Abstract
Purpose
Kartagener syndrome (KS), also known as visceral inversion-nasosinusitis-bronchiectasis syndrome, or familial bronchiectasis, is an autosomal recessive inherited disease. In this study, through two cases of KS, we aimed to assess the clinical and genetic characteristics of KS caused by DNAH5 mutations.
Methods
The two cases of KS from the same family underwent extensive clinical assessments, with next-generation DNA sequencing and bioinformatics analysis to identify pathogenic genes. In addition, Sanger sequencing was used to verify the pedigrees.
Results
The present study employed a directional capture strategy for hereditary disease screening, which correctly identified the virulence sites in the pedigree, and facilitated the differential diagnosis among multiple genes. Two novel mutations were detected in DNAH5: c.7778C>T (missense mutation) and c.13729G>A (nonsense mutation). They were not found in dbSNP, 1000 Genomes, and ExAC.
Conclusions
These findings demonstrated that new DNAH5 mutations could be used for molecular diagnosis of KS, providing families with genetic counseling and prenatal diagnosis.
Keywords: Kartagener syndrome, DNAH5, Gene sequencing, Visceral inversion, Mutation, Precision medicine
Introduction
Kartagener syndrome (KS), also known as visceral inversion-nasosinusitis-bronchiectasis syndrome, or familial bronchiectasis, is an autosomal recessive inherited disease. To date, cases with X-linked inheritance have been identified [1].
The first case of bronchiectasis accompanied with visceral inversion was reported by Siewart in 1904. Further in 1933, four cases with the triad of complete visceral inversion, bronchiectasis, and paranasal sinusitis were identified by Kartagener, after whom the disease was named. Afzelius demonstrated by electron microscopy that the disease is caused by immotile cilia due to congenital ultrastructural abnormalities and named this disease immotile cilia syndrome. However, Sleigh et al. showed in 1981 that the cilia are not absolutely immotile; on the contrary, they have abnormal movements, and secretions could not be effectively expelled, consequently inducing bronchiectasis. Therefore, this congenital disease was termed primary ciliary dyskinesia (PCD) or dyskinesia cilia syndrome (DSC). KS is a subtype of PCD, referring to PCD cases accompanied with visceral ectopia. KS tends to run in families, and could occur in the same or different generations, and the parents of most patients are close relatives. The incidence of PCD is about 1/16,000 [2]; meanwhile, studies have reported an incidence of birth defects patients with PCD is about 1/20,000–1/60,000 [3].
KS is pathologically based on movement disorders of the ciliated respiratory epithelium; thus, the transportation function of mucosal cilia is decreased, and secretions cannot be effectively expelled. This induces repeated, long-term chronic infection and causes bronchiectasis and paranasal sinusitis. As cilia are abundant in the respiratory tract, middle ear, oviduct, spermatic flagellum, and brain and spinal ependyma, KS could be accompanied with pneumonia, conductive deafness, ectopic pregnancy, infertility, and hydrocephalus. KS onset mainly occurs in childhood, but this disease is easily misdiagnosed [4]. The major clinical presentations of KS are repeated cough, expectoration, hemoptysis, nasal congestion, running nose, dizziness, and headache. In addition, some patients may seek healthcare services for infertility. Imaging could provide important evidence for the diagnosis of KS. Indeed, chest X-ray, CT scanning, and ultrasound examinations could identify visceral inversion.
KS is a hereditary disease, which is associated with multiple virulence genes. Therefore, KS diagnosis requires not only clinical data but also evidence of molecular diagnosis. The clinical presentations of KS vary greatly, and multiple virulence genes are involved; this makes its diagnosis and differential diagnosis difficult [5]. Currently, genetic testing is used for precise diagnosis; for instance, Sanger sequencing, amplification refractory mutation system-polymerase chain reaction (ARMS-PCR), and Droplet Digital PCR (ddPCR) measure the sequences of certain genes. However, the number of genes is very high, and the above methods are costly, with lengthy measurement times; therefore, it is very difficult to use them in clinical practice, especially for diagnosing diseases with similar characteristics. Next-generation sequencing (NGS) could provide high throughput sequencing of multiple genes, including all genes associated with a given disease, thus reducing cost and measurement time. Therefore, NGS is suitable for gene screening of hereditary diseases with similar clinical characteristics [6]. In the present study, we used the chip capturing high-throughput sequencing method for whole exon sequencing of monogenic pulmonary disease-related genes and successfully identified a virulence gene site in one pedigree. To our knowledge, this novel mutation is the first report about DNAH5, which has a certain value for the epidemiological investigation of KS in China. In addition, the identification of this gene site could further complement the Hereditary Disease Library of China, facilitate KS diagnosis, and promote the development of precision medicine, including preimplantation genetic screening (PGS), preimplantation genetic diagnosis (PGD), prenatal screening, and prenatal diagnosis.
Patients and methods
Patients
Between October 2014 and September 2015, two children (a 2-month old boy and his 9.5-year-old sister) from the same family suspected with KS were enrolled from the 7th Pediatric Department of our hospital of Hunan Province and Pediatric Outpatient Department, respectively. The present study was approved by the Ethics Committee of our hospital; informed consent was obtained from the children’s families. Medical history, physical examination data, and related examination data of the two children were collected.
Measuring of virulence genes
Sample collection
A total of 4 mL blood was collected from the proposituses and their parents into EDTA anticoagulant tubes, and BloodGen Midi Kit (Kangwei Century biological technology co., LTD, Beijing, China) was used for genomic DNA extraction.
Target sequence capturing and high-throughput sequencing
According to previous references and the OMIM database, the NimbleGencapture probe (Roche) was customized for the exon regions of over 4000 hereditary diseases included in the OMIM database and used for full exon capture of target genes.
Library preparation
First, genomic DNA was fragmented to about 200 bp by the Cavoris system. Then, end-filling repair of the fragmented DNA was performed using Klenow Fragment, T4 DNA polymerase, and T4 PNK. For 3′- adenylation, an A base was added to the 3′ end of the repaired product with the polymerase system, which was used for subsequent connection. Afterwards, the T4 DNA ligase reaction system was prepared, and the adapter and products with the A base added were ligated on a Thermo mixer under appropriate temperature. The ligation products were amplified for 4–6 cycles by LM-PCR. For hybridization, the library and probes were mixed in a hybridization system at 65 °C for 60–68 min. After magnetic bead washing, streptomycin labeled magnetic beads were incubated with the hybridized samples, and then eluted. Finally, elution products were amplified for 10 cycles by LM-PCR.
Illumina sequencing
Sequencing was performed on a standard Illumina hiseq2500 sequencing system; original sequencing images were obtained and analyzed by the official Illumina base call analysis software; BclToFastq was used to generate raw data.
Data analysis
For analyzing basic information, raw data were processed by removing connection contamination, ruling out low-quality data. Final data were compared with reference sequences using the BWA software, with the hg38 genome as the reference genome. For SNP detection and annotation, the Samtools software was used for analysis. The Pindel software was used for Indel detection and annotation; false positive mutations were ruled out according to sequencing depth and mutation quality, with the Indel used for screening high-quality, reliable mutations. Mutations were annotated according to the locations of SNP and Indel on gene sequences; the effects on amino acid changes, gene splicing, UTR, and intron mutations were analyzed. Algorithms such as homologous alignment and conservativeness of protein structure were applied using SIFT to predict the effects of the screened mutations on proteins, and splicing harmfulness of the mutation close to splicing sites was predicted. Further data analysis was performed to explore the hereditary mode and identify the mutations with the clinical presentations matching the children’s symptoms.
Verification with Sanger sequencing
Primers were designed according to DNAH5 sequences confirmed beforehand. PCR was used for amplification, with the ABI 3730XL sequencer used for sequencing, with the original PCR primers. The DNASTA software was used for sequence analysis and comparison, with NM_003560 employed as the mRNA comparison template. Sanger sequencing was performed for the samples from the children’s parents for verification.
Results
Clinical data
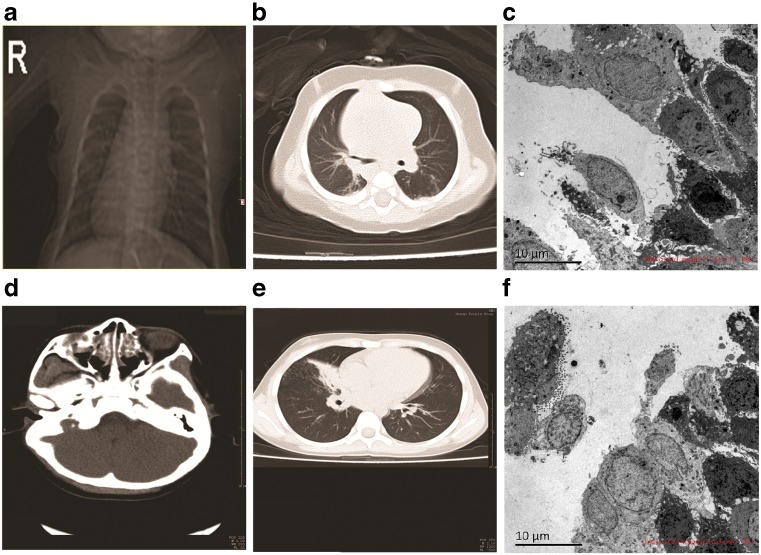
Case report: Case 1 was a 2-month-old boy (referred to as the younger brother) hospitalized due to cough and nasal congestion for 3 days. He was diagnosed with pneumonia, cytomegalovirus infection, and complete visceral inversion. Physical examination showed red throat, coarse respiratory sounds in bilateral lungs; moderate to coarse moist rale was noticed, while no pleural friction sound was found. The heart was on the right side; no protrusion of precordium was found, with no abnormality in auscultation. The liver was touched on the left side of the abdomen, and no other abnormality was found. The boy was hospitalized for 7 days, received treatments such as anti-infection therapy, sputum suction, and antiasthma by aerosol inhalation, and discharged after disease improvement. However, intermittent cough, wheezing, and nasal congestion remained after discharge, and milk choking occurred occasionally. The boy was the second birth on second pregnancy, delivered by full-term cesarean section, with no birth injury or history of asphyxia recorded. He weighed 3.2 kg at birth and received mixed feeding; growth and development were in agreement with the children of the same. In laboratory examinations, normal results were obtained for routine blood and urine, liver function, renal function, and myocardial enzyme assessments. C-reactive protein levels were normal; antibodies against pneumonia and mycoplasma were negative, and CMV-IgM was positive. Chest X-ray revealed that the lung markings in bilateral lungs were increased, disarranged, and slightly unclear, while no evident patchy shadow was found. The sizes of bilateral lung hilum were normal; the cardiac apex was rightward, with heart size and shape in normal ranges (Fig. 1a). Chest CT scanning showed that the heart shadow was evident at the right side; thoracic aorta was on the right side of the thoracic cavity, and lung markings in bilateral lungs were increased and unclear. Diffused high-density patchy unclear shadows were found in bilateral lungs. No enlarged lymph nodes were found at the bilateral lung hilum and mediastinum, and openings of the segmental and lobe bronchus were unobstructed. No pleural effusion was found, while evident liver and stomach inversions were noticed. These imaging presentations suggested bronchopneumonia and indicated complete visceral inversion (Fig. 1b). Bronchofiberscopy suggested epiglottic cartilage softening, endobronchitis, and left and right lung inversion. Electron microscopy of the bronchial mucous membrane showed that mucosal tissues were mainly composed of columnar epithelium; columnar epithelial cell hyperplasia, increased layers, and irregular arrangement were found. In addition, inflammatory cells (mainly neutrophils) were mixed among the columnar epithelial cells, and microvilli at the epithelial surface were sparse and fine, with no cilia found. Collagenous hyperplasia was found under the mucosa, with the infiltration of large amounts of inflammatory cells (Fig. 1c).
Fig. 1.
a Chest X-ray image of the boy. b Chest CT image of the boy. c Electron micrograph of the bronchial mucosa of the boy. d Nasosinus CT image of the girl. e Chest CT image of the girl. f Electron micrograph of the bronchial mucosa of the girl
Case 2 was the elder sister of the above boy. She was 9.5 years old and received treatment in the outpatient department of our hospital in September 2015, for repeated pneumonia and nasosinusitis for 5 years. The girl was diagnosed with pneumonia, mycoplasma infection, and nasosinusitis, and received treatments of anti-inflammation, phlegm dispersion, and aerosol inhalation for 5 days, with symptoms improvement. This girl was the first born on first pregnancy and delivered by full-term natural birth, with no birth injury or history of asphyxia. She weighed 3.4 kg at birth and received breast feeding; growth and development were in agreement with children of the same age. She had relatively poor health conditions since birth, with a history of pneumonia and nasosinusitis. Physical examination showed red throat, grade II tonsil enlargement, and pressing pain in the nasosinus area. Lung auscultation showed coarse respiratory sounds and coarse moist rale in bilateral lungs; respiratory sounds in the right lung were relatively low. Laboratory examinations showed normal routine blood and urine results; kidney and liver functions were abnormal; C-reactive protein was 32 mg/L, and antibodies against pneumonia and mycoplasma were positive. Nasosinus CT scanning showed paranasal sinusitis, bilateral inferior turbinate hypertrophy, and nasal septum deviation (Fig. 1d). Chest CT scanning showed inflammation of the lower lobe of right lung, consolidation and atelectasis of the middle lobe of right lung (right middle lobe syndrome) (Fig. 1e). Mucosal fibers (bronchial mucosa) were biopsied for electronic microscopy and showed large amounts of bronchial gland and epithelial cells with large amounts of secretory granules (mucous) in the cytoplasm; these cells were arranged into a lumen-shaped structure, surrounded by large amounts of collagen fibers. Small amounts of tall columnar cells were also found, with no cilia on the surface. Small amounts of inflammatory and red blood cells were also observed, with no typical alveolar epithelial cells or ciliary alveoli. In addition, no ciliated cells were found (Fig. 1f).
Family history
The children’s parents had good health conditions. No similar disorders were reported, and they were not close relatives.
Genetic testing
Exon capturing high-throughput sequencing, bioinformatics analysis, and clinical data revealed two suspicious mutation sites in the DNAH5 gene in the boy. After verification by first generation sequencing and pedigree analysis, the following results were obtained:
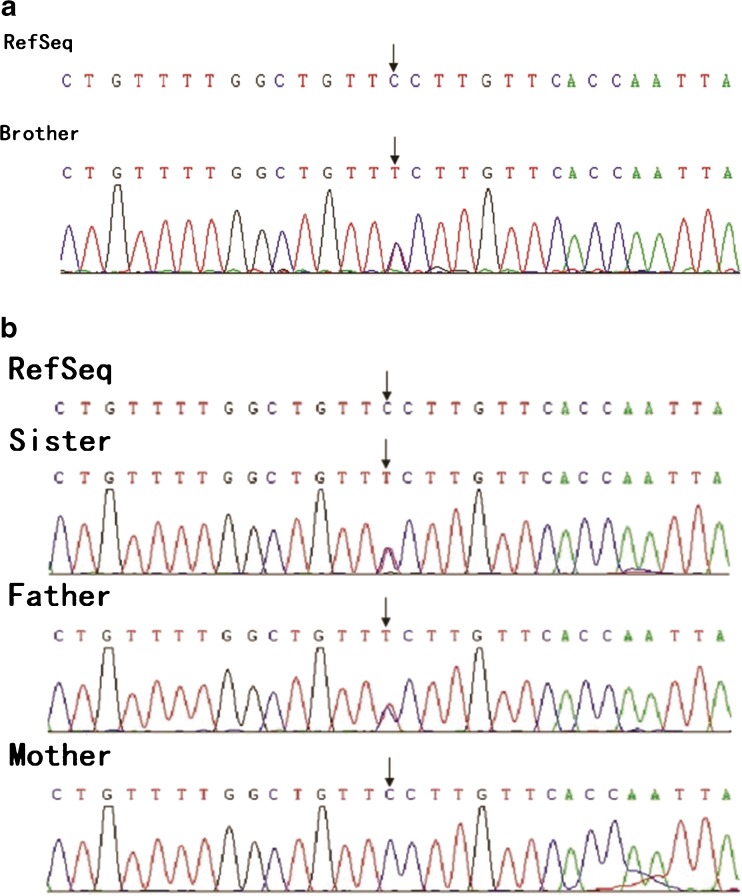
A missense mutation c.7778C>T (p.G2593E) was found in the DNAH5 gene in both children, who carried heterozygous mutations predicted to be harmful by the software (Fig. 2a). Pedigree verification showed that the father harbored a heterozygous mutation in the gene, while the mother had the wild-type gene (Fig. 2b).
Fig. 2.
a c.7778C>T (p.G2593E) mutation in the DNAH5 gene in the brother. Black arrows show the heterozygous mutation. b c.7778C>T (p.G2593E) mutation in the DNAH5 gene in the sister. Black arrows show the heterozygous mutation. The father carried the heterozygous mutation, and the mother had the wild-type gene
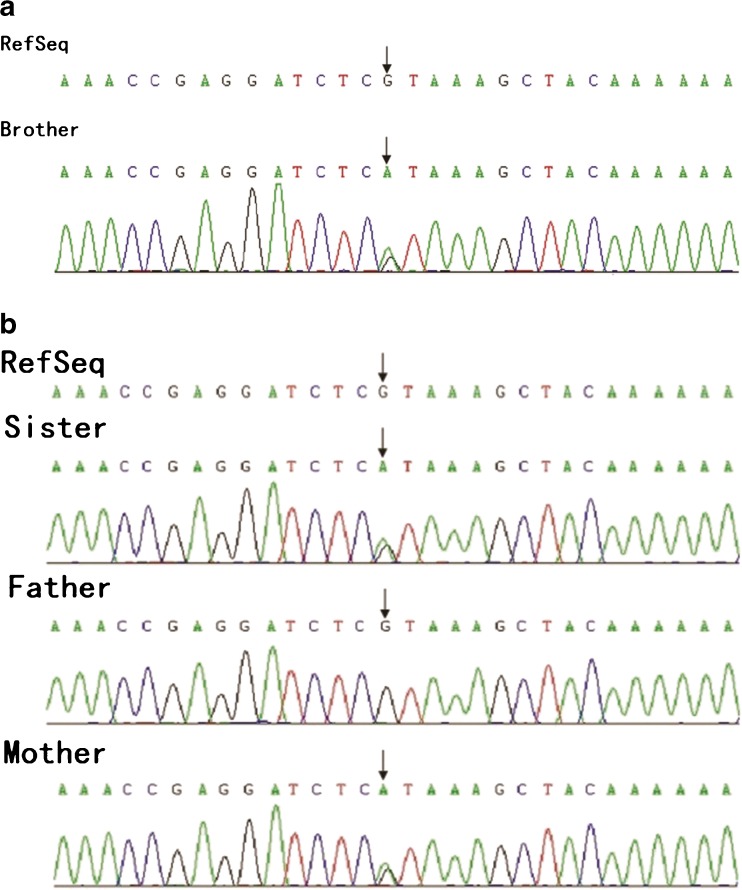
A nonsense mutation c.13729G>A (p.R4577X) was found in the DNAH5 gene in both children, who carried heterozygous mutations. This nonsense mutation could result in early termination of protein translation (Fig. 3a). Pedigree verification showed that the mother harbored heterozygous mutation, with the father having the wild-type gene (Fig. 3b).
Fig. 3.
a c.13729G>A (p.R4577X) mutation in the DNAH5 gene of the brother. Black arrows show the heterozygous mutation. b c.13729G>A (p.R4577X) mutation in the DNAH5 gene of the sister. Black arrows show the heterozygous mutation. The father carried the wild-type gene, with the mother harboring the heterozygous mutation
Both genetic testing and pedigree verification showed that the above children carried compound heterozygous mutations; parents were carriers, in accordance with the hereditary mode of KS. Therefore, both children were diagnosed with KS. A distribution frequency for these two mutations of 0 was obtained in the dbSNP database, 1000 Genomes, and ExAC. To our knowledge, this is the first report describing these two mutations to date.
Discussion
KS, a type of PCD, is characterized by the triad of visceral inversion, bronchiectasis, and paranasal sinusitis. Current studies demonstrated that PCD is a series of monogenic diseases. Multiple subtypes of PCD have been reported, with various subtypes associated with different genes. DNAH5 and DNAI1 were shown to be virulence genes in about 1/3 of all PCD patients, i.e., they were found in 28 and 10% of all PCD patients, respectively; thus, these two genes are considered the major virulence genes [7–10]. In the present study, multiple KS-related genes (Table 1) were measured simultaneously; the wide coverage could effectively improve the positive rate of disease screening.
Table 1.
List of the genes tested in the study
| DNAH5 | DNAH1 | SPAG1 | RSPH4A | DNAI2 |
| CCDC65 | DRC1 | MCIDAS | CCNO | CCDC151 |
| DNAH8 | RSPH9 | RSPH4A | RSPH3 | CCDC114 |
| DNAI1 | SPAG1 | LRRC6 | INSL6 | DNAAF3 |
| ARMC4 | DNAAF2 | HYDIN | DNAAF1 | C21orf59 |
| DNAL1 | GAS8 | CCDC103 | SLC38A8 | RSPH1 |
| AK7 | PRKAR1A | … |
KS is a rare autosomal recessive disorder that occurs in infancy; however, no specific treatment method is available for this ailment. The most common abnormality of KS is caused by complete or partial deletion of outer dynein arms (ODAs); indeed, disease severity depends on the degree of deletion [3]. Other causes include deletion of inner dynein arms and microtubule malposition [11]. Defects of the ciliary ultrastructure composed of dynein arms could affect the movement of cilia, and thus cause cough, pneumonia, infertility, and even anosphrasia and deafness [12–16]. ODAs are composed of protein products encoded by multiple genes, among which the DNAH5 gene has 79 exons [17] that encode the heavy chain of ODA. DNAH5 is associated with the activities of ATPase and microtubule motor proteins; defects in the DNAH5 gene could cause ODA deletion, inducing the presentations of KS.
In the present study, a compound heterozygous mutation in the DNAH5 gene was identified and was shown to cause KS in this family. There are several similarities and differences between these findings and previous studies. DNAH5 mutations were similar to previously reported data; for instance, mutations are easy to occur in the DNAH5 gene, which could cause KS. No presentations were found in carriers, indicating that KS is an autosomal recessive disorder. Patients in the same family could carry the same mutations; however, the disease occurred earlier in males than females [18]. In addition, the severity of KS in males is higher than in females, with most male patients potentially losing fertility [18–21]. An important finding of this study is that although the boy and his sister carried the same mutated gene, no typical KS triad symptoms were found in the girl (e.g., no visceral inversion). These findings suggest that even in the same pedigree, the same gene mutation could result in different clinical presentations of KS. Almost 50% of PCD individuals have a situs inversus totalis whereas the other patients have situs solitus or (rarely) heterotaxia. Therefore, these findings indicate that diagnosing KS is very complex, requiring molecular genetic analyses.
The c.7778C>T and c.13729G>A mutations reported in this study were verified by pedigree analyses to be compound heterozygous mutations. Although not reported in previous studies, these mutations could help in molecular diagnosing, genetic counseling, and prenatal screening. NGS has become an important method for molecular investigations, including disease pathogenesis and classification, due to the advantages of high-throughput and low cost. Currently, NGS has been successfully applied in screening and auxiliary diagnosis of hereditary diseases, as well as in precision medicine, including non-invasive prenatal testing (NIPT), PGS, PGD, individualized medication for tumors, and response monitoring. The present study employed a directional capture strategy for hereditary disease screening, which correctly identified the virulence sites in the pedigree and facilitated the differential diagnosis among multiple genes. With the development of NGS and cost decrease, full exon sequencing would be universally used for disease screening. In addition, application of whole genome sequencing would also be possible in the future.
Currently, dozens of monogenic diseases could be screened in China, which would effectively reduce the risk of birth defects. In addition, precise classification diagnosis provides the foundation for disease assessment and a basis for prenatal screening, prenatal diagnosis, and genetic counseling. We believe that with the accumulation of cases and genetic data in China, birth defects, including KS, could be effectively avoided in the future.
Compliance with ethical standards
Research involving human participants
The present study was approved by the Ethics Committee of People’s Hospital. All procedures performed in studies involving human participants were in accordance with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Capsule These findings demonstrated that new DNAH5 mutations could be used for molecular diagnosis of KS, providing families with genetic counseling and prenatal diagnosis.
References
- 1.Moore A, Escudier E, Roger G, Tamalet A, Pelosse B, Marlin S, et al. RPGR is mutated in patients with a complex X linked phenotype combining primary ciliary dyskinesia and retinitis pigmentosa. J Med Genet. 2006;43(4):326–33. doi: 10.1136/jmg.2005.034868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Knowles MR, Leigh MW, Ostrowski LE, Huang L, Carson JL, Hazucha MJ, et al. Exome sequencing identifies mutations in CCDC114 as a cause of primary ciliary dyskinesia. Am J Hum Genet. 2013;92(1):99–106. doi: 10.1016/j.ajhg.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Afzelius B, Mossberg B, Bergstrom S. Immotile-cilia syndrome (primary ciliary dyskinesia), including Kartagener syndrome. Metabol Molecul Base Inherit Dis. 1995;3:3943–54. [Google Scholar]
- 4.Kennedy MP, Omran H, Leigh MW, Dell S, Morgan L, Molina PL, et al. Congenital heart disease and other heterotaxic defects in a large cohort of patients with primary ciliary dyskinesia. Circulation. 2007;115(22):2814–21. doi: 10.1161/CIRCULATIONAHA.106.649038. [DOI] [PubMed] [Google Scholar]
- 5.Hogg C, Bush A. Genotyping in primary ciliary dyskinesia: ready for prime time, or a fringe benefit? Thorax. 2012;67(5):377–8. doi: 10.1136/thoraxjnl-2011-201320. [DOI] [PubMed] [Google Scholar]
- 6.O’Roak BJ, Deriziotis P, Lee C, Vives L, Schwartz JJ, Girirajan S, et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat Genet. 2011;43(6):585–9. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geremek M, Schoenmaker F, Zietkiewicz E, Pogorzelski A, Diehl S, Wijmenga C, et al. Sequence analysis of 21 genes located in the Kartagener syndrome linkage region on chromosome 15q. Euro J Hum Genet: EJHG. 2008;16(6):688–95. doi: 10.1038/ejhg.2008.5. [DOI] [PubMed] [Google Scholar]
- 8.Djakow J, Svobodova T, Hrach K, Uhlik J, Cinek O, Pohunek P. Effectiveness of sequencing selected exons of DNAH5 and DNAI1 in diagnosis of primary ciliary dyskinesia. Pediatr Pulmonol. 2012;47(9):864–75. doi: 10.1002/ppul.22520. [DOI] [PubMed] [Google Scholar]
- 9.Failly M, Bartoloni L, Letourneau A, Munoz A, Falconnet E, Rossier C, et al. Mutations in DNAH5 account for only 15% of a non-preselected cohort of patients with primary ciliary dyskinesia. J Med Genet. 2009;46(4):281–6. doi: 10.1136/jmg.2008.061176. [DOI] [PubMed] [Google Scholar]
- 10.Fliegauf M, Olbrich H, Horvath J, Wildhaber JH, Zariwala MA, Kennedy M, et al. Mislocalization of DNAH5 and DNAH9 in respiratory cells from patients with primary ciliary dyskinesia. Am J Respir Crit Care Med. 2005;171(12):1343–9. doi: 10.1164/rccm.200411-1583OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jorissen M, Willems T, Van der Schueren B, Verbeken E, De Boeck K. Ultrastructural expression of primary ciliary dyskinesia after ciliogenesis in culture. Acta Otorhinolaryngol Belg. 2000;54(3):343–56. [PubMed] [Google Scholar]
- 12.Afzelius BA. A human syndrome caused by immotile cilia. Science. 1976;193(4250):317–9. doi: 10.1126/science.1084576. [DOI] [PubMed] [Google Scholar]
- 13.Geremek M, Witt M. Primary ciliary dyskinesia: genes, candidate genes and chromosomal regions. J Appl Genet. 2004;45(3):347–61. [PubMed] [Google Scholar]
- 14.Noone PG, Leigh MW, Sannuti A, Minnix SL, Carson JL, Hazucha M, et al. Primary ciliary dyskinesia: diagnostic and phenotypic features. Am J Respir Crit Care Med. 2004;169(4):459–67. doi: 10.1164/rccm.200303-365OC. [DOI] [PubMed] [Google Scholar]
- 15.Halbert SA, Patton DL, Zarutskie PW, Soules MR. Function and structure of cilia in the fallopian tube of an infertile woman with Kartagener’s syndrome. Hum Reprod. 1997;12(1):55–8. doi: 10.1093/humrep/12.1.55. [DOI] [PubMed] [Google Scholar]
- 16.Fischer TJ, McAdams JA, Entis GN, Cotton R, Ghory JE, Ausdenmoore RW. Middle ear ciliary defect in Kartagener’s syndrome. Pediatrics. 1978;62(4):443–5. [PubMed] [Google Scholar]
- 17.Guichard C, Harricane MC, Lafitte JJ, Godard P, Zaegel M, Tack V, et al. Axonemal dynein intermediate-chain gene (DNAI1) mutations result in situs inversus and primary ciliary dyskinesia (Kartagener syndrome) Am J Hum Genet. 2001;68(4):1030–5. doi: 10.1086/319511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eliasson R, Mossberg B, Camner P, Afzelius BA. The immotile-cilia syndrome. A congenital ciliary abnormality as an etiologic factor in chronic airway infections and male sterility. N Engl J Med. 1977;297(1):1–6. doi: 10.1056/NEJM197707072970101. [DOI] [PubMed] [Google Scholar]
- 19.Hornef N, Olbrich H, Horvath J, Zariwala MA, Fliegauf M, Loges NT, et al. DNAH5 mutations are a common cause of primary ciliary dyskinesia with outer dynein arm defects. Am J Respir Crit Care Med. 2006;174(2):120–6. doi: 10.1164/rccm.200601-084OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Afzelius BA, Eliasson R. Male and female infertility problems in the immotile-cilia syndrome. Eur J Respir Dis Suppl. 1983;127:144–7. [PubMed] [Google Scholar]
- 21.Munro NC, Currie DC, Lindsay KS, Ryder TA, Rutman A, Dewar A, et al. Fertility in men with primary ciliary dyskinesia presenting with respiratory infection. Thorax. 1994;49(7):684–7. doi: 10.1136/thx.49.7.684. [DOI] [PMC free article] [PubMed] [Google Scholar]