of pregnancy loss. Yin et al. found that DNMT1 expression and DNA global methylation levels were significantly down-regulated in villous of early pregnancy loss [32]. Here, we found that the −448A > G polymorphism of DNMT3A was associated with a significantly increased risk of pregnancy loss. Individuals bearing the AA genotype had a 16-fold increased risk of pregnancy loss than GG genotype carriers, suggesting that polymorphism −448A > G in the human DNMT3A promoter may affect the promoter activity of DNMT3A gene and DNA methylation levels, being involved in the progression of pregnancy loss. Besides, this SNP could be a predictor of SA for those women <35 years in the first trimester. No associations were observed between the SNP and risk of SA among those whose gestational weeks ≥12 or maternal age ≥35 years. However, the A variant genotype still had a 3–4-fold increased risk of pregnancy loss among the advanced age, owing to the small number of these samples after stratification; the differences were not significant and further studies are required. Advanced age is a risk factor for female infertility, pregnancy loss, fetal anomalies, and stillbirth, and age-related aneuploidy is the most common reason for pregnancy loss [33]. In the current study, we merely analyzed the prevalence of DNMT3A promoter polymorphism in euploid embryos, and the age of the patients with SA/stillbirth was comparable to that of women with normal pregnancy. It thus suggested that the A variant genotype of −448A > G in the DNMT3A promoter region in pregnancy loss was not related to maternal age.
ART interventions are often implemented during the phase of reprogramming of gene imprinting, which is required for generating functional germ cells, and failure to do this appropriately usually results in infertility or developmentally abnormal embryos that die during gestation. Therefore, it was suggested that ART procedures might affect the methylation process in gametes and embryos [34, 35]. However, more recent studies failed to support these findings and could not demonstrate an association between imprinting disorders and ARTs [36, 37]. In the present study, no association was found between the DNMT3A polymorphism −448A > G with conception modes, suggesting that ART might not change the expression of DNMT3A gene and the methylation patterns of imprinting genes. Previously, Shi presented that methylation errors of H19 differentially methylated region (DMR), PEG1 DMR, and KvDMR1 were, respectively, found in 8.0 %, 16.9 %, and 10.4 % day 3 embryos of poor quality [38]. Thus, it cannot be neglected that the ART group represents selected embryos that developed normally during in vitro culture and were suitable for transfer to patients, according to morphological criteria, while SAs after natural conceptions do not undergo any artificial selection. Since imprinting disorders are so rare, further multi-center randomized studies of larger sample size are needed to address the question whether ART is associated with an increased risk of imprinting disorders [39].
The ICF syndrome (for immunodeficiency, centromere instability, and facial anomalies), the only genetic disorder known to involve constitutive abnormalities of genomic methylation patterns, was found to have mutations in both alleles of the gene that encodes DNMT3B [40]. Okano et al. analyzed immortalized lymphoblasts derived from an affected individual and confirmed that the intronic mutation and the altered DNMT3B transcript are responsible for ICF syndrome. Inactivation of Dnmt3b in mouse embryonic fibroblasts results in DNA hypomethylation, chromosomal instability, and spontaneous immortalization [41]. The above studies suggest that DNMT3B gene plays an important role in normal growth and development of disease. Several previous studies have shown that polymorphism of DNMT3B −149C > T is associated with cancer development in a variety of tumors [21, 42, 43]. We observed genotype frequencies of 100 % (TT) in all 307 tissues. Since no papers have reported the SNPs of DNMT3B in embryonic or extraembryonic tissues, this observed TT genotype of −149C > T might represent the predominant genotype and not be involved in the pathogeny of SA, which needs further research.
Several limitations persist when interpreting the present findings. First, we merely measured one single polymorphism of DNMT3A and 3B, and thus, further study covering more SNPs is needed. Second, the exact effects of the genetic variants of DNMT3A −448A > G or DNMT3B −149G > T on the expression of their individual enzymatic activities could not be determined. Additional investigations on the underlying molecular mechanism of the polymorphisms and the expression of DNMT3s are warranted. Finally, the sample size of the present study was small, particularly for the analysis of gene polymorphisms and stratification.
In summary, our study provides the first evidence that the −448A > G polymorphism of the DNMT3A promoter is significantly associated with an increased risk of pregnancy loss in the Chinese population, especially for early pregnancy loss (<12 weeks) in younger women (<35 years). The potential mechanism for the higher risk associated with the variants may be an increased activity of G > A. These results suggest that the polymorphism of DNMT3A could be used as an important marker of genetic susceptibility to SA, although additional studies using larger sample sizes are required to confirm our findings. ART may not affect the distribution of −448A > G in SA or normal pregnancy, suggesting that ART might not affect the DNA methylation of imprinted genes. Meanwhile, it is possible that the observed TT genotype of DMNT3B is the predominant genotype of this population.
Electronic supplementary material
Below is the link to the electronic supplementary material.
PCR-based restriction fragment length polymorphism genotyping of DNMT3A (a) and DNMT3B (b). a The wild-type G allele consists of a TaaI restriction site that results in two bands (153, 94, and 87 bp; 87 and 94 bp cannot be distinguished by electrophoresis, being combined into one band), the variant A allele also produces two bands (247 and 87 bp). Lanes 3–5, 7, 9, and 12: GG wild-type; lanes 1, 2, 6, 8, 11, and 13: AG heterozygotes; and lanes 10: AA variant (lanes 1–6, samples from the study group; lanes 7–13, samples from the control group). b The variant T allele had an AvrII restriction site that results in two bands (207 and 173 bp), and the wild-type C allele lacked the AvrII restriction site and therefore produced a single 380-bp band. Lanes 1–16: TT variant (lanes 1–8, samples from the study group; lanes 9–16, samples from the control group). (GIF 37 kb)
Acknowledgments
The authors gratefully acknowledge the technical support and valuable suggestions of all members from the Research Center of Clinical Medicine of Nanfang Hospital, and the help of the staff from the Department of Gynecology and Obstetrics in Nanfang Hospital in sample collection.
Compliance with ethical standards
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional research committee and with the 1964 Helsinki declaration and its later amendments.
Funding
This study was funded by the National Natural Science Foundation of China (no. 81170574 and no. 31371517) and High-level Project Matching Foundation of Nanfang Hospital, Southern Medical University (G201206).
Conflict of Interest
The authors declare that they have no competing interests.
Footnotes
Capsule The DNMT3A −448A > G polymorphism is associated with the risk of human spontaneous abortion in Chinese population and assisted reproduction techniques may not affect the distribution of −448A > G in pregnancy loss and normal pregnancy.
References
- 1.Miller JF, Williamson E, Glue J, Gordon YB, Grudzinskas JG, Sykes A. Fetal loss after implantation. A prospective study. Lancet. 1980;2(8194):554–6. doi: 10.1016/S0140-6736(80)91991-1. [DOI] [PubMed] [Google Scholar]
- 2.Ono R, Nakamura K, Inoue K, Naruse M, Usami T, Wakisaka-Saito N, et al. Deletion of Peg10, an imprinted gene acquired from a retrotransposon, causes early embryonic lethality. Nat Genet. 2006;38(1):101–6. doi: 10.1038/ng1699. [DOI] [PubMed] [Google Scholar]
- 3.Adalsteinsson BT, Ferguson-Smith AC. Epigenetic control of the genome-lessons from genomic imprinting. Genes (Basel) 2014;5(3):635–55. doi: 10.3390/genes5030635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293(5532):1089–93. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 5.Santos F, Hendrich B, Reik W, Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol. 2002;241(1):172–82. doi: 10.1006/dbio.2001.0501. [DOI] [PubMed] [Google Scholar]
- 6.Guo H, Zhu P, Yan L, Li R, Hu B, Lian Y, et al. The DNA methylation landscape of human early embryos. Nature. 2014;511(7511):606–10. doi: 10.1038/nature13544. [DOI] [PubMed] [Google Scholar]
- 7.Benoff S, Hurley IR. Epigenetic and experimental modifications in early mammalian development: part I. Preface Hum Reprod Update. 2001;7(2):211–6. doi: 10.1093/humupd/7.2.211. [DOI] [PubMed] [Google Scholar]
- 8.Khosla S, Dean W, Brown D, Reik W, Feil R. Culture of preimplantation mouse embryos affects fetal development and the expression of imprinted genes. Biol Reprod. 2001;64(3):918–26. doi: 10.1095/biolreprod64.3.918. [DOI] [PubMed] [Google Scholar]
- 9.Zechner U, Pliushch G, Schneider E, El HN, Tresch A, Shufaro Y, et al. Quantitative methylation analysis of developmentally important genes in human pregnancy losses after ART and spontaneous conception. Mol Hum Reprod. 2010;16(9):704–13. doi: 10.1093/molehr/gap107. [DOI] [PubMed] [Google Scholar]
- 10.Zheng HY, Shi XY, Wu FR, Wu YQ, Wang LL, Chen SL. Assisted reproductive technologies do not increase risk of abnormal methylation of PEG1/MEST in human early pregnancy loss. Fertil Steril. 2011;96(1):84–9. doi: 10.1016/j.fertnstert.2011.04.021. [DOI] [PubMed] [Google Scholar]
- 11.Zheng HY, Tang Y, Niu J, Li P, Ye DS, Chen X, et al. Aberrant DNA methylation of imprinted loci in human spontaneous abortions after assisted reproduction techniques and natural conception. Hum Reprod. 2013;28(1):265–73. doi: 10.1093/humrep/des358. [DOI] [PubMed] [Google Scholar]
- 12.Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet. 2002;3(9):662–73. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- 13.Bestor TH. The DNA, methyltransferases of mammals. Hum Mol Genet. 2000;9(16):2395–402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 14.Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet. 1998;19(3):219–20. doi: 10.1038/890. [DOI] [PubMed] [Google Scholar]
- 15.Bourc’His D, Xu GL, Lin CS, Bollman B, Bestor TH. Dnmt3L and the establishment of maternal genomic imprints. Science. 2001;294(5551):2536–9. doi: 10.1126/science.1065848. [DOI] [PubMed] [Google Scholar]
- 16.Robertson KD, Uzvolgyi E, Liang G, Talmadge C, Sumegi J, Gonzales FA, et al. The human DNA methyltransferases (DNMTs) 1, 3a and 3b: coordinate mRNA expression in normal tissues and overexpression in tumors. Nucleic Acids Res. 1999;27(11):2291–8. doi: 10.1093/nar/27.11.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99(3):247–57. doi: 10.1016/S0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 18.Suetake I, Shinozaki F, Miyagawa J, Takeshima H, Tajima S. DNMT3L stimulates the DNA methylation activity of Dnmt3a and Dnmt3b through a direct interaction. J Biol Chem. 2004;279(26):27816–23. doi: 10.1074/jbc.M400181200. [DOI] [PubMed] [Google Scholar]
- 19.Hata K, Okano M, Lei H, Li E. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development. 2002;129(8):1983–93. doi: 10.1242/dev.129.8.1983. [DOI] [PubMed] [Google Scholar]
- 20.Fan H, Liu D, Qiu X, Qiao F, Wu Q, Su X, et al. A functional polymorphism in the DNA methyltransferase-3A promoter modifies the susceptibility in gastric cancer but not in esophageal carcinoma. BMC Med. 2010;8:12. doi: 10.1186/1741-7015-8-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen H, Wang L, Spitz MR, Hong WK, Mao L, Wei Q. A novel polymorphism in human cytosine DNA-methyltransferase-3B promoter is associated with an increased risk of lung cancer. Cancer Res. 2002;62(17):4992–5. [PubMed] [Google Scholar]
- 22.Zhang W, Xu Y, Ma G, Qi W, Gu H, Jiang P. Genetic polymorphism of DNA methyltransferase 3A rs1550117 A > G and risk of cancer: a meta-analysis. J Investig Surg. 2015;28(6):346–53. doi: 10.3109/08941939.2015.1010024. [DOI] [PubMed] [Google Scholar]
- 23.Chung CJ, Chang CH, Liu CS, Huang CP, Chang YH, Chien SN, et al. Association of DNA methyltransferases 3A and 3B polymorphisms, and plasma folate levels with the risk of urothelial carcinoma. PLoS One. 2014;9(8) doi: 10.1371/journal.pone.0104968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Plasschaert RN, Bartolomei MS. Genomic imprinting in development, growth, behavior and stem cells. Development. 2014;141(9):1805–13. doi: 10.1242/dev.101428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Piedrahita JA. The role of imprinted genes in fetal growth abnormalities. Birth Defects Res A Clin Mol Teratol. 2011;91(8):682–92. doi: 10.1002/bdra.20795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaneda M, Okano M, Hata K, Sado T, Tsujimoto N, Li E, et al. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature. 2004;429(6994):900–3. doi: 10.1038/nature02633. [DOI] [PubMed] [Google Scholar]
- 27.Takashima S, Takehashi M, Lee J, Chuma S, Okano M, Hata K, et al. Abnormal DNA methyltransferase expression in mouse germline stem cells results in spermatogenic defects. Biol Reprod. 2009;81(1):155–64. doi: 10.1095/biolreprod.108.074708. [DOI] [PubMed] [Google Scholar]
- 28.Tadokoro Y, Ema H, Okano M, Li E, Nakauchi H. De novo DNA methyltransferase is essential for self-renewal, but not for differentiation, in hematopoietic stem cells. J Exp Med. 2007;204(4):715–22. doi: 10.1084/jem.20060750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Najafi S, Hadinedoushan H, Eslami G, Aflatoonian A. Association of IL-17A and IL-17 F gene polymorphisms with recurrent pregnancy loss in Iranian women. J Assist Reprod Genet. 2014;31(11):1491–6. doi: 10.1007/s10815-014-0294-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sazegari A, Kalantar SM, Pashaiefar H, Mohtaram S, Honarvar N, Feizollahi Z, et al. The T657C polymorphism on the SYCP3 gene is associated with recurrent pregnancy loss. J Assist Reprod Genet. 2014;31(10):1377–81. doi: 10.1007/s10815-014-0272-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Su MT, Lin SH, Chen YC, Kuo PL. Gene-gene interactions and gene polymorphisms of VEGFA and EG-VEGF gene systems in recurrent pregnancy loss. J Assist Reprod Genet. 2014;31(6):699–705. doi: 10.1007/s10815-014-0223-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yin LJ, Zhang Y, Lv PP, He WH, Wu YT, Liu AX, et al. Insufficient maintenance DNA methylation is associated with abnormal embryonic development. BMC Med. 2012;10:26. doi: 10.1186/1741-7015-10-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sauer MV. Reproduction at an advanced maternal age and maternal health. Fertil Steril. 2015;103(5):1136–43. doi: 10.1016/j.fertnstert.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 34.van Montfoort AP, Hanssen LL, de Sutter P, Viville S, Geraedts JP, de Boer P. Assisted reproduction treatment and epigenetic inheritance. Hum Reprod Update. 2012;18(2):171–97. doi: 10.1093/humupd/dmr047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.White CR, Denomme MM, Tekpetey FR, Feyles V, Power SG, Mann MR. High frequency of imprinted methylation errors in human preimplantation embryos. Sci Rep. 2015;5:17311. doi: 10.1038/srep17311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Camprubi C, Iglesias-Platas I, Martin-Trujillo A, Salvador-Alarcon C, Rodriguez MA, Barredo DR, et al. Stability of genomic imprinting and gestational-age dynamic methylation in complicated pregnancies conceived following assisted reproductive technologies. Biol Reprod. 2013;89(3):50. doi: 10.1095/biolreprod.113.108456. [DOI] [PubMed] [Google Scholar]
- 37.Laprise SL. Implications of epigenetics and genomic imprinting in assisted reproductive technologies. Mol Reprod Dev. 2009;76(11):1006–18. doi: 10.1002/mrd.21058. [DOI] [PubMed] [Google Scholar]
- 38.Shi X, Chen S, Zheng H, Wang L, Wu Y. Abnormal DNA methylation of imprinted loci in human preimplantation embryos. Reprod Sci. 2014;21(8):978–83. doi: 10.1177/1933719113519173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Horsthemke B, Ludwig M. Assisted reproduction: the epigenetic perspective. Hum Reprod Update. 2005;11(5):473–82. doi: 10.1093/humupd/dmi022. [DOI] [PubMed] [Google Scholar]
- 40.Xu GL, Bestor TH, Bourc’His D, Hsieh CL, Tommerup N, Bugge M, et al. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature. 1999;402(6758):187–91. doi: 10.1038/46052. [DOI] [PubMed] [Google Scholar]
- 41.Dodge JE, Okano M, Dick F, Tsujimoto N, Chen T, Wang S, et al. Inactivation of Dnmt3b in mouse embryonic fibroblasts results in DNA hypomethylation, chromosomal instability, and spontaneous immortalization. J Biol Chem. 2005;280(18):17986–91. doi: 10.1074/jbc.M413246200. [DOI] [PubMed] [Google Scholar]
- 42.Wang C, Jia Z, Cao D, You L, Jin M, Wu X, et al. Polymorphism of DNA methyltransferase 3b and association with development and prognosis in gastric cancer. PLoS One. 2015;10(8) doi: 10.1371/journal.pone.0134059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iacopetta B, Heyworth J, Girschik J, Grieu F, Clayforth C, Fritschi L. The MTHFR C677T and DeltaDNMT3B C-149T polymorphisms confer different risks for right- and left-sided colorectal cancer. Int J Cancer. 2009;125(1):84–90. doi: 10.1002/ijc.24324. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PCR-based restriction fragment length polymorphism genotyping of DNMT3A (a) and DNMT3B (b). a The wild-type G allele consists of a TaaI restriction site that results in two bands (153, 94, and 87 bp; 87 and 94 bp cannot be distinguished by electrophoresis, being combined into one band), the variant A allele also produces two bands (247 and 87 bp). Lanes 3–5, 7, 9, and 12: GG wild-type; lanes 1, 2, 6, 8, 11, and 13: AG heterozygotes; and lanes 10: AA variant (lanes 1–6, samples from the study group; lanes 7–13, samples from the control group). b The variant T allele had an AvrII restriction site that results in two bands (207 and 173 bp), and the wild-type C allele lacked the AvrII restriction site and therefore produced a single 380-bp band. Lanes 1–16: TT variant (lanes 1–8, samples from the study group; lanes 9–16, samples from the control group). (GIF 37 kb)