Significance
Gram-negative pathogens use a specialized secretion system to inject effector proteins into host cells to facilitate infection. Members of the IpaH family are a conserved group of effector proteins that function in the host cell as novel E3 ubiquitin ligases. How these ligases carry out their enzymatic function and how they autoregulate remain unclear. Using targeted mutagenesis, we uncover the specific function of IpaH conserved residues in E3 Ub ligase activity, either directly in the catalytic mechanism or in the recruitment of the upstream E2 enzyme. Comparison of autoinhibitory properties between IpaH members and the HECT-class E3 ligase Smurf2 reveals that IpaH enzymes short circuit the levels of charged E2 in vitro, which has the potential to subvert host ubiquitination during pathogenesis.
Keywords: IpaH family, bacterial E3 ubiquitin ligase, ubiquitin, host-pathogen
Abstract
IpaH enzymes are secreted bacterial effectors that function within host cells as E3 ubiquitin (Ub) ligases. Catalytic activity is imparted by a conserved novel E3 ligase (NEL) domain that is unique to Gram-negative pathogens and whose activity is repressed by a flanking substrate-binding leucine-rich repeat (LRR) domain when substrate is absent. How the NEL domain catalyzes the conjugation of Ub onto substrates, recognizes host E2s, and maintains its autoinhibited state remain poorly understood. Here we used mutagenesis and enzyme kinetic analyses to address these gaps in knowledge. Mutagenesis of conserved residues on two remote surfaces of the NEL domain identified functional clusters proximal to and distal to the active site cysteine. By analyzing the kinetics of Ub charging and discharging, we identified proximal active site residues that function as either the catalytic acid or catalytic base for aminolysis. Further analysis revealed that distal site residues mediate the direct binding of E2. In studying the full-length protein, we also have uncovered that IpaH family autoinhibition is achieved by a short-circuiting mechanism wherein the LRR domain selectively blocks productive aminolysis, but not the nonproductive discharge of Ub from the E3 to solvent. This mode of autoinhibition, which is not shared by the HECT domain ligase Smurf2, leads to the unanticipated depletion of E2∼Ub and thus a concomitant dominant-negative effect on other E3s in vitro, raising the possibility that short circuiting also may serve to restrict the function of host E3s in cells.
Posttranslational modification by the covalent attachment of the small protein ubiquitin (Ub) onto lysine side chains of target proteins is an essential eukaryotic modification that imparts a wide range of consequences to the substrate, from targeting to the proteasome for degradation to the assembly of signaling complexes (1, 2). Ubiquitination begins with an E1 enzyme, which activates Ub through the consumption of ATP to form a thioester E1∼Ub adduct on its catalytic cysteine. The E1∼Ub conjugate then transthiolates Ub to the active-site cysteine of an E2 enzyme to form an E2∼Ub adduct. Finally, the charged E2∼Ub then collaborates with a diverse group of E3 ligases to catalyze the stable attachment of Ub to the ε-amino group of side-chain lysine residues or, in some cases, the N-terminal amino group of protein substrates via an aminolysis reaction.
E3 ligases can be divided into two groups based on whether or not they use a catalytic cysteine to form an E3∼Ub intermediate. The HECT (homologous to E6AP C-terminus), RBR (RING-in-between-RING), and IpaH (invasion-plasmid antigen H) families of structurally distinct E3 ligases use a catalytic cysteine residue to accept activated Ub in the form of an E3∼Ub thioester adduct. The E3 itself then catalyzes the aminolysis reaction to form an isopeptide bond between Ub and the acceptor lysine substrate. In contrast, the RING (really interesting new gene)-related E3s lack a catalytic cysteine and instead stimulate the ability of the E2 itself to catalyze aminolysis between the E2-conjugated Ub and the acceptor lysine substrate through an allosteric mechanism (3). Thus, the process of ubiquitination can be visualized as a series of activation and transthiolation reactions that terminate with a common catalytic aminolysis step.
Because Ub has seven lysine residues (and a free amino terminus), iterative action of the Ub cascade can form poly-Ub chains of various linkage topologies and lengths (1, 2), resulting in a diverse repertoire of biological outcomes. Recent structures of various enzymatic intermediates of the Ub cascade for E1, E2, RING, HECT, and RBR proteins provide a strong groundwork for understanding how these classes of eukaryotic enzymes function normally and how they are dysregulated during disease (4–7).
Ub and Ub-like modifications are heavily involved in proper immune function and regulation (8, 9). Therefore, preventing or diverting Ub signaling is a common strategy used by invasive pathogens to facilitate infection (10). Toward this end, pathogenic Gram-negative bacteria use specialized secretion systems to deliver effector proteins into the host cell as a means of manipulating Ub signaling in diverse ways. For example, the protein kinase OspG binds to and is activated by E2∼Ub conjugates, thereby competing for the availability of charged E2 (11, 12); the deamidase OspI modifies UBC13, hindering its ability to function as an E2 for the immune-related E3 ligase TRAF6 (13); the methyltransferase NleE modifies zinc-coordinating cysteine residues on TAB2 and TAB3, preventing its ability to bind K63-linked Ub chains and promote NF-κB signaling (14); and the E3 ligases SopA, a structural mimic of HECT-class E3 ligases (15), and AvrPtoB, a structural mimic of RING-class E3 ligases (16), both subvert the Ub proteasome pathway to target host proteins for destruction. Along these lines, the IpaH family is a group of conserved effector proteins found in numerous Gram-negative pathogens that subvert the host Ub pathway through their ability to function as an E3 ligase (17). Identification of in vivo targets for several Shigella IpaH members have revealed a recurring theme of suppression of the host inflammasome and innate immune responses (18) through, for example, ubiquitination of GLMN by IpaH7.8 (19) or of NEMO by IpaH9.8 (20).
IpaH enzymes catalyze aminolysis via a novel E3 ligase (NEL) domain (17, 21–23), which, despite the lack of any structural similarity to the HECT and RBR classes of E3 ligases, proceeds through an analogous IpaH∼Ub thioester intermediate to generate Ub chains (17). IpaH members have a conserved two-domain architecture consisting of (i) a C-terminal NEL domain, which fully recapitulates E3 ligase activity in its isolated form, and (ii) an N-terminal leucine-rich repeat (LRR) domain, which serves the dual function of binding specific substrates for ubiquitination and autoinhibiting the constitutive activity of the NEL domain in the absence of a bound substrate (23, 24). Structures of isolated IpaH1.4 NEL domain (22), full-length autoinhibited IpaH members IpaH3 (21) and SspH2 (23), and substrate-bound complexes of SspH1-PKN1 (25) and SlrP-Trx (26) have provided an initial understanding of substrate recognition and the mechanism underlying IpaH activation. However, many functional questions remain, particularly for how IpaH E3 ligases recognize host E2 enzymes, how they catalyze aminolysis, and how precisely autoinhibition of NEL domain activity is achieved. Laying the groundwork for future structural and functional studies, here we report the mechanism of IpaH autoinhibition and provide a systematic interrogation of conserved regions within the NEL domain, elucidating the function of specific residues underlying catalysis and E2 recognition.
Results
Surface Conservation Identifies Two Functionally Relevant Interfaces.
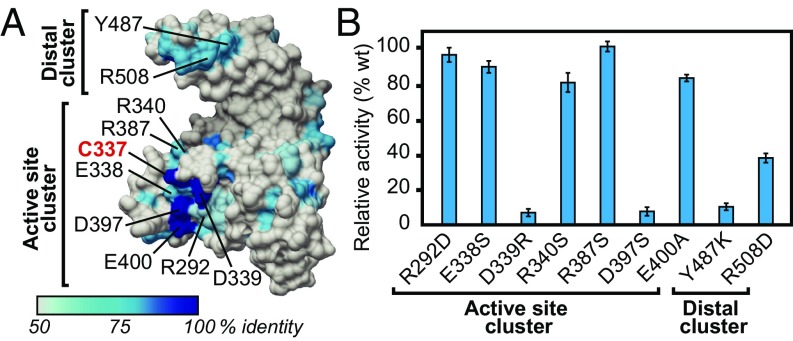
To identify functionally relevant residues involved in NEL domain E3 ligase activity, we mapped conservation onto the surface of the representative Shigella IpaH1.4 NEL domain structure (22) (Fig. 1A), substituting the better functionally characterized IpaH9.8 (20) residue numbering scheme (Fig. S1; 99% identity to the IpaH1.4 NEL domain). This exercise highlighted two clusters of surface conservation, one surrounding the active site cysteine (C337), and another distal to the active site (Fig. 1A). Conserved residues in the vicinity of the catalytic cysteine included R292, E338, D339, R340, R387, D397, and E400. The most highly conserved of these (D339 and D397) were previously validated as essential for IpaH function (21, 22), but their specific functions and those of surrounding residues in catalysis have not been addressed. Conserved residues at the distal region included Y487 and R508, neither of which has been functionally probed by mutagenesis.
Fig. 1.
Mutational analysis of conserved residues on the IpaH9.8 NEL domain. (A) Surface representation of the IpaH1.4 NEL domain (PDB ID code 3CKD) (22), numbered using the IpaH9.8 sequence, color-coded by conservation according to the key. Residues mutated in this study are indicated in black with the catalytic cysteine indicated in red. Conserved interfaces are grouped into active site and distal site clusters. (B) Catalytic E3 Ub ligase activity of IpaH9.8 NEL domain mutants relative to the WT enzyme in vitro (n = 3).
Fig. S1.
Protein sequence alignment of NEL domain homologs from Salmonella (SspH1, SspH2, and SlrP), Shigella (IpaH9.8), Rhizobium (NopM), Bradyrhizobium (Blr1904), Yersinia (y3397), and Pseudomonas (PSPTO_1492, Pfl01_4099) species. For each gene, the UniProt identifier is indicated in brackets. Sequences are shaded by similarity with a 50% identity cutoff, with residues grouped based on their side chain character using ALINE software (49). Sequence numbering for IpaH9.8 and SspH1 are shown above their respective sequences. A red star highlights the catalytic cysteine, and green triangles highlight conserved surface residues targeted for mutagenesis in this study.
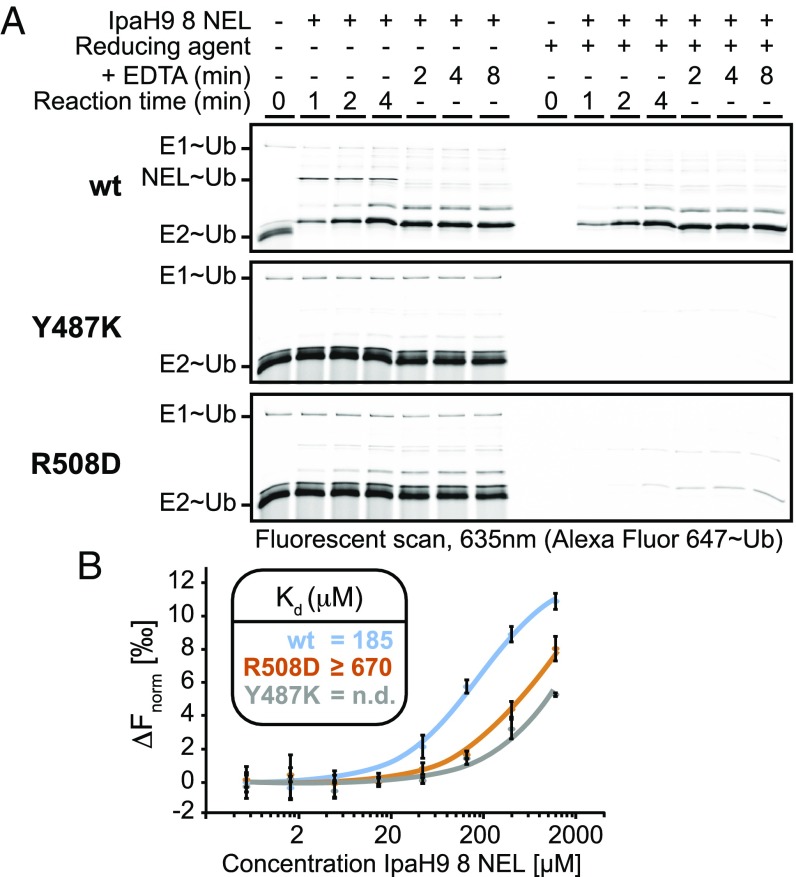
To ascertain the functional importance of conserved residues at both spatially distinct regions, we generated point mutations within the IpaH9.8 NEL domain and quantitatively tested each point mutant for its effect on in vitro polyubiquitination activity using fluorescein-labeled Ub (Fig. 1B). Mutations of D339 and D397 caused a dramatic loss of activity, as expected. Surprisingly, however, mutations at surrounding conserved positions, including R292, E338, R340, R387, and E400, showed no striking effect on ubiquitination activity (Fig. 1B; active site cluster). We reasoned that D339 and D397 play direct roles in catalyzing aminolysis, given their proximity to the catalytic cysteine. Mutations at positions on the distal site cluster also showed defects in poly-Ub formation, with the Y487K mutation having a more pronounced effect relative to the R508D mutation (Fig. 1B; distal cluster). Based on the large separation in space between the distal and catalytic clusters (∼30 Å), we hypothesized that the distal cluster residues play an indirect role in aminolysis, possibly by composing the E2 binding site.
D339 Functions as a Catalytic Acid and D397 Functions as a Catalytic Base in IpaH9.8 Aminolysis.
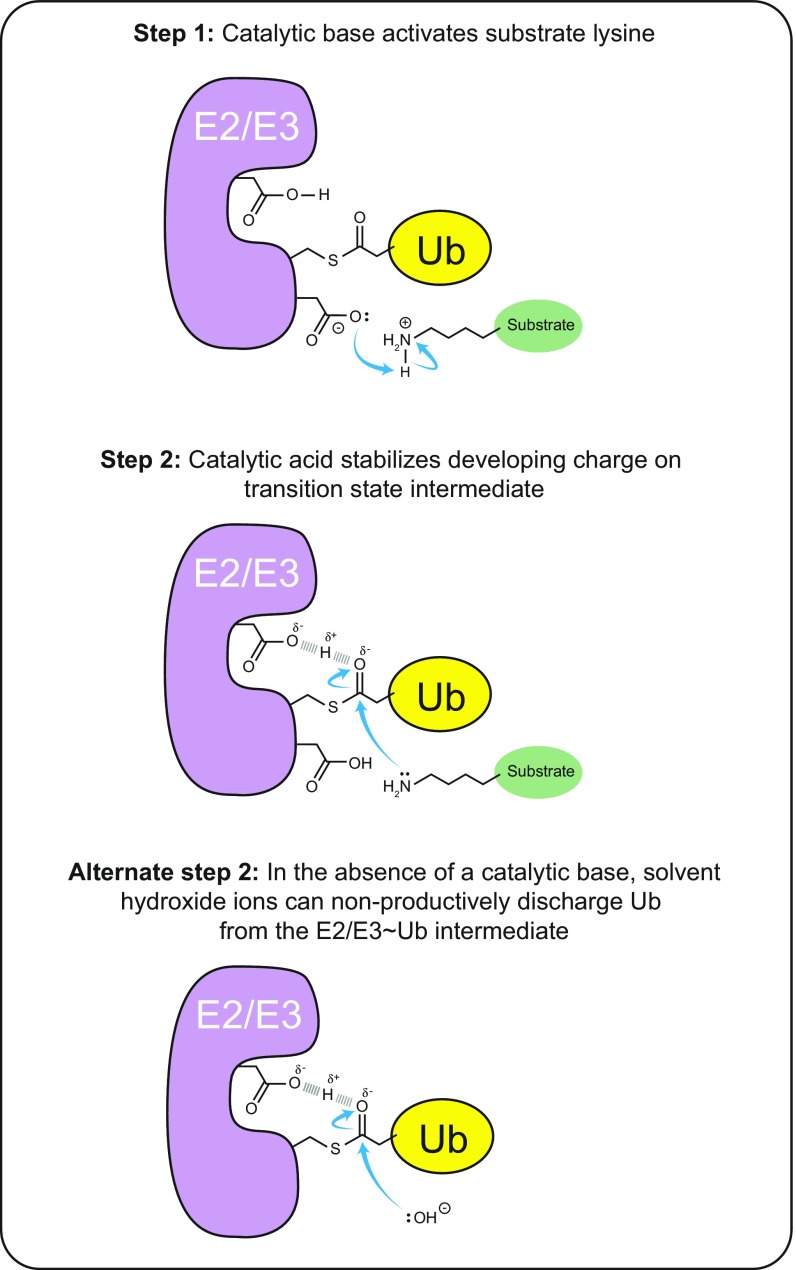
In general (and as best worked out for E2 enzymes), aminolysis between Ub charged onto an E2 or E3 enzyme and a lysine acceptor involves general acid-base catalysis (27–31). Within the active site, a catalytic base acts to deprotonate the incoming substrate lysine to promote its nucleophilic potential. Then, during the nucleophilic attack on the Ub∼enzyme thioester bond, a catalytic acid acts to stabilize the developing negative charge of the tetrahedral transition-state intermediate (Fig. S2). The participation of specific residues in either function can be discerned by assaying the effect of mutation on the penultimate steps of Ub thioester charging and discharging of the E2/E3 enzyme and on Ub–acceptor bond formation. For example, mutation of the catalytic base can allow apparently normal E3∼Ub charging and discharging, but with a failure to form Ub chains owing to a lack of nucleophilic activation of the substrate lysine (27–30). In this context, an outwardly normal Ub discharge reaction in the absence of a deprotonated lysine acceptor can be achieved via nucleophilic attack by solvent molecules, yielding nonproductive hydrolysis. Importantly, the resultant defect in aminolysis can be partially rescued by the nonspecific deprotonation of an attacking lysine side chain through an increase in the solution pH (30). In contrast, mutation of the catalytic acid produces an exaggeratedly more stable E3∼Ub/E2∼Ub intermediate owing to impaired discharge function resulting from insufficient stabilization of the tetrahedral intermediate formed by an attacking nucleophile (27–29). This defect is apparent in the presence of an efficiently deprotonated lysine (in the case of substrate attack) or water (in the case of nonproductive hydrolysis).
Fig. S2.
Summary of the generalizable roles of catalytic acid and catalytic base in aminolysis as inferred from representative E2 and E3 enzymes. Step 1: Lysine ubiquitination requires a catalytic base to activate its nucleophilic potential by deprotonating the positively charged nitrogen atom. Step 2: Subsequent nucleophilic attack by the activated lysine on the active site thioester-bound Ub requires stabilization of the developing negative charge on the carbonyl oxygen by a catalytic acid, which donates a proton to stabilize the transition state intermediate. Alternate step 2: Mutation of the catalytic base leads to accumulation of the Ub-charged state of the E2/E3 enzyme, which can be offset by nonproductive discharge of Ub to solvent by the nucleophilic attack of hydroxide ions. The choice of pathway is affected by the bulk pH of the solution and is viable because the intact catalytic acid stabilizes the transition state intermediate. Mutation of the catalytic acid leads to accumulation of a Ub-charged state of the E2/E3 enzyme, because neither an attacking lysine nor a hydroxide nucleophile can proceed through the transition state intermediate.
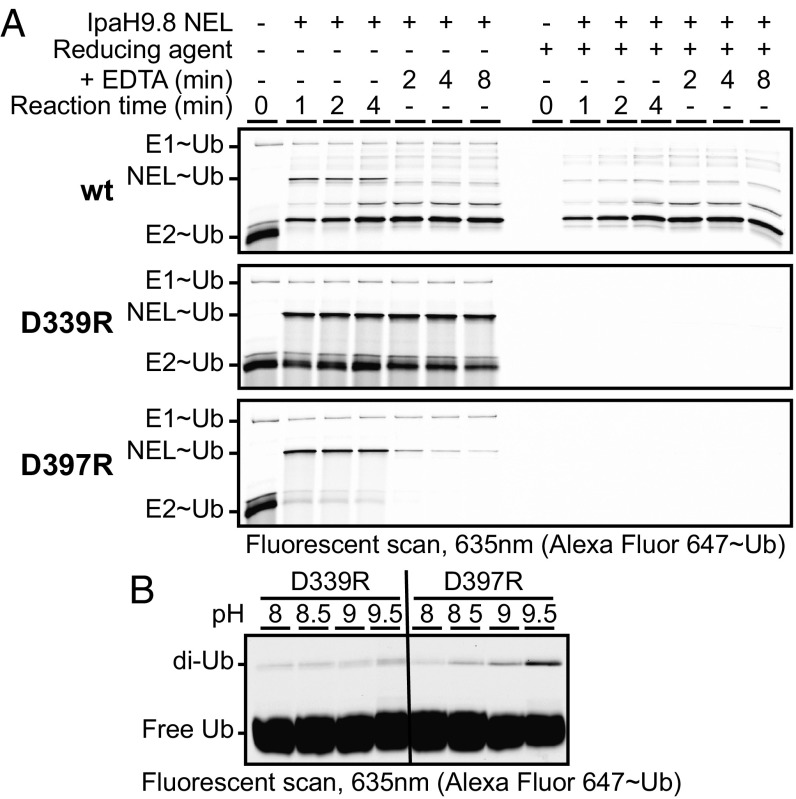
To discern the specific function of conserved active site residues in IpaH9.8 E3 ligase activity, we developed NEL domain Ub charging and discharging assays. To this end, we premixed E1 and E2 with ATP and Ub to enable steady-state formation of covalently charged E1∼Ub and E2∼Ub intermediates (Fig. 2A, Top, lane 1). We then assessed IpaH charging by adding wild-type (WT) IpaH9.8 NEL domain to the prepared mixture and visualizing the appearance of new molecular weight species corresponding to the NEL∼Ub intermediate and poly-Ub products over time (Fig. 2A, Top, lanes 2–4). Ubiquitinated products were distinguished from thioester intermediates by running equal reaction volumes in the presence and absence of reducing agent, which selectively breaks down thioester but not isopeptide bonds, thereby allowing identification of the NEL∼Ub intermediate (Fig. 2A, Top). Coinciding with the appearance of the NEL∼Ub intermediate, we observed a concomitant loss of E2∼Ub species, indicating that transthiolation from E2 to E3 occurs more rapidly than Ub transthiolation from E1 to E2 under our reaction conditions (indicating that E1 is limiting). Furthermore, the NEL∼Ub intermediate did not accumulate beyond the first time point (1 min), indicating that the NEL∼Ub intermediate rapidly achieved steady state.
Fig. 2.
Functional analysis of active site mutants. (A) Kinetics of Ub charging and discharging for the indicated WT (wt) and mutant IpaH9.8 NEL domains. (B) Effect of pH on the residual catalytic activity of indicated IpaH9.8 NEL domain mutants in vitro.
Discharge analysis was then initiated by spiking EDTA into the reaction mix to inhibit E1 function, thereby preventing further activation of free Ub. The addition of EDTA led to loss of the NEL∼Ub intermediate within the first time point tested (2 min), indicating that the NEL∼Ub conjugate is rapidly discharged/consumed when using a WT IpaH NEL domain (Fig. 2A, Top, lanes 5–7). In contrast to the WT enzyme, the IpaH active site mutant D339R (Fig. 2A, Middle) showed rapid and efficient NEL∼Ub charging, but was severely compromised for discharge function. This correlated with stabilization of the E2∼Ub conjugate at all time points, possibly owing to saturation of the NEL domain in its Ub-charged state, which would prevent further transthiolation from E2∼Ub. In contast, the active site mutant D397R behaved more like the WT protein (Fig. 2A, Bottom), displaying outwardly normal charging and discharging activities despite a lack of aminolysis activity.
Taken together, these mutational results support a model in which D397 functions as a catalytic base and D339 functions as a catalytic acid. To further validate these inferences, we measured the effect of pH on the residual activity of D339 and D397 mutants. Consistent with only D397 functioning as a catalytic base, the D397R mutant, but not the D339R mutant, showed a partial rescue of aminolysis with an increase in solution pH (Fig. 2B).
Distal Residues Mediate E2 Binding.
The clustering of functionally important conserved residues at a position distal from the active site strongly suggested an indirect contribution toward catalytic chemistry, such as by mediating interaction with the E2 enzyme or positioning of substrate. To elucidate the specific function of distal site residues, we tested both Y487K and R508D mutants in our NEL∼Ub charging and discharging assay. In contrast to the behavior of WT enzyme (Fig. 3A, Top), both Y487K and R508D mutants (Fig. 3A, Middle and Bottom) yielded little to no detectable NEL∼Ub species, which was further correlated by the findings of no appreciable change in E2∼Ub levels. These results are reminiscent of the behavior of HECT domain mutants defective for E2 binding, which exhibit a loss of E2-E3 transthiolation and thus stabilization of the E2∼Ub conjugate in charging assays (32).
Fig. 3.
Functional analysis of distal site mutants. (A) Kinetics of Ub charging and discharging for the indicated WT (wt) and mutant IpaH9.8 NEL domains. (B) Binding analysis of UBCH5 to the indicated WT or mutant IpaH9.8 NEL domains using MST. (Inset) Calculated Kd values (n = 3).
To directly test whether binding of the IpaH9.8 NEL domain with its cognate E2, UBCH5B, was hindered by either Y487K or R508D mutations, we tested both WT and NEL mutants for interaction with fluorescent-labeled UBCH5B by microscale thermophoresis (MST) (Fig. 3B). The WT NEL domain produced significant thermophoretic shifts, yielding an estimated dissociation constant (Kd) of 185 μM (Fig. 3B, blue curve). This is comparable to previous reports measuring the binding of free NEL domain to the E2 enzyme by fluorescent polarization (22). Directly supporting a functional role for the conserved distal site in E2 binding, the R508D mutant displayed a ≥4-fold reduction in affinity for E2 (Fig. 3B, orange curve; Kd ≥670 μM), whereas the Y487K mutant showed even greater loss of affinity (Fig. 3B, gray curve; Kd not determined). Importantly, the relative severity of the R508D and Y487K mutants in E2 binding correlated well with their respective loss of ubiquitination activity (Fig. 1B, distal cluster). We conclude that the conserved distal site on the NEL domain is an E2-binding surface.
The Mechanism of Action of IpaH9.8 Is Generalizable to Other IpaH Family Members.
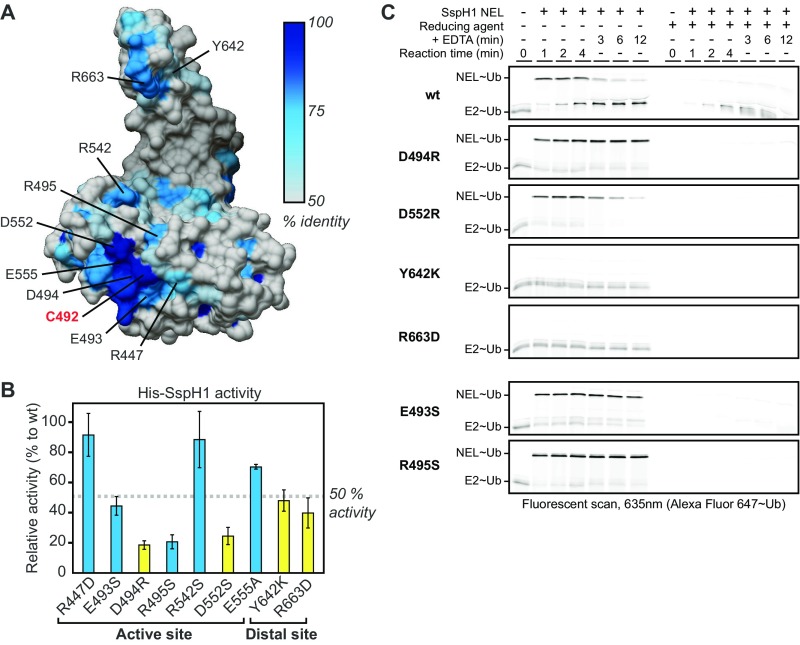
The IpaH family member SspH1 from Salmonella shares only ∼40% identity of its NEL domain with that of IpaH members from Shigella. To determine whether our insight into the catalytic mechanism of IpaH9.8 extends to SspH1 as well, we generated and characterized the corresponding mutations in full-length SspH1 (Fig. S3). Assessment of poly-Ub chain formation revealed that four residues in SspH1 (D494, D552, Y642, and R663, corresponding to residues D339, D397, Y487, and R508 in IpaH9.8) and two conserved residues in the vicinity of the catalytic cysteine (E493 and R495) are critical for function (Fig. S3B). To assess how these residues contribute to catalytic function, we tested all six SspH1 mutants for NEL∼Ub charging and discharging kinetics. The mutants D494R, D552R, Y642K, and R663D in SspH1 demonstrated charging and discharging defects comparable to those of the corresponding mutants D339R, D397R, Y487K, and R508D, respectively, in IpaH9.8 (Fig. S3C, panels 2–5). The E493S and R495S mutants (Fig. S3C, panels 6 and 7) behaved similarly to the D494R mutant in this assay, suggesting that they cooperate with D494 in the stabilization of the tetrahedral intermediate formed during nucleophilic attack. Taken together, these findings confirm that our insight into the catalytic mechanism of IpaH9.8 extends to SspH1 and likely, more generally, to all IpaH family members as well.
Fig. S3.
Mutational analysis of conserved residues in SspH1 on E3 ligase activity and Ub charging and discharging kinetics. (A) Surface representation of SspH2 NEL domain color-coded according to conservation (PBD ID code 3G06; 78% identity to the SspH1 NEL domain) (23) with residues mutated in this study labeled according to the SspH1 numbering. (B) Relative E3 Ub ligase activity of His-tagged SspH1 mutants in vitro (n = 3). Mutated residues corresponding to those that perturb IpaH9.8 activity are in yellow. (C) In vitro Ub charging and discharging kinetics for WT (wt) and mutant SspH1 NEL domains.
Autoregulation Short Circuits the NEL∼Ub Charged Intermediate.
Intramolecular interactions between the substrate-binding LRR domain and catalytic NEL domain restrict unbridled IpaH Ub ligase activity (23, 24). Engagement of substrate by the LRR domain relieves autoinhibition by competitively displacing interactions between the LRR and NEL domains (25). Comparing the structures of autoinhibited and isolated NEL domains suggests that autoinhibition is achieved by limiting accessibility of the catalytic cysteine, thereby preventing charging to form the NEL∼Ub intermediate (21–24). Direct biochemical evidence supporting this model is lacking, however.
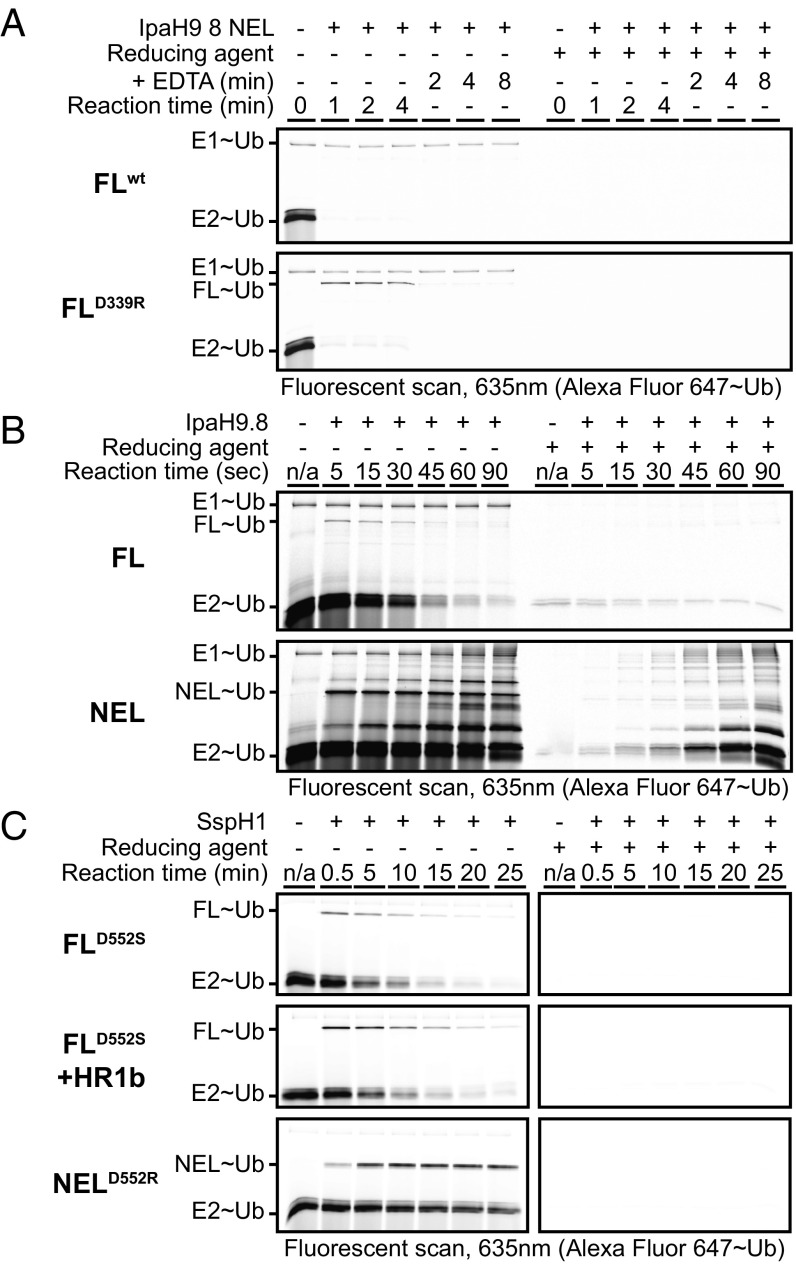
To shed light on the mechanism of autoinhibition, we purified full-length IpaH9.8 and tested Ub charging and discharging kinetics. Consistent with expectations, when full-length IpaH9.8 (IpaH9.8FL) was added to a mixture of precharged E1∼Ub and E2∼Ub, no charged IpaH9.8FL∼Ub intermediate was detected (Fig. 4A, Top). Unexpectedly, however, the addition of IpaH9.8FL to the reaction mix coincided with a wholesale loss of the E2∼Ub intermediate (Fig. 4A, Top), raising the possibility that transthiolation of Ub from E2 to E3 was in fact occurring.
Fig. 4.
IpaH autoinhibition involves a short-circuit mechanism. (A) Kinetics of Ub charging and discharging for WT (wt) or discharge mutant (D339R) full-length (FL) autoinhibited IpaH9.8. (B) Kinetics of Ub charging and discharging for WT FL and isolated NEL domain of IpaH9.8. (C) Kinetics of Ub charging and discharging for catalytic base (D552) mutants of FL and isolated NEL domain of SspH1 in the presence or absence of the substrate, PKN1 fragment HR1b (HR1b).
We reasoned that the absence of a detectable IpaH9.8FL∼Ub intermediate might be due to a rapid, nonproductive discharge of Ub to solvent. To explore this possibility, we tested the effect of the discharge-deficient mutant D339R in the context of the full-length enzyme. In this experiment, the D339R mutant caused the same instability of the E2∼Ub intermediate, but now with detectable formation of the elusive IpaH9.8FL∼Ub intermediate (Fig. 4A, Bottom). Surprisingly, the D339R IpaH9.8FL∼Ub intermediate was more unstable than the D339R IpaH9.8NEL∼Ub intermediate (Fig. 2A, Middle), suggesting that autoinhibitory interactions involving the LRR domain may potentiate the nonproductive discharge of Ub.
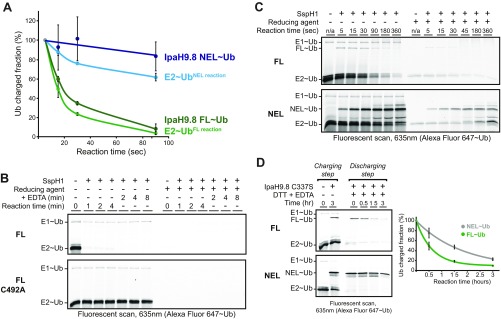
To first confirm that the WT full-length IpaH enzyme forms an E3∼Ub thioester intermediate followed by rapid hydrolysis, we used experimental conditions reoptimized to favor detection of the WT E3∼Ub thioester intermediate, i.e., higher (E2) and lower temperatures (Fig. 4B; quantification in Fig. S4A). Under these conditions, we observed rapid formation and depletion of the IpaH9.8FL∼Ub intermediate (Fig. 4B, Top) and rapid formation but slow depletion of the IpaH9.8 NEL∼Ub intermediate (Fig. 4B, Bottom). These results establish that full-length IpaH9.8 is fully competent for charging, but that the thioester-linked Ub intermediate is rapidly discharged by a short-circuiting mechanism involving nonproductive hydrolysis. Similar analyses demonstrated that the short-circuiting mechanism applies to SspH1 as well (Fig. S4 B and C).
Fig. S4.
Comparison of full-length (FL) versus isolated NEL domain in Ub charging and discharging reactions. (A) Quantification of Ub-charged intermediates over time for reactions involving the FL or isolated NEL domain of IpaH9.8 shown in Fig. 4B. (B) In vitro Ub charging and discharging kinetics for WT SspH1 FL and a catalytic cysteine mutant (C492A). (C) Kinetics of Ub charging and discharging for WT SspH1 FL and isolated NEL domain in conditions favoring visualization of the thioester intermediate. (D) Ub discharging kinetics for C337S mutants of the IpaH9.8 FL or isolated NEL domain. Quantification of Ub-charged intermediates is shown at the right.
In the foregoing experiments, the full-length enzyme consistently displayed a faster rate of thioester discharge compared with the isolated NEL domain (Fig. 4B; quantification in Fig. S4A). This difference in E3∼Ub stability was recapitulated when we used a catalytic Cys-to-Ser mutational strategy to enhance detection of the E3∼Ub intermediate (Fig. S4D). We reasoned that the apparent increase in Ub thioester discharge of full-length enzyme can be explained by the fact that the autoinhibitory state promotes E3∼Ub hydrolysis. To explore this possibility, we compared the rate of hydrolysis in the presence and absence of substrate, because substrate will competitively displace autoinhibitory interactions and thus mimic the status of an isolated NEL domain with a low hydrolysis rate. To avoid the complicating effect of substrate addition on the stimulation of aminolysis, we introduced the catalytic base mutation (D552S or R) into the full-length and isolated NEL domains of SspH1 to disable aminolysis activity while leaving the hydrolysis reaction intact (Fig. S3C, panel 3). As expected, full-length SspH1D552S demonstrated a much more rapid rate of hydrolysis than the isolated NELD552R domain (Fig. 4C, Top vs. Bottom). On the addition of substrate (PKN1 HR1b subdomain), no significant change in the rate of hydrolysis was observed for the full-length enzyme (Fig. 4C, Middle), indicating that autoinhibition does not promote hydrolysis per se, but rather a high rate of hydrolysis is a constitutive feature of both the active and autoinhibited states of the full-length IpaH enzyme. We conclude that autoinhibition is achieved by the selective blockade of aminolysis activity in a manner that leaves Ub charging function and the discharge function by nonproductive hydrolysis unperturbed. Furthermore, we attribute the apparent difference in hydrolysis rates of the IpaH full-length and isolated NEL domain proteins to an intrinsic difference in the overall catalytic efficiency of the two enzyme constructs. Providing a possible explanation for the selective blocking of aminolysis over Ub charging and nonproductive hydrolysis functions, inspection of autoinhibited IpaH structures identified the loop region harboring the catalytic base as the primary point of contact for the LRR domain (Fig. S5). Thus, selective perturbation of the catalytic base would allow for an off-state that blocks aminolysis while leaving the hydrolysis function intact.
Fig. S5.
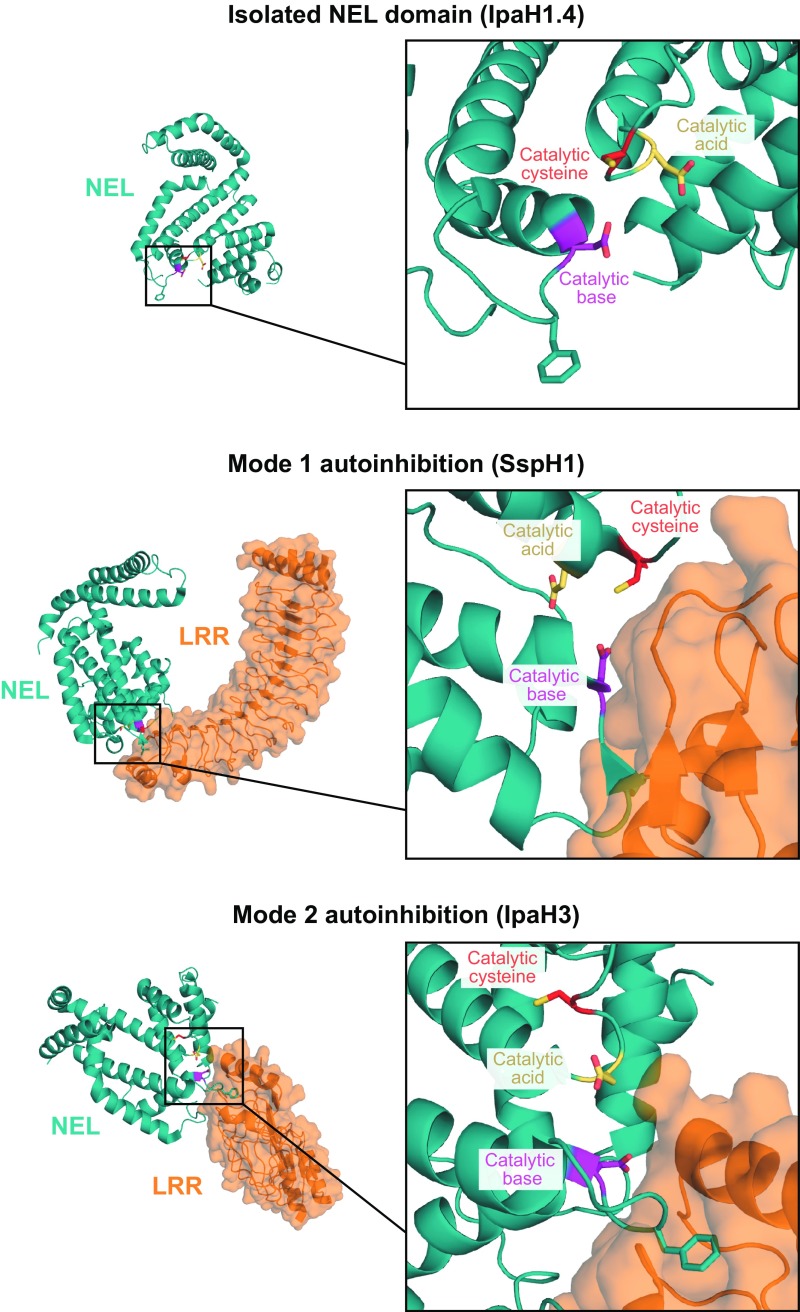
Structural comparison of the isolated IpaH1.4 NEL domain (PDB ID code 3CKD), autoinhibited SspH2 (PDB ID code 3G06), and autoinhibited IpaH3 (PDB ID code 3CVR) (21–23). NEL domains are colored in teal, and LRR domains are in orange. Side chains, highlighted as sticks, correspond to the catalytic cysteine (red), catalytic base (magenta), or catalytic acid (yellow).
Comparison of IpaH Autoregulation with the Eukaryotic HECT Class E3 Ligase Smurf2.
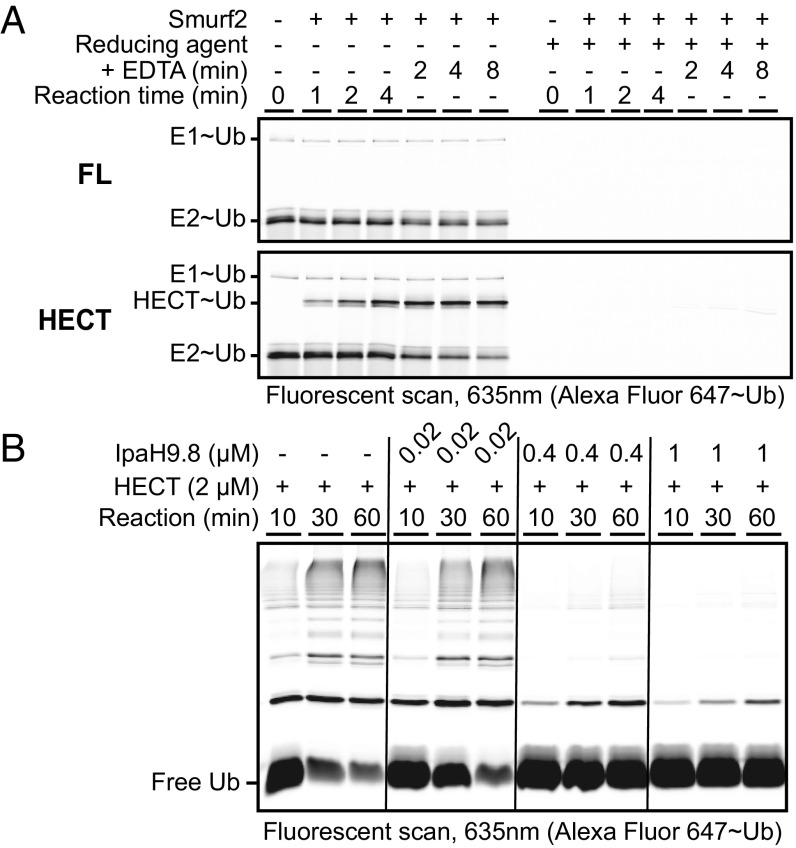
Smurf2 is a HECT class E3 ligase that shares several characteristics with the IpaH family: (i) both form Ub charged intermediates via a catalytic cysteine (1, 17), and (ii) both demonstrate autoinhibition by the intermolecular association of flanking regulatory domains (23, 24, 33, 34). To examine how autoregulation differs between these two classes of E3 ligases, we characterized the kinetic properties of autoinhibited full-length Smurf2 and its constitutively active isolated HECT domain. Using a similar Ub charging and discharging assay as that used for IpaH enzymes in this study, we found that full-length Smurf2 had no significant effect on E1∼Ub or E2∼Ub levels and showed no evidence of charging to form a Smurf2∼Ub intermediate (Fig. 5A, Top). As expected, the isolated HECT domain showed evidence of charging, but without apparent perturbation of the steady-state levels of E2∼Ub (Fig. 5A, Bottom). The latter observation suggests that unlike what was observed for IpaH proteins, one or more aspects of Smurf2 HECT domain function were rate-limiting (e.g., transthiolation from the E2, catalysis of aminolysis, or Ub hydrolysis from the E3). These results indicate that the short-circuit mechanism of autoinhibition used by IpaH9.8 and SspH1 is not used by the HECT ligase Smurf2.
Fig. 5.
The short-circuit mechanism of autoinhibition used by IpaH9.8 is not used by Smurf2 and produces a dominant-negative effect on other E3s. (A) Kinetics of Ub charging and discharging for full-length Smurf2 (FL) or its isolated HECT domain. (B) Dominant-negative effect of autoinhibited IpaH9.8 on the Ub ligase activity of constitutively active Smurf2 HECT domain in vitro.
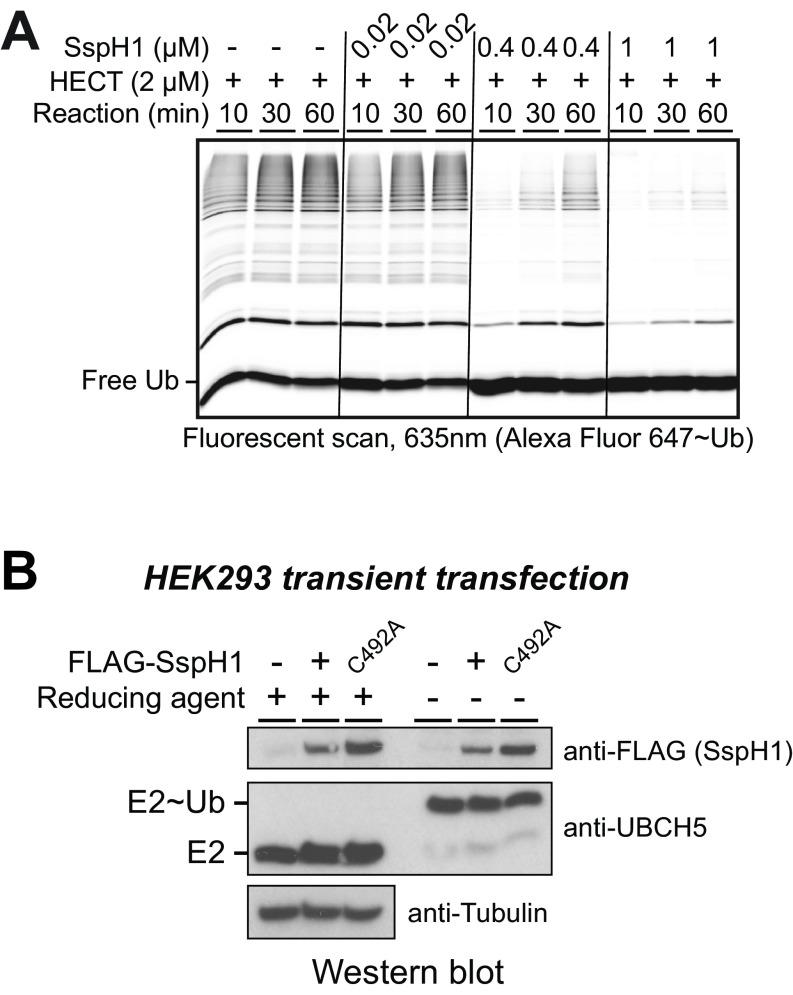
The observation that IpaH9.8, but not Smurf2, can efficiently deplete steady-state levels of E2∼Ub conjugates in our in vitro ubiquitination reactions suggested that autoinhibited IpaH9.8 might be able to exert a dominant-negative effect on Smurf2 function if the two enzymes were mixed in reactions lacking IpaH substrate. To explore this possibility, we mixed increasing amounts of full-length autoinhibited IpaH9.8 with a fixed level (2 μM) of isolated Smurf2 HECT domain. On its own, the Smurf2 HECT domain displayed robust ubiquitination activity (Fig. 5B, lanes 1–3); however, as the level of autoinhibited IpaH9.8 was increased, a potent concomitant loss of Ub chain formation by the Smurf2 HECT domain was observed (Fig. 5B, lanes 7–9). This dominant-negative effect was also observed when using SspH1 (Fig. S6A) and is consistent with an autoinhibitory mechanism for IpaH enzymes involving a continuous discharge of Ub from the E3 after transthiolation from E2.
Fig. S6.
SspH1 short-circuiting and functional characterization in cells. (A) Dominant-negative effect of autoinhibited SspH1 on the Ub ligase activity of the constitutively active Smurf2 HECT domain in vitro. (B) Western blot analysis of HEK-293 lysates with the indicated antibodies to the FLAG epitope (SspH1) and UBCH5. HEK-293 cells were transiently transfected with empty plasmid vector control or plasmids expressing WT or the catalytic mutant (C492A) of SspH1. UBCH5 charging was assessed by comparing reduced and nonreduced samples.
The observation that IpaH members potently deplete UBCH5∼Ub pools in vitro raised the possibility that autoinhibited IpaH enzymes might suppress host ubiquitination through depletion of UBCH5∼Ub pools in cells. To test this possibility, we assessed the levels of UBCH5 and UBCH5∼Ub in whole-cell lysates from HEK-293 cells transfected with plasmid alone or with plasmid expressing WT or a catalytically dead mutant of SspH1 (C492A). No discernable effect on the total level of Ub-charged UBCH5 was observed across all treatment groups (Fig. S6B), indicating that short circuiting does not influence the global level of UBCH5∼Ub in cells.
Discussion
A worldwide increase in microbial drug resistance, coupled with stagnant growth in the discovery of new antibiotics, underscores the need for novel therapeutic approaches to directly target pathogenesis (35–37). The IpaH family, with its high conservation across numerous pathogenic Gram-negative bacteria, is an attractive candidate for the development of mechanism-based inhibitors with possibly broad applicability (18). Here we used mutational analysis of residues conserved through evolution to examine the mechanisms underlying catalysis and autoregulation for IpaH family members from Shigella and Salmonella. The insights gained may help inform future therapeutic strategies to fight bacterial infections. In addition, our study has uncovered an unanticipated mechanism by which IpaH enzymes may modulate the host Ub machinery beyond their ability to target host proteins for ubiquitination, namely by depleting the pool of charged E2s.
Eukaryotic E2 and E3 ligases share many similarities in catalyzing aminolysis despite their significant differences in structure. By analyzing the effect of mutations within the vicinity of the active site cysteine on catalysis, Ub charging, and Ub discharging kinetics, specific residues for E2 enzymes (acting in conjunction with RING E3 ligases) have been assigned functional roles as either catalytic bases or catalytic acids in aminolysis (27, 28). Although the catalytic mechanisms of the RBR (30, 38, 39) and HECT (29, 40) classes of E3 ligases are less well understood, the mutagenic results accumulated to date are consistent with specific side chains functioning analogously. Our mutational, biochemical, and enzymologic analyses of conserved residues in the IpaH family NEL domain are suggestive of a similar approach to aminolysis despite overt differences in catalytic domain structure and flanking domain architecture. Atomic structures of catalytic intermediates, such as the NEL∼Ub conjugate, will be highly instructive for explaining how exactly the residues characterized in this study support enzyme function. Our work also has revealed that the full-length enzyme has greater hydrolytic activity than the isolated NEL domain, suggesting that the LRR domain or the LRR-NEL interdomain linker may contribute in some way to NEL domain catalytic function, possibly by providing supporting contacts to the conjugated donor Ub or by affecting NEL domain stability and dynamics. Additional structures of full-length catalytic intermediates may explain the basis for this effect.
A previous study probing the binding interaction between the E2∼Ub intermediate and IpaH NEL domain identified an extended contact surface on the E2∼Ub conjugate spanning both the E2 and the covalently attached Ub (41). This contact surface differed considerably from the contact surface on the E2∼Ub conjugate used for engaging HECT domains (32). Our study has identified likely residues on the reciprocal contact surface of the NEL domain for E2∼Ub interaction that combines the catalytic cysteine and a remote conserved surface located 24–28 Å away. Elucidation of the atomic structure of an E2-NEL domain complex will provide valuable information on how these two reciprocal interaction surfaces come together to support the transthiolation reaction.
E3 ligases are regulated through diverse mechanisms; for example, the multi-subunit Cullin RING E3 ligases are regulated by posttranslational modification with NEDD8 (42), the HECT-class E3 ligase Smurf2 is regulated by intramolecular interactions between the HECT domain and its flanking N-terminal C2 domain (33), and the RBR-class E3 ligase HHARI is regulated by intramolecular interactions between the RBR module and its flanking C-terminal Ariadne domain (43). Not surprisingly, the IpaH family of E3 ligases, many of which are also regulated, have a completely different mechanism of autoregulation. Our work reveals that autoinhibited IpaH enzymes are not static entities completely deficient in all catalytic functions, but instead represent an active “short-circuited” state whereby Ub is efficiently transferred from E2 to the NEL domain and rapidly discharged nonproductively to solvent. This mode of autoinhibition may reflect a bacterial adaptation to achieve the dual goal of suppressing unwanted formation of poly-Ub chains in the absence of substrate while also depleting the local pool of charged E2s to subvert the cellular function of other E3 enzymes. Although we found that the IpaH family member SspH1 did not short circuit the global cellular pool of charged UBCH5 in cells, we cannot rule out the possibility that short-circuiting occurs more locally in the cell, for example, in the immediate vicinity of the bacterial vacuole (44) or in smaller subcellular compartments to which specific IpaH enzymes localize, such as at the apical plasma membrane in the case of SspH2 (23) and within the nucleus in the case of IpaH9.8 (45) or SspH1 (46). Further work is needed to prove that this described function of IpaH enzymes in vitro is exploited for virulence in vivo.
Materials and Methods
Detailed information is provided in SI Materials and Methods. Recombinant proteins were expressed from Escherichia coli and purified as described previously (25, 47, 48). Fluorescent labeling of indicated proteins was adapted from a previous report (47). Ubiquitination reactions were performed largely as described previously (25), with proteins visualized by fluorescence using a Typhoon FLA 9500 imager (GE Healthcare). Ub charging and discharging assays were performed similarly to the ubiquitination assays, but at 4 °C or 16 °C. Analysis of E2 charging in cells was adapted from previous work (25) by treating HEK-293 lysate with 1.35 M β-mercaptoethanol or not, as indicated, and running samples on BIS-Tris SDS/PAGE gels under neutral pH. MST-binding assays were carried out using fluorescent-labeled UBCH5B mixed with indicated amounts of IpaH9.8 NEL domain, with measurements made with a Monolith NT.Automated device (NanoTemper Technologies).
SI Materials and Methods
Bacterial Expression Plasmids.
Bacterial expression vectors for pETM30 UBA1, pProEx UBCH5B, pProEx Ub, pProEx UbCys0, pETM30 IpaH9.8 NEL254–545, pGEX IpaH9.8, pProEx SspH1 LRR-NEL162–700, pProEx Smurf2 HECT366–748, pGEX Smurf2, and pGEX PKN1 HR1b122–199 were used as reported previously (25, 47, 48). Because the isolated His-tagged SspH1 NEL domain is poorly expressed in bacteria, the pProEx SspH1 LRR-NEL162–700 vector was modified to remove the original N-terminal tobacco etch virus (TEV) protease site (to make the His-tag uncleavable), followed by insertion of a seven-residue TEV cleavage site (ENLYFQG) between the LRR and NEL domains preceding position Glu403. A modified vector of pGEX HR1b122–199 was used to generate cysteine-conjugated fluorescein∼HR1b by mutation in a Cys residue at position −4 just after the TEV cleavage site following the N-terminal GST tag. Mammalian expression vectors for SspH1 were reported previously (25). All mutants tested were generated by site-directed mutagenesis using the QuikChange method (Stratagene) and confirmed by sequencing.
Expression and Purification of Recombinant Proteins.
Bacterial expression and purification of GST-tagged proteins (UBA1, IpaH9.8 NEL254–545, IpaH9.8, Smurf2, and PKN1 HR1b122–199) and His-tagged proteins (UBCH5B, Ub, UbCys0, SspH1 LRR-NEL162–700, and Smurf2 HECT366–748) were performed as described previously (24, 25, 48). His-tagged (SspH1 LRR-NELTEV linker) and GST-tagged (PKN1 HR1bCys-4) proteins generated in this study were similarly produced in E. coli BL21 (DE3)-RIL CodonPlus cells (Stratagene) grown to an OD600 of 0.6–0.8 before induction with 0.25 mM isopropyl-β-d-thiogalactopyranoside and a change in temperature to 17 °C overnight. Tagged proteins were affinity-purified with a HisTrap HP nickel column (GE Healthcare) or glutathione Sepharose 4B resin (GE Healthcare). Where indicated, affinity tags were removed with TEV protease overnight at 4 °C and proteins were further purified by nickel subtraction to remove His-tagged TEV protease, followed by gel filtration chromatography to exchange buffer (20 mM Hepes pH 7.5, 150 mM NaCl, and 2 mM DTT). Proteins destined for conjugation with cysteine-reactive fluorescent dyes (UBCH5B, PKN1 HR1bCys-4, and UbCys0) used a modified sizing buffer, with 2 mM DTT replaced by 1 mM TCEP.
Fluorescent Labeling of Recombinant Proteins.
Fluorescent labeling of proteins with cysteine-reactive fluorescein [5-(iodoacetamido)fluorescein; Sigma-Aldrich], Alexa Fluor 647 (Alexa Fluor C2 647 maleimide; Thermo Fisher Scientific), or an Alexa Fluor 647 variant (NT-647-maleimide; NanoTemper Technologies) was performed as described previously (47) with minor modifications. In brief, 0.2–0.25 mM protein was incubated with 3 molar excess (for UbCys0 and HR1bCys-4) or 8 molar substoichiometric (for UBCH5B) dye in conjugation buffer (20 mM Hepes pH 7.5, 150 mM NaCl, and 0.2 mM TCEP) at room temperature overnight. Reactions were then quenched with 5 mM DTT and diluted to lower the concentration of NaCl to 50 mM. The resultant mixture was then loaded onto a HiTrap Q HP (for HR1b) or SP HP column (for Ub and UBCH5B) (GE Healthcare) and eluted with a linear gradient (50–500 NaCl), collecting peak fractions. Purified proteins were then concentrated and buffer-exchanged by S75 gel filtration chromatography.
Sequence Alignment and Surface Mapping.
Sequences of selected NEL domains were accessed from UniProt, and alignments were prepared using Aline (49). For conservation mapping, alignment was carried out using Clustal Omega with the default settings (50) and mapped onto the surface of the NEL domain of IpaH1.4 (22) or SspH2 (23) using UCSF Chimera (51).
E2 Binding Assay Using MST.
MST binding assays were performed using 330 nM NT-647-maleimide–labeled UBCH5B diluted in binding buffer consisting of 20 mM Hepes pH 7.5, 0.12% Brij-35, 0.3 mg/mL BSA, and 1 mM DTT. Purified IpaH9.8 NEL C337A, IpaH9.8 NEL C337A/Y487K, and IpaH9.8 NEL C337A/R508D were titrated into fluorescently labeled UBC5B and incubated for 30 min at room temperature before MST scans using an Monolith NT.Automated device (NanoTemper Technologies). Binding curves were fit using NT Analysis Software.
In Vitro Ubiquitination Activity Assays.
Ubiquitination assays were carried out in a common reaction condition consisting of 6.5 μM total Ub (19:1 ratio of unlabeled to Alexa Fluor 647-labeled Ub), 0.15 μM UBA1, 0.25 μM UBCH5B, 28 mM Hepes pH 7.5, 50 mM NaCl, 10 mM MgCl2, 5 mM ATP pH∼7, and 1 mM TCEP. Reactions were initiated by addition of 0.5 μM IpaH9.8 NEL domain or 0.5 μM His-tagged SspH1 LRR-NEL and 2 μM PKN1 fluorescein-labeled HR1bCys-4 substrate at 25 °C. After 15 min, the reactions were quenched by the addition of SDS/PAGE loading dye, followed by boiling for 10 s. Reactions were resolved by SDS/PAGE. Without removing the glass plates, the gel cassettes were rinsed and dried, followed by visualization of fluorescence using a Typhoon FLA 9500 scanner (GE Healthcare) at 500 V power and 50-μm resolution. Quantification was carried out using ImageQuant software, with calculation of the ratio of higher molecular weight species vs. the total (high molecular weight species + free Ub) signals, normalized to a loaded standard curve. To assess the effect of pH on the residual activity of D339R and D397R IpaH9.8 NEL domain mutants, reactions were adjusted to 0.1 μM UBA1 and 1 μM IpaH9.8 NEL, with Hepes replaced by 75 mM BIS-Tris propane (pH 8–9) or CAPS (pH 9.5) for higher-pH analysis. Samples were incubated at 25 °C for 80 min before quenching reactions were performed.
Ubiquitination reactions performed with full-length Smurf2 and isolated HECT domain were performed as for IpaH enzymes but with 13 μM total Ub, 0.5 μM UBA1, 1 μM UBCH5B, 2 μM Smurf2 HECT domain, and indicated concentrations of full-length IpaH or SspH1 enzyme (1, 0.4, or 0.02 μM). Reactions were incubated at 37 °C and harvested at indicated reaction times (10, 30, or 60 min).
Ub Charging and Discharging Kinetics.
Ub charging and discharging kinetics were performed in a common reaction buffer consisting of 6.5 μM total Ub (19:1 mixture of cold Ub and fluorescently labeled Ub), 0.5 μM UBA1, 1 μM UBCH5B, 28 mM Hepes pH 7.5, 200 mM NaCl, 10 mM MgCl2, 5 mM ATP pH∼7, and 1 mM TCEP, at 16 °C. NEL charging reactions were initiated by addition of 2 µM E3 and then quenched at the indicated time points by adding SDS/PAGE loading buffer with or without reducing agent (1.35 mM β-mercaptoethanol) to discriminate thioester from isopeptide bond linked products and heating on a 95 °C block for 15 s. NEL∼Ub discharging was assessed by adding EDTA to a final concentration of 30 mM at the 4 min time-point of a charging reaction and harvesting volume-adjusted samples at 2, 4, and 8 min after the addition of EDTA. All charging and discharging harvested samples were run on SDS/PAGE and visualized as described for the E3 ligase activity assays.
To detect the WT IpaH FL∼Ub intermediates in Ub charging and discharging reactions, conditions were optimized by preparing an initial pool of Ub-charged E1 and E2 (mixing 3.25 µM fluorescent Ub, 0.25 µM E1, and 2 µM E2 in adjusted reaction buffer containing 8.5 mM Hepes pH 7.5, 200 mM NaCl, 2 mM MgCl2, 1 mM ATP pH∼7, and 1 mM TCEP) at 25 °C for 12 min. Further Ub charging was terminated by the addition of EDTA to a final concentration of 30 mM, after which the reaction mixture was cooled on ice for 2 min. Charging was initiated by the addition of E3 enzyme to a concentration of 0.5 µM. Aliquots at the indicated subsequent time points were analyzed by SDS/PAGE.
To follow the discharge of Ub from C337S mutants of IpaH9.8 FL or NEL domain enzymes, an initial pool of Ub-charged E3∼Ub intermediate was prepared by mixing 3.25 µM fluorescent Ub, 0.5 µM E1, 2 µM E2, and 2 µM E3 in charging buffer (8.5 mM Hepes pH 7.5, 200 mM NaCl, 2 mM MgCl2, 1 mM ATP pH∼7, and 1 mM TCEP) at 25 °C for 3 h. Discharge was initiated by the addition of EDTA to a final concentration of 30 mM and DTT to a final concentration of 15 mM. Aliquots at the indicated subsequent time points were analyzed by SDS/PAGE.
In-Cell Experiment for Analyzing E2∼Ub Levels.
HEK-293 cells were seeded into six-well plates and grown to 80% confluence in complete medium [DMEM, high glucose, supplemented with 10% (vol/vol) FBS; Invitrogen]. Before transfection, the medium was changed to Opti-MEM (1 mL per well) for 2.5 h before the transfection reagent was added. The transfection reagent was prepared according to the manufacturer’s protocols using 5 µL Lipofectamine 3000 (Invitrogen) and 2.5 mg of total DNA per well. Cells were incubated for 4 h after transfection, after which the medium was replaced with 2 mL of complete medium. Lysates were prepared at 2 d after transfection in 100 µL of lysis buffer [50 mM TrisCl pH 7.5, 150 mM NaCl, 1% Nonidet P-40, 0.1% SDS, 1 mM EDTA, 10 mM N-ethylmaleimide, 1 mM PMSF, and 1× cOmplete EDTA-free inhibitor mixture (Roche)] per well, scraping cells and nutating at 4 °C for 10 min, followed by centrifugation for 10 min (20,800 × g) and harvesting of supernatant. Total protein was quantified by Bradford’s reagent, and equivalent total protein was loaded per lane using 6× loading dye supplemented with or without 1.35 M β-mercaptoethanol. Samples were run over a neutral BIS-Tris pH 6.5 SDS/PAGE gel in neutral running buffer (50 mM Mes, 50 mM Tris base, 0.5% SDS, and 5 mM EDTA) before being transferred by a semidry method onto PVDF (20 V, 35 min). PVDF membranes were blocked with 5% (wt/vol) BSA in PBST buffer for 2 h at 4 °C before blotting with primary antibodies in 3% (wt/vol) BSA overnight.
The primary antibodies used in this work were anti-UBCH5 (Santa Cruz Biotechnology; C-6, 1:1,000 dilution) and anti-Flag (Sigma-Aldrich; F3165, 1:2,500 dilution). Secondary goat anti-mouse HRP (Invitrogen; 1:9,000 dilution) was blotted in 3% (wt/vol) BSA for 3 h at 4 °C. Western blots were visualized with Clarity Western ECL reagent (Bio-Rad) on Amersham hyperfilm (GE Healthcare).
Acknowledgments
This work was supported by Canadian Institutes of Health Research Grants FDN-143277 and MOP-57795 (to F.S.) and by Natural Sciences and Engineering Research Council Postgraduate Scholarship PGSD3-426371-2012 (to A.F.A.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1611595114/-/DCSupplemental.
References
- 1.Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81(1):203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 2.Yau R, Rape M. The increasing complexity of the ubiquitin code. Nat Cell Biol. 2016;18(6):579–586. doi: 10.1038/ncb3358. [DOI] [PubMed] [Google Scholar]
- 3.Pruneda JN, et al. Structure of an E3:E2∼Ub complex reveals an allosteric mechanism shared among RING/U-box ligases. Mol Cell. 2012;47(6):933–942. doi: 10.1016/j.molcel.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Streich FC, Jr, Lima CD. Structural and functional insights to ubiquitin-like protein conjugation. Annu Rev Biophys. 2014;43:357–379. doi: 10.1146/annurev-biophys-051013-022958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berndsen CE, Wolberger C. New insights into ubiquitin E3 ligase mechanism. Nat Struct Mol Biol. 2014;21(4):301–307. doi: 10.1038/nsmb.2780. [DOI] [PubMed] [Google Scholar]
- 6.Popovic D, Vucic D, Dikic I. Ubiquitination in disease pathogenesis and treatment. Nat Med. 2014;20(11):1242–1253. doi: 10.1038/nm.3739. [DOI] [PubMed] [Google Scholar]
- 7.Buetow L, et al. Activation of a primed RING E3-E2-ubiquitin complex by non-covalent ubiquitin. Mol Cell. 2015;58(2):297–310. doi: 10.1016/j.molcel.2015.02.017. [DOI] [PubMed] [Google Scholar]
- 8.Park Y, Jin H-S, Aki D, Lee J, Liu Y-C. The ubiquitin system in immune regulation. Adv Immunol. 2014;124:17–66. doi: 10.1016/B978-0-12-800147-9.00002-9. [DOI] [PubMed] [Google Scholar]
- 9.Fritah S, et al. Sumoylation controls host anti-bacterial response to the gut invasive pathogen Shigella flexneri. EMBO Rep. 2014;15(9):965–972. doi: 10.15252/embr.201338386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tanner K, Brzovic P, Rohde JR. The bacterial pathogen-ubiquitin interface: Lessons learned from Shigella. Cell Microbiol. 2015;17(1):35–44. doi: 10.1111/cmi.12390. [DOI] [PubMed] [Google Scholar]
- 11.Grishin AM, et al. Structural basis for the inhibition of host protein ubiquitination by Shigella effector kinase OspG. Structure. 2014;22(6):878–888. doi: 10.1016/j.str.2014.04.010. [DOI] [PubMed] [Google Scholar]
- 12.Pruneda JN, et al. E2∼Ub conjugates regulate the kinase activity of Shigella effector OspG during pathogenesis. EMBO J. 2014;33(5):437–449. doi: 10.1002/embj.201386386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanada T, et al. The Shigella flexneri effector OspI deamidates UBC13 to dampen the inflammatory response. Nature. 2012;483(7391):623–626. doi: 10.1038/nature10894. [DOI] [PubMed] [Google Scholar]
- 14.Zhang L, et al. Cysteine methylation disrupts ubiquitin-chain sensing in NF-κB activation. Nature. 2011;481(7380):204–208. doi: 10.1038/nature10690. [DOI] [PubMed] [Google Scholar]
- 15.Diao J, Zhang Y, Huibregtse JM, Zhou D, Chen J. Crystal structure of SopA, a Salmonella effector protein mimicking a eukaryotic ubiquitin ligase. Nat Struct Mol Biol. 2008;15(1):65–70. doi: 10.1038/nsmb1346. [DOI] [PubMed] [Google Scholar]
- 16.Janjusevic R, Abramovitch RB, Martin GB, Stebbins CE. A bacterial inhibitor of host programmed cell death defenses is an E3 ubiquitin ligase. Science. 2006;311(5758):222–226. doi: 10.1126/science.1120131. [DOI] [PubMed] [Google Scholar]
- 17.Rohde JR, Breitkreutz A, Chenal A, Sansonetti PJ, Parsot C. Type III secretion effectors of the IpaH family are E3 ubiquitin ligases. Cell Host Microbe. 2007;1(1):77–83. doi: 10.1016/j.chom.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 18.Ashida H, Sasakawa C. Shigella IpaH family effectors as a versatile model for studying pathogenic bacteria. Front Cell Infect Microbiol. 2016;5:100. doi: 10.3389/fcimb.2015.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suzuki S, et al. Shigella IpaH7.8 E3 ubiquitin ligase targets glomulin and activates inflammasomes to demolish macrophages. Proc Natl Acad Sci USA. 2014;111(40):E4254–E4263. doi: 10.1073/pnas.1324021111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ashida H, et al. A bacterial E3 ubiquitin ligase IpaH9.8 targets NEMO/IKKgamma to dampen the host NF-kappaB-mediated inflammatory response. Nat Cell Biol. 2010;12(1):66–73, 1–9. doi: 10.1038/ncb2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu Y, et al. Structure of a Shigella effector reveals a new class of ubiquitin ligases. Nat Struct Mol Biol. 2008;15(12):1302–1308. doi: 10.1038/nsmb.1517. [DOI] [PubMed] [Google Scholar]
- 22.Singer AU, et al. Structure of the Shigella T3SS effector IpaH defines a new class of E3 ubiquitin ligases. Nat Struct Mol Biol. 2008;15(12):1293–1301. doi: 10.1038/nsmb.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quezada CM, Hicks SW, Galán JE, Stebbins CE. A family of Salmonella virulence factors functions as a distinct class of autoregulated E3 ubiquitin ligases. Proc Natl Acad Sci USA. 2009;106(12):4864–4869. doi: 10.1073/pnas.0811058106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chou Y-C, Keszei AFA, Rohde JR, Tyers M, Sicheri F. Conserved structural mechanisms for autoinhibition in IpaH ubiquitin ligases. J Biol Chem. 2012;287(1):268–275. doi: 10.1074/jbc.M111.316265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keszei AFA, et al. Structure of an SspH1-PKN1 complex reveals the basis for host substrate recognition and mechanism of activation for a bacterial E3 ubiquitin ligase. Mol Cell Biol. 2014;34(3):362–373. doi: 10.1128/MCB.01360-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zouhir S, et al. The structure of the Slrp-Trx1 complex sheds light on the autoinhibition mechanism of the type III secretion system effectors of the NEL family. Biochem J. 2014;464(1):135–144. doi: 10.1042/BJ20140587. [DOI] [PubMed] [Google Scholar]
- 27.Plechanovová A, Jaffray EG, Tatham MH, Naismith JH, Hay RT. Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature. 2012;489(7414):115–120. doi: 10.1038/nature11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dou H, Buetow L, Sibbet GJ, Cameron K, Huang DT. BIRC7-E2 ubiquitin conjugate structure reveals the mechanism of ubiquitin transfer by a RING dimer. Nat Struct Mol Biol. 2012;19(9):876–883. doi: 10.1038/nsmb.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kamadurai HB, et al. Mechanism of ubiquitin ligation and lysine prioritization by a HECT E3. eLife. 2013;2:e00828. doi: 10.7554/eLife.00828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stieglitz B, et al. Structural basis for ligase-specific conjugation of linear ubiquitin chains by HOIP. Nature. 2013;503(7476):422–426. doi: 10.1038/nature12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dou H, Buetow L, Sibbet GJ, Cameron K, Huang DT. Essentiality of a non-RING element in priming donor ubiquitin for catalysis by a monomeric E3. Nat Struct Mol Biol. 2013;20(8):982–986. doi: 10.1038/nsmb.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kamadurai HB, et al. Insights into ubiquitin transfer cascades from a structure of a UbcH5B approximately ubiquitin-HECTNEDD4L complex. Mol Cell. 2009;36(6):1095–1102. doi: 10.1016/j.molcel.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wiesner S, et al. Autoinhibition of the HECT-type ubiquitin ligase Smurf2 through its C2 domain. Cell. 2007;130(4):651–662. doi: 10.1016/j.cell.2007.06.050. [DOI] [PubMed] [Google Scholar]
- 34.Mari S, et al. Structural and functional framework for the autoinhibition of Nedd4-family ubiquitin ligases. Structure. 2014;22(11):1639–1649. doi: 10.1016/j.str.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 35.World Health Organization . Antimicrobial Resistance: Global Report on Surveillance. World Health Organization; Geveva, Switzerland: 2014. [Google Scholar]
- 36.Bassetti M, Merelli M, Temperoni C, Astilean A. New antibiotics for bad bugs: Where are we? Ann Clin Microbiol Antimicrob. 2013;12:22. doi: 10.1186/1476-0711-12-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Allen RC, Popat R, Diggle SP, Brown SP. Targeting virulence: Can we make evolution-proof drugs? Nat Rev Microbiol. 2014;12(4):300–308. doi: 10.1038/nrmicro3232. [DOI] [PubMed] [Google Scholar]
- 38.Dove KK, Stieglitz B, Duncan ED, Rittinger K, Klevit RE. Molecular insights into RBR E3 ligase ubiquitin transfer mechanisms. EMBO Rep. 2016;17(8):1221–1235. doi: 10.15252/embr.201642641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lechtenberg BC, et al. Structure of a HOIP/E2∼ubiquitin complex reveals RBR E3 ligase mechanism and regulation. Nature. 2016;529(7587):546–550. doi: 10.1038/nature16511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maspero E, et al. Structure of a ubiquitin-loaded HECT ligase reveals the molecular basis for catalytic priming. Nat Struct Mol Biol. 2013;20(6):696–701. doi: 10.1038/nsmb.2566. [DOI] [PubMed] [Google Scholar]
- 41.Levin I, et al. Identification of an unconventional E3 binding surface on the UbcH5∼Ub conjugate recognized by a pathogenic bacterial E3 ligase. Proc Natl Acad Sci USA. 2010;107(7):2848–2853. doi: 10.1073/pnas.0914821107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Read MA, et al. Nedd8 modification of cul-1 activates SCF(beta(TrCP))-dependent ubiquitination of IkappaBalpha. Mol Cell Biol. 2000;20(7):2326–2333. doi: 10.1128/mcb.20.7.2326-2333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Duda DM, et al. Structure of HHARI, a RING-IBR-RING ubiquitin ligase: autoinhibition of an Ariadne-family E3 and insights into ligation mechanism. Structure. 2013;21(6):1030–1041. doi: 10.1016/j.str.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang J, Brumell JH. Bacteria-autophagy interplay: A battle for survival. Nat Rev Microbiol. 2014;12(2):101–114. doi: 10.1038/nrmicro3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Toyotome T, et al. Shigella protein IpaH(9.8) is secreted from bacteria within mammalian cells and transported to the nucleus. J Biol Chem. 2001;276(34):32071–32079. doi: 10.1074/jbc.M101882200. [DOI] [PubMed] [Google Scholar]
- 46.Haraga A, Miller SI. A Salmonella enterica serovar typhimurium translocated leucine-rich repeat effector protein inhibits NF-κ B-dependent gene expression. Infect Immun. 2003;71(7):4052–4058. doi: 10.1128/IAI.71.7.4052-4058.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang H, et al. E2 enzyme inhibition by stabilization of a low-affinity interface with ubiquitin. Nat Chem Biol. 2014;10(2):156–163. doi: 10.1038/nchembio.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ogunjimi AA, et al. Regulation of Smurf2 ubiquitin ligase activity by anchoring the E2 to the HECT domain. Mol Cell. 2005;19(3):297–308. doi: 10.1016/j.molcel.2005.06.028. [DOI] [PubMed] [Google Scholar]
- 49.Bond CS, Schüttelkopf AW. ALINE: A WYSIWYG protein-sequence alignment editor for publication-quality alignments. Acta Crystallogr D Biol Crystallogr. 2009;65(Pt 5):510–512. doi: 10.1107/S0907444909007835. [DOI] [PubMed] [Google Scholar]
- 50.Sievers F, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pettersen EF, et al. UCSF Chimera: A visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]