Abstract
Recent studies reveal that mitochondria take substantial responsibility in cellular functions that are closely related to aging diseases caused by degeneration of neurons. These studies emphasize that the membrane and crista morphology of a mitochondrion should receive attention in order to investigate the link between mitochondrial function and its physical structure. Electron microscope tomography (EMT) allows analysis of the inner structures of mitochondria by providing highly detailed visual data from large volumes. Computerized segmentation of mitochondria with minimum manual effort is essential to accelerate the study of mitochondrial structure/function relationships. In this work, we improved and extended our previous attempts to detect and segment mitochondria from transmission electron microcopy (TEM) images. A parabolic arc model was utilized to extract membrane structures. Then, curve energy based active contours were employed to obtain roughly outlined candidate mitochondrial regions. Finally, a validation process was applied to obtain the final segmentation data. 3D extension of the algorithm is also presented in this paper. Our method achieved an average F-score performance of 0.84. Average Dice Similarity Coefficient and boundary error were measured as 0.87 and 14 nm respectively.
Keywords: Image processing, Electron tomography, Curve fitting, Active contour, Mitochondrion
1. Introduction
Mitochondrial structural correlates to function are attracting increased attention as the relation between mitochondrial function and degenerative disorders related to aging such as Alzheimer’s and Parkinson’s diseases is becoming empowered by recent studies (Burté et al., 2014; Franco-Iborra et al., 2015; Wang et al., 2014). Three dimensional (3D) visualization of mitochondria is coming into prominence because these studies expose the need for detailed analysis of high-resolution physical alterations in mitochondria.
Investigations of subcellular structures are being made more effective by means of advances in electron microscopy. The physical formation of mitochondria emerges in detail as new imaging techniques have been developed. Serial block-face scanning electron microscopy (SBEM) is one of the emerging methods for volumetric mitochondrial imaging that provides 3D datasets consisting of voxels with a size typically of 5–10 nm in x- and y-axes and 20–80 nm in z-axis (Chavan et al., 2015; Wanner et al., 2015). Transmission electron microscope (TEM) tomography is another powerful technique that renders possible the visualization of structures down to a few nm resolution (Davies et al., 2014; Harapin et al., 2013). Such a resolution offers a clear separation of the double membrane structure of mitochondria and distinctive arrangement of cristae as well as the visualization of other subcellular structures such as synaptic vesicles (SV) and the endoplasmic reticulum (ER). Considering that the size of mitochondria varies between 0.3 and 10 µm (Scheffler, 2007), large 3D tomographic volumes are often required to study the structure of whole mitochondria.
In a study by Perkins et al. (2003) various structural features, such as the width of the peripheral inner and outer membranes of mitochondria and cristae, the number of crista segments, crista junctions and contact site diameters were measured and hypothesized to have potential effect on mitochondrial function. Visualization details depend on the resolution of the tomographic volume as well as the structural preservation of the sample. To perform a detailed mitochondria segmentation in such volumes, both peripheral and cristae membranes are required to have high contrast with respect to the background. By heavy-metal staining, TEM tomography highlights the two mitochondrial membrane systems. Cristae segmentation provides a basis for our motivation to develop an automatized segmentation method for mitochondrial boundary to be used in this imaging modality.
Visual investigation of tomographic volumes of mitochondria are currently carried out by manual segmentation utilizing specialized software tools such as IMOD (Kremer et al., 1996) and Amira. However, hand segmentation of features of a volume may produce flawed results because of human error even with highly trained segmenters. Because mitochondria are pleomorphic and appear in many different forms based on cell type, respiration or disease state, and sample preparation, a perfect automated segmentation is still an unsolved problem.
Various attempts have been made to automatically segment membranes and mitochondria from transmission electron microscope (TEM) tomography images. A confidence-connected and level-set based segmentation scheme was realized to extract contours of membranes of mitochondria (Bazán et al., 2009). The method produced successful segmentation results. However, it requires a manual connected-component removal and clear visualization of membranes.
Mitochondria segmentation based on classification of random forest patches and contour pairs (Giuly et al., 2012), utilization of supervoxel segmentation (Ghita et al., 2014; Lucchi et al., 2012) and spectral clustering techniques (Dietlmeier et al., 2013) which basically use intensity distribution of mitochondria and differentiation from background were also asserted. However, these methods are not suitable for specimens specially prepared for cristae analysis as mentioned earlier since the differentiation of the intensity distribution is lost in the preparation process.
Several generic methods to segment membrane-like thin structures from electron tomography images were developed as well. A membrane segmentation method based on Hessian-based ridge detection was developed for extraction of membrane-like structures such as mitochondria boundary, SV and ER (Martinez-Sanchez et al., 2014, 2013, 2011). Bartesaghi et al. (2005) proposed a semi-automated 3D segmentation method based on minimal surface of closed geodesic curves. Another approach was introduced by Sandberg and Brega (2007) in which a contour tracing mechanism utilized local features extracted by line and orientation filter transforms. A bilateral edge filtering method was presented by Pantelic et al. (2007) for 2D segmentation of noisy electron microscopy and cryo-electron microscopy images. A 3D extension of this method was later proposed by Ali et al. (2012). A cascaded hierarchical model (CHM) by Seyedhosseini et al. (2013b) was used for membrane segmentation in SBEM and TEM images and provided promising results. Another intriguing semi-automated procedure was established by Page et al. (2015) for cell segmentation which is based on watersheds exploiting the differentiation of structures inside and outside of the cell. Although notable results have been obtained, these proposed methods have not been capable of separating mitochondria from each other or from membranes of other subcellular structures, or segmenting the more morphologically complex mitochondrial inner membrane.
In our previous attempt (Mumcuoglu et al., 2012), we employed a double membrane detection based on kernel pairs and an ellipse fitting approach for 2D detection and separation of mitochondria followed by active contours and a modified livewire method for 2D accurate segmentation of mitochondria. This algorithm depends on the successful removal of cristae in one of the intermediate steps to locate the peripheral mitochondrial membranes. Our current work describes a better mechanism to separate peripheral membranes from cristae membranes.
In another study (Seyedhosseini et al., 2013a), algebraic curve based segmentation was applied to 2D mitochondrion images which is reasonable to obtain a curve which tracks the boundary of a mitochondrion. However, it can easily be attracted by cristae and further, it is not sufficient to separate mitochondria from each other.
In our preliminary study (Tasel et al., 2014), a 2D detection method for mitochondria performed by active contours using a parabolic arc based membrane detection process was proposed. Although the method was tested on a limited dataset, the study revealed that the curve based membrane detection approach is useful to extract mid-level features which are relatively easy to manage and capable to differentiate mitochondria peripheral membranes and cristae membranes.
In this study, we have improved the aforementioned parabolic arc model fitting algorithm and presented the separation of the two peripheral membranes and cristae membranes. In our new scheme, an active contour model driven by a curve energy image is utilized to obtain candidate mitochondrial regions. Finally, a validation process in order to filter false shapes is applied by using features extracted from continuity, curvature and signature characteristics of boundary and additional curve energy of cristae. Instead of removing cristae (as in our previous attempt), we adopted a mechanism which uses existence of cristae as a sign of the presence of a mitochondrion, because mitochondria are the only structures inside a cell that possess cristae. We have tested our method on a much larger volume of datasets that were used in our preliminary study. The major contribution of the work is 3D extension of the algorithms. Detection and segmentation performances of 2D and 3D method are also compared and discussed.
The subsequent sections of this paper present information about the datasets used, the acquisition of ground truth, the proposed methods, experiments and results.
2. Datasets and ground truth
The datasets used for the experiments in this work were collected from the Cell Centered Database (CCDB) supported by the National Center for Microscopy and Imaging Research (NCMIR) (Martone et al., 2008, 2003, 2002). We chose eight TEM datasets which include tomograms having diversity in image contrast and variety in mitochondrial membrane and crista characteristics. The collection comprises mitochondria appearing in various shapes and sizes. Some properties of the datasets such as image size or voxel size are given in Table 1.
Table 1.
Properties of datasets used in the experiments.
| Dataset basename | Accession number | Image size (width × height) | Number of slices used | Voxel size (nm) (X × Y × Z) |
|---|---|---|---|---|
| 6_22.sub | 5,274,878 | 1960 × 2560 | 91 | 1.1 × 1.1 × 1.1 |
| bclpb-d.sub | 5,274,930 | 720 × 878 | 61 | 2.4 × 2.4 × 2.4 |
| cone.sub | 54 | 736 × 1010 | 97 | 2.4 × 2.4 × 2.4 |
| gap18_sub | 8747 | 350 × 600 | 54 | 2.2 × 2.2 × 2.2 |
| mac_serial_sub | 5,274,996 | 907 × 1172 | 111 | 2.4 × 2.4 × 2.4 |
| od.sub | 8752 | 1960 × 2560 | 91 | 1.1 × 1.1 × 1.1 |
| pedicle | 5,274,970 | 950 × 1280 | 31 | 2.4 × 2.4 × 2.4 |
| spherule24mos1 | 8495 | 1996 × 1996 | 86 | 1.67 × 1.67 × 1.67 |
Ground truth data were prepared via hand segmentation outlining the outer membrane of a total of 96 mitochondria appearing in 622 slices by using IMOD software. Since the images corresponding to the very top and bottom sections of the volume become very blurry, only the useful slices were processed for ground truth. It is noteworthy to emphasize that creating such a ground truth database requires considerable amount of human effort and it is a considerably time consuming process which justifies that computerized segmentation is obligatory to accelerate tomographic analysis.
3. Methods
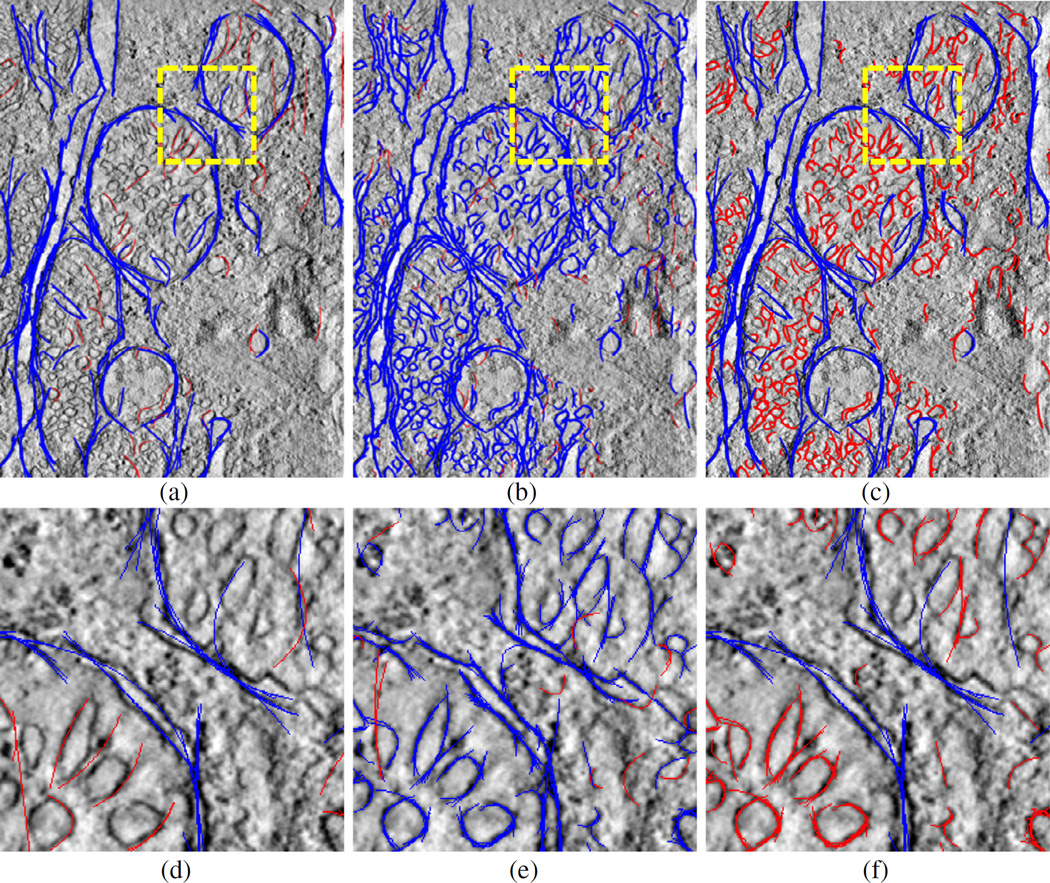
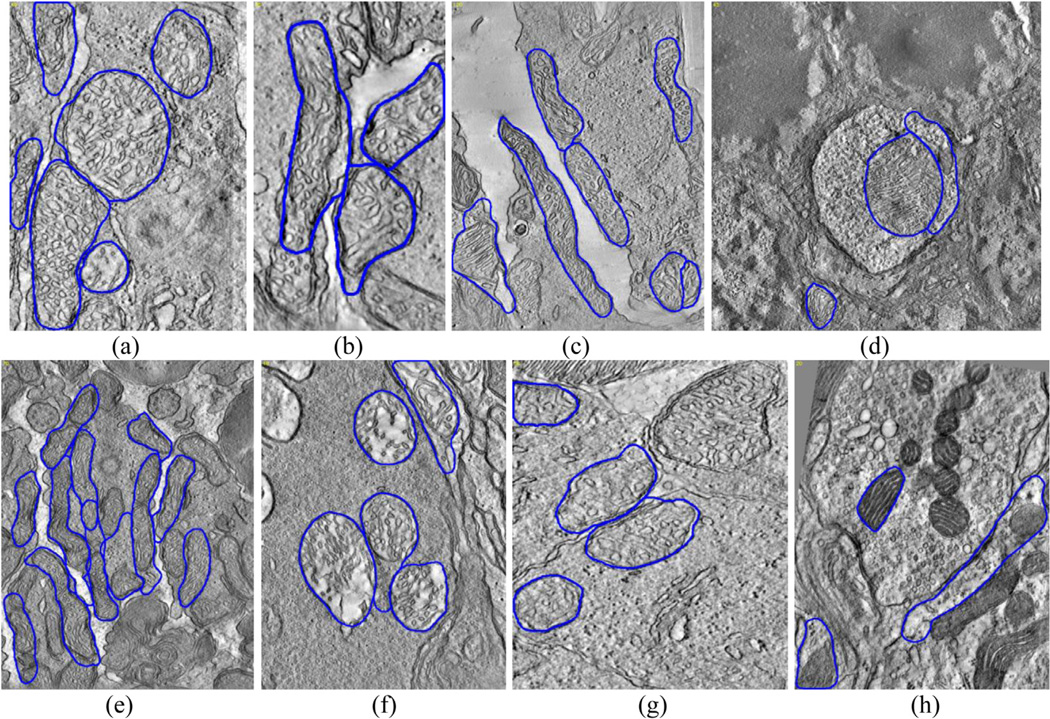
Our mitochondria detection and segmentation method comprises preprocessing, ridge detection, energy mapping, curve fitting and filtering, snake-based shape extraction, validation and post-processing steps. A flowchart of the algorithm and sample images regarding several intermediate steps is illustrated in Fig. 1.
Fig. 1.
Flowchart of the mitochondria detection and segmentation algorithm (top) and sample images (a–g) showing the output of intermediate steps: (a) input image; (b) preprocessed and (c) ridge energy image; (d) low frequency ridge energy map (LFREM) image and (e) high frequency ridge energy map (HFREM) image for the region indicated by the red square shown in (c); (f) Detected curves after filtering (blue: large-scale curves, red: small-scale curves); (g) Validated snake output; (h) Output boundary after post-processing.
The following subsections describe each step of the 2D method and extension of the algorithm to 3D.
3.1. 2D Method
3.1.1. Preprocessing
The potential effect of contrast and membrane strength characteristics of the input image on the output of any further step is handled in the preprocessing step. It also deals with the noise of TEM images. Moreover, it dissipates parameter tuning difficulty originating from the difference in pixel size of the datasets. Fig. 1(b) shows the preprocessed form of the image shown in Fig. 1(a). The preprocessing step consists of three parts described below:
Auto-contrast adjustment: Reconstructed ET datasets may contain extreme values due to the presence of artifacts originating from the gold markers utilized or X-rays and other flaws in CCD camera images that produce extreme low or high values that skew the contrast. Normalization of gray values for such datasets cause a substantial degradation in image contrast. Auto-contrast adjustment re-normalizes gray values of pixels of the image into a certain range (such as 0–1 or 0–255) assuming that the gray value distribution has some extreme points. A small portion (e.g. 0.5%) from the lowest and highest gray values of the histogram area are set to the minimum and the maximum possible value (i.e. 0 and 255) respectively. The other pixel values are linearly scaled and normalized between these maximum and minimum values.
Resampling: In order to ease the parameter tuning, the contrast adjusted images are converted to a fixed pixel size by interpolation. Considering that the typical membrane thickness of mitochondria is in the range of 4–6 nm (Róg et al., 2009), interpolating the image to 2 nm pixel size does not generate data loss problems for the mitochondrial membrane as we justified in our pilot study (Mumcuoglu et al., 2012).
Smoothing: Bilateral filtering (Tomasi and Manduchi, 1998) which is an edge-preserving smoothing technique is applied as a noise removal process as it is recommended for electron microscopy (Bazán et al., 2009; Jiang et al., 2003). In bilateral filtering, the contribution of each pixel depends on both the Gaussian kernel and the gray value differences of pixels in a neighborhood. This technique is able to reduce the effect of the noise as well as the non-membrane-like structures while preserving membranes.
3.1.2. Ridge detection
In the ridge detection step, membrane-like structures are extracted. Since bright-dark-bright transition occurs in membrane profile, membranes exist at the locations where intensity increases in opposite directions and remains stable in orthogonal directions. In order to detect membranes, we constructed a Hessian-based ridge detector applied to the preprocessed image which is sensitive to valley-like shapes. Let λ1 and λ2 be eigenvalues of Hessian matrix defined as:
| (1) |
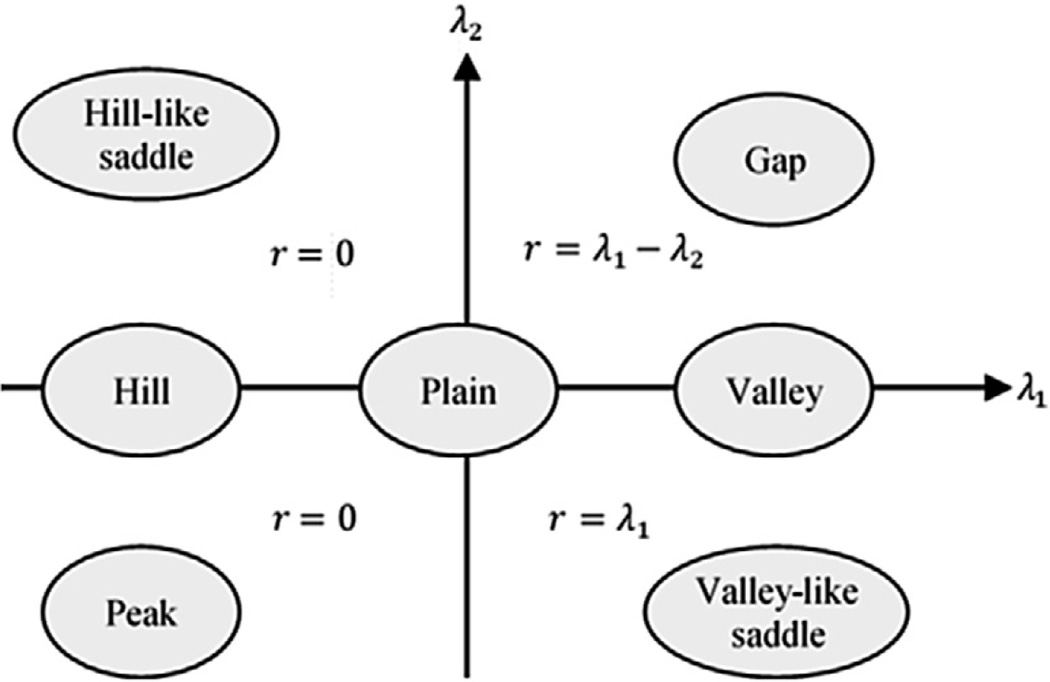
where Gxx, Gyy, Gxy and Gyx correspond to second order Gaussian derivatives of the image intensity with respect to the axis specified by the subscript and the eigenvalues satisfy |λ1| ≥ |λ2|. As illustrated in Fig. 2, a valley is obtained at a location where λ1 ≫ |λ2|. On the other hand, the shape becomes a gap (i.e. a dark blob) when λ1 ≈ λ2 ≫ 0 as shown in the first quadrant of the coordinate system in Fig. 2. In the fourth quadrant, a saddle point on a valley is achieved when λ1 ≫ 0 and λ2 ≪ 0 which corresponds to a relatively weak point on a membrane. By considering the potential discontinuity in membranes, we define ridge energy as:
| (2) |
Fig. 2.
Topographic shapes to be detected with respect to eigenvalues of Hessian matrix and assigned ridge energy.
In the above formulation, the ridge energy increases while λ1 is increasing and λ2 is getting close to zero when both eigenvalues are positive. In case of discontinuity of the membrane, λ2 becomes negative and the energy only depends on the valley depth that is proportional to λ1. Hence, it favors objects which are in the form of elongated dark stripes. The ridge energy is zero in the second and third quadrant since we are not interested in hill-like (i.e. bright) structures. Fig. 1(c) depicts the ridge image in which bright points correspond to the locations such as peripheral membrane and crista membrane where the ridge energy is high. We also extract ridge direction from the second eigenvector.
3.1.3. Energy mapping
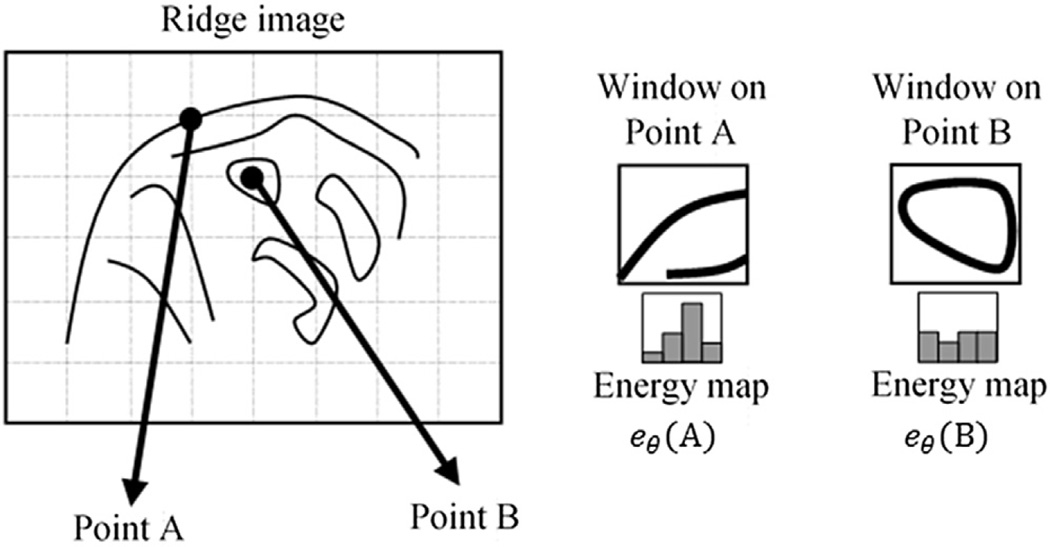
Although the ridge image is useful to detect membrane-like structures, it carries local information and is not sufficient to classify mitochondria peripheral and cristae membranes. Therefore, we constitute a ridge energy map based on the fact that peripheral membranes are longer structures having relatively low curvature in general which indicates clues pertaining to the big picture.
A window with a size ω is scanned through each point of the ridge image in order to inspect the ridge distribution over a region and avoid misleading local information. To reduce computation and provide smoothness to ridges having excessive curvature, the ridge direction is quantized to four angles such that the intervals (−π/8, π/8) and (7π/8, 9π/8) are assigned to 0; (π/8, 3π/8) and (−5π/8, −7π/8) are assigned to π/4; (3π/8, 5π/8) and (−3π/8, −5π/8) are assigned to π/2 and (5π/8, 7π/8) and (−π/8, −3π/8) are assigned to 3π/4. We then compute the total ridge energy within the window by using the points having the same ridge direction. The process is repeated for all quantized angles. The window is passed through all the pixels in the image in order to form the energy map eθ(s) that stores the total ridge energy of angle θ around the point s.
Fig. 1(d) and (e) depict ridge energy map images for the same segment of the ridge image by adjusting the window size as ω = 30 nm and ω = 8 nm respectively. Dashes indicate the major direction which has the maximum energy. Subtle (high frequency) information is extracted by using a small value of ω whereas rough structures (low frequency) are provided by a large value of ω. In the high frequency ridge energy map (HFREM) (see Fig. 1(e)), both cristae and mitochondrion boundary appear in detail. On the other hand, in the low frequency ridge energy map (LFREM) (see Fig. 1 (d)), the visualization of the structure of the cristae is mostly eliminated due to the use of a large window. On the contrary, boundaries of mitochondria are still preserved. Fig. 3 illustrates these two cases by delineating the energy map as histogram for two points. A uniform histogram is expected for cristae compared to peripheral mitochondrial membranes in the low frequency energy map.
Fig. 3.
Sample points A and B on the ridge image (left); corresponding windows placed on A and B (right); energy maps (eθ(A) and eθ(B)) (bottom-right). Each point on the energy map corresponds to a histogram indicating the total ridge energy with respect to each ridge angle within the window.
3.1.4. Parabolic arc model
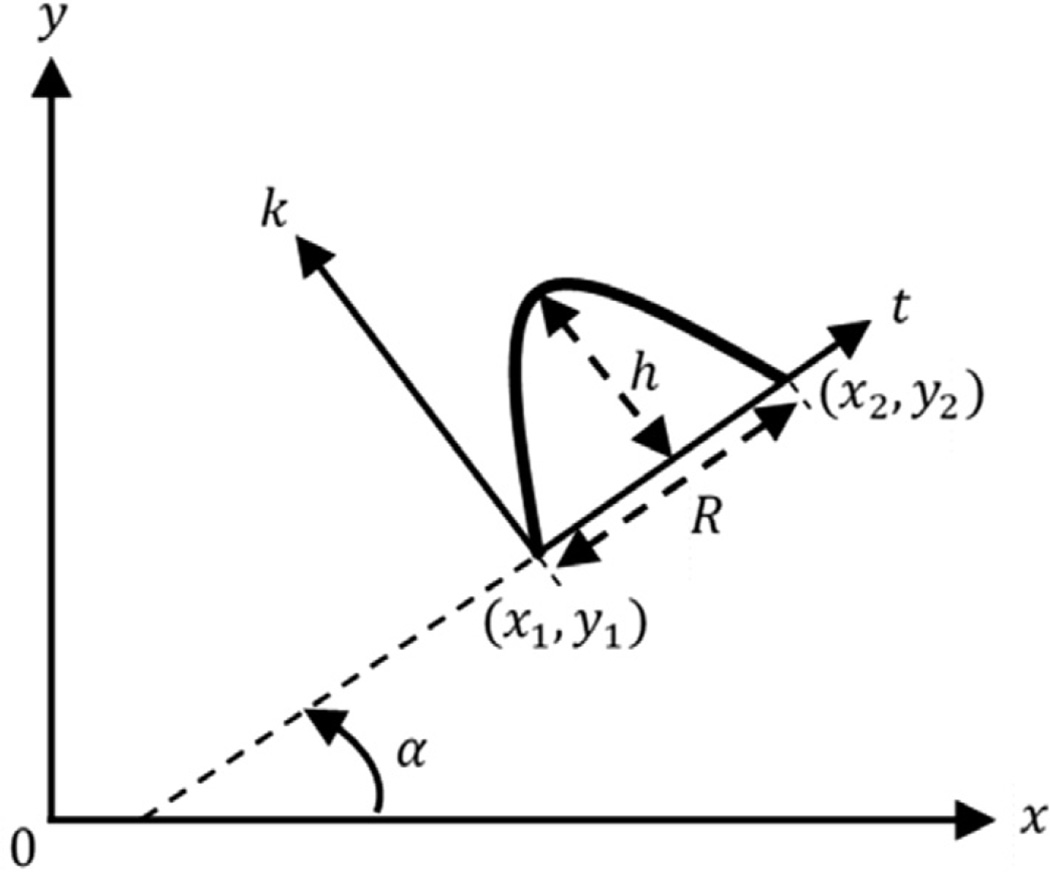
Peripheral mitochondrial and cristae membranes may be represented by a combination of curve segments. In general, the peripheral membranes are long and smooth curves whereas cristae membranes are relatively short curves with high curvature compared to peripheral membranes. We have adopted a parabolic arc model as delineated in Fig. 4 in order to extract membrane-like patterns by utilizing the ridge energy maps. A 2D parabolic arc model is represented by two tip points (x1, y1), (x2, y2) and height (sagitta length) h.
Fig. 4.
A Parabolic arc model represented by two tip points (x1, y1), (x2, y2) and height h.
A parabolic arc passing through the origin of the kt-plane with height h and width (chord length) R is given by:
| (3) |
where and with
| (4) |
and
| (5) |
Then, a point s on the rotated parabolic arc in xy-plane is given by:
| (6) |
where and . The tangential angle φs of the parabola on the point s is approximated as:
| (7) |
where Δt is step size on the t-axis which can be arranged to scan the equidistant points on the curve. A smaller step size forms a smoother curve but increases the computation time. In order to achieve the same smoothness for curves with different parameters, we use a dynamic step size which is given by:
| (8) |
where S denotes the desired distance (e.g. 1 pixel) between each consecutive point on the curve. In addition to the model described, we also defined a scale invariant curvature of the parabolic arc as:
| (9) |
The next section describes the curve fitting algorithm developed for the parabolic arc model.
3.1.5. Curve fitting and filtering algorithm
In the curve fitting step, membrane-like structures are extracted by utilizing the parabolic arc model and ridge energy maps described in the previous sections. We define an energy function of the curve as:
| (10) |
where Ω is the set of points on the parabolic arc obtained by Eq. (6) and φs represents the tangential angle of the parabola at the point s. The weight function w is defined as:
| (11) |
The contribution of the energy associated to the point s in Eq. (10) is controlled by the weight function based on proximity of tangential angle of the parabola (φs) and the ridge direction (θ). If they are similar (i.e. if the difference is close to zero), the weight function gets close to one, thus allows the maximum contribution. If the difference is high (i.e. close to ±π/2), it tends to be closer to minus one and causes a negative contribution. Therefore, the curve is forced to pass through points where the ridges are strong and have a suitable direction in order to gather high energy. According to this framework, the curve fitting process becomes an energy maximization problem. A brute-force approach like searching for all possible curves is infeasible. Hence, an iterative curve growing method is employed in which the candidate curves are initialized as single points on local maxima of the ridge energy map where the total ridge energy associated to the major direction is sufficiently high (i.e. maxθeθ(s) > Tmap where Tmap is a threshold).
The curve fitting algorithm is summarized below:
Initialization: Initialize a curve as a single point for each local maximum of maxθ eθ(s) satisfying maxθ eθ (s) > Tmap such that (x1, y1) = (x2, y2) = s and h = 0.
- Growing phase 1:
- Let , h(0) = h and i = 0.
-
Compute E(Ω) by using Eq. (10) for each parameter set candidate: and height h(i+1) ∈ {a|h(i) − τh ≤ a ≤ h(i) + τh} where the set of curve points Ω is formed by using Eq. (6) for the current values of parameters and t = 0, Δt, 2Δt, 3Δt, …, R.Select the parameter set which maximizes E(Ω).
- If , increment i and go to step ii.
- Set and h = h(i).
Growing phase 2: This phase is the repetition of the first growing phase. However, the search operation is done by altering the parameters (x2, y2, h) this time and keeping (x1, y1) fixed.
The update parameters τx, τy and τh determine the search neighborhood at each iteration and control growing speed. Model parameters (x1, y1), (x2, y2) and h are updated within the range specified by the update parameters to maximize the energy iteratively.
The curve fitting process is repeated for two different values of window size ω used in the energy mapping step. Hence, two different sets of curve segments are obtained. Fig. 5(a) and (b) shows obtained curve segments (in blue) by using LFREM (ω = 30 nm) and HFREM (ω = 8 nm) respectively. The curve segments obtained from HFREM appear relatively short in general since the parabolic arc model cannot perfectly fit in large-scales. They are considered as small-scale curves in this paper. These curves cover all membrane-like structures (cristae, boundary etc.). On the other hand, longer curve segments having relatively high energy can be acquired by using LFREM. In this case, the curve segments generally correspond to the mitochondrion boundary and endoplasmic reticulum and they are considered as large-scale curves.
Fig. 5.
Curve segments obtained by using low frequency energy map (a); high frequency energy map (b) and filtered curves (c). In (a) and (b), blue curves show successful detections and red curves are eliminated curves due to weakness and shortness. In (c), blue and red curves show the accepted detections in (a) and the obtained curves from (b) respectively after the elimination and filtering process are applied; (d), (e) and (f) are close-up images taken from the yellow region placed on (a), (b) and (c) respectively.
Some of the detected curves are considered as unreliable and thus filtered out in the next step. Although a parabolic arc with excessive height does not perfectly fit into the mitochondrial membrane, such curve segments can reasonably fit into the energy map. Therefore, the curves which unexpectedly have high curvature (i.e. κ > 1 which means |h| > R such that the parabola has a sharp bend at the sagitta point) are removed. The curve segments initiated at irrelevant locations (e.g. cytoplasm) are not expected to grow and have high energy (E(Ω)). Therefore, the curves that are too short and too weak are also eliminated. For this purpose, the large- and small-scale curves which have an arc-length shorter than 100 nm and 20 nm respectively or which have an average energy less than 30% of the maximum energy obtained in the map were removed in our experiments. The removed curves are shown in red in Fig. 5(a) and (b).
It is clearly seen that small-scale curves (Fig. 5(b)) cover large-scale curves (Fig. 5(a)). In the last step of the curve filtering process, the remaining curve segments are combined on a single image by removing small-scale curves which overlap a large-scale curve. We assume that a small-scale curve overlaps a large-scale curve if 70% of the former lies within ω neighborhood of the latter (ω is the window size of LFREM). As illustrated in Figs. 5(c) and 1(f), blue curves are large-scale curves which were detected by using LFREM. Such curves mostly appear on the boundary of mitochondria and long membrane-like structures. The red ones are the curve segments which can be extracted from only HFREM and usually detected on cristae and small scale membrane-like structures.
3.1.6. Snake-based shape extraction
Curve fitting itself is not competent to segment mitochondria since they may not appear as fully surrounded by a membrane. Moreover, curve fitting may fail as a result of inaccurate ridge detection due to weak appearance of the membrane. A curve segment may also overlap multiple mitochondria which reside close to each other. Therefore, separation of mitochondrial regions require further processing in which the polymorphic characteristic of mitochondria are also taken into consideration.
We employed balloon snake which is a variant of the active contour model in order to overcome the challenges encountered in the segmentation problem. Active contour models are versatile in segmenting irregular shapes and have a gap filling property (Kass et al., 1988). Balloon snake has an additional inflation feature to segment the object by approaching from the inside. It has been shown that balloon snakes exhibit a significant stability when initialized on a plain region where there exists no sufficient nearby contours (Cohen, 1991).
The snake method is implemented by energy minimization of the snake model:
| (12) |
where ν denotes the boundary of the object (i.e. mitochondrion) to be segmented that is represented by x and y coordinates of the snake contour along a trajectory t. Eint is a smoothness term that consists of the first and second order derivatives of ν:
| (13) |
where wa controls the contribution of tension on the contour created by the first derivative and wb controls the curvature energy generated by the second derivative. Eext is the external energy term which motivates the snake towards the boundary to be segmented and Einf creates an outwards inflation force. In our snake-based shape extraction scheme, the snake contour is driven by an energy image which is constituted by a curve energy function E(Ω) composed of average curve energy:
| (14) |
where x is a point on the image,Ωp is the set of points on the curve segment p and the cardinality term |Ωp| gives the number of points on the curve. Eext is defined as the accumulation of curve energy images:
| (15) |
where wc is the weight parameter to regulate the effect of curves on the snake contour and CL denotes the set of large-scale curve segments. The energy function in Eq. (15) is a negative valued function since the snake is interested in minimum energy and should converge on the points where the curve energy is maximized. Moreover, it depends on the total energy so that overlapping curves make the contour stronger.
The snake energy (Eq. (12)) is minimized iteratively by updating the snake contour which is initialized as a unit circle near to the vertex of a parabola until it is converged. The snake initialization process is discussed in detail in Section 3.2.2. The update equation is defined as:
| (16) |
where γ is step size and should be set to a suitable value (e.g. 1/|∇Esnake|) in order to prevent the snake from oversighting the mitochondrion boundary. During the update operations, the distance between consecutive points of the snake may become unequal due to the arrangement of the vector field formed by ∇Esnake(νi). In order to fix this problem, an equidistant correction process is applied in addition to the snake model (Valdés et al., 1996). The term ∇Esnake is given by:
| (17) |
where the internal energy update term can be found as:
| (18) |
Eq. (18) can be computed numerically by using five consecutive points on the snake contour. On the other hand, the term ∇Eext is computed by taking the derivative of Eext with respect to the x and y axes individually. The gradients of energy terms can be considered as the inversely oriented vectors of the internal and external forces that push the snake to desired locations. Accordingly, ∇Einf is defined as the inversely weighted outwards normal vector:
| (19) |
where N⃗ν is the unit normal vector at the boundary point ν of the snake and wd adjusts the magnitude of the inflation force. This force pulls the snake boundary outwards and prevents the snake from vanishing in the absence of external forces. Thus, the snake will grow when it is initialized on plain regions far from curves. Besides, the internal and inflation forces will cancel out each other and the snake will stay in balance after it grows a while, since the internal force increases as the snake is being inflated. Moreover, a strong inflation force can make the snake continue to grow when weak external forces are acting on the snake. In this respect, the snake may converge into different regions depending on the strength of curves surrounding the region and the size of the region. A small value of wd will be sufficient to extract the boundary of a small mitochondrion. In case of a large mitochondrion, a relatively high of wd is required. If wd is excessively high, the mitochondrion boundary may not hold the snake if the curves detected on the boundary are weak. On the other hand, the possibility of false-positive large-scale curve detections on cristae must be taken into account. To handle such cases, wd should be set to a high value.
We set up the following algorithm in order to extract candidate mitochondrial regions with different size, boundary strength and cristae arrangement:
- Repeat the following sub-steps for each parabolic arc:
- Initialize wd with a small value.
- Initialize the snake nearby the vertex of the parabolic arc as a unit circle.
- Execute the snake algorithm.
- If a different boundary is obtained, save the snake boundary νi.
- Increase wd.
- If wd is smaller than a threshold (Tinf), go to step ii.
Use the saved snake boundaries as candidate shapes for the next stage.
The algorithm described above extracts snake boundaries for ascending values of the inflation weight parameter wd iteratively. Fig. 6 depicts sample output of the algorithm. The whole image is scanned by executing the algorithm for each curve located on different regions. Note that weak curve segments should be ignored since misleading initialization would occur due to false detection. Hence, curves having sufficiently high energy are considered in the process as indicated in the preceding curve filtering step. In case of an initialization of a snake outside a mitochondrion, the process would yield a non-mitochondrial region if the snake encounters appropriate enclosing curves. Therefore, we adopt a validation procedure in the next step in which awkward and irrelevant segmentations are discarded.
Fig. 6.
Sample output of the shape extraction algorithm: (a) 1st iteration; (b) 2nd iteration and (c) 3rd iteration.
3.1.7. Validator function
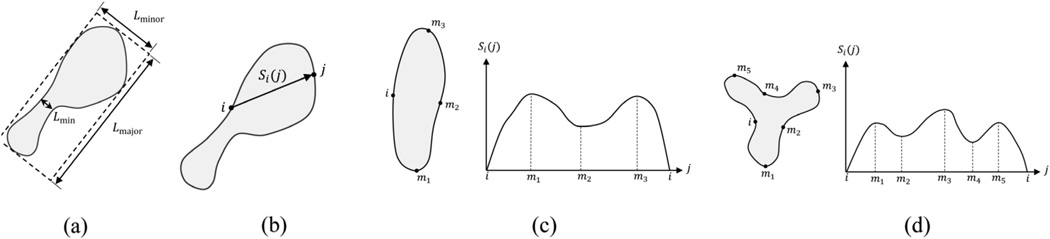
A mitochondrion is differentiated from the background by an enclosing membrane and cristae inside. Although mitochondria cannot be strictly represented by a particular shape, the vast majority appear as a form of distorted, bended or laterally compressed form of an ellipsoid (Tasel et al., 2014) as illustrated in Fig. 7. We built a validation mechanism for candidate shapes extracted in the previous step in order to sort out potential outliers. The validator function F(ν) is a binary valued function that returns a decision towards the acceptance or rejection of the shape ν. A set of descriptors and conditions are verified by the validator function. The shape ν is accepted as a mitochondrial region if all of the conditions are satisfied. In order to obtain scale-invariant features, circumference and area of the shape are used to normalize quantities and denoted by C and A respectively in the equations presented in this section. The descriptors involved in F(ν) are explained in five categories below:
- Energy: A shape having strong curves on the boundary and cristae inside indicates the presence of a mitochondrion. The average boundary energy of the shape ν is given by:
Note that CL is the set of the large-scale curves. In order to compute the average energy corresponding to internal region of a given shape, we can similarly arrange Eq. (20) to operate on the small-scale curves. Hence, the average region energy is given by:(20)
where Sν is the set of points lying inside the region formed by ν and CS denotes the set of small-scale curves. Thus, the acceptance criteria associated to shape energy may be found as Eboundary > Tenergy_b and Ecrista > Tenergy_c where right-hand side terms denote energy thresholds for boundary and crista respectively.(21) Area: The area of the candidate shape is used to determine whether the size of the shape is acceptable. Therefore, the area is expected to be in a range specified by the physical limitations of mitochondria. Although the cross-sectional area of a mitochondrion may drop down to zero due to orientation and location of the cross-section, extremely small snippets of mitochondria can be unreliable. Hence, the corresponding acceptance criterion is Tarea_min < A < Tarea_max.
Discontinuity: The gap filling property of the snake model may lead to a false segmentation in which the shape boundary has some large segments on a region where the external energy is too weak. In such cases, it is necessary to query whether the shape is dependable. We expect the snake boundary to be located partially on a gap by considering that the mitochondrion may appear as a roughly closed region or it may be located on the image border and seen partially. Moreover, no large-scale curve may be detected on the weak boundary. Nevertheless, allowing shapes with too large gaps would also increase the false positive rate. Therefore, we adopt a gap detection mechanism in order to reject the shape according to the gap length. A gap is detected on the boundary point t of the candidate shape if the boundary energy measurement ∑p∈CL Ecurve(t, p) is too small (i.e. less than a small threshold value Tgap_energy). We measure the total gap length (Lgap_total), the maximum gap length (Lgap_max), the ratio of the total gap length to circumference (Lgap_ratio) and the ratio of the length of the total gap on the image border to circumference (Lgap_border). Herewith, four additional acceptance criteria associated to boundary continuity descriptors are specified by setting an upper limit to each descriptor: (1) Lgap_total < Tgap_total, (2) Lgap_max < Tgap_max, (3) Lgap_ratio < Tgap_ratio and finally (4) Lgap_border < Tgap_border. Hence, a shape is rejected if one of the gap descriptors indicates extremely high discontinuity of shape boundary.
- Curvature: The shape boundary is expected to have a reasonable smoothness. In this category, local and average curvature of the boundary is investigated. A shape is accepted if the curvature is suitable for the contour of a mitochondrion. In this sense, sharp or frequent changes in direction of the boundary path is inadmissible. The local curvature of a curve is defined as (Stewart, 1998):
where T⃗ is the unit tangent vector and l denotes the arc length. In the discrete case, the local curvature at a sample point of ν(t) can be approximated by using the equation below:(22)
where d(t) = ν(t + 1) − ν(t). The average curvature is given by computing the average of Eq. (23) over all boundary points:(23)
where n is the number of points on the boundary. The acceptance criteria concerning curvature descriptors are maxt κL(t) < Tcurv_max and κA(ν) < Tcurv_ave. Note that setting up a limitation to the maximum local curvature guarantees that smoothness is satisfied by every point whereas restricting average curvature precludes tortuosity.(24) - Signature: Circular, elliptical or elongated patterns are frequently observed in the cross-sections of mitochondria. An example for an elongated shape is demonstrated in Fig. 7(c). On the other hand, a mitochondrion appearing like an asterisk shape (see Fig. 7(d)) is an unusual case in tomographic images. In order to expose such morphologic attributes, we extract indicative features via signature functions. The signature function of the boundary point j with respect to reference point i for shape ν is defined as the distance between i and j as illustrated in Fig. 7(b) and formulized below:
(25)
Fig. 7.
(a) Major axis length (Lmajor), minor axis length (Lminor) of a shape and the minimum thickness (Lmin); (b) Signature function Si(j) from the reference point i to boundary point j; (c and d) The reference point i and local extremum points mj on the shape boundary and sketch of corresponding signature function.
Local extrema count Ni pertaining to the signature function is a useful feature to determine the extension count of the object. As delineated in Fig. 7(c and d), local extrema count (within a single period) of Si(j) is proportional to the extension count (e.g. Ni ≤ 2 for a circular object and Ni ≤ 4 for an elliptic object). For the shape given in Fig. 7(d), Ni is equal to 6. Note that the number of local extrema depends on the chosen reference point and it should be checked for all possible reference points placed on the boundary of the shape. We expect that maxi Ni ≤ 4 must be satisfied for a valid mitochondrial shape.
The minimum cross-sectional thickness Lmin (see Fig. 7(a)) is another descriptor which can also be extracted from Si(j). Lmin is equal to the least local extrema achievable given by:
| (26) |
where mk denotes local extrema points. In our validation algorithm, a valid shape is not allowed to have too short Lmin. Hence, the acceptance criterion is given by Lmin > Tsig_min. We also use major and minor axes’ lengths as two additional descriptors. The length of major axis (Lmajor) is given by maxi,jSi(j) and the length of minor axis (Lminor) is determined by the width of the bounding rectangle placed along the major axis as shown in Fig. 7(a). Lmajor and Lminor are used to inspect whether the size of candidate shapes does not exceed the expectations based on the physical limitations of mitochondria. Hence, corresponding acceptance criteria are specified as Lmajor < Tmajor and Lminor > Tminor.
3.1.8. Post-processing
A mitochondrion may not be fully segmented using a single initialization depending on the false-positive curves detected on cristae and the shape of the mitochondrion. For example, narrow and curvy mitochondria create strong internal forces that prevent the snake from converging to the correct boundary. Instead, multiple snakes are initialized for all the curves which surround the same mitochondrion in our approach. Each snake is expected to cover a different portion of the mitochondrial region as it can be seen from the image showing validated shapes in Fig. 1(g). In order to achieve a better segmentation, the resulting contours after execution of snakes are merged if the ratio of overlapping area is greater than a threshold. The merged shapes in Fig. 1(h) were obtained by continually joining contours when the area of intersection (with another contour) is larger than 30% of the area of the entire shape. The major drawback of such an automatized merging mechanism is that it may erroneously join more than one mitochondrion together when a snake spreading onto multiple mitochondria due to over-inflation is found to be valid. Nevertheless, it can be beneficial when integrated into semi-automatic systems.
3.2. 3D extension of the algorithm
3.2.1. A 2.5D approach to snake model
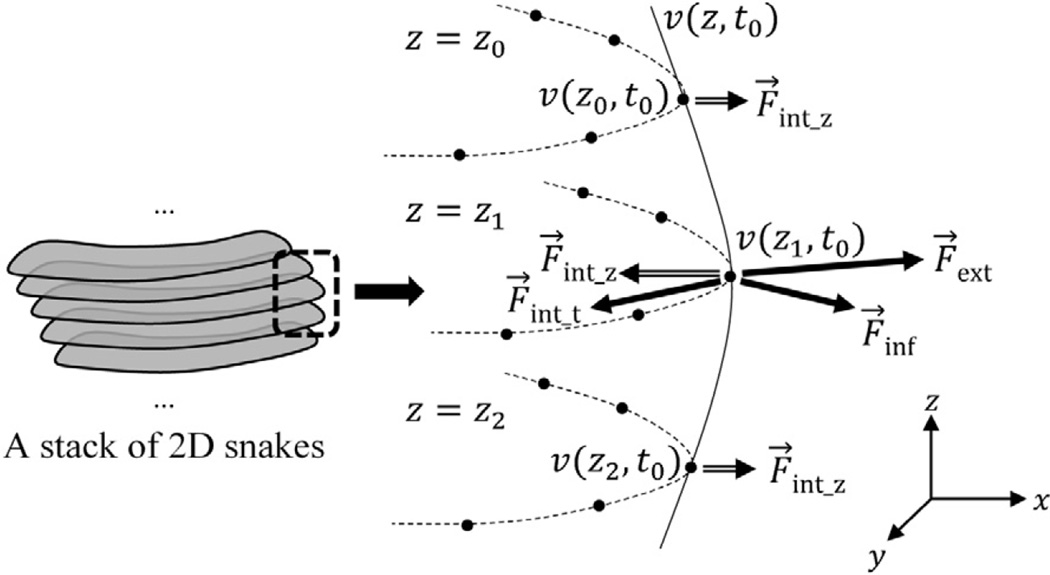
The standard 2D snake method involves evolution of 2D boundary vertices under the effect of internal, external and inflation forces. The 3D extension of the model is achieved by denoting each vertex as a point (x, y, z) in 3D space and redefining the active forces in terms of several vectors which are computed along the path on both xy-plane and z-axis (Ahlberg, 1996). This scheme allows the forces to shift the z-position of the vertices and make the snake boundary grow through the z-axis as well as the xy-plane.
In our 2D balloon snake method, the external forces are supplied by the curves detected on membranes that appear as elongated ridge structures. Since a membrane that is parallel to the xy-plane does not appear as a ridge-like structure in the tomogram, no curve is detected for such a membrane and consequently, no force can be created. Therefore, the aforementioned method is not directly applicable to our 2D system. Instead, we propose a quasi-3D (2.5D) approach in which the snake model is constructed in 3D space as a stack of 2D snakes. The internal forces acting at vertices on each slice depend on the neighboring vertices along the paths on both xy-plane and z-axis as depicted in Fig. 8. However, a vertex is not allowed to change its initial z-position. The external forces are obtained in the same way as we proposed in the 2D approach.
Fig. 8.
A stack of snakes and acting forces on a vertex of the snake.
Let ν(z, t) = (xz,t, yz,t) represent a 2D point on the snake boundary for the slice z. The internal energy term is redefined as follows:
| (27) |
The control parameters wat and waz are weights for tension on the xy-plane and along the z-axis respectively, wbt and wbz are corresponding weights for curvature. Thus, the internal energy term is minimized by using the update vector given by:
| (28) |
Note that Eq. (28) produces a 2D vector since the vertex ν is defined as a 2D point. Hence, each 2D snake is allowed to evolve on its own slice. Considering the contribution of external and inflation energy, we can rewrite the boundary update term as below:
| (29) |
where is the gradient of external energy image acquired for slice z and ∇Einf is the inflation energy update term which is computed on a slice-by-slice basis in a 2D manner. These update terms are visualized in Fig. 8 as the forces which are acting on the object and given by inversely oriented vectors (i.e. F⃗ = −∇E). The internal forces F⃗int_t and F⃗int_z shown in Fig. 8 correspond to the vectors which are formed by the first two and the last two terms of Eq. (28) respectively. The internal force emerged from the derivation over z-axis (F⃗int_z) works in favor of aligning points along z-axis. Hence, the evolution of the snake boundary is carried out by indirect interaction of forces via minimization of energy among neighboring slices.
3.2.2. Snake initialization
In the 2D approach, we adopted an algorithm in which snakes are initialized as small circles (e.g. unit circle) near to the curves that are placed on several locations and have sufficiently high energy. Although this scheme is useful to scan all mitochondria in the image, initialization of the snakes may differ due to displacement of curves extracted from the other slices. Furthermore, the output of the algorithm is independently evaluated by the validator function with respect to a single slice. Consequently, inconsistent contours can be obtained according to the initial locations and validation of snakes at each slice. Therefore, 2D initialization is not suitable to produce a smooth 3D mesh. In order to reduce the inconsistency problems, we propose a new initialization method that uses the curves in multiple slices by considering the possibility of movement of membranes and variability in detection. According to our new method, the initial locations of snakes are determined by a modified version of Density-Based Spatial Clustering of Applications with Noise (DBSCAN) algorithm (Ester et al., 1996).
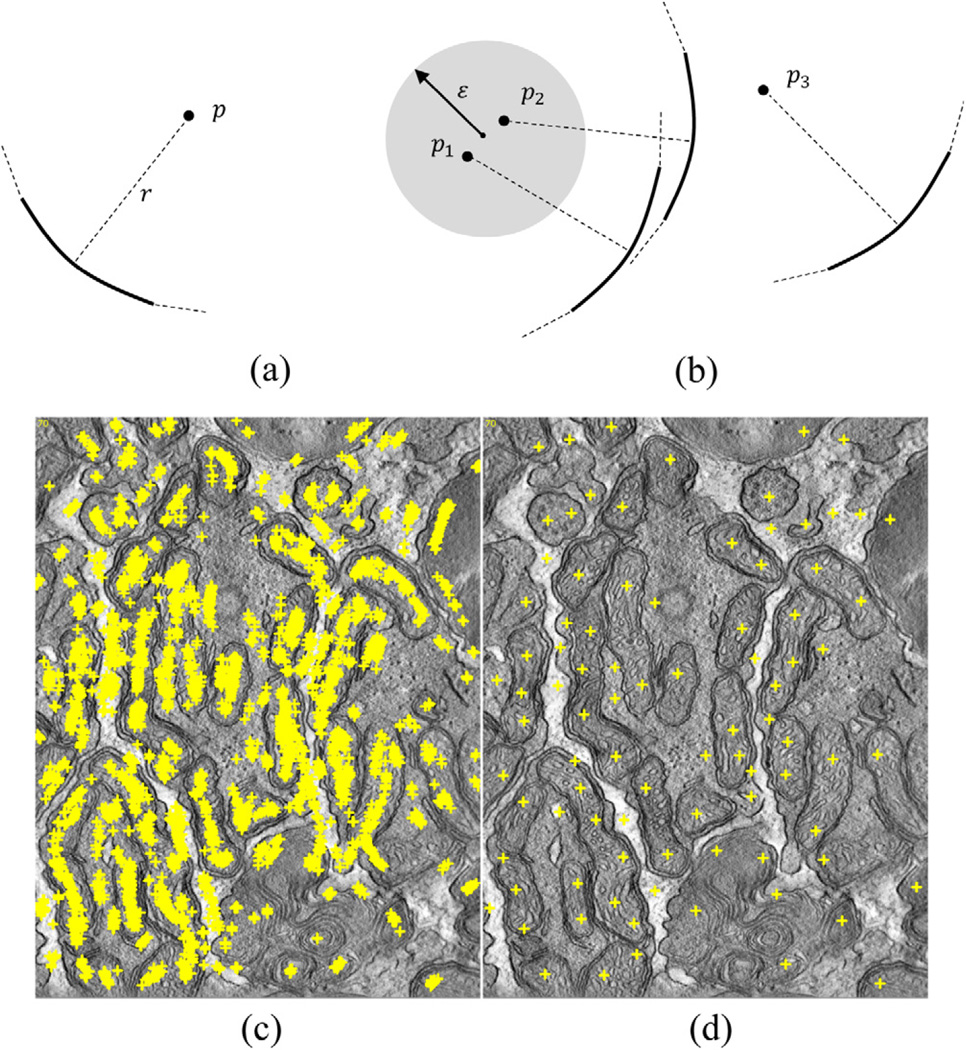
We defined the cue point of a curve segment as the point in the concave side of the parabolic arc with distance r in the orthogonal direction to the vertex point as shown in Fig. 9(a). Cue points within a particular neighborhood support each other if all of them reside in the concave sides of the corresponding parabolic arcs. For example, the points p1 and p2 support each other whereas p3 is blackballed as illustrated in Fig. 9(b). Sufficiently populated cluster centers which are formed by the supportive cue points within the neighborhood are used as actual initial points. Thus, snakes are initialized on the cluster centers as a cylinder (or a circle in the 2D case). In order to guarantee to catch the mitochondrion boundary, the distance parameter r and diameter of the cylinder should be set to a value smaller than the size of minimum cross-sectional thickness of the mitochondrion to be detected. On the other hand, height of the cylinder depends on z-thickness of the mitochondrion appearing in the volume. A larger height is expected to extract a better surface since more information is utilized. However, a snake with a too large height that exceeds the size of the mitochondria may not converge successfully. In order to initialize snakes only at reliable locations, the clusters having fewer cue points than a threshold are ignored. Hence, the effect of false positively detected curves is reduced.
Fig. 9.
(a) A cue point p of a curve segment and (b) a cluster formed by the cue points; (c) Cue points extracted from the dataset “od_sub” (slice range: 70–89) and (d) corresponding cluster centers.
The algorithm that is used to initialize snakes is given below:
Determine all of the cue points for each curve which is detected in a given z-interval.
Find all supportive points for each cue point within ε neighborhood.
Constitute a cluster for each neighborhood having sufficient number of points.
Initialize a snake as a cylinder at each cluster center (i.e. the center of mass formed by the cue point population in the cluster).
Curve segments on a strongly detected membrane overlap each other. Similarly, the curves on a strong membrane continuing along the z-axis should appear at close-range. Therefore, the cue points are expected to support each other for such curves. Fig. 9(c) portrays every cue point gathered from 20 slices of a dataset. The corresponding cluster centers are displayed in Fig. 9(d). Since the initialization procedure utilizes each curve from a range of tomograms, it is robust against false detection and noise.
The original DBSCAN algorithm is used to create dense clusters satisfying the criterion that each point in the cluster is sufficiently close to at least one point in the cluster. This method aims to make clusters as wide as possible. However, converting a pervasive distribution to a single cluster is not a desired property since a better segmentation could be achieved by trying multiple initializations inside the mitochondrion. In our algorithm, the extent of clusters is limited to ε neighborhood such that the cluster centers are found to be at least 2ε distant.
3.2.3. Adaptation of the validator function
In the 2D method, the validator function decides acceptance or rejection of a given 2D snake. Formally, it can be defined as F:ν → {0, 1} where ν is a 2D shape. The function returns 1 in case of acceptance and returns 0 otherwise. In order to validate 2.5D snakes, we extend the definition of the validator function to a validity measurement formulated as:
| (30) |
where νz is a 2D cross section of snake ν at z-level and k is the z-thickness of the snake satisfying 1 ≤ z ≤ k. The validity V is a real-valued function which ranges over [0, 1] and returns the probability of the shape being valid. In a sense, measurement of validity performs a voting process and yields the acceptance rate of all voters. Thus, the snake ν is accepted as valid if the validity V(ν) is greater than a threshold (TV) or rejected otherwise.
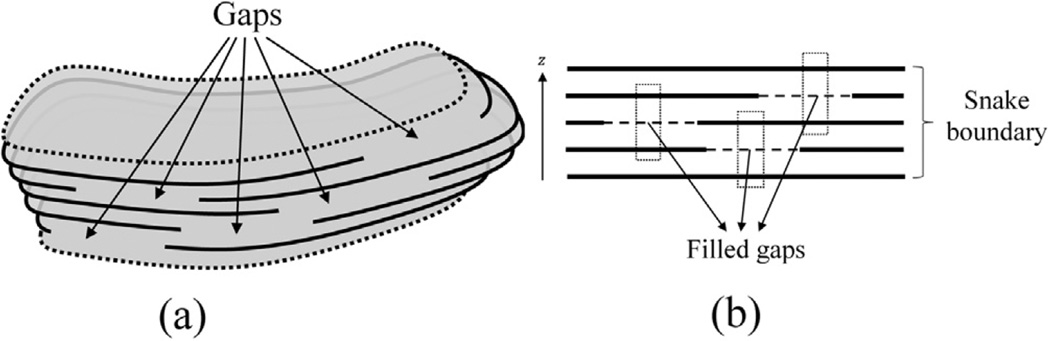
The 2.5D model may have some cross-sections which appear to be invalid due to a large amount of gaps where inflation of the boundary is suppressed despite lack of external energy and convergence is enforced by the effect of internal energy along the z-axis that is revealed by means of neighboring slices. In such cases, the validity value may be found to be unexpectedly low although the mitochondrion seems to be successfully segmented. Fig. 10(a) depicts an explanatory example in which each cross-section of snake has substantial amount of gaps making them invalid. Hence, the validity value of the whole shape is reduced. In order to dissipate such a potential problem, a window is passed through the z-axis and the median filter is applied to the boundary energy measurement ∑p∈CLEcurve(t, p) within the window. Therefore, a gap is artificially filled by neighboring slices if there exists a sufficient number of neighbors with high boundary energy as shown in Fig. 10(b). Conversely, a false boundary point may become a gap if it is not supported by its neighbors along the z-axis.
Fig. 10.
(a) A 2.5D snake is assumed to have gaps where boundary energy is weak; (b) gap filling mechanism.
3.2.4. Adaptation of the post-processing
In the 3D case, the merging operation is done considering overlapping volumes of 3D segmented regions. In this manner, valid 2.5D snakes are joined iteratively until the ratio of the overlapping volume of no region is greater than the threshold.
4. Experimental results
Both 2D and 2.5D algorithms were tested by using every slice of the eight datasets listed in Table 1. The parameter sets used in the system are presented in Table 2. Adjustments of these parameters are discussed in Section 4.3. The candidate regions obtained from the 2.5D snake algorithm were subjected to the validation process with and without the gap filling mechanism enabled (as described in Section 3.2.3). The size of the window (Wgap) to fill gaps was adjusted as 3, 5, 7 and 9 separately. The z-thickness (k) of the 2.5D snake model was set to 10, 20, 30 and full (equal to dataset thickness) separately. The datasets have been split into subsections such that each sub-section contains consecutive k slices. For example, five sections are obtained for a dataset consisting of 100 slices when k = 20. The initialization and execution of snakes have been individually performed in each section. Additionally, the snake output has been merged in post-processing if the ratio of overlapping area (or volume) to entire region was greater than 30%. Then, the whole process was repeated for three different settings of gap parameters given in Table 2.
Table 2.
List of parameters and settings used in the proposed system.
| Parameter description | Used setting | ||
|---|---|---|---|
| Preprocessing parameters | |||
| Auto-contrast histogram cut | 0.5% | ||
| Resampling pixel-size | 2 nm | ||
| Bilateral filtering spatial kernel size | 60 nm | ||
| Bilateral filtering gray sigma | 0.2 | ||
| Ridge detection parameter | |||
| Gaussian derivative sigma | 3 nm | ||
| Energy mapping parameter | |||
| Window size (ω) | 30 nm (LFREM) | ||
| 8 nm (HFREM) | |||
| Curve fitting parameters | |||
| Threshold energy (Tmap) for initialization | 40% of max eθ(s) | ||
| Curve update parameters (τx, τy, τh) | 8 nm | ||
| Curve filtering parameters | |||
| Max. allowed value for curvature (κ) | 1 | ||
| Min. arc length | 100 nm (large-scale curves) | ||
| 20 nm (small-scale curves) | |||
| Min. average energy | 30% of max eθ(s) | ||
| Shape extraction/snake parameters | |||
| Tension weight (wa, wat) | 1.0 | ||
| Curvature weight (wb, wbt) | 200 | ||
| External energy weight (wc) | 0.5 (2D) | ||
| 1.0 (2.5D) | |||
| Inflation weights (wd) | 0.5, 1.0, 1.5 … 3.0 | ||
| Tension weight along z-axis (waz) | 5.0 | ||
| Curvature weight along z-axis (wbz) | 5.0 | ||
| z-thickness (k) | 1 (2D), 10, 20, 30, max | ||
| Snake initialization parameters | |||
| Distance to the vertex point of arc (r) | 40 nm | ||
| Cluster neighborhood (ε) | 100 nm | ||
| Min. cluster size | [1.5xz – thickness] | ||
| Validator function parameters | |||
| Min. average boundary energy (Tenergy_b) | 20 | ||
| Min. average region energy (Tenergy_c) | 0.1 | ||
| Max. local curvature (Tcurv_max) | 1/45 nm−1 | ||
| Max. average curvature (Tcurv_ave) | 1/180 nm−1 | ||
| Max. number of local extrema of the signature function (maxiNi) |
4 | ||
| Min. thickness (Tsig_min) | 70 nm | ||
| Max. major axis length (Tmajor) | 2000 nm | ||
| Min. minor axis length (Tminor) | 140 nm | ||
| Min. area (Tarea_min) | 0.02 µm2 | ||
| Max. area (Tarea_max) | 0.7 µm2 | ||
| Gap-filling window size (Wgap) | 3, 5, 7, 9 | ||
| 1st setting |
2nd setting |
3rd setting |
|
| Boundary energy measurement threshold (Tgap_energy) |
5 | 10 | 15 |
| Total gap length (Tgap_total) | 500 nm | 500 nm | 600 nm |
| Max. gap length (Tgap_max) | 400 nm | 400 nm | 600 nm |
| Max. gap ratio (Tgap_ratio) | 0.4 | 0.4 | 0.4 |
| Max. gap ratio on border (Tgap_border) | 0.3 | 0.3 | 0.4 |
| Post-processing parameter | |||
| Merging threshold | 30% of area (or volume) overlapped |
||
Comparison of convergence of 2D and 2.5D snakes is illustrated in Fig. 11. Fig. 11(a) exhibits that 2D snake may over-inflate due to inaccurate curve detection on membranes. On the other hand, the 2.5D model is capable to compensate false-negative error as shown in Fig. 11(b). Similarly, the 2D approach may fail to inflate the balloon properly in case of the existence of false-positive curves on cristae whereas such problems are eliminated with the aid of neighboring slices in the 2.5D model. The 2D model also suffers from instability problems among consecutive slices where false-positive and false-negative contours appear to be valid. On the contrary, the 2.5D model attains robust results compared to the other model.
Fig. 11.
Boundary of an identically initialized (a) 2D snake output (performed in slice-by-slice manner) and (b) 2.5D snake output for the dataset “bclpb-d.sub” (slice range: 20–39).
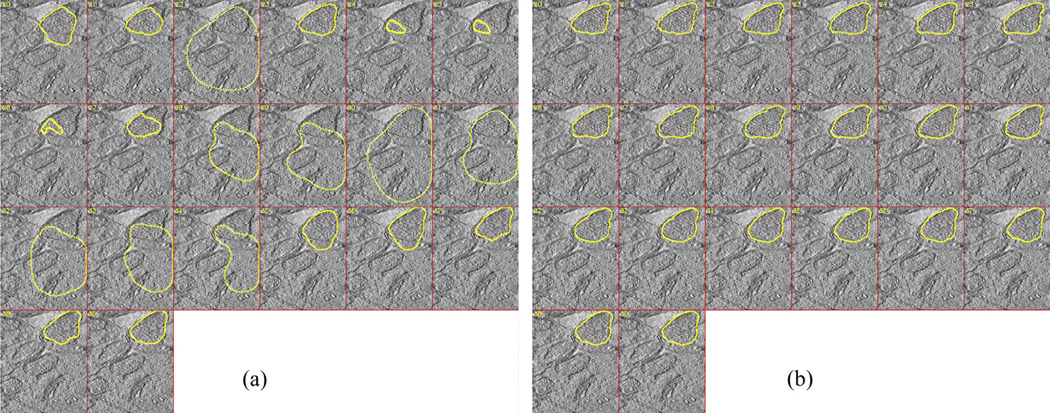
Another significant issue is the adjustment of z-thickness (k) of the snake. A relatively thin snake (having a few slices) does not effectively utilize most of the curves regarding each slice. On the other hand, an excessively thick snake partially converges and it is possibly rejected by the validator function if the mitochondrion to be segmented does not span the z-range of the snake. Mitochondria can be segmented in all slices by executing the algorithm in all sections individually. Fig. 12(a) demonstrates a sample output for four snakes for k = 20. A smooth transition among sections may not be observed in some cases due to the discontinuity between the bottom and top slices of the sections. In this case, it would be better to re-run the snake algorithm with total z-thickness of previously segmented sections in order to segment the whole mitochondrion as illustrated in Fig. 12(b). Segmentation outputs of several mitochondria which were obtained by using the maximum achievable z-thickness are shown in Fig. 12(c). Fig. 13 exhibits segmentation boundaries in sample tomograms taken from each dataset.
Fig. 12.
A sample merged shape with (a) k = 20 and (b) k = 80; (c) Merged shape samples for maximum valid n. The outputs were visualized by MeshLab tool (Cignoni et al., 2008).
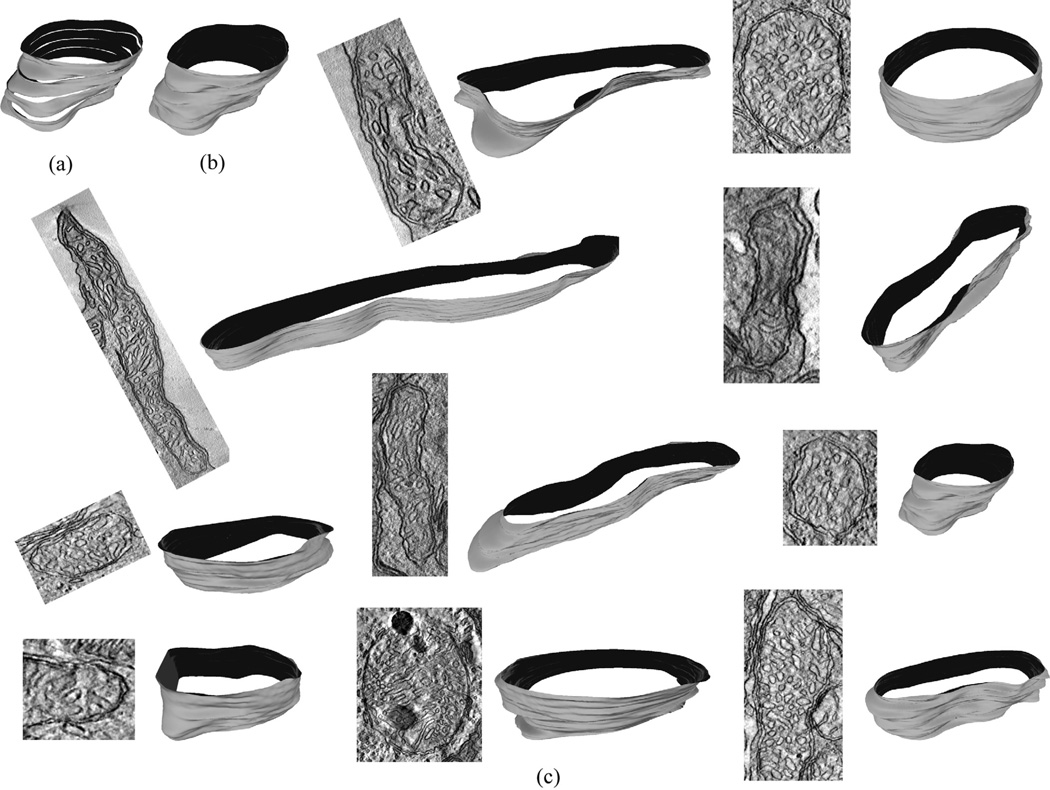
Fig. 13.
Mitochondria segmentation results (Image basename and slice number): (a) cone.sub (slice #: 40); (b) gap18_sub (slice #: 35); (c) 6_22.sub (slice #: 120); (d) spherule24mos1 (slice #: 45); (e) od.sub (slice #: 70); (f) pedicle (slice #: 40); (g) bclpb-d.sub (slice #: 50); (h) mac_serial_sub (slice #: 20).
4.1. Performance evaluation
Precision and recall are two fundamental performance indicators that are used for measuring detection accuracy. Precision is the ratio of the detection outcome being correct (true positives/all positives). On the other hand, recall gives the ratio of true elements being successfully detected (true positives/all true elements). In image segmentation problems in which pixels are used as the elements to be detected, precision is given by the ratio of the size (i.e. area or volume) of correctly detected regions to the size of all detected regions whereas recall is given by the ratio of the size of correctly detected regions to the size of true regions according to basic definitions. However, the definition can also be modified depending on what is to be detected, nature of the domain and how the results are interpreted.
In our preceding studies (Mumcuoglu et al., 2012; Tasel et al., 2014), a region was accepted as a correct detection if at least 70% of area of the region overlapped a single mitochondrion. Then, precision and recall were defined based on the number of regions (not the size of regions). Such a definition ensures an accuracy measurement such that each region is evaluated depending on whether it segments a single mitochondrion or not. On the other hand, the definition also entails that a region should be accepted as a correct detection even if it segments only a small portion of the mitochondrion. Note that the whole mitochondrion is assumed to be successfully detected even if it is not covered by multiple regions in this scheme. Indeed, even a single small region can make the whole mitochondrion detected successfully.
In this study, a new approach has been applied to measure precision and recall by considering potential segmentation boundaries among algorithm output and ground truth. In the first step, each detected shape is matched to the mitochondrion boundary where the Dice Similarity Coefficient (DSC) is maximized. The DSC is defined as:
| (31) |
where G and S denote the ground truth shape of a mitochondrion and the detected shape obtained by the algorithm respectively. The cardinality operator |·| denotes the area of 2D shapes and the volume of 3D shapes. Let Si denote the ith detected shape and let Gi denote the ground truth of the ith mitochondrion and denote the best matching ground truth region for Si. Then, the precision and recall measurements are formulized as follows:
| (32) |
| (33) |
Note that the numerator of the fractions in Eqs. (32) and (33) focus on the union of intersections of each shape with the best matching ground truth shape. Hence, detection performance is evaluated by considering the criterion that each shape or union of a set of shapes should segment a single mitochondrion. Additionally, recall and precision reflect more reasonable values since the area (or volume in the 3D case) of the region of intersection proportionally affects the fractions. The performance measurements have also been empowered by evaluation of segmentation accuracy. Two metrics have been utilized:
Dice Similarity Coefficient (DSC)
The average DSC (see Eq. (31)) of all of the snakes together with corresponding best matching mitochondrion boundary has been computed.
Median Symmetric Boundary Error (MSBE)
The median of the set composed of the minimum Euclidean distance from each snake boundary point to mitochondrion boundary and from each mitochondrion boundary point to the snake boundary has been computed. The formulation is given below:
| (34) |
where Ĝ(j) and Ŝ(i) are boundary points, N and M are the number of points on the boundary of ground truth and snake output respectively.
4.2. Quantitative results
The precision and recall values have been calculated for every possible validity threshold (TV). The recall has been presented for fully seen mitochondria and all of the mitochondria separately. DSC and MSBE have been measured for both original snake output and merged snake-output. Additionally, we utilized F-scores in order to facilitate the comparison of algorithm performance. The F-score is equal to the harmonic mean of precision and recall. It provides a balanced measurement considering both precision and recall (Powers, 2011).
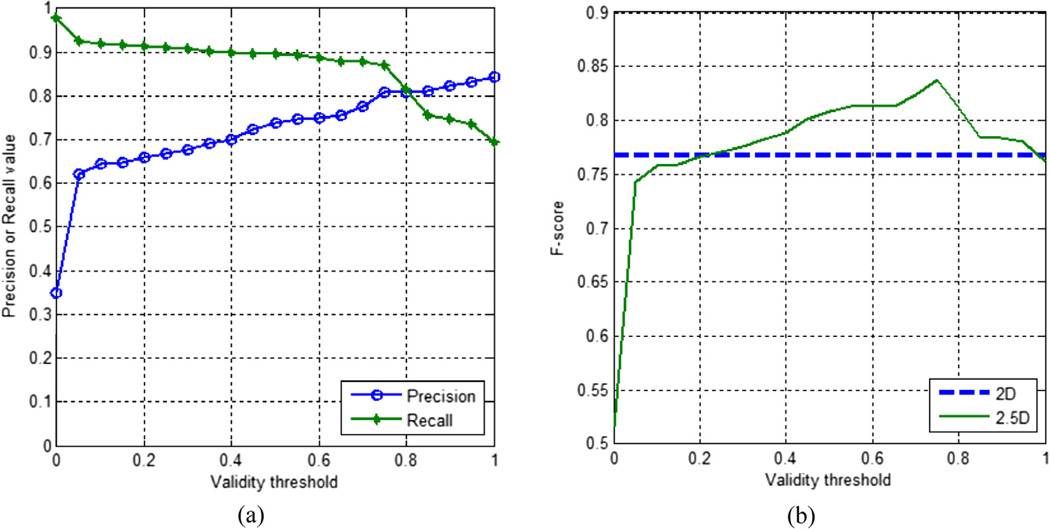
Table 3 contains snake and validation parameters, corresponding average precision and recall that maximize F-score. Precision of the 2D method is 0.71 whereas the 2.5D method achieves a value up to 0.81. A slight improvement was also obtained for the recall value that was calculated for fully seen mitochondria. The recall was found as 0.84 for 2D method and raised up to 0.87 by the 2.5D approach. The best results were obtained when the snake thickness (k) was 20 and the validity threshold (TV) was 0.75. Henceforth, the best-case results of the 2D and 2.5D methods with respect to k and TV are presented and discussed below unless stated otherwise.
Table 3.
The best achievements of the algorithms and corresponding parameters.
| Snake thickness (k) | Gap window (Wgap) | Gap setting | Validity threshold (TV) | Precision | Recall | F-score | ||
|---|---|---|---|---|---|---|---|---|
| Fully seen | All | Fully seen | All | |||||
| 1 (2D) | N/A | 2nd | N/A | 0.71 | 0.84 | 0.71 | 0.77 | 0.71 |
| 10 (2.5D) | 5 | 1st | 0.5 | 0.74 | 0.87 | 0.76 | 0.80 | 0.75 |
| 20 (2.5D) | 5 | 3rd | 0.75 | 0.81 | 0.87 | 0.71 | 0.84 | 0.76 |
| 30 (2.5D) | 5 | 3rd | 0.7 | 0.80 | 0.86 | 0.69 | 0.82 | 0.74 |
| Full (2.5D) | 3 | 3rd | 0.6 | 0.79 | 0.87 | 0.71 | 0.83 | 0.75 |
Fig. 14(a) demonstrates the precision-recall graph with respect to TV. The precision increases and the recall decreases while TV is increasing. In other words, the voting process is more reliable when consensus achieved is stronger. However, precise decisions lead to loss of some of the correct detections (false negative error). Fig. 14(b) shows the F-score versus TV graph and the achievement border of the 2D method. It is observed that the 2.5D method provides better results for a wide range of TV.
Fig. 14.
(a) The detection performance of 2.5D snake (k = 20; Wgap = 5; gap setting: 3rd) and comparison with 2D snake (gap setting: 2nd): (a) precision-recall (fully seen) vs. validity threshold (TV) graph; (b) F-score (fully seen) vs. validity threshold (TV) graph.
Table 4 compares the detection performance of 2D and 2.5D methods in eight datasets. Both methods fail in the dataset “mac_serial_sub” due to lack of accurate ridge detection since the mitochondria boundaries do not appear as ridge structures. Cristae and separation of peripheral membranes are not clearly visible since this particular dataset contains condensed mitochondria. As this is not a rare case, datasets which can be used for cristae segmentation are expected to have high contrast between membranes and background. Considering the failure in this dataset, the average precision and recall are given when it is both included and excluded. When the failed dataset is ignored, an improvement of 10% in precision and 3% in recall was observed on average with respect to the 2D model by utilizing the 2.5D approach. In the datasets “od_sub” and “6_22.sub”, the 2D method appears to produce a better recall compared to the 2.5D method. However, the 2D algorithm performance was measured on a slice-by-slice basis due to the nature of the algorithm where the segmentation of each slice is independently performed. Particularly, 2D snake boundaries that were successfully extracted from the slices where the 2.5D method failed led to an illusory increase in recall value. In such cases, boundaries of 2D snakes extracted from the consecutive slices are unconformable and the segmentation result is not continuous along the z-direction. Note that a high recall value does not mean a useful 3D segmentation in the 2D approach. On the other hand, a considerable amount of eligible slices must be segmented to obtain a valid snake in the 2.5D method. Otherwise, the validation process would be inclined to reject the shape in order to avoid reduction in precision. Furthermore, it is always possible to tune the algorithm behavior by adjusting the TV parameter (see Fig. 14(a)).
Table 4.
The detection performance of 2.5D snake (k = 20; Wgap = 5; gap setting: 3rd; TV = 0.75) and 2D snake (gap setting: 2nd) algorithms for eight datasets.
| Dataset basename | 2D | 2.5D | ||||
|---|---|---|---|---|---|---|
| Precision | Recall | Precision | Recall | |||
| Fully seen |
All | Fully seen |
All | |||
| 6_22.sub | 0.76 | 0.86 | 0.65 | 0.84 | 0.80 | 0.58 |
| bclpb-d.sub | 0.82 | 0.73 | 0.59 | 0.88 | 0.95 | 0.57 |
| cone.sub | 0.87 | 0.93 | 0.73 | 0.92 | 0.93 | 0.81 |
| gap18_sub | 0.79 | 0.79 | 0.60 | 0.85 | 0.84 | 0.60 |
| mac_serial_sub | 0.31 | 0.23 | 0.20 | 0.45 | 0.06 | 0.11 |
| od.sub | 0.60 | 0.75 | 0.64 | 0.65 | 0.62 | 0.52 |
| pedicle | 0.68 | 0.86 | 0.83 | 0.90 | 0.95 | 0.92 |
| spherule24mos1 | 0.43 | 0.95 | 0.95 | 0.62 | 0.99 | 0.99 |
| Average | 0.66 | 0.76 | 0.65 | 0.76 | 0.77 | 0.64 |
| Average (excluding mac_serial_sub) |
0.71 | 0.84 | 0.71 | 0.81 | 0.87 | 0.71 |
The effect of window size (Wgap) of the gap filling mechanism to detection performance is given in Table 5. The gap filling mechanism decreases precision but increases recall. Specifically, a larger window causes a larger decrease in precision and a larger increase in recall. Since the gain in recall is more than the loss in precision, it is reasonable to activate the gap filling mechanism. Experiments show that setting Wgap = 5 (in terms of the number of slices) is usually a good choice.
Table 5.
The detection performance of 2.5D snake (k = 20; gap setting: 3rd; TV = 0.75) with respect to different values of gap window size (Wgap).
| Gap window size (Wgap) | Precision | Recall | F-score | ||
|---|---|---|---|---|---|
| Fully seen | All | Fully seen | All | ||
| Disabled | 0.82 | 0.75 | 0.59 | 0.78 | 0.68 |
| 3 | 0.81 | 0.82 | 0.66 | 0.82 | 0.73 |
| 5 | 0.81 | 0.87 | 0.71 | 0.84 | 0.76 |
| 7 | 0.78 | 0.87 | 0.71 | 0.82 | 0.75 |
| 9 | 0.77 | 0.88 | 0.72 | 0.82 | 0.75 |
Table 6 lists the segmentation accuracy measured by two metrics (DSC and MSBE) with respect to the snake thickness (k). MSBE values are given in nanometers. The original and merged forms of snake outputs have been evaluated separately. The best results have been obtained for the 2D snake with the 2nd gap setting and for the 2.5D snake with the 3rd gap setting when k = 20, Wgap = 5 and TV = 0.75 (see Table 2 for the used parameter list). The average DSC was found as 0.79 for the 2D method and 0.84 for the 2.5D method. The merged snakes usually performed finer segmentation since segmentation of some of the mitochondria was achieved by the union of snakes. Note that multiple mitochondria may also be accidentally merged which causes a false detection. Considering such cases, falsely merged snakes were removed for the measurement of segmentation accuracy. The average of MSBE indicates that boundary segmentation is not as accurate as in the previous study (Mumcuoglu et al., 2012) since the double membrane tracing process has not been applied. Nevertheless, it is sufficient to extract rough annotations of mitochondria considering that total thickness of the double membrane of the vast majority of mitochondria is about 18–48 nm.
Table 6.
Segmentation accuracy of 2D snake (gap setting: 2nd) and 2.5D snake (k = 20; Wgap = 5; gap setting: 3rd; TV = 0.75) algorithms for different thickness values and datasets.
| Metric | DSC | |||||||||
| Snake state Dataset |
Original Snake thickness (k) |
Merged Snake thickness (k) |
||||||||
| 1 (2D) | 10 | 20 | 30 | Full | 1 (2D) | 10 | 20 | 30 | Full | |
| 6_22.sub | 0.68 | 0.68 | 0.72 | 0.74 | 0.73 | 0.72 | 0.76 | 0.75 | 0.77 | 0.82 |
| bclpb-d.sub | 0.85 | 0.89 | 0.92 | 0.92 | 0.91 | 0.86 | 0.90 | 0.91 | 0.91 | 0.89 |
| cone.sub | 0.86 | 0.86 | 0.93 | 0.93 | 0.94 | 0.88 | 0.87 | 0.90 | 0.90 | 0.90 |
| gap18_sub | 0.82 | 0.84 | 0.82 | 0.82 | 0.85 | 0.85 | 0.89 | 0.87 | 0.86 | 0.87 |
| mac_serial_sub | 0.68 | 0.71 | 0.73 | 0.77 | 0.35 | 0.65 | 0.69 | 0.70 | 0.77 | 0.35 |
| od.sub | 0.75 | 0.77 | 0.78 | 0.78 | 0.75 | 0.70 | 0.73 | 0.77 | 0.76 | 0.72 |
| pedicle | 0.73 | 0.72 | 0.79 | 0.77 | 0.74 | 0.78 | 0.78 | 0.91 | 0.83 | 0.84 |
| spherule24mos1_ | 0.85 | 0.82 | 0.92 | 0.87 | 0.98 | 0.92 | 0.95 | 0.97 | 0.97 | 0.97 |
| Average | 0.78 | 0.79 | 0.83 | 0.82 | 0.78 | 0.79 | 0.82 | 0.85 | 0.85 | 0.80 |
| Average (excluding mac_serial_sub) | 0.79 | 0.80 | 0.84 | 0.83 | 0.84 | 0.81 | 0.84 | 0.87 | 0.86 | 0.86 |
| Metric | MSBE (in nanometers) | |||||||||
| Snake state Dataset |
Original Snake thickness (k) |
Merged Snake thickness (k) |
||||||||
| 1 (2D) | 10 | 20 | 30 | Full | 1 (2D) | 10 | 20 | 30 | Full | |
| 6_22.sub | 33 | 30 | 24 | 24 | 18 | 32 | 23 | 32 | 34 | 14 |
| bclpb-d.sub | 16 | 11 | 8 | 10 | 9 | 17 | 11 | 9 | 11 | 9 |
| cone.sub | 18 | 17 | 10 | 10 | 9 | 14 | 14 | 10 | 10 | 10 |
| gap18_sub | 12 | 9 | 9 | 8 | 9 | 12 | 9 | 9 | 9 | 10 |
| mac_serial_sub | 26 | 19 | 28 | 21 | 110 | 31 | 23 | 42 | 21 | 110 |
| od.sub | 20 | 19 | 18 | 18 | 22 | 31 | 26 | 22 | 24 | 31 |
| pedicle | 48 | 49 | 41 | 49 | 50 | 32 | 32 | 11 | 21 | 21 |
| spherule24mos1_ | 30 | 31 | 18 | 28 | 8 | 15 | 9 | 7 | 7 | 8 |
| Average | 25 | 23 | 20 | 21 | 29 | 23 | 18 | 18 | 17 | 27 |
| Average (excluding mac_serial_sub) | 25 | 24 | 18 | 21 | 18 | 22 | 18 | 14 | 17 | 15 |
Table 7 presents average execution durations of major steps in our system. Computations and measurements were made on a computer with Intel Core 2.4 GHz microprocessor by using a single thread. The implementation was realized by using C++ programming language and OpenCV library. The 2.5D snake algorithm utilizes approximately 33% less CPU time compared to the 2D method since the new initialization procedure is capable of performing even better segmentation by using 33% less initial points. Although the 2.5D method requires additional computation, no significant change in CPU time per snake slice was attained.
Table 7.
Average execution time of major steps in the system.
| Step name | Scale | Duration (s) |
|---|---|---|
| Preprocessing | Per slice | 0.6 |
| Curve detection | Per slice | 18.1 |
| Snake algorithm (2D) | Per slice | 293.7 |
| Per snake-slice | 0.5 | |
| Snake algorithm (2.5D) | Per slice | 194.1 |
| Per snake-slice | 0.5 |
4.3. Parameter analysis
The response of the algorithms to different parameter values were investigated and discussed for each step below:
Preprocessing
The specified values of preprocessing parameters in Table 2 were tested with both low and high contrast tomograms and provided good and stable results. The auto-contrast histogram cut parameter should be kept as small as possible to eliminate only extreme gray values. High values may cause data loss in the intensity distribution. Resampling of the image to a fixed pixel size is useful for the parameter tuning process (especially for validator function parameters) and a 2 nm pixel size is adequate to visualize membranes. In our experiments, the bilateral filtering was capable of reducing the noise and the effect of non-membrane ridges without losing weak membrane structures when the spatial kernel size was set to a value between 30 and 60 nm. However, values lower than 30 nm precipitated detection of false positive large-scale curves on cristae. Conversely, true curves were lost when excessively large kernels were used. The gray sigma parameter provided suitable smoothing of membranes and background when it was set to 0.1–0.2 assuming that the gray value ranges between 0 and 1. Values greater than 0.2 give rise to smoothing of the edges (between membrane and background).
Ridge detection
The sigma parameter of Gaussian derivatives controls the expected thickness of membranes to be detected as ridges. This value is adjusted in conformity with the membrane thickness of mitochondria (4–6 nm). Experimental results show that relatively strong ridges are obtained when the sigma value is set to 2–3 nm.
Energy mapping
Peripheral membranes of mitochondria can be discriminated from cristae when the energy mapping window covers the double membrane structure of mitochondrion. Since total energy is expected to be doubled, the ω parameter can be set to the maximum thickness of the double membrane (~48 nm). Considering the distance among cristae membranes, we preferred to use the average double membrane thickness (~30 nm). If crista detection is desired, the window size is arranged in order to cover a single membrane (>6 nm). Note that it should be kept below the minimum double membrane thickness (<18 nm) to avoid the effect of a double membrane.
Curve fitting
Our experiments show that using local maxima points in LFREM/HFREM which have at least 30–50% of the maximum energy provides a good curve extraction covering both peripheral and crista membranes and avoiding initialization on non-membrane regions. The update parameters τx, τy and τh should be set to a reasonably high value in order to avoid local maxima of the energy function E(Ω) where a better curve is available in a small neighborhood in the model parameter space (x1, y1, x2, y2, h). On the other hand, excessively high values can make the curve span disjoint membranes. Suitable curves were obtained for a range of 4–8 nm.
Curve filtering
Curve filtering parameters (arc-length and curve energy thresholds) were adjusted empirically such that large-scale curves were preserved at the peripheral membrane of mitochondria and similarly, small-scale curves were preserved at both peripheral and crista membranes whereas a considerable amount of undesired curves at irrelevant locations were eliminated. Note that the expected curve energy and length are affected by the window size (ω) of the energy map. Therefore, we recommend to search these thresholds by starting from linearly scaled values with respect to ω when it is necessary to tune thresholds empirically. These parameters introduced acceptable results within a range of ±70%.
Snake-based shape extraction
The behavior of the system is not sensitive to small changes in energy weights of the snake model (wa, wat, wb, wbt, wc, wd, waz, wbz). Our parameter settings do not require further tuning according to the input dataset or shape of mitochondria. Nonetheless, the ratio of parameters with respect to each other has some significance. We preferred to set the tension weights (wa, wat) to a fixed value (i.e. 1.0) and tune other parameters accordingly. Our experiments showed that strong stiffness increased stability and the capability of gap filling and tolerance to false positive large-scale curves. Therefore, we adopted a strategy to set the curvature weights to a value satisfying wb, wbt ≫ wc, wd. The external energy weight (wc) was doubled for 2.5D snakes considering the effect of internal energy on the xy-plane and along the z-axis. The inflation weight (wd) should scan a wide range of values (i.e. 0.5, 1.0, 1.5…3.0) due to the presence of false-positive large-scale curves and diverse membrane strength. However, the step size can be further decreased to provide more robustness to membrane weakness at the expense of increased computation time. Since the z-thickness of the 2.5D snake is much shorter than its circumference on the xy-plane, tension and curvature weights along the z-axis (waz, wbz) were set to a higher value than wat and wbt in order to boost the effect of neighboring slices.
Snake initialization
The snake initialization may be unstable if the snake is initialized at a location too close to the vertex point of arc due to failure of proper inflation under the strong effect of external forces. The distance between the initial location and the arc (r) can be adjusted according to snake weight parameters. The snake boundary usually progressed without problems while r > 20 nm with our weight settings. The upper bound of r and the cluster neighborhood size (ε) can be adjusted according to the minimum expected diameter of mitochondria (~250 nm). In order to create at least one cluster in such a small mitochondrion, ε was set to 100 nm. Larger values may make the cluster be affected by nearby cue points which are located outside of the mitochondrion. We specified the threshold concerning the minimum number of elements in each cluster with respect to the z-thickness of the snake. Small numbers indirectly improve recall by increasing the number of initializations. On the other hand, the clusters supported by more cue points are expected to be more reliable. We obtained acceptable results when this number was 1.0–2.0 times the z-thickness.
Validator function
Validator function parameters were roughly tuned according to the morphology of mitochondria. Determination of the parameters Tmajor, Tminor, Tarea_min, Tarea_max are conducted by the range of physical size of mitochondria and arranged to establish a coarse restriction for candidate shapes. However, they can be refined when it is necessary to segment mitochondria of a particular size. Similarly, the curvature parameters (Tcurv_max and Tcurv_ave) are upper-bounds for radial movement of the peripheral membrane in terms of the radius of a circle (used to estimate the degree of curvature). These parameters serve to regulate the amount of exclusion of false positive detections and can be decreased to improve precision if required. The energy constraints (Tenergy_b and Tenergy_c) were adjusted to a value as small as possible to cover mitochondria which exhibit low-contrast peripheral membranes and vaguely appearing cristae. Since the refinement of such parameters may vary with respect to the input dataset, they must be altered by taking potential overfitting problems into consideration. The validation step is further discussed in Section 6.
Post-processing
In our experiments, feasible results were obtained when the merging threshold is set to a value between 15% and 35%. Separate mitochondria may be erroneously merged if the value is too low. On the contrary, snakes lying inside a single mitochondrion may not be merged for high threshold values.
5. Conclusion
In this study, we focused on detection and segmentation of both fully and partially seen mitochondria from 2D and 3D transmission electron microscopy images. Our segmentation method can be used to outline the peripheral membrane of mitochondria. The first step in the system is preprocessing. In this step, the image is resampled, smoothed and its contrast is adjusted. Then, membrane strength is measured by a Hessian-based ridge detection method. The local energy sums within two different neighborhoods are computed to create energy maps. The next step is the curve extraction process which utilizes the energy maps to locate peripheral membranes and cristae of mitochondria. Then, a snake algorithm driven by the extracted curves is employed. Finally, a validator function is used to refine the results.
Despite the various computerized segmentation attempts for mitochondria, this problem does not seem satisfactorily solved yet since the appearance of mitochondria is multifarious depending on cell type, condition and preparation of specimens. Considering that the subsequent problem is to deal with demands for high throughput automatic cristae segmentation, we preferred to use mostly datasets with clear settlement of cristae (as well as the mitochondrial boundary membranes). We want to remind the readers that for the other types of images (which exhibit dark blob-like mitochondrial regions), the problem of automatic detection and segmentation of mitochondria is a much simpler problem (using intensity-based features) which has been mostly successfully solved by the previous studies.
6. Discussion
A manual effort is still needed to obtain a flawless segmentation. Thus, designing some user-interaction tools to augment the automatic tools developed here will replace the hard work required by the user for the full hand-segmentation. These tools include validation, rejection, merging, splitting and dragging operations (Cocelli, 2015). False-negative error can be reduced by validating snakes that have been erroneously labeled as invalid under the control of the user. Similarly, a faulty portion of redundantly auto-merged shapes can be rejected by the user to deal with a false-positive error. Since such tools work on pre-determined snakes, the interaction with the user can be realized in real-time. Merging and splitting operations can be also used to fix auto-merging errors. In case of insufficient snake detection, these tools also allow the user to run the snake algorithm at a proper location. A partially consistent segmentation may also be corrected by dragging the defective boundary to the correct location.
The current implementation has not been fully optimized yet. Hence, it is not capable of responding to users’ needs in real time. Fortunately, the algorithms developed are highly parallelizable and adaptable to be run on a GPU in order to gain a tremendous speedup. GPU parallelization of curve fitting and snake algorithms are in our future plans. The current software is planned to be made available by the end of the second quarter of 2016. Additionally, we will build an easy to use application as soon as we complete optimization and parallelization of algorithms.
Since the Hessian ridge detector is not sufficiently capable of detecting peripheral and crista membranes of condensed mitochondria (see Fig. 13(h)), one can replace the Hessian matrix (Eq. (1)) with an appropriate tensor matrix based on the first derivative to extract edges instead of ridges. Besides, it is advantageous to take this approach since the first derivative is less sensitive to noise.
The snake contours produced by the proposed snake algorithm tend to pass through mid-points of double membrane structure of mitochondria since the external energy formulation is based on LFREM. This energy map indicates the total ridge energy within a large neighborhood (30 nm) which usually covers both membranes. Hence, the energy is maximized between membranes. Since the snake algorithm in our system utilizes parabolic arcs based on the energy maps, a perfect segmentation is not expected. However, the accuracy can be significantly increased by refining techniques described in a study by Jorstad and Fua (Jorstad and Fua, 2015) or a modified live-wire algorithm proposed in our previous study (Mumcuoglu et al., 2012).
We tested our system under different signal-to-noise ratio (SNR) conditions by using a mitochondrion phantom (see Fig. S1) taken from Fig. 7 in (Fernández et al., 2002). Note that the phantom was not sufficiently elaborated in order to test the whole system. Nevertheless, we ran our algorithm to see how the curves and snakes fit into the synthetic boundary. Fig. S2 depicts a sample tomogram of the phantom with additive Gaussian noise having different SNR values (∞, 20, 10, 4, 2, 1.33, 1.0, 0.80, 0.66 and 0.5). Fig. S3 shows curve fitting results for the same tomogram. For SNR ≥ 4, there was no curve extracted from HFREM (red curves/small-scale curves). The reason is that the phantom does not have realistic curvy cristae structures. For SNR ≤ 2, some false-positive curves were detected. The curves extracted from LFREM (blue curves/large-scale curves) covered the whole boundary in each tomogram for SNR ≥ 0.80. However, small-scale curves were too few to validate the segmented boundary. In order to see the final results, we lowered the minimum average region energy threshold (Tenergy_c) to zero (i.e. disabled its effect) while leaving the remaining parameters values unchanged. For SNR = 0.66, the boundary was partially detected. The large-scale curves were usually lost due to very weak ridge response for SNR = 0.5. In Fig. S4, the initial points of snakes were illustrated. No initial points were located outside of the mitochondrion. The algorithm failed for SNR = 0.5, since there were not enough large-scale curves detected in the volume. Fig. S5 exhibits the segmentation results. For SNR ≥ 0.80, we obtained good results. However, we noticed very small (a few pixels) fluctuations on the boundary along the z-direction for SNR ≤ 1. For SNR = 0.66, all of the candidate shapes were refused by the validator function due to weak boundary energy. However, as shown in Fig. S6, the obtained snake boundary was actually useful. To handle such cases, the user can be oriented to use the method with a parameter set which is arranged for low SNR conditions.
Although our method is specialized for mitochondria detection, it involves a Hessian-based membrane detection step which can be compared to generic methods used for membrane detection (aforementioned in Section 1). According to our elementary experiments, our simple ridge detector shows considerable success when compared to generic methods. Furthermore, our method can be integrated with membrane detection methods in order to increase accuracy.
We additionally tested our algorithm on a cryotomography (cryoET) dataset (ID: 6471) in EMDataBank (Wang et al., 2015). This dataset is quite large and has very low SNR. We cropped small sections (consisting of 20 tomograms with a size of 500 × 500). A tomogram (slice #: 310) from the dataset is provided in Fig. S7. Fig. S8(a) shows the snake initial points, Fig. S8(b) shows the curve fitting results obtained from LFREM (blue) and HFREM (red) and Fig. S8(c) shows the final segmentation result. Our method managed to locate and segment the mitochondrion with the same parameter sets given in Table 2. However, such datasets (not being plentifully populated by mitochondria) may require an additional preprocessing step that highlights the double membrane structure in order to attain a good detection performance. Parameter sets can be additionally revised for cryoET datasets. In the current form of the method, the most of the segmented regions are expected to be invalidated by the validator function due to weak ridge response. One example is given in Fig. S9. In this example, a candidate shape was extracted successfully but rejected by the validator function since the no strong small-scale curve was detected inside the candidate shape (low Ecrista).
The validation scheme in our system corresponds to a dichotomy in pattern classification problems. Hence, each valid snake can be considered as a point in a hyper-rectangular region. For future work, we are planning to build an improved validation function in which supervised learning techniques are utilized based on the validation descriptors in order to achieve better filtering results. In addition, integration of our system into a semiautomatic segmentation software package through the implementation of the aforementioned tools and the development of automatic cristae segmentation methods are also among the items considered for our future work.
Supplementary Material
Footnotes
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.jsb.2016.03.002.
References
- Ahlberg J. Active Contours in Three Dimensions. Linköping University; 1996. [Google Scholar]
- Ali RA, Landsberg MJ, Knauth E, Morgan GP, Marsh BJ, Hankamer B. A 3D image filter for parameter-free segmentation of macromolecular structures from electron tomograms. PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0033697. http://dx.doi.org/10.1371/journal.pone.0033697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartesaghi A, Sapiro G, Subramaniam S. An energy-based three-dimensional segmentation approach for the quantitative interpretation of electron tomograms. IEEE Trans. Image Process. 2005;14:1314–1323. doi: 10.1109/TIP.2005.852467. http://dx.doi.org/10.1109/TIP.2005.852467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazán C, Miller M, Blomgren P. Structure enhancement diffusion and contour extraction for electron tomography of mitochondria. J. Struct. Biol. 2009;166:144–155. doi: 10.1016/j.jsb.2009.02.009. http://dx.doi.org/10.1016/j.jsb.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burté F, Carelli V, Chinnery PF, Yu-Wai-Man P. Disturbed mitochondrial dynamics and neurodegenerative disorders. Nat. Rev. Neurol. 2014;11:11–24. doi: 10.1038/nrneurol.2014.228. http://dx.doi.org/10.1038/nrneurol.2014.228. [DOI] [PubMed] [Google Scholar]
- Chavan V, Willis J, Walker SK, Clark HR, Liu X, Fox MA, Srivastava S, Mukherjee K. Central presynaptic terminals are enriched in ATP but the majority lack mitochondria. PLoS ONE. 2015;10:e0125185. doi: 10.1371/journal.pone.0125185. http://dx.doi.org/10.1371/journal.pone.0125185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cignoni P, Callieri M, Corsini M, Dellepiane M, Ganovelli F, Ranzuglia G. MeshLab: an open-source mesh processing tool; Sixth Eurographics Italian Chapter Conference; 2008. pp. 129–136. [Google Scholar]
- Cocelli M. Semi-Automatic/User-Guided Segmentation of Mitochondria on Transmission Electron Microscopy Images. Middle East Technical University; 2015. [Google Scholar]
- Cohen LD. On active contour models and balloons. CVGIP Image Underst. 1991;53:211–218. http://dx.doi.org/10.1016/1049-9660(91)90028-N. [Google Scholar]
- Davies KM, Daum B, Gold VAM, Mühleip AW, Brandt T, Blum TB, Mills DJ, Kühlbrandt W. Visualization of ATP synthase dimers in mitochondria by electron cryo-tomography. J. Vis. Exp. 2014:51228. doi: 10.3791/51228. http://dx.doi.org/10.3791/51228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietlmeier J, Ghita O, Duessmann H, Prehn JHM, Whelan PF. Unsupervised mitochondria segmentation using recursive spectral clustering and adaptive similarity models. J. Struct. Biol. 2013;184:401–408. doi: 10.1016/j.jsb.2013.10.013. http://dx.doi.org/10.1016/j.jsb.2013.10.013. [DOI] [PubMed] [Google Scholar]
- Ester M, Kriegel HP, Sander J, Xu X. A density-based algorithm for discovering clusters in large spatial databases with noise; Second International Conference on Knowledge Discovery and Data Mining; 1996. pp. 226–231. [Google Scholar]
- Fernández J-J, Lawrence AF, Roca J, García I, Ellisman MH, Carazo J-M. High-performance electron tomography of complex biological specimens. J. Struct. Biol. 2002;138:6–20. doi: 10.1016/s1047-8477(02)00017-5. http://dx.doi.org/10.1016/S1047-8477(02)00017-5. [DOI] [PubMed] [Google Scholar]
- Franco-Iborra S, Vila M, Perier C. The parkinson disease mitochondrial hypothesis: where are we at? Neuroscience. 2015 doi: 10.1177/1073858415574600. 1073858415574600. http://dx.doi.org/10.1177/1073858415574600. [DOI] [PubMed] [Google Scholar]
- Ghita O, Dietlmeier J, Whelan P. Automatic segmentation of mitochondria in EM data using pairwise affinity factorization and graph-based contour searching. IEEE Trans. Image Process. 2014;23:4576–4586. doi: 10.1109/TIP.2014.2347240. http://dx.doi.org/10.1109/TIP.2014.2347240. [DOI] [PubMed] [Google Scholar]
- Giuly RJ, Martone ME, Ellisman MH. Method: automatic segmentation of mitochondria utilizing patch classification, contour pair classification, and automatically seeded level sets. BMC Bioinformatics. 2012;13:29. doi: 10.1186/1471-2105-13-29. http://dx.doi.org/10.1186/1471-2105-13-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harapin J, Eibauer M, Medalia O. Structural analysis of supramolecular assemblies by cryo-electron tomography. Structure. 2013;21:1522–1530. doi: 10.1016/j.str.2013.08.003. http://dx.doi.org/10.1016/j.str.2013.08.003. [DOI] [PubMed] [Google Scholar]
- Jiang W, Baker ML, Wu Q, Bajaj C, Chiu W. Applications of a bilateral denoising filter in biological electron microscopy. J. Struct. Biol. 2003;144:114–122. doi: 10.1016/j.jsb.2003.09.028. http://dx.doi.org/10.1016/j.jsb.2003.09.028. [DOI] [PubMed] [Google Scholar]
- Jorstad A, Fua P. Refining mitochondria segmentation in electron microscopy imagery with active surfaces. Lect. Notes Comput. Sci. 2015;8928:367–379. http://dx.doi.org/10.1007/978-3-319-16220-1_26 (including Subser. Lect. Notes Artif. Intell. Lect. Notes Bioinformatics) [Google Scholar]
- Kass M, Witkin A, Terzopoulos D. Snakes: active contour models. Int. J. Comput. Vis. 1988;1:321–331. http://dx.doi.org/10.1007/BF00133570. [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. http://dx.doi.org/10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Lucchi A, Smith K, Achanta R, Knott G, Fua P. Supervoxel-based segmentation of mitochondria in em image stacks with learned shape features. IEEE Trans. Med. Imaging. 2012;31:474–486. doi: 10.1109/TMI.2011.2171705. http://dx.doi.org/10.1109/TMI.2011.2171705. [DOI] [PubMed] [Google Scholar]
- Martinez-Sanchez A, Garcia I, Fernandez JJ. A differential structure approach to membrane segmentation in electron tomography. J. Struct. Biol. 2011;175:372–383. doi: 10.1016/j.jsb.2011.05.010. http://dx.doi.org/10.1016/j.jsb.2011.05.010. [DOI] [PubMed] [Google Scholar]
- Martinez-Sanchez A, Garcia I, Fernandez JJ. A ridge-based framework for segmentation of 3D electron microscopy datasets. J. Struct. Biol. 2013;181:61–70. doi: 10.1016/j.jsb.2012.10.002. http://dx.doi.org/10.1016/j.jsb.2012.10.002. [DOI] [PubMed] [Google Scholar]
- Martinez-Sanchez A, Garcia I, Asano S, Lucic V, Fernandez JJ. Robust membrane detection based on tensor voting for electron tomography. J. Struct. Biol. 2014;186:49–61. doi: 10.1016/j.jsb.2014.02.015. http://dx.doi.org/10.1016/j.jsb.2014.02.015. [DOI] [PubMed] [Google Scholar]
- Martone ME, Gupta A, Wong M, Qian X, Sosinsky G, Ludäscher B, Ellisman MH. A cell-centered database for electron tomographic data. J. Struct. Biol. 2002;138:145–155. doi: 10.1016/s1047-8477(02)00006-0. http://dx.doi.org/10.1016/S1047-8477(02)00006-0. [DOI] [PubMed] [Google Scholar]
- Martone ME, Zhang S, Gupta A, Qian X, He H, Price DL, Wong M, Santini S, Ellisman MH. The cell-centered database: a database for multiscale structural and protein localization data from light and electron microscopy. Neuroinformatics. 2003;1:379–395. doi: 10.1385/NI:1:4:379. http://dx.doi.org/10.1385/NI-1-4-379. [DOI] [PubMed] [Google Scholar]
- Martone ME, Tran J, Wong WW, Sargis J, Fong L, Larson S, Lamont SP, Gupta A, Ellisman MH. The Cell Centered Database project: an update on building community resources for managing and sharing 3D imaging data. J. Struct. Biol. 2008;161:220–231. doi: 10.1016/j.jsb.2007.10.003. http://dx.doi.org/10.1016/j.jsb.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumcuoglu EU, Hassanpour R, Tasel SF, Perkins G, Martone ME, Gurcan MN. Computerized detection and segmentation of mitochondria on electron microscope images. J. Microsc. 2012;246:248–265. doi: 10.1111/j.1365-2818.2012.03614.x. http://dx.doi.org/10.1111/j.1365-2818.2012.03614.x. [DOI] [PubMed] [Google Scholar]
- Page C, Hanein D, Volkmann N. Accurate membrane tracing in three-dimensional reconstructions from electron cryotomography data. Ultramicroscopy. 2015;155:20–26. doi: 10.1016/j.ultramic.2015.03.021. http://dx.doi.org/10.1016/j.ultramic.2015.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantelic RS, Ericksson G, Hamilton N, Hankamer B. Bilateral edge filter: photometrically weighted, discontinuity based edge detection. J. Struct. Biol. 2007;160:93–102. doi: 10.1016/j.jsb.2007.07.005. http://dx.doi.org/10.1016/j.jsb.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Perkins GA, Ellisman MH, Fox DA. Three-dimensional analysis of mouse rod and cone mitochondrial cristae architecture: bioenergetic and functional implications. Mol. Vis. 2003;9:60–73. v9/a10 [pii] [PubMed] [Google Scholar]
- Powers D. Evaluation: from precision, recall and F-measure to ROC, informedness, markedness and correlation. J. Mach. Learn. Technol. 2011;2:37–63. [Google Scholar]
- Róg T, Hector MS, Munck N, Orešič M, Karttunen M, Vattulainen I. Role of cardiolipins in the inner mitochondrial membrane: insight gained through atom-scale simulations. J. Phys. Chem. B. 2009;113:3413–3422. doi: 10.1021/jp8077369. http://dx.doi.org/10.1021/jp8077369. [DOI] [PubMed] [Google Scholar]
- Sandberg K, Brega M. Segmentation of thin structures in electron micrographs using orientation fields. J. Struct. Biol. 2007;157:403–415. doi: 10.1016/j.jsb.2006.09.007. http://dx.doi.org/10.1016/j.jsb.2006.09.007. [DOI] [PubMed] [Google Scholar]
- Scheffler IE. Mitochondria. second. Hoboken, NJ, USA: John Wiley & Sons Inc; 2007. http://dx.doi.org/10.1002/9780470191774. [Google Scholar]
- Seyedhosseini M, Ellisman MH, Tasdizen T, et al. Segmentation of mitochondria in electron microscopy images using algebraic curves; 2013 IEEE 10th International Symposium on Biomedical Imaging ({ISBI}); 2013a. pp. 860–863. http://dx.doi.org/10.1109/ISBI.2013.6556611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyedhosseini M, Sajjadi M, Tasdizen T. Image segmentation with cascaded hierarchical models and logistic disjunctive normal networks; Proceedings of the IEEE International Conference on Computer Vision; 2013b. pp. 2168–2175. http://dx.doi.org/10.1109/ICCV.2013.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart J. Calculus, Concepts and Contexts. Cole, Pacific Grove, CA: 1998. [Google Scholar]
- Tasel SF, Hassanpour R, Mumcuoglu EU, Perkins GC, Martone M. Automatic detection of mitochondria from electron microscope tomography images: a curve fitting approach. SPIE Medical Imaging 2014. 2014:903449. http://dx.doi.org/10.1117/12.2043517. [Google Scholar]
- Tomasi C, Manduchi R. Bilateral filtering for gray and color images. Int. Conf. Comput. Vis. 1998:839–846. http://dx.doi.org/10.1109/ICCV.1998.710815. [Google Scholar]
- Valdés I, Nomura Y, Mitsuya Y. Equidistant snakes: accurate irregular shaped contour detection. Mach. Vis. Appl. 1996:285–288. [Google Scholar]
- Wang X, Wang W, Li L, Perry G, Lee H, Zhu X. Oxidative stress and mitochondrial dysfunction in Alzheimer’s disease. Biochim. Biophys. Acta – Mol. Basis Dis. 2014;1842:1240–1247. doi: 10.1016/j.bbadis.2013.10.015. http://dx.doi.org/10.1016/j.bbadis.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Stone RL, Kaelber JT, Rochat RH, Nick AM, Vijayan KV, Afshar-Kharghan V, Schmid MF, Dong J-F, Sood AK, Chiu W. Electron cryotomography reveals ultrastructure alterations in platelets from patients with ovarian cancer. Proc. Natl. Acad. Sci. U.S.A. 2015;112 doi: 10.1073/pnas.1518628112. 201518628. http://dx.doi.org/10.1073/pnas.1518628112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner AA, Kirschmann MA, Genoud C. Challenges of microtome-based serial block-face scanning electron microscopy in neuroscience. J. Microsc. 2015;259:137–142. doi: 10.1111/jmi.12244. http://dx.doi.org/10.1111/jmi.12244. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.