Abstract
It is clear that viral entry, replication, and spread is a complex process involving a dialog between the virus and the targeted host cell. Viruses have evolved highly specific strategies to hijack cellular factors to promote their internalization, initiate their replication, and facilitate their eventual spread. However, the identification of many of these host cell molecules has been hindered by the requirement for robust genome-scale loss-of-function assays that are capable of targeting a wide variety of host factors. The more recent use of genome-scale or genome-wide RNA interference (RNAi) screens have extended our knowledge of the complex interplay between a virus and host and have implicated a wide variety of cellular factors required for infection of a number of viruses. Here, we describe an approach to target mammalian host cell factors involved in regulating viral infections by the use of a genome-scale RNAi library screen.
Keywords: High-throughput screening, RNAi, Host factors
1. Introduction
Most viruses have evolved strategies to coopt host cell factors to promote a variety of events throughout their infectious life cycles. These events can be associated with modification(s) of the host cell that facilitate endocytic uptake, initiate viral replication, and/ or manipulate apoptotic and innate immune signaling. The identification of these host cell factors has remained elusive largely in part due to the lack of robust high-throughput screening assays that target a wide variety of host genes simultaneously. The advent of genome-scale RNAi screening approaches has provided a substantial step forward in our ability to identify host cell factors involved in regulating viral infections in mammalian cells. Several genome-wide RNAi screens have implicated a wide variety of cellular factors required for infection of a number of viruses (1–7). Although the functions of many of these signaling molecules remains to be defined, the results clearly implicate intracellular cell signaling as a key strategy used by viruses during infection.
Here, we present a generalized protocol for a high-throughput RNAi screen to identify mammalian cellular factors involved in viral infection using Coxsackievirus B3 and human osteosarcoma (U2OS) cells as examples.
2. Materials
2.1. siRNA and Transfections
384-Well plates, black with clear bottom (Corning 3712).
12 pin vacuum manifold (Drummond).
Automated liquid handling device (Well-mate, Thermo Fisher).
Genome-scale library of duplex siRNAs (Druggable Genome, Ambion) (see Note 1).
siRNA duplex optimized to reduce viral receptor expression by >80% (Ambion) (see Note 2).
Opti-MEM reduced serum medium (Invitrogen).
Hiperfect Transfection reagent (Qiagen).
2.2. Cell Culture
U2OS cells (ATCC) are a human osteosarcoma cell line that serves as an ideal cell type for high-throughput screening strategies as they are highly adherent and are highly transfectable by most commercially available transfection reagents (see Note 3).
Dulbecco’s Modified Eagle Medium High Glucose, containing 4,500 mg/L D-glucose, and sodium pyruvate (DME-H, Invitrogen). Supplement with 10% fetal bovine serum (FBS, Invitrogen), 100 μg/mL penicillin, and 100 μg/mL streptomycin (Invitrogen).
Trypsin/EDTA: 0.25% trypsin, 1 mM ethylenediamine tetraacetic acid (EDTA).
2.3. Infections
1. Coxsackievirus B3 is a member of the picornavirus family of small positive sense RNA viruses. This virus is commercially available (ATCC) and can be expanded and titred in HeLa cells as described previously (8).
2.4. Immunofluorescent Staining
Ice cold methanol/acetone (3:1 ratio).
Phosphate buffered saline (PBS), 137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.47 mM.
KH2PO4, adjust to a final pH of 7.4.
0.1% Triton-X 100 in PBS.
Primary antibody recognizing the enterovirus viral coat protein VP1 (anti-VP1) monoclonal antibody (NCL-ENTERO, Novocastra Laboratories) (see Note 4).
Secondary antibody: Alexa Fluor-488 Goat anti-mouse IgG (Invitrogen).
4′,6-Diamidino-2-phenylindole (DAPI), a blue-fluorescent nucleic acid stain to identify nuclei. Dissolve 10 mg in 2 mL of deionized water for a final concentration of 5 mg/mL. This stock can be stored at <20°C for long-term storage. To counterstain nuclei, dilute the 5 mg/mL stock to make a 300 nM working solution in PBS.
Adhesive sticker (Fisher Scientific).
2.5. Microscope and Software for Automated Image Acquisition and Analysis
ImageXpress Micro (Molecular Devices) with a 10× objective.
Automated image analysis software (MetaXpress, Leica).
3. Methods
3.1. Complexing siRNAs and Transfection Reagent
Using an automated liquid handling station (which is beneficial to avoid well to well variability), array a pool of siRNAs (three siRNAs per target gene) in 384-well plates (25 nM final concentration) (see Note 5).
Create a mix of Hiperfect transfection reagent with Opti-MEM (0.5 μL Hiperfect and 9.5 μL Opti-MEM per well). To limit errors in pipetting and to account for void volume in Well-Mate tubing, mix sufficient reagent for 400 wells/plate (thus, 200 μL Hiperfect in 3.8 mL of Opti-MEM per 384-well plate). This solution is stable for ~1 h at room temperature.
Add 10 μL of the Hiperfect/Opti-MEM mix to each well.
Spin plates at 800 × g for 1 min (see Note 6).
Incubate for 10 min at room temperature to form siRNA:Hiperfect complexes.
3.2. Preparation and Transfection of Cells
Grow U2OS cells at 37°C and 5% CO2 in complete DMEM-H medium. Cells should be grown to approximately 80% confluence.
Wash cells briefly with PBS and dislodge with trypsin/EDTA.
Pellet cells (800 × g, 3 min) and resuspend in medium at a concentration of 80,000 cells/mL.
Add 25 μL of the cell suspension (2,000 cells/well) to wells containing precomplexed siRNAs.
Spin plates (800 × g, 1 min).
Incubate at 37°C and 5% CO2 for 48 h (see Note 7).
3.3. Infections
Prepare virus stocks (Coxsackievirus B3-RD strain, ~1 × 1011 pfu/mL) (see Note 8).
Aspirate medium from the wells using a vacuum manifold.
Infect cells by adding 25 μL virus suspension in DMEM-H at an MOI of 0.5–1 (see Note 8) which leads to ~20–30% infection level.
Spin plates at 800 × g for 1 min.
Incubate for 8 h at 37°C and 5% CO2 (see Note 7).
3.4. Immunofluorescent Staining
Aspirate virus-containing medium from wells using a vacuum manifold.
Wash wells briefly with 30 μL PBS to remove residual medium.
Add 30 μL ice cold 3:1 methanol:acetone to each well to fix the cells.
Spin plates at 800 × g for 1 min.
Incubate for 5 min at room temperature.
Aspirate methanol:acetone using vacuum manifold and add 50 μL PBS per well. At this point, the plates can be stored at 4°C for up to 1 week or stained immediately.
Aspirate PBS using a vacuum manifold and add 25 μL of primary antibody diluted 1:500 in PBS.
Spin plates at 800 × g for 1 min.
Incubate at room temperature for 1 h.
Aspirate and wash wells with 50 μL of PBS. Repeat this wash step three times (for a total of four washes).
Add 20 μL of secondary antibody diluted 1:2,000 in PBS containing 300 nM DAPI.
Spin plates at 800 × g for 1 min.
Aspirate and wash wells with 50 μL of PBS. Repeat this wash step three times.
After the final wash, add 30 μL of PBS for storage and cover the plate with adhesive sticker. The fluorescent signal is stable for several weeks if the plates are stored at 4°C.
3.5. Image Acquisition and Analysis
Capture at least three images per well in both the DAPI channel and the virus channel (488 nm).
Use automated image analysis software to calculate the number of cells (Dapi+) and the number of infected cells (VP1+). The ratio of these values (VP1+/DAPI+) is the level of infection.
Calculate robust Z-scores for each well (see Note 9).
“Hits” can be identified as those wells that exhibited a change in infection (either decrease or increase) by ≥2 standard deviations from calculated Z-scores (see Note 9).
Analysis of cell number can be used to remove siRNAs with cytotoxic effects.
Toxic siRNAs are excluded based upon decreased cell viability as measured by a robust Z-score ≤–2. In general, this corresponds to a decrease in cell number of more that 30% (Fig. 1).
Fig. 1.
Identification of toxic siRNAs. Representative fluorescence images of cytotoxic siRNAs. DAPI-stained nuclei of nontoxic (left ) and toxic (right ) siRNAs. Corresponding Z-scores are shown in white. Blue, DAPI-stained nuclei and green, VP1.
3.6. Limiting Wellto-Well and Plate-to-Plate Variability
Plate-to-plate and day-to-day variability are minimized via the use of automated liquid handling.
Cell number should be optimized prior to RNAi screening in order to eliminate any cell piling that may occur throughout the course of the screen (there should be a single monolayer of cells to ensure proper image acquisition).
The level of infection should also be optimized (MOI and time of infection) in order to achieve 30–50% infection. This will allow for the identification of hits that have both an inhibitory and stimulatory effect on virus infection levels.
To control for variability between plates/wells (and to control for siRNA transfection efficiency between plates/wells), it is ideal to include preoptimized siRNAs that reduce expression of viral receptors, if possible. We previously used siRNAs that reduce the expression of the coxsackievirus and adenovirus receptor (CAR) by >75% (8, 9). These siRNAs are spotted in three wells of each plate.
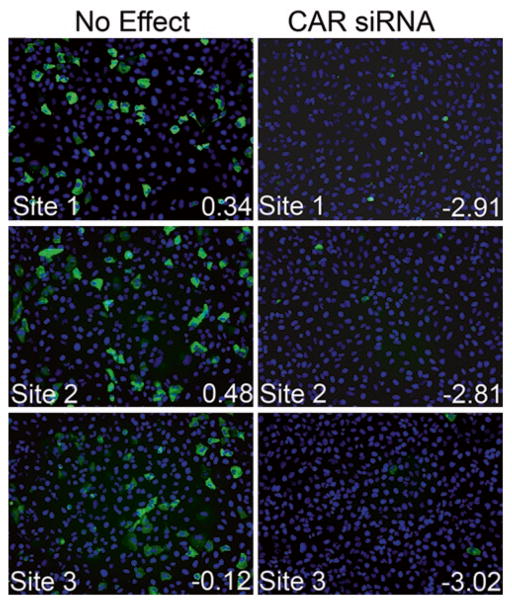
These wells should be identified as hits in the screen and should exhibit minimal variability in Z-scores between plates (Fig. 2).
Fig. 2.
Receptor siRNAs serve as effective internal controls. Shown are three independent immunofluorescence images from a single well of CVB-infected U2OS cells transfected with an siRNA which had no effect (left ) or transfected with positive control CAR siRNA (right ). Corresponding Z-scores (bottom right ) are shown in white text. Blue, DAPI-stained nuclei, and green, VP1.
3.7. Validation and Follow-Up
Positive candidates identified in the initial screen (primary screen) should be validated by performing a secondary screen analysis using independent siRNAs designed against another region of the mRNA (this will minimize effects induced due to off-target events). A gene is included in follow-up studies when it has been identified as a hit in both primary and secondary screens.
Additional follow-up experiments to confirm positive candidates include pharmacological inhibitors and/or dominant-negative mutants. Multiple methods to validate a given hit will ensure that a hit is genuine.
Once a positive candidate is validated by the above-described means, the function of that gene in regulating virus infection can begin to be dissected.
Acknowledgments
This work was supported by grants from the NIH [R01AI081759 (CBC) and R01AI074951, U54AI057168 (SC)].
Footnotes
. The Ambion Druggable Genome RNAi library is a commercially available library containing siRNAs targeting a varying amount of human genes (V3 targets ~7,000 genes whereas V4 targets ~9,000 genes), with four individual siRNAs per target. Many of these siRNAs have been tested for efficacy. However, some siRNAs are not specific to given genes owing to redundancy and sequence homology (generally within members of related gene families). The library targets a wide variety of molecules involved in metabolism, intracellular signaling (including serine/threonine and tyrosine kinases), small GTP-binding proteins and their effector molecules, phosphatases, proteases, a variety of ion channels, caspase and caspase-related molecules, and molecular motor-related targets. The library is shipped as 0.25 nmol/siRNA lyophilized powder in 96-well plates (267 total plates for V3 and 356 total plates for V4). SiRNAs are reconstituted in nuclease-free water at the desired final concentration (an automated website from Ambion for this calculation can be found at: http://www.ambion.com/techlib/append/oligo_dilution.html). For example, to achieve a 1 μM stock of siRNA, each well would be reconstituted with 250 μL of nuclease-free water. Plates are then stored at −80°C.
. CAR siRNA sequence: sense, 5′-GGUGGAUCAAGUGA UUAUU-3′ and antisense, 5′-AAUAAUCACUUGAUCC ACC-3′.
. It is imperative to begin any RNAi screen with cells that are healthy and have been maintained properly. We test our cells for mycoplasma contamination biweekly by PCR to ensure that they do not exhibit any growth or infection anomalies.
. As the screening strategy relies on immunofluorescent means to detect viral replication, it is important to optimize staining conditions (including optimal fixation conditions and dilution of primary and secondary antibodies) prior to performing any screen.
. Individual siRNAs (reconstituted as described in Note 1) should be pooled together using an arrayer robot to avoid errors (such as those associated with manual pipetting) that might alter siRNA concentrations (for example, TekBench from TekCel). The final volume of each pooled set of siRNAs should be 25 nM (thus, 6.25 nM final concentration for each individual siRNA).
. To ensure that there is no solution remaining on the walls of the well, it is helpful to perform a “tap spin” for a brief period at a low g-force to ensure that all media/solution covers the bottom of the well (this is particularly critical when a low volume is added to each well). These “tap spins” appear at a variety of steps throughout the screening protocol. This is most easily accomplished in a swinging bucket rotor cell culture centrifuge with the appropriate multiwell plate adaptors (such as the Eppendorf 5804).
. To avoid loss of medium due to evaporation throughout the experiment, it is ideal to maintain plates under high humidity. We accomplish this by using large Tupperware containers lined with water soaked paper towels.
. CVB3 can be expanded, purified, and titered as described (8). It is best to prepare a “master mix” of media: virus at MOI = 1 to avoid pipetting areas that could contribute to plate-to-plate variability in infection levels. For example, as each well is infected with 25 μL of media containing virus (MOI = 1), each plate will require 9.6 mL of virus-containing media (assuming 384-well plates). If the RNAi screen totals 50 384-well plates, a total of 480 mL of virus-containing media would be required.
. A robust Z-score is a statistical measure that quantifies the distance (in standard deviations) that a data point is from the median (and median absolute deviation) of a data set. In RNAi high-throughput screening, the calculation of Z-scores does not rely on control siRNAs, but instead assumes that the majority of siRNAs will have no effect (on virus infection, in this case) and can thus serve as controls. Thus, to calculate Z-scores, first perform a log transformation of all infection levels from each site of each well of a plate [log (VP1/DAPI)] (using Excel or another equivalent statisical software) and transform. Then, calculate the median and interquartile ranges (which represents the lowest 25% and the highest 25% of infection levels) of these data. Then, calculate Z-scores based on these data. A Z-score equal to 0 has the exact same value as the mean of the plate (although in a large data set, this is a rare value), whereas a Z-score equal to 1 is exactly one standard-deviation above the mean (and −1 is therefore one standard-deviation below the mean). Malo et al. (10) provide an excellent review describing the use of robust statistics in high-throughput screening and the benefits and pitfalls of these techniques.
References
- 1.Pelkmans L, Fava E, Grabner H, Hannus M, Habermann B, et al. Genome-wide analysis of human kinases in clathrin- and caveolae/raft-mediated endocytosis. Nature. 2005;436:78–86. doi: 10.1038/nature03571. [DOI] [PubMed] [Google Scholar]
- 2.Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, et al. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 3.Krishnan MN, Ng A, Sukumaran B, Gilfoy FD, Uchil PD, et al. RNA interference screen for human genes associated with West Nile virus infection. Nature. 2008;455:242–245. doi: 10.1038/nature07207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hao L, Sakurai A, Watanabe T, Sorensen E, Nidom CA, et al. Drosophila RNAi screen identifies host genes important for influenza virus replication. Nature. 2008;454:890–893. doi: 10.1038/nature07151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tai AW, Benita Y, Peng LF, Kim SS, Sakamoto N, et al. A functional genomic screen identifies cellular cofactors of hepatitis C virus replication. Cell Host Microbe. 2009;5:298–307. doi: 10.1016/j.chom.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sessions OM, Barrows NJ, Souza-Neto JA, Robinson TJ, Hershey CL, et al. Discovery of insect and human dengue virus host factors. Nature. 2009;458:1047–1050. doi: 10.1038/nature07967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cherry S, Kunte A, Wang H, Coyne C, Rawson RB, et al. COPI activity coupled with fatty acid biosynthesis is required for viral replication. PLoS Pathog. 2006;2:e102. doi: 10.1371/journal.ppat.0020102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coyne CB, Bergelson JM. Virus-induced Abl and Fyn kinase signals permit coxsackievirus entry through epithelial tight junctions. Cell. 2006;124:119–131. doi: 10.1016/j.cell.2005.10.035. [DOI] [PubMed] [Google Scholar]
- 9.Coyne CB, Kim KS, Bergelson JM. Poliovirus entry into human brain microvascular cells requires receptor-induced activation of SHP-2. EMBO J. 2007;26:4016–4028. doi: 10.1038/sj.emboj.7601831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malo N, Hanley JA, Cerquozzi S, Pelletier J, Nadon R. Statistical practice in high-throughput screening data analysis. Nat Biotechnol. 2006;24:167–175. doi: 10.1038/nbt1186. [DOI] [PubMed] [Google Scholar]