Significance
Phenotypic plasticity is common in nature, yet the mechanisms by which environmental inputs are detected and alter developmental programs are not well understood. Even less explored are the mechanisms underlying transgenerational plasticity. Here we address the problem of how asexual aphids produce winged or wingless offspring in response to external cues. We find that gene expression patterns suggest the importance of the hormone ecdysone in this polyphenism. We show that aphids injected with ecdysone or its analog produce fewer winged offspring, and aphids with inhibited ecdysone signaling produce more winged offspring. Our results provide insight into the hormonal control of alternative aphid morphs, which have long been a textbook example of polyphenism, but whose control mechanisms have long proven elusive.
Keywords: wing polyphenism, phenotypic plasticity, pea aphid, Acyrthosiphon pisum, ecdysone signaling
Abstract
The wing polyphenism of pea aphids is a compelling laboratory model with which to study the molecular mechanisms underlying phenotypic plasticity. In this polyphenism, environmental stressors such as high aphid density cause asexual, viviparous adult female aphids to alter the developmental fate of their embryos from wingless to winged morphs. This polyphenism is transgenerational, in that the pea aphid mother experiences the environmental signals, but it is her offspring that are affected. Previous research suggested that the steroid hormone ecdysone may play a role in this polyphenism. Here, we analyzed ecdysone-related gene expression patterns and found that they were consistent with a down-regulation of the ecdysone pathway being involved in the production of winged offspring. We therefore predicted that reduced ecdysone signaling would result in more winged offspring. Experimental injections of ecdysone or its analog resulted in a decreased production of winged offspring. Conversely, interfering with ecdysone signaling using an ecdysone receptor antagonist or knocking down the ecdysone receptor gene with RNAi resulted in an increased production of winged offspring. Our results are therefore consistent with the idea that ecdysone plays a causative role in the regulation of the proportion of winged offspring produced in response to crowding in this polyphenism. Our results also show that an environmentally regulated maternal hormone can mediate phenotype production in the next generation, as well as provide significant insight into the molecular mechanisms underlying the functioning of transgenerational phenotypic plasticity.
One of the clearest examples of the effects of organism–environment interactions is phenotypic plasticity, where a single genotype can give rise to multiple phenotypes, depending on environmental cues (1). Polyphenism is the most extreme form of phenotypic plasticity, with only a few discontinuous phenotypes produced. Despite considerable interest, known control mechanisms of polyphenic development remain limited to a handful of taxa (e.g., refs. 2–6) in which environmental factors act directly on the individuals that go on to display the corresponding phenotype. In contrast, aphids exhibit a transgenerational wing polyphenism in which external cues act maternally and are transmitted to offspring, which are the individuals that develop into the alternative winged and wingless morphs. Wing polyphenism in aphids has served as a powerful evolutionary model for the study of trade-offs between dispersal and reproduction (7–9), but the molecular mechanisms underlying the induction of winged vs. wingless aphids remain unknown. More generally, little is known about the molecular mechanisms that control transgenerational polyphenism. The goal of our study was to illuminate these mechanisms.
The aphid wing polyphenism occurs during the asexual portion of the aphid life cycle. Wingless asexual females parthenogenetically and viviparously produce genetically identical wingless and winged offspring. Winged aphids are produced in response to stressful environmental conditions such as crowding, poor food quality, or the presence of predators (10). The overall success of a particular aphid genotype likely depends on a balance between the high fecundity of wingless morphs and the superior dispersal ability of the winged morph. Wing morph determination in pea aphids occurs prenatally, during embryogenesis (11). Only during this sensitive period can embryos receive the maternal signal that determines the wing phenotype. Once born, a nymph’s developmental trajectory, and thus its adult phenotype, is set.
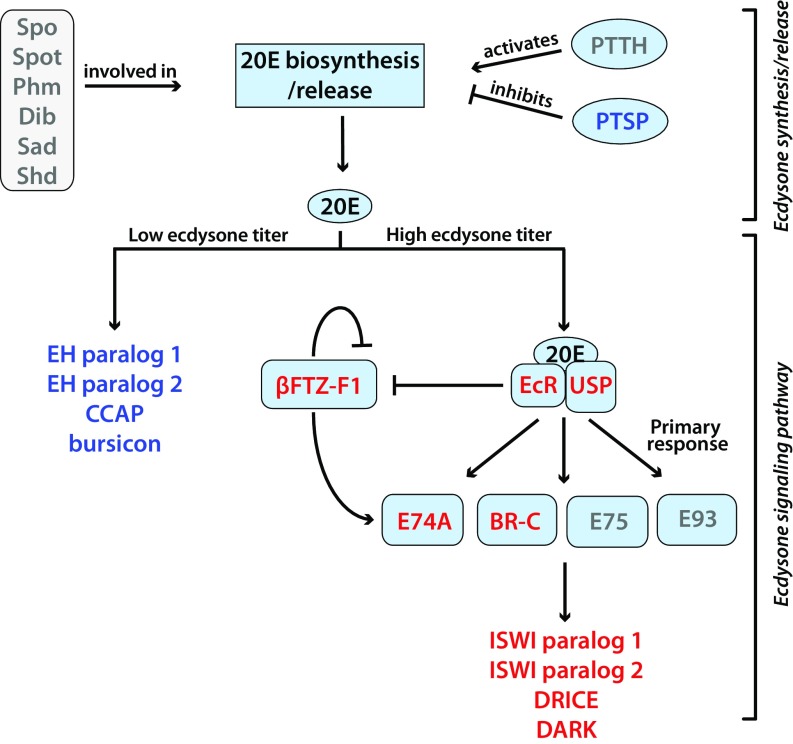
We previously performed a genome-wide transcriptional profiling study on pea aphids (Acyrthosiphon pisum) during prenatal wing determination to identify the key molecular pathways involved in this process (12). We found that the genes differentially expressed between aphid mothers producing winged or wingless progeny suggested that ecdysone signaling may play a role in controlling wing morph determination. For example, the prothoracicostatic peptide gene, whose gene product inhibits ecdysone synthesis (13), was at higher levels in females producing winged offspring. This indicated that ecdysone levels were lower in these females. Consistent with this inference, genes that are activated in response to dropping ecdysone levels [eclosion hormone, crustacean cardioactive peptide, bursicon (14)] were at higher levels in female adults producing winged offspring. In contrast, genes that are ecdysone inducible, or are coactivators of the ecdysone receptor (15–17), were at higher levels in adult females producing wingless offspring. These include the two genes whose products form the heterodimeric ecdysone receptor (EcR and ultraspiracle), along with other well-known ecdysone-inducible genes such as broad and ftz-f1. Combined, these data showed that genes associated with ecdysone signaling were differentially expressed in aphid mothers that were induced to produce winged offspring compared with their uninduced counterparts (summarized in Fig. S1; gene identities listed in Table S1).
Fig. S1.
Expression of ecdysone-associated genes correlates with offspring phenotypes. Genes associated with key aspects of 20-OH ecdysone (20E) synthesis and signaling are shown. Gene names in red are those that are at significantly higher levels in wingless offspring producing females, blue are those that are at higher levels in winged offspring producing females, and gray genes are those that were not differentially expressed. βFtz-f1, fushi tarazu 1; BR-C, broad-complex core protein isoform; DARK, apoptotic protease-activating factor 1; Dib, disembodied; DRICE = caspase-1; EcR, ecdysone receptor; EH Paralog 1&2, eclosion hormone paralog1&2; ISWI, ISWI chromatin remodeling complex; Phm, phantom; PTSP, prothoracicostatic peptide; Sad, shadow; Shd, shade; Spo, spook; Spot, Spookiest; Usp, ultraspiracle.
Table S1.
Ecdysone-related genes with significant differential expression between winged offspring producing (induced) and wingless offspring producing (uninduced) pea aphid mothers
| Gene | Putative identity | Relation to ecdysone | P value* | Fold change & direction† |
| ACYPI085492 | Eclosion hormone (EH, paralog 2) | Activated by dropping ecdysone levels (14) | 0.0001 | 2.9× up in induced |
| ACYPI062598 | Crustacean cardioactive peptide (CCAP) | Activated by dropping ecdysone levels (14) | 0.0003 | 2.2× up in induced |
| ACYPI42083 | Eclosion hormone (EH, paralog 1) | Activated by dropping ecdysone levels (14) | 0.0004 | 2.6× up in induced |
| ACYPI005281 | Bursicon | Activated by dropping ecdysone levels (14) | 0.01 | 1.9× up in induced |
| ACYPI007510 | Prothoracicostatic peptide (PTSP) | Inhibits ecdysone release (13) | 0.01 | 1.9× up in induced |
| ACYPI001692 | Ecdysone receptor (EcR) | Ecdysone receptor (15) | 0.0001 | 3.3× up in uninduced |
| ACYPI005934 | Ultraspiracle protein (Usp) | 2.2× up in uninduced | ||
| ACYPI004047 | Chromatin-remodeling complex atpase chain ISWI (paralog 2) | Coactivator of EcR (53) | 0.001 | 2.5× up in uninduced |
| ACYPI40836 | Broad-complex core protein isoform | Ecdysone-inducible, primary response gene (16) | 0.009 | 3.0× up in uninduced |
| ACYPI083237 | Nuclear hormone receptor ftz-f1 | Coactivator of EcR (17) | 0.01 | 3.4× up in uninduced |
| ACYPI001312 | Chromatin-remodeling complex atpase chain ISWI (paralog 1) | Coactivator of EcR (53) | 0.046 | 3.7× up in uninduced |
| ACYPI25933 | E74a | Ecdysone-inducible, primary response gene (54) | 0.0001 | 2.1× up in uninduced |
| ACYPI49157 | DARK (apoptotic protease-activating factor 1) | Ecdysone inducible (55) | 0.0003 | 2.6× up in uninduced |
| ACYPI003094 | DRICE (Caspase-1) | Ecdysone inducible (55) | 0.048 | 1.7× up in uninduced |
| ACYPI007773 | E75 | Ecdysone-inducible, primary response gene (56) | ns | |
| ACYPI002959 | E93 | Ecdysone-inducible, primary response gene (57) | ns | |
| ACYPI37989 | Prothoracicotropic hormone (PTTH) | Activates ecdysone release (58) | ns | |
| ACYPI001519 | Spook (Spo) | Ecdysone biosynthesis (59) | ns | |
| ACYPI000716 | Spookiest (Spot) | Ecdysone biosynthesis (59) | ns | |
| ACYPI006623 | Phantom (Phm) | Ecdysone biosynthesis (59) | ns | |
| ACYPI006729 | Disembodied (Dib) | Ecdysone biosynthesis (59) | ns | |
| ACYPI000973 | Shadow (Sad) | Ecdysone biosynthesis (59) | ns | |
| ACYPI008228 | Shade (Shd) | Ecdysone biosynthesis (59) | ns |
P value from RNA-Seq experiment (12), false discovery rate corrected.
Bold genes exhibited higher expression in aphids induced to produce winged offspring, underlined genes had higher expression in uninduced aphids, and the remaining genes were not significantly differentially expressed.
Ecdysone (and its bioactive derivative 20-OH ecdysone) is a well-studied insect steroid hormone, primarily known for its role in molting and metamorphosis (18). Ecdysone binds to its nuclear receptor, the ecdysone receptor, a heterodimer comprising the EcR protein and the Ultraspiracle protein (15). Upon association with its hormone, the receptor binds cis-regulatory DNA (19) elements and recruits cofactors to regulate transcription (20–23). Ecdysone-mediated signaling therefore has the ability to mediate the expression of the many phenotypes that differ between the two morphs by acting as a transcriptional modulator. Ecdysone-mediated control of seasonal morphs has been described before in the butterflies Araschnia levana, Precis coenia, and Bicyclus anynana (24–26), and the ecdysone receptor gene product (EcR) mediates the quantitative differences in plasticity between fore and hindwings in Bicyclus anynana (27). Here we demonstrate a causative role for ecdysone signaling in the transgenerational pea aphid wing polyphenism. This study describes a role for ecdysone in polyphenism outside of the Lepidoptera.
Results and Discussion
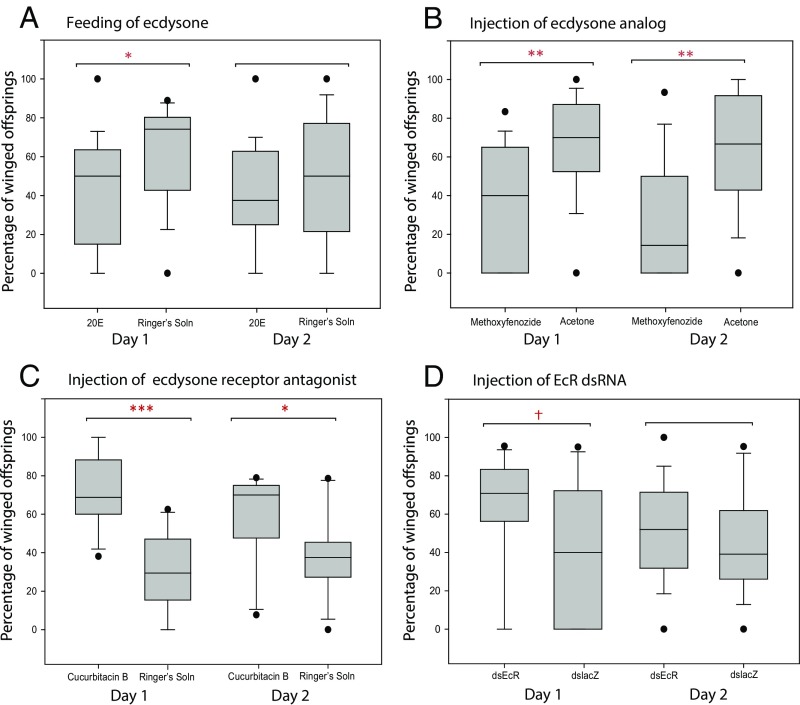
To determine whether ecdysone’s role in the aphid wing polyphenism was correlative or causative, we tested whether ecdysone application affected winged offspring production. On the basis of the direction of differential expression of ecdysone-related genes from our previous study (12) (Table S1 and Fig. S1), we hypothesized that higher levels of ecdysone would result in lower percentages of winged offspring. To test this, we raised aphids at moderate density (20 per plant) so that they would produce both winged and wingless offspring. After reaching adulthood, aphids were placed on an artificial liquid diet (diet “A” of ref. 28) containing 250 µg/mL 20-hydroxyecdysone (20E) or on a control without 20E for 24 h. As anticipated, 20E-treated females produced a significantly lower proportion of winged offspring compared with control aphids when ingesting 20E [Fig. 1A, day 1; Mann–Whitney U test (MW) test, offspring counted from n = 18 groups of three adult aphids for treatment, n = 19 groups of three aphids for control; P = 0.014]. This effect dissipated after the aphids were returned to plants posttreatment; offspring produced in the second 24 h did not have significantly different percentages of winged offspring (Fig. 1A, day 2; MW, n = 18 groups of three aphids for treatment, n = 19 groups of three aphids for control; P = 0.197).
Fig. 1.
The addition of ecdysone causes the production of fewer winged offspring, whereas interference with ecdysone receptor activity causes the production of more winged offspring. (A) Adult, wingless aphids were fed on artificial media containing 20E or control media without 20E (n = 18 sets of three aphids for the treatment, n = 19 for the control), or (B) were injected with methoxyfenozide dissolved in acetone or acetone only (n = 20 sets of three aphids for the treatment, n = 19 for the control). (C) Adult, wingless aphids were injected with cucurbitacin B dissolved in Ringer’s solution or a Ringer’s solution control (n = 15 sets of three aphids each for treatment and control), or (D) with dsRNA against EcR or EcR compared with control aphids (n = 15 sets of three aphids each for treatment and control). Each data point is the percentage of winged offspring produced by sets of three aphids. Boxes show the interquartile range, and the line is the median value of each group. Black circles are outliers. Significant differences between treatments are represented by asterisks: †P < 0.10; *P < 0.05; ***P < 0.001.
To corroborate that ecdysone could cause a shift in the wing polyphenism response, we also treated the aphids with an ecdysone agonist, methoxyfenozide (RH-2485) (19). Because methoxyfenozide is not water soluble, we could not administer it via feeding. Instead, we used abdominal injections. We again raised aphids at moderate density so that they produced offspring of both phenotypes. Note that raising aphids at moderate density does not control the exact proportion of winged and wingless offspring they produce, so relevant comparisons are within experiments (treatment vs. control for a particular compound), rather than between experiments. As with the 20E treatment, methoxyfenozide-treated aphids produced a significantly lower proportion of winged offspring than controls. This effect persisted, strongly, for 2 d (Fig. 1B; MW, n = 20 groups of three aphids for treatment, n = 19 groups of three aphids for control; P = 0.008, day 1; P = 0.003, day 2). We conclude that the addition of ecdysone or its analog can affect the production of winged vs. wingless offspring.
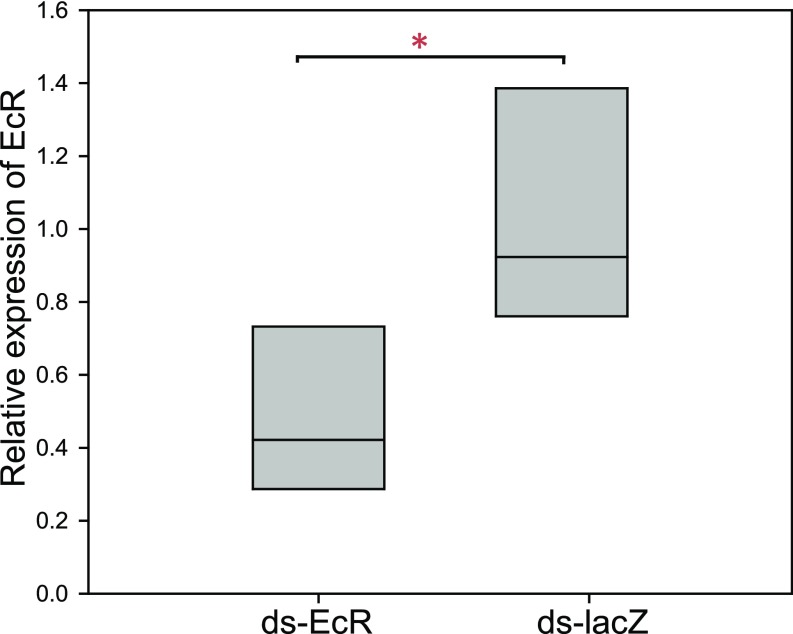
To further investigate the role of ecdysone in the pea aphid wing polyphenism, we reasoned that reduced sensitivity to circulating ecdysone might be sufficient to cause adult females to produce more winged offspring, the opposite of the outcomes observed above. We thus interfered with ecdysone signaling activity, using cucurbitacin B, an ecdysone receptor antagonist that prevents the binding of ecdysone (29). This compound has previously been used in butterflies to inhibit ecdysone signaling (27). As expected, treatment with cucurbitacin B caused a significantly higher production of winged offspring over the 2 dd posttreatment (Fig. 1C; MW, n = 15 groups of three aphids; P < 0.001, day 1; P = 0.036, day 2). We also interfered with ecdysone signaling activity, using EcR RNAi. Double-stranded EcR RNA injected into wingless aphid adults raised at moderate densities resulted in a slightly higher percentage of winged offspring, although the effect was not significant at the P < 0.05 level (Fig. 1D; MW, n = 15 groups of three aphids; P = 0.092 day 1; P = 0.237 day 2). As with other dsRNA studies in aphids (e.g., ref. 30), and indeed many other organisms, we were only able to achieve partial and variable knockdown of the transcript (∼50% knockdown 24 h after injection; Fig. S2). The comparative weakness of these results likely reflects this partial knockdown, as well as autoregulation of the EcR gene (31). Importantly, however, the trend on day 1 mirrors the cucurbitacin B results and supports the idea that decreasing ecdysone signaling affects the offspring phenotypes.
Fig. S2.
Injection with double-stranded RNA against EcR (dsEcR) results in lower relative expression of EcR compared with control aphids (dslacZ). Data shown are the relative expression levels of Ecr in dsEcr and dslacZ injected aphids (n = 5 sets of three aphids each for treatment and control; each data point consists of RNA collected from three injected aphids). Boxes are the interquartile range, and the line is the median value of each group. *P < 0.05.
Treatments also had an effect on offspring production. For 20E ingestion, the number of nymphs produced per female was significantly lower in treated vs. control aphids in the first 24 h, an effect that was no longer present in the second 24 h (MW, offspring counted from n = 18 groups of three adult aphids for treatment, n = 19 for control; P < 0.001 day 1; P = 0.274 day 2). Similarly, injection with the ecdysone analog, methoxyfenozide, significantly repressed the number of offspring produced by the mothers on both days (MW, n = 20; P = 0.025, day 1; P = 0.038, day 2). Conversely, treatment with the ecdysone receptor antagonist, cucurbitacin B, resulted in a significantly higher number of total offspring relative to controls (MW, n = 15; P = 0.003, day 1; P < 0.001, day 2), and the number of offspring was higher in the EcR dsRNA samples on day 1, although not significantly higher; no differences were seen on day 2 (MW, n = 15; P = 0.118 day 1; P = 0.272 day 2). The feeding of ecdysone or ecdysone analogs inhibits ovarian maturation and egg production in houseflies and other insects (18, 32), so this repressive effect on offspring production in the first two experiments is likely mediated by the excess ecdysone, with the opposite effect in the latter two experiments. This expected reproductive effect is an independent indication that the aphids are, in fact, sensing the ecdysone manipulations.
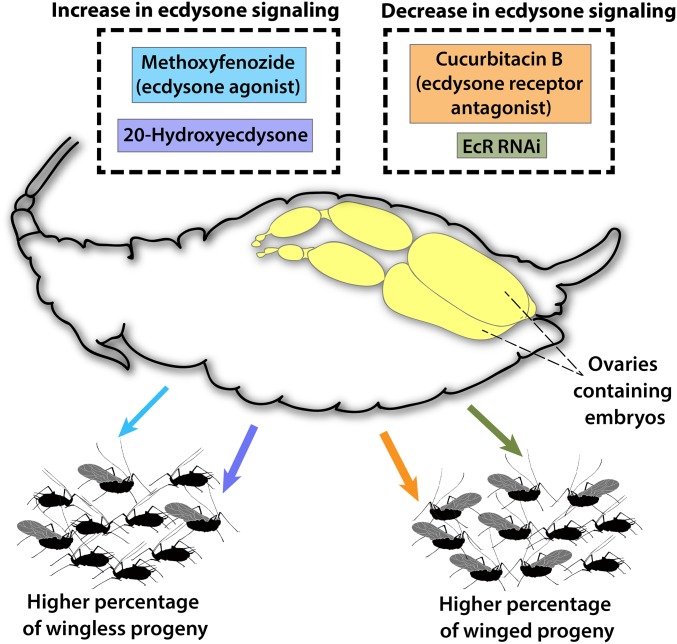
In summary, using transcript level differences and functional studies, we have shown that ecdysone-mediated signaling is a critical pathway underlying the transgenerational pea aphid wing polyphenism. Treatment with ecdysone or an analog resulted in higher proportions of wingless offspring, whereas treatment with an ecdysone receptor antagonist or EcR RNAi resulted in more winged offspring (Fig. 2). Our results suggest that higher aphid densities result in lower levels of ecdysone pathway signaling in aphid mothers, which causes embryos in their ovaries to be fated to become winged. In adult insects generally, including in aphids, the primary organ that secretes ecdysone is the ovaries (33, 34). Ecdysone released by the mother’s ovaries could directly signal the embryos, which could then respond by changing their developmental program. This hypothesized mechanism could connect environmental perception by the mother with a phenotypic response by the offspring.
Fig. 2.
Summary of ecdysone signaling manipulation experiments. All treatments, indicated along the top, were applied to adult wingless females. The relative effect of each treatment (i.e., the shift in the median of the experimental group relative to the control) is represented by the width of arrows shown along the bottom, and the direction of morph shift in the form of the offspring phenotypes. Arrows correspond to the treatments, coded by different colors. Treatments that manipulated ecdysone signaling in a similar manner are represented within each dotted box.
Our results provide significant insight into the molecular determination of the aphid wing polyphenism, which has long been a textbook example of transgenerational polyphenism, but whose control mechanisms up until now have proven elusive (35, 36). This study provides strong support for a causative factor in this process of prenatal morph determination. Most previous molecular studies of the aphid wing polyphenism profiled gene expression changes far downstream of morph determination, such as differences between winged and wingless adults (37–39; although, see ref. 40); these studies examined the transcriptional basis of morph development and function, but not morph determination. Previous physiological studies primarily focused on the possible role of another major insect hormone, juvenile hormone, with the hypothesis that a wingless adult is a juvenilized winged adult. A role for juvenile hormone in the wing polyphenism is not ruled out by this study, especially given the complex crosstalk that can occur between the two hormone signaling pathways (41–43), but to date, no definitive role for juvenile hormone has emerged (7, 35, 44), although it probably functions in the process of wing development in some aphid species (45, 46).
Polyphenisms vary greatly in the timing of when development diverges. Earlier determination allows morphs to have more significant changes in morphology, and thus become more fine-tuned to their morph’s function, whereas later determination ensures the correct matching of morph to the environment (7, 47). For the pea aphid wing polyphenism, morph determination is embryonic and maternally mediated, with the resulting adult morphs highly differentiated from one another; winged and wingless morphs differ not only in the complete presence or absence of wings and wing musculature but also in a suite of other morphological, behavioral, and developmental characteristics (reviewed in ref. 35). Other polyphenisms are determined later in development, are intragenerational, and result in less differentiated forms that may only differ in one particular trait (7). For example, the brown plant hopper (Nilaparvata lugens) produces long-winged and reduced-winged morphs in response to population density and host plant quality, and determination occurs at later nymphal instars (48, 49). The morphs differ primarily in their wing size, and consequently their flight capability. Different developmental divergence times likely require different molecular mechanisms. Insulin receptor signaling, known for its conserved role in growth (50), controls wing morph differences in the plant hopper wing polyphenism, primarily through action in the wing buds (51). In contrast, in the pea aphid wing polyphenism, we would predict that insulin signaling controls differential growth of the two morphs, but that this differential growth is downstream of morph determination. In other words, insulin receptor signaling may not solely be able to control the larger range of traits that differ between winged and wingless pea aphids compared with long-winged and reduced-winged plant hopper morphs. In general, whether or not there are physiological mechanisms that are optimal for earlier relative to later morph determination awaits information from even more polyphenic systems.
Materials and Methods
The pea aphid line used for all experiments in this study was ROC-1, collected from Rochester, New York, in 2008. Aphids were reared in cages on fava (Vicia fabae) seedlings.
Gene Expression Analysis.
We previously performed an RNA-Seq study using RNA collected from adult aphids with ovaries and embryos removed to compare two treatments: females that had been crowded and were known to be producing primarily winged offspring or females that had not been crowded and were known to be producing primarily wingless offspring (12). That global analysis of transcriptional differences revealed enrichment of genes involved in the ecdysone receptor-mediated signaling pathway or hormonal activity, among many other categories. Here we focused specifically on these genes and also included other known ecdysone-related genes we mined from the literature (Table S1). Methods of calculating significant differential expression can be found in ref. 12.
Feeding of 20E Using Artificial Media.
20E (250 µg/mL, Sigma) was resuspended in Ringer’s solution and added to 175 µL artificial media (28). An equal amount of Ringer’s without 20E was added to the control media. This media was placed in film canisters between two layers of Parafilm stretched over one end. Aphids were raised at moderate density, with ∼20 aphids per plant. At day three of adulthood, aphids were randomly placed into the treatment or control canisters in groups of three, for a total of 48 aphids in the treatment group and 51 in the control (n = 18 and n = 19, respectively). After 24 h, nymphs were removed and placed on plants; adults were also placed on plants in groups of three for an additional day of nymph deposition. Nymph phenotypes (winged or wingless) were determined after they reached adulthood.
Injection of Methoxyfenozide and Cucurbitacin B.
For the remaining experiments (pharmaceutical and RNAi injections), aphids were raised at low density (five per fava plant) for several generations. From a low-density stock, sets of 20 first instar aphids were moved onto plants to create a single generation of moderate density. These aphids, as adults, were then removed from plants and randomized as to whether they were used for treatments or controls.
Methoxyfenozide was a gift from S. R. Palli (University of Kentucky) and was resuspended in acetone at a concentration of 100 µg/µL. Cucurbitacin B was obtained from Sigma. Aphids at day three of adulthood were injected in their abdomens with ∼0.2 µL 100 µg/µL methoxyfenozide or an acetone control (60 aphids each), or 0.2 µL 1 µg/µL cucurbitacin B or a Ringer’s solution control (45 aphids each). After injection, aphids were placed in groups of three on plants for 24 h. Injected aphids were moved to new plants after another 24 h to monitor the nymphs produced each day and then killed. Offspring were phenotyped as winged or wingless after maturation.
RNAi Injections and Knockdown Verification.
Using T7 containing primers (TGTACCTGAAGTTCAATGTGCAG and AGCTTCACTTGAGCAAGCCT), 467 bp of EcR (ACYPI001692) was amplified from cDNA. Four hundred sixty-six base pairs EcR, ACYPI001692, was amplified using T7 containing primers (TGTACCTGAAGTTCAATGTGCAG and AGCTTCACTTGAGCAAGCCT) from cDNA. A similar-sized lacZ DNA fragment was a gift from J. H. Werren (University of Rochester). Two micrograms PCR product was used to make double-stranded RNA using the MEGAscript T7 Kit (Ambion), according to manufacturer’s instructions. dsRNA was used at concentrations of 2.3 µg/µL for EcR and 2.7 µg/µL for lacZ. Forty-five aphids were used in each of the treatment and control groups. After injection, aphids were placed on plants in groups of three and moved every 24 h for 3 d of nymph production. EcR mRNA knockdown by RNAi injection was validated by qRT-PCR in a separate set of aphids. For this, adult aphids (15 each for treatment and control) were treated with double-stranded EcR or lacZ RNA (dsEcR or dslacZ) and placed in groups of three on plants. After 24 h, the adults were flash frozen in liquid nitrogen. Total RNA was extracted from five groups of three aphids for each treatment, using the TRIzol Reagent (Ambion), following manufacturer’s instructions. First-strand cDNA was synthesized using iScript cDNA synthesis kit (Bio-Rad) from 1 µg RNA. PCR amplifications were carried out for each set, using iTaq Universal SYBR Green Supermix (Bio-Rad). Along with the EcR gene-specific primers (GTTACCATTACAACGCGCTGA and ACTTCCGCCTCATGTACATGT), primers for NADH dehydrogenase (TTGGTACACTGGTGAAGGTATG and AGCGGTAGCTTCTTGGTATTG) were used as a reference gene to amplify each set of samples because of its stability of expression. Reactions were performed in a CFX96 Touch Real-Time PCR Detection System (Bio-Rad). Each biological replicate was measured with three technical replicates, and fold change was calculated using the 2−ΔΔCT method (52).
Acknowledgments
We gratefully thank Ryan D. Bickel and Tony J. Zera for valuable discussions, Jen Keister for technical assistance, and two reviewers for comments that improved the manuscript. This research was supported by Award R01GM116867 from the National Institute of General Medical Sciences (to J.A.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1617640114/-/DCSupplemental.
References
- 1.Whitman DW, Agrawal AA. What is phenotypic plasticity and why is it important? In: Whitman DW, Ananthakrishnan TN, editors. Phenotypic Plasticity of Insects. CRC Press; Boca Raton, FL: 2009. pp. 1–63. [Google Scholar]
- 2.Anstey ML, Rogers SM, Ott SR, Burrows M, Simpson SJ. Serotonin mediates behavioral gregarization underlying swarm formation in desert locusts. Science. 2009;323(5914):627–630. doi: 10.1126/science.1165939. [DOI] [PubMed] [Google Scholar]
- 3.Kucharski R, Maleszka J, Foret S, Maleszka R. Nutritional control of reproductive status in honeybees via DNA methylation. Science. 2008;319(5871):1827–1830. doi: 10.1126/science.1153069. [DOI] [PubMed] [Google Scholar]
- 4.Kijimoto T, Moczek AP, Andrews J. Diversification of doublesex function underlies morph-, sex-, and species-specific development of beetle horns. Proc Natl Acad Sci USA. 2012;109(50):20526–20531. doi: 10.1073/pnas.1118589109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ragsdale EJ, Müller MR, Rödelsperger C, Sommer RJ. A developmental switch coupled to the evolution of plasticity acts through a sulfatase. Cell. 2013;155(4):922–933. doi: 10.1016/j.cell.2013.09.054. [DOI] [PubMed] [Google Scholar]
- 6.Ma Z, Guo W, Guo X, Wang X, Kang L. Modulation of behavioral phase changes of the migratory locust by the catecholamine metabolic pathway. Proc Natl Acad Sci USA. 2011;108(10):3882–3887. doi: 10.1073/pnas.1015098108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zera AJ, Denno RF. Physiology and ecology of dispersal polymorphism in insects. Annu Rev Entomol. 1997;42:207–230. doi: 10.1146/annurev.ento.42.1.207. [DOI] [PubMed] [Google Scholar]
- 8.Zera AJ, Brisson JA. Quantitative, physiological, and molecular genetics of dispersal/migration. In: Clobert J, Baguette M, Benton T, Bullock J, editors. Dispersal: Causes and Consequences. Oxford Univ. Press; Oxford, United Kingdom: 2012. [Google Scholar]
- 9.Dixon AFG. H.M.T., Dispersal in aphids, a problem in resource allocation. In: Danthanarayana W, editor. Insect Flight: Dispersal and Migration. Springer-Verlag; Berlin, Germany: 1986. pp. 145–151. [Google Scholar]
- 10.Müller CB, Williams IS, Hardie J. The role of nutrition, crowding and interspecific interactions in the development of winged aphids. Ecol Entomol. 2001;26(3):330–340. [Google Scholar]
- 11.Sutherland ORW. The role of crowding in the production of winged forms by two strains of the pea aphid, Acyrthosiphon pisum. J Insect Physiol. 1969;15:1385–1410. [Google Scholar]
- 12.Vellichirammal NN, Madayiputhiya N, Brisson JA. The genomewide transcriptional response underlying the pea aphid wing polyphenism. Mol Ecol. 2016;25(17):4146–4160. doi: 10.1111/mec.13749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hua YJ, et al. Identification of a prothoracicostatic peptide in the larval brain of the silkworm, Bombyx mori. J Biol Chem. 1999;274(44):31169–31173. doi: 10.1074/jbc.274.44.31169. [DOI] [PubMed] [Google Scholar]
- 14.Davis MM, O’Keefe SL, Primrose DA, Hodgetts RB. A neuropeptide hormone cascade controls the precise onset of post-eclosion cuticular tanning in Drosophila melanogaster. Development. 2007;134(24):4395–4404. doi: 10.1242/dev.009902. [DOI] [PubMed] [Google Scholar]
- 15. Riddiford LM, Cherbas P, JW Truman. 2001. Ecdysone receptors and their biological actions. Vitam Horm 60:1–73. [DOI] [PubMed]
- 16.DiBello PR, Withers DA, Bayer CA, Fristrom JW, Guild GM. The Drosophila Broad-Complex encodes a family of related proteins containing zinc fingers. Genetics. 1991;129(2):385–397. doi: 10.1093/genetics/129.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu J, Chen L, Sun G, Raikhel AS. The competence factor beta Ftz-F1 potentiates ecdysone receptor activity via recruiting a p160/SRC coactivator. Mol Cell Biol. 2006;26(24):9402–9412. doi: 10.1128/MCB.01318-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nijhout HF. Insect Hormones. Princeton University Press; Princeton, NJ: 1998. [Google Scholar]
- 19.Ishaaya I, Yablonski S, Horowitz AR. Comparative toxicology of two ecdysteroid agonists, RH-2485 and RH-5992, on susceptible and pyrethroid-resistant strains of the Egyptian cotton leafworm, Spodoptera littoralis. Phytoparasitica. 1995;23(2):139–145. [Google Scholar]
- 20.Bai J, Uehara Y, Montell DJ. Regulation of invasive cell behavior by taiman, a Drosophila protein related to AIB1, a steroid receptor coactivator amplified in breast cancer. Cell. 2000;103(7):1047–1058. doi: 10.1016/s0092-8674(00)00208-7. [DOI] [PubMed] [Google Scholar]
- 21.Sedkov Y, et al. Methylation at lysine 4 of histone H3 in ecdysone-dependent development of Drosophila. Nature. 2003;426(6962):78–83. doi: 10.1038/nature02080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Biddie SC, et al. Transcription factor AP1 potentiates chromatin accessibility and glucocorticoid receptor binding. Mol Cell. 2011;43(1):145–155. doi: 10.1016/j.molcel.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Privalsky ML. The role of corepressors in transcriptional regulation by nuclear hormone receptors. Annu Rev Physiol. 2004;66:315–360. doi: 10.1146/annurev.physiol.66.032802.155556. [DOI] [PubMed] [Google Scholar]
- 24.Koch PB, Buckmann D. Hormonal control of seasonal morphs by the timing of ecdysteroid release in Araschnia levana L. (Nymphalidae: Lepidoptera) J Insect Physiol. 1987;33(11):823–829. [Google Scholar]
- 25.Rountree DB, Nijhout HF. Hormonal control of a seasonal polyphenism in Precis coenia (Lepidoptera: Nymphalidae) J Insect Physiol. 1995;41(11):987–992. [Google Scholar]
- 26.Nijhout HF. Control mechanisms of polyphenic development. Bioscience. 1999;49(3):181–192. [Google Scholar]
- 27.Monteiro A, et al. Differential expression of ecdysone receptor leads to variation in phenotypic plasticity across serial homologs. PLoS Genet. 2015;11(9):e1005529. doi: 10.1371/journal.pgen.1005529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Febvay G, Rahbe Y, Rynkiewicz M, Guillaud J, Bonnot G. Fate of dietary sucrose and neosynthesis of amino acids in the pea aphid, acyrthosiphon pisum, reared on different diets. J Exp Biol. 1999;202(Pt 19):2639–2652. doi: 10.1242/jeb.202.19.2639. [DOI] [PubMed] [Google Scholar]
- 29.Dinan L, et al. Cucurbitacins are insect steroid hormone antagonists acting at the ecdysteroid receptor. Biochem J. 1997;327(Pt 3):643–650. doi: 10.1042/bj3270643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jaubert-Possamai S, et al. Gene knockdown by RNAi in the pea aphid Acyrthosiphon pisum. BMC Biotechnol. 2007;7:63. doi: 10.1186/1472-6750-7-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thummel CS. From embryogenesis to metamorphosis: The regulation and function of Drosophila nuclear receptor superfamily members. Cell. 1995;83(6):871–877. doi: 10.1016/0092-8674(95)90203-1. [DOI] [PubMed] [Google Scholar]
- 32.Robbins WE, et al. Ecdysones and analogs: Effects on development and reproduction of insects. Science. 1968;161(3846):1158–1160. doi: 10.1126/science.161.3846.1158. [DOI] [PubMed] [Google Scholar]
- 33.Johnson B. A histological study of neurosecretion in aphids. J Insect Physiol. 1963;9(5):727–739. [Google Scholar]
- 34.Delbecque JP, Weidner K, Hoffmann KH. Alternative sites for ecdysteroid production in insects. Invertebr Reprod Dev. 1990;18(2):29–42. [Google Scholar]
- 35.Braendle C, Davis GK, Brisson JA, Stern DL. Wing dimorphism in aphids. Heredity (Edinb) 2006;97(3):192–199. doi: 10.1038/sj.hdy.6800863. [DOI] [PubMed] [Google Scholar]
- 36.Zera AJ, Brisson JA. Induction and function of polyphenic morphs: Proximate regulatory mechanisms and evolutionary implications. In: Martin LB, Ghalambor CK, Woods HA, editors. Integrative Organismal Biology. John Wiley & Sons; Hoboken, NJ: 2014. pp. 71–90. [Google Scholar]
- 37. Brisson JA, Davis GK, Stern DL (2007) Brisson JA, Davis GK, Stern DL (2007) Common genome-wide patterns of transcript accumulation underlying the wing polyphenism and polymorphism in the pea aphid (Acyrthosiphon pisum). Evol Dev 9(4):338–346. [DOI] [PubMed]
- 38.Ghanim M, Dombrovsky A, Raccah B, Sherman A. A microarray approach identifies ANT, OS-D and takeout-like genes as differentially regulated in alate and apterous morphs of the green peach aphid Myzus persicae (Sulzer) Insect Biochem Mol Biol. 2006;36(11):857–868. doi: 10.1016/j.ibmb.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 39.Yang X, et al. Gene expression profiling in winged and wingless cotton aphids, Aphis gossypii (Hemiptera: Aphididae) Int J Biol Sci. 2014;10(3):257–267. doi: 10.7150/ijbs.7629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ishikawa A, et al. Screening of upregulated genes induced by high density in the vetch aphid Megoura crassicauda. J Exp Zool A Ecol Genet Physiol. 2012;317(3):194–203. doi: 10.1002/jez.1713. [DOI] [PubMed] [Google Scholar]
- 41.Fang F, Xu Y, Jones D, Jones G. Interactions of ultraspiracle with ecdysone receptor in the transduction of ecdysone- and juvenile hormone-signaling. FEBS J. 2005;272(7):1577–1589. doi: 10.1111/j.1742-4658.2005.04578.x. [DOI] [PubMed] [Google Scholar]
- 42.Jones G, et al. Juvenile hormone action through a defined enhancer motif to modulate ecdysteroid-activation of natural core promoters. Comp Biochem Physiol B Biochem Mol Biol. 2012;161(3):219–225. doi: 10.1016/j.cbpb.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Riddiford LM. How does juvenile hormone control insect metamorphosis and reproduction? Gen Comp Endocrinol. 2012;179(3):477–484. doi: 10.1016/j.ygcen.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 44.Schwartzberg EG, Kunert G, Westerlund SA, Hoffmann KH, Weisser WW. Juvenile hormone titres and winged offspring production do not correlate in the pea aphid, Acyrthosiphon pisum. J Insect Physiol. 2008;54(9):1332–1336. doi: 10.1016/j.jinsphys.2008.04.025. [DOI] [PubMed] [Google Scholar]
- 45.Lees AD. The development of juvenile hormone sensitivity in alatae of the aphid Megoura viciae. J Insect Physiol. 1980;26(2):143–151. [Google Scholar]
- 46.Ishikawa A, Gotoh H, Abe T, Miura T. Juvenile hormone titer and wing-morph differentiation in the vetch aphid Megoura crassicauda. J Insect Physiol. 2013;59(4):444–449. doi: 10.1016/j.jinsphys.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 47.Dixon AFG. Aphid ecology. Chapman & Hall; London, United Kingdom: 1985. [Google Scholar]
- 48.Kisimoto R. Effect of crowding during the larval period on the determination of the wing-form of an adult plant-hopper. Nature. 1956;178:641–642. [Google Scholar]
- 49.Syobu S, Mikuriya H, Yamaguchi J, Matsuzaki M, Matsumura M. Fluctuations and factors affecting the wing-form ratio of the brown planthopper, Nilaparvata lugens Stål in rice fields. Jap J Appl Entomol Zool. 2002;46(3):135–143. [Google Scholar]
- 50.Brogiolo W, et al. An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr Biol. 2001;11(4):213–221. doi: 10.1016/s0960-9822(01)00068-9. [DOI] [PubMed] [Google Scholar]
- 51.Xu H-J, et al. Two insulin receptors determine alternative wing morphs in planthoppers. Nature. 2015;519(7544):464–467. doi: 10.1038/nature14286. [DOI] [PubMed] [Google Scholar]
- 52.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 53.Badenhorst P, et al. The Drosophila nucleosome remodeling factor NURF is required for Ecdysteroid signaling and metamorphosis. Genes Dev. 2005;19(21):2540–2545. doi: 10.1101/gad.1342605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Burtis KC, Thummel CS, Jones CW, Karim FD, Hogness DS. The Drosophila 74EF early puff contains E74, a complex ecdysone-inducible gene that encodes two ets-related proteins. Cell. 1990;61(1):85–99. doi: 10.1016/0092-8674(90)90217-3. [DOI] [PubMed] [Google Scholar]
- 55.Cakouros D, Daish T, Martin D, Baehrecke EH, Kumar S. Ecdysone-induced expression of the caspase DRONC during hormone-dependent programmed cell death in Drosophila is regulated by Broad-Complex. J Cell Biol. 2002;157(6):985–995. doi: 10.1083/jcb.200201034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Segraves WA, Hogness DS. The E75 ecdysone-inducible gene responsible for the 75B early puff in Drosophila encodes two new members of the steroid receptor superfamily. Genes Dev. 1990;4(2):204–219. doi: 10.1101/gad.4.2.204. [DOI] [PubMed] [Google Scholar]
- 57.Baehrecke EH, Thummel CS. The Drosophila E93 gene from the 93F early puff displays stage- and tissue-specific regulation by 20-hydroxyecdysone. Dev Biol. 1995;171(1):85–97. doi: 10.1006/dbio.1995.1262. [DOI] [PubMed] [Google Scholar]
- 58.Bollenbacher WE, Agui N, Granger NA, Gilbert LI. In vitro activation of insect prothoracic glands by the prothoracicotropic hormone. Proc Natl Acad Sci USA. 1979;76(10):5148–5152. doi: 10.1073/pnas.76.10.5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rewitz KF, Rybczynski R, Warren JT, Gilbert LI. The Halloween genes code for cytochrome P450 enzymes mediating synthesis of the insect moulting hormone. Biochem Soc Trans. 2006;34(Pt 6):1256–1260. doi: 10.1042/BST0341256. [DOI] [PubMed] [Google Scholar]