Significance
SHORT VALVE 1 (STV1), a ribosomal protein, is required for the development of Arabidopsis. However, its functional mechanism remains to be identified. This research shows that STV1 binds the stem-loop flanked by a short 5′ arm within primary miRNAs and facilitates the recruitment of primary miRNAs to the DICER-LIKE1 complex. Consequently, this study provides insights into the mechanisms controlling miRNA production and identifies an extraribosomal function of STV1. Because STV1 is a conserved protein in eukaryotes, the results may produce a broader impact.
Keywords: miRNA biogenesis, Arabidopsis, STV1 RNA binding, pri-miRNAs
Abstract
MicroRNAs (miRNAs) are key regulators of gene expression. They are processed from primary miRNA transcripts (pri-miRNAs), most of which are transcribed by DNA-dependent polymerase II (Pol II). miRNA levels are precisely controlled to maintain various biological processes. Here, we report that SHORT VALVE 1 (STV1), a conserved ribosomal protein, acts in miRNA biogenesis in Arabidopsis. A portion of STV1 localizes in the nucleus and binds pri-miRNAs. Using pri-miR172b as a reporter, we show that STV1 binds the stem-loop flanked by a short 5′ arm within pri-miRNAs. Lack of STV1 reduces the association of pri-miRNAs with their processing complex. These data suggest that STV1 promotes miRNA biogenesis through facilitating the recruitment of pri-miRNAs to their processing complex. Furthermore, we show that STV1 indirectly involves in the occupancy of Pol II at the promoters of miRNA coding genes (MIR) and influences MIR promoter activities. Based on these results, we propose that STV1 refines the accumulation of miRNAs through its combined effects on pri-miRNA processing and transcription. This study uncovers an extraribosomal function of STV1.
MicroRNAs (miRNAs) are noncoding RNAs that regulate multiple biological processes by inhibiting gene expression at posttranscriptional levels (1–3). The majority of miRNA-coding genes (MIR) are transcribed by the DNA-dependent RNA polymerase II (Pol II) to produce primary miRNA transcripts (pri-miRNAs) (4). In Arabidopsis, a complex containing DICER-LIKE1 (DCL1, an RNase III enzyme), HYPONASTIC LEAVES 1 (HYL1, a double-stranded RNA-binding protein), SERRATE (SE, a Zinc-finger protein), and TOUGH (TGH, an RNA-binding protein) process the imperfect stem-loops residing in the pri-miRNAs to release the miRNA/miRNA* (passenger strand) duplex in the nucleus (3, 5–9). The miRNA duplex is then methylated by HUA1 ENHANCER1 (HEN1) to prevent uridylation and degradation (3). Following biogenesis, miRNAs are loaded into the effector protein called ARGONAUTE 1 (AGO1), which requires HSP90 and the Cyclophilin 40 (CYP-40), to repress the expression of genes containing their homolog sequences through mRNA cleavage and translational inhibition in Arabidopsis (10–13).
Plant miRNA biogenesis is regulated through pri-miRNA transcription, stability, and processing (14). In Arabidopsis, the transcription factors, including the mediator complex, the elongator complex, Negative on TATA less 2 (NOT2), and CELL DIVISION CYCLE 5 (CDC5) have been shown to positively regulate MIR transcription through recruiting Pol II to MIR promoters (15–17). The transcription of some MIRs, such as MIR172 and MIR156, is also subject to physiological, temporal, or spatial regulation by specific transcription factors (18, 19). The RNA-binding proteins DAWDLE (DDL) and PLEIOTROPIC REGULATORY LOCUS 1 (PRL1) are proposed to stabilize pri-miRNAs following their transcription (14, 20). Several protein factors, including DDL, CDC5, PRL1, and CBP20/80 (two cap-binding proteins), associate with the DCL1 complex to promote pri-miRNA processing (14, 17, 20–22), whereas the elongator complex, NOT2 and MODIFIER OF SNC1, 2 (MOS2, an RNA-binding protein), are required for the correct assembly of the DCL1 complex (16, 23). Proteins including SICKLE (SIC, a proline-rich protein), RECEPTOR FOR ACTIVATED C KINASE 1 (RACK1), STABILIZED1 (STA1, a pre-mRNA processing factor 6 homolog), HOS5 and GRP7 (a glycine-rich RNA-binding protein), act outside of the DCL1 complex to participate in pri-miRNA processing (24–29). Protein modifications also regulate the activity of the DCL1 complex. For example, HYL1 activity is controlled by phosphorylation and dephosphorylation (30, 31). Furthermore, the expression levels of the DCL1 complex are regulated at transcriptional and posttranscriptional levels to influence miRNA accumulation (32–34). The structure and length of the stem-loop also impact pri-miRNA processing. For example, the presence of an internal loop following a ∼15-bp stem below the miRNA/miRNA* is required for efficient processing of some pri-miRNAs (35–39). Despite these progresses, it is still enigmatic how the DCL1 complex recognizes pri-miRNAs, because the stem loops of pri-miRNAs are diversified in structures, lengths, and sequences.
Ribosomal proteins are ribosome components responsible for ribosome assembly and protein translation. Because of their ability to interact with RNAs and proteins, their extraribosomal functions have been proposed (40). However, most extraribosomal functions of ribosomal proteins are still related to the ribosome or its synthesis. Thus, the real extraribosomal functions of ribosomal proteins still remain to be uncovered (40). The ribosomal protein L24 (RPL24) exists in archaebacteric and eukaryotic, but not prokaryotic, ribosomes (41); It facilitates the interaction between the ribosomal large subunit and the small subunit in archaebacteria (41). However, in yeast, RPL24 is not essential for cell viability (42). In Arabidopsis, SHORT VALVE 1 (STV1), an RPL24 homolog, is required for the translation initiation of some proteins including ETT and MP, two auxin response factors, involved in gynoecium patterning (43). However, whether it functions outside ribosome is not known.
Here, we report that STV1 is required for miRNA accumulation in Arabidopsis. Lack of STV1 in stv1-1 alters the occupancy of Pol II at MIR promoters and influence MIR transcription. However, STV1 does not interact with Pol II and MIR promoters, indicating that STV1 may indirectly regulate MIR transcription. A portion of STV1 localizes in the nucleus and directly binds pri-miRNAs in vivo. STV1 interacts with the stem-loop flanked by a short 5′ arm within pri-miRNAs. In addition, both the stem-loop and the short 5′ arm are required for STV1 binding. In stv1, the association of pri-miRNAs with HYL1 is reduced. These results suggest that STV1 directly acts in miRNA biogenesis through facilitating the recruitment of pri-miRNA to the processing complex.
Results
STV1 Is Required for Proper miRNA Accumulation.
Besides abnormal vascular patterning and gynoecium structures (43), the stv1 mutants displayed other developmental defects, such as delayed growth, smaller plant size, clustered inflorescence, shorter siliques, and reduced fertility (Fig. S1 A and B). We suspected that the developmental defects of stv1 could result from the impaired translation at global levels, because STV1 is a ribosomal protein. To test this possibility, we compared the polysomes profile between Columbia (Col) and stv1-3 (Salk_045401, in Col genetic background) (Fig. S1C). Both 80S monosomes and polysomes were largely intact in stv1-3, although their relative abundance was slightly different with that in Col (Fig. S1C). This result indicates that like in other organisms, STV1 may not play essential roles in protein translation in Arabidopsis.
Fig. S1.
Phenotypes of stv1-1. (A and B) stv1-1 causes developmental defects. (Scale bar, 2 cm.) (C) Polysome sedimentation of Col and stv1-3. Ribosome profiles were determined by the absorbance (A254 nm) of sucrose density gradient fractionated ribosomes.
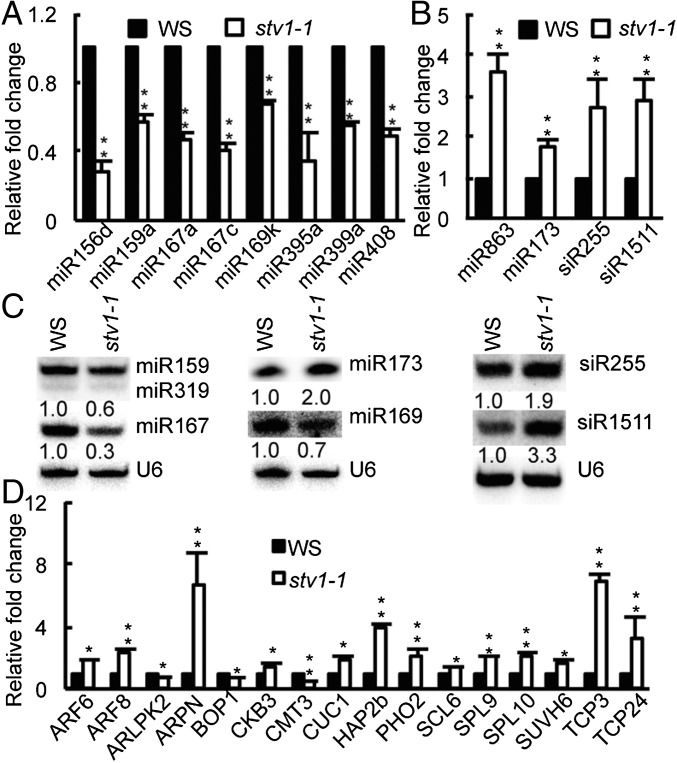
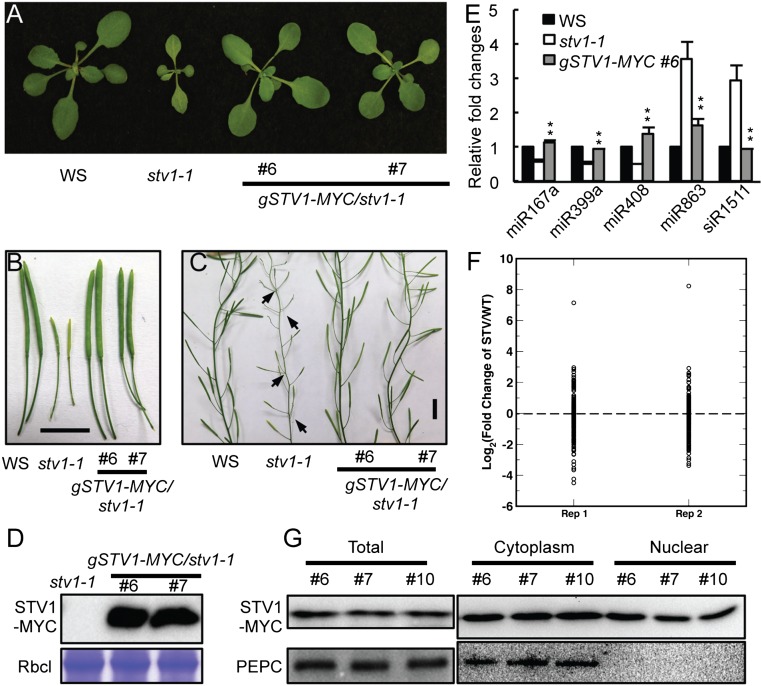
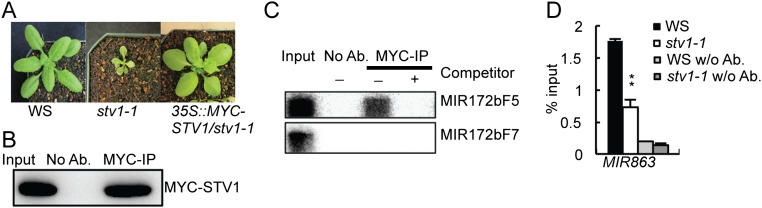
We next tested miRNA accumulation in inflorescences of stv1-1 because impaired miRNA biogenesis is known to cause pleiotropic developmental defects. Indeed, quantitative RT-PCR (qRT-PCR) analyses showed that several examined miRNAs displayed increased or decreased abundance in stv1-1 [Wassilewskija (WS) genetic background] relative to WS (WT) (Fig. 1 A and B). Northern blot analyses further confirmed reduced accumulation of miR167 and miR169 and elevated abundance of miR173 in stv1-1 relative to WS (Fig. 1C). To demonstrate that loss-of-function of STV1 is responsible for the alteration of miRNA accumulation in stv1, we introduced an STV1 genomic fragment under the control of its native promoter (pSTV1::STV1-MYC) into stv1-1. The expression of STV1-MYC rescued the developmental defects of stv1-1 and fully recovered miRNA levels (Fig. S2 A–E). Next, we analyzed the miRNA profile of inflorescences from WS and stv1-1 through deep sequencing. The abundance of most miRNAs was altered in stv1-1 relative to WS in two biological replicates (Fig. S2F and Dataset S1). We further asked whether STV1 could affect the accumulation of transacting siRNAs (ta-siRNAs). We found that siR255 and 1511, two examined ta-siRNAs, were increased in abundance in stv1-1 relative to WS (Fig. 1 B and C). However, the increased levels of siR255 and 1511 are likely caused by elevated abundance of miR173, which triggers the production of siR255 and 1511.
Fig. 1.
stv1-1 influences the accumulation of miRNAs and ta-siRNAs. (A and B) miRNA level in stv1-1 and WS detected by qRT-PCR. miRNA levels in stv1-1 were normalized to those of U6 RNA and compared with WS.Values of WS were set as 1. Error bars indicate SEs (SD) of three replicates (**P < 0.01). (C) The expression levels of small RNAs in WS and stv1-1 detected by Northern blot. U6 RNA was used for the loading control. The relative abundance of small RNAs is shown below the picture and represents the mean of three repeats (*P < 0.05). For miR159/319, the upper band was miR159 and the lower band showed miR319. (D) The expression levels of miRNA target genes in WS and stv1-1 detected by qRT-PCR. The expression levels of miRNA targets were normalized to those of UBQ5 and compared with WS. Error bars indicate SEs (SD) of three replicates (*P < 0.05; **P < 0.01).
Fig. S2.
STV1 affects miRNA accumulation. (A–C) Expression of STV1 complements the developmental defects of stv1-1. (Scale bars, 1 cm.) (D) Detection of the MYC-STV1 protein in stv1-1 harboring pSTV1::STV1-MYC (gSTV1-MYC) by Western blot. The large subunit of Rubisco (RbcL) serves as a protein loading control. (E) Expression of STV1 rescues the defection of miRNAs in stv1-1. miRNA levels in stv1-1 and WS were detected by qRT-PCR, normalized to those of U6 RNA, and compared with WS (WT). Values of WS were set as 1. Error bars indicate SEs (SD) of three replicates. **P < 0.01. (F) miRNA profile in stv1-1 and WT. RNAs extracted from inflorescences were used to prepare small RNA libraries. The miRNA abundance was calculated as reads per million and a log2-transformed ratio of stv1/WT were plotted. Each circle indicates one miRNA. (G) The STV1 protein localizes at the cytoplasm and nucleus. MYC-STV1 and PEPC from nuclear and cytoplasmic fractions were detected with anti-MYC and anti-PEPC antibodies, respectively, and indicated on the right.
We then examined the transcript levels of miRNA targets in stv1-1 and WS using qRT-PCR. The result showed that ARF6/8, ARPN, CKB3, CUC1, HAP2b, PHO2, SCL6, SPL9/10, SUVH6, and TCP3/24, which are the target of miR167, miR408, miR397, miR164, miR168, miR399, miR171, miR156, miR778, and miR319 (reduced levels in stv1-1), respectively, were increased in abundance (Fig. 1D). In contrast, the accumulation of ARLPK2, BOP1, and CMT3, which are the targets of miR863, miR836 and miR823 (elevated levels in stv1-1), was reduced (Fig. 1D). These results suggest that alterations of miRNA abundance may influence the expression levels of some targets.
A Portion of STV1 Localizes in the Nucleus.
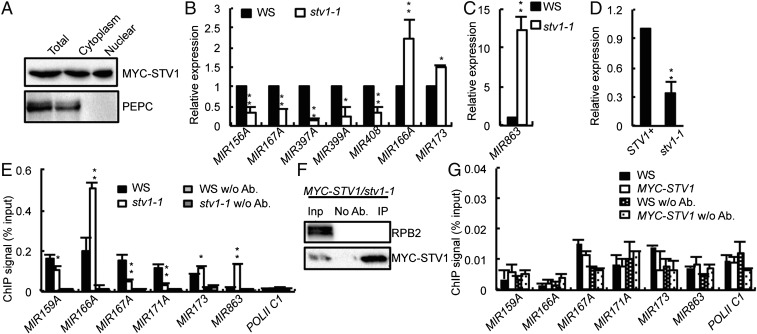
Because plant miRNA biogenesis occurs in the nucleus, STV1 should localize in the nucleus if it had a direct role in miRNA generation. To test this possibility, we expressed GFP-STV1 and STV1-GFP driven by the native STV1 promoter in stv1-1, respectively. However, both transgenes could not complement the defection caused by stv1, which is likely because the GFP protein (∼26 kDa) interfered the function of STV1 (∼19 kDa). Thus, to determine STV1 localization, we prepared proteins from nuclear and cytoplasmic fractions of three independent complementation lines harboring pSTV1::STV1-MYC and examined the presence of STV1-MYC in each fraction by Western blot. As expected, the majority of STV1 localized in the cytoplasm (Fig. 2A and Fig. S2G). However, a portion of STV1 did exist in the nucleus (Fig. 2A and Fig. S2G). The presence of STV1 in the nucleus was unlikely to be caused by the contamination of cytoplasmic proteins, because phosphoenolpyruvate carboxylase (PEPC), a cytoplasmic marker protein, could not be detected in the nuclear fraction (Fig. 2A and Fig. S2G). These results suggest that STV1 may function in the nucleus and participate in miRNA biogenesis.
Fig. 2.
STV1 influences the transcription of genes encoding miRNAs. (A) The STV1 protein localizes at the cytoplasm and nucleus. MYC-STV1 and PEPC from nuclear and cytoplasmic fractions were detected with anti-MYC and anti-PEPC antibodies, respectively, and indicated on the right. (B and C) stv1-1 alters the expression levels of pri-miRNAs. The expression levels of pri-miRNAs in stv1-1 were analyzed by qRT-PCR, normalized to those of those of UBQ5, and compared with WS (values were set as 1). Error bars indicate SEs (SD) of three replicates (*P < 0.05; **P < 0.01). (D) GUS transcript levels in STV1+ and stv1-1 harboring the pMIR167a:GUS transgene detected by qRT-PCR. GUS transcript levels were normalized to those of UBQ5 and compared with WS (values were set as 1). Error bars indicate SEs (SD) of three replicates (**P < 0.01). STV+: STV1/STV1 or STV1/stv1-1. (E) The occupancy of Pol II at MIR promoters in WS and stv1-1 detected by ChIP with anti-RPB2 antibodies. The intergenic region between At2g17470 and At2g17460 (Pol II C1) serves as a negative control for Pol II occupancy. Means and standard derivation of three technical repeats are presented. (**P < 0.01). (F) STV1 does not associate with RPB2 by co-IP. MYC-STV1 was immunoprecipitated with anti-MYC antibodies. MYC-STV1 and RPB2 were detected with anti-MYC and anti-RPB2 antibodies, respectively. (G) STV1 does not associate with MIR promoters detected by a ChIP assay. Pol II C1 was used as a control.
STV1 Indirectly Influences MIR Transcription.
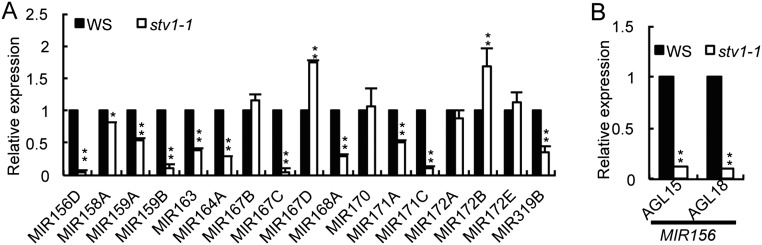
There are at least two possible ways by which STV1 acts in miRNA biogenesis. It may regulate the transcription of pri-miRNAs or participate in pri-miRNA processing. We first evaluated pri-miRNA levels in stv1-1 using qRT-PCR. Among 25 examined pri-miRNAs, 21 pri-miRNAs were either decreased or increased in abundance in stv1-1 compared with those in WS, whereas the remaining 4 were not affected by stv1 (Fig. 2 B and C and Fig. S3A). Because transcription is one of several factors affecting pri-miRNA levels, we examined the effect of STV1 on MIR promoter activity using a GUS reporter gene driven by the MIR167a promoter (pMIR167a::GUS), which has been used to study the functions of mediator complex and PRL1 in miRNA biogenesis (15, 39). We crossed the transgenic line harboring pMIR167a::GUS to stv1-1. In the F2 generation, we obtained the STV1+(STV1/STV1 or STV1/stv1) and stv1-1 genotypes containing the pMIR167a::GUS transgene, respectively. The GUS mRNA level was ∼40% lower in the stv1-1 mutant than that in STV+ background (Fig. 2D), indicating that STV1 might affect miRNA promoter activity. To validate this result, we checked the occupancy of Pol II on MIR promoters using ChIP assay with anti-RNA polermerase II (RPB2, the second largest subunit of Pol II) antibodies. As expected, the ChIP signals of MIR promoters were enriched in Pol II immunoprecipitates relative to “no-antibody” controls in WS (Fig. 2E). Compared with WS, stv1-1 reduced Pol II occupancy at the promoters of MIR159A, MIR167A, and MIR171A, but increased Pol II occupancy at the promoters of MIR166A, MIR173 and MIR863 (Fig. 2E). These results were consistent with reduced levels of pri-miR159a, pri-miR167a, and pri-miR171a and increased levels of pri-miR166a, pri-miR173, and pri-miR863, which support the potential role of STV1 in regulating MIR transcription.
Fig. S3.
STV1 affects the accumulation of pri-miRNAs. (A) stv1-1 alters the expression levels of pri-miRNAs. (B) stv1-1 reduces the expression of AGL15 and AGL18. The expression levels of various RNAs in stv1-1 were analyzed by qRT-PCR, normalized to those of those of UBQ5, and compared with WS (values were set as 1). Error bars indicate SEs (SD) of three replicates (*P < 0.05; **P < 0.01).
Next, we asked if STV1 has a direct role in promoting MIR transcription. First, the association of Pol II with STV1 in the stv1 complementation line was examined using a coimmunoprecipitation (co-IP) assay. However, STV1 did not coprecipitate with RPB2 (Fig. 2F). Next, we tested if STV1 occupied at MIR promoter using ChIP assay. In the stv1 transgenic line harboring pSTV1::STV1-MYC, the occupancy of STV1 in the promoters of MIR159A, MIR66A, MIR167A, MIR171A, MIR173, and MIR863 was not detected (Fig. 2G). These results indicate that STV1 may indirectly regulate MIR transcription.
STV1 Does Not Associate with the DCL1 Complex.
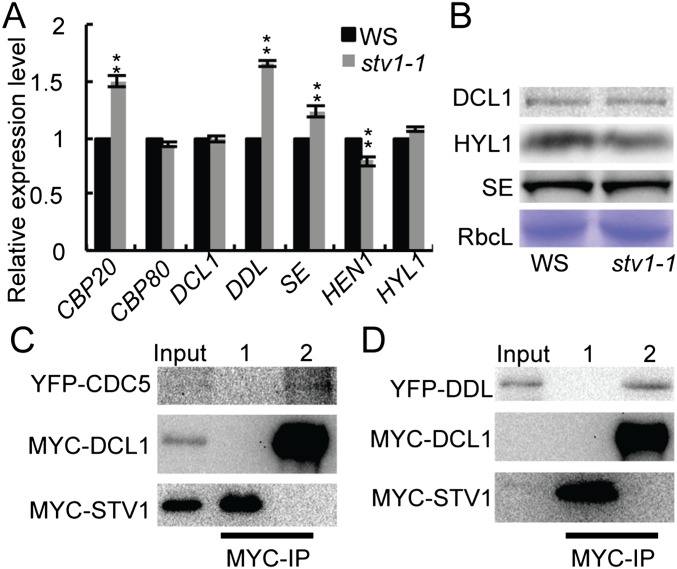
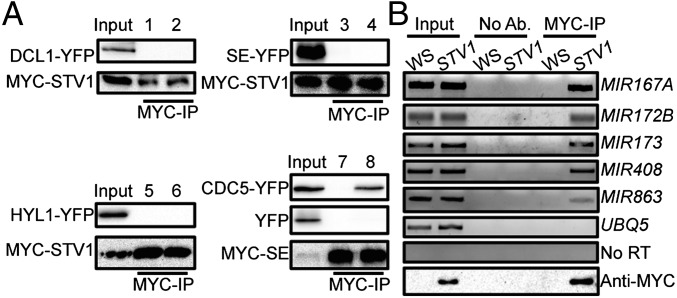
Next we asked if STV1 has a role in pri-miRNA processing. We tested if STV1 affected the expression levels of genes that are involved in pri-miRNA processing using qRT-PCR. stv1-1 did not significantly change the transcript levels of DCL1, HYL1, HEN1, and CPB80, although it slightly increased the expression levels of DDL, SE, and CPB20 (Fig. S4A). Consistent with these results, stv1-1 did not show effect on the protein levels of HYL1, SE, and DCL1 (Fig. S4B). We next examined if STV1 acted as a component of the DCL1 complex to participate in pri-miRNA processing because its homologs in other organisms can function through protein–protein interactions. To test the interaction of STV1 with DCL1, HYL1, SE, DDL, and CDC5, we coexpressed MYC-STV1 with DCL1-YFP, SE-YFP, HYL1-YFP, YFP-DDL, or YFP-CDC5 in Nicotiana benthamiana and performed co-IPs assay using anti-MYC antibodies. STV1 did not interact with DCL1, HYL1, SE, DDL, and CDC5 (Fig. 3A and Fig. S4 C and D), indicating that STV1 may not associate with the DCL1 complex.
Fig. S4.
The function of STV1 in the miRNA pathway. (A) The transcript levels of several genes required for miRNA biogenesis detected by qRT-PCR. The transcript levels of examined genes were normalized to those of UBQ5 and compared with WS (value set as 1). Means of three replicates were shown. **P < 0.01. (B) Protein levels of DCL1, HYL1, and SE in WS and stv1-1 detected by Western blot. The large subunit of Rubisco (RbcL) was stained as a loading control. (C) STV1 does not interact with CDC5. 1, protein extracts containingYFP-CDC5 and MYC-STV1; 2, protein extracts containing YPF-CDC5 and MYC-DCL1. The DCL1-CDC5 interaction is shown as a positive control. (D) STV1 does not interact with DDL. MYC-STV1 was coexpressed with YFP-CDC5 or YFP-DDL in N. benthamiana. 1, protein extracts containing YFP-DDL and MYC-STV1; 2, protein extracts containing YPF-DDL and MYC-DCL1. MYC-STV1 was IPed with anti-MYC antibodies. MYC-STV1 and YFP-tagged proteins were detected with anti-MYC and anti-YFP antibodies, respectively, and indicated on the left. The DCL1–DDL interaction is shown as a positive control.
Fig. 3.
STV1 associates with pri-miRNAs but not the DCL1 complex. (A) STV1 does not interact with DCL1, HYL1, and SE. MYC-STV1 was coexpressed with DCL1-YFP, SE-YFP, or HYL1-YFP in N. benthamiana. After protein extraction, MYC-STV1 was immunoprecipitated with anti-MYC antibodies. MYC-STV1 and YFP-tagged proteins were detected with anti-MYC and anti-YFP antibodies, respectively, and indicated on the left. The interaction of SE and CDC5 is shown as a positive control. Numbers on top of the pictures indicate protein extracts containing: 1, YFP and MYC-STV1; 2, DCL1-YFP and MYC-STV1; 3, YFP and MYC-STV1; 4, SE-YFP and MYC-STV1; 5, YFP and MYC-STV1; 6, HYL1-YFP and MYC-STV1; 7, YFP and MYC-SE (Middle); 8, YFP-CDC5 and MYC-SE (Top). (B) STV1 binds pri-miRNAs in vivo detected by RIP. Inflorescences from WS and stv1-1 harboring MYC-STV1 were used for RIP assay with anti-MYC antibodies. No antibody (No Ab.) was used as a negative control. Ten-percent of IPs and 2% input proteins were used for Western blot. Five percent RNAs were used as input RNA. STV1 on the top of the picture indicates MYC-STV1.
STV1 Binds pri-miRNAs in Vivo.
The facts that STV1 homologs associate with ribosome RNAs (41), and that a portion of STV1 exists in the nucleus, prompted us to test if STV1 binds pri-miRNAs in vivo. RNA immunoprecipitation assay (RIP) was performed on the seedlings of stv1-1 harboring the pSTV1::STV1-MYC transgene. After cross-linking, nuclear isolation, and IP, RT-PCR analyses showed that pri-miR167a, pri-miR172b, pri-miR173, pri-miR408, and pri-miR863 were enriched in the MYC-STV1 immunoprecipitates, but not in the no-antibody control or IPs from the control plants (Fig. 3B). In addition, the control UBIQUITIN 5 (UBQ5) mRNA was not detected in the STV1 complex (Fig. 3B). These results show that STV1 binds pri-miRNAs in vivo.
STV1 Binds a pri-miR172b Fragment Containing the Stem-Loop Flanked by a 39-nt 5′ Arm.
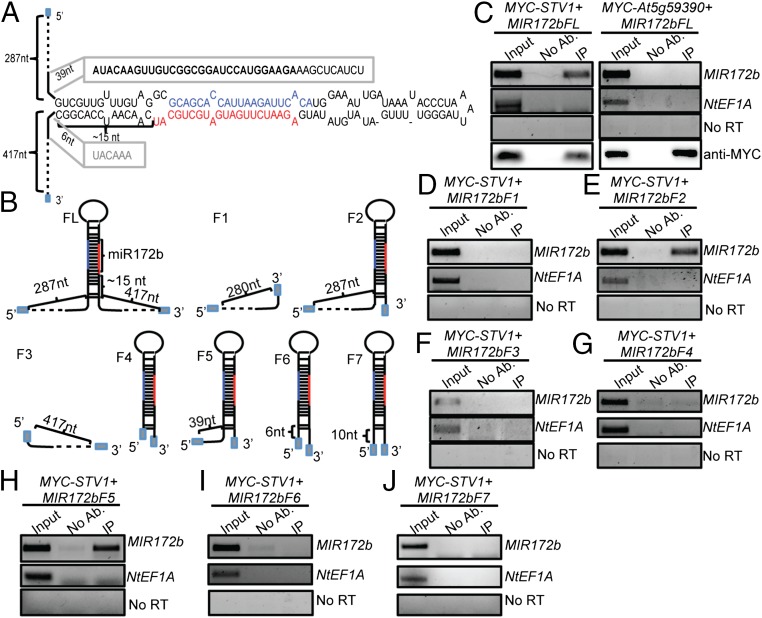
Next, we sought to identify the potential STV1-binding region within pri-miRNAs. Because the recombinant STV1 protein could not be expressed in Escherichia coli, an in vivo RNA-binding assay was used to examine the STV1–pri-miRNA interaction. In this assay, we coexpressed 35S::MYC-STV1, which complements the defects of stv1 (Fig. S5A) with 35S::MIR172b in N. benthamiana and checked the STV1-MIR172b interaction by RIP. Transcription of 35S::MIR172b generated a transcript containing an ∼120-nt upstream vector sequence, MIR172b that composes of the stem-loop flanked by an ∼287-nt upstream arm and a 417-nt downstream arm, and an ∼354-nt downstream vector sequence (MIR172bFL) (Fig. 4 A and B). MIR172bFL, but not NtEF1A that is an endogenous mRNA from N. benthamiana, was enriched in the MYC-STV1 IPs relative to the “no antibody” control (Fig. 4C). As a negative control, we coexpressed MIR172b with an unrelated RNA-binding protein (URP, At5g59390). This protein did not pull down MIR172bFL. These results suggest that this assay could be used to determine the STV1-binding position within MIR172b. We first examined the interaction of STV1 with the 5′ arm (MIR172bF1; ∼287nt), a truncated MIR172b fragment (MIR172bF2) containing a 5′ arm (∼287 nt) and a short 3′ arm (6 nt; MIR172bF2), and the 3′ arm (MIR172bF3; ∼417 nt) (Fig. 4B). STV1 bound MIR172bF2, but not MIR172bF1 and MIR172bF3 (Fig. 4 D–F), indicating that STV1 may bind the stem-loop of MIR172b. To confirm this result, we tested the association of STV1 with the stem-loop of MIR172b with the 6-nt 3′ arm (MIR172bF4) (Fig. 4B). However, STV1 did not bind MIR172bF4 (Fig. 4G). These results suggest that STV1 requires both the 5′ arm and stem-loop to bind pri-miR172b. To further determine sequence features of the 5′ arm required for STV1 binding, we checked the effect of 5′ arm truncation (MIR172bF5-F7) on the interaction of STV1 with MIR172b. STV1 bound MIR172bF5 (containing a 39-nt 5′ arm) (Fig. 4 B and H), but not MIR172bF6 (containing a 6-nt 5′ arm) (Fig. 4 B and I) and MIR172bF7 (containing a 10-nt 5′ arm) (Fig. 4 B and J). To rule out the effect of the vector sequence on STV1-binding, we in vitro-transcribed MIR172bF5 and MIR172bF7 and tested their interactions with IPed MYC-STV1 using an RNA pull-down assay (7). MYC-STV1 binds MIR172bF5, but not MIR172bF7 (Fig. S5 B and C). These results show that STV1 bind the stem-loop of pri-miR172b with the 39-nt 5′ arm and the sequence between 39 nt and 10 nt upstream of the stem loop is required for STV1-binding.
Fig. S5.
STV1 interacts with pri-miRNAs. (A) Four-week-old seedlings of WS, stv1-1, and stv1-1 harboring 35S::MYC-STV1. (Scale bar, 1 cm.) (B) IP of MYC-STV1 detected by Western blot. (C) STV1 binds pri-miR172b in vitro. MIR172bF5 and MIR172bF7 were produced by in vitro transcription. NoAb: No antibody control. RNA pull down was performed using IPed MYC-STV1. (D) The association of HYL1 with pri-miR863 is reduced in stv1-1. Pri-miR863 associated with HYL1 was examined by qRT-PCR and normalized to the input. UBQ5 serves as a negative control. **P < 0.01.
Fig. 4.
The interaction of STV1 with MIR172b. (A) Diagram of the stem-loop and flanking sequences of MIR172b. (B) Diagrams of various MIR172b constructs used for the STV1-binding assay. Gray box indicate the sequences from vectors. miR172 is shown in red; miR172b* is shown in dark blue. (C–J) The interaction of STV1 with full-length and truncated MIR172b RNAs. MYC-STV1 and MIR172b were transiently coexpressed in N. benthamiana. IP was performed with anti-MYC antibodies. No Ab: IP without antibody. No RT was performed with primers recognizing MIR172b. Input RNA was 5%.
stv1-1 Reduces the Interaction of pri-miRNAs with HYL1.
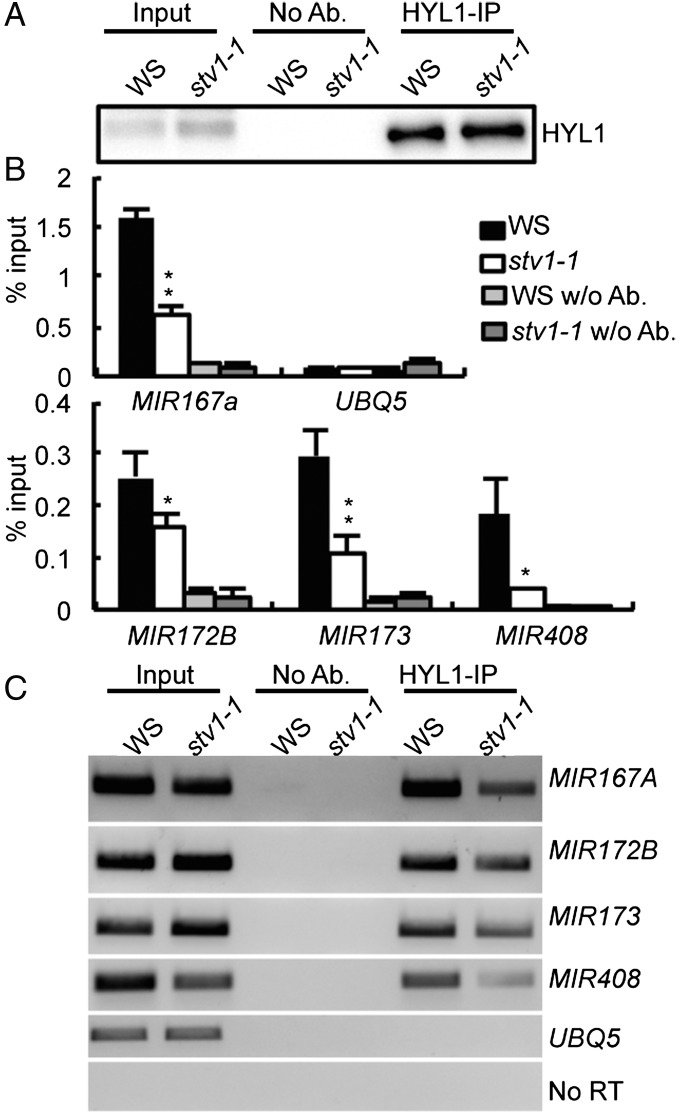
In yeast, RPL24 acts in the 60S subunit-joining step during translation initiation (42). By analog, we suspected that STV1-binding might enhance the recruitment of pri-miRNAs to the DCL1 complex. If so, we would expect a reduced association of HYL1 with pri-miRNAs in stv1. We examined the HYL1–pri-miRNA interaction in WS and stv1-1 by RIP using antibodies against HYL1. The amount of HYL1 protein in the IPs from stv1-1 was similar to that in WS (Fig. 5A). RT-PCR and qRT-PCR analyses showed that the levels of several examined pri-miRNAs (MIR167a, MIR172b, MIR173, MIR408, and MIR863) in the HYL1 IPs were lower in stv1-1 than those in WS (Fig. 5 B and C and Fig. S5D), demonstrating that STV1 contributes to the loading of pri-miRNAs to the DCL1 complex.
Fig. 5.
STV1 affects the interaction between HYL1 and pri-miRNAs. (A) Detection of HYL1. IP was performed with the anti-HYL1 antibodies. Ten percent of IPs and 2% input proteins were detected by Western blot. (B and C) The association of HYL1 with pri-miRNAs is reduced in stv1-1. Pri-miRNAs associated with HYL1 were examined by qRT-PCR and normalized to the input. UBQ5 serves as a negative control. *P < 0.05; **P < 0.01.
Discussion
In conclusion, we show here that STV1, a ribosomal protein, is required for miRNA biogenesis. This finding is supported by the association of STV1 with pri-miRNAs and the altered accumulation of miRNAs and pri-miRNAs in stv1. Altered miRNA accumulation may partially contribute to the pleiotropic developmental defects of stv1, given the essential roles of miRNAs in regulating development. However, STV1 must have other functions, as its effect on miRNA accumulation is less than DCL1, HYL1, and SE. It is still possible that STV1 influences the translation of some proteins. In fact, STV1 modulates the translation initiation of ETTIN, an auxin response factor (43), and influences the activity of the cauliflower mosaic virus transactivator, TAV, which controls the translation reinitiation of polycistronic RNAs encoded by virus (44). Furthermore, some ribosomal proteins are involved in the formation of the preribosome in the nucleus (45). It is reasonable to speculate that STV1 may participate in the process.
In miRNA biogenesis, STV1 may facilitate the interaction of pri-miRNAs with the processing complex, which is supported by the association of STV1 with pri-miRNAs and the reduced amount of pri-miRNAs in the HYL1 complex. Because STV1 does not interact with the DCL1 complex, we propose that STV1 may bind pri-miRNAs before the loading of the DCL1 complex to pri-miRNAs. STV1 also affects the transcription of many pri-miRNAs, based on the observations that the occupancy of Pol II at the MIR promoters and the levels of many pri-miRNAs are altered in stv1. However, this effect maybe indirect, as STV1 itself does not interact with Pol II and MIR promoters. How does STV1 indirectly regulate activities of these MIR promoters? We suspect that STV1 may indirectly impact the expression of transcription factors of MIRs. For example, stv1 reduces the transcript levels of AGL15 and AGL18 (Fig. S3B), which encode positive transcription factors of MIR156 (46), but increases the abundance of SPL9 and 10 (Fig. 2B), which are targets of miR156 and promote the expression of MIR172B (47, 48). These results are consistent with decreased pri-miR156 levels and elevated pri-miR172b abundance in stv1 (Fig. 2B and Fig. S3A). How STV1 affects MIR transcription factors is not clear, but may partially attribute to its effect on miRNA levels. Additionally, STV1 may influence the translation of some proteins that may play roles in regulating the transcription of MIR transcription factors. STV1 may also affect pri-miRNA stability, which clearly needs further investigation.
The assembly of RNA–protein complex often involves the conformation changes of RNAs (41). Many ribosomal proteins bind rRNAs to alter their structures for ribosome assembly (41). Thus, STV1 may alter the structure of pri-miRNAs to enable them more accessible for the processing complex. STV1 binds the stem-loop with a short 5′ arm with MIR172b, and this binding requires the presence of both stem-loop and the 39-nt 5′ arm. Because the presence of 120-nt vector sequence in various forms of MIR172b transcripts, it is unlikely the length of 5′ arm is required for STV1 binding. Rather, the sequence or structure features within the 39-nt 5′ arm sequence together with the stem-loop enables STV1 binding.
In summary, we discovered a previously unknown extraribosomal activity of STV1. STV1 binds pri-miRNAs and promotes miRNA biogenesis through facilitating the interaction of pri-miRNAs with the processing complex. Besides this function, STV1 may indirectly modulate pri-miRNA accumulation through negatively or positively influencing the levels of transcription factors that regulate MIR transcription. Thus, the accumulation of miRNAs in stv1 reflects the effect of STV1 on pri-miRNA loading and transcription. Through these combined effects, STV1 may be able to fine-tune the levels of miRNAs to ensure normal development and physiology of Arabidopsis. STV1 is a conserved protein in eukaryotes. It is possible that STV1 homologs act in miRNA biogenesis in other organisms. Indeed, lack of STV1 homolog in mice causes lethality, consistent with the crucial role of miRNA in development (49).
Materials and Methods
Plant materials, plasmid construction, polysome profile analysis, deep sequencing, and RNA analysis are described in SI Materials and Methods. Data generated from deep sequencing of small RNA libraries were deposited in the National Center for Biotechnology Information (accession no. GSE91013).
Nuclear-Cytoplasmic Fractionation.
Inflorescences of stv1-1 harboring the pSTV1::STV1-MYC transgene were used to perform nuclear-cytopasmic fractionation according to Park et al. (50). The isolated nuclear pellets were further sonicated in the nuclear lysis buffer for 1 min on ice, as described previously (51). Following this step, the supernatants were collected as nuclear extracts through centrifugation (51). Anti-PEPC antibody was used to detect the cytoplasmic fraction.
ChIP and RIP Analyses.
Inflorescences of stv1-1 harboring pSTV1:STV1-MYC were used for RIP analyses. RIP assay was performed as described previously (7). After cross-linking, the nuclear fractions were extracted, and then incubated with anti-MYC antibodies overnight at 4 °C. STV1-associate RNAs were then extracted and analyzed with RT-PCR. To analyze the interaction of STV1 with various MIR172b fragments, 35S::MYC-STV1 and MIR172b constructs were transiently coexpressed in N. benthaminana leaves. Three grams of leaves were used for RIP analysis.
ChIP assay was performed as previously described (17). Three biological replicates were performed. Anti-RPB2 and anti-MYC antibodies were used for IP. qPCR was performed using the primers listed in Table S1.
Table S1.
DNA oligos used in this study
| Name | Sequence | Application |
| Primers for constructs | ||
| pSTV1-1F | CACC CTTGCGTGGCATCATCCCAAGA | STV1 genomic amplication |
| STV1CDS-1R | GCGTTTGCCACCACCACC | STV1 genomic amplication |
| STV1CDS-1F | ATGGTTCTCAAGACGGAGCTT | cDNA amplification |
| delMIR172B-1F | CACC ATATAACAAACATCGTATTCT | MIR172b-F1, F2 |
| delMIR172B-2F | CACC ATACAAGTTGTCGGCGGATCCA | MIR172b-F5 |
| delMIR172B-3F | CACC GAGAAAGAAACTTGAAGATAT | MIR172b-F3 |
| delMIR172B-6F | CACC GTCGTTGTTTGTAGGCGCAG | MIR172b-F4 |
| delMIR172B-7F | CACC TCATCTGTCGTTGTTTGTAGGC | MIR172b-F6 |
| delMIR172B-8F | CACC AAGCTCATCTGTCGTTGTTTGTA | MIR172b-F7 |
| delMIR172B-1R | TGGATCCGCCGACAACTTGTAT | MIR172b-F1 |
| delMIR172B-2R | AAGCTTTAGGTATTTGTAGCC | MIR172b-F2, F4, F5, F6, F7 |
| delMIR172B-3R | ACCAATAGAAGTGTACTTACTC | MIR172b-F3 |
| Primers for stem-loop PCR | ||
| miR156d- stemloop RT | GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCAC CAGAGCCA AC GTTATG | Stem-loop PCR |
| miR156d-Forward | CGGCGG GCTCACTCTCTTTTT | Stem-loop PCR |
| miR159a stemloop RT | GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACTAGAGC | Stem-loop PCR |
| miR159a stemloop forward | CGGCGGTTTGGATTGAAGGGA | Stem-loop PCR |
| miR167a stem-loop RT | GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCAC CAGAGCCA ACTAGATC | Stem-loop PCR |
| miR167a Forward | GGCGTC TGAAGCTGCCAGCAT | Stem-loop PCR |
| miR167c-stemloop RT | GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCAC CAGAGCCA AC CAAGAT | Stem-loop PCR |
| miR167c-Forward | GTTGGC TAAGCTGCCAGCATG | Stem-loop PCR |
| miR169k stem-loop RT | GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCAC CAGAGCCA AC CAGGCA | Stem-loop PCR |
| miR169k forward | GGCGTC TAGCCAAGGATGACT | Stem-loop PCR |
| miR173 stem-loop RT | GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCAC CAGAGCCA ACGTGATT | Stem-loop PCR |
| miR173 forward | GTTGGC TTCGCTTGCAGAGAG | Stem-loop PCR |
| miR395a stem-loop RT | GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCAC CAGAGCCA ACGACTTC | Stem-loop PCR |
| miR395a Forward | TCGCGT CTCAAGGGGTTTGTG | Stem-loop PCR |
| miR399a stem-loop RT | GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCAC CAGAGCCA ACCAGGGC | Stem-loop PCR |
| miR399a forward | CGGCGG TGCCAAAGGAGATTT | Stem-loop PCR |
| miR408 stem-loop RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGCCAGG | Stem-loop PCR |
| miR408 Forward | GACGTCATGCACTGCCTCTTC | Stem-loop PCR |
| miR863 stem-loop RT | GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCAC CAGAGCCA AC | Stem-loop PCR |
| miR863 Forward | CGGCGG TTATGTGTTGTTGAT | Stem-loop PCR |
| siR255 stem-loop RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTACGCT | Stem-loop PCR |
| siR255 Forward | CCGTCGTTCTAAGTCCAACAT | Stem-loop PCR |
| siR1511 stem-loop RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAAGTAT | Stem-loop PCR |
| siR1511 Forward | GGTCGTCCAAGCGAATGATG | Stem-loop PCR |
| U6 stem-loop RT | GTGCAGGGTCCGAGGTTTTGGACCATTTCTCGAT | Stem-loop PCR |
| U6 Forward | GGAACGATACAGAGAAGATTAGCA | Stem-loop PCR |
| Universal | GTGCAGGGTCCGAGGT | Stem-loop PCR |
| Probe sequence for Northern blot | ||
| miR319/159 | GGG+AGC+TCC+CTT+CAG+TCC+AA | Northern blot |
| miR167 | T+AGA+TCA+TGT+TGG+CAG+TTT+CA | Northern blot |
| miR169 | T+CGG+CAA+GTC+ATC+CTT+GGC+TG | Northern blot |
| miR173 | G+TGA+TTT+CTC+TCT+CGA+AGC+GAA | Northern blot |
| siR255 | T+ACG+CTA+TGT+TGG+ACT+TAG+AA | Northern blot |
| siR1511 | AAGTATCATCATTCGCTTGGA | Northern blot |
| U6 | TCATCCTTGCGCAGGGGCCA | Northern blot |
| Primers for RT-PCR | ||
| qAGL15-1F | TTGAAGAATCACGCCTCAAGGAAC | RT-PCR |
| qAGL15-1R | AACCTGTCTACGCAAGGTCTCG | RT-PCR |
| qAGL18-1F | TGACGCCGAGGTTGCTCTTATC | RT-PCR |
| qAGL18-1R | TTGCTCCATACAGACGCTGGAG | RT-PCR |
| qAGL15-1F | TTGAAGAATCACGCCTCAAGGAAC | RT-PCR |
| qARF6-1F | GGGGTCCTTTGGTAGGTCGC | RT-PCR |
| qARF6-1R | CGGCCAGGGGTCATCACCAA | RT-PCR |
| qARF8-1F | CAGGGTCGGTCGGGCGATCA | RT-PCR |
| qARF8-1R | CCCCTGCTCCCCCATCTTTT | RT-PCR |
| qARLPK2-F | TGAGGAATGCTGTTCTTGATC | RT-PCR |
| qARLPK2-R | CTAAAATTTCCCAATCTCTTCAA | RT-PCR |
| qARPN-1F | AAGGCAAACACTTTAGAG | RT-PCR |
| qARPN-1R | GTTGTAACTTCCGCTATC | RT-PCR |
| qBOP1-1F | CTCTTTCCAGGCTTCCAGTT | RT-PCR |
| qBOP1-1R | AGTCCCAAGTTCAGTGTTCAG | RT-PCR |
| qCKB3-1F | ATGTACAAGGAACGTAGTGG | RT-PCR |
| qCKB3-1R | CTAGATGTGGTGGTGGAAGT | RT-PCR |
| qCMT3-1F | ATGCTGAAGATGGCTAAG | RT-PCR |
| qCMT3-1R | CATTCCTCACTTGGTAATTC | RT-PCR |
| CUC1qF | CGCCTTGACGGCAAATTCTCTTAC | RT-PCR |
| CUC1qR | GATGATCGGAGCAATTGCAGAACC | RT-PCR |
| HAP2b-qF | TGCTGCAATTTCAAAACCTG | RT-PCR |
| HAP2b-qR | TCACCTCACGGCGAAGTTAC | RT-PCR |
| qSPL9-F | CAAGGTTCAGTTGGTGGAGGA | RT-PCR |
| qSPL9-R | TGAAGAAGCTCGCCATGTATTG | RT-PCR |
| qSUVH6-1F | CGTTAGCAACCAGCATAG | RT-PCR |
| qSUVH6-1R | CACAAGATATAATCCGTCGTAA | RT-PCR |
| TCP3-QF | CATCCAGTTTATAGCCAAA | RT-PCR |
| TCP3-QR | ATGGCGAGAATCGGATGAA | RT-PCR |
| TCP10-QF | GTTTCTTGGTGGCCAACAA | RT-PCR |
| TCP10-QR | GAGGTGTGAGTTTGGAGGA | RT-PCR |
| qPHO2-1F | GTGAAGGACCATTTTACGCACC | RT-PCR |
| qPHO2-1R | CCATATAAGCCTTGCACGCAG | RT-PCR |
| DCL1 qF | CGTTGTTATGCGTTTCGACCTTGC | RT-PCR |
| DCL1 qR | AACGCTGCGTGAGATACATTTCCTC | RT-PCR |
| HYL1 qF | TTGCCTGGATTCTTCAATCGTAAGG | RT-PCR |
| HYL1 qR | TAGGTTCTTGCATAATCCCGTTTCG | RT-PCR |
| SE qF | CCACCGCCTCGTAGGGATTACA | RT-PCR |
| SE qR | CCACCATGGTCATACCCAAATCTTC | RT-PCR |
| DDLqF | ATGAGCCCCCAGAGGCTAGAAAAC | RT-PCR |
| DDLqR | CTGCAAGATGGGTGATCCGTAGGAA | RT-PCR |
| CBP20qF | ACCGGCCTATTCGTGTGGATTTTG | RT-PCR |
| CBP20qR | TGCCTTTGTGCTTCGAGTTCCTTC | RT-PCR |
| CBP80qF | TCTGGCAACTGCAACAGTATCCGTA | RT-PCR |
| CBP80qR | GGCAGCAGATGATAGCAATGTTTCG | RT-PCR |
| HEN1 qF | TTAGGATGACACCCCCTGATGCTG | RT-PCR |
| HEN1 qR | AAAAGCCGCCTCCATTCGTTCTTC | RT-PCR |
| MIR159a-F | TCAGGAGCTTTAACTTGCCCTTT | RT-PCR |
| MIR159a-R | CACGCTAAACATTGCTTCGGAAT | RT-PCR |
| pri-miR166a-F | GACTCTGGCTCGCTCTATTCA | RT-PCR |
| pri-miR166a-R | TGGTCCGAAGACGCTAAAAC | RT-PCR |
| MIR167a qF | TGTTGTGTTTCATGACGATGG | RT-PCR/RIP |
| MIR167a qR | AGCTCACAAAATCAGACTGAAGA | RT-PCR/RIP |
| pri-miR168a-QF | AGTAGAGTCTCACCATCGGGCT | RT-PCR |
| pri-miR168a-QR | TTACACCTCGAGGATCCGATT | RT-PCR |
| MIR172b-qF | GTAGGCGCAGCACCATTAAG | RT-PCR/RIP |
| MIR172b-qR | TTTGTAGCCGTCGATTGTTG | RT-PCR/RIP |
| MIR173-qF | CTTCTTCTCACAAATAAACCCA | RT-PCR/RIP |
| MIR173-qR | AAGATCTCTAACATTAAATCAT | RT-PCR/RIP |
| MIR397A-qF | GCAGCGTTGATGTAATTTCG | RT-PCR |
| MIR397A-qR | GCATACCTGTTTAAGTGTTCCT | RT-PCR |
| MIR399A-qF | CCGTGGAATATGCTTCTT | RT-PCR |
| MIR399A-qR | TTGGCAGATAACTCACTAAT | RT-PCR |
| MIR408-qF | GTAAGTAGCCACCAGTAA | RT-PCR/RIP |
| MIR408-qR | ATATCTGTTGCCATCCTT | RT-PCR/RIP |
| MIR863-qF | CTGACCAGAAGGAGTTAA | RT-PCR/RIP |
| MIR863-qR | GAGATCAACAAGACATAAGAA | RT-PCR/RIP |
| GUS-F | CGATGTCACTCCGTATGTTATTG | RT-PCR |
| GUS-R | CAGTTCTTTCGGCTTGTTGC | RT-PCR |
| UBQ5-N | GGTGCTAAGAAGAGGAAGAAT | RT-PCR |
| UBQ5-C | CTCCTTCTTTCTGGTAAACGT | RT-PCR |
| Primers for ChIP-PCR | ||
| Pol II-C1-F | AGTTCAATGGAGAGATGTCGAAATATG | ChIP-PCR |
| Pol II-C1-R | AAGAGGAAAAGAAAGAGATGGAGAGA | ChIP-PCR |
| cMIR166a-F | TGGCTCTCTCCACTACTCAA | ChIP-PCR |
| cMIR166a-R | GACAACAGTCCCCTCAAAA | ChIP-PCR |
| cMIR167a-F | CGACCCTTAAACTCTCCATAA | ChIP-PCR |
| cMIR167a-R | ACTTCACCGTAGCAGATCAA | ChIP-PCR |
| cMIR171a-F | TGCTTTGGTAGTAGATGAGGTT | ChIP-PCR |
| cMIR171a-R | CGTGTGTGGTCAGGTAAGAT | ChIP-PCR |
| cMIR863-F | GATGGGGTCATCAAGTCAGCAT | ChIP-PCR |
| cMIR863-R | GGGTTGGGCTGGTATGCTTCTT | ChIP-PCR |
| Primers for in vitro RNA binding | ||
| T7miR172F5-F | TAATACGACTCACTATAGGG ATACAAGTTGTCGGCGGATCCA | RNA binding |
| T7miR172F7-F | TAATACGACTCACTATAGGG AAGCTCATCTGTCGTTGTTTGTA | RNA binding |
SI Materials and Methods
Plant Materials and Growth Condition.
The stv1-1 (CS6957) mutant is in the Wassilewskija (WS) genetic background and stv1-3 (Salk_045401) is in Columbia (Col) background. The two mutants were obtained from the Arabidopsis Biological Resources Center. The transgenic line harboring pMIR167a::GUS was crossed to stv1-1. In the F2 generation, STV1+ (STV1/STV1 and STV1/stv1-1) and stv1-1 were isolated through PCR genotyping.
Plasmid Construction.
An ∼3.1-kb DNA fragment covering the promoter and coding region of STV1 was PCR-amplified and cloned into pEarleyGate303 binary vector to generate the pSTV1::STV1-MYC plasmid. The STV1 full-length cDNA was generated through RT-PCR and cloned pEarleyGate203 binary vector to generate the 35S::MYC-STV1 plasmid. To construct MIR172b expression vectors, a series of MIR172b derivatives were PCR amplified, cloned into the pENTRY/d-TOPO vector, and subsequently cloned into the binary vector pMDC32. The primers are listed in Table S1.
Polysome Profile Analysis.
Nine-day-old seedlings of Col and stv1-3 were used for polysome analysis. Polysomes were isolated from aerial tissues as described previously (52). Polysomes were sedimented through 4.5 mL 15–60% (wt/vol) sucrose gradients containing 400 mM Tris pH 8.4, 200 mM KCl, 100 mM MgCl2, 50 μg/mL cycloheximide, and 50 μg/mL chloramphenicol. Gradients were then analyzed using a BR 188 Density Gradient Fractionation System (Brandel) to generate continuous A254-nm profiles.
Small RNA Sequencing.
Small RNA libraries were prepared using total RNAs extracted from inflorescences following standard protocol. After sequencing, miRNAs are analyzed according to Ren et al. (7). After removing reads aligned to t/r/sn/snoRNA, the total numbers of perfectly aligned reads were used for normalization (53). miRNA abundance was compared by using EdgeR with trimmed mean of M values normalization method (54). The dataset was deposited into the National Center for Biotechnology Information Gene Expression Omnibus (accession no. GSE91013).
RNA Analyses.
For qRT-PCR analyses of pri-miRNAs, miRNA targets and genes required for miRNA biogenesis, RNA was reverse-transcribed using M-MLV (Promega) and an oligo (dT)18 primer to generate cDNA. qPCR was then performed using an SYBR green kit (Bio-Rad). Detection of small RNAs by Northern blot was performed according to (7). qRT-PCR analyses of miRNAs were performed as described previously (55). Probes and primers used for RNA analyses are listed in Table S1.
Co-IP Analyses.
Co-IP was performed as described previously (7). MYC-STV1 was coexpressed with DCL1-YFP, SE-YFP, or HYL1-YFP in Nicotiana benthamiana leaves. Total protein extracts were incubated with anti-MYC antibodies coupled to protein G agarose beads overnight at 4 °C. After washing five times, the precipitates subjected to SDS/PAGE and probed with anti-GFP or anti-MYC antibodies.
RNA Pull-Down Assay.
MYC-STV1 was in N. benthamiana. After protein extraction, MYC-STV1 was immunoprecipitated with anti-MYC antibodies coupled to protein G agarose beads. Before RNA pull-down, the beads were treated with nuclease to remove RNAs and DNAs. MIR172bF5 and MIR172bF7 were produced by in vitro transcription. RNA pull-down was performed as according to Ren et al. (7).
Supplementary Material
Acknowledgments
This work was supported by Nebraska Soybean Board Award 16R-05-3/3 #1706 (to B.Y.); National Science Foundation Award OIA-1557417 (to B.Y.); and National Natural Science Foundation of China Award 31471221 (to G.R.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE91013).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1613069114/-/DCSupplemental.
References
- 1.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Chen X. MicroRNA biogenesis and function in plants. FEBS Lett. 2005;579(26):5923–5931. doi: 10.1016/j.febslet.2005.07.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136(4):669–687. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 4.Baulcombe D. RNA silencing in plants. Nature. 2004;431(7006):356–363. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- 5.Dong Z, Han MH, Fedoroff N. The RNA-binding proteins HYL1 and SE promote accurate in vitro processing of pri-miRNA by DCL1. Proc Natl Acad Sci USA. 2008;105(29):9970–9975. doi: 10.1073/pnas.0803356105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fang Y, Spector DL. Identification of nuclear dicing bodies containing proteins for microRNA biogenesis in living Arabidopsis plants. Curr Biol. 2007;17(9):818–823. doi: 10.1016/j.cub.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ren G, et al. Regulation of miRNA abundance by RNA binding protein TOUGH in Arabidopsis. Proc Natl Acad Sci USA. 2012;109(31):12817–12821. doi: 10.1073/pnas.1204915109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rogers K, Chen X. Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell. 2013;25(7):2383–2399. doi: 10.1105/tpc.113.113159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xie M, Zhang S, Yu B. microRNA biogenesis, degradation and activity in plants. Cell Mol Life Sci. 2015;72(1):87–99. doi: 10.1007/s00018-014-1728-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vaucheret H. Plant ARGONAUTES. Trends Plant Sci. 2008;13(7):350–358. doi: 10.1016/j.tplants.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 11.Baumberger N, Baulcombe DC. Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci USA. 2005;102(33):11928–11933. doi: 10.1073/pnas.0505461102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith MR, et al. Cyclophilin 40 is required for microRNA activity in Arabidopsis. Proc Natl Acad Sci USA. 2009;106(13):5424–5429. doi: 10.1073/pnas.0812729106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Earley KW, Poethig RS. Binding of the cyclophilin 40 ortholog SQUINT to Hsp90 protein is required for SQUINT function in Arabidopsis. J Biol Chem. 2011;286(44):38184–38189. doi: 10.1074/jbc.M111.290130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang S, Liu Y, Yu B. New insights into pri-miRNA processing and accumulation in plants. Wiley Interdiscip Rev RNA. 2015;6(5):533–545. doi: 10.1002/wrna.1292. [DOI] [PubMed] [Google Scholar]
- 15.Kim YJ, et al. The role of Mediator in small and long noncoding RNA production in Arabidopsis thaliana. EMBO J. 2011;30(5):814–822. doi: 10.1038/emboj.2011.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang L, et al. NOT2 proteins promote polymerase II-dependent transcription and interact with multiple MicroRNA biogenesis factors in Arabidopsis. Plant Cell. 2013;25(2):715–727. doi: 10.1105/tpc.112.105882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang S, Xie M, Ren G, Yu B. CDC5, a DNA binding protein, positively regulates posttranscriptional processing and/or transcription of primary microRNA transcripts. Proc Natl Acad Sci USA. 2013;110(43):17588–17593. doi: 10.1073/pnas.1310644110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poethig RS. Small RNAs and developmental timing in plants. Curr Opin Genet Dev. 2009;19(4):374–378. doi: 10.1016/j.gde.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fouracre JP, Poethig RS. The role of small RNAs in vegetative shoot development. Curr Opin Plant Biol. 2016;29:64–72. doi: 10.1016/j.pbi.2015.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu B, et al. The FHA domain proteins DAWDLE in Arabidopsis and SNIP1 in humans act in small RNA biogenesis. Proc Natl Acad Sci USA. 2008;105(29):10073–10078. doi: 10.1073/pnas.0804218105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gregory BD, et al. A link between RNA metabolism and silencing affecting Arabidopsis development. Dev Cell. 2008;14(6):854–866. doi: 10.1016/j.devcel.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 22.Laubinger S, et al. Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2008;105(25):8795–8800. doi: 10.1073/pnas.0802493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu X, et al. A role for the RNA-binding protein MOS2 in microRNA maturation in Arabidopsis. Cell Res. 2013;23(5):645–657. doi: 10.1038/cr.2013.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ben Chaabane S, et al. STA1, an Arabidopsis pre-mRNA processing factor 6 homolog, is a new player involved in miRNA biogenesis. Nucleic Acids Res. 2013;41(3):1984–1997. doi: 10.1093/nar/gks1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhan X, et al. Arabidopsis proline-rich protein important for development and abiotic stress tolerance is involved in microRNA biogenesis. Proc Natl Acad Sci USA. 2012;109(44):18198–18203. doi: 10.1073/pnas.1216199109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Köster T, et al. Regulation of pri-miRNA processing by the hnRNP-like protein AtGRP7 in Arabidopsis. Nucleic Acids Res. 2014;42(15):9925–9936. doi: 10.1093/nar/gku716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Speth C, Willing EM, Rausch S, Schneeberger K, Laubinger S. RACK1 scaffold proteins influence miRNA abundance in Arabidopsis. Plant J. 2013;(3):433–445. doi: 10.1111/tpj.12308. [DOI] [PubMed] [Google Scholar]
- 28.Chen T, Cui P, Xiong L. The RNA-binding protein HOS5 and serine/arginine-rich proteins RS40 and RS41 participate in miRNA biogenesis in Arabidopsis. Nucleic Acids Res. 2015;43(17):8283–8298. doi: 10.1093/nar/gkv751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karlsson P, et al. KH domain protein RCF3 is a tissue-biased regulator of the plant miRNA biogenesis cofactor HYL1. Proc Natl Acad Sci USA. 2015;112(45):14096–14101. doi: 10.1073/pnas.1512865112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raghuram B, Sheikh AH, Rustagi Y, Sinha AK. MicroRNA biogenesis factor DRB1 is a phosphorylation target of mitogen activated protein kinase MPK3 in both rice and Arabidopsis. FEBS J. 2015;282(3):521–536. doi: 10.1111/febs.13159. [DOI] [PubMed] [Google Scholar]
- 31.Manavella PA, et al. Fast-forward genetics identifies plant CPL phosphatases as regulators of miRNA processing factor HYL1. Cell. 2012;151(4):859–870. doi: 10.1016/j.cell.2012.09.039. [DOI] [PubMed] [Google Scholar]
- 32.Kim W, et al. Histone acetyltransferase GCN5 interferes with the miRNA pathway in Arabidopsis. Cell Res. 2009;19(7):899–909. doi: 10.1038/cr.2009.59. [DOI] [PubMed] [Google Scholar]
- 33.Cho SK, Ben Chaabane S, Shah P, Poulsen CP, Yang SW. COP1 E3 ligase protects HYL1 to retain microRNA biogenesis. Nat Commun. 2014;5:5867. doi: 10.1038/ncomms6867. [DOI] [PubMed] [Google Scholar]
- 34.Fang X, Shi Y, Liu X, Chen Z, Qi Y. CMA33/XCT regulates small RNA production through modulating the transcription of dicer-like genes in Arabidopsis. Mol Plant. 2015;8(8):1227–1236. doi: 10.1016/j.molp.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 35.Mateos JL, Bologna NG, Chorostecki U, Palatnik JF. Identification of microRNA processing determinants by random mutagenesis of Arabidopsis MIR172a precursor. Curr Biol. 2010;20(1):49–54. doi: 10.1016/j.cub.2009.10.072. [DOI] [PubMed] [Google Scholar]
- 36.Song L, Axtell MJ, Fedoroff NV. RNA secondary structural determinants of miRNA precursor processing in Arabidopsis. Curr Biol. 2010;20(1):37–41. doi: 10.1016/j.cub.2009.10.076. [DOI] [PubMed] [Google Scholar]
- 37.Werner S, Wollmann H, Schneeberger K, Weigel D. Structure determinants for accurate processing of miR172a in Arabidopsis thaliana. Curr Biol. 2010;20(1):42–48. doi: 10.1016/j.cub.2009.10.073. [DOI] [PubMed] [Google Scholar]
- 38.Bologna NG, et al. Multiple RNA recognition patterns during microRNA biogenesis in plants. Genome Res. 2013;23(10):1675–1689. doi: 10.1101/gr.153387.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu H, et al. Bidirectional processing of pri-miRNAs with branched terminal loops by Arabidopsis Dicer-like1. Nat Struct Mol Biol. 2013;20(9):1106–1115. doi: 10.1038/nsmb.2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Warner JR, McIntosh KB. How common are extraribosomal functions of ribosomal proteins? Mol Cell. 2009;34(1):3–11. doi: 10.1016/j.molcel.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289(5481):905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 42.Dresios J, Derkatch IL, Liebman SW, Synetos D. Yeast ribosomal protein L24 affects the kinetics of protein synthesis and ribosomal protein L39 improves translational accuracy, while mutants lacking both remain viable. Biochemistry. 2000;39(24):7236–7244. doi: 10.1021/bi9925266. [DOI] [PubMed] [Google Scholar]
- 43.Nishimura T, Wada T, Yamamoto KT, Okada K. The Arabidopsis STV1 protein, responsible for translation reinitiation, is required for auxin-mediated gynoecium patterning. Plant Cell. 2005;17(11):2940–2953. doi: 10.1105/tpc.105.036533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park HS, Himmelbach A, Browning KS, Hohn T, Ryabova LA. A plant viral “reinitiation” factor interacts with the host translational machinery. Cell. 2001;106(6):723–733. doi: 10.1016/s0092-8674(01)00487-1. [DOI] [PubMed] [Google Scholar]
- 45.de la Cruz J, Karbstein K, Woolford JL., Jr Functions of ribosomal proteins in assembly of eukaryotic ribosomes in vivo. Annu Rev Biochem. 2015;84:93–129. doi: 10.1146/annurev-biochem-060614-033917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Serivichyaswat P, et al. Expression of the floral repressor miRNA156 is positively regulated by the AGAMOUS-like proteins AGL15 and AGL18. Mol Cells. 2015;38(3):259–266. doi: 10.14348/molcells.2015.2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu G, et al. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell. 2009;138(4):750–759. doi: 10.1016/j.cell.2009.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang JW, Czech B, Weigel D. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell. 2009;138(4):738–749. doi: 10.1016/j.cell.2009.06.014. [DOI] [PubMed] [Google Scholar]
- 49.Oliver ER, Saunders TL, Tarlé SA, Glaser T. Ribosomal protein L24 defect in belly spot and tail (Bst), a mouse Minute. Development. 2004;131(16):3907–3920. doi: 10.1242/dev.01268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Park MY, Wu G, Gonzalez-Sulser A, Vaucheret H, Poethig RS. Nuclear processing and export of microRNAs in Arabidopsis. Proc Natl Acad Sci USA. 2005;102(10):3691–3696. doi: 10.1073/pnas.0405570102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ye R, et al. Cytoplasmic assembly and selective nuclear import of Arabidopsis Argonaute4/siRNA complexes. Mol Cell. 2012;46(6):859–870. doi: 10.1016/j.molcel.2012.04.013. [DOI] [PubMed] [Google Scholar]
- 52.Juntawong P, Girke T, Bazin J, Bailey-Serres J. Translational dynamics revealed by genome-wide profiling of ribosome footprints in Arabidopsis. Proc Natl Acad Sci USA. 2014;111(1):E203–E212. doi: 10.1073/pnas.1317811111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nobuta K, McCormick K, Nakano M, Meyers BC. Bioinformatics analysis of small RNAs in plants using next generation sequencing technologies. Methods Mol Biol. 2010;592:89–106. doi: 10.1007/978-1-60327-005-2_7. [DOI] [PubMed] [Google Scholar]
- 54.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen C, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33(20):e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.