Significance
The survival and function of insulin-secreting β cells is critical for the prevention of diabetes. We identify a previously unknown function for activating transcription factor 5 (ATF5) in the apoptotic susceptibility of β cells. ATF5 expression in β cells is stress inducible, and ATF5 deficiency results in a significant increase in β-cell apoptosis in response to stress. Further, eukaryotic translation initiation factor 4E-binding protein 1 (EIF4EBP1), a component of the mammalian target of rapamycin (mTOR) pathway that inhibits protein translation through interaction with EIF4E, is regulated by ATF5 and likely is involved in apoptotic susceptibility through regulation of global translation. Understanding the role of ATF5 deepens our understanding of the complex mechanisms governing β-cell fate decisions, providing a potential pathway for the prevention of diabetes.
Keywords: ATF5, 4EBP1, pancreatic β cell, stress, apoptosis
Abstract
The stress response and cell survival are necessary for normal pancreatic β-cell function, glucose homeostasis, and prevention of diabetes. The homeodomain transcription factor and human diabetes gene pancreas/duodenum homeobox protein 1 (Pdx1) regulates β-cell survival and endoplasmic reticulum stress susceptibility, in part through direct regulation of activating transcription factor 4 (Atf4). Here we show that Atf5, a close but less-studied relative of Atf4, is also a target of Pdx1 and is critical for β-cell survival under stress conditions. Pdx1 deficiency led to decreased Atf5 transcript, and primary islet ChIP-sequencing localized PDX1 to the Atf5 promoter, implicating Atf5 as a PDX1 target. Atf5 expression was stress inducible and enriched in β cells. Importantly, Atf5 deficiency decreased survival under stress conditions. Loss-of-function and chromatin occupancy experiments positioned Atf5 downstream of and parallel to Atf4 in the regulation of eIF4E-binding protein 1 (4ebp1), a mammalian target of rapamycin (mTOR) pathway component that inhibits protein translation. Accordingly, Atf5 deficiency attenuated stress suppression of global translation, likely enhancing the susceptibility of β cells to stress-induced apoptosis. Thus, we identify ATF5 as a member of the transcriptional network governing pancreatic β-cell survival during stress.
Reduced pancreatic β-cell number and function characterize all forms of diabetes. Insulin-secreting β cells are notoriously susceptible to stress, including endoplasmic reticulum (ER), cytokine, and oxidative stress (1–4). Thus, understanding apoptotic cell-fate decisions during stress could provide new targets that could be exploited for the prevention or amelioration of diabetes.
In secretory cells such as the β cell, the unfolded protein response (UPR) and regulation of translation, particularly in response to stress, are key factors in maintaining cellular homeostasis, as clearly demonstrated in mouse models with deficiencies of critical regulators such as protein kinase R-like ER kinase (PERK) and EIF2α (5–7). In humans, PERK mutation causes Wolcott–Rallison syndrome, a rare autosomal recessive disorder characterized by permanent neonatal diabetes (8, 9). Downstream of PERK, the basic leucine zipper (bZIP) transcription factor activating transcription factor 4 (ATF4) regulates the expression of 4ebp1, a member of the eukaryotic translation initiation factor 4E (eIF4E)-binding protein family (4EBPs). 4EBP1 is the most abundant mammalian isoform in the pancreas (10) and functions as an inhibitor of translation initiation by binding the cap-binding protein eIF4E, thereby preventing formation of the eIF4F translational initiation complex (11, 12). Expression of 4EBP1 is induced by stress, and 4ebp1 deficiency results in deregulated translational control and increased susceptibility to ER stress-mediated apoptosis in β cells (13).
We previously demonstrated that the homeodomain transcription factor and human diabetes gene pancreas/duodenum homeobox protein 1 (Pdx1) regulates pancreatic β-cell susceptibility to ER stress. Although PDX1 expression itself is not induced by stress, PDX1 does function to orchestrate islet compensation for insulin resistance, at least in part through direct transcriptional regulation of the UPR mediators ATF4 and Wolfram syndrome 1 (Wfs1) (14–16). A cDNA microarray of Pdx1+/− islets indicated Atf5 as a possible downstream target of Pdx1 (15). Atf5 is a close but less well-studied homolog of Atf4 that also contains a bZIP domain. The interconnected nature of their functions, homology, and expression make it necessary to elucidate the overlapping and independent functions of ATF4 and ATF5 in cell survival. ATF5 binds to CCAAT/enhancer-binding protein (C/EBP)-ATF response element (CARE) sites (17) and is itself a direct transcriptional target of ATF4 and C/EBP homologous protein (CHOP) in response to ER stress (18). Similar to ATF4, ATF5 expression is regulated by both transcriptional and translational mechanisms that allow selective translation in response to ER stress controlled by upstream ORFs (19, 20). ATF4 has established roles in cell susceptibility to ER stress through the regulation of CHOP, whereas ATF5 has been found to have antiapoptotic roles in several tissues, including cartilage, hematopoietic cells, cancers, and olfactory sensory neurons (18–25). Interestingly, ATF5 regulates expression and promotes cell death downstream of CHOP in mouse embryonic fibroblasts (18, 26). Two direct transcriptional targets of ATF5 have been identified in transformed cells, the antiapoptotic factor B-cell chronic lymphocytic leukemia/lymphoma 2 (BCL-2) and mammalian target of rapamycin (mTOR), a conserved serine/threonine kinase that controls cell growth (22, 23). The tissue-specific duality of both adaptive and maladaptive roles in cell survival and stress response position ATF5 as an interesting target for study in the β cell, which requires precise regulation of protein homeostasis and stress response for survival.
Here, we examine the β-cell–specific role of ATF5, independent of ATF4, in cell survival. Our results position ATF5 in a cross-regulatory transcriptional network governing protein translation and survival of pancreatic β cells during stress.
Results
Stress- and PDX1-Regulated Expression of ATF5 in Murine β Cells.
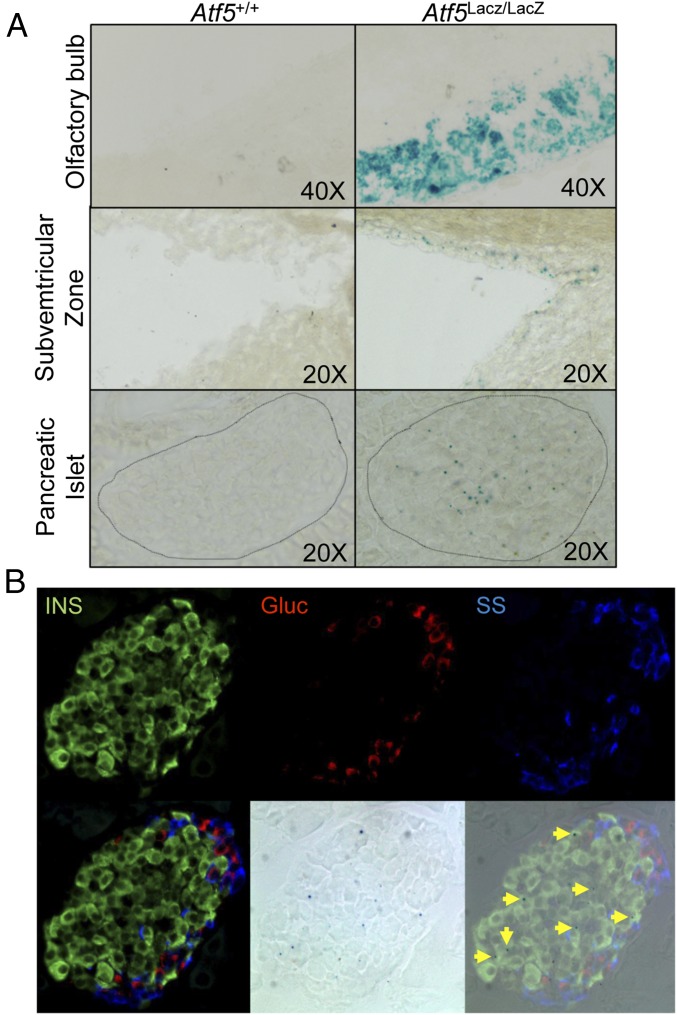
We assessed β-gal enzymatic activity in tissues of Atf5LacZ/LacZ mice in which the LacZ reporter was homologously recombined into the endogenous Atf5 locus (25). As previously reported, β-gal activity was observed in the olfactory bulb and the subventricular zone in the brain of Atf5LacZ/LacZ mice but not in Atf5+/+ littermate controls (Fig. 1A) (21, 25). We observed a similar level of β-gal activity in a subset of insulin-positive cells in the pancreatic islets of Atf5LacZ/LacZ mice, which was not seen in Atf5+/+ littermate control islets (Fig. 1). This pattern of β-gal activity was unchanged after treatment with thapsagargin (Tg) (Fig. S1). The expression of ATF5 in β cells of the murine pancreatic islet is consistent with several single-cell transcriptomic studies that identified Atf5 transcript in murine and human β cells (27–29).
Fig. 1.
ATF5 is expressed in pancreatic islets. (A) β-Gal activity assessed by X-Gal staining of the olfactory bulb (Top), subventricular zone (Center), and islets (Bottom) from Atf5+/+ (Left) and Atf5LacZ/LacZ (Right) mice. (B) β-Gal and immunofluorescent staining of parallel sections from Atf5LacZ/LacZ pancreata for insulin (Upper Left), glucagon (Upper Center), and somatostatin (Upper Right). The lower row shows merged (Lower Left), β-gal bright-field (Lower Center), and merged IF and X-Gal (Lower Right) images. Arrows indicate insulin-positive, β-gal+ cells.
Fig. S1.
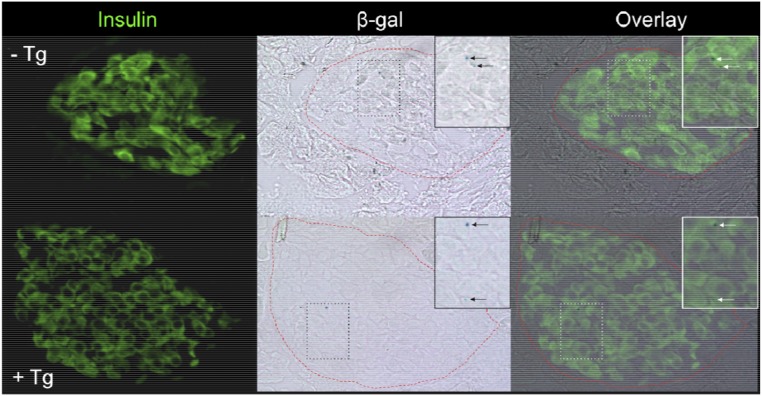
Insulin and β-gal staining of pancreatic sections from Atf5LacZ/LacZ mice treated with Tg. Immunofluorescent staining of insulin and β-gal activity was assessed by X-Gal staining of pancreatic sections from Atf5LacZ/LacZ mice treated with vehicle control (PBS) (−Tg) (Upper Row) or Tg (1 μg/kg) (+Tg) (Lower Row) by i.p. injection for 6 h before tissue collection (magnification: 20×). The Insets (magnification: 40×) show insulin-positive, β-gal+ cells.
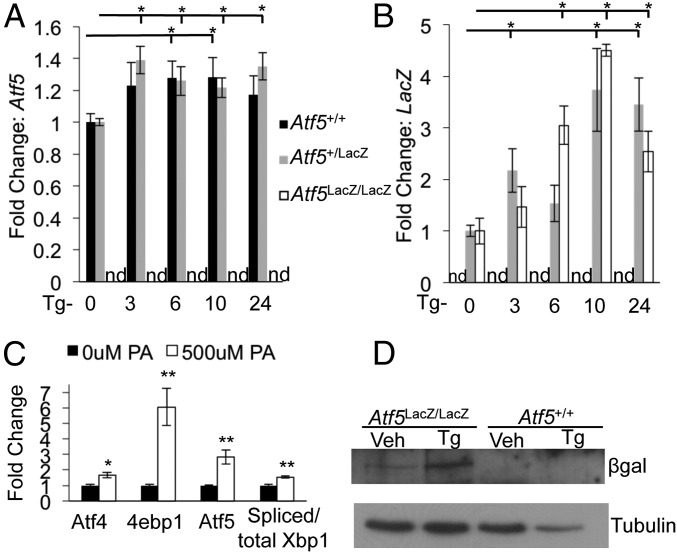
ER stress has an established role in β-cell apoptosis and diabetes development (2, 7, 30, 31). Signs of ER stress have been observed in the β cells of patients with type 2 diabetes, and ER stress is thought to contribute to the β-cell failure and insulin resistance leading to disease progression (32–34). A previous study demonstrated stress induction of ATF5 by arsenite (35). To determine whether ATF5 expression is stress responsive in the β cell, we induced ER stress by Tg or the fatty acid palmitate. Both Atf5 and LacZ mRNA showed a significant increase in Atf5+/+ and Atf5LacZ/+ islets during the time course of Tg administration, and the protein encoded by the ATF5–LacZ fusion, expressed under the control of endogenous Atf5 regulatory elements, was induced also (Fig. 2 A, B, and D). Atf5LacZ/+ islets expressed wild-type levels of Atf5 and LacZ transcript, suggesting the possibility of autoregulation. Palmitate treatment significantly induced Atf5 mRNA in Min6 cells (Fig. 2C). Taken together, these results indicate that ATF5 is expressed in the murine pancreatic islet and that its expression is induced by stress.
Fig. 2.
ATF5 expression is induced by stress. (A and B) Murine pancreatic islets were isolated from Atf5+/+, Atf5+/LacZ, and Atf5LacZ/LacZ mice, cultured and treated with Tg (1 uM) over time (expressed in hours), and then were collected and harvested for RNA. qPCR results are shown for Atf5 (A) and LacZ (B). n = 3; nd, not detected. (C) Min6 cells were treated with 500 uM palmitate (PA) for 48 h and then were harvested for RNA. qPCR results are shown for Atf4, 4ebp1, Atf5, and spliced/total Xbp1. n = 3. (D) Western blot analysis of whole-cell lysate from isolated islets cultured and treated with Tg (1 μM) for 6 h and then harvested for protein. n = 3. Bars in A–C show the mean, and error bars indicate the SEM. P values were calculated with Student’s t test; *P ≤ 0.02, **P ≤ 0.01.
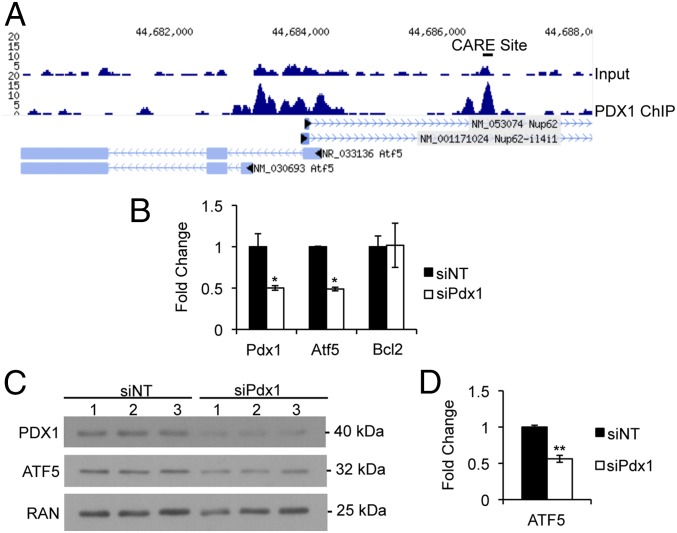
A cDNA microarray of Pdx1+/− islets revealed Atf5 as downstream of Pdx1 (15). ChIP-sequencing (ChIP-seq) analysis of isolated murine islets demonstrated enrichment of PDX1 near the ATF5 transcriptional start site at an upstream CARE site previously shown to be enriched for ATF4 and CHOP (Fig. 3A) (18), suggesting that Pdx1 might regulate Atf5 expression directly. Indeed, siRNA-mediated reduction of Pdx1 in Min6 cells reduced the levels of Atf5 transcript (Fig. 3B) and protein (Fig. 3 C and D). These results are consistent with ATF5 being a direct transcriptional target of PDX1. It is important to note that it has been shown previously that PDX1 expression is not induced by ER stress (16). However, ATF4 and CHOP are selectively translated in response to stress, so it is possible, particularly because they are enriched at similar loci, that PDX1 cooperates with these or other transcription factors to regulate ATF5 expression in the context of stress stimuli.
Fig. 3.
Atf5 is a transcriptional target of PDX1. (A) Mouse islet PDX1 ChIP-seq profile for the Atf5 locus. (B and C) Min6 cells nucleofected with siRNA duplexes targeting PDX1 were harvested 96 h after nucleofection for RNA (B) and protein with loading control ras-related nuclear protein (RAN) (C). qPCR results are shown for Pdx1, Atf5, and Bcl2. *P < 0.05. (D) Quantification of Western blot analysis. n = 6. Bars in B and D show the mean, and error bars indicate the SEM. P values were calculated with Student’s t test; **P < 0.01.
ATF5 Regulates the Susceptibility of β Cells to Stress-Induced Apoptosis.
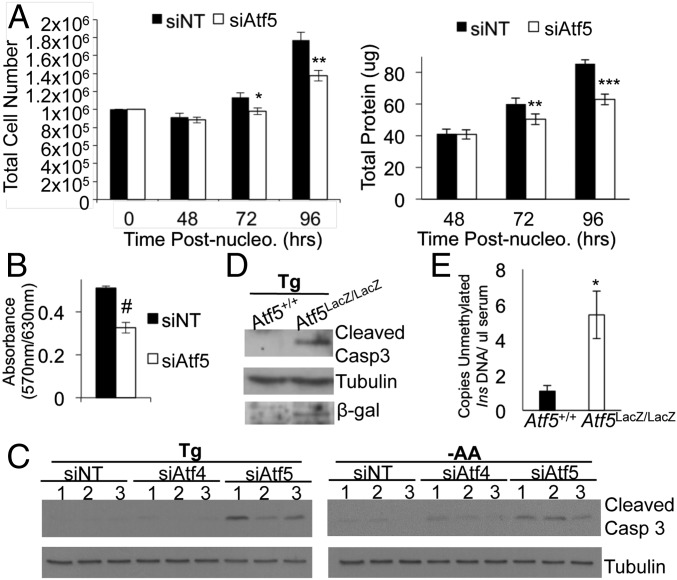
We observed a significant reduction in cell number and total protein in Min6 cells 72 h after siRNA-mediated reduction of Atf5 (Fig. 4A), and this reduction was corroborated by an MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] reduction assay (Fig. 4B). As compared with nonimmortalized cell lines, immortalized cell lines can be susceptible to stresses induced by high cell density (36). Therefore we hypothesized that ATF5 regulates β-cell viability in the immortalized Min6 insulinoma cell line in response to stress caused by high cell density. Indeed, we observed a significant increase in the cleavage of caspase-3, which normally is increased during apoptosis, in Atf5-deficient Min6 cells over that seen in control cells in the setting of ER stress induced by Tg or amino acid deprivation (Fig. 4C). The degree of caspase-3 cleavage in these cells was notably greater than that seen with Atf4 deficiency (Fig. 4C). These observations suggest a role for ATF5 independent of ATF4 in susceptibility to apoptosis.
Fig. 4.
ATF5 deficiency decreases cell viability and increases stress-induced susceptibility to apoptosis independent of ATF4. (A) Growth curve assay. Min6 cells were nucleofected with siRNA targeting Atf5 (siAtf5) or with nontargeting control siRNA (siNT) and were harvested for cell counting (Left) and protein (Right). n = 3. (B) MTT assay performed 96 h after nucleofection. n = 3. (C) Min6 cells were nucleofected with siRNA targeting Atf5 or Atf4 or with control siRNA, were treated at 96 h after nucleofection with either Tg (1 μM) or amino acid deprivation (−AA) for 6 h, and then were harvested for protein and analyzed by Western blot for cleaved caspase-3 and tubulin. Lanes were run on same gel but were noncontiguous. n = 3. (D) Islets isolated from Atf5+/+ or Atf5LacZ/LacZ mice were cultured, treated with 1 μM Tg for 6 h, and then were harvested for protein and analyzed by Western blot for cleaved caspase-3, tubulin, and β-gal. n = 3. (E) Blood serum was collected from Atf5LacZ/LacZ mice and from Atf5+/+ controls. Levels of circulating unmethylated mouse insulin DNA levels were measured by droplet PCR and are denoted as copies per microliter. n = 6. Bars in A, B, and E show the mean, and error bars indicate the SEM. P values were calculated with Student’s t test; *P < 0.02; **P < 0.0007; ***P = 6.1 × 10−8; #P < 0.001.
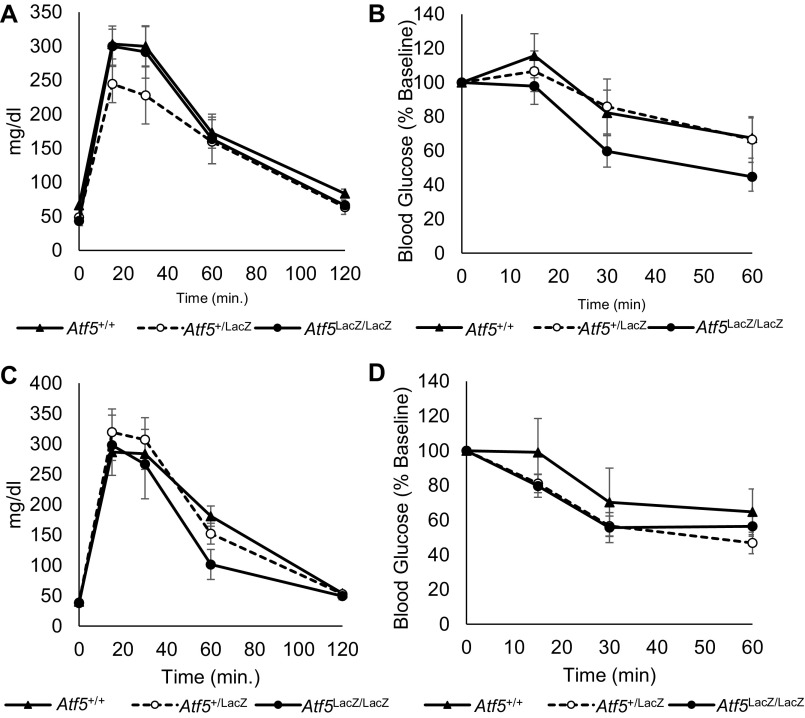
To assess the potential role of ATF5 in primary β-cell survival, we examined Atf5LacZ/LacZ mice. No overt glucose homeostasis phenotype was observed; the results of glucose and insulin tolerance tests of Atf5LacZ/LacZ mice were indistinguishable from those of control littermates (Fig. S2). Although we previously demonstrated the prosurvival role of Pdx1 in the context of a C57Bl6 genetic background and diet-induced insulin resistance (15), Atf5LacZ/LacZ mice on a C57Bl6 background exhibit high neonatal lethality (25), and the strain of Atf5LacZ/LacZ mice on the Balb/C genetic background used in our study is resistant to a range of metabolic stresses (37–40), including diet-induced metabolic phenotypes (37). These factors precluded the establishment of an in vivo stress model. Therefore, we challenged isolated Atf5LacZ/LacZ islets with Tg to induce ER stress. Atf5LacZ/LacZ islets exhibited a significant enhancement of caspase-3 cleavage compared with Atf5+/+ islets after Tg exposure, indicating an increased susceptibility to ER stress-induced apoptosis (Fig. 4D). To assess β-cell apoptosis in vivo further, droplet digital PCR was used to measure circulating levels of unmethylated cell-free insulin 2 (Ins2) DNA in serum (41–43). Importantly, Atf5LacZ/LacZ mice had significantly more circulating unmethylated Ins2 DNA than Atf5+/+ controls (Fig. 4E), strongly supporting a role for ATF5 in reducing β-cell susceptibility to apoptosis.
Fig. S2.
Glucose and insulin tolerance of Atf5+/+, Atf5+/LacZ, and Atf5LacZ/LacZ mice. (A) Results of i.p. glucose tolerance tests in 6-wk-old male Atf5+/+ (n = 6), Atf5+/LacZ (n = 6), and Atf5LacZ/LacZ (n = 5) mice after 16 h fasting. (B) Results of insulin tolerance tests in 8-wk-old male Atf5+/+ (n = 6), Atf5+/LacZ (n = 6), and Atf5LacZ/LacZ (n = 5) mice after 6 h fasting. (C) Results of i.p. glucose tolerance tests in 6-wk-old female Atf5+/+ (n = 6), Atf5+/LacZ (n = 8), and Atf5LacZ/LacZ (n = 6) mice after 16 h fasting. (D) Results of insulin tolerance tests in 8-wk-old female Atf5+/+ (n = 6), Atf5+/LacZ (n = 8), and Atf5LacZ/LacZ (n = 6) mice after 6 h fasting.
Identification of 4ebp1 as an ATF5 Transcriptional Target.
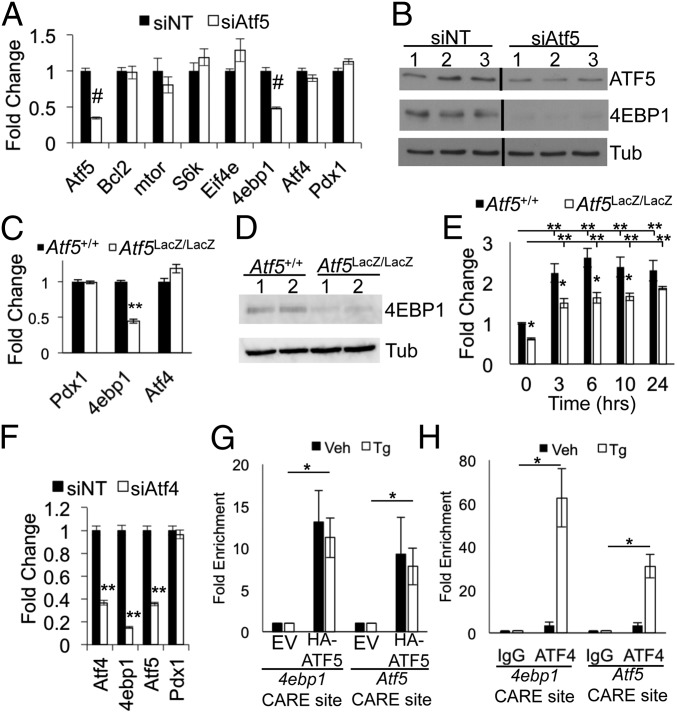
BCL-2 and mTOR have been identified as direct targets of ATF5 regulation in glioblastoma cells and in BCR-ABL–transformed myeloid progenitor cells, respectively (22, 23). In contrast, Atf5 deficiency in Min6 cells was not associated with a change in the transcript levels of Bcl2, mTor, or the mTOR pathway components S6K and Eif4e (Fig. 5A). We did, however, observe a significant reduction in the level of 4ebp1 transcript and protein (Fig. 5 A and B). Similarly, quantitative PCR (qPCR) and Western blot analysis of Atf5LacZ/LacZ isolated islets demonstrated a significant decrease in 4ebp1 transcript and 4EBP1 protein, respectively (Fig. 5 C and D). Tg treatment of Atf5LacZ/LacZ islets still induced 4ebp1 transcript; however, the degree of induction was significantly less than in Atf5+/+ control islets (Fig. 5E). Notably, the reduction of 4ebp1 occurred in the absence of any change in Atf4 expression in Atf5-deficient Min6 cells and primary islets (Fig. 5 A and C). siRNA-mediated silencing of Atf4 resulted in a significant decrease in Atf5 transcript, indicating that ATF4 regulates Atf5 in Min6 cells, in agreement with previous findings in mouse embryonic fibroblasts (Fig. 5F) (18). Deficiency of Atf4 led to a decrease in Atf5 and 4ebp1, whereas Atf5 deficiency affected 4ebp1 but not Atf4, thus positioning Atf5 both downstream of and parallel to Atf4 in the regulation of 4ebp1.
Fig. 5.
ATF5 regulates 4ebp1. (A and B) Min6 cells were nucleofected with siRNA targeting Atf5 (siAtf5) or with nontargeting control siRNA (siNT) and were harvested 96 h later for RNA (A) and protein (B). Lanes were run on same gel but were noncontiguous. qPCR results are shown for Atf5, mTor, S6k, Eif4e, 4ebp1, Atf4, and Pdx1. n = 6. (C and D) Islets isolated from Atf5+/+ and Atf5LacZ/LacZ mice were harvested for RNA (C) and protein (D). qPCR results are shown for Pdx1, 4ebp1, and Atf4. (n = 5) (E) Islets isolated from Atf5+/+ and Atf5LacZ/LacZ mice were cultured, treated with Tg (1 μM) for the denoted times and then were harvested for RNA. qPCR results are shown for 4ebp1. n = 5. (F) Min6 cells were nucleofected with siRNA targeting Atf4 or with nontargeting control siRNA and were harvested for RNA. qPCR results are shown for ATF4, 4ebp1, Atf5, and Pdx1. n = 3. (G and H) Min6 cells were infected with lentivirus expressing C-terminal HA-tagged ATF5 or empty vector control. ChIP PCR was performed on chromatin isolated from the cells expressing HA-ATF5 (G) and from wild-type Min6 cells (H), with or without treatment with Tg (1 μM) for 6 h using anti-HA, anti-ATF4, or anti-IgG. qPCR results are shown for CARE sites near the transcriptional start sites of 4ebp1 and Atf5. P values compare TG-treated empty vector (EV) and TG-treated HA-ATF5 cells. n = 3. Bars in A, C, and E–H show the mean, and error bars indicate the SEM. P values were calculated with Student’s t test; *P ≤ 0.03; **P < 2 × 10−4; #P < 1 × 10−7.
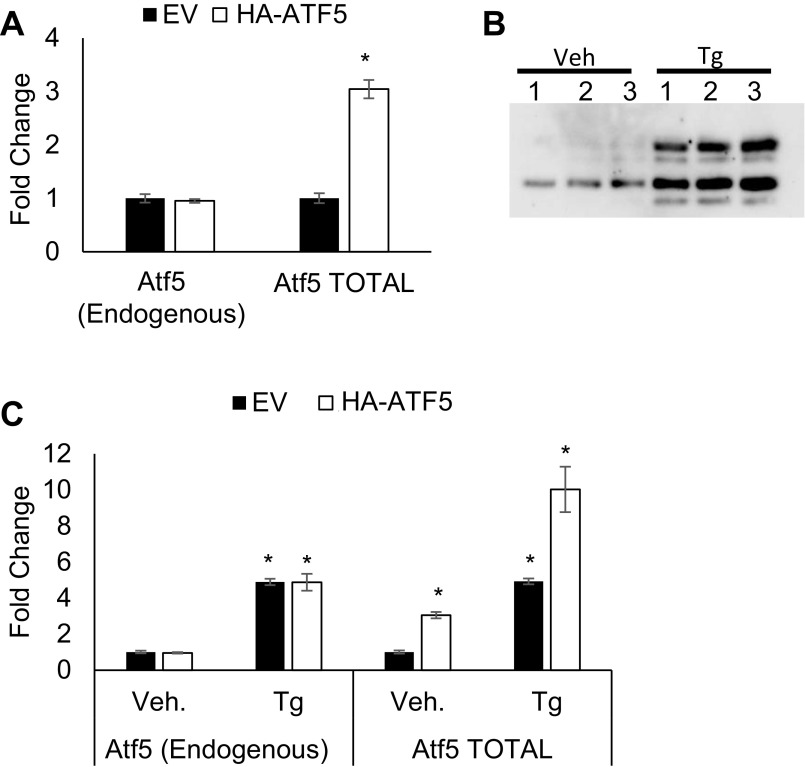
To determine whether ATF5 could regulate 4ebp1 directly, we performed ChIP. Because ATF5 antisera suitable for ChIP are not available, we generated a Min6 cell line stably expressing a C-terminally epitope-tagged form of ATF5 (HA-ATF5) at approximately twofold above endogenous levels (Fig. S3 A and B). Protein levels expressed by the CMV-driven HA-ATF5 construct were induced by Tg. The induction of HA-ATF5 protein expression by Tg correlated with induction of the HA-Atf5 transcript, suggesting transcriptional regulation of the expression construct by Tg. Indeed, the CMV promoter can be induced by Tg through the JNK pathway (19, 44, 45). We also noted a significant induction of endogenous Atf5, similar to our observations in primary islets (Fig. 3 and Fig. S3 B and C). HA-ATF5 was enriched at the CARE site near the transcriptional start site of 4ebp1 by ChIP PCR (Fig. 5G). Enrichment also was observed at the CARE site near the Atf5 transcriptional start site, consistent with autoregulation. HA-ATF5 enrichment at these CARE sites was not altered by Tg exposure, despite the induction of protein expression, implying that the role of ATF5 at these loci is in basal, and not induced, expression. In contrast, ATF4 was significantly enriched in a Tg-induced manner at the CARE sites of both 4ebp1 and Atf5, consistent with previous reports (Fig. 5H) (13, 18). Taken together, these findings suggest that ATF5 directly regulates the transcription of 4ebp1 and that ATF5 recruitment is not ATF4 dependent.
Fig. S3.
Expression of Atf5 transcript after lentiviral infection with EV-GFP or HA-ATF5 in stable Min6 cell lines. (A) qPCR of endogenous and total Atf5 transcript in Min6 cells stably expressing EV-GFP or HA-ATF5. n = 3. *P < 0.001. Endogenous Atf5 transcript was measured using primers in the Atf53′ UTR (Table S2); total Atf5 (endogenous + HA-ATF5) transcript was measured using primers in the Atf5 coding sequence (Table S2). (B and C) Min6 cells stably expressing HA-ATF5 were treated or not treated with Tg (1 μM) for 6 h, harvested for protein, and blotted with anti-HA (B) or were harvested for RNA and qPCR for endogenous Atf5 transcript and total Atf5 (endogenous + HA-ATF5) transcript (C). n = 3.
Atf5 Deficiency Increases Global Protein Translation.
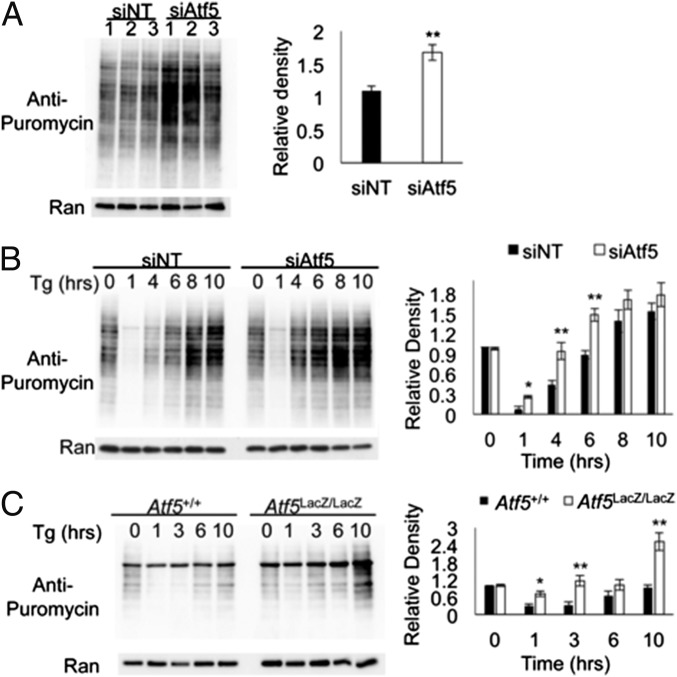
Several studies have elucidated the importance of 4E-BP1 expression and translational regulation for β-cell survival, particularly in response to stress (5, 6, 13). A central role for the PERK arm of the UPR response is the down-regulation of global translation to allow recovery by reducing the protein load on the ER while simultaneously and selectively translating transcripts required for recovery, e.g., Atf4 (5, 19). Perturbations of these translational regulation mechanisms are detrimental to cell survival, particularly in highly secretory cells such as β cells that normally function at augmented levels of protein synthesis (5–8). To assess global translation rates under basal and stressed conditions, we treated Atf5-deficient and control Min6 cells with Tg over a time course and measured global translation by puromycin incorporation followed by Western blot analysis. Under basal conditions, ATF5 deficiency induced an increase in basal global translation (Fig. 6A). After Tg exposure, rates of translation are initially suppressed at 1 h and then recover over 8–10 h in control cells; in Atf5-deficient Min6 cells a notable enhancement of translational recovery was observed beginning at 4 h, and the maximal level of translation at 10 h also was visibly increased (Fig. 6B). Similarly, an increased rate of translational recovery after Tg was seen in primary isolated Atf5LacZ/LacZ islets compared with Atf5+/+ islets, with global translation at 10 h surpassing that observed under basal conditions (Fig. 6C). The results indicate that Atf5 deficiency abrogates the reduced protein load in the ER that normally allows the restoration of cellular homeostasis under stress conditions.
Fig. 6.
Atf5 deficiency enhances the recovery of global translation under stress conditions. (A) Min6 cells were nucleofected with siRNA targeting Atf5 (siAtf5) or with nontargeting control siRNA (siNT). At 96 h after nucleofection, cells were treated with puromycin (1 μg/mL) for 30 min and then were harvested for protein. (Left) Western blot analysis with anti-puromycin for the basal level of global translation. (Right) Measurement of the relative density of blots. n = 3. (B) Min6 cells were nucleofected with siAtf5 or with siNT. At 96 h after nucleofection, cells were treated with Tg (1 μM) for the denoted time (in hours) and with puromycin (1 μg/mL) for the last 30 min and were harvested for protein. (Left) Western blot analysis with anti-puromycin for global translation by puromycin incorporation. (Right) Quantitation of relative density. n = 3. (C, Left) Islets harvested from Atf5+/+ and Atf5LacZ/LacZ mice were cultured, treated with Tg (1 μM) for the denoted time, and harvested for protein and Western blot analysis. (Right) Quantitation of relative density. n = 3. Bars show the mean, and error bars indicate the SEM. P values were calculated with Student’s t test; *P ≤ 0.05; **P ≤ 0.01.
Discussion
ER stress is particularly detrimental to highly secretory cells such as pancreatic β cells, because they are dependent on optimal protein synthesis for survival and function. In normal cells, stress and perturbations in protein synthesis lead to the activation of the UPR and eventual restoration of homeostasis. We find that ATF5 is induced by stress and is critical for the survival of β cells during stress. Loss-of-function studies and ChIP assays further positioned Atf5 in the transcriptional network governing β-cell survival during stress and suggest that 4EBP1 is a direct transcriptional target of ATF5 in this context. Deficiency of ATF5 resulted in an enhancement of global translation, explaining, at least in part, the increase in apoptotic susceptibility of β cells during stress.
Loss of ATF5 appears to “prime” the β cell for apoptotic susceptibility by impairing its ability to down-regulate translation in response to stress (Fig. 7). ER stress has been observed in the islets of patients with both type 1 and type 2 diabetes, and multiple studies have demonstrated the importance of translational regulation for β-cell survival (2, 7, 13, 30–34, 46). 4EBP1 is an important negative regulator of translation that reduces the ER protein load during ER stress, allowing the cell an opportunity for recovery and restoration of homeostasis. Droplet digital PCR results measuring circulating cell-free unmethylated Ins2 DNA demonstrated a significant increase in β-cell apoptosis in Atf5LacZ/LacZ mice as compared with Atf5+/+ controls even before stress exposure. The preservation of Atf5LacZ/LacZ β-cell mass under basal conditions is likely related to the Balb/C genetic background, possibly through a small adaptive increase in proliferation. We speculate that on a susceptible genetic background such as C57BL6, β-cell mass and function would deteriorate under stress, analogous to the normal glucose homeostasis of 4ebp1−/− mice that deteriorates under stress imposed by Wfs1 deficiency or expression of the Akita misfolding mutation in Ins2 (13). This experiment is not yet possible because of the neonatal lethality of Atf5LacZ/LacZ mice on a C57Bl6 genetic background and the unavailability of a conditional allele to date. Overall, our findings indicate that Atf5 deficiency leads to reduced 4ebp1 expression and impaired adaptive translational suppression, which likely contribute to enhanced β-cell susceptibility to stress-induced apoptosis.
Fig. 7.
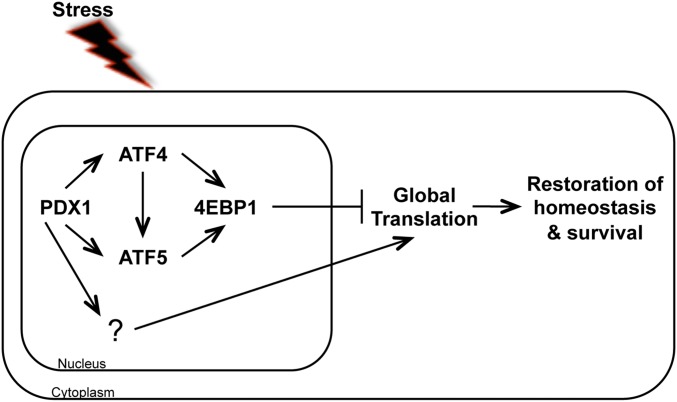
Role of ATF5 in the survival of β cells during stress. Our findings place ATF5 within the PDX1–ATF4–4EBP1 regulatory pathway, downstream of and parallel to ATF4. ATF5 regulates 4ebp1, a negative regulator of translation. In response to stress, 4EBP1 binds to EIF4E, preventing it from joining the cap-dependent translation initiation complex, thereby inhibiting global translation. Global translation and protein load on the ER decrease, allowing the cell to restore homeostasis and survive.
The close ATF5 homolog ATF4 has been studied extensively in cellular stress responses, and its downstream targets include both negative and positive regulators of translation and protein synthesis (Fig. 5G) (13, 26, 47). There appear to be distinct temporal dynamics and thresholds of ATF4 expression, and thereby protein synthesis, that are beneficial to cell survival. ATF4 is one member of a set of mRNAs specifically translated in the context of stress when there is global translation reduction of most other mRNAs. Translational regulation of ATF4 is controlled by the presence of an inhibitory ORF in the 5′ UTR, which is skipped through delayed ribosomal reinitiation during stress (19). Examination of the ATF5 mRNA sequence demonstrates that its mRNA transcript has a similar dual upstream ORF that could regulate translation in a similar stress-dependent manner. The complexity and overlapping levels of translational regulation highlight the importance of tightly controlled protein synthesis and the specific expression of stress-response components. Our results, consistent with findings in other systems, suggest that Atf5 regulation of 4ebp1 downstream of Atf4 may serve as a threshold checkpoint for protein synthesis during cellular stress recovery.
Our chromatin occupancy and loss-of-function experiments position ATF5 both downstream of and parallel to ATF4 in the regulation of 4e-bp1. Overlapping chromatin occupancy of the CARE sites of Atf5 and 4ebp1 strongly suggests a level of stress-induced cooperation between ATF4 and ATF5, but the molecular mechanism remains to be determined. It is not yet clear whether they interact in a complex or have a sequential role in DNA binding and transcriptional regulation. One defining characteristic of the bZIP domains is the ability to heterodimerize with other bZIP domain-containing proteins. Although the exact mechanisms of bZIP-domain specificity are not defined, this ability for protein–protein interaction and the plethora of possible bZIP partners creates the potential for diverse interactions. In particular, the bZIP domain containing the C/EBP transcription factor C/EBPβ is highly expressed in β cells (48) and can be induced by ER stress in other cell types (49). Further investigation will be required to determine whether and how C/EBPβ and/or other bZIP and related factors participate with ATF4 and ATF5 in the transcriptional network governing protein translation and cell survival in β cells.
Although the stresses may be distinct, the survival of pancreatic β cells is impaired in both type 1 and type 2 diabetes, and islet inflammation has been observed during the progression of both forms of diabetes (50, 51). In addition to the prosurvival roles we identify here, ATF5 has been identified as a transcriptional regulator of the proapoptotic CHOP transcription factor as well as TXNIP, a proinflammatory component that increases the expression of IL-1β in response to oxidative stress and ER stress (52, 53). The effect of stress on the β cell has been reported to depend on the length and severity of the stresses (2, 54, 55). Reduction of the global translational load allows recovery and return to homeostasis, but long-term suppression of translation would not be beneficial to the β cell. Our findings suggest that ATF5 is necessary for survival in the setting of acute stress through suppression of translation; however, the overall function of ATF5 in the regulation of the proinflammatory and proapoptotic components may be to serve as a threshold checkpoint for chronic stresses that cannot be overcome. The involvement of ATF5 in both inflammatory signaling and translational regulation in response to environmental stresses suggests a nuanced role at the apex of β-cell survival that requires further investigation. The identification of ATF5 as a potential gatekeeper of decisions regarding β-cell fate during stress introduces a potential node for therapeutic intervention to prevent or ameliorate diabetes by improving β-cell durability.
Materials and Methods
Mice.
The Atf5-knockout allele (herein referred to as Atf5LacZ/LacZ) was generated by inserting a LacZ-Neo cassette in frame, replacing most of exon 1 and all of exons 2 and 3 (26). To assess β-cell apoptosis, droplet digital PCR (Bio-Rad QX200 Droplet Digital PCR System) of serum was used to measure unmethylated insulin DNA (44). All animal procedures were approved by the University of Pennsylvania Institutional Animal Care and Use Committee.
Cell Culture and siRNA Nucleofection.
Passage 25-35 Min6 cells were cultured in DMEM (Invitrogen) supplemented with 10% (vol/vol) FBS. For siRNA experiments, cells were nucleofected by AMAXA with predesigned siRNAs targeting Pdx1, Atf5, or Atf4 or with a nontargeting control (Ambion) and were harvested 72–96 h after nucleofection for protein or RNA.
Islet Isolation and Culture.
Islets were isolated from 8- to 10-wk-old mice by collagenase digestion (56). After hand-picking and overnight culture in RPMI 1640, size-matched islets were treated with vehicle (DMSO), 1 μM Tg (Sigma), or 500 μM palmitate for the denoted times.
Immunofluorescence and β-Gal Staining.
Immunofluorescence (IF) was carried out as previously described (57). Additional information is given in SI Materials and Methods.
Western Blot Analysis.
Protein lysates were prepared using Nonidet P-40 lysis buffer (0.4% Nonidet P-40, 150 mM NaCl, 50 mM Tris base, pH 7.6). Western blot analysis was carried out as previously described (15). Use and dilution information for antisera is found in Table S1. Additional information is given in SI Materials and Methods.
Table S1.
Antisera source, dilution, and application
| Antisera | Source | Dilution | Application |
| Anti–β-gal | Santa Cruz | 1:1,000 | WB |
| Anti-4EBP1 | Cell Signaling | 1:1,000 | WB |
| Anti-ATF4 | Santa Cruz | 10 μg, 1:500 | ChIP, WB |
| Anti-ATF5 | Sigma | 1:1,000 | WB |
| Anti–cleaved caspase-3 | Cell Signaling | 1:1,000 | WB |
| Anti-glucagon | Santa Cruz | 1:500 | IF |
| Anti-HA HRP | Roche | 1:1,000 | WB |
| Anti-HA agarose | Sigma | 30 μL | ChIP |
| Anti-insulin | Abcam | 1:1,000 | IF |
| Anti-puromycin | Millipore | 1:10,000 | WB |
| Anti-ran | BD | 1:10,000 | WB |
| Anti-somatostatin | Santa Cruz | 1:500 | IF |
| Anti-tubulin | Sigma | 1:10,000 | WB |
ChIP.
ChIP was carried out as previously described (15). Primer sequences are found in Table S2. Additional information is given in SI Materials and Methods.
Table S2.
Primer sequences
| Transcript/gene locus | Sequence |
| 4ebp1 | F-TGCTACTGCAGGGGAATGACT |
| R-TGCTACTGCAGGGGAATGACT | |
| 4ebp1 CARE site (ChIP) | F-TGCTACTGCAGGGGAATGACT |
| R-TGCTACTGCAGGGGAATGACT | |
| Atf4 | F-TGCTACTGCAGGGGAATGACT |
| R-TGCTACTGCAGGGGAATGACT | |
| Atf5 (3′ UTR) | F-TGCTACTGCAGGGGAATGACT |
| R-TGCTACTGCAGGGGAATGACT | |
| Atf5 (coding region) | F-TGCTACTGCAGGGGAATGACT |
| R-TGCTACTGCAGGGGAATGACT | |
| Atf5 CARE site (ChIP) | F-TGCTACTGCAGGGGAATGACT |
| R-TGCTACTGCAGGGGAATGACT | |
| Bcl2 | F-TGCTACTGCAGGGGAATGACT |
| R-TGCTACTGCAGGGGAATGACT | |
| Eif4e | F-TGCTACTGCAGGGGAATGACT |
| R-TGCTACTGCAGGGGAATGACT | |
| Hprt | F-TGCTCGAGATGTCATGAAGGA |
| R-CCAGCAGGTCAGCAAAGAACT | |
| LacZ | F-ATACTGTCGTCGTCCCCTCAAAC |
| R-CGGATTCTCCGTGGGAACAA | |
| mTOR | F-TCCCAGGACAGTGAAACACA |
| R-GGTTGAGAGGACCAACTGGA | |
| Pdx1 | F-GAACCCGAGGAAAACAAGAGG |
| R-GTTCAACATCACTGCCAGCTC | |
| Spliced Xbp1 | F-CTGAGTCCGAATCAGGTGCAG |
| R-GTCCATGGGAAGATGTTCCTGG | |
| Total Xbp1 | F-TGGCCCGGGTCTGCTGAGTCCG |
| R-GTCCATGGGAAGATGTTCCTGG | |
| S6k | F-CCTCTAGGACACACCCTCCA |
| R-ATCAACCCTGTCTGGATTCG |
RNA Isolation and qPCR.
Min6 cells and isolated islets were harvested and stored in TRIzol (Invitrogen). Min6 RNA was extracted using ethanol precipitation. Islet RNA was extracted using the RNeasy Mini Kit (QIAGEN). Min6 samples were reverse transcribed using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) and random primers. Islet samples were reverse transcribed using SuperScript III (Invitrogen) and oligo(dT). Transcript was analyzed by qPCR (Bio-Rad CFX384) and normalized to hypoxanthine-guanine phosphoribosyltransferase (Hprt). Primer sequences are given in Table S2.
Measurement of Global Translation.
Min6 cells were nucleofected with siRNA targeting Atf5 or with nontargeting (NT) control siRNA. Nucleofected cells or primary Atf5+/+ and Atf5LacZ/LacZ islets were treated with 1 μM Tg for the denoted times or were left untreated. Puromycin (1 μg/mL) was applied during the last 30 min of incubation. Cells were washed with 1× PBS and collected for protein.
Statistics.
Data are presented as mean ± SEM. Differences between compared groups were achieved by two-tailed Student’s t tests and were considered significant when P values were less than 0.05.
SI Materials and Methods
IF and β-Gal Staining.
Tissues were dissected from 8- to 10-wk-old male mice, fixed on ice for 45 min in 4% paraformaldehyde, and washed three times with cold 1× PBS. Pancreata were incubated overnight at 4 °C in 30% sucrose and then were embedded in optimal cutting temperature (O.C.T.) embedding medium before cryosectioning. For β-gal staining, slides were incubated overnight at 37 °C with X-Gal staining solution [5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6, 2 mM MgCl2, 0.01% Na deoxycholate, 0.02% Nonidet P-40, 1 mg/mL X-Gal] (Fisher). For β-gal/fluorescence double staining, slides were washed in 1× PBS, blocked (1× PBS + 5% donkey serum + 1% BSA) for 1 h at 4 °C, and then incubated in primary antibody overnight at 4 °C. Slides then were washed in 1× PBS, incubated for 1 h at room temperature with secondary Cy2-, Cy3-, or Cy5-conjugated antibodies, washed, and cover-slipped. For IF and double IF, the following primary antibodies were used at the following dilutions: rabbit anti-glucagon (1:500; Santa Cruz), guinea pig anti-insulin (1:500; Abcam), and goat anti-somatostatin (1:500; Santa Cruz).
Western Blot Analysis.
Protein lysates were prepared using Nonidet P-40 lysis buffer [0.4% Nonidet P-40, 150 mM NaCl, 50 mM Tris base (pH 7.6)]. Protein concentration was determined using BCA (Fisher). Proteins were separated by SDS/PAGE and immunoblotted with the following primary antibodies: rabbit anti-ATF5 (Sigma Aldrich), mouse anti-Ran (BD), rabbit anti–cleaved caspase-3 (Cell Signaling), mouse anti-tubulin (Sigma Aldrich), rabbit anti-4ebp1 (Cell Signaling), rabbit anti–β-gal (Molecular Probes), mouse anti-puromycin (Millipore), and HRP-conjugated anti-HA (Roche). HRP-conjugated antibodies were incubated for 2 h at room temperature. Luminata Crescendo HRP substrate (Millipore) was used to visualize blots with film or a ChemiDoc Touch Imaging system (Bio-Rad).
ChIP.
Min6 cells (∼1 × 107 cells) were cross-linked with 1% formaldehyde in 1× PBS for 10 min at room temperature and were quenched with glycine to a final concentration of 0.125 M. Chromatin was sonicated in a Bioruptor (Diagenode) and precleared with normal mouse IgG (Santa Cruz) overnight at 4 °C. After removal of an aliquot for input, precleared chromatin was divided equally for immunoprecipitation with rabbit anti-ATF4 (Santa Cruz), anti–HA-conjugated agarose (Sigma), or normal IgG (Santa Cruz) for 3 h at 4 °C. Data were analyzed quantitatively in triplicate by real-time PCR.
Acknowledgments
We thank Sarah Tersey and Kieran Mather of the Translation Core, Center for Diabetes and Metabolic Diseases, Indiana University School of Medicine for performing digital droplet PCR for unmethylated DNA. This work was supported by National Institutes of Health Grants P01 DK49210 and R01 DK068157 (to D.A.S.) and F32-DK103454 (to C.A.J.). M.R.G. is an investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1620705114/-/DCSupplemental.
References
- 1.Chan JY, et al. The balance between adaptive and apoptotic unfolded protein responses regulates β-cell death under ER stress conditions through XBP1, CHOP and JNK. Mol Cell Endocrinol. 2015;413:189–201. doi: 10.1016/j.mce.2015.06.025. [DOI] [PubMed] [Google Scholar]
- 2.Eizirik DL, Cardozo AK, Cnop M. The role for endoplasmic reticulum stress in diabetes mellitus. Endocr Rev. 2008;29(1):42–61. doi: 10.1210/er.2007-0015. [DOI] [PubMed] [Google Scholar]
- 3.Hodish I, et al. In vivo misfolding of proinsulin below the threshold of frank diabetes. Diabetes. 2011;60(8):2092–2101. doi: 10.2337/db10-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharma RB, et al. Insulin demand regulates β cell number via the unfolded protein response. J Clin Invest. 2015;125(10):3831–3846. doi: 10.1172/JCI79264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harding HP, Zhang Y, Bertolotti A, Zeng H, Ron D. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol Cell. 2000;5(5):897–904. doi: 10.1016/s1097-2765(00)80330-5. [DOI] [PubMed] [Google Scholar]
- 6.Harding HP, et al. Diabetes mellitus and exocrine pancreatic dysfunction in perk-/- mice reveals a role for translational control in secretory cell survival. Mol Cell. 2001;7(6):1153–1163. doi: 10.1016/s1097-2765(01)00264-7. [DOI] [PubMed] [Google Scholar]
- 7.Scheuner D, et al. Control of mRNA translation preserves endoplasmic reticulum function in beta cells and maintains glucose homeostasis. Nat Med. 2005;11(7):757–764. doi: 10.1038/nm1259. [DOI] [PubMed] [Google Scholar]
- 8.Julier C, Nicolino M. Wolcott-Rallison syndrome. Orphanet J Rare Dis. 2010;5:29. doi: 10.1186/1750-1172-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wolcott CD, Rallison ML. Infancy-onset diabetes mellitus and multiple epiphyseal dysplasia. J Pediatr. 1972;80(2):292–297. doi: 10.1016/s0022-3476(72)80596-1. [DOI] [PubMed] [Google Scholar]
- 10.Tsukiyama-Kohara K, et al. Adipose tissue reduction in mice lacking the translational inhibitor 4E-BP1. Nat Med. 2001;7(10):1128–1132. doi: 10.1038/nm1001-1128. [DOI] [PubMed] [Google Scholar]
- 11.Pause A, et al. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature. 1994;371(6500):762–767. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- 12.Yanagiya A, et al. Translational homeostasis via the mRNA cap-binding protein, eIF4E. Mol Cell. 2012;46(6):847–858. doi: 10.1016/j.molcel.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamaguchi S, et al. ATF4-mediated induction of 4E-BP1 contributes to pancreatic beta cell survival under endoplasmic reticulum stress. Cell Metab. 2008;7(3):269–276. doi: 10.1016/j.cmet.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 14.Sachdeva MM, Stoffers DA. Minireview: Meeting the demand for insulin: Molecular mechanisms of adaptive postnatal beta-cell mass expansion. Mol Endocrinol. 2009;23(6):747–758. doi: 10.1210/me.2008-0400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sachdeva MM, et al. Pdx1 (MODY4) regulates pancreatic beta cell susceptibility to ER stress. Proc Natl Acad Sci USA. 2009;106(45):19090–19095. doi: 10.1073/pnas.0904849106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Templin AT, Maier B, Tersey SA, Hatanaka M, Mirmira RG. Maintenance of Pdx1 mRNA translation in islet β-cells during the unfolded protein response. Mol Endocrinol. 2014;28(11):1820–1830. doi: 10.1210/me.2014-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fawcett TW, Martindale JL, Guyton KZ, Hai T, Holbrook NJ. Complexes containing activating transcription factor (ATF)/cAMP-responsive-element-binding protein (CREB) interact with the CCAAT/enhancer-binding protein (C/EBP)-ATF composite site to regulate Gadd153 expression during the stress response. Biochem J. 1999;339(Pt 1):135–141. [PMC free article] [PubMed] [Google Scholar]
- 18.Teske BF, et al. CHOP induces activating transcription factor 5 (ATF5) to trigger apoptosis in response to perturbations in protein homeostasis. Mol Biol Cell. 2013;24(15):2477–2490. doi: 10.1091/mbc.E13-01-0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vattem KM, Wek RC. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc Natl Acad Sci USA. 2004;101(31):11269–11274. doi: 10.1073/pnas.0400541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Watatani Y, et al. Stress-induced translation of ATF5 mRNA is regulated by the 5′-untranslated region. J Biol Chem. 2008;283(5):2543–2553. doi: 10.1074/jbc.M707781200. [DOI] [PubMed] [Google Scholar]
- 21.Arias A, et al. Regulated ATF5 loss-of-function in adult mice blocks formation and causes regression/eradication of gliomas. Oncogene. 2012;31(6):739–751. doi: 10.1038/onc.2011.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dluzen D, Li G, Tacelosky D, Moreau M, Liu DX. BCL-2 is a downstream target of ATF5 that mediates the prosurvival function of ATF5 in a cell type-dependent manner. J Biol Chem. 2011;286(9):7705–7713. doi: 10.1074/jbc.M110.207639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sheng Z, Ma L, Sun JE, Zhu LJ, Green MR. BCR-ABL suppresses autophagy through ATF5-mediated regulation of mTOR transcription. Blood. 2011;118(10):2840–2848. doi: 10.1182/blood-2010-12-322537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song B, Scheuner D, Ron D, Pennathur S, Kaufman RJ. Chop deletion reduces oxidative stress, improves beta cell function, and promotes cell survival in multiple mouse models of diabetes. J Clin Invest. 2008;118(10):3378–3389. doi: 10.1172/JCI34587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang SZ, Ou J, Zhu LJ, Green MR. Transcription factor ATF5 is required for terminal differentiation and survival of olfactory sensory neurons. Proc Natl Acad Sci USA. 2012;109(45):18589–18594. doi: 10.1073/pnas.1210479109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma Y, Brewer JW, Diehl JA, Hendershot LM. Two distinct stress signaling pathways converge upon the CHOP promoter during the mammalian unfolded protein response. J Mol Biol. 2002;318(5):1351–1365. doi: 10.1016/s0022-2836(02)00234-6. [DOI] [PubMed] [Google Scholar]
- 27.Benner C, et al. The transcriptional landscape of mouse beta cells compared to human beta cells reveals notable species differences in long non-coding RNA and protein-coding gene expression. BMC Genomics. 2014;15:620. doi: 10.1186/1471-2164-15-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li J, et al. Single-cell transcriptomes reveal characteristic features of human pancreatic islet cell types. EMBO Rep. 2016;17(2):178–187. doi: 10.15252/embr.201540946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xin Y, et al. Use of the Fluidigm C1 platform for RNA sequencing of single mouse pancreatic islet cells. Proc Natl Acad Sci USA. 2016;113(12):3293–3298. doi: 10.1073/pnas.1602306113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herbach N, et al. Dominant-negative effects of a novel mutated Ins2 allele causes early-onset diabetes and severe β-cell loss in Munich Ins2C95S mutant mice. Diabetes. 2007;56(5):1268–1276. doi: 10.2337/db06-0658. [DOI] [PubMed] [Google Scholar]
- 31.Wang J, et al. A mutation in the insulin 2 gene induces diabetes with severe pancreatic β-cell dysfunction in the Mody mouse. J Clin Invest. 1999;103(1):27–37. doi: 10.1172/JCI4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laybutt DR, et al. Endoplasmic reticulum stress contributes to beta cell apoptosis in type 2 diabetes. Diabetologia. 2007;50(4):752–763. doi: 10.1007/s00125-006-0590-z. [DOI] [PubMed] [Google Scholar]
- 33.Ozcan U, et al. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science. 2004;306(5695):457–461. doi: 10.1126/science.1103160. [DOI] [PubMed] [Google Scholar]
- 34.Huang CJ, et al. High expression rates of human islet amyloid polypeptide induce endoplasmic reticulum stress mediated beta-cell apoptosis, a characteristic of humans with type 2 but not type 1 diabetes. Diabetes. 2007;56(8):2016–2027. doi: 10.2337/db07-0197. [DOI] [PubMed] [Google Scholar]
- 35.Zhou D, et al. Phosphorylation of eIF2 directs ATF5 translational control in response to diverse stress conditions. J Biol Chem. 2008;283(11):7064–7073. doi: 10.1074/jbc.M708530200. [DOI] [PubMed] [Google Scholar]
- 36.Brezden CB, Rauth AM. Differential cell death in immortalized and non-immortalized cells at confluency. Oncogene. 1996;12(1):201–206. [PubMed] [Google Scholar]
- 37.Montgomery MK, et al. Mouse strain-dependent variation in obesity and glucose homeostasis in response to high-fat feeding. Diabetologia. 2013;56(5):1129–1139. doi: 10.1007/s00125-013-2846-8. [DOI] [PubMed] [Google Scholar]
- 38.Gurley SB, et al. Impact of genetic background on nephropathy in diabetic mice. Am J Physiol Renal Physiol. 2006;290(1):F214–F222. doi: 10.1152/ajprenal.00204.2005. [DOI] [PubMed] [Google Scholar]
- 39.Hayashi K, Kojima R, Ito M. Strain differences in the diabetogenic activity of streptozotocin in mice. Biol Pharm Bull. 2006;29(6):1110–1119. doi: 10.1248/bpb.29.1110. [DOI] [PubMed] [Google Scholar]
- 40.Montgomery MK, et al. Disparate metabolic response to fructose feeding between different mouse strains. Sci Rep. 2015;5:18474. doi: 10.1038/srep18474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Akirav EM, et al. Detection of β cell death in diabetes using differentially methylated circulating DNA. Proc Natl Acad Sci USA. 2011;108(47):19018–19023. doi: 10.1073/pnas.1111008108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fisher MM, Perez Chumbiauca CN, Mather KJ, Mirmira RG, Tersey SA. Detection of islet β-cell death in vivo by multiplex PCR analysis of differentially methylated DNA. Endocrinology. 2013;154(9):3476–3481. doi: 10.1210/en.2013-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fisher MM, et al. Elevations in circulating methylated and unmethylated preproinsulin DNA in new-onset type 1 diabetes. Diabetes. 2015;64(11):3867–3872. doi: 10.2337/db15-0430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodova M, Jayini R, Singasani R, Chipps E, Islam MR. CMV promoter is repressed by p53 and activated by JNK pathway. Plasmid. 2013;69(3):223–230. doi: 10.1016/j.plasmid.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spohn D, et al. Thapsigargin induces expression of activating transcription factor 3 in human keratinocytes involving Ca2+ ions and c-Jun N-terminal protein kinase. Mol Pharmacol. 2010;78(5):865–876. doi: 10.1124/mol.110.067637. [DOI] [PubMed] [Google Scholar]
- 46.Marhfour I, et al. Expression of endoplasmic reticulum stress markers in the islets of patients with type 1 diabetes. Diabetologia. 2012;55(9):2417–2420. doi: 10.1007/s00125-012-2604-3. [DOI] [PubMed] [Google Scholar]
- 47.Han J, et al. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat Cell Biol. 2013;15(5):481–490. doi: 10.1038/ncb2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu M, Seufert J, Habener JF. Pancreatic beta-cell-specific repression of insulin gene transcription by CCAAT/enhancer-binding protein beta. Inhibitory interactions with basic helix-loop-helix transcription factor E47. J Biol Chem. 1997;272(45):28349–28359. doi: 10.1074/jbc.272.45.28349. [DOI] [PubMed] [Google Scholar]
- 49.Hayakawa K, et al. ER stress depresses NF-kappaB activation in mesangial cells through preferential induction of C/EBP beta. J Am Soc Nephrol. 2010;21(1):73–81. doi: 10.1681/ASN.2009040432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Atkinson MA, et al. How does type 1 diabetes develop?: The notion of homicide or β-cell suicide revisited. Diabetes. 2011;60(5):1370–1379. doi: 10.2337/db10-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Butler AE, et al. Beta-cell deficit and increased beta-cell apoptosis in humans with type 2 diabetes. Diabetes. 2003;52(1):102–110. doi: 10.2337/diabetes.52.1.102. [DOI] [PubMed] [Google Scholar]
- 52.Oslowski CM, et al. Thioredoxin-interacting protein mediates ER stress-induced β cell death through initiation of the inflammasome. Cell Metab. 2012;16(2):265–273. doi: 10.1016/j.cmet.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamazaki T, et al. Regulation of the human CHOP gene promoter by the stress response transcription factor ATF5 via the AARE1 site in human hepatoma HepG2 cells. Life Sci. 2010;87(9-10):294–301. doi: 10.1016/j.lfs.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 54.Fonseca SG, Gromada J, Urano F. Endoplasmic reticulum stress and pancreatic β-cell death. Trends Endocrinol Metab. 2011;22(7):266–274. doi: 10.1016/j.tem.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scheuner D, Kaufman RJ. The unfolded protein response: A pathway that links insulin demand with beta-cell failure and diabetes. Endocr Rev. 2008;29(3):317–333. doi: 10.1210/er.2007-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scharp DW, Kemp CB, Knight MJ, Ballinger WF, Lacy PE. The use of ficoll in the preparation of viable islets of langerhans from the rat pancreas. Transplantation. 1973;16(6):686–689. doi: 10.1097/00007890-197312000-00028. [DOI] [PubMed] [Google Scholar]
- 57.Ediger BN, et al. Islet-1 Is essential for pancreatic β-cell function. Diabetes. 2014;63(12):4206–4217. doi: 10.2337/db14-0096. [DOI] [PMC free article] [PubMed] [Google Scholar]