Abstract
Background
The genus Thermus, which has been considered for a long time as a fruitful source of biotechnological relevant enzymes, has emerged more recently as suitable host to overproduce thermozymes. Among these, α-galactosidases are widely used in several industrial bioprocesses that require high working temperatures and for which thermostable variants offer considerable advantages over their thermolabile counterparts.
Results
Thermus thermophilus HB27 strain was used for the homologous expression of the TTP0072 gene encoding for an α-galactosidase (TtGalA). Interestingly, a soluble and active histidine-tagged enzyme was produced in larger amounts (5 mg/L) in this thermophilic host than in Escherichia coli (0.5 mg/L). The purified recombinant enzyme showed an optimal activity at 90 °C and retained more than 40% of activity over a broad range of pH (from 5 to 8).
Conclusions
TtGalA is among the most thermoactive and thermostable α-galactosidases discovered so far, thus pointing to T. thermophilus as cell factory for the recombinant production of biocatalysts active at temperature values over 90 °C.
Electronic supplementary material
The online version of this article (doi:10.1186/s12934-017-0638-4) contains supplementary material, which is available to authorized users.
Keywords: α-Galactosidase, Thermus thermophilus, Thermozymes, Recombinant expression, Themostability
Background
After cellulose, the second most abundant biopolymer on earth is hemicellulose, an heterogeneous polymer of pentoses (xylose and arabinose) and hexoses (glucose, galactose, mannose) [1]. Among these, mannans comprise linear or branched polymers derived from sugars such as d-mannose, d-galactose and d-glucose. Moreover, they represent the major source of secondary cell wall found in conifers (softwood) and leguminosae [2]. Based on their sugar composition, mannans are classified in four subfamilies: i.e. mannans, glucomannans, galactomannans and galactoglucomannans. The concerted action of different hydrolytic enzymes such as β-glucosidases (EC 3.2.1.21), endo-mannanases (EC 3.2.1.78), mannosidases (EC 3.2.1.25) and α-galactosidases (EC 3.2.1.22) is needed to achieve the degradation of galactoglucomannans, the most complex mannans subfamily [3].
α-Galactosidases (α-d-galactoside galactohydrolase EC 3.2.1.22) are exoglycosidases that catalyse the cleavage of the terminal non-reducing α-1,6-linked galactose residues present in different galactose-containing oligo- and polysaccharides. α-Galactosidases are widely distributed in microorganisms, plants, animals and mammalians, including humans [4]. The localization of α-galactosidases can be cytoplasmic (e.g. Escherichia coli), lysosomal (e.g. Homo sapiens) or extracellular (e.g. yeast) [5, 6].
α-Galactosidases have a great potential in both biotechnological and medical applications. For instance, these enzymes are used for the treatment of Fabry’s disease [7], in xenotransplantation [8] and in blood group transformation for safety transfusion [9]. Furthermore, α-galactosidases offer a promising solution for the degradation of raffinose in beet sugar industry [10], pulp and paper manufacturing [11] as well as in soy food [12] and animal feed processing [13].
The interest towards enzymes hydrolysing hemicellulose, such as α-galactosidases, has particularly increased in recent years. Indeed, they are extensively used in synergic combination with cellulases [14] for the production of bioethanol from lignocellulose [15]. In this regard, it is noteworthy that the pretreatment of raw lignocellulosic material requires elevated temperatures; therefore, thermostable and thermoactive enzymes are suitable for this purpose [16], since they can withstand high temperature, extreme pH and pressure, as well as the presence of some inhibitors, including toxic metals. Furthermore, as catalysts, thermostable enzymes might provide additional advantages, since higher temperatures often promote: (1) better enzyme penetration, (2) cell wall disorganisation of the raw materials, (3) increase of the substrate solubility and (4) reduction of contamination. Over all, these features may improve the global yield of the process [17].
In recent years, the thermophilic microorganism T. thermophilus has emerged has a rich source of polymer-degrading enzymes (e.g. α-amylases, xylanases, esterases, lipases, proteases and pullulanases). Indeed, this thermophilic bacterium is able to grow on different organic sources such as various proteinaceous and carbohydrates substrates [18].
Although E. coli has been successfully employed to produce thermophilic recombinant proteins/enzymes [19–24], in some cases their expression level can be too low to perform functional and structural characterization as well as to exploit their biotechnological potential [25–27]. For this reason, efficient and reliable “hot” expression systems are needed.
Some genetic systems for both archaeal and bacterial thermophilic microorganisms have been designed [28–32]. However, the amount of the recombinant proteins is in general lower than that achieved through conventional mesophilic expression systems [33].
A thermophilic expression system for T. thermophilus HB27 was previously developed and proved to be suitable to achieve high expression levels of heterologous proteins [34, 35]. Therefore, we have used this system to produce a novel α-galactosidase (named TtGalA) from T. thermophilus HB27. In particular, the coding gene was over-expressed in the native host and the recombinant enzyme was purified and characterized. TtGalA exhibits an optimal hydrolytic activity at high temperature (90 °C and pH 6.0), a good catalytic efficiency (kcat = 709.7/s) and a significant thermostability (30 h at 70 °C), which are all interesting features for industrial applications.
Results and discussion
The genus Thermus comprises thermophilic and hyperthermophilic bacteria [18], which represent genetic reservoirs of several thermozymes potentially useful for industrial bioprocesses at high temperature [36, 37]. This aspect, along with other intrinsic features such as the natural competence of many strains, high growth rates and biomass yields, make these thermophiles suitable models for the production of thermozymes [27, 38].
In this work we have used T. thermophilus HB27::nar as host for the homologous expression of a novel α-galactosidase (TtGalA), one of the most thermoactive α-galactosidases identified so far.
Sequence analysis
An in-depth survey of the carbohydrate-active enzymes database CAZy (http://www.cazy.org/) was carried out to identify putative α-galactosidases in the genome of T. thermophilus HB27. The only protein sequence retrieved is encoded by TTP0072 (Accession No. AAS82402) and shows high sequence identity with previously characterized α-galactosidases from Thermus sp. strain T2 (75%; Accession No. BAA76597), Thermus brockianus (73%; Accession No. AF135398), Thermotoga neapolitana (36%; Accession No. AF011400), Thermotoga maritima (35%; Accession No. AJ001776), Sulfolobus solfataricus (39%; Accession No. Q97U94) and H. sapiens (23%; Accession No. CAA29232.1). In general, sequence identities among the analysed proteins fall into regions that are “signature” of the catalytic activity and/or of the structural properties of α-galactosidases. A multiple alignment (Fig. 1a) revealed the presence of a consensus motif (FEVFQIDDGW) in the TTP0072 translated sequence characteristic of α-galactosidases. This is generally localised within the central region of bacterial enzymes (CAZy family GH36) or at the amino-terminal region of eukaryotic variants (CAZy family GH27). The presence of such a sequence points out to a common reaction mechanism and/or a similar substrate binding site [39]. Two aspartic residues i.e. D193–194 underlined in the above consensus sequence are followed by a cysteine or a glycine in the GH27 or GH36 members, respectively. Moreover, GH27 and GH36 families share another common feature, which is the presence of two additional aspartic acids involved in the acid–base catalytic mechanism (D301 and D355 in the protein from T. thermophilus HB27, Fig. 1). At consensus level, whereas the motif including the catalytic nucleophile (D301) is fully conserved [K(Y/V/L/W)D], the A/B aspartic acid motif (D355) is more variable (RXXXD) (Fig. 1c). The co-presence of these two motifs defines the sub-group identity of GH36bt (where “bt” stands for bacterial thermophilic) [40] that also includes the thermophilic Thermotoga and Thermus α-galactosidases. Another characteristic of the GH36bt enzymes is the presence of two conserved cysteine residues (C161 and C336) (Fig. 1b) whose functional, structural and stabilization role is questioned.
Fig. 1.

Sequence homology of α-galactosidases from different sources. The amino acid sequences of Thermus thermophilus HB27 (Accession No. AAS82402), Thermus sp. strain T2 (75%), Thermus brockianus (73%), Thermotoga neapolitana (36%), Thermotoga maritima (35%), Sulfolobus solfataricus (39%) and Homo sapiens (23%) were aligned for optimal sequence similarity using the program CLUSTAL W. a The consensus motif ([LIVMFY]-x(2)-[LIVMFY]-x-[LIVM]-D-D-x-[WY]) is characteristic of α-galactosidases; b amino acid stretches including two conserved cysteine residues; c amino acid stretches surrounding the aspartic acids responsible for the nucleophilic and acid–base catalytic mechanism. Conserved amino acids are highlighted with asterisk in T. thermophilus HB27 sequence
The TTP0072 gene is located on the megaplasmid pTT27 upstream of a gene encoding for the galactose-1-phosphate uridylyltransferase gene (galT) and partially overlapping it. This operon-like structure closely resembles that found in other Thermus spp. [41], thus suggesting that their functional association might be important for galactose metabolism.
Cloning, expression and purification of TtGalA
The TTP0072 gene was cloned both in pET28(b) and in pMKE2 vectors to compare the expression levels of the N-terminal His-tagged enzymes EcGalA and TtGalA in the heterologous (E. coli) as well as in the homologous (T. thermophilus HB27::nar) hosts. In the case of E. coli, after a two-step purification procedure i.e. a thermal precipitation at 70 °C and an affinity chromatography through His-trap column, the final protein amount was very low (0.5 mg/L) (Fig. 2a) (Additional file 1: Table S1).
Fig. 2.
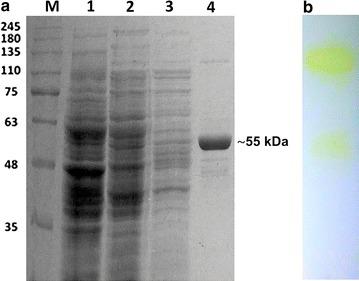
SDS-PAGE of the recombinant α-galactosidase TtGalA fused to a His-tag at it N-terminal sequence. a M, molecular mass markers; 1, T.thermophilus::nar cellular extract not transformed; 2, T.thermophilus::nar pMKE2-TtgalA cellular extract after overnight growth; 3, anionic exchange chromatography; 4, His-Trap affinity chromatography; b zimography
On the other hand, the enzyme TtGalA was expressed at higher level in T. thermophilus HB27::nar with a final amount of 5 mg/L (tenfold higher than EcGalA). Moreover, its specific activity (338 U/mg) was twofold higher than that of EcGalA (Additional file 1: Tables S1, S2). As previously observed, the over-production of soluble and active enzymes in mesophilic hosts is in some cases limited by: (1) differences in the codon usage [42]; (2) domain misfolding during protein synthesis at a temperature (37 °C), which is far lower than the optimal growth temperature of the native host; (3) requirement of specific chaperone(s), cofactors, metals as well as genus- or species-specific post-translational modifications [27].
Given the significant higher amount of the soluble active enzyme produced by the thermophilic host, all the subsequent characterizations were performed on the TtGalA enzyme. The nar promoter, which drives the expression of the genes cloned in the pMKE2 vector, is induced by the combined action of nitrate and anoxia in the facultative anaerobic derivatives of T. thermophilus HB27 [34]. In our tested conditions, the biomass yield of the culture achieved under anaerobic growth was rather low, thus negatively affecting the amount of enzyme produced by the cells. To overcome this limitation, the expression of TtGalA was carried out by growing T. thermophilus HB27::nar cells aerobically to reach an higher biomass yields. A total protein amount, similar to that of the cells cultured anaerobically, was achieved.
We resolved to set up a purification protocol from crude extracts of aerobically cultured cells based on anionic exchange followed by affinity His-trap chromatography. The recombinant enzyme purified to homogeneity displays a single band on SDS-PAGE with an estimated molecular weight (MW) of 55 kDa (Fig. 2a), which is consistent with the predicted MW of a his-tagged monomer (55.8 kDa). The identity of the recombinant TtGalA was verified by mass spectrometry (data not shown). It is important to note that the presence of the His-tag at the N-terminus of the recombinant enzyme ensures that the reported experiments are not affected by the presence of the endogenous enzyme. Furthermore, zymography revealed the presence of two bands with hydrolytic activity indicating that: (1) TtGalA adopts an oligomeric structure; (2) the oligomer is far more active than the monomer and (3) it is partially resistant to the denaturing conditions employed in the SDS-PAGE. These results prompted us to further investigate on the quaternary structure of TtGalA (Fig. 2b).
Characteristics of the recombinant TtGalA
Determination of the molecular weight
To assess the quaternary structure, size-exclusion chromatography coupled with a triple-angle light scattering-QELS, was performed. This analysis showed a molecular weight of about 320 kDa ± 0.2% (RH = 8.1 nm ± 3%), thus indicating that TtGalA is a hexamer in solution. This oligomeric structure is in agreement with that of some previously characterised α-galactosidases, which adopt dimeric (Thermotoga maritima) [43], trimeric (Sulfolobus solfataricus) [40], tetrameric (T. brockianus) [39], octameric (Thermus sp. strain T2) structure [44]. This complex oligomeric state might correlate with the stability at higher temperature of TtGalA, as it was showed for B. stearothermophilus α-galactosidases [45].
To understand if C161 and C336 had a structural role (Fig. 1b), the purified TtGalA was analysed on SDS-PAGE with and without a reducing agent. In the sample containing β-mercaptoethanol TtGalA is present mainly in monomeric form, while in the absence of this reducing agent the protein forms essentially high MW oligomers (data not shown). Indeed, the widespread stabilizing role of intracellular disulphide bonds in thermophiles and hyperthermophiles, has been already established as a strategy for protein stabilization [46].
Catalytic and stability properties
TtGalA is able to hydrolyze pNP-α-d-galactopyranoside, but shows negligible activity on both pNP-α-substituted (d-glucose, d-mannose, l-rhamnose) and pNP-β-substituted (d-galactose, d-glucose, and d-mannose) (Table 1). In particular, the enzyme has a barely detectable activity for β anomer of galactose (6.75 U/mg), which is 50-fold lower than activity over pNP-α-galactose (338.0 U/mg). Therefore, the kinetic enzymatic properties were determined, using pNP-α-d-galactopyranoside as substrate, at optimal pH and temperature (Table 2). TtGalA shows higher affinity towards its substrate (KM = 0.69 mM) compared to α-galactosidase from Thermus sp. strain T2 (KM = 4.7 mM) [44] and T. brockianus (KM = 2.5 mM) [39]. However, comparing the kinetic parameters between the most known thermoactive (Topt 105 °C) α-galactosidase of Thermotoga neapolitana (TnGalA) and the TtGalA, the KM value is very similar. Interestingly, the different catalytic constant of 152.5/s towards 709.7/s, for TnGalA and TtGalA respectively [47] (Table 2) reflects a great efficiency of TtGalA on pNP-α-d-galactopyranoside substrate. This aspect constitutes an important criterion for employing different enzyme variants for industrial purposes. For istance, the high catalytic efficiency of TtGalA makes it a suitable candidate to enhance pulp bleachability in combination with other hemicellulases exhibiting different catalytic specificity [11].
Table 1.
Substate specificity of TtGalA
| Substrate | Specific activity (U/mg) |
|---|---|
| pNP-α-d-mannose | 0 |
| pNP-β-glucose | 0.65 |
| pNP-α-l-rhamnose | 1.5 |
| pNP-α-d-glucose | 2.16 |
| pNP-β-galactose | 6.75 |
| pNP-β-mannose | 7.13 |
| pNP-α-d-galactose | 338.0 |
Table 2.
Kinetic parameters for the hydrolysis pNPG hydrolysis at 90 °C by the TtGalA
| KM (mM) | Vmax (U/mg) | kcat (/s) | kcat/KM (/s M) |
|---|---|---|---|
| 0.69 ± 0.017 | 338.0 ± 7.9 | 709.7 ± 17.7 | 1.03 × 104 ± 0.025 × 104 |
TtGalA is among the most thermoactive and pH tolerant α-galactosidases known so far. Indeed, when assayed at different pH and temperatures, it exhibited an optimal hydrolytic activity at 90 °C and pH 6.0 (Figs. 3, 4). As reported for an α-galactosidase from Bacillus megaterium VHM1 (optimal pH 7–7.5) [48], it is possible to foresee the employment of TtGalA for the removal of oligosaccharides from soya based foods, thus improving their nutritive value. Noteworthy, TtGalA might be a better catalyst for this process, since its pH optimum (pH 6.0) is even closer to that of the soymilk hydrolysis process (pH 6.2–6.4).
Fig. 3.
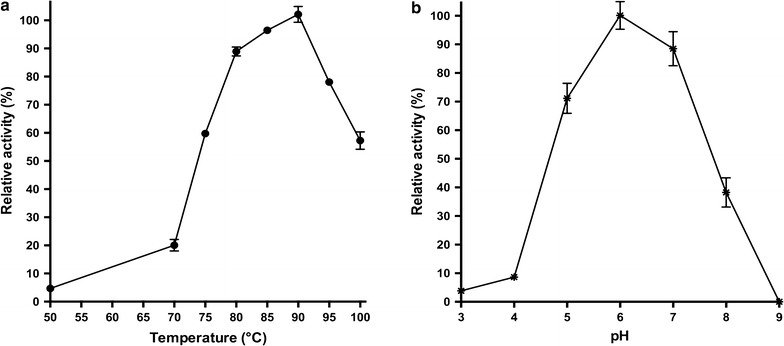
a Determination of the TtGalA catalytic activity in the temperature range of 50 to 100 °C. pNP-α-d-galactopyranoside was used as substrate dissolved in 50 mM sodium phosphate buffer pH 6.0. Relative activity at 90 °C was considered as 100%. b The relative activity was measured between pH 3.0 and pH 9.0, at 90 °C considering as 100% the activity at pH 6.0
Fig. 4.
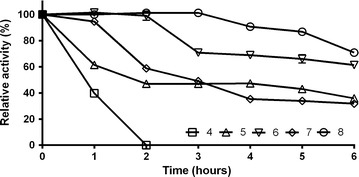
pH stability of TtGalA. The enzyme was incubated in various buffers (pH 4–8) and aliquots of different time intervals were used for the residual activity assay
Interestingly, the retained activity was greater than 40% within the pH range from 5.0 to 8.0, which is a quite wide tolerance range compared to other characterised thermostable α-galactosidases [40]. Therefore, pH shifts during on-going enzymatic reaction in industrial processes could have a minor impact on its activity (Fig. 3). The recombinant enzyme did not lose activity after 2 h of incubation at pH 6.0 and 8.0, and it retained up to 60% of residual activity after 6 h (Fig. 4). Approximately 50–60% loss in activity was recorded on either side of the pH optimum after 6 h incubation, while at pH 4.0, the activity rapidly dropped, as expected from the pH-dependence data (Fig. 4). Because of its considerable tolerance towards neutral/slightly alkaline pH values, TtGalA could be employed in hydrothermal processing in which water in liquid phase or in vapour phase is used to pretreat lignocellulosic biomasses [49]. In this process, controlling the pH around neutral values minimizes the formation of fermentation inhibitors [50]. Typically, the optimal temperature for catalysis of thermophilic enzymes mirrors the growing temperature of the native host, like the α-galactosidase from Thermus sp. strain T2 (75 °C) [44], whereas their activity is limited at lower temperatures. TtGalA is unusual since its optimum was set at 90 °C and its activity was lower at temperature ≤70 °C, which is the optimal growth temperature for most of Thermus species. Moreover, TtGalA is among the most thermophilic α-galactosidases so far characterised, such as that of T. neapolitana (105 °C) [47], T. brockianus (94 °C) and T. maritima (95 °C) [39, 43].
The thermal inactivation data indicate that the TtGalA has a half-life of 60 min (Fig. 5) at its optimal temperature (90 °C). Moreover, residual activity higher than 90% was detected up to 6 h of incubation at 70 °C (Fig. 5), retaining 50% of its activity after 30 h (not shown).
Fig. 5.
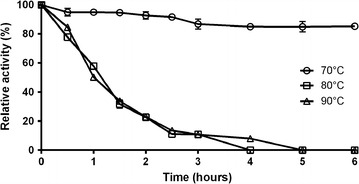
Thermal inactivation of TtGalA. The purified enzyme from T. thermophilus was incubated in 20 mM sodium-phosphate buffer pH 6.0 at 90, 80 and 70 °C for different period of time and then assayed for residual activity at 90 °C
It has already been reported that previously characterised enzymes from Thermus species display in vitro an optimal catalytic temperature higher than the growing temperature of the native host [51], thus making this microorganism a fruitful source of enzyme catalytically active at temperature above 70 °C. Due to the elevated temperatures used during some industrial applications (such as sugar manufacturing processes and/or raw material pretreatments in bioethanol production), stability at high temperatures is an important feature for the utilization of α-galactosidases, since it prevents the loss of ternary/quaternary structures that leads to enzyme inactivation as for the mesophilic counterparts [36, 37].
Effect of metal ions
Metal ions can be released during processing of biomass as consequence of corrosion of pretreatment equipment, resulting in the liberation of heavy metal ions, which can be inhibitory to biocatalysts [52]. Moreover, other cations can derive from chemicals used to adjustment the pH. Noteworthy, several divalent cations are potent inhibitors of α-galactosidases; therefore, we tested their effect on the enzyme activity over a range of concentration from 0.5 to 5 mM (Table 3). Interestingly, the enzyme turned to be slightly activated in the presence of Co+2, Mn+2, Zn+2 at 1 mM concentration suggesting that it is a metalloenzyme or requires divalent cations as cofactors. On the other hand, this effect is negligible with monovalent cations (Table 3). The inhibition effect occurred only at the highest concentration of metal ions tested (5 mM). To further investigate on the possible role of metal ions, the enzyme activity was assayed in the presence of EDTA (5 mM) and a 20% reduction of its catalytic activity was observed. The inhibitory effect of EDTA was studied up to 40 mM concentration (not shown) resulting in a linear decrease of the enzymatic activity, thus indirectly confirming the role as cofactor of the metal ions (Co+2, Mn+2, Zn+2) (Table 3). Similar to these findings, EDTA slightly inhibited also the α-galactosidase activity from Lenzites elegans [53], whereas it has no effect on the activity of α-galactosidases from B. megaterium and Ganoderma lucidum [48, 54].
Table 3.
Influence of metal ions on the relative activity of TtGalA
| Metal ions | 0.5 mM | 1 mM | 2.5 mM | 5 mM |
|---|---|---|---|---|
| Mg+2 | 99.3 ± 0.7 | 91.7 ± 0.3 | 89.9 ± 2.5 | 55.6 ± 0.5 |
| Ca+2 | 95.9 ± 0.7 | 95.8 ± 0.1 | 90.1 ± 2.4 | 27.8 ± 0.4 |
| Cu+2 | 87. 7 ± 3.1 | 77.2 ± 1.6 | 63.9 ± 0. 97 | 22.2 ± 0.3 |
| Li+ | 106.3 ± 0.5 | 113.9 ± 3.6 | 114.8 ± 3.1 | 93.6 ± 2.5 |
| Zn+2 | 100.6 ± 2.2 | 110.3 ± 0.6 | 70.4 ± 0.6 | 15.1 ± 0.3 |
| Mn+2 | 103.4 ± 1.0 | 128.5 ± 1.9 | 114.8 ± 3.0 | 20.2 ± 0.6 |
| Co+2 | 101.8 ± 2.6 | 136.2 ± 0.4 | 92.8 ± 0.9 | 30.1 ± 0.7 |
Inhibition of activity
Since detergents are reported to strongly inhibit the α-galactosidase activity in Glycine max and Pencillium griseoroseum [55, 56], we assayed TtGalA in the presence of 5 mM detergents. The enzyme turned out to be very sensitive to the common anionic detergent SDS, which leads to a complete loss of function possibly due to the disruption of enzyme native structure. By contrast, the non-ionic detergents Tween 20 and Triton X-100 had a less marked effect with reduction of the enzyme activity to ~40 and ~19%, respectively (Table 4).
Table 4.
Influence of additives on the activity of TtGalA
| Compound (5 mM) | Relative activity (%) |
|---|---|
| EDTA | 79.9 |
| d-Galactose | 27.9 |
| d-Saccarose | 44.6 |
| d-Arabinose | 54.7 |
| Urea | 60.0 |
| Guanidine chloride | 36.8 |
| SDS | 1.6 |
| Tween 20 | 39.8 |
| Triton X100 | 18.8 |
During pretreatment and saccharification of lignocellulosic biomasses, several sugars are released. These can inhibit the activity of glycoside hydrolases during the saccharification phase [49, 50]. Moreover, various sugars were also reported to inhibit the α-galactosidase activity, for instance, an α-galactosidase from Aspergillus nidulans is competitively inhibited by d-galactose and d-glucose [57]. Accordingly, the activity of TtGalA turned out to be inhibited by the presence of several saccharides, such as d-galactose, d-saccarose and d-arabinose (Table 4). Nevertheless, TtGalA might have a potential use in the sugar beet industry for raffinose hydrolysis, because it retains 44.6% of its activity in presence of sucrose [10].
Finally, TtGalA was assayed in presence of caotropic agents such as urea and guanidine chloride. The partial reduction of the catalytic activity is in agreement with its intrinsic stability as thermozyme [20].
Conclusions
In this work, we report the biochemical characterization of a thermoactive and thermostable α-galactosidase from T. thermophilus HB27 (TtGalA). Moreover, the drawbacks of using a heterologous mesophilic host (E. coli) for the production of this thermozyme have been highlighted. Indeed, “hot” expression systems are in some cases indispensable to get functional thermozymes in reasonable amounts.
Interestingly, the long-term retained activity of TtGalA (30 h at 70 °C) might pave the way to its utilization after thermal pretreatment of lignocellulosic biomass (pre-saccharification), when the temperature is still too high for the fungal enzymes currently used for the hydrolysis of the biomass.
Despite its already interesting catalytic features, a fine-tuning of TtGalA enzymatic properties, through genetic engineering, will be attempted to make it even more suitable for industrial applications.
Methods
Bacterial strains and growth conditions
Thermus thermophilus HB27 strain was purchased from DSMZ. A frozen (−80 °C) stock culture was streaked on a Thermus Medium (TM) solidified by the addition of 0.8% Gelrite® (Sigma) and incubated at 70 °C overnight [58]. T. thermophilus HB27::nar strain, kindly provided by Prof. J. Berenguer (Universidad Autónoma de Madrid) was used for the homologous expression of TtGalA. E. coli strains were grown in Luria–Bertani (LB) medium at 37 °C with 50 μg/ml kanamycin, 33 μg/ml chloramphenicol as required. E. coli DH5α and BL21-CodonPlus (DE3)-RIL (Stratagene, La Jolla, CA, USA) strains were used for DNA manipulations and for the heterologous expression of the recombinant α-galactosidase, respectively.
Cloning and sequencing of TTP0072 gene
A single colony of T. thermophilus HB27 was inoculated into TM liquid medium and genomic DNA was isolated using DNeasy® 124 Tissue kit, (Qiagen), according to the instruction manufacturer. TTP0072 gene, encoding for a putative α-galactosidase, was amplified by PCR from T. thermophilus HB27 genomic DNA using the primers 5′GGAGGGCATATGAGGCTGAA3′ (NdeI site underlined) and 5′CGGTGGAAGCTTTTATAGAAGG3′ (HindIII site underlined) and Phusion Taq Polymerase (NEB). The amplification was carried out at 94 °C for 1 min, 58 °C for 1 min and 72 °C for 1 min, for 35 cycles. The PCR product was purified with QIAquick PCR purification kit (Qiagen Spa, Milan, Italy) and cloned in pCR4-TOPO_vector using the TOPO TA CLONING Kit (Invitrogen). The identity of the cloned DNA fragment was confirmed by DNA sequencing (BMR Genomics). Then, the insert was sub-cloned into the NdeI/HindIII digested pET28(b) and for pMKE2 [34] vectors for E. coli and T. thermophilus HB27::nar expression, respectively.
Expression and purification of recombinant EcGalA and TtGalA
The recombinant α-galactosidases expressed in E. coli (EcGalA) and T. thermophilus HB27::nar (TtGalA) bear an His-tag at their N-terminus. To express EcGalA, E. coli BL21-CodonPlus (DE3)-RIL was transformed with the recombinant plasmid pET28(b)-EcgalA. Protein expression was induced by adding 0.5 mM of isopropil-β-d-1-thiogalactopyranoside (IPTG) to exponentially growing cells (0.5 OD600) and culturing them for 12 h. Since EcGalA was poorly expressed, different approaches were attempted to achieve a sufficient amount of soluble recombinant protein: (1) varying the induction time (2, 4, 6, and 8 h and overnight induction); (2) lowering the temperature during induction (down to 20 °C); (3) decreasing the IPTG concentration (0.01–0.1 mM). At every conditions, the expression levels were monitored by SDS-PAGE and enzymatic assays. However, none of these strategies turned out to reasonably increase the final amount of the recombinant protein.
For the homologous expression 200 ng of pMKE2-TtgalA plasmid were added to exponentially growing (0.4 OD600nm) T. thermophilus HB27::nar cells and transformants were selected on TM plates at 70 °C containing 50 μg/ml kanamycin [59]. The induction of TtGalA expression was performed as previously described [34].
Crude extracts from both E. coli BL21-CodonPlus (DE3)-RIL and T. thermophilus HB27::nar cells were prepared following a similar procedure. Cell pellets were collected from 1-L cultures by centrifugation at 4000×g for 15 min at 4 °C and resuspended in buffer A (50 mM Tris–HCl, pH 7.5 and 500 mM NaCl) for EcGalA and in buffer B (50 mM Tris–HCl, pH 7.5) for TtGalA purification, respectively. Protease inhibitor cocktail tablets (Roche) were added in both cases. The cells were homogenized by sonication (Sonicator:Heat System Ultrasonic, Inc.) for 5 min, alternating 30 s of pulse-on and 30 s of pulse-off and clarification of the cell extract was obtained by centrifugation at 40,000×g for 20 min at 4 °C. Purification of EcGalA from the soluble fraction was performed through a two-step procedure, i.e. (1) thermal precipitation at 70 °C for 10 min followed by centrifugation at 5000×g for 20 min at 4 °C; (2) affinity chromatography on a His-Trap column (1 ml, GE Healthcare) connected to an AKTA Explorer system (GE Healthcare). The affinity chromatography was equilibrated with buffer A and the EcGalA was eluted with the same buffer A supplemented with a linear gradient of imidazole (0–250 mM).
TtGalA was purified through a similar procedure, except that the first thermal precipitation step was substituted with an anionic exchange chromatography on a Hi-trap Q HP column (5 ml, GE Healthcare). The column was equilibrated in buffer B and elution was performed in the same buffer through a linear gradient from 0 to 500 mM NaCl. The affinity chromatography was carried out under the same conditions described above. Protein identity was further verified by Western blot analysis using anti-His antibodies and LC–MS/MS analysis.
Protein concentration was estimated by using bovine serum albumin as standard according to Bradford [60]. Protein fractions displaying α-galactosidase hydrolysing activity toward p-nitrophenyl-α-d-galactopyranoside (herein named pNP-α-d-galactopyranoside, Sigma) were pooled, dialyzed against 20 mM Tris–HCl pH 7.5 and analysed by 12% SDS-PAGE [61]. The TtGalA activity was detected through zimography in 12% SDS-PAGE under not reducing conditions. After electrophoresis, the gel was soaked in 2.5% Triton X-100 for 30 min at 4 °C and then was incubated with 20 mM of pNP-α-d-galactopyranoside solution at 90 °C for 10 min.
Molecular weight determination
The native molecular weight of the purified TtGalA was analysed by gel-filtration chromatography connected to Mini DAWN Treos light-scattering system (Wyatt Technology) equipped with a QELS (quasi-elastic light scattering) module mass value and hydrodynamic radius (Rh) measurements [62]. Five hundreds micrograms of protein (1 mg/ml) were loaded on a S200 column (16/60, GE Healthcare) with a flow-rate of 0.5 ml/min and equilibrated in buffer A. Data were analysed using Astra 5.3.4.14 software (Wyatt Technology).
Determination of the α-galactosidase activity
The TtGalA standard assay was performed by using pNP-α-d-galactopyranoside as substrate (2.0 mM) in 50 mM sodium phosphate buffer (pH 6.0) at 90 °C in 160 μl of the reaction mixture using 50 ng of the enzyme. All assays were performed in triplicate in a 96-well microplate reader (Synergy H4, Biotek), on at least 3 different enzyme preparations. The reaction was stopped, after 10 min, by addition of 1 volume of cold 0.5 M Na2CO3 and the concentration of the released p-nitrophenol (molar extinction coefficient, 18.5/mM cm) was determined by measuring A405nm. One unit of enzyme activity (U) was defined as the amount of enzyme required to release 1 μmol p-nitrophenol per minute, under the above assay conditions.
Catalytic and stability properties
The optimal pH and temperature were determined by performing the pNP-α-d-galactopyranoside assay between pH 3.0–9.0 using the following buffers (each 50 mM): citrate phosphate (pH 3.0–5.0), sodium phosphate (pH 6.0–9.0). Thermal inactivation assays were performed by incubating 50 ng of enzyme at different temperatures (70, 80, 90 °C) in buffer sodium phosphate at pH 6.0 and taking aliquots at regular time intervals to measure the residual enzyme activity under standard assay conditions (90 °C for 10 min, pH 6.0). To test enzyme stability to pH, the assays were performed by incubating TtGalA at 70 °C in various buffers (pH 4.0–8.0). The residual activity was measured at different time intervals following the α-galactosidase standard assay.
Inhibition of activity
The effect of MgCl2, CaCl2, CuCl2, LiCl, ZnSO4, MnCl2, CoSO4 on the α-galactosidase activity was studied over a range of concentrations (0.5–5.0 mM) by pre-incubating the enzyme (50 ng) for 5 min at room temperature with the specific metal ions and by measuring the residual activity under standard assay conditions.
The influence of reducing (DTT, β-mercaptoethanol) and chelating (EDTA) agents as well as of sugars (d-galactose, sucrose, l-arabinose, and d-fucose), each tested at 5 mM concentration, was studied under the same pre-incubation and assay conditions as above.
Substrate specificity
TtGalA was tested for the hydrolysis of pNP-α-substituted hexoses (d-glucose, d-mannose, l-rhamnose) and of pNP-β-substituted hexoses (d-galactose, d-glucose, and d-mannose) at a concentration of 2 mM under standard conditions (90 °C and pH 6.0 for 10 min). The kinetic parameters were determined using different pNP-α-d-galactopyranoside concentration (ranging from 0.0125 to 2 mM) with 50 ng of TtGalA for 3 min. The Michaelis constant (KM) and Vmax were calculated by non-linear regression analysis using GraphPad 6.0 Prism software.
Authors’ contributions
MA and SF performed experiments. SF, GF, DL, EP, SB and PC supervised the project. MA and PC drafted the manuscript. All authors read and approved the final manuscript.
Acknowledgements
We thank Dr. Andrea Carpentieri for performing the LC–MS/MS analysis and Prof. José Berenguer who kindly provided us with the pMKE2 plasmid.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
Recombinant strains described in this work are made available upon request to the corresponding author. Data sharing not applicable to this article as no datasets were generated or analysed during the current study.
Funding
This work was supported by BIOPOLIS: PON03PE_00107_1 CUP: E48C14000030005.
Abbreviations
- bt
bacterial thermophilic
- CAZy
carbohydrate-active enzymes
- DTT
dithiothreitol
- EcGalA
Thermus thermophilus α-galactosidase expressed in E. coli
- EDTA
ethylenediaminetetraacetic acid
- GH
glycoside hydrolase
- g
gravity
- h
hour(s)
- IPTG
isopropil-β-d-1-tiogalattopiranoside
- kcat
catalytic constant
- KM
michaelis constant
- LB
Luria-Bertani broth
- LC–MS/MS
liquid chromatography-tandem mass spectrometry
- pNP
P-nitrophenol
- QELS
quasi-elastic light scattering
- Rh
hydrodynamic radius
- s
second(s)
- TM
thermus medium
- TnGalA
Thermotoga neapolitana α-galactosidase
- TtGalA
Thermus thermophilus α-galactosidase
- SDS-PAGE
sodium dodecyl sulphate-polyacrylamide gel electrophoresis
Additional file
Additional file 1: Table S1. Purification table of EcGalA. Table S2. Purification table of TtGalA.
Contributor Information
Martina Aulitto, Email: martina.aulitto@unina.it.
Salvatore Fusco, Email: fuscos@chalmers.se.
Gabriella Fiorentino, Email: fiogabri@unina.it.
Danila Limauro, Email: limauro@unina.it.
Emilia Pedone, Email: empedone@unina.it.
Simonetta Bartolucci, Email: bartoluc@unina.it.
Patrizia Contursi, Phone: (++39) 081 679 166, Email: contursi@unina.it.
References
- 1.Pauly M, Keegstra K. Plant cell wall polymers as precursors for biofuels. Curr Opin Plant Biol. 2010;13:304–311. doi: 10.1016/j.pbi.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 2.Malgas S, van Dyk JS, Pletschke BI. A review of the enzymatic hydrolysis of mannans and synergistic interactions between β-mannanase, β-mannosidase and α-galactosidase. World J Microbiol Biotechnol. 2015;31:1167–1175. doi: 10.1007/s11274-015-1878-2. [DOI] [PubMed] [Google Scholar]
- 3.Chauhan PS, Puri N, Sharma P, Gupta N. Mannanases: microbial sources, production, properties and potential biotechnological applications. Appl Microbiol Biotechnol. 2012;93:1817–1830. doi: 10.1007/s00253-012-3887-5. [DOI] [PubMed] [Google Scholar]
- 4.Marraccini P, Rogers WJ, Caillet V, Deshayes A, Granato D, Lausanne F, Lechat S, Pridmore D, Pétiard V. Biochemical and molecular characterization of alpha-d-galactosidase from coffee beans. Plant Physiol Biochem. 2005;43:909–920. doi: 10.1016/j.plaphy.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 5.Pourcher T, Bassilana M, Sarkar HK, Kaback HR, Leblanc G. Melibiose permease and alpha-galactosidase of Escherichia coli: identification by selective labeling using a T7 RNA polymerase/promoter expression system. Biochemistry. 1990;29:690–696. doi: 10.1021/bi00455a014. [DOI] [PubMed] [Google Scholar]
- 6.Yang D, Tian G, Du F, Zhao Y, Zhao L, Wang H, Ng TB. A fungal alpha-galactosidase from Pseudobalsamia microspora capable of degrading raffinose family oligosaccharides. Appl Biochem Biotechnol. 2015;176:2157–2169. doi: 10.1007/s12010-015-1705-0. [DOI] [PubMed] [Google Scholar]
- 7.Linthorst GE, Hollak CE, Donker-Koopman WE, Strijland A, Aerts JM. Enzyme therapy for Fabry disease: neutralizing antibodies toward agalsidase alpha and beta. Kidney Int. 2004;66:1589–1595. doi: 10.1111/j.1523-1755.2004.00924.x. [DOI] [PubMed] [Google Scholar]
- 8.Lim HG, Kim GB, Jeong S, Kim YJ. Development of a next-generation tissue valve using a glutaraldehyde-fixed porcine aortic valve treated with decellularization, α-galactosidase, space filler, organic solvent and detoxification. Eur J Cardiothorac Surg. 2015;48:104–113. doi: 10.1093/ejcts/ezu385. [DOI] [PubMed] [Google Scholar]
- 9.Olsson ML, Hill CA, De La Vega H, Liu QP, Stroud MR, Valdinocci J, Moon S, Clausen H, Kruskall MS. Universal red blood cells—enzymatic conversion of blood group A and B antigens. Transfus Clin Biol. 2004;11:33–39. doi: 10.1016/j.tracli.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 10.Linden JC. Immobilized α-d-galactosidase in the sugar beet industry. Enzym Microb Technol. 1982;4:130–136. doi: 10.1016/0141-0229(82)90103-X. [DOI] [Google Scholar]
- 11.Clarke JH, Davidson K, Rixon JE, Halstead JR, Fransen MP, Gilbert HJ, Hazlewood GP. A comparison of enzyme-aided bleaching of softwood paper pulp using combinations of xylanase, mannanase and alpha-galactosidase. Appl Microbiol Biotechnol. 2000;53:661–667. doi: 10.1007/s002530000344. [DOI] [PubMed] [Google Scholar]
- 12.Pessela BC, Fernández-Lafuente R, Torres R, Mateo C, Fuentes M, Filho M, Vian A, García JL, Guisán JM, Carrascosa AV. Production of a thermoresistant alpha-galactosidase from Thermus sp. strain T2 for food processing. Food Biotechnol. 2007;21:91–103. doi: 10.1080/08905430701191221. [DOI] [Google Scholar]
- 13.Prashanth SJ, Mulimani V. Soymilk oligosaccharide hydrolysis by Aspergillus oryzae α-galactosidase immobilized in calcium alginate. Process Biochem. 2005;40:1199–1205. doi: 10.1016/j.procbio.2004.04.011. [DOI] [Google Scholar]
- 14.Rosgaard L, Pedersen S, Langston J, Akerhielm D, Cherry JR, Meyer AS. Evaluation of minimal Trichoderma reesei cellulase mixtures on differently pretreated Barley straw substrates. Biotechnol Prog. 2007;23:1270–1276. doi: 10.1021/bp070329p. [DOI] [PubMed] [Google Scholar]
- 15.Sun Y, Cheng J. Hydrolysis of lignocellulosic materials for ethanol production: a review. Bioresour Technol. 2002;83:1–11. doi: 10.1016/S0960-8524(01)00212-7. [DOI] [PubMed] [Google Scholar]
- 16.Linares-Pasten JA, Andersson M, Karlsson NE. Thermostable glycoside hydrolases in biorefinery technologies. Current Biotechnol. 2014;3:26–44. doi: 10.2174/22115501113026660041. [DOI] [Google Scholar]
- 17.Paës G, O’Donohue MJ. Engineering increased thermostability in the thermostable GH-11 xylanase from Thermobacillus xylanilyticus. J Biotechnol. 2006;125:338–350. doi: 10.1016/j.jbiotec.2006.03.025. [DOI] [PubMed] [Google Scholar]
- 18.Henne A, Brüggemann H, Raasch C, Wiezer A, Hartsch T, Liesegang H, Johann A, Lienard T, Gohl O, Martinez-Arias R, et al. The genome sequence of the extreme thermophile Thermus thermophilus. Nat Biotechnol. 2004;22:547–553. doi: 10.1038/nbt956. [DOI] [PubMed] [Google Scholar]
- 19.Limauro D, D’Ambrosio K, Langella E, De Simone G, Galdi I, Pedone C, Pedone E, Bartolucci S. Exploring the catalytic mechanism of the first dimeric Bcp: functional, structural and docking analyses of Bcp4 from Sulfolobus solfataricus. Biochimie. 2010;92:1435–1444. doi: 10.1016/j.biochi.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 20.Contursi P, D’Ambrosio K, Pirone L, Pedone E, Aucelli T, She Q, De Simone G, Bartolucci S. C68 from the Sulfolobus islandicus plasmid-virus pSSVx is a novel member of the AbrB-like transcription factor family. Biochem J. 2011;435:157–166. doi: 10.1042/BJ20101334. [DOI] [PubMed] [Google Scholar]
- 21.Prato S, Vitale RM, Contursi P, Lipps G, Saviano M, Rossi M, Bartolucci S. Molecular modeling and functional characterization of the monomeric primase–polymerase domain from the Sulfolobus solfataricus plasmid pIT3. FEBS J. 2008;275:4389–4402. doi: 10.1111/j.1742-4658.2008.06585.x. [DOI] [PubMed] [Google Scholar]
- 22.Fiorentino G, Del Giudice I, Bartolucci S, Durante L, Martino L, Del Vecchio P. Identification and physicochemical characterization of BldR2 from Sulfolobus solfataricus, a novel archaeal member of the MarR transcription factor family. Biochemistry. 2011;50:6607–6621. doi: 10.1021/bi200187j. [DOI] [PubMed] [Google Scholar]
- 23.Contursi P, Farina B, Pirone L, Fusco S, Russo L, Bartolucci S, Fattorusso R, Pedone E. Structural and functional studies of Stf76 from the Sulfolobus islandicus plasmid-virus pSSVx: a novel peculiar member of the winged helix-turn-helix transcription factor family. Nucleic Acids Res. 2014;42:5993–6011. doi: 10.1093/nar/gku215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Contursi P, Fusco S, Limauro D, Fiorentino G. Host and viral transcriptional regulators in Sulfolobus: an overview. Extremophiles. 2013;17:881–895. doi: 10.1007/s00792-013-0586-9. [DOI] [PubMed] [Google Scholar]
- 25.Francis DM, Page R. Strategies to optimize protein expression in E. coli. Curr Protoc Protein Sci. 2010;Chapter 5:Unit 5.24.21–29. doi: 10.1002/0471140864.ps0524s61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosano GL, Ceccarelli EA. Recombinant protein expression in Escherichia coli: advances and challenges. Front Microbiol. 2014;5:172. doi: 10.3389/fmicb.2014.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hidalgo A, Betancor L, Moreno R, Zafra O, Cava F, Fernández-Lafuente R, Guisán JM, Berenguer J. Thermus thermophilus as a cell factory for the production of a thermophilic Mn-dependent catalase which fails to be synthesized in an active form in Escherichia coli. Appl Environ Microbiol. 2004;70:3839–3844. doi: 10.1128/AEM.70.7.3839-3844.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Contursi P, Cannio R, She Q. Transcription termination in the plasmid/virus hybrid pSSVx from Sulfolobus islandicus. Extremophiles. 2010;14:453–463. doi: 10.1007/s00792-010-0325-4. [DOI] [PubMed] [Google Scholar]
- 29.Fusco S, She Q, Bartolucci S, Contursi P. T(lys), a newly identified Sulfolobus spindle-shaped virus 1 transcript expressed in the lysogenic state, encodes a DNA-binding protein interacting at the promoters of the early genes. J Virol. 2013;87:5926–5936. doi: 10.1128/JVI.00458-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Contursi P, Fusco S, Cannio R, She Q. Molecular biology of fuselloviruses and their satellites. Extremophiles. 2014;18:473–489. doi: 10.1007/s00792-014-0634-0. [DOI] [PubMed] [Google Scholar]
- 31.Prato S, Cannio R, Klenk HP, Contursi P, Rossi M, Bartolucci S. pIT3, a cryptic plasmid isolated from the hyperthermophilic crenarchaeon Sulfolobus solfataricus IT3. Plasmid. 2006;56(35–45):31. doi: 10.1016/j.plasmid.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 32.Bartolucci S, Contursi P, Fiorentino G, Limauro D, Pedone E. Responding to toxic compounds: a genomic and functional overview of Archaea. Front Biosci (Landmark Ed) 2013;18:165–189. doi: 10.2741/4094. [DOI] [PubMed] [Google Scholar]
- 33.Suzuki H, Yoshida K-I, Ohshima T. Polysaccharide-degrading thermophiles generated by heterologous gene expression in Geobacillus kaustophilus HTA426. Appl Environ Microbiol. 2013;79:5151–5158. doi: 10.1128/AEM.01506-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moreno R, Haro A, Castellanos A, Berenguer J. High-level overproduction of His-tagged Tth DNA polymerase in Thermus thermophilus. Appl Environ Microbiol. 2005;71:591–593. doi: 10.1128/AEM.71.1.591-593.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu WL, Liao JH, Lin GH, Lin MH, Chang YC, Liang SY, Yang FL, Khoo KH, Wu SH. Phosphoproteomic analysis reveals the effects of PilF phosphorylation on type IV pilus and biofilm formation in Thermus thermophilus HB27. Mol Cell Proteomics. 2013;12:2701–2713. doi: 10.1074/mcp.M113.029330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Viikari L, Alapuranen M, Puranen T, Vehmaanperä J, Siika-Aho M. Thermostable enzymes in lignocellulose hydrolysis. Adv Biochem Eng Biotechnol. 2007;108:121–145. doi: 10.1007/10_2007_065. [DOI] [PubMed] [Google Scholar]
- 37.Elleuche S, Schäfers C, Blank S, Schröder C, Antranikian G. Exploration of extremophiles for high temperature biotechnological processes. Curr Opin Microbiol. 2015;25:113–119. doi: 10.1016/j.mib.2015.05.011. [DOI] [PubMed] [Google Scholar]
- 38.Sarmiento F, Peralta R, Blamey JM. Cold and hot extremozymes: industrial relevance and current trends. Front Bioeng Biotechnol. 2015;3:148. doi: 10.3389/fbioe.2015.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fridjonsson O, Watzlawick H, Gehweiler A, Rohrhirsch T, Mattes R. Cloning of the gene encoding a novel thermostable alpha-galactosidase from Thermus brockianus ITI360. Appl Environ Microbiol. 1999;65:3955–3963. doi: 10.1128/aem.65.9.3955-3963.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brouns SJ, Smits N, Wu H, Snijders AP, Wright PC, de Vos WM, van der Oost J. Identification of a novel alpha-galactosidase from the hyperthermophilic archaeon Sulfolobus solfataricus. J Bacteriol. 2006;188:2392–2399. doi: 10.1128/JB.188.7.2392-2399.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fridjonsson O, Watzlawick H, Mattes R. The structure of the alpha-galactosidase gene loci in Thermus brockianus ITI360 and Thermus thermophilus TH125. Extremophiles. 2000;4:23–33. doi: 10.1007/s007920050134. [DOI] [PubMed] [Google Scholar]
- 42.Jenney FE, Adams MW. Hydrogenases of the model hyperthermophiles. Ann NY Acad Sci. 2008;1125:252–266. doi: 10.1196/annals.1419.013. [DOI] [PubMed] [Google Scholar]
- 43.Liebl W, Wagner B, Schellhase J. Properties of an α-galactosidase, and structure of its gene galA, within an α- and β-galactoside utilization gene cluster of the hyperthermophilic bacterium Thermotoga maritima. Syst Appl Microbiol. 1998;21:1–11. doi: 10.1016/S0723-2020(98)80002-7. [DOI] [PubMed] [Google Scholar]
- 44.Ishiguro M, Kaneko S, Kuno A, Koyama Y, Yoshida S, Park GG, Sakakibara Y, Kusakabe I, Kobayashi H. Purification and characterization of the recombinant Thermus sp. strain T2 alpha-galactosidase expressed in Escherichia coli. Appl Environ Microbiol. 2001;67:1601–1606. doi: 10.1128/AEM.67.4.1601-1616.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gote M, Khan M, Gokhale D, Bastawde K, Khire J. Purification, characterization and substrate specificity of thermostable α-galactosidase from Bacillus stearothermophilus (NCIM-5146) Process Biochem. 2006;41:1311–1317. doi: 10.1016/j.procbio.2006.01.003. [DOI] [Google Scholar]
- 46.Beeby M, O’Connor BD, Ryttersgaard C, Boutz DR, Perry LJ, Yeates TO. The genomics of disulfide bonding and protein stabilization in thermophiles. PLoS Biol. 2005;3:e309. doi: 10.1371/journal.pbio.0030309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Duffaud GD, McCutchen CM, Leduc P, Parker KN, Kelly RM. Purification and characterization of extremely thermostable beta-mannanase, beta-mannosidase, and alpha-galactosidase from the hyperthermophilic eubacterium Thermotoga neapolitana 5068. Appl Environ Microbiol. 1997;63:169–177. doi: 10.1128/aem.63.1.169-177.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Patil A, Praveen Kumar S, Mulimani VH, Veeranagouda Y, Lee K. α-Galactosidase from Bacillus megaterium VHM1 and its application in removal of flatulence-causing factors from soymilk. J Microbiol Biotechnol. 2010;20:1546–1554. doi: 10.4014/jmb.0912.12012. [DOI] [PubMed] [Google Scholar]
- 49.Hu F, Ragauskas A. Pretreatment and lignocellulosic chemistry. Bioenergy Research. 2012;5:1043–1066. doi: 10.1007/s12155-012-9208-0. [DOI] [Google Scholar]
- 50.Jönsson LJ, Martín C. Pretreatment of lignocellulose: formation of inhibitory by-products and strategies for minimizing their effects. Bioresour Technol. 2016;199:103–112. doi: 10.1016/j.biortech.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 51.Blank S, Schröder C, Schirrmacher G, Reisinger C, Antranikian G. Biochemical characterization of a recombinant xylanase from Thermus brockianus, suitable for biofuel production. JSM Biotechnol Biomed Eng. 1027;2014:2. [Google Scholar]
- 52.Turner P, Mamo G, Karlsson EN. Potential and utilization of thermophiles and thermostable enzymes in biorefining. Microb Cell Fact. 2007;6:9. doi: 10.1186/1475-2859-6-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sampietro D, Quiroga E, Sgariglia M, Soberón J, Vattuone MA. A thermostable α-galactosidase from Lenzites elegans (Spreng.) ex Pat. MB445947: purification and properties. Antonie Van Leeuwenhoek. 2012;102:257–267. doi: 10.1007/s10482-012-9734-y. [DOI] [PubMed] [Google Scholar]
- 54.Sripuan T, Aoki K, Yamamoto K, Tongkao D, Kumagai H. Purification and characterization of thermostable α-galactosidase from Ganoderma lucidum. Biosci Biotechnol Biochem. 2003;67:1485–1491. doi: 10.1271/bbb.67.1485. [DOI] [PubMed] [Google Scholar]
- 55.Falkoski DL, Guimarães VM, Callegari CM, Reis AP, de Barros EG, de Rezende ST. Processing of soybean products by semipurified plant and microbial alpha-galactosidases. J Agric Food Chem. 2006;54:10184–10190. doi: 10.1021/jf0617162. [DOI] [PubMed] [Google Scholar]
- 56.Mi S, Meng K, Wang Y, Bai Y, Yuan T, Luo H, Yao B. Molecular cloning and characterization of a novel α-galactosidase gene from Penicillium sp. F63 CGMCC 1669 and expression in Pichia pastoris. Enzym Microb Technol. 2007;40:1373–1380. doi: 10.1016/j.enzmictec.2006.10.017. [DOI] [Google Scholar]
- 57.Ríos S, Pedregosa AM, Fernández Monistrol I, Laborda F. Purification and molecular properties of an alpha-galactosidase synthesized and secreted by Aspergillus nidulans. FEMS Microbiol Lett. 1993;112:35–41. doi: 10.1016/0378-1097(93)90534-9. [DOI] [PubMed] [Google Scholar]
- 58.Del Giudice I, Limauro D, Pedone E, Bartolucci S, Fiorentino G. A novel arsenate reductase from the bacterium Thermus thermophilus HB27: its role in arsenic detoxification. Biochim Biophys Acta Proteins Proteom. 2013;1834:2071–2079. doi: 10.1016/j.bbapap.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 59.Koyama Y, Hoshino T, Tomizuka N, Furukawa K. Genetic transformation of the extreme thermophile Thermus thermophilus and of other Thermus spp. J Bacteriol. 1986;166:338–340. doi: 10.1128/jb.166.1.338-340.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 61.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 62.Limauro D, De Simone G, Pirone L, Bartolucci S, D’Ambrosio K, Pedone E. Sulfolobus solfataricus thiol redox puzzle: characterization of an atypical protein disulfide oxidoreductase. Extremophiles. 2014;18:219–228. doi: 10.1007/s00792-013-0607-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Recombinant strains described in this work are made available upon request to the corresponding author. Data sharing not applicable to this article as no datasets were generated or analysed during the current study.


