Abstract
Primary ciliary dyskinesia (PCD) is a genetically heterogeneous recessive disorder caused by several distinct defects in genes responsible for ciliary beating, leading to defective mucociliary clearance often associated with randomization of left/right body asymmetry. Individuals with PCD caused by defective radial spoke (RS) heads are difficult to diagnose owing to lack of gross ultrastructural defects and absence of situs inversus. Thus far, most mutations identified in human radial spoke genes (RSPH) are loss-of-function mutations, and missense variants have been rarely described. We studied the consequences of different RSPH9, RSPH4A, and RSPH1 mutations on the assembly of the RS complex to improve diagnostics in PCD. We report 21 individuals with PCD (16 families) with biallelic mutations in RSPH9, RSPH4A, and RSPH1, including seven novel mutations comprising missense variants, and performed high-resolution immunofluorescence analysis of human respiratory cilia. Missense variants are frequent genetic defects in PCD with RS defects. Absence of RSPH4A due to mutations in RSPH4A results in deficient axonemal assembly of the RS head components RSPH1 and RSPH9. RSPH1 mutant cilia, lacking RSPH1, fail to assemble RSPH9, whereas RSPH9 mutations result in axonemal absence of RSPH9, but do not affect the assembly of the other head proteins, RSPH1 and RSPH4A. Interestingly, our results were identical in individuals carrying loss-of-function mutations, missense variants, or one amino acid deletion. Immunofluorescence analysis can improve diagnosis of PCD in patients with loss-of-function mutations as well as missense variants. RSPH4A is the core protein of the RS head.
Keywords: cilia, primary ciliary dyskinesia, human radial spoke protein 9, human radial spoke protein 4A, human radial spoke protein 1
Clinical Relevance
We demonstrate that mutations in the human radial spoke genes RSPH1, RSPH4A, and RSPH9 cause distinct defects of the radial spoke (RS) head in ciliary axonemes detectable by immunofluorescence (IF) analysis. IF can be used to diagnose primary ciliary dyskinesia with RS defects and help determine the pathogenicity of DNA variants detected by mutational analysis.
Primary ciliary dyskinesia (PCD; MIM 244400) is a rare inherited recessive disorder caused by defective ciliary motility with an incidence of 1:20,000 (1). Defective mucociliary clearance results in recurrent airway inflammation (2) and bronchiectasis, which can progress to lung failure (3). In half of the affected individuals, PCD occurs with situs inversus, and is then referred to as Kartagener syndrome (1, 4), with an increased risk for congenital heart defects (5). In males with PCD, dysfunctional sperm flagella frequently cause infertility (4). Diagnosis is often delayed, especially when clinical signs, such as situs inversus, are missing (6). Due to the heterogeneity of the encountered defects, there are no simple diagnostic tests available.
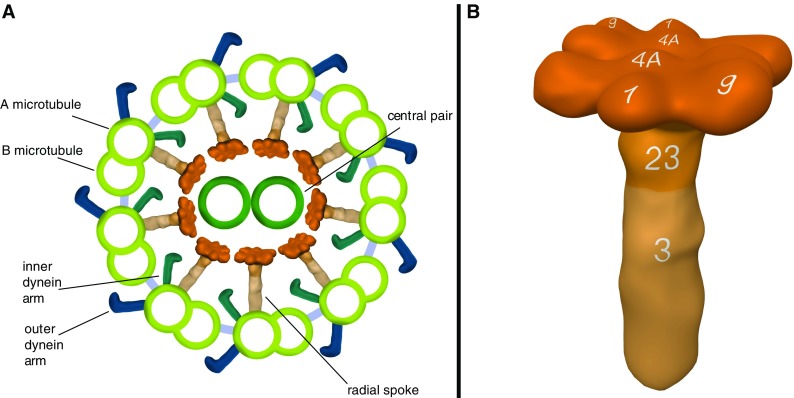
Respiratory cilia are composed of more than 200 proteins (7) that form 9 microtubular doublets arranged around a central microtubulus pair (CP) (Figure 1A). Dynein chains attached to the A-microtubule, referred to as outer dynein arms (ODA) and inner dynein arms (IDA), promote microtubular sliding through ATPase activity (8).
Figure 1.
Model of a ciliary cross-section and the radial spoke (RS) complex based on immunofluorescence (IF) findings and Chlamydomonas orthology. Ciliary cross-section showing the typical 9 + 2 ultrastructure (A) of nine outer microtubulus doublets arranged around the central pair. Outer dynein arms, inner dynein arms, and RSs emerge from each A-microtubule. As illustrated, the RS complexes connect the outer microtubulus doublets to the central pair. The RS proteins, RSPH9, RSPH4A, and RSPH1, are located in the head complex and arranged in a twofold rotational symmetry, whereas RSPH23 is found in the neck and RSPH3 in the stalk compartment. The location of these proteins is indicated by the numbers 9, 1, 4A, 23, 3 (B). Model of the RS complex was created based on published findings in Chlamydomonas reinhardtii (31, 33, 43, 44, 47–49).
Mutations in multiple genes can cause PCD. Among these are autosomal recessive mutations in: DNAH5 (MIM 603335), DNAH11 (MIM 611884), and DNAI1 (MIM 604366), TXNDC3 (MIM 607421), and DNAI2 (MIM 605483) encoding ODA components (9–13). Mutations in CCDC114 (MIM 615038), ARMC4 (MIM 615408), and CCDC151 (MIM 615956) affect the ODA docking complex (14–17), whereas in cytoplasmatic preassembly factors, such as DNAAF1/LRRC50 (MIM 613190 [18]), DNAAF2/KTU (MIM 612517 [19]), DNAAF3 (MIM 614566 [20]), and DYX1C1 (MIM 608706 [21]), as well as in genes encoding for other factors, cause PCD with abnormal ODA and IDA complex composure (22–27). Nexin–dynein regulatory complexes (N-DRC) and IDAs can be affected by mutations in CCDC39 (MIM 613798) and CCDC40 (MIM 613799) (28, 29), whereas isolated defects of the N-DRC are caused by mutations in CCDC65 (MIM 611088) and CCDC164 (MIM 615288) (27, 30).
Radial spokes (RSs) are T-shaped structures aligned between the CP and the peripheral A-microtubule, transmitting regulatory signals between the CP and the dynein arms (1) (Figure 1A). Based on Chlamydomonas reinhardtii orthology, it is composed of at least 23 proteins and divided in a head, neck, and stalk compartment (Figure 1B) (31–33). Mutations in RSPH9, RSPH4A, and RSPH1 encoding RS head components (33–37), as well as a CP defect due to HYDIN (MIM 610812 [38]) mutations, affect the interaction between the RS head and the central pair and result in PCD.
Genetic analysis can be used for diagnosis of PCD with RS head defects (37). However, pathogenetic assessment of detected missense or small in-frame variants is difficult. In this study we sequenced RSPH9, RSPH4A, and RSPH1 in individuals with classical PCD symptoms with situs solitus, frequently normal transmission electron microscopy (TEM) findings and subtle changes in the ciliary beating pattern (Table 1). In addition, we evaluated the impact of the identified mutations on the RS assembly. We first studied the impact of loss-of-function mutations, and then tested whether the observed axonemal RS head abnormalities were also present in individuals with PCD with missense variants in genes encoding for RS head proteins or a small in-frame deletion so far only rarely reported in PCD with RS head defects.
Table 1.
Overview of Clinical Details of the Affected Individuals with Mutations in the Human Radial Spoke Genes RSPH1, RSPH4A and RSPH9
| Individual Code | Mutated Gene | DNA Change | Protein Change | Sex (Consang.) | Age* (Yr) | Origin | nNO Production Rate (nl/min) | Situs | nRDS | BE | Chronic Cough, Wheeze, Bronchitis, rRI | Sinusitis, Rhinorrhea | OM | TEM | HSVM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OP-1428 II1 | RSPH1 | [c.275-2A>C]; [c.433C>T] | [?]; [p.Gln145*] | M (N.A.) | 4 | Germany | 18,53† | SS | Y | N.A. | Y | Y | Y | 9 + 2, rarely 9 + 0 | Reduced amplitude, stiff beating pattern |
| OP-562 | RSPH1 | [c.275-2A>C]; [c.275-2A>C] | [?]; [?] | M (Y) | N.A. | Belgium | N.A. | N.A. | Y | Y | Y | N.A. | N.A. | 9 + 2, rarely 9 + 0 | N.A. |
| OI-162 | RSPH4A | [c.1393C>T]; [c.1393C>T] | [p.Arg465*]; [p.Arg465*] | F (Y) | N.A. | Israel | 11,22† | SS | N.A. | Y | Y | Y | Y | 9 + 2, rarely 9 + 0 | N.A. |
| OP-225 II2 | RSPH4A | [c.1129delG]; [c.1129delG] | [p.Glu377Lysfs11*]; [p.Glu377Lysfs11*] | F (Y) | 7 | Germany | N.A. | SS | Y | N.A. | Y | Y | Y | N.A. | Reduced amplitude, stiff beating pattern |
| OP-1603 II1 | RSPH4A | [c.1105G>C]; [c.1105G>C] | [p.Ala369Pro]; [p.Ala369Pro] | F (Y) | 13 | Turkey | N.A. | SS | N | Y | Y | Y | Y | 9 + 2, rarely 9 + 0 | Reduced amplitude, stiff beating pattern |
| OP-1603 II2 | RSPH4A | [c.1105G>C]; [c.1105G>C] | [p.Ala369Pro]; [p.Ala369Pro] | F (Y) | 3 | Turkey | N.A. | SS | N | Y | Y | Y | Y | N.A. | Reduced amplitude, stiff beating pattern |
| OP-943 IV1 | RSPH4A | [c.1105G>C]; [c.1105G>C] | [p.Ala369Pro]; [p.Ala369Pro] | F (Y) | 13 | Germany | N.A. | SS | Y | Y | Y | Y | Y | 9 + 2, rarely 9 + 0 | Reduced amplitude, stiff and rotatory beating pattern |
| OP-708 | RSPH4A | [c.1105G>C]; [c.1105G>C] | [p.Ala369Pro]; [p.Ala369Pro] | F (N.A.) | 9 | Denmark | 16.2‡ | SS | N.A. | Y | Y | Y | Y | 9 + 2, rarely 9 + 0 | N.A. |
| OP-1606 II2 | RSPH4A | [c.1105G>C]; [c.1105G>C] | [p.Ala369Pro]; [p.Ala369Pro] | M (Y) | 18 | Germany | 9,57 | SS | Y | Y | Y | Y | Y | N.A. | Reduced amplitude, stiff beating pattern |
| OP-89 II2 | RSPH4A | [c.1391G>A]; [c.1391G>A] | [p.Gly464Glu]; [p.Gly464Glu] | F (Y) | 12 | Germany | N.A. | SS | N.A. | Y | Y | Y | Y | 9 + 2, rarely 9 + 0 or 8 + 1 (transposition defect) | Reduced amplitude, stiff rotatory beating pattern |
| OP-1795 II1 | RSPH4A | [c.1391G>A]; [c.1391G>A] | [p.Gly464Glu]; [p.Gly464Glu] | F (N.A.) | 34 | Morocco | 7,63† | SS | Y | Y | Y | Y | Y | 9 + 2, rarely 9 + 0 or 8 + 1 (transposition defect) | Reduced amplitude, stiff rotatory beating pattern |
| OP-1795 II2 | RSPH4A | [c.1391G>A]; [c.1391G>A] | [p.Gly464Glu]; [p.Gly464Glu] | M (N.A.) | 22 | Morocco | 2,77† | SS | Y | N | Y | Y | Y | 9 + 2, rarely 9 + 0 or 8 + 1 (transposition defect) | Reduced amplitude, stiff rotatory beating pattern |
| OP-1795 II3 | RSPH4A | [c.1391G>A]; [c.1391G>A] | [p.Gly464Glu]; [p.Gly464Glu] | F (N.A.) | 39 | Morocco | 10,77† | SS | Y | Y | Y | Y | Y | 9 + 2, rarely 9 + 0 or 8 + 1 (transposition defect) | Reduced amplitude, stiff rotatory beating pattern |
| OP-1135 | RSPH9 | [c.466C>T]; [c.466C>T] | [p.Arg156*]; [p.Arg156*] | M (N) | 14 | Germany | N.A. | SS | N | Y | Y | Y | Y | 9 + 2, rarely 9 + 0 or 8 + 1 (transposition defect) | N.A. |
| OI-4 II2 | RSPH9 | [c.2T>C]; [c.2T>C] | [p.Met1?]; [p.Met1?] | F (N) | N.A. | Israel | 13,2† | SS | Y | Y | Y | N | N | 9 + 2, rarely 9 + 0 | N.A. |
| OI-107 | RSPH9 | [c.752A>G]; [c.752A>G] | [p.His251Arg]; [p.His251Arg] | M (N.A.) | N.A. | Israel | N.A. | SS | N.A. | N | Y | Y | Y | 9 + 2, rarely 9 + 0 | N.A. |
| OP-658 | RSPH9 | [c.523-1G>C]; [c.610A>T] | [?]; [p.Lys204*] | F (N.A.) | 21 | Denmark | 15,9‡ | SS | N | Y | Y | Y | N | 9 + 2, rarely 9 + 0 | N.A. |
| OP-671 | RSPH9 | [c.523-1G>C]; [c.610A>T] | [?]; [p.Lys204*] | M (N.A.) | 16 | Denmark | 4,5‡ | SS | Y | Y | Y | Y | Y | 9 + 2, rarely 9 + 0 | N.A. |
| OP-1665 II1 | RSPH9 | [c.2T>C]; [c.610A>T] | [p.Met1?]; [p.Lys204*] | F (N.A.) | 13 | Germany | N.A. | N.A. | N.A. | N.A. | Y | Y | N.A. | 9 + 2, rarely 9 + 0 | N.A. |
| OP-601 II1 | RSPH9 | [c.801_803delGAA]; [c.801_803delGAA] | [p.268delLys]; [p.268delLys] | F (N.A.) | 3 | Denmark | 52,5‡ | SS | Y | N.A. | Y | Y | Y | 9 + 2, rarely 9 + 0 | Reduced amplitude, stiff beating pattern |
| OP-601 II2 | RSPH9 | [c.801_803delGAA]; [c.801_803delGAA] | [p.268delLys]; [p.268delLys] | M (N.A.) | 2 | Denmark | 15,3‡ | SS | Y | N.A. | Y | N.A. | Y | 9 + 2, rarely 9 + 0 | Reduced amplitude, stiff beating pattern |
Definition of abbreviations: BE, bronchiectasis; Consang., consanguinity status; F, female; HSVM, high-speed video microscopy; M, male; N, no; N.A., data not available; nNO, nasal nitric oxide; nRDS, neonatal respiratory distress syndrome; OM, otitis media; rRI, recurrent respiratory infections; PCD, primary ciliary dyskinesia; RSPH, human radial spoke gene; SS, situs solitus; TEM, transmission electron microscopy; Y, yes.
Age at which PCD was diagnosed.
nNO measurements were obtained with the EcoMedicsCLD88Chemiluminescence nitric oxide analyzer (Duernten, Switzerland).
nNO measurements were obtained with the NIOX Flex equipment (Aerocrine, Solna, Sweden). The splice site mutation c.275-2A>C has been demonstrated to cause aberrant splicing (36).
Materials and Methods
Whole-Exome Sequencing
Whole-exome sequencing was performed at the Wellcome Trust Sanger Institute. Genomic DNA fragmented to an average size of 150 bp was subjected to DNA library creation using established Illumina paired-end protocols (Agilent Technologies, Santa Clara, CA). Adapter-ligated libraries were amplified and indexed via PCR. A portion of each library was used to create an equimolar pool comprising eight indexed libraries. The exomes were produced using Illumina HiSeq and SureSelect Human All Exon v5 (Agilent Technologies) SeqCap pulldown technology. Two callers were used and their results merged for each sample separately: an all-sites Binary Call Format was created with samtools mpileup (samtools version 0.1.18; https://github.com/samtools/) and then variants (single nucleotide polymorphisms and indels) were called by bcftools; also variant sites (single nucleotide polymorphisms and indels) were called using the GATK Unified Genotyper (GATK version 1.3-21; https://www.broadinstitute.org/gatk/index.php). The variants called by each of the callers were (soft) filtered separately and annotated using vcf-annotate (VCFtools; http://vcftools.sourceforge.net/). 1000Genomes population allele frequencies were applied (http://www.1000genomes.org). The Integrative Genomics Viewer (https://www.broadinstitute.org/igv/) was used to visually study the sequencing data. All identified mutations were confirmed by Sanger sequencing in the index case as well as parents and/or siblings, if DNA was available (see Figures E1 and E2 in the online supplement).
Immunofluorescence Microscopy
Immunofluorescence (IF) analyses were performed as described previously (39). Monoclonal mouse anti–acetylated-α-tubulin (T7451) was obtained from Sigma (Taufkirchen, Germany). We used the commercially available antibodies (Atlas Antibodies, Stockholm, Sweden) rabbit polyclonal anti-RSPH1 (HPA016816) directed against RSPH1 amino acids 227–309, anti-RSPH3 (HPA039109) directed against RSPH3 amino acids 268–354, anti-RSPH4A (HPA031196) directed against RSPH4A amino acids 22–94, anti-RSPH9 (HPA031703) directed against RSPH9 amino acids 96–187, and anti-RSPH23 (HPA044555) directed against RSPH23 amino acids 141–207 all diluted 1:400. An overview of the epitopes used to generate the antibodies directed against RSPH9, RSPH4A, and RSPH1 is given in Figure E3. Anti-mouse Alexa Fluor 488 and anti-rabbit Alexa Fluor 546 were used as secondary antibodies (Molecular Probes, Invitrogen, Darmstadt, Germany). DNA was stained with Hoechst 33342 (Sigma). To evaluate the antibodies, we stained respiratory cells from disease control subjects as well as from patients carrying homozygous mutations in HYDIN (c.922A>T, p.Lys307*) (38) and CCDC40 (c.2630 delG, p.Glu877Argfs*) (Figure E4). Disease control subjects with recurrent respiratory infections showed normal ciliary beating patterns (high-speed video microscopy [HSVM]), normal dynein arms and normal N-DRC (IF). Images were taken with a Zeiss Apotome Axiovert 200 (Munich, Germany) and processed with AxioVision 4.8 (http://www.zeiss.de/microscopy/de_de/downloads/axiovision.html) and Adobe Creative Suite 4 (http://www.adobe.com). A total of 10–20 cells per staining were analyzed and stainings were performed at least two times. Exposure time was determined using cilia from healthy control subjects. Patient cilia were first analyzed with the same exposure time as determined for the control subjects and then overexposed to confirm absence of the analyzed protein.
Western Blot Analyses
Western blots were performed as previously described (21, 40). Axonemal pig lysates provide highly concentrated samples for Western blots and were used for anti-RSPH1, anti-RSPH9, and anti-RSPH23, whereas human respiratory cell lysates were used for anti-RSPH4A and anti-RSPH3. Human respiratory cells from healthy German control subjects were used to prepare lysates. The same primary antibodies as for IF experiments were used for immunoblotting, diluted 1:500 or 1:1,000.
Additional methods are provided in the online supplement.
Results
Mutational Status of RSPH9, RSPH4A, and RSPH1 in Individuals with PCD
Whole-exome sequencing results: In RSPH1 we found in the individual OP-562 a homozygous mutation of the obligatory splice site (c.275–2A>C); In RSPH4A we found in the individual OP-89 II2 a homozygous missense variant (c.1391G>A, p.Gly464Glu). In RSPH9 bi-allelic homozygous loss-of-function mutations were identified in the individuals OP-1135 (c.466C>T, p.Arg156*), OI-4 II2 (c.2T>C, p.Met1?), OP-658 and OP-671 (c.523–1G>C, mutation of the obligatory canonical splice site; c.610A>T, p.Lys204*). In OP-601 II1 and OP-601 II2 we found in RSPH9 homozygous in-frame deletions predicting the loss of one amino acid (c.801_803 delGAA, p.268 delLys). All mutations were confirmed by Sanger sequencing.
Sanger sequencing results: In RSPH4A we found in the individual OI-162 a homozygous loss-of-function mutation (c.1393C>T, p.Arg465*). In the individuals OP-1603 II1, OP-1603 II2, OP-943 IV1, OP-1606 II2 and OP-708 bi-allelic missense variants in RSPH4A were identified (c.1105G>C, p.Ala369Pro). In OP-1795 II1, OP-1795 II2 and OP-1795 II3 another bi-allelic missense variant was identified (c.1391G>A, p.Gly464Glu). ClustalW alignment analysis demonstrated that both of these missense variants are localized in evolutionary conserved regions of RSPH4A. In RSPH9 we found a bi-allelic loss-of-function mutation in OP-1665 II1 (c.2T>C, p.Met1?) and a homozygous missense variant in OI-107 (c.752A>G, p.His251Arg).
Overall we identified 7 novel mutations not reported previously and not described in the 1,000 Genomes database (in RSPH4A c.1393C>T, p.Arg465*, c.1105G>C, p.Ala369Pro and c.1391G>A, p.Gly464Glu; in RSPH9 c.2T>C, p.Met1?, c.523–1G>C, splicing mutation, c.752A>G, p.His251Arg and c.610A>T, p.Lys204*).
The variation c.466C>T, p.Arg156* (rs2295947) is listed in the 1,000 Genomes database (TMP_ESP_6_43623371) with an allele frequency of 0.0005 in the European population. We confirm that it is disease-causing in our patient OP-1135, because it is a low frequent homozygous loss-of-function mutation and we demonstrated the absence of RSPH9 by IF analysis. The homozygous in-frame deletion c.801_803 delGAA, p.268 delLys (rs397515340) is also listed in the 1,000 Genomes database (TMP_ESP_6_43638655_43638657) with an allele frequency of 0.0001 in the European population and has been previously described (33). Our data strongly indicates that the variant is disease causing in OP-601 II1 and OP-602 II2 because we demonstrated by IF analysis that RSPH9 is absent from the ciliary axonemes. Additionally, the variant co-segregates with the disease phenotype, the allele frequency is very low and the deletion appears in an evolutionary conserved region of the protein.
The mutations in the individuals OP-225 II2 (c.1129 delG, p.Glu377Lysfs11* in RSPH4A) and OP-1428 II1 (c.275–2A>C, rs151107532; mutation of the obligatory canonical splice site; c.433C>T, p.Gln145* in RSPH1) have been reported previously (35–37, 41). The mutation c.275–2A>C [p.?] was also found homozygous in OP-562. This mutation has been previously reported in 20 patients from three different studies (35–37). It is described in the 1,000 Genomes database and has an allele frequency of 0,001 in the European population (Table 1).
Clinical Findings in the Affected Individuals
All of the affected individuals included in the screening suffered from classical PCD symptoms, including recurrent respiratory infections, chronic cough, wheeze, and bronchitis. None showed laterality defects (situs solitus). We analyzed 21 individuals originating from 16 families. Out of 21, 12 displayed neonatal respiratory distress, 14 had documented bronchiectasis, all 21 had recurrent respiratory infections and chronic cough, 18 suffered from chronic sinusitis and rhinorrhea, and 17 had otitis media (Table 1). In 12 patients, the nasal nitric oxide production rate was well below the threshold of 77 nl/min (42). TEM analyses showed mostly cross-sections with the normal 9 + 2 composition; occasionally, 9 + 0 cilia were observed. HSVM analyses revealed reduced amplitude and stiff and sometimes rotatory beating patterns. Clinical and diagnostic findings are depicted in Table 1. Dynein arm defects and N-DRC defects were ruled out by IF. Consanguinity is known in the families OP-562, OI-162, OP-225, OP-1603, OP-1606, OP-943, and OP-89, whereas others are nonconsanguineous (OP-1135 and OI-4) or with unknown consanguinity status (OP-1428, OP-708, OP-1795, OP-658, OP-671, OP-1665, OP-601, and OI-107). Healthy German control individuals were included in the study.
RSPH9, RSPH4A, and RSPH1 Loss-of-Function Mutations Cause Deficiency of the Assembly of the RS Head Protein, RSPH9
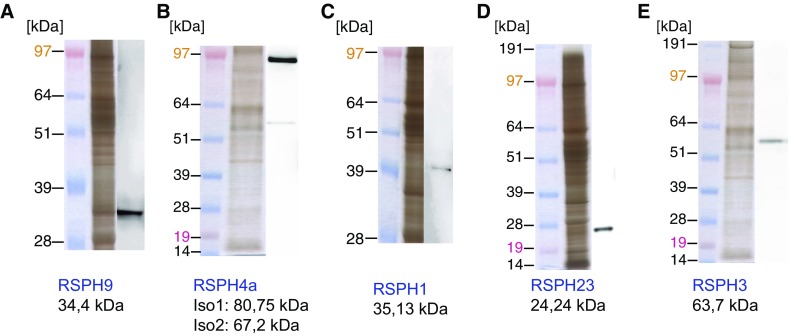
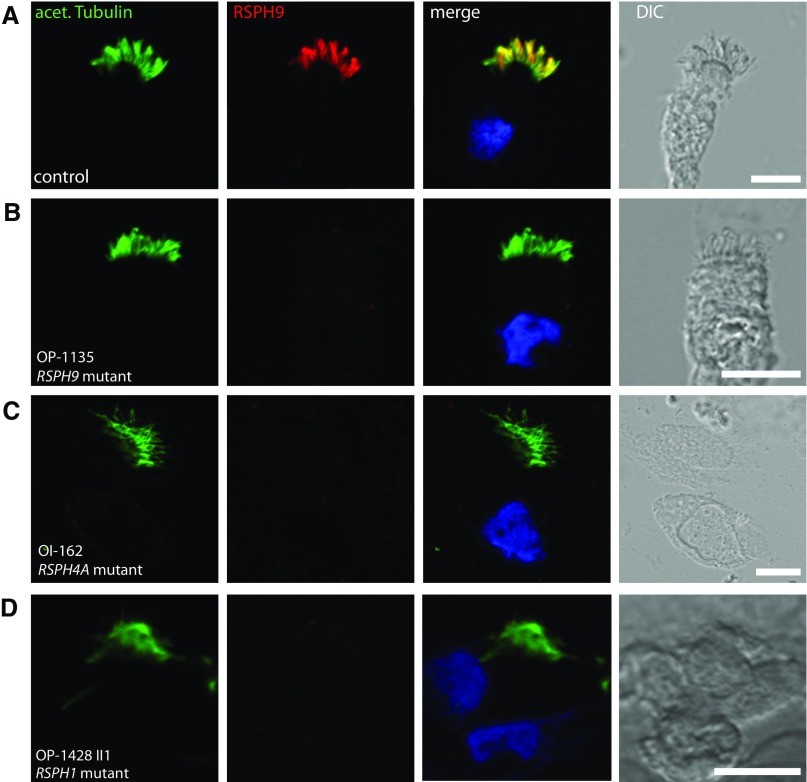
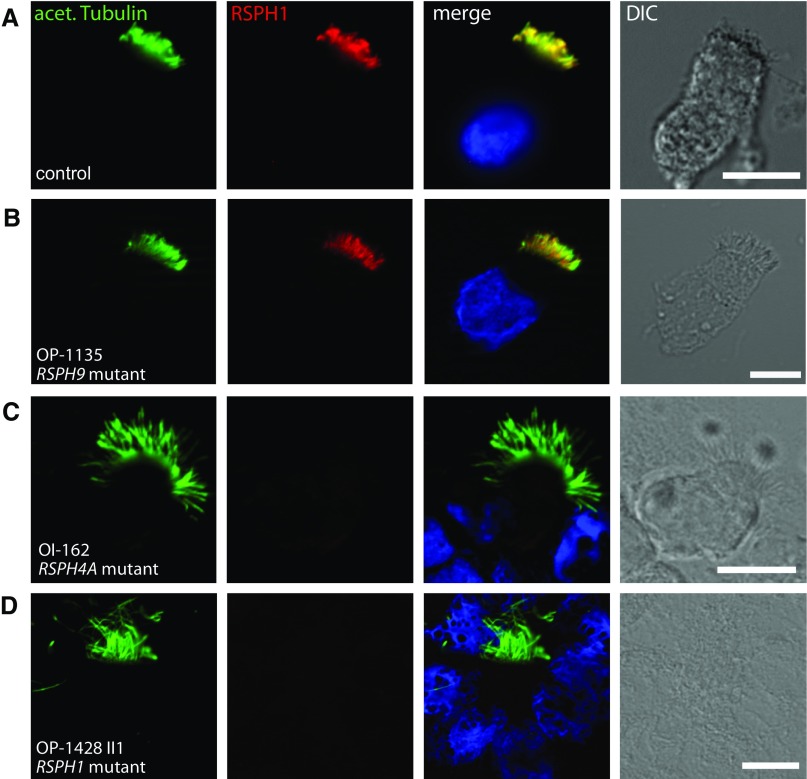
To analyze the structure of the RS on the subcellular level, we performed IF microscopy of respiratory epithelial cells from the affected individuals, OI-4 II2, OP-1135, OP-658, OP-671, and OP-1665 II1, carrying biallelic loss-of-function mutations in RSPH9 using rabbit polyclonal antibodies directed against the RS head protein, RSPH9. Western blot analysis proved specificity of the antibody targeting RSPH9 (34.4 kD; Figure 2A). Although RSPH9 localized to the entire axonemal length in cells from control subjects (Figure 3A), it was undetectable in the cells of the patients (Figures 3B and Figure E5B, shown for OP-1135), consistent with recessive loss-of-function mutations that result in failure to produce a functional RSPH9 protein. In addition, we analyzed the RSPH9 localization in RSPH4A-deficient cells (OI-162) and RSPH1-deficient cells (OP-1428 II1). RSPH9 was undetectable in the mutant respiratory cells (Figures 3C and 3D and Figure E5C, shown for OI-162), indicating that the assembly of RSPH9 into respiratory axonemes is RSPH4A and RSPH1 dependent. Interestingly, these findings are consistent with data from interaction experiments performed using different Chlamydomonas RSP mutants (43, 44).
Figure 2.
Western blot analyses of antibodies directed against human radial spoke (RSPH) proteins. The antibodies used in this study, directed against RSPH9 (A), RSPH4A (B), RSPH1 (C), RSPH23 (D), and RSPH3 (E), were validated by Western blot and specifically detect bands with the predicted molecular weight or similar to the predicted molecular weight of the proteins.
Figure 3.
Loss-of-function mutations in the human radial spoke genes RSPH9, RSPH4A, and RSPH1 disrupt the assembly of the RS head protein, RSPH9. Respiratory epithelial cells from control subjects (A) and individuals affected by primary ciliary dyskinesia (PCD) carrying biallelic loss-of-function mutations in RSPH9 (OP-1135 [B]), RSPH4A (OI-162 [C]), or RSPH1 (OP-1428 II1 [D]) were double labeled with antibodies directed against acetylated α-tubulin (green) and RSPH9 (red). Both proteins colocalize (yellow) along the ciliary axonemes in cells from the unaffected control subjects (A). In contrast, in respiratory cilia of individuals with loss-of-function mutations in RSPH9, RSPH4A, and RSPH1, RSPH9 is undetectable in ciliary axonemes (B–D), indicating that the ciliary localization of RSPH9 is RSPH4A and RSPH1 dependent. Nuclei were stained with Hoechst 33342 (blue). acet. Tubulin, acetylated α-tubulin; DIC, differential interference contrast. Scale bars, 10 μm.
RSPH4A Loss-of-Function Mutations Cause Deficiency of the Assembly of the RS Head Protein RSPH4A; Loss-of-Function Mutations in RSPH1 and RSPH9 Do Not Affect the RSPH4A Localization into Axonemes
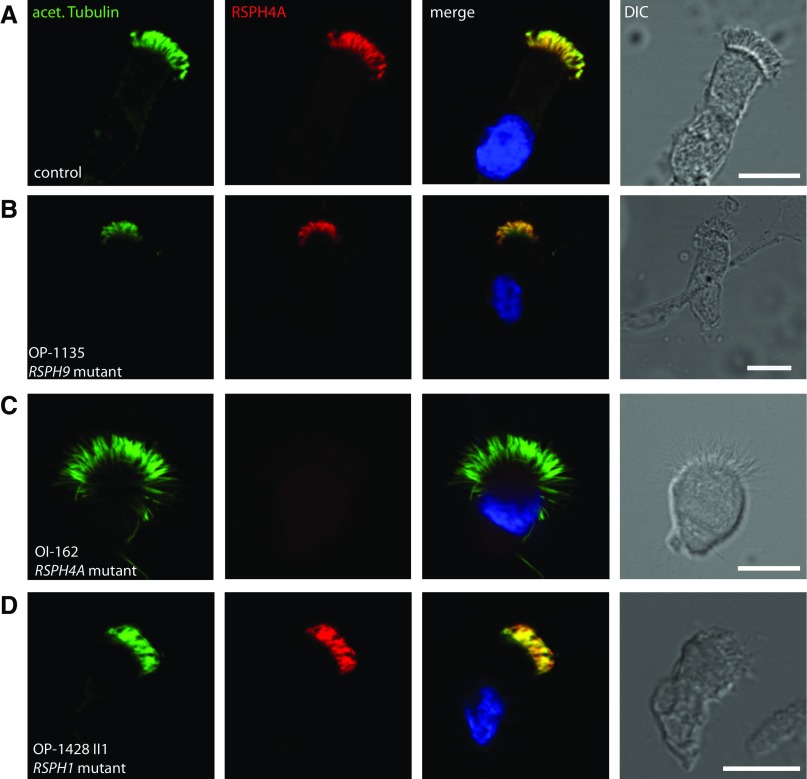
Furthermore, we analyzed the RSPH4A localization in respiratory epithelial cells from the affected patients, OP-225 II2 and OI-162, carrying biallelic loss-of-function mutations in RSPH4A using rabbit polyclonal antibodies directed against the RS head protein, RSPH4A. Western blot analysis showed that the antibodies specifically recognize both isoforms of RSPH4A, detecting two protein bands of the predicted molecular weight (80.75 and 67.2 kD; Figure 2B). Although RSPH4A localized to the entire axonemal length in wild-type cells (Figure 4A), it was undetectable in the cells of the affected individuals (Figures 4C, shown for OI-162), consistent with recessive loss-of-functions mutations that result in failure to produce a functional RSPH4A protein. Interestingly, by analyzing the RSPH4A localization in RSPH1-deficient cells (OP-1428 II1) or RSPH9-deficient cells (OP-1135), we showed that RSPH4A is still present in the axonemes of both mutants (Figures 4B and 4D), indicating that the assembly of RSPH4A into respiratory axonemes is neither RSPH1 nor RSPH9 dependent.
Figure 4.
Assembly of the RS head protein, RSPH4A, into ciliary axonemes is affected by loss-of-function mutations in RSPH4A but not by mutations in RSPH1 and/or RSPH9. Respiratory epithelial cells from control (A) and individuals affected by PCD carrying biallelic loss-of-function mutations in RSPH9 (OP-1135 [B]), RSPH4A (OI-162 [C]), or RSPH1 (OP-1428 II1 [D]) were double labeled with antibodies directed against acetylated α-tubulin (green) and RSPH4A (red). Both proteins colocalize (yellow) along the ciliary axonemes in cells from the unaffected control subjects and RSPH9 or RSPH1 mutant individuals (A, B, and D). In contrast, in respiratory cilia of RSPH4A mutant individuals, RSPH4A is undetectable in the ciliary axonemes (C), indicating that the ciliary localization of RSPH4A is affected by mutations in RSPH4A but not by mutations in RSPH9 (B) or RSPH1 (D). Nuclei were stained with Hoechst 33342 (blue). Scale bars, 10 μm.
RSPH4A and RSPH1 Loss-of-Function Mutations Cause Deficiency of the Assembly of the RS Head Protein, RSPH1, whereas Loss-of-Function Mutations in RSPH9 Do Not
We performed IF of respiratory epithelial cells from the affected patients OP-1428 II1 and OP-562 carrying biallelic loss-of-function mutations in RSPH1 using rabbit polyclonal antibodies directed against the RS head protein, RSPH1. Western blot analysis proved specificity of the antibody targeting RSPH1 (35.13 kD; Figure 2C). Although RSPH1 localized to the entire axonemal length in cells from control subjects (Figure 5A), it was undetectable in the cells of the affected individuals (Figure 5D, shown for OP-1428 II1), consistent with recessive loss-of-function mutations that result in the failure to produce a functional RSPH1 protein. In addition, we analyzed the RSPH1 localization in RSPH4A-deficient cells (OI-162) or RSPH9-deficient cells (OP-1135). Interestingly, RSPH1 was also undetectable in the RSPH4A mutant (Figure 5C), but still present in the RSPH9 mutant (Figure 5B), indicating that the assembly of RSPH1 into respiratory axonemes is RSPH4A dependent, but RSPH9 independent.
Figure 5.
RSPH4A and RSPH1 loss-of-function mutations disrupt the assembly of the RS head protein, RSPH1, whereas mutations in RSPH9 do not. Respiratory epithelial cells from control subjects (A) and individuals affected by PCD carrying biallelic loss-of-function mutations in RSPH9 (OP-1135 [B]), RSPH4A (OI-162 [C]), or RSPH1 (OP-1428 II1 [D]) were double labeled with antibodies directed against acetylated α-tubulin (green) and RSPH1 (red). Both proteins colocalize (yellow) along the ciliary axonemes in cells from the unaffected control subjects and the RSPH9 mutant individual (A and B). In contrast, in respiratory cilia of individuals with loss-of-function mutations in RSPH4A or RSPH1, RSPH1 is undetectable in the ciliary axonemes (C and D), indicating that the ciliary localization of RSPH1 is also affected by loss-of-function mutations in RSPH4A but not in RSPH9. Nuclei were stained with Hoechst 33342 (blue). Scale bars, 10 μm.
RSPH9, RSPH4A, and RSPH1 Mutations Do Not Affect the Assembly of the RS Neck Projection, RSPH23, or the RS Stalk Projection, RSPH3
Analyses of the localization of the RS neck protein, RSPH23, and the RS stalk protein, RSPH3, in respiratory epithelial cells from affected patients OI-162 (RSPH4A mutant), OP-1428 II1 (RSPH1 mutant), and OP-671 (RSPH9 mutant), using polyclonal antibodies directed against RSPH23 or RSPH3, revealed that both proteins still assembled to the axonemes of the different RS head mutants, the same as in control cells (Figures E6A–E6H). Western blot analysis showed that the antibodies specifically recognize RSPH23 and RSPH3 (24.24 and 63.7 kD; Figures 2D and 2E).
Missense Variants in RSPH4A and RSPH9 and a Small In-Frame Deletion in RSPH9 Affect the RS Head Proteins in the Same Way as Loss-of-Function Mutations in RSPH4A and RSPH9
To assess the significance of the detected missense variants and in-frame deletion, we performed identical IF analysis in respiratory cells. First, we analyzed the localization of the three head proteins, RSPH9, RSPH4A, and RSPH1, in individuals carrying different homozygous missense variants in RSPH4A. IF analysis with polyclonal antibodies revealed that RSPH4A, RSPH1, and RSPH9 are absent in ciliary axonemes of the RSPH4A mutants (OP-1603 II1, OP-1603 II2, OP-943 IV1, OP-708, OP-1606 II2, OP89 II2, OP-1795 II1, OP-1795 II2, and OP-1795 II3; Figures E7B, E7F, and E7J, shown for OP-89 II2). Second, we demonstrated that the in-frame deletion in RSPH9 (OP-601 II1 and II2), as well as the missense mutation in RSPH9 (OI-107), solely affected the assembly of RSPH9 (Figures E7K and E7L), but not of RSPH4A or RSPH1 in the ciliary axonemes (Figures E7C, E7D, E7G, and E7H). Because the results are identical to findings obtained in individuals with RSPH4A and RSPH9, classical loss-of-function mutations, we assume that assembly of the RS head is disturbed, possibly resulting in protein degradation and, subsequently, absence of the mutant protein in axonemes.
Interestingly, there are two isoforms described for RSPH9 encoding 276 amino acids (NM_152732) and 306 amino acids (NM_001193341). The identified missense variant, c.752A>G, results in an amino acid change from histidine to arginine (p.His251Arg) in the RSPH9 isoform encoded by transcript NM_152732 and a synonymous amino acid change from proline to proline (p.Pro268Pro) in the isoform encoded by transcript NM_001193341, which should not affect the protein. However, because no signal was obtained for RSPH9 in cilia of OI-107, we conclude that the isoform encoded by transcript NM_001193341 is probably not functionally present or expressed in respiratory cells.
Discussion
PCD due to defects in RS function are very difficult to diagnose, because ultrastructural analyses of cross-sections studied by TEM can be normal (33–37, 45), and some RS complexes can still be observed in TEM sections of patients with RSPH1 and RSPH4A mutations (35, 36, 46). Furthermore, ciliary beating abnormalities are very subtle and only detectable by experienced HSVM examiners (33, 35–37, 45). Here, we searched for mutations in RSPH9, RSPH4A, and RSPH1 in individuals with classical PCD symptoms with situs solitus, frequently normal TEM findings, and subtle changes in the ciliary beating pattern, as evaluated by HSVM (Table 1). We characterized the molecular defects of all currently genetically known PCD variants with RS defects at the protein level to better understand the disease mechanisms and to exploit this knowledge for novel diagnostic strategies.
RS multiprotein complexes form a signal-transduction scaffold between the CP and the dynein arms localized on the outer microtubuli doublets, and thus play a major role in regulating the ciliary beating (47). It is reported that the CP–RS interactions determine the waveform and the bend direction, whereas RS–IDA interactions influence the velocity of ciliary beating (48). In the alga, C. reinhardtii, the RSs are composed of 23 proteins that form a T-shaped complex, including a stalk, neck, and head compartment (31). Cryo-electron tomography studies revealed that there are three types of RSs, RS1, RS2, and RS3 (49). Interestingly, the third RS is structurally divergent between Chlamydomonas and humans (49). At least 12 RS proteins have apparent human orthologs, including RSPH9, RSPH4A, RSPH1, RSPH23, and RSPH3, underlining the high conservation throughout evolution (32).
So far, only a few studies address PCD with RS defects, and it has been suggested that genetic testing might be a powerful tool for diagnosis (37). Here, we report the largest cohort to date of genetically characterized individuals with PCD (n = 21 individuals from 16 families) with biallelic mutations in either RSPH9, RSPH4A, or RSPH1 (Table 1). We identified, in eight individuals, classical loss-of-function mutations and confirmed absence of the respective protein by IF analysis in mutant respiratory cells. Furthermore, we observed that c.2T>C (p.Met1?) and c.610A>T (p.Lys204*) in RSPH9 represent recurrent mutations. The mutation, c.2T>C, has arisen in the two unrelated families, OI-4 and OP-1665, both of different ethnic origin, as well as the mutation, c.610A>T, in the unrelated families, OP-658/OP-671 (siblings) and OP-1665. In addition, the splice site acceptor mutation, c.275–2A>C, in RSPH1 (OP-1428 II1) is also a recurrent mutation previously reported in 20 individuals from three different studies (35–37). This indicates that these mutations represent hotspot mutations.
We used respiratory cells from individuals with loss-of-function mutations in RSPH9, RSPH4A, or RSPH1, as well as control cells to characterize the RS defect on the molecular level using high-resolution IF microscopy. First, we confirmed specific expression of RS components by Western blot analysis in respiratory cell protein extracts (Figure 2). Using high-resolution IF analysis, we found that, in wild-type respiratory cilia, the RS head proteins, RSPH9, RSPH4A, and RSPH1 (Figures 3A, 4A, 5A and Figure E5A), the RS neck protein, RSPH23, as well as the RS stalk protein, RSPH3, are all present throughout the entire length of the ciliary axonemes (Figures E6A and E6E). In respiratory cells from individuals carrying homozygous RSPH4A loss-of-function mutations, RSPH4A was not detectable (Figure 4C), consistent with RSPH4A deficiency. Interestingly, the other RS head proteins, RSPH9 and RSPH1, were also absent from the ciliary axonemes (Figures 3C and 5C). The presence of the neck protein, RSPH23, and the stalk protein, RSPH3, in RSPH4A-deficient axonemes (Figures E6C and E6G) indicates that RSPH4A mutations affect solely the assembly of the RS head and have no impact on assembly of the neck or stalk complexes. Described findings obtained by cryo-electron tomography studies support this observation (49). In addition, our results provide the first evidence that RSPH4A mutations result in disruption of the RS head complex and axonemal loss of the components, RSPH1 and RSPH9, in humans. Although our results indicate interaction between RSPH9, RSPH4A, and RSPH1 in humans, additional interaction experiments (i.e., coimmunoprecipitation) have to be performed to confirm our findings. In Chlamydomonas, the RS head is composed of two identical subdomains aligned in twofold rotational symmetry and interconnecting to the neck (43, 44). Findings from 3D ultrastructural analysis of Chlamydomonas mutants lacking RSP4 orthologous to human RSPH4A (43, 44) are consistent with our findings in human respiratory cilia of RSPH4A-deficient individuals, where the head complex proteins RSPH9, RSPH4A and RSPH1 are absent (Figure 6). Our results are also consistent with the sequential assembly of flagellar RSs in Chlamydomonas where mutants lacking RSP4, do not partially preassemble a 12S complex that usually contains all the RS head proteins. Consequently Chlamydomonas flagella lack the RS head while a residual assembly of a stalk precursor form can still be observed. In addition, three-dimensional ultrastructural analysis of RSP4-deficient flagella of Chlamydomonas lack the head complex, leaving neck and stalk unaffected (31), which is also consistent with our observations (Figure E6). Thus, the molecular mechanisms of RS head assembly, including the functional role of RSP4/RSPH4A, has been evolutionary conserved from the unicellular green alga, C. reinhardtii, to man.
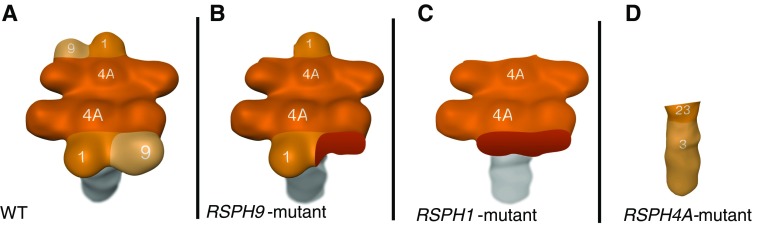
Figure 6.
Summary of the defects caused by mutations in RSPH9, RSPH4A, and RSPH1. Compared with wild-type (WT) RSs (A), mutation of RSPH9 solely leads to deficiency of the RSPH9 projection (B). Based on our IF findings, we conclude that deficiency of the RSPH1 projection leads to the lack of RSPH9 and RSPH1 (C), whereas RSPH4A mutants lack the entire head complex, just leaving the neck and stalk unaffected (D). Therefore, we conclude the following ladder of importance for the correct assembly of the RS head, starting with the most significant: RSPH4A, RSPH1, and RSPH9.
Respiratory cells from the affected individuals, OP-1428 II1 and OP-562, carrying biallelic RSPH1 loss-of-function mutations showed absence of the proteins, RSPH9 (Figure 3D) and RSPH1 (Figure 5D), from the ciliary axonemes, but RSPH4A remained detectable in the ciliary axonemes (Figure 4D). These findings are consistent with the normal RSPH4A localization in RSPH1 mutant cilia reported previously (36), and are supported by the fact that, in Chlamydomonas, RSP4, the ortholog of RSPH4A, is assembled before RSP1, orthologous to RSPH1 (44). Thus, the RSPH1 assembly is apparently RSPH4A dependent and not vice versa. Consequently, our findings contradict the abnormal RSPH4A localization in RSPH1 mutant cilia reported by others (35). Moreover, deficiency of the assembly of the RS head protein, RSPH9, in RSPH1 mutant cilia (Figure 3D) is consistent with previous findings (35).
Concerning RSPH9 loss-of-function mutants, our findings indicate that cilia deficient for RSPH9 still correctly assemble the RSPH4A and RSPH1 projections (Figures 4B and 5B), and demonstrate that RSPH9 does not play a crucial role in the assembly of the other RS head proteins, RSPH4A and RSPH1, but depends on their presence.
Interestingly, by genetic testing, we identified nine individuals with homozygous missense variants in RSPH4A, one individual with a homozygous RSPH9 missense variant, as well as two individuals with a homozygous in-frame deletion in RSPH9 predicting the loss of one amino acid residue. These variants affect evolutionarily conserved amino acid residues and were either not reported in the 1,000 genome database or were only present at very low frequency. However, it is difficult to prove that amino acid exchanges or the loss of one amino acid are indeed disease causing or represent very rare DNA polymorphisms. Therefore, we used respiratory cells from those individuals to test if IF analysis can be used to clarify whether or not the observed mutations are causative. Our IF results indicate that the homozygous missense variants in RSPH4A c.1105G>C, p.Ala369Pro in OP-1603 II1, OP-1603 II2, OP-943 IV1, OP-1606 II2, and OP-708, and the RSPH4A missense variant c.1391G>A, p.Gly464Glu in OP-89 II2, OP-1795 II1, OP-1795 II2, and OP-1795 II3 are indeed disease causing and result in absence of the RS head proteins, RSPH9, RSPH4A, and RSPH1 (Figures E7B, E7F, and E7J), indistinguishable from defects observed in RSPH4A loss-of-function mutations. The homozygous in-frame deletion mutation in RSPH9 (c.801_803 delGAA, p.268 delLys) in the siblings, OP-601 II1 and OP-601 II2, as well as the RSPH9 missense variant in OI-107 (c.752A>G, p.His251Arg) are also disease causing, because RSPH9 was not detectable in the ciliary axonemes (Figures E7K and E7L). Identical findings were obtained in RSPH9 loss-of-function mutations. Thus, we provide strong evidence that IF analysis can be used to diagnose PCD with RS head defects, and can be helpful to clarify the functional consequences of missense variants or other small in-frame mutations. However, additional functional studies could help ascertain the pathogenicity of missense variants. Interestingly, in our cohort missense variants represented a common type of mutation, but have rarely been reported in PCD with RS defects. We suspect that, due to the lack of confirmative measures, such as IF, some missense variants could not be further validated for their pathogenic significance.
Nodal cilia have a 9 + 0 microtubular pattern lacking the central pair, which interacts with the RSs to regulate the ciliary beating pattern (30). Although it has been described that nodal cilia contain RS proteins (33), RS complexes might not have a specific function, and lack of RS complexes does not affect nodal ciliary beating pattern. Consistent with the reported absence of left–right asymmetry defects in patients with RS defects (33), all the affected individuals presented here have situs solitus. This is an important fact that has to be considered, because the absence of laterality abnormalities makes early diagnosis of PCD less likely (6). Our findings will be especially beneficial for individuals with PCD without laterality defects, because we strongly believe that patients with deficient RS components can easily be overlooked by conventional TEM and HSVM.
We confirm that genetic testing is useful for PCD diagnostics of RS defects, especially the sequencing of hotspot mutations in RSPH1 and RSPH9. In addition, we illustrate that IF studies of respiratory cells obtained by simple nasal brushing using antibodies targeting RS head components could possibly be used as a diagnostic tool for PCD with RS defects, recognizing loss-of-function mutations as well as missense variants. We conclude that RSPH9 stainings might be useful for PCD diagnostics of individuals with typical PCD symptoms without situs inversus, and abnormal HSVM and/or TEM findings, as illustrated in Table 1. In addition, RSPH9 staining might be helpful to clarify the consequence of detected variants in RS head genes. Thus, we recommend performing IF with RSPH9 antibodies, because RSPH9 was absent from the ciliary axonemes in all studied RS mutant cells. In case abnormal results are obtained, further analysis using antibodies targeting RSPH1 and RSPH4A should be performed. In this way, genetic screening can be tailored to the relevant gene.
In conclusion, detailed knowledge of the interaction between RSPH9, RSPH4A, and RSPH1 and their IF analysis will possibly aid the future diagnostics of PCD. This is especially important, because there is currently no simple test available for PCD diagnosis in cases in which TEM analysis appears normal in individuals with RSPH9, RSPH4A, and RSPH1-mutations.
Acknowledgments
Acknowledgments
The authors thank the individuals affected by primary ciliary dyskinesia and their families for their participation and acknowledge the German patient support group “Kartagener Syndrom und Primaere Ciliaere Dyskinesie e.V.” They thank D. Ernst, M. Herting, L. Overkamp, F. J. Seesing, and Maria Philipsen for excellent technical work and Dr. P. Pennekamp for scientific support. They greatly appreciate the contribution of Robin Selbach, who provided technical assistance for three-dimensional figure construction, and they acknowledge Dr. Seithe (Nürnberg), Dr. Griese (Munich), Dr. Grychtol (Freiburg), and Dr. Zeidler for providing patient samples and thank the DNA pipelines team at the Wellcome Trust Sanger Institute for sequencing data generation.
Footnotes
This work was supported by a fellowship of the Nordrhein-Westfalen Research School “Cell Dynamics and Disease, CEDAD” to H. Omran, by Deutsche Forschungsgemeinschaft grants OM 6/4 and OM 6/7 (H. Omran), by European Commission (FP7/2007–2013) grant agreement (GA) number 262,055 (H. Omran), as a Transnational Access project of the European Sequencing and Genotyping Infrastructure and the Interdisziplinäres Zentrum für Klinische Forschung (Om2/009/12) Muenster to H. Omran as well as funding from European Union seventh FP under GA Nr. 241955, project SYStems biology approach to dissect CILIA function (SYSCILIA) (H. Omran) and GA Nr. 305404 project Better Experimental Screening and Treatment for Primary Ciliary Dyskinesia (BESTCILIA) (K.G.N. and H. Omran) and by the Deutsche Forschungsgemeinschaft grant OL450/1 to H. Olbrich.
Author Contributions: A.F. characterized the antibodies RSPH4A (human radial spoke protein 4A), RSPH23, and RSPH3 by Western blot (WB) and all studied antibodies by immunofluorescence (IF); performed IF experiments in OP-89 II2, OP-225 II2, OP-601 II1, OP-601 II2, OP-943 IV1, OP-1603 II1, OP-1603 II2, OP-1795 II1, OP-1795 II2, OP-1795 II3, prepared the clinical table, prepared the figures, assisted in sequencing experiments, and wrote the manuscript; R.H. identified mutations in OI-162 and OP-1665 II1, confirmed the mutations from WES data by Sanger sequencing (OP-1428 II1, OP-89 II2, OP-1135, OI-4 II2, OP-658, OP-671), performed IF stainings, prepared the figures, and wrote the manuscript; N.T.L. established the antibodies RSPH1 and RSPH9 in WB, supervised A.F.’s experiment, wrote the manuscript, and prepared the figures; C.E. sequenced OP-1603 II1, OP-1603 II2, OP-1606 II2, OP-943 IV, OP-1795 II1, OP-1795 II2, OP-1795 II3, OP-601 II1, OP-601 II2 and OP-708, prepared IF stainings of OP-708, prepared the clinical table, wrote the manuscript, and prepared the figures; C.J. identified mutations in OP-225 II2; J.R., J.W., C.W., and J.G.-O. provided clinical data for OP-1428 II1 and OP-1795, and critically assessed and corrected the manuscript; H. Olbrich identified mutations in OP-89 II2, OP-943 IV1 and OI-107, supervised C.J.’s work, prepared the figures, and critically assessed and corrected the manuscript; S.C. did IF screening experiments for radial spoke mutants; M.J. and M.B. provided patient samples and the clinical details of OP-562; Y.M. performed whole-exome data analysis; R.D., A.K.-K., and S.S. performed whole-exome project coordination and data production; J.K.M. and K.G.N. provided the patients OP-658, OP-671, OP-601 II1, OP-601 II2, OP-708 and their clinical details, and critically assessed and corrected the manuscript; I.A., N.E., E.K., and D.S. provided patients OI-4 II2, OI-107, and OI-162 and their clinical details, and critically assessed and corrected the manuscript; K.H. provided patients OP-89 II2, OP-1665, OP-1603, OP-943 IV and their clinical details, and critically assessed and corrected the manuscript; H. Omran designed the study, analyzed the data, supervised the work, and wrote the manuscript.
Originally Published in Press as DOI: 10.1165/rcmb.2014-0483OC on March 19, 2015
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Ibañez-Tallon I, Heintz N, Omran H. To beat or not to beat: roles of cilia in development and disease. Hum Mol Genet. 2003;12:R27–R35. doi: 10.1093/hmg/ddg061. [DOI] [PubMed] [Google Scholar]
- 2.Marthin JK, Petersen N, Skovgaard LT, Nielsen KG. Lung function in patients with primary ciliary dyskinesia: a cross-sectional and 3-decade longitudinal study. Am J Respir Crit Care Med. 2010;181:1262–1268. doi: 10.1164/rccm.200811-1731OC. [DOI] [PubMed] [Google Scholar]
- 3.Knowles MR, Daniels LA, Davis SD, Zariwala MA, Leigh MW. Primary ciliary dyskinesia: recent advances in diagnostics, genetics, and characterization of clinical disease. Am J Respir Crit Care Med. 2013;188:913–922. doi: 10.1164/rccm.201301-0059CI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zariwala MA, Knowles MR, Omran H. Genetic defects in ciliary structure and function. Annu Rev Physiol. 2007;69:423–450. doi: 10.1146/annurev.physiol.69.040705.141301. [DOI] [PubMed] [Google Scholar]
- 5.Kennedy MP, Omran H, Leigh MW, Dell S, Morgan L, Molina PL, Robinson BV, Minnix SL, Olbrich H, Severin T, et al. Congenital heart disease and other heterotaxic defects in a large cohort of patients with primary ciliary dyskinesia. Circulation. 2007;115:2814–2821. doi: 10.1161/CIRCULATIONAHA.106.649038. [DOI] [PubMed] [Google Scholar]
- 6.Kuehni CE, Frischer T, Strippoli MF, Maurer E, Bush A, Nielsen KG, Escribano A, Lucas JSA, Yiallouros P, Omran H, et al. Factors influencing age at diagnosis of primary ciliary dyskinesia in European children. Eur Respir J. 2010;36:1248–1258. doi: 10.1183/09031936.00001010. [DOI] [PubMed] [Google Scholar]
- 7.Pazour GJ, Agrin N, Leszyk J, Witman GB. Proteomic analysis of a eukaryotic cilium. J Cell Biol. 2005;170:103–113. doi: 10.1083/jcb.200504008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Becker-Heck A, Loges NT, Omran H. Dynein dysfuntion as a cause of primary ciliary dyskinesia and other ciliopathies. In: King SM, editor. Dyneins, 1st ed. Waltham, MA: Academic Press; 2011. pp. 602–627. [Google Scholar]
- 9.Olbrich H, Häffner K, Kispert A, Völkel A, Volz A, Sasmaz G, Reinhardt R, Hennig S, Lehrach H, Konietzko N, et al. Mutations in DNAH5 cause primary ciliary dyskinesia and randomization of left–right asymmetry. Nat Genet. 2002;30:143–144. doi: 10.1038/ng817. [DOI] [PubMed] [Google Scholar]
- 10.Pennarun G, Escudier E, Chapelin C, Bridoux AM, Cacheux V, Roger G, Clément A, Goossens M, Amselem S, Duriez B. Loss-of-function mutations in a human gene related to Chlamydomonas reinhardtii dynein IC78 result in primary ciliary dyskinesia. Am J Hum Genet. 1999;65:1508–1519. doi: 10.1086/302683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mazor M, Alkrinawi S, Chalifa-Caspi V, Manor E, Sheffield VC, Aviram M, Parvari R. Primary ciliary dyskinesia caused by homozygous mutation in DNAL1, encoding dynein light chain 1. Am J Hum Genet. 2011;88:599–607. doi: 10.1016/j.ajhg.2011.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duriez B, Duquesnoy P, Escudier E, Bridoux A, Escalier D, Rayet I, Marcos E, Vojtek A, Bercher J, Amselem S. A common variant in combination with a nonsense mutation in a member of the thioredoxin family causes primary ciliary dyskinesia. Proc Natl Acad Sci USA. 2007;104:3336–3341. doi: 10.1073/pnas.0611405104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loges NT, Olbrich H, Fenske L, Mussaffi H, Horvath J, Fliegauf M, Kuhl H, Baktai G, Peterffy E, Chodhari R, et al. DNAI2 mutations cause primary ciliary dyskinesia with defects in the outer dynein arm. Am J Hum Genet. 2008;83:547–558. doi: 10.1016/j.ajhg.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knowles MR, Leigh MW, Ostrowski LE, Huang L, Carson JL, Hazucha MJ, Yin W, Berg JS, Davis SD, Dell SD, et al. Exome sequencing identifies mutations in CCDC114 as a cause of primary ciliary dyskinesia. Am J Hum Genet. 2013;92:99–106. doi: 10.1016/j.ajhg.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Onoufriadis A, Paff T, Antony D, Shoemark A, Micha D, Kuyt B, Schmidts M, Petridi S, Dankert-Roelse JE, Haarman EG, et al. Splice-site mutations in the axonemal outer dynein arm docking complex gene CCDC114 cause primary ciliary dyskinesia. Am J Hum Genet. 2013;92:88–98. doi: 10.1016/j.ajhg.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hjeij R, Lindstrand A, Francis R, Zariwala MA, Liu X, Li Y, Damerla R, Dougherty GW, Abouhamed M, Olbrich H, et al. ARMC4 mutations cause primary ciliary dyskinesia with randomization of left/right body asymmetry. Am J Hum Genet. 2013;93:357–367. doi: 10.1016/j.ajhg.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hjeij R, Onoufriadis A, Watson CM, Slagle CE, Klena NT, Dougherty GW, Kurkowiak M, Loges NT, Diggle CP, Morante NF, et al. CCDC151 mutations cause primary ciliary dyskinesia by dysruption of the outer dynein arm docking complex formation. Am J Hum Genet. 2014;95:257–274. doi: 10.1016/j.ajhg.2014.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loges NT, Olbrich H, Becker-Heck A, Häffner K, Heer A, Reinhard C, Schmidts M, Kispert A, Zariwala MA, Leigh MW, et al. Deletions and point mutations of LRRC50 cause primary ciliary dyskinesia due to dynein arm defects. Am J Hum Genet. 2009;85:883–889. doi: 10.1016/j.ajhg.2009.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Omran H, Kobayashi D, Olbrich H, Tsukahara T, Loges NT, Hagiwara H, Zhang Q, Leblond G, O’Toole E, Hara C, et al. Ktu/PF13 is required for cytoplasmic pre-assembly of axonemal dyneins. Nature. 2008;456:611–616. doi: 10.1038/nature07471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitchison HM, Schmidts M, Loges NT, Freshour J, Dritsoula A, Hirst RA, O’Callaghan C, Blau H, Al Dabbagh M, Olbrich H, et al. Mutations in axonemal dynein assembly factor DNAAF3 cause primary ciliary dyskinesia. Nat Genet. 2012;44:381–389, S1–S2. doi: 10.1038/ng.1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tarkar A, Loges NT, Slagle CE, Francis R, Dougherty GW, Tamayo JV, Shook B, Cantino M, Schwartz D, Jahnke C, et al. DYX1C1 is required for axonemal dynein assembly and ciliary motility. Nat Genet. 2013;45:995–1003. doi: 10.1038/ng.2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Panizzi JR, Becker-Heck A, Castleman VH, Al-Mutairi DA, Liu Y, Loges NT, Pathak N, Austin-Tse C, Sheridan E, Schmidts M, et al. CCDC103 mutations cause primary ciliary dyskinesia by disrupting assembly of ciliary dynein arms. Nat Genet. 2012;44:714–719. doi: 10.1038/ng.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kott E, Duquesnoy P, Copin B, Legendre M, Dastot-Le Moal F, Montantin G, Jeanson L, Tamalet A, Papon J, Siffroi J, et al. Loss-of-function mutations in LRRC6, a gene essential for proper axonemal assembly of inner and outer dynein arms, cause primary ciliary dyskinesia. Am J Hum Genet. 2012;91:958–964. doi: 10.1016/j.ajhg.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zariwala MA, Gee HY, Kurkowiak M, Al-Mutairi DA, Leigh MW, Hurd TW, Hjeij R, Dell SD, Chaki M, Dougherty GW, et al. ZMYND10 is mutated in primary ciliary dyskinesia and interacts with LRRC6. Am J Hum Genet. 2013;93:336–345. doi: 10.1016/j.ajhg.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horani A, Druley TE, Zariwala MA, Patel AC, Levinson BT, Van Arendonk LG, Thornton KC, Giacalone JC, Albee AJ, Wilson KS, et al. Whole-exome capture and sequencing identifies HEATR2 mutation as a cause of primary ciliary dyskinesia. Am J Hum Genet. 2012;91:685–693. doi: 10.1016/j.ajhg.2012.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knowles MR, Ostrowski LE, Loges NT, Hurd T, Leigh MW, Huang L, Wolf WE, Carson JL, Hazucha MJ, Yin W, et al. Mutations in SPAG1 cause primary ciliary dyskinesia associated with defective outer and inner dynein arms. Am J Hum Genet. 2013;93:711–720. doi: 10.1016/j.ajhg.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Austin-Tse C, Halbritter J, Zariwala MA, Gilberti RM, Gee HY, Hellman N, Pathak N, Liu Y, Panizzi JR, Patel-King RS, et al. Zebrafish ciliopathy screen plus human mutational analysis identifies C21orf59 and CCDC65 defects as causing primary ciliary dyskinesia. Am J Hum Genet. 2013;93:672–686. doi: 10.1016/j.ajhg.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Becker-Heck A, Zohn IE, Okabe N, Pollock A, Lenhart KB, Sullivan-Brown J, McSheene J, Loges NT, Olbrich H, Haeffner K, et al. The coiled-coil domain containing protein CCDC40 is essential for motile cilia function and left–right axis formation. Nat Genet. 2011;43:79–84. doi: 10.1038/ng.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Merveille A, Davis EE, Becker-Heck A, Legendre M, Amirav I, Bataille G, Belmont J, Beydon N, Billen F, Clément A, et al. CCDC39 is required for assembly of inner dynein arms and the dynein regulatory complex and for normal ciliary motility in humans and dogs. Nat Genet. 2011;43:72–78. doi: 10.1038/ng.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wirschell M, Olbrich H, Werner C, Tritschler D, Bower R, Sale WS, Loges NT, Pennekamp P, Lindberg S, Stenram U, et al. The nexin–dynein regulatory complex subunit DRC1 is essential for motile cilia function in algae and humans. Nat Genet. 2013;45:262–268. doi: 10.1038/ng.2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pigino G, Ishikawa T. Axonemal radial spokes: 3D structure, function and assembly. BioArchitecture. 2012;2:50–58. doi: 10.4161/bioa.20394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang P, Diener DR, Yang C, Kohno T, Pazour GJ, Dienes JM, Agrin NS, King SM, Sale WS, Kamiya R, et al. Radial spoke proteins of Chlamydomonas flagella. J Cell Sci. 2006;119:1165–1174. doi: 10.1242/jcs.02811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Castleman VH, Romio L, Chodhari R, Hirst RA, de Castro SCP, Parker KA, Ybot-Gonzalez P, Emes RD, Wilson SW, Wallis C, et al. Mutations in radial spoke head protein genes RSPH9 and RSPH4A cause primary ciliary dyskinesia with central-microtubular-pair abnormalities. Am J Hum Genet. 2009;84:197–209. doi: 10.1016/j.ajhg.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ziętkiewicz E, Bukowy-Bieryłło Z, Voelkel K, Klimek B, Dmeńska H, Pogorzelski A, Sulikowska-Rowińska A, Rutkiewicz E, Witt M. Mutations in radial spoke head genes and ultrastructural cilia defects in East-European cohort of primary ciliary dyskinesia patients. PLoS One. 2012;7:e33667. doi: 10.1371/journal.pone.0033667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Onoufriadis A, Shoemark A, Schmidts M, Patel M, Jimenez G, Liu H, Thomas B, Dixon M, Hirst RA, Rutman A, et al. Targeted NGS gene panel identifies mutations in RSPH1 causing primary ciliary dyskinesia and a common mechanism for ciliary central pair agenesis due to radial spoke defects. Hum Mol Genet. 2014;23:3362–3374. doi: 10.1093/hmg/ddu046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kott E, Legendre M, Copin B, Papon J, Dastot-Le Moal F, Montantin G, Duquesnoy P, Piterboth W, Amram D, Bassinet L, et al. Loss-of-function mutations in RSPH1 cause primary ciliary dyskinesia with central-complex and radial-spoke defects. Am J Hum Genet. 2013;93:561–570. doi: 10.1016/j.ajhg.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knowles MR, Ostrowski LE, Leigh MW, Sears PR, Davis SD, Wolf WE, Hazucha MJ, Carson JL, Olivier KN, Sagel SD, et al. Mutations in RSPH1 cause primary ciliary dyskinesia with a unique clinical and ciliary phenotype. Am J Respir Crit Care Med. 2014;189:707–717. doi: 10.1164/rccm.201311-2047OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Olbrich H, Schmidts M, Werner C, Onoufriadis A, Loges NT, Raidt J, Banki NF, Shoemark A, Burgoyne T, Al Turki S, et al. Recessive HYDIN mutations cause primary ciliary dyskinesia without randomization of left–right body asymmetry. Am J Hum Genet. 2012;91:672–684. doi: 10.1016/j.ajhg.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Omran H, Loges NT. Immunofluorescence staining of ciliated respiratory epithelial cells. Methods Cell Biol. 2009;91:123–133. doi: 10.1016/S0091-679X(08)91007-4. [DOI] [PubMed] [Google Scholar]
- 40.Fliegauf M, Olbrich H, Horvath J, Wildhaber JH, Zariwala MA, Kennedy M, Knowles MR, Omran H. Mislocalization of DNAH5 and DNAH9 in respiratory cells from patients with primary ciliary dyskinesia. Am J Respir Crit Care Med. 2005;171:1343–1349. doi: 10.1164/rccm.200411-1583OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raidt J, Wallmeier J, Hjeij R, Große Onnebrink J, Pennekamp P, Loges NT, Olbrich H, Häffner K, Dougherty GW, Omran H, et al. Ciliary beat pattern and frequency in genetic variants of primary ciliary dyskinesia. Eur Respir J. 2014;44:1579–1588. doi: 10.1183/09031936.00052014. [DOI] [PubMed] [Google Scholar]
- 42.Leigh MW, Hazucha MJ, Chawla KK, Baker BR, Shapiro AJ, Brown DE, Lavange LM, Horton BJ, Qaqish B, Carson JL, et al. Standardizing nasal nitric oxide measurement as a test for primary ciliary dyskinesia. Ann Am Thorac Soc. 2013;10:574–581. doi: 10.1513/AnnalsATS.201305-110OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pigino G, Bui KH, Maheshwari A, Lupetti P, Diener D, Ishikawa T. Cryoelectron tomography of radial spokes in cilia and flagella. J Cell Biol. 2011;195:673–687. doi: 10.1083/jcb.201106125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Diener DR, Yang P, Geimer S, Cole DG, Sale WS, Rosenbaum JL. Sequential assembly of flagellar radial spokes. Cytoskeleton (Hoboken) 2011;68:389–400. doi: 10.1002/cm.20520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Daniels ML, Leigh MW, Davis SD, Armstrong MC, Carson JL, Hazucha M, Dell SD, Eriksson M, Collins FS, Knowles MR, et al. Founder mutation in RSPH4A identified in patients of Hispanic descent with primary ciliary dyskinesia. Hum Mutat. 2013;34:1352–1356. doi: 10.1002/humu.22371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burgoyne T, Lewis A, Dewar A, Luther P, Hogg C, Shoemark A, Dixon M. Characterizing the ultrastructure of primary ciliary dyskinesia transposition defect using electron tomography. Cytoskeleton (Hoboken) 2014;71:294–301. doi: 10.1002/cm.21171. [DOI] [PubMed] [Google Scholar]
- 47.Smith EF, Yang P. The radial spokes and central apparatus: mechano-chemical transducers that regulate flagellar motility. Cell Motil Cytoskeleton. 2004;57:8–17. doi: 10.1002/cm.10155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kohno T, Wakabayashi K, Diener DR, Rosenbaum JL, Kamiya R. Subunit interactions within the Chlamydomonas flagellar spokehead. Cytoskeleton (Hoboken) 2011;68:237–246. doi: 10.1002/cm.20507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin J, Yin W, Smith MC, Song K, Leigh MW, Zariwala MA, Knowles MR, Ostrowski LE, Nicastro D. Cryo-electron tomography reveals ciliary defects underlying human RSPH1 primary ciliary dyskinesia. Nat Commun. 2014;5:5727. doi: 10.1038/ncomms6727. [DOI] [PMC free article] [PubMed] [Google Scholar]