Abstract
A novel high throughput method for synthesis and screening of customized protein-resistant surfaces was developed. This method is an inexpensive, fast, reproducible and scalable approach to synthesize and screen protein-resistant surfaces appropriate for a specific feed. The method is illustrated here by combining a high throughput platform (HTP) approach together with our patented photo-induced graft polymerization (PGP) method developed for facile modification of commercial poly(aryl sulfone) membranes. We demonstrate that the HTP–PGP approach to synthesize and screen fouling-resistant surfaces is general, and thus provides the capability to develop surfaces optimized for specific feeds. Surfaces were prepared via graft polymerization onto poly(ether sulfone) (PES) membranes and were evaluated using a protein adsorption assay followed by pressure-driven filtration. We have employed the HTP–PGP approach to confirm previously reported successful monomers and to develop new antifouling surfaces from a library of 66 monomers for four different challenges of interest to the biotechnology community: hen egg-white lysozyme, supernatant from Chinese Hamster Ovary (CHO) cells in phosphate buffered saline (PBS) solution as a model cell suspension, and immunoglobulin G (IgG) precipitated in the absence and presence of bovine serum albumin (BSA) in high salt solution as a model precipitation process.
1. Introduction
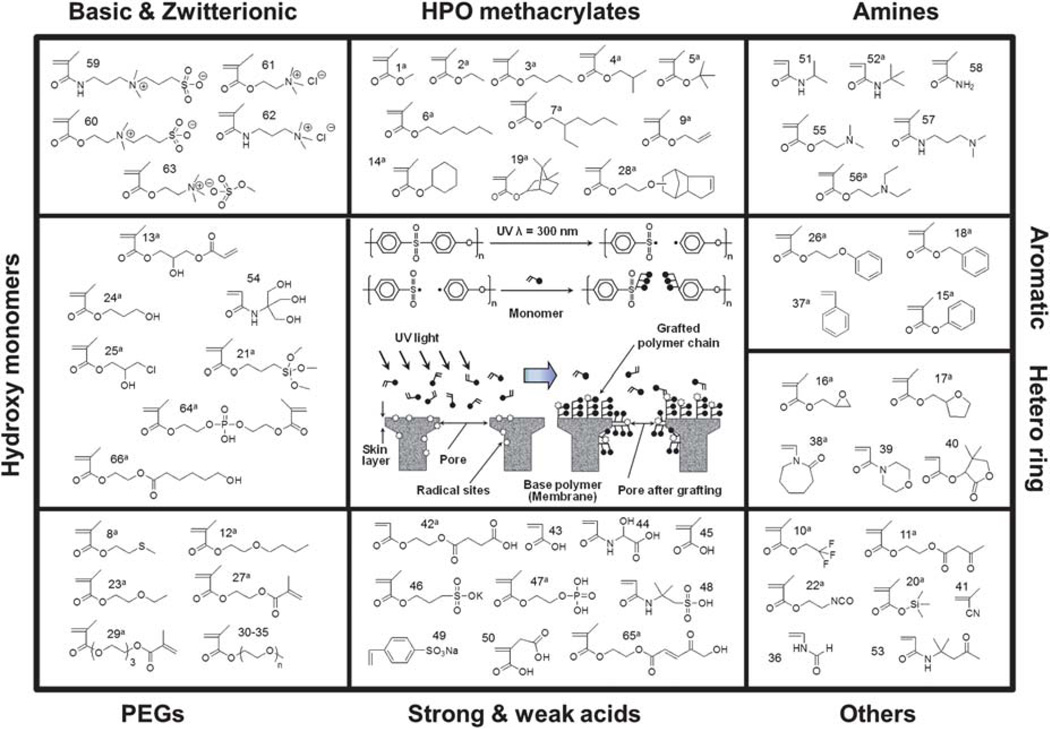
Both rational and trial-and-error methods have been used to search for low fouling surfaces for marine,1 medical,2 separations technology3 and other applications. Although these approaches to-date have produced some successes,2 we still do not fully understand how to rationally choose a low fouling surface. Unfortunately, surface science has not yet developed to the point that allows prediction of the surface or functional characteristics needed to minimize undesirable interactions with solution components, and thus to control fouling. Previous work with plasma treatment and grafting in our laboratory and that of other groups in academia and industry has approached the protein resistance or fouling problem by first choosing a priori a few “attractive” monomers (based mostly on trial and error approaches reported in the literature, intuition and the assumption that hydrophilic monomers with hydroxyl or ethylene glycol groups are best),4–6 conducting confirmatory studies to verify grafting and then testing filtration efficacy.7 The process has been slow, with a low probability of success and offering little mechanistic insight. Here, we offer a new approach to rapidly, efficiently, and reproducibly select optimal polymeric surfaces for a particular application, and subsequently analyze its mechanism of action, to gain understanding for future design of surfaces for reduction of such fouling. The new method adapts—for the first time—high throughput platform (HTP) approaches successfully used in chemistry (e.g. combinatorial spot/well analysis)8 and biology (e.g. phage display9 and SELEX10) to the facile modification of a base polymer, poly- (ether sulfone) (PES), ultrafiltration membrane. We combine HTP with our patented photo-induced graft polymerization (PGP) method11,12 using 66 commercially available vinyl monomers to produce testable polymeric synthetic membrane surfaces. We call this method HTP–PGP. We have tested these newly modified PES membrane surfaces in a 96-filter well format in quadruplicate using ultrafiltration of phosphate buffered saline (PBS) or deionized (DI) water as a post-challenge assay after solute adhesion. We use the HTP–PGP method to modify poly(ether sulfone), in part because it has excellent physical and transport characteristics, although the approach could be applied to other materials. During photo-induced graft polymerization, poly(aryl sulfone) membranes are UV-irradiated, cleaving trunk polymer chains and forming reactive radical sites.11,12 Either water or ethanol-soluble vinyl monomers covalently bond to these radical sites and undergo free-radical polymerization. A schematic illustration of the mechanism is shown in Fig. 1. In contrast to some other free-radical polymerization methods such as atom transfer radical polymerization, no initiator or catalyst is required. The novel method described here proffers an inexpensive, fast, simple, reproducible and scalable modification procedure for testing hundreds if not thousands of surfaces in relatively short periods (weeks to months). Hence, our new approach has substantially increased the chances of finding surfaces with superior anti-fouling characteristics.
Fig. 1.
The PGP method (center box) illustrating radical formation from UV radiation on PES membranes and subsequent graft polymerization of vinyl monomers. Sixty-six vinyl monomers (in nine classes based on functionality, surrounding boxes) were grafted onto PES using the HTP–PGP platform and tested for protein adhesion and filtration. In the text the following abbreviations in parenthesis are used for the monomers in the outer 9 boxes in clockwise direction: charged (Basic and Zwitterionic or Zwit), hydrophobic methacrylates (HPO MA), amines (Amines), aromatic (Aromatic), hetero ring (Hetero ring), monomers that do not easily fit into the other categories (Others), strong and weak acids (Acid), polyethylene glycols (PEGs) and hydroxyl monomers (Hydroxy).
The strategy for membrane material and process development using HTP–PGP has been discussed elsewhere.13 Briefly, an initial monomer library was chosen from a pool of likely candidates. For photo-graft polymerization using PGP, monomers that had pendent vinyl groups were needed. The initial monomer library employed in this work, shown in Fig. 1, represents 66 vinyl monomers that are commercially available (Sigma-Aldrich Co., St Louis, MO) (Table S1, ESI†). For convenience we have categorized them into nine classes based on chemical functionality. These monomers were then employed to modify surfaces in quadruplicate using the HTP–PGP approach. Candidate surfaces were prepared, screened and characterized in terms of buffer or DI water permeation flux prior to and after exposure to the four challenge solutions in the same multi-well filter plate.
To our knowledge this is the first work employing an HTP approach to synthesize and screen fouling-resistant surfaces. We have previously confirmed the excellent reproducibility of the HTP–PGP method by measuring the resistance change after graft modification, Rmod (m−1), by 66 monomers in two separate experiments, which yielded a correlation coefficient R2 = 0.95.13 We have also validated the HTP–PGP approach by comparison with previous results of PES grafted with six different monomers using a bench-scale low-throughput (LT) protein solution filtration protocol previously used by our group as described by Taniguchi and Belfort.14 HTP–PGP approach yielded similar trends in membrane resistance after modification, total resistance after fouling, and bovine serum albumin (BSA) rejection,13 demonstrating the scalability of the HTP approach.
A protein adsorption protocol was also evaluated by comparing the resistance measured during the protein solution filtration assay with that measured during filtration of a protein-free buffer feed after static protein adsorption.13,15 This alternative approach to assess protein/surface interactions avoids complications arising from the convective transport of protein to the membrane surface during the assay, which can result in the formation of a protein cake. The filtration and static adsorption screening assays exhibited similar trends in fouling resistance, confirming the scalability of the static adsorption protocol.13,15 In this work we have chosen to employ the adsorption protocol as an assay to search for new anti-foulant membrane chemistries.
The HTP–PGP approach was applied to identify surfaces for their ability to resist interaction with BSA, as a model protein.13 Surfaces were prepared and screened from a monomer library of 66 commercial vinyl monomers. Some of the monomers, such as poly(ethylene glycol) (PEG), are previously known protein resistant polymeric surfaces, i.e. those reported in the literature by various research groups.5,6,16,17 These surfaces were also protein-resistant in HTP–PGP experiments, thus fulfilling an important criterion for gaining confidence in the HTP–PGP method. Several new surfaces were also identified, confirming that the HTP–PGP method offers new opportunities for choosing new membrane chemistries that minimize fouling.
Several of the most promising monomers identified in the high throughput experiments (and one that was not among the best) were tested in bench-scale filtration experiments with mixing to assess the scalability of the results.13 We found that surfaces identified by the HTP–PGP method, having low protein interactions after adsorption, were also favorable for filtration applications. The performance of the monomers at the two different scales correlated reasonably well, even though the membranes used at the two scales were not identical, and despite differences in hydrodynamics and fouling mechanisms.13
We have also employed the HTP–PGP method to identify surfaces that resist fouling by Eliott soil humic acid, as model for natural organic matter (NOM).15 As with BSA protein solutions, new and previously known low-fouling surfaces for NOM were identified. A comparison of surfaces having the ability to mitigate either NOM or BSA adsorption reveals that some surfaces work well for both feeds, such as diacetone acrylamide and N-isopropylacrylamide, a neutral monomer containing a secondary amine. On the other hand, surfaces made from 2-ethylhexyl methacrylate performed well for BSA feeds but poorly for NOM feeds. These findings demonstrate that different surface chemistries are optimal for different challenges, and suggest that the HTP approach can identify surfaces that perform well for specific feeds (“feed specific surfaces”).
The objective of this paper is to demonstrate that the HTP–PGP approach to synthesize and screen fouling-resistant surfaces provides the capability to develop surfaces optimized for specific feeds of interest to the biotechnology community. This paper presents the method and the new chemistry findings—surfaces having optimal surface chemistry—for four different challenges or assays, none of which have been tested or reported previously. We demonstrate that exceptions to general guidelines for fouling-resistant surfaces may be found. The challenges include exposure of the modified surfaces to hen egg-white lysozyme solution, supernatant from Chinese Hamster Ovary (CHO) cells in PBS as a model cell suspension, and immunoglobulin G (IgG) precipitated in the absence and presence of BSA in high salt solution as a model precipitation process. These solutions offer a range of properties to challenge the HTP–PGP approach. Note that we also report a control (BSA), which we have published previously, for comparison purposes.
2. Experimental
2.1 Materials
Polypropylene 96-well filter plates (Seahorse Labware, Chicopee, MA) were used in HTP–PGP experiments. A 100 kDa cut-off PES membrane coupon (effective area 19.35 mm2) was mounted by the manufacturer on the bottom of each 400 µL well. The hydraulic resistance of the 96 membranes ranged from 8.12 × 109 to 9.49 × 109 cm−1 with a coefficient of variation (σ/μ) equal to 4.0%. Prior to use, the filter plates were washed several times with DI water and then soaked in DI water overnight to remove surfactant from the membrane coupons. Commercial vinyl monomers (66 total, Fig. 1) were purchased from Sigma-Aldrich (Saint Louis, MO) and were used as-received without further purification. The name, structure, and formula weight (FW) of these monomers are shown in Table S1†. These monomers were either dissolved in reagent grade water or ethanol depending on their solubility. A monomer concentration of 0.2 mol l−1 was employed for grafting experiments. Lysozyme was chosen as the model protein to assess membrane fouling. Lysozyme is a small globular protein (Mw = 14.7 kDa, pI = 11), which is positively charged under our experimental conditions. Solutions were prepared by dissolving single protein into PBS to yield a protein concentration of 1 mg ml−1. Lysozyme and PBS tablets were purchased from Sigma-Aldrich (Saint Louis, MO). Chinese hamster ovary cell culture supernatant was provided by Biogen-Idec (San Diego, CA). The monoclonal antibody (Mw = 155 kDa) titer was 1.67 mg ml−1 with an isoelectric point (pI) between pH 8.3 and 8.9. The pH and conductivity of the solution (measured at 19 °C) were 7.4 and 17.09 mS cm−1, respectively. Testing solution was prepared by diluting the cell culture supernatant to an antibody concentration of 1 mg ml−1 and a total protein concentration of ~4 mg ml−1. A stock solution of IgG (5 mg ml−1) was prepared in phosphate buffer (50 mM, pH 7.0). IgG was precipitated by controlled addition of ammonium sulfate salt (1.75 M). Ammonium sulfate was slowly added to the feed container with constant stirring. Care was taken such that all unstable precipitate from a previous addition of salt was re-dissolved before addition of more salt. Salt addition was continued until a stable protein precipitate was obtained. The precipitated protein suspension was diluted with phosphate buffer (50 mM) containing ammonium sulfate (1.75 M), to a concentration of 0.5 mg ml−1. A stock solution (5 mg ml−1) of BSA (Mw = 66.43 kDa, pI 4.7) was also prepared in phosphate buffer (50 mM, pH 7.0). BSA was diluted to 0.5 mg ml−1 concentration in a similar manner. Equal volumes of the IgG and BSA solutions were then mixed to obtain a total protein concentration of 0.5 mg ml−1 for IgG precipitate/BSA testing solution.
2.2 Preparation of modified surfaces using HTP–PGP
The membranes on the 96-well filter plates were modified using the UV-induced graft polymerization method; mechanisms have been described in our previous publications.11 UV irradiation was done in a chamber (F300S, Fusion UV Systems, Inc., Gaithersburg, MD) containing an electrodeless microwave lamp (~7% of the energy was at <280 nm). A band-pass UV filter (UG-11, Newport Corporation, Franklin, MA) was placed between the 96-well filter plate and the UV lamp to reduce the energy at wavelengths below 280 nm to <1%. The membrane modification consisted of the following steps. After washing, the hydraulic permeability of each well was measured simultaneously with DI water. The membranes were then modified by adding monomer solution (200 µl) to each well, shaking the plates on an orbital shaker at 100 rpm for 1 h, purging with N2 for 15 min to remove O2, and irradiating plates in the UV chamber for 30 s. After modification, the plates were washed by shaking in water for 1 h. Each 96-well plate allowed evaluation of 22 monomers with 4 replicates for each monomer, and 8 controls: 4 membrane coupons were treated with ethanol without UV irradiation to serve as a control for membranes grafted with the monomers dissolved in ethanol and the remaining 4 wells were used as-received to serve as a control for the membranes grafted with monomers dissolved in DI water.
2.3 Evaluation of modified membranes by static adsorption challenge
The resistance of modified and control membranes was evaluated using a static challenge (adsorption) protocol. DI water or buffer permeation flux prior to and after adsorption was measured as criteria to evaluate membrane performance. This is a physical characterization of newly synthesized surfaces with regard to adsorption of feed components, and subsequent impact on filtration. In this method, 300 µl of feed challenge solution (four types) were added to each well, and the plate was sealed with a piece of adhesive film to eliminate evaporation. The plate was then placed on a shaker (as above) for a fixed period (i.e. 44 h for lysozyme and CHO supernatant, and 2 h for IgG and IgG/BSA). After equilibration, the wells were then gently emptied, and DI (for lysozyme) or buffer (for others) added to measure permeation flux, J (cm s−1). The membrane resistance was calculated using flux values. The resistance, R (cm−1), was calculated as R = ΔP/µJ, where ΔP (g cm−1 s−2) is the transmembrane pressure and µ (g cm−1 s−1) is the kinematic viscosity of the solution at 22 ± 2 °C. Resistance usually increased for the modified membranes relative to the as-received membrane due to the presence of the grafted polymer. In order to compare the relative permeation fluxes or resistances between the modified and the as-received (control) membrane (unmodified), we used a ratio of differences of R before and after fouling (challenges) for the modified to the control membrane, ℛ = (Rfouled − R)mod/(Rfouled − R)control = ΔRmod/ΔRcontrol, and plotted this value for different monomers.
2.4 Analytical methods
A Microplate Spectrophotometer (PowerWave XS, BioTek Instruments Inc., Winooski, VT) was used to measure the volume of permeate solution in the receiver plate wells. The acrylic 96-well receiver plates are UV transparent. This facilitates permeate analysis by light absorbance in both UV and near infrared regions. The volume of permeate in each receiver well was measured at 977 nm. Proteins do not absorb at this wavelength, whereas water exhibits an absorbance peak. Volumetric flux, J, was calculated as J = V/At, where V (cm3) is the cumulative permeate volume, A (cm2) is the membrane area, and t (s) is the filtration time.
3. Results and discussion
3.1 Trends identified using the HTP–PGP technique
The resistance to water permeation after modification (but prior to fouling), Rmod, relative to the resistance of the as-received membrane, RAR, represents the factor by which membrane resistance increased after modification, and is a rough indicator of the amount of grafted material. To assess foulant/surface interactions, a fouling index, ℛ, was defined as the resistance increase of grafted membranes caused by fouling normalized by that of the ungrafted membrane control, ℛ = ΔRmod/ΔRcontrol, where ΔRmod = (Rfouled − R)mod and ΔRcontrol = (Rfouled − R)control. The control was the as-received membrane treated with either water (in which case Rcontrol = RAR) or ethanol, depending on which solvent was used to dissolve the monomer. The fouling index was chosen as a meaningful and practical way to characterize results from a fouling protocol that mimics a specific filtration application, assuring the relevance of new fouling resistant membranes. The increase in the modified membrane resistance after protein adsorption should be lower than that of the control when the modified surface resists protein interactions. For filtration applications involving permeation, the resistance of the modified membranes should be near that of the as-received membrane (Rmod ≈ RAR), although a higher resistance may be acceptable when it correlates with higher rejection, and high rejection is a goal.
The fouling index data were fitted to 24 different distributions having a lower bound equal to zero and yielding only positive values, to reflect the properties of the fouling index. The General Extreme Value distribution was chosen to represent the data because it described the data well, as indicated by Kolmogorov–Smirnov, Anderson–Darling and χ-squared goodness-of-fit statistics. It is a flexible three-parameter model that combines the Gumbel, Fréchet, and Weibull maximum extreme value distributions. It uses readily interpretable scale (standard deviation, σ) and location (mean, µ) parameters, as well as a shape parameter, k. For k ≠ 0 the probability density function is:
| (1) |
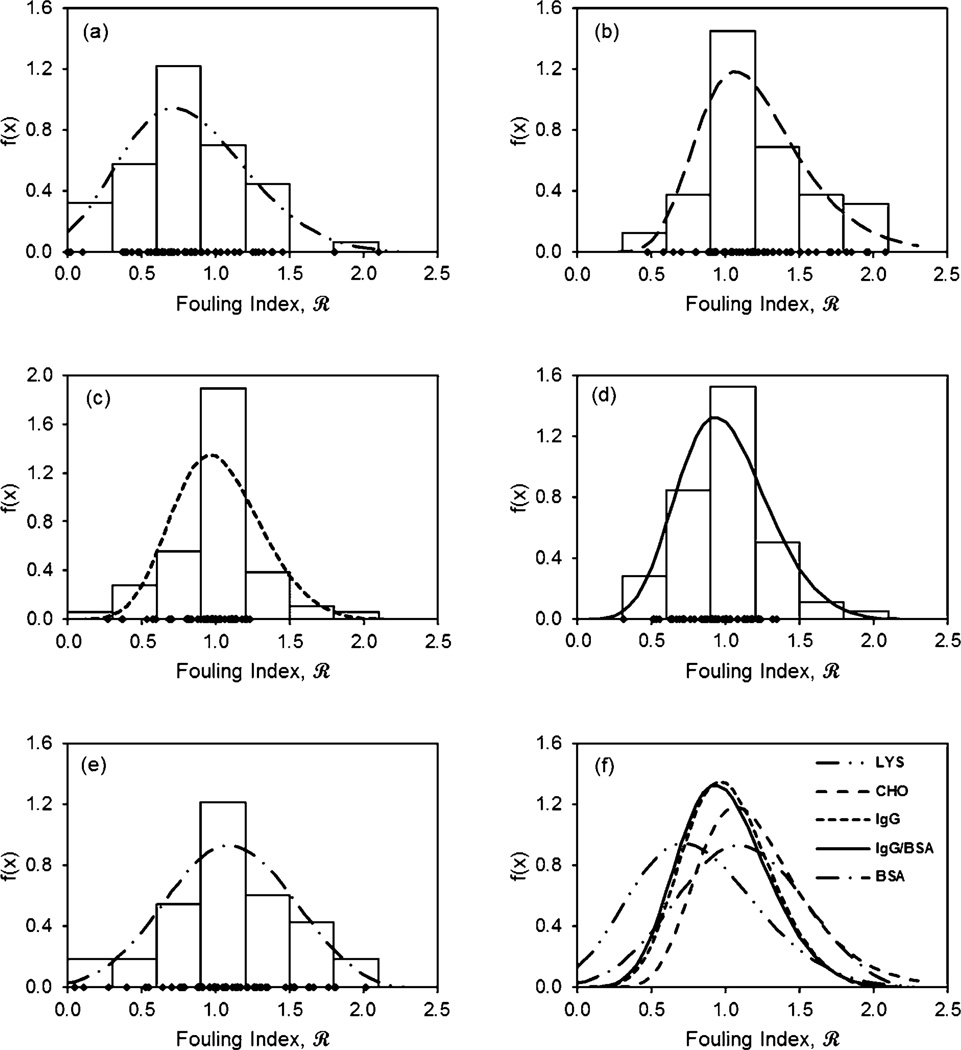
where z = (x − µ)/σ and where the range of definition of the distribution for k ≠ 0 is positive, i.e., 1 + kz > 0. The best-fit General Extreme Value distributions for different feeds are shown in Fig. 2, and the corresponding parameters are shown in Table 1. Note that in all cases, k < 0, corresponding to a reversed Weibull distribution. Other parameters being equal, larger negative values of k yield a distribution skewed toward lower values of the fouling index, indicating successful surface modification.
Fig. 2.
Measured (histogram) and best fit General Extreme Value (lines) fouling index distributions for (a) hen egg lysozyme solution in PBS at 1 mg ml−1; (b) Chinese hamster ovary cell supernatant in PBS at ~4 mg ml−1 total protein with 1 mg ml−1 IgG; (c) IgG precipitate with 0.5 mg ml−1 IgG; (d) IgG precipitate with 0.25 mg ml−1 IgG in the presence of 0.25 mg ml−1 of BSA; (e) BSA solution in PBS at 1 mg ml−1; and (f) comparison of fitted distributions. Points on the x-axis show location of measured fouling indices. Histogram bin size shown corresponds approximately to one standard deviation in the fouling index data over the range 0 to ±2.
Table 1.
Best fit General Extreme Value distribution parameters for different feeds
| Param. | LYS | CHO | IgG | IgG/BSA | BSA |
|---|---|---|---|---|---|
| μ | 0.639 | 1.049 | 0.903 | 0.878 | 0.948 |
| σ | 0.396 | 0.311 | 0.278 | 0.282 | 0.416 |
| k | −0.187 | −0.060 | −0.195 | −0.177 | −0.305 |
For all feeds except CHO, the location parameter (mean) is below one, indicating less fouling than the as-received membrane. The mean for the lysozyme feed is the lowest, equal to only 0.64. The standard deviations range from 0.278 to 0.416 for protein feeds, with the higher values corresponding to feeds having the largest number of low fouling surfaces (BSA and lysozyme).
The fouling index distributions provide a measure of the fouling intensity of the feed solutions. They reveal that the highest proportion of anti-fouling surfaces were synthesized for lysozyme; this is reflected in the distribution parameters as a negative value of k and a low value of the mean fouling index. The CHO supernatant is the most challenging feed, with the highest mean fouling index and lowest proportion of surfaces having a fouling index less than 0.50. Although both CHO supernatant and BSA have mean values near one, the BSA distribution has a larger negative k value and a larger standard deviation, yielding a much higher proportion of surfaces having a fouling index less than 0.50. The distributions for IgG in the presence and absence of BSA are quite similar, suggesting that the presence of BSA does not significantly affect IgG interactions with the surfaces studied. However, as will be discussed below, although the distribution of fouling index is similar, several of the selected surfaces differ.
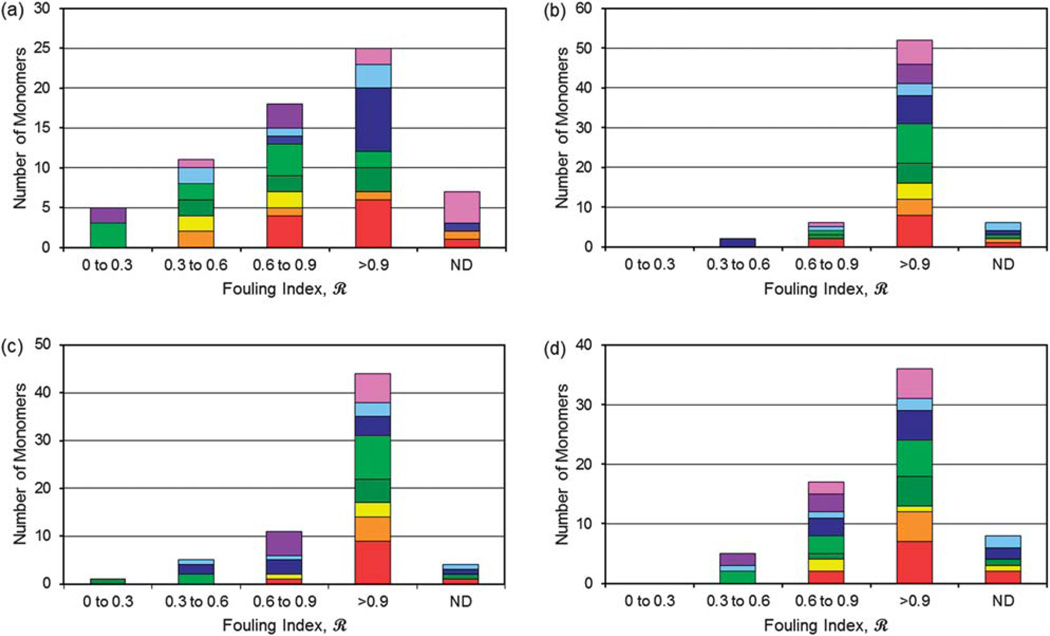
In order to search for trends of ratings for the four challenges, we examined the number of monomers with specific ratings for different monomer classes. The monomers were rated according to the fouling index ℛ, where “3+” = excellent, 0 < ℛ < 0.3; “2+” = very good, 0.3 < ℛ < 0.6; and “1+” = good, 0.6 < ℛ < 0.9. Results are shown in Fig. 3. For the lysozyme challenge, only PEG and Basic and Zwitterionic monomers produced surfaces rated excellent (0 < ℛ < 0.3). However, representatives from these categories also had lower rated surfaces; indeed, having representatives from a given monomer category in multiple fouling index ranges was the rule. It appears that whereas only a few monomer categories produce anti-fouling surfaces, being a member of such a category is not sufficient to ensure a high performing surface. The Acid category performed the worst—most of the acid monomers made surfaces that fell primarily in the highest (ℛ > 0.9) fouling index range. This is likely caused by electrostatic interactions between negative (deprotonated) acid groups and positively charged moieties on the lysozyme surface at the pH of these experiments (pH = 7.4), because lysozyme is net positively charged below its pI of 11.
Fig. 3.
Number of monomers with specific ratings for different monomer classes for fouling-resistant surfaces due to challenges from (a) hen egg lysozyme solution in PBS at 1 mg ml−1; (b) Chinese hamster ovary cell supernatant in PBS at ~4 mg ml−1 total protein with 1 mg ml−1 IgG; (c) IgG precipitate with 0.5 mg ml−1 IgG; (d) IgG precipitate with 0.25 mg ml−1 IgG in the presence of 0.25 mg ml−1 of BSA. Monomers: purple: charged (Basic and Zwitterionic or Zwit), red: hydrophobic methacrylates (HPO MA), light blue: amines (Amines), yellow: aromatic (Aromatic), orange: hetero ring (Hetero ring), rose: monomers that do not easily fit into the other categories (Others), dark blue: strong and weak acids (Acid), light green: polyethylene glycols (PEGs) and dark green: hydroxyl monomers (Hydroxy). ND = not determined due to low modified membrane permeability.
For the stringent CHO cell supernatant in PBS challenge, there were no excellent ratings; in contrast to lysozyme, the best performing surfaces were two very good (2+) monomers from the Acid group. Again, there were several representatives from every monomer category, including PEG, in the highest (ℛ > 0.9) fouling index range. For the IgG precipitate challenge, there was one excellent (3+) rating from the hydroxyl group, although five surfaces synthesized from other hydroxy monomers had ℛ > 0.9. Very good (2+) surfaces were found using monomers from the PEG, Amine and Acid categories. There were several representatives from every monomer category, including PEG, in the highest (ℛ > 0.9) fouling index category. For IgG in the presence of BSA, there were no surfaces with an excellent rating. Very good (2+) surfaces were found using monomers from the PEG and Amine categories; in contrast to IgG precipitate alone, no acid monomers received a 2+ rating, but two quaternary amines from the Basic and Zwitterion group did.
3.2 Discovery of new surfaces using the HTP–PGP technique
In Tables 2–4, the best ten surfaces synthesized from a total of 66 commercial monomers relative to the as-received poly(ether sulfone) membrane using the HTP–PGP method for each feed are tabulated. Success is measured in terms of the fouling index, ℛ, based on either DI water (lysozyme), PBS buffer (CHO) or high salt buffer (IgG) filtration after exposure to the protein feed. Monomer class is defined according to the groups listed in Fig. 1. Each monomer was run in quadruplet. It is clear that the HTP approach has identified many new and previously reported surfaces that perform significantly better than the as-received membrane, offering significantly lower resistance due to fouling (ℛ ≪ 1).
Table 2.
Optimized (ten best monomers, rank-ordered) selection of hen egg lysozyme-resistant surfaces
| # | ℛa | Name | Structure | Classc |
|---|---|---|---|---|
| 60 | ~0.00 ± 0.081 | [2-(Methacryloyloxy) ethyl]dimethyl- (3-sulfopropyl) ammonium hydroxide |
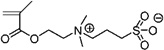 |
Zwit |
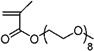
| 33 | ~0.00 ± 0.015 | Poly(ethylene glycol) methyl ether methacrylate |
 |
PEG |
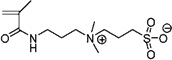
| 59 | ~0.00 ± 0.045 | [3-(Methacryloylamino) propyl]dimethyl (3-sulfopropyl) ammonium hydroxide inner salt |
 |
Zwit |
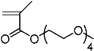
| 32 | 0.022 ± 0.004 | Poly(ethylene glycol) methyl ether methacrylate |
 |
PEG |
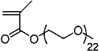
| 34 | 0.103 ± 0.001 | Poly(ethylene glycol) methyl ether methacrylate |
 |
PEG |
| 39 | 0.373 ± 0.003 | 4-Acryloylmorpholine | 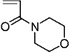 |
Hetero ring |
| 35 | 0.387 ± 0.024 | Poly(ethylene glycol) methyl ether methacrylate |
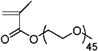 |
PEG |
| 38b | 0.398 ± 0.041 | N-Vinylcaprolactam | 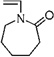 |
Hetero ring |
| 37b | 0.436 ± 0.003 | Styrene | Aromatic | |
| 55 | 0.478 ± 0.003 | 2-(Dimethylamino)ethyl methacrylat |
Amine |
Feed: hen egg lysozyme solution in PBS at 1 mg ml−1 at 22 ± 2 °C.
90% ethanol was used as the solvent. All others used DI water.
Zwit: zwitterionic monomers, PEG: monomers having ethylene glycol groups, Hetero ring: hetero ring group monomers, Amine: amine monomers, Hydroxy: hydroxyl monomers, and Others: other monomers.
Table 4.
Optimized (ten best monomers, rank-ordered) selection of Chinese hamster ovary cell supernatant-resistant surfaces
| # | ℛa | Name | Structure | Classc |
|---|---|---|---|---|
| 50 | 0.474 ± 0.093 | Itaconic acid | 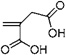 |
Acid |
| 44 | 0.582 ± 0.013 | 2-Acrylamidoglycolic acid | 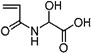 |
Acid |
| 32 | 0.649 ± 0.153 | Poly(ethylene glycol) methyl ether methacrylate |
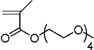 |
PEG |
| 6 | 0.702 ± 0.241 | Hexyl methacrylate | HPO MAs | |
| 4b | 0.801 ± 0.145 | Isobutyl methacrylate | 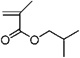 |
HPO MAs |
| 21b | 0.807 ± 0.136 | 3-(Trimethoxysilyl)propyl methacrylate |
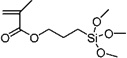 |
Hydroxy |
| 52b | 0.885 ± 0.014 | N-tert-Butylacrylamide | Amine | |
| 41 | 0.897 ± 0.068 | Methacrylonitrile | Others | |
| 3b | 0.903 ± 0.043 | Butyl methacrylate | HPO MAs | |
| 8b | 0.915 ± 0.106 | 2-(Methylthio)ethyl methacrylate | PEG |
Feed: Chinese hamster ovary cell supernatant in PBS at ~4 mg ml−1 total protein with 1 mg ml−1 IgG at 22 ± 2 °C.
90% ethanol was used as the solvent. All others used DI water.
Acid: strong and weak acid monomers, PEG: monomers having ethylene glycol groups, HPO MAs: methacrylates with hydrophobic side chains, Hydroxy: hydroxyl monomers, and Amine: amine monomers.
Many of our results are generally consistent with results from studies of protein interactions with surfaces having a variety of functionalities created using self-assembled monolayers (SAMs) of alkanethiolates on gold as a model substrate.6,17–23 Such studies have identified general features of surfaces having low affinity for proteins: (i) they are hydrophilic (wettable), (ii) they contain hydrogen bond acceptors, (iii) they lack hydrogen bond donors, and (iv) they are electrically neutral.6,18,19 The poly- (ethylene glycol)monomers, which are water soluble, electrically neutral polyethers, represent the “standard” for protein resistance and satisfy all four criteria. PEGs were one of two monomer categories (with amines) to appear in the top 10 of all feeds considered. Many researchers have observed that PEG surfaces resist non-specific adsorption of proteins; this property has resulted in their wide use in biomedical devices.16,24 Although protein resistance has been observed to increase with density and chain length of surface grafted PEGs,17 a finding consistent with our work with BSA,13 results of this work suggest that the optimal chain length depends on the feed characteristics.
The favorable properties of PEGs have motivated attempts to use them to improve membrane surface chemistry.25–27 The ability of such membranes to resist fouling was attributed to hydrogen bonding between water and the ether oxygen groups. We have shown here that PEG monomers performed quite well at the HTP scale, and in previous work we have shown that they also performed well at the bench scale.13 These results provide verification for the HTP approach—it was able to identify surfaces that are known to resist protein fouling.
Because potential limitations of PEGs include their propensity to degrade both in the presence of dioxygen and transition metal ions, and their inability to retain anti-fouling properties above 35 °C,5,28–30 there is increasing interest in identifying alternate low-fouling materials other than PEG. It is likely that successful alternative surfaces influence water structure and its impact on protein stability. The role of water in the folding of globular proteins is critical and connected to minimizing the non-polar surface, while simultaneously providing hydrogen bonding interactions for buried backbone groups, usually in the form of secondary structures (α-helices, β-sheets, turns and random segments).31 This process is exacerbated when pseudo-stable globular proteins are exposed to solid substrates such as hydrophobic or hydrophilic (water covered) surfaces.32–34 How water structure impacts adsorption and subsequent stability of proteins is of great interest.35–37
Amines were the second monomer category to appear in the top 10 of all feeds considered. Several of the monomers identified in this work have been studied for their ability to resist protein adsorption or cell adhesion, but to our knowledge have not been evaluated for their ability to reduce fouling or feed component adhesion during membrane filtration. The monomer 2-(dimethylamino) ethyl methacrylate (#55) performed well for lysozyme (ℛ = 0.478 ± 0.003), and also performed well for BSA (ℛ = 0.410 ± 0.008).13 The brushes formed by 2-(dimethylamino)ethyl methacrylate have been shown to be sensitive to solution pH and ionic strength; increasing either leads to a conformation switch from a stretched brush to a collapsed state. In contrast to polymerized N-isopropylacrylamide, the collapsed state enhances hydrophilicity and protein-resistance of the grafted surfaces, due to a higher surface enrichment of ester groups.38–41 A weak polyelectrolyte ultrafiltration membrane based on poly- (acrylonitrile and 2-dimethylamino ethyl methacrylate) copolymer was reported.40 X-Ray photoelectron spectroscopy confirmed enrichment of poly(2-dimethylamino ethyl methacrylate) on the membrane surface, which made water flux tunable by switching from the stretched to collapsed state. Surface enrichment of ester groups improved water flux;40 however, effects on fouling were not examined.
Monomer 2-(diethylamino)ethyl methacrylate (#56) successfully resisted fouling of IgG precipitate in the presence (ℛ = 0.554 ± 0.057) and absence (ℛ = 0.533 ± 0.136) of BSA. This monomer has been used to form pH-responsive smart copolymers. 42–44 It was found that the polyampholyte microgels consisting of poly(methacrylic acid) and poly(2-(diethylamino)ethyl methacrylate) showed enhanced hydrophilic behavior in aqueous medium at low and high pH but become hydrophobic and compact between pH 4 and 6 near the isoelectric point.42,43 Monomer N-tert-butylacrylamide (#52) (ℛ = 0.885 ± 0.014) resisted fouling by CHO supernatant, but was not a top performer. It has been used to form thermoresponsive surfaces to control bioadhesion of protein and bacterial, and cell attachment and growth.45–48
Two zwitterions, [3-(methacryloylamino)propyl]dimethyl (3-sulfopropyl)ammonium hydroxide inner salt (#59) and [2-(methacryloyloxy)ethyl]dimethyl-(3-sulfopropyl)ammonium hydroxide (#60), were among the top performers for the lysozyme feed (ℛ ≈ 0.0 ± 0.045 for #59, ℛ ≈ 0.0 ± 0.081 for #60), and monomer #60 also yielded surfaces that resisted IgG fouling in the presence (ℛ = 0.629 ± 0.118) and absence (ℛ = 0.804 ± 0.144) of BSA. Zwitterions also performed well for BSA feed, yielding surfaces with a fouling index as low as 0.650 (±0.030).13 However, no zwitterions performed well for the CHO supernatant feed.
Whitesides’ group was one of the first to show that by combining oppositely charged polymers in a self-assembled monolayer (mixed), they were able to demonstrate excellent protein-resistant properties.17 Azzaroni et al.49 have shown that graft polymerization of sulfobetaines (i.e. Zwit #60) as a brush on silicon and gold surfaces lead to a reversible collapsed state with increase of brush thickness. The first use of this monomer (Zwit #60) as a surface coating on a synthetic membrane was by Nabe et al.50 From their study it is unclear whether the grafted polymer was in a collapsed state. It did, however, perform relatively well as a protein-resistant surface. Cho et al.51 showed that the surface grafted with zwitterionic monomer #59 exhibited resistance to the non-specific adsorption of proteins, comparable to that of the best known systems such as PEG-like films.
The zwitterion [3-(methacryloylamino)propyl]dimethyl(3-sulfopropyl) ammonium hydroxide inner salt (#59) conforms to the net neutrality criterion, but also contains a secondary amine in an amide group. Others have noted that primary and secondary amines adsorb more protein than structurally similar groups in the form of amides.19 Furthermore, it should also be noted that other molecules containing hydrogen bond donors, such as mannitol, have exhibited protein resistance.5
Hetero-ring monomer #39, 4-acryloylmorpholine, yielded a surface that successfully resisted lysozyme fouling (ℛ = 0.373 ± 0.003). Although it has been studied for its metal binding properties as a co-polymer with, e.g., cellulose and 2-acrylamido glycolic acid,52,53 it has not been investigated for its surface properties or for applications involving protein adsorption or cell adhesion, or filtration. Therefore, there was no basis in the literature for anticipating how this monomer would perform in terms of protein adsorption or filtration, demonstrating the ability of the HTP platform to identify new high-performance surfaces and assess them for protein interaction. N-Vinylcaprolactam (#38) is another hetero-ring monomer that yielded a surface that successfully resisted lysozyme fouling (ℛ = 0.398 ± 0.041). Poly(N-vinylcaprolactam) bonded to aminopropyl silica was shown to be temperature responsive, exhibiting a transition from hydrophilic to hydrophobic interaction between 30 and 40 °C.54 Lequieu et al.55 modified the surface of poly(ethylene terephthalate) track etched membranes with poly(N-vinylcaprolactam) and showed that the water permeation dramatically increased when the cloud point of the grafted chains was reached. However, the fouling behavior of such membranes was not reported.
A rather surprising finding in the present work is the success of surfaces grafted with acidic monomers, including the two surfaces that showed the lowest fouling by CHO supernatant, itaconic acid (#50) and 2-acrylamidoglycolic acid (#44), and three surfaces that resisted fouling by IgG, ethylene glycol methacrylate phosphate (#47),methacrylic acid (#45), and monomer #44. This is in contrast to our findings with lysozyme and BSA,13 but consistent with our findings for humic acid, a naturally occurring negatively charged polyelectrolyte.15 In that work, the efficacy of acid monomers was attributed to charge repulsion effects. Itaconic acid (#50) has been used in graft polymerization of membranes to improve the separation properties of various organic and inorganic chemicals, such as urea, NaCl, saccharose, acetic acid, etc.56–59 2-Acrylamidoglycolic acid (#44) has been applied to membrane modification to reduce protein fouling by our group.60–62 Monomer #47 is an acid–PEG monomer with ionic group. It has been used to form a mineral phase throughout a PEG hydrogel to improve cell adhesion,63 to produce proton conducting polymer materials,64,65 and to produce cation-exchange stationary phase.66 It has also been used to enhance the surface bioactivity of polytetrafluoroethylene (PTFE) membranes.67 To our knowledge it has not been used to synthesize anti-fouling surfaces.
3.3 Potential limitations of HTP–PGP technique
It is important to point out potential limitations of the HTP–PGP method. First, the data reported here were obtained under well-defined experimental conditions of pH, temperature, and monomer concentration. In addition, the grafting conditions were chosen based on our previous 14 years experience using photo-induced graft polymerization with 14 monomers. 11,12,14,60,62,68–71 It is likely that the grafting conditions (e.g., monomer concentration and UV irradiation time) for many previously untested monomers were sub-optimal. As mentioned above, the HTP–PGP method is being used to optimize these conditions. Actual irradiation energy received by the membrane surface is difficult to assess due to absorption/scattering by the well walls. Although mixing can be controlled during the feed solution adhesion step, it is difficult to implement during the filtration assay. As a result, we may expect some difference between rankings obtained at HTP scale and those identified under bench-scale or pilot-scale conditions with cross-flow, pressure gradients, etc. The HTP approach identifies surface chemistries that minimize interactions with feed components as a way to mitigate the initial stages of fouling. The resistance after static adsorption likely represents the fouling potential of membrane surfaces in terms of feed component affinity, chemistry and structure, and may also incorporate pore blockage and pore constriction that result from solute adsorption to the membrane surface and pore walls. However, this approach does not incorporate all fouling mechanisms, nor does it predict the effects of hydrodynamics. What is especially significant is that in spite of these limitations, many previously identified monomers and several new ones were selected by the HTP–PGP method.
4. Conclusions
In this paper, we have demonstrated the application of a new high throughput synthesis and testing method to obtain a fast, efficient, reproducible, and economic method to select the best polymeric surface for four biotechnology applications. Not only has the HTP–PGP method identified previously reported successful monomers (such as PEG and zwitterionic materials), it has identified previously unreported surfaces that exhibit excellent fouling resistance to the four challenges. These include hetero-ring monomer #39, 4-acryloylmorpholine, and amine monomer #55, 2-(dimethylamino)ethyl methacrylate, for the lysozyme challenge; hydroxyl monomer #64, bis[2-(methacryloyloxy) ethyl] phosphate, acid monomer #47, ethylene glycol methacrylate phosphate, and amine monomer #56, 2-(diethylamino)ethyl methacrylate, for the IgG challenge. Future work will involve using the membrane-based approach to determine, in the HT platform, performance parameters specific to membrane applications, including permeation flux, flux decline due to feed interactions with the membrane, feed component sieving, and cleanability. The buffer/DI water filtration assay will also be evaluated for other applications including marine and medical fouling of surfaces which often occur in the presence of fluid flow across a surface. With the recent development of methods to resolve and analyze spatially resolved micro-patterned array surfaces via X-ray photoelectron spectroscopy, time-of-flight secondary ion mass spectroscopy and water contact angle measurements,72 we are working on methods to characterize individual filter surfaces.
Supplementary Material
Table 3.
Optimized (ten best monomers, rank-ordered) selection of IgG precipitate-resistant surfaces. Also shown are IgG precipitate-resistant surfaces in the presence of BSA and BSA-resistant surfaces
| # | ℛa IgG | ℛb IgG/BSA | ℛc BSA | Name | Structure | Classe |
|---|---|---|---|---|---|---|
| 64d | 0.274 ± 0.056 | 0.839 ± 0.104 (16) | 2.024 ± 0.024 (55) | Bis[2-(methacryloyloxy)ethyl] phosphate |
Hydroxy | |
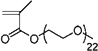
| 34 | 0.365 ± 0.041 | 0.310 ± 0.076 (1) | 0.558 ± 0.021 (6) | Poly(ethylene glycol) methyl ether methacrylate |
 |
PEG |
| 47d | 0.371 ± 0.041 | 0.914 ± 0.194 (23) | 1.822 ± 0.015 (54) | Ethylene glycol methacrylate phosphate |
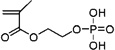 |
Acid |
| 56d | 0.533 ± 0.136 | 0.554 ± 0.057 (5) | 2.700 ± 0.146 (58) | 2-(Diethylamino)ethyl methacrylate |
Amine | |
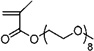
| 33 | 0.574±0.116 | 0.519±0.087 (3) | 0.712±0.014 (8) | Poly(ethylene glycol) methyl ether methacrylate |
 |
PEG |
| 45 | 0.594 ± 0.084 | 0.672 ± 0.108 (9) | 3.179 ± 0.044 (59) | Methacrylic acid | Acid | |
| 44 | 0.683 ± 0.087 | ND | ND | 2-Acrylamidoglycolic acid | 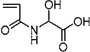 |
Acid |
| 61 | 0.695 ± 0.038 | 0.537 ± 0.055 (4) | 1.542 ± 0.017 (48) | [2-(Methacryloyloxy)ethyl] trimethylammonium chloride |
Basic | |
| 63 | 0.704 ± 0.024 | 0.513 ± 0.060 (2) | 0.901 ± 0.018 (16) | [2-(Methacryloyloxy)ethyl] trimethylammonium methyl sulfate |
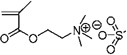 |
Basic |
| 60 | 0.804 ± 0.144 | 0.629 ± 0.118 (6) | 0.803 ± 0.027 (13) | [2-(Methacryloyloxy) ethyl]dimethyl- (3-sulfopropyl) ammonium hydroxide |
Zwit |
Feed: IgG precipitate with 0.5 mg ml−1 IgG in the absence of bovine serum albumin; monomers are rank-ordered.
Feed: IgG precipitate with 0.25 mg ml−1 IgG in the presence of 0.25 mg ml−1 of bovine serum albumin at 22 ± 2 °C. The number in parentheses following the fouling index is the monomer ranking.
Feed: bovine serum albumin solution in PBS at 1 mg ml−1 at 22 ± 2 °C. The number in parentheses following the fouling index is the monomer ranking.
90% ethanol was used as the solvent. All others used DI water.
PEG: monomers having ethylene glycol groups, Hydroxy: hydroxyl monomers, Basic: basic monomers, Amine: amine monomers, Acid: strong and weak acid monomers, and Zwit: zwitterionic monomers.
Not determined.
Acknowledgments
The authors acknowledge the US Environmental Protection Agency (EPA grant RD83090901-0), US Department of Energy (DOE DE-FG02-90ER14114 and DOE DE-FG02-07ER46429), the NIH (Grant RO1 DE016516) and the NSF (Grant # CBET-0730449) for financial support. They also thank Ron Lipsky and Seahorse Labware (Chicopee, MA) for filter plates, Jorge Thommes and Biogen-Idec (San Diego, CA) and Dr Susan Sharfstein and Duan Shen for the plate reader.
Footnotes
Electronic supplementary information (ESI) available: A Table providing the names, structures, and formula weights of the monomers employed in this study. See DOI: 10.1039/c0jm01266a
Supporting information is available online from Wiley InterScience or from the authors.
References
- 1.Dobretsov S, Dahms H-U, Qian P-Y. Biofouling. 2006;22:43–54. doi: 10.1080/08927010500504784. [DOI] [PubMed] [Google Scholar]
- 2.Castner DG, Ratner BD. Surf. Sci. 2002;500:28–60. [Google Scholar]
- 3.Belfort G, Zydney AL. In: Biopolymers at Interfaces. Malmste M, editor. New York: Marcel Dekker; 2003. [Google Scholar]
- 4.Kane RS, Deschatelets P, Whitesides GM. Langmuir. 2003;19:2388–2391. [Google Scholar]
- 5.Luk Y-Y, Kato M, Mrksich M. Langmuir. 2000;16:9604–9608. [Google Scholar]
- 6.Ostuni E, Chapman RG, Holmlin RE, Takayama S, Whitesides GM. Langmuir. 2001;17:5605–5620. doi: 10.1021/la0015258. [DOI] [PubMed] [Google Scholar]
- 7.Leach AR, Gillet VJ. An Introduction to Chemoinformatics. Dordrecht, The Netherlands: Kluwer Academic Publishers; 2003. [Google Scholar]
- 8.Bannwarth W, Felder E. Combinatorial Chemistry: a Practical Approach. Germany: Wiley-VCH Weinheim; 2000. [Google Scholar]
- 9.Clackson T, Hoogenboom HR, Griffiths AD, Winter G. Nature. 1991;352:624. doi: 10.1038/352624a0. [DOI] [PubMed] [Google Scholar]
- 10.Gold L, Brown D, He Y-Y, Shtatland T, Singer BS, Wu Y. Proc. Natl. Acad. Sci. U. S. A. 1997;94:59. doi: 10.1073/pnas.94.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamagishi H, Crivello JV, Belfort G. J. Membr. Sci. 1995;105:237–247. [Google Scholar]
- 12.Crivello JV, Belfort G, Yamagishi H. 5468390. Troy, NY, USA: Rensselaer Polytechnic Institute; US Pat. 1995
- 13.Zhou M, Liu H, Kilduff JE, Langer R, Anderson DG, Belfort G. AIChE J. 2010;56:1932–1945. [Google Scholar]
- 14.Taniguchi M, Belfort G. J. Membr. Sci. 2004;231:147–157. [Google Scholar]
- 15.Zhou M, Liu H, Kilduff JE, Langer R, Anderson DG, Belfort G. Environ. Sci. Technol. 2009;43:3865–3871. doi: 10.1021/es9003697. [DOI] [PubMed] [Google Scholar]
- 16.Harris JM, Zalipsky S. Poly(ethylene glycol): Chemistry and Biological Applications. Washington, DC: American Chemical Society Publication; 1997. [Google Scholar]
- 17.Holmlin RE, Chen X, Chapman RG, Takayama S, Whitesides GM. Langmuir. 2001;17:2841–2850. doi: 10.1021/la0015258. [DOI] [PubMed] [Google Scholar]
- 18.Chapman RG, Ostuni E, Takayama S, Holmlin RE, Yan L, Whitesides GM. J. Am. Chem. Soc. 2000;122:8303. [Google Scholar]
- 19.Chapman RG, Ostuni E, Liang MN, Meluleni G, Kim E, Yan L, Pier G, Warren HS, Whitesides GW. Langmuir. 2001;17:1225–1233. [Google Scholar]
- 20.Mrksich M, Whitesides GM. Annu. Rev. Biophys. Biomol. Struct. 1996;25:55–78. doi: 10.1146/annurev.bb.25.060196.000415. [DOI] [PubMed] [Google Scholar]
- 21.Ostuni E, Yan L, Whitesides GM. Colloids Surf., B. 1999;15:3–30. [Google Scholar]
- 22.Vutukuru S, Bethi SR, Kane RS. Langmuir. 2006;22:10152–10156. doi: 10.1021/la062093p. [DOI] [PubMed] [Google Scholar]
- 23.Chen S, Zheng J, Li L, Jiang S. J. Am. Chem. Soc. 2005;127:14473–14478. doi: 10.1021/ja054169u. [DOI] [PubMed] [Google Scholar]
- 24.McPherson T, Kidane A, Szleifer I, Park K. Langmuir. 1998;14:176–186. [Google Scholar]
- 25.Chen Y, Ying L, Yu W, Kang ET, Neoh KG. Macromolecules. 2003;36:9451–9457. [Google Scholar]
- 26.Akthakul A, Salinaro RF, Mayes AM. Macromolecules. 2004;37:7663–7668. [Google Scholar]
- 27.Chen Y, Deng Q, Xiao J, Nie H, Wu L, Zhou W, Huang B. Polymer. 2007;48:7604–7613. [Google Scholar]
- 28.Leckband D, Sheth S, Halpern A. J. Biomater. Sci. Polym. Ed. 1999;10:1125–1147. doi: 10.1163/156856299x00720. [DOI] [PubMed] [Google Scholar]
- 29.Shen M, Martinson L, Wagner MS, Castner DG, Ratner BD, Horbett TA. J. Biomater. Sci. Polym. Ed. 2002;13:367–390. doi: 10.1163/156856202320253910. [DOI] [PubMed] [Google Scholar]
- 30.Gerhardt W, Wartens C. Z. Chem. 1985;25:143–143. [Google Scholar]
- 31.Dyson HJ, Wright PE, Scheraga HA. Proc. Natl. Acad. Sci. U. S. A. 2006;103:13057–13061. doi: 10.1073/pnas.0605504103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sethuraman A, Belfort G. Biophys. J. 2005;88:1322–1333. doi: 10.1529/biophysj.104.051797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sethuraman A, Han M, Kane RS, Belfort G. Langmuir. 2004;20:7779–7788. doi: 10.1021/la049454q. [DOI] [PubMed] [Google Scholar]
- 34.Sethuraman A, Vedantham G, Przybyccien T, Belfort G. Proteins: Struct., Funct., Bioinf. 2004;56:669–678. doi: 10.1002/prot.20183. [DOI] [PubMed] [Google Scholar]
- 35.Vogler EA. Adv. Colloid Interface Sci. 1998;74:69–117. doi: 10.1016/s0001-8686(97)00040-7. [DOI] [PubMed] [Google Scholar]
- 36.Harder P, Grunze M, Dahint R, Whitesides GM, Laibinis PE. J. Phys. Chem. B. 1998;102:426–436. [Google Scholar]
- 37.Li L, Chen S, Zheng J, Ratner BD, Jiang S. J. Phys. Chem. B. 2005;109:2934–2941. doi: 10.1021/jp0473321. [DOI] [PubMed] [Google Scholar]
- 38.Kusumo A, Bombalski L, Lin Q, Matyjaszewski K, Schneider JW, Tilton RD. Langmuir. 2007;23:4448–4454. doi: 10.1021/la063660b. [DOI] [PubMed] [Google Scholar]
- 39.Su Y, Li C. J. Colloid Interface Sci. 2007;316:344–349. doi: 10.1016/j.jcis.2007.08.017. [DOI] [PubMed] [Google Scholar]
- 40.Su Y, Li C. J. Membr. Sci. 2007;305:271–278. [Google Scholar]
- 41.Sun Q, Su Y, Ma X, Wang Y, Jiang Z. J. Membr. Sci. 2006;285:299–305. [Google Scholar]
- 42.Tan BH, Ravi P, Tam KC. Macromol. Rapid Commun. 2006;27:522–528. [Google Scholar]
- 43.Tan BH, Ravi P, Tan LN, Tam KC. J. Colloid Interface Sci. 2007;309:453–463. doi: 10.1016/j.jcis.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 44.Baquey G, Sakai K, Perrier S, Biggs S. 10th Annual NSTI Nanotechnology Conference and Trade Show. Cambridge, MA 02139, United States, Santa Clara, CA, United States: Nano Science and Technology Institute; 2007. pp. 41–44. [Google Scholar]
- 45.Cunliffe D, De Las Heras Alarcon C, Peters V, Smith JR, Alexander C. Langmuir. 2003;19:2888–2899. [Google Scholar]
- 46.Gilcreest VP, Carroll WM, Rochev YA, Blute I, Dawson KA, Gorelov AV. Langmuir. 2004;20:10138–10145. doi: 10.1021/la0487996. [DOI] [PubMed] [Google Scholar]
- 47.Lynch I, Blute IA, Zhmud B, MacArtain P, Tosetto M, Allen LT, Byrne HJ, Farrell GF, Keenan AK, Gallagher WM, Dawson KA. Chem. Mater. 2005;17:3889–3898. [Google Scholar]
- 48.Rochev Y, O’Halloran D, Gorelova T, Gilcreest V, Selezneva I, Gavrilyuk B, Gorelov A. J. Mater. Sci.: Mater. Med. 2004;15:513–517. doi: 10.1023/b:jmsm.0000021130.11711.16. [DOI] [PubMed] [Google Scholar]
- 49.Azzaroni O, Brown AA, Huck WTS. Angew. Chem., Int. Ed. 2006;45:1770–1774. doi: 10.1002/anie.200503264. [DOI] [PubMed] [Google Scholar]
- 50.Nabe A, Staude E, Belfort G. J. Membr. Sci. 1997;133:57–72. [Google Scholar]
- 51.Cho WK, Kong B, Choi IS. Langmuir. 2007;23:5678–5682. doi: 10.1021/la063737w. [DOI] [PubMed] [Google Scholar]
- 52.Rivas BL, Maureira A, Geckeier KE. J. Appl. Polym. Sci. 2006;101:180–185. [Google Scholar]
- 53.Rivas BL, Maureira A. Eur. Polym. J. 2008;44:523–533. [Google Scholar]
- 54.Miserez B, Lynen F, Wright A, Euerby M, Sandra P. Chromatographia. 2010;71:1–6. [Google Scholar]
- 55.Lequieu W, Shtanko NI, Prez FED. J. Membr. Sci. 2005;256:64–71. [Google Scholar]
- 56.Pulat M, Babayigit D. Polym. Test. 2001;20:209–216. [Google Scholar]
- 57.Isiklan N, Sanli O. J. Appl. Polym. Sci. 2004;93:2322–2333. [Google Scholar]
- 58.Higuchi A, Iijima T. J. Appl. Polym. Sci. 1986;32:3229–3237. [Google Scholar]
- 59.Higuchi A, Iijima T. J. Appl. Polym. Sci. 1986;31:419–428. [Google Scholar]
- 60.Taniguchi M, Kilduff JE, Belfort G. J. Membr. Sci. 2003;222:59–70. [Google Scholar]
- 61.Kaeselev B, Kingshott P, Jonsson G. Desalination. 2002;146:265–271. [Google Scholar]
- 62.Kaeselev B, Pieracci J, Belfort G. J. Membr. Sci. 2001;194:245–261. [Google Scholar]
- 63.Nuttelman CR, Benoit DSW, Tripodi MC, Anseth KS. Biomaterials. 2006;27:1377–1386. doi: 10.1016/j.biomaterials.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 64.Kufaci M, Bozkurt A, Tulu M. Solid State Ionics. 2006;177:1003–1007. [Google Scholar]
- 65.Zukowska G, Williams J, Stevens JR, Jeffrey KR, Lewera A, Kulesza PJ. Solid State Ionics. 2004;167:123–130. [Google Scholar]
- 66.Wang F, Dong J, Jiang X, Ye M, Zou H. Anal. Chem. 2007;79:6599–6606. doi: 10.1021/ac070736f. [DOI] [PubMed] [Google Scholar]
- 67.Wentrup-Byrne E, Suzuki S, Grondahl L, Leavesley D. 7th World Biomaterials Congress Biomaterials 2004 Congress Managers; Sydney, NSW 2001, Australia, Sydney, Australia. 2004. p. 1425. [Google Scholar]
- 68.Pieracci J, Crivello JV, Belfort G. J. Membr. Sci. 1999;156:223–240. [Google Scholar]
- 69.Pieracci J, Crivello JV, Belfort G. Chem. Mater. 2002;14:256–265. [Google Scholar]
- 70.Yamagishi H, Crivello JV, Belfort G. J. Membr. Sci. 1995;105:249–259. [Google Scholar]
- 71.Pieracci J, Crivello JV, Belfort G. J. Membr. Sci. 2002;202:1–16. [Google Scholar]
- 72.Urquhart AJ, Taylor M, Anderson DG, Langer R, Davies MC, Alexander MR. Anal. Chem. 2008;80:135–142. doi: 10.1021/ac071560k. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.