 CBT-Cys click condensation reaction has a high second-order reaction rate constant and has found wide applicability in recent years.
CBT-Cys click condensation reaction has a high second-order reaction rate constant and has found wide applicability in recent years.
Abstract
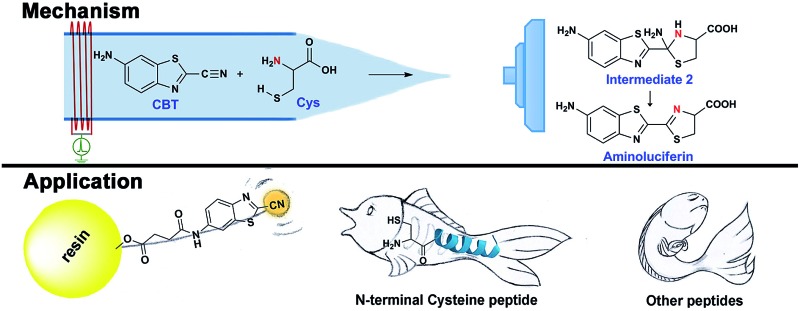
CBT-Cys click condensation reaction has a high second-order reaction rate constant and has found wide applicability in recent years. However, its reaction mechanism has not been experimentally validated and its application for identifying bioactive N-terminal Cys peptides in real clinical samples has not been reported. Herein, firstly, by employing induced nanoelectrospray ionization-mass spectrometry (InESI-MS) and a home-built micro-reactor, we successfully intercepted and structurally characterized the crucial intermediate in this click reaction for the first time. With the intermediate, the proposed mechanism of this reaction was corroborated. Moreover, we also applied this MS setup to monitor the reaction in real time and obtained the second-order reaction rate constants of this reaction at different pH values. After mechanistic study, we applied this click reaction for identifying bioactive N-terminal cysteine peptides in amniotic fluid (AF). Eight unique N-terminal Cys peptides in AF, three of which are located in the functional domain regions of their corresponding proteins, were identified with a false positive rate less than 1%. One of the three peptides was found able to inhibit the growth of uterine endometrial cancer HEC-1-B cells but not the endometrial normal cells via a typical apoptotic pathway. With its mechanism satisfactorily elucidated, the kinetic parameters obtained, as well as its application for fishing bioactive N-terminal Cys peptides from vast complex clinical samples, we anticipate that this CBT-Cys click reaction could be applied more widely for the facile isolation, site-specific identification, and quantification of N-terminal Cys-containing peptides in complex biological samples.
Introduction
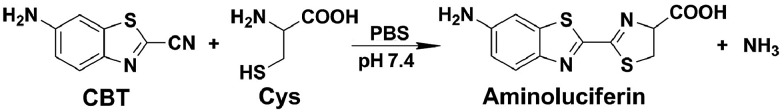
The click reaction is rapidly becoming an essential tool in medicinal chemistry, combinatorial chemistry, material science, and chemical biology.1–5 Due to the inherent advantages of a click reaction, including high specificity, quantitative yield, and fidelity under physiological conditions, more efforts have been made on the development and evaluation of more reaction classes possessing these characteristics.6–8 Recently, based on the proposed regeneration pathway of d-luciferin in firefly, Rao and co-workers developed a novel click reaction with high biocompatibility and controllability.9 Briefly, as shown in Scheme 1, this click reaction involves the efficient condensation between l-cysteine (Cys) (or d-cysteine) and the cyano group of 2-cyano-6-aminobenzothiazole (CBT) (or 2-cyano-6-hydroxybenzothiazole) to yield Aminoluciferin (or luciferin) in d- or l-form. The second-order rate constant of this click condensation reaction was reported to be 26.8 M–1 s–1,10 which is 300 times larger than that of the well defined copper-free azide–alkyne cycloaddition (AAC) click reaction (7.6 × 10–2 M–1 s–1).11 To date, this click condensation reaction has been successfully employed to design smart imaging probes (optical, magnetic resonance, or nuclear),12–16 synthesize cyclic superstructures,17 overcome multidrug resistance,18 and prepare oligomeric hydrogels.19 Although this CBT-Cys click reaction has been widely used for several years and its reaction equation looks simple, its underlying mechanism has remained less explored. Recently, Liang et al. proposed a mechanism for this reaction,20 however, up to now, there has been no experimental result to validate the mechanism (e.g., direct isolation or characterization of the intermediates in this reaction).
Scheme 1. Reaction between 2-cyano-6-aminobenzothiazole (CBT) and cysteine (Cys) to yield the product Aminoluciferin.
Protein cysteine, serving as an important mediator of redox signaling and regulation, is one of the most reactive amino acid residues in proteins.21 Remarkably, many of the biologically active peptides contain one or multiple Cys for each, which are believed to contribute to the stability and activity of the peptides.22–24 However, our overall knowledge of Cys-containing peptides with bioactivity potential is still quite limited owing to the challenges associated with the isolation, site-specific identification, and quantification of these peptides. Conventional methods for the enrichment of Cys-containing peptides are known to use affinity or covalent capture with suitable resins or magnetic beads. For affinity-based methods, affinity capture tags have to be introduced to the Cys-containing peptides through the derivatization of the Cys residues. With the labeled affinity tags, these peptides can be selectively captured by resins (or beads) and then eluted for identification.25 Thiol–disulfide exchange and thiol–thioester exchange are commonly employed in covalent capture approaches. For example, via thiol–disulfide exchange with thiopropyl sepharose on the solid support, Cys-containing peptides can be selectively separated by covalent chromatography.26,27 Although these methods are able to enrich Cys-containing peptides in a peptide sample pool, the sites of Cys on the enriched peptides (N-terminal, middle, or C-terminal) are not differentiated.
Owing to its capabilities for structural characterization, as well as high specificity, sensitivity, and speed, mass spectrometry (MS) is advantageous over other established spectroscopic techniques which require a conventional periodic sampling–quenching–concentrating–analyzing process for reaction monitoring. Moreover, through interception of reactive intermediates, MS can additionally provide a wealth of mechanistic information.28–31 Among the MS techniques, electron spray ionization mass spectrometry (ESI-MS) and tandem MS are advantageous in solution-phase and are rapidly becoming the techniques of choice for mechanistic studies in chemistry.32,33 For example, following the pioneering study by Chen et al. on the mechanistic investigation of organometallic chemistry with ESI-MS,34 De Angelis et al. recently uncovered the mechanism underlying the Cu+-catalyzed AAC (CuAAC) click reaction using ESI-MS.35 Moreover, MS (either ESI-MS or matrix-assisted laser desorption ionization (MALDI) MS) has also been widely used for peptide (or protein) identification.36,37 Terminal labeling of peptide (or protein) can either simplify the mass spectra or help to distinguish the fragmentation ion series of the peptide (or protein), thus facilitating the direct readout of the amino acid sequence of the peptide (or protein) from the spectra. Therefore, it is of broad interest to develop new approaches to specifically label peptide termini for mass spectrometric analysis.
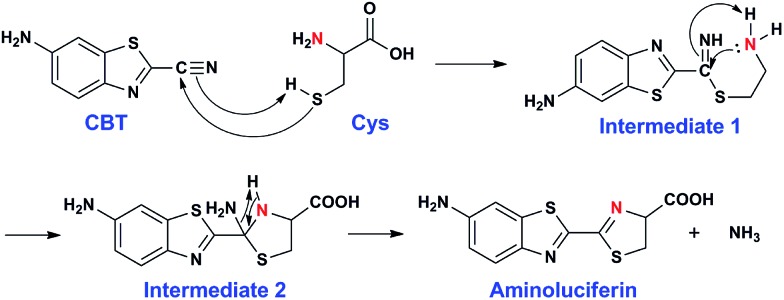
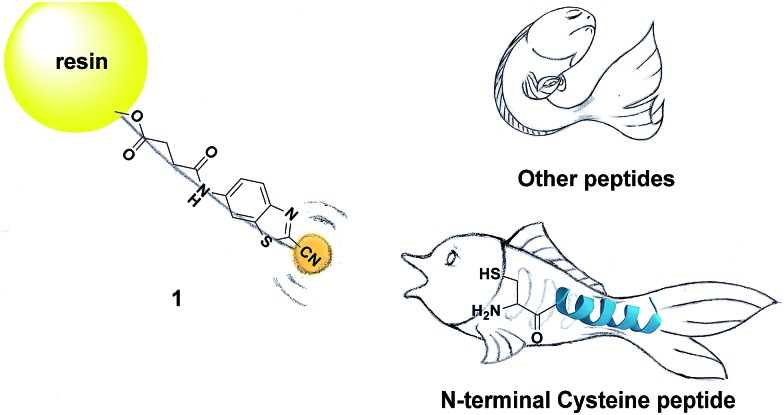
Inspired by the pioneering studies above, in this work, we firstly aim to investigate the mechanism of CBT-Cys click reaction by using induced nano-electrospray ionization mass spectrometry (InESI-MS), a new emerging MS technique which has shown good performance in matrix tolerance and thus facilitated the direct identification of intermediates in raw reaction mixtures.38–40 Using a 15N-labeled Cys as one reactant and InESI-MS, we firstly validated that the N-atom in the second thiazole ring of Aminoluciferin originated from Cys but not from cyano group of CBT (Scheme 1). Then, we assembled a micro-reactor for InESI-MS analysis and successfully intercepted the crucial Intermediate 2, with which the mechanism of this CBT-Cys click reaction was satisfactorily interpreted, as shown in Scheme 3. Finally, a series of CBT-Cys click reactions at different pH values were monitored with our InESI-MS in real time, which gave according kinetic parameters of this reaction. With the mechanism of this CBT-Cys click reaction elucidated, we further employed it for identifying bioactive N-terminal cysteine peptides in amniotic fluid with MALDI-MS. As illustrated in Scheme 2, we rationally designed a CBT derivative, CBT succinic amide (1), which has a free carbonic acid group to covalently conjugate to 2-chlorotrityl chloride resin and a light molecular weight (MW) of 275.28 daltons. In the presence of a reducing agent (e.g., dithiothreitol, DTT) and at pH 7.4, the N-terminal Cys peptide in the vast complex peptide sample (e.g., Cys(SEt)-Glu-Tyr(H2PO3)-Phe-Phe-Gly-OH, 2 in this work) click reacts with 1 to yield the condensation product which can be cleaved from the resin for mass spectrometric analysis. Employing this strategy and MALDI-MS, we successfully identified eight unique N-terminal Cys peptides from human amniotic fluid (AF). Proteomic analyses indicated that three of the eight peptides have C-coil structures and coincidentally are located in the functional domain regions of their corresponding proteins. The cell cytotoxicity study indicated that one of the three peptides showed an obvious inhibitory effect on endometrial cancer HEC-1-B cells. To the best of our knowledge, this is the first time that the N-terminal Cys peptides in real clinic samples are selectively enriched for proteomic analysis.
Scheme 3. Proposed reaction mechanism between CBT and Cys.
Scheme 2. Schematic illustration of using solid phase CBT to fish N-terminal cysteine (Cys) peptides in the vast complex peptide sample.
Results and discussion
Assignment of the origin of the nitrogen atom
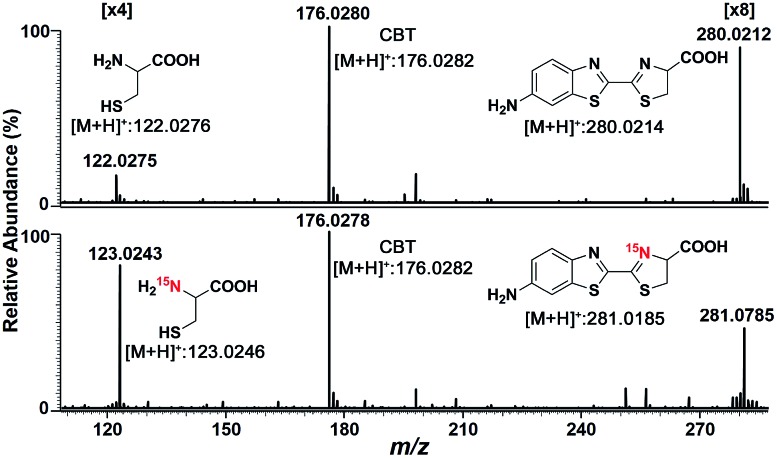
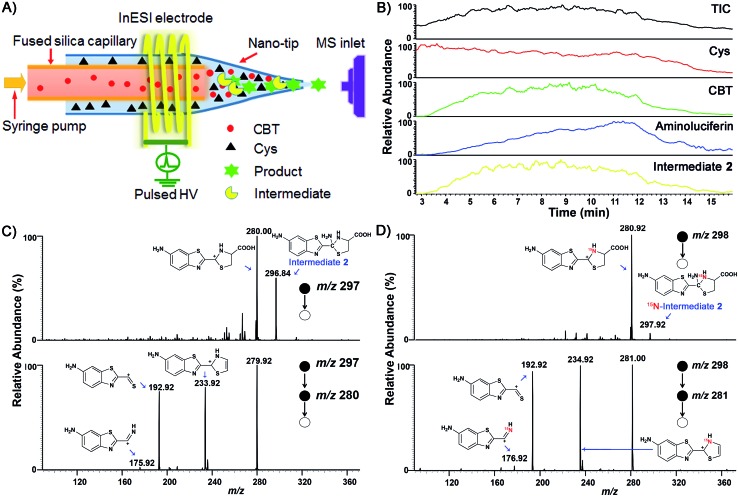
We began the study by comparing the InESI-MS and conventional nESI-MS to choose one more suitable means for studying the mechanism of our CBT-Cys click reaction. Fig. S7 and S8† indicated that the molecular ion peak of the product Aminoluciferin (m/z 280) was totally suppressed in conventional nESI-MS due to the interference from the high background peak of the plasticizer (m/z 278) while that in InESI-MS was clearly observed under the same condition. Direct InESI-MS and corresponding tandem MS, together with high resolution InESI-MS, clearly showed the starting materials, the product Aminoluciferin, as well as their characteristic fragment ions (Fig. S9 and S10†). All these results above indicated that InESI-MS had better matrix tolerance and was more suitable than conventional nESI-MS for studying our CBT-Cys click reaction. Therefore, InESI was chosen to investigate the mechanism of CBT-Cys click reaction. As shown in Scheme 1, the CBT-Cys click condensation reaction involves the release of one unit of ammonia gas (NH3) and the formation of a new thiazole ring (i.e., the second thiazole ring) in the product Aminoluciferin. Therefore, whether the N atom in the second thiazole ring of Aminoluciferin originated from the cyano group of CBT or the amino group of Cys is critical for resolving the mechanism of this reaction. To assign the origin of this N atom, we employed a stable isotope-labeling method to trace the N atom. In detail, the reaction mixture of CBT with natural Cys or 15N-Cys was separately subjected to InESI-MS and their corresponding MS spectra were recorded for comparative analysis. Fig. 1 showed that Aminoluciferin from the reaction mixture of natural Cys and CBT was observed at m/z 280, while m/z of Aminoluciferin from 15N-Cys and CBT shifted to 281. This suggests that the N-atom in the second thiazole ring of Aminoluciferin originated from Cys but not CBT.
Fig. 1. InESI(+)-MS spectra of the CBT-Cys reaction using natural Cys (top) or 15N-labeled Cys (bottom). Reaction conditions: 50 μM CBT with 50 μM Cys under pH 7.4, CH3OH : H2O = 1 : 1.
Proposal and validation of the reaction mechanism
After assignment of the N atom in the second thiazole ring of the product Aminoluciferin, the mechanism underlying this CBT-Cys click condensation reaction can be roughly drawn, as depicted in Scheme 3. In detail, since the thiol group has a much higher nucleophilicity than the amino group, we propose that the S atom on Cys first attacks the C atom on the cyano group of CBT, causing the N atom on the cyano group to become negatively charged which thereafter attracts a H-atom on the –SH group to yield Intermediate 1. Then the amino group on the cysteine attacks the same C atom, while in the meantime the N atom on the enamine of Intermediate 1 draws a H-atom from the amino group, to form a new thiazolidin structure of Intermediate 2. Then the N atom originating from the cyano group of CBT acquires its third hydrogen from thiazolidin to yield NH3 which is released.
To validate the proposed mechanism above, we need to intercept and characterize the transient intermediates in this click reaction. Obviously, Intermediate 2 is structurally more stable than Intermediate 1 but both intermediates have an m/z 297 (for natural Cys reaction). Structural analysis with the obtained MS data indicated that the intermediate we intercepted in this work was Intermediate 2. Taking their very short lifetimes into consideration, interception of the intermediates challenges most of the conventional MS methods. In this study, as shown in Fig. 2A, we introduced a thin fused silica capillary (loaded with the solution of 100 μM CBT) into the nanospray emitter (pre-loaded with the solution of 100 μM Cys) to initiate the reaction in the spray emitter for immediate InESI-MS analysis. The mixing rate of the two reactants was controlled by a syringe pump connected to that fused silica capillary. Once the reactants were mixed, the reaction started and the mixture was immediately subjected to InESI-MS analysis. By this method, the reaction was monitored in real time and transient species were trapped and analyzed. Using this home-built micro-reactor, for the first time, we successfully intercepted Intermediate 2 and characterized its structure by tandem MS. Fig. 2B showed the selected ion chronograms of reactants Cys (red, m/z 122†) and CBT (green, m/z 176), product Aminoluciferin (blue, m/z 280), and the ion m/z 297 (yellow, the Intermediate 2) which recorded their dynamic changes in ion abundance. Note here that the ion chronogram of m/z 297 was generated simultaneously with the injection of CBT by the springe pump (100 nL min–1 for 1 min), suggesting that the intermediate was successfully captured in its rapid occurrence. Further tandem MS analyses structurally resolved that the captured m/z 297 was the transient Intermediate 2. As shown by the MS/MS spectrum in the top panel of Fig. 2C, the ion at m/z 297 produces a fragment at m/z 280, which is equal to that of the parent ion of Aminoluciferin, as the first prominent product ion. Further MS3 fragmentation on the m/z 280 ion yielded the ion fragments at m/z 234, 193, and 176 (bottom panel in Fig. 2C), which also appeared in the MS2 spectrum of Aminoluciferin (Fig. S9†). These data above suggest that the ion m/z 297 is Intermediate 2 whose structure is depicted in Scheme 3. Similarly, we also used 15N-Cys and CBT for real time InESI-MS analyses and further confirmed the captured intermediate was Intermediate 2. As shown in the top panel of Fig. 2D, the ion at m/z 298 produces a fragment at m/z 281 as the first prominent product ion, which is equal to that of the parent ion of 15N-Aminoluciferin. Consistently, further MS3 fragmentation on the m/z 281 ion yielded the fragments at m/z 235, 193, and 177 which were corresponding fragment ions of 15N-Aminoluciferin (bottom panel in Fig. 2D). These showed that the ion m/z 298 is 15N-labeled Intermediate 2. Interestingly, the ion fragment m/z 193 appeared in both MS3 spectra of Intermediate 2 and 15N-labeled Intermediate 2 because the fragment ion does not contain the N-atom from either natural Cys or 15N-Cys. This additionally verified our structural analysis.
Fig. 2. (A) Instrument illustration to show online CBT-Cys click reaction in the micro-reactor. (B) Ion chronograms of the CBT-Cys click reaction. InESI-(+)-MS/MS (top) and InESI-(+)-MS/MS/MS (bottom) spectra of the intermediates of the CBT-Cys click reaction using natural Cys (C) or 15N-Cys (D) as the reactant. Reaction conditions: pH 7.4, CH3OH : H2O = 1 : 1.
Kinetic study of the CBT-Cys click reaction
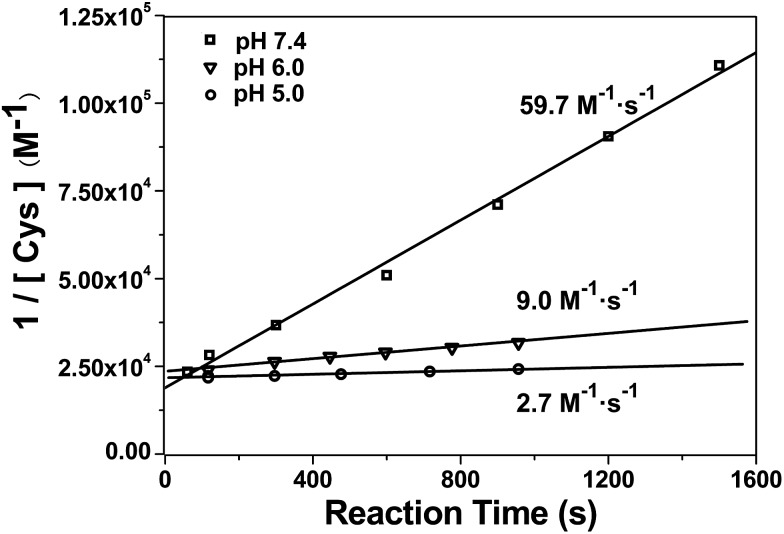
After mechanistic study, we also applied our system to investigate the kinetics of this click reaction. The nucleophilicity of the thiol group of Cys is pH-dependent. Therefore, the kinetics of this click condensation reaction at different pH values should be different. To prove this, an online method of uninterrupted analysis of the reaction mixture was designed to monitor the dynamic processes of this reaction in real-time. When the reactants were progressively transformed into the product Aminoluciferin, the composition of the reaction mixture was monitored by InESI-MS over time. Since this reaction originally occurs in the firefly body, we firstly studied its kinetics at a physiological condition of pH 7.4. Selected ion chronograms of the reactant Cys (m/z 122) and the product Aminoluciferin (m/z 280) recorded the dynamic changes of their ion abundances, as shown in Fig. S11 (ESI†). As the initial concentration of Cys was known, we thus used the concentration change of Cys but not Aminoluciferin for the determination of the second-order reaction rate constant. Applying the similar kinetic analysis reported by Ren et al.,41 we plotted 1/[Cys] vs. reaction time and the linear regression analysis of the plot gave the second-order reaction rate constant of 59.7 M–1 s–1 at pH 7.4, as shown in Fig. 3. This result is consistent in order of magnitude with that recently reported, calculated by the conventional high performance liquid chromatography (HPLC) method (26.8 M–1 s–1).10 Selected ion chronograms of the reactant Cys (m/z 122) and the product Aminoluciferin (m/z 280) in this click reaction at pH 6 and pH 5 are shown in Fig. S12 and S13 (ESI†). Similarly, their reaction rate constants were calculated to be 9.0 M–1 s–1 for pH 6 and 2.7 M–1 s–1 for pH 5, respectively, as shown in Fig. 3. We did not conduct the kinetic study on this click reaction at pH higher than 8 because we found that the reactant CBT was hydrolysed at these conditions.42,43
Fig. 3. Linear regression analysis of 1/[Cys] vs. time of the CBT-Cys click reaction under different pH values. Reaction conditions: 50 μM CBT and 50 μM Cys, CH3OH : H2O = 1 : 1.
Using solid phase CBT derivative 1 to fish N-terminal Cys peptide 2
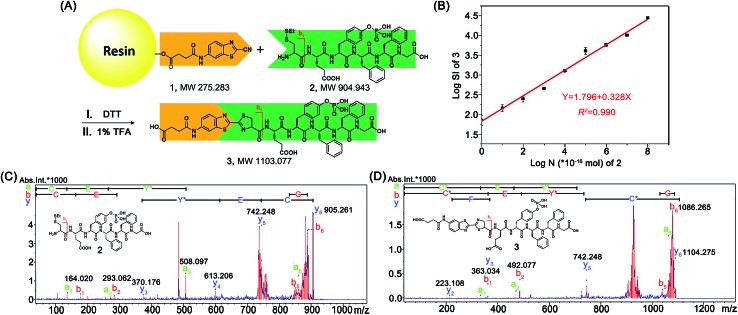
After mechanistic study of this CBT-Cys click reaction, we further employed it for identifying bioactive N-terminal cysteine peptides with MALDI-MS. We began the study with the syntheses of compound 1 and peptide 2. The syntheses are simple and straightforward. Briefly, in the presence of 4-methylmorpholine, CBT reacts with succinic anhydride to yield 1 after high performance liquid chromatography (HPLC) purification (Scheme S1,† Fig. 4A, S1–S3†). Peptide 2 was synthesized with solid phase peptide synthesis (SPPS), followed by the deprotection of the tBu and Boc groups and purified with HPLC (Scheme S2 and Fig. S4–S6†). After syntheses, we conjugated 500 nmol 1 to the resin. Then, different amounts of 2 (from 100 amol to 10 nmol), together with 20 equiv. of DTT, in N,N-dimethylformamide (DMF) were shaken with the resin overnight at room temperature. After the reaction was completed, the resin was thoroughly washed with DMF and the title product 3 was cleaved from the resin for matrix-assisted laser desorption ionization/time of flight mass spectrometry (MALDI/TOF MS) analyses (Fig. 4A). As shown in Fig. S14 (ESI†), the MS signal of product 3 could be detected with the loading amount of peptide 2 as low as 100 amol. Quantitative analysis indicated that there was a high linearity between the log value of the relative mass signal intensity (SI) of 3 and the substance amount (N) of peptide 2 over the range of 100 amol to 10 nmol (Y = 1.796 + 0.328X, R 2 = 0.990, Fig. 4B). The recovery test showed that, at the amount of 1–10 nmol of 2, our method had a recovery rate of 86.8–105.7%, indicating good reliability of the method (Fig. S15 and Table S1†). This suggests that, in combination with MS, our solid phase CBT derivative 1 could be applied to fish and quantitatively detect N-terminal Cys peptides within a wide amount range with very high sensitivity. In addition, both the quantity and sequence identity of the fished peptides were determined by MALDI TOF/TOF MS. MW of the N-terminal Cys peptide was measured by a TOF-MS scan as the parent ion and fragment ions of the parent ion were obtained by the TOF/TOF MS in a LIFT mode.44 In this work, fished N-terminal Cys peptides were quantified by measuring the relative signal intensities of the peptide's parent ion tagged with the CBT derivative (i.e., 1) in TOF MS mode, respectively. For example, the standard peptide 2 was fragmented in TOF/TOF MS, and the fragmentation of the b1 ion did start from the N-terminal disulfided Cys (Cys(SEt)), whose MW is 164.020 (Fig. 4C). Fig. 4D showed that, after reaction with 1 on the resin, the Cys b1 ion of 2 shifted to 363.034 which corresponds to the b1 ion of 3. Therefore in this work, we used m/z 363.034 as a characteristic fragment ion for qualitative analysis of the N-terminal Cys peptides which conjugated to the “fishhook” (i.e., 1) on the resin. Combinational analyses of the TOF MS and TOF/TOF MS results of 1-labeled peptides therefore determined their relative quantities as well as the sequence identities in a single automated operation. Due to the specificity of the CBT-Cys click reaction, every peptide fished by our CBT derivative 1 incontrovertibly starts with an amino acid of Cys from the N-terminal.
Fig. 4. (A) Schematic illustration of using solid phase CBT derivative 1 to fish N-terminal Cys peptide 2. (B) The fitted calibration line between the log value of the relative mass signal intensity (SI) of product 3 and the substance amount (N) of peptide 2 (×10–16 mol). (C) TOF/TOF MS spectrum of 2 showing the pairs of its fragment ions. (D) TOF/TOF MS spectrum of 3 showing the pairs of its fragment ions.
Fishing and identification of N-terminal peptides in amniotic fluid
After validation of the feasibility of our method, we applied our solid phase CBT to fish and identify N-terminal peptides in AF for clinical research. AF fills the amniotic cavity that is lined by amnion epithelial cells. The cells are of fetal origin and hence reflect the genotypic constitution of the fetus.45,46 During pregnancy of the mother, the composition of AF changes, and its protein profile reflects the physiological and pathological changes of the placenta that affect both the fetus and the mother. All the proteins and peptides in the AF, each of which has a MW less than 10 kD, were extracted by centrifugal ultrafiltration and then applied to the CBT resin. The N-terminal Cys peptides reacted with 1 and conjugated to the resin while the unreacted proteins (peptides) were eluted. After being cleaved from the resin and lyophilized, the labeled N-terminal peptides were subjected to MALDI-TOF/TOF MS analysis immediately (Fig. S16†). The acquired MS data of the peptides were analyzed by biotools software Sequence Editor for the determination of their denovo sequences. Then the sequences were searched and validated with software Mascot using the Swiss-Prot human peptide database (none trypsin-digested peptide). In this work, 27 N-terminal Cys peptides in total were “fished” out by our CBT derivative 1. However, only 17 of them, whose parent ion peaks in the mass spectra showed intensity over 5000 and S/N > 15 to ensure reliable fragment ion peaks, were selected for secondary mass analyses. Based on the parent ions and fragment ions in their MALDI TOF/TOF MS spectra, these 17 peptides were automated denovo sequenced. However, only eight of these 17 N-terminal Cys peptides that were statistically meaningful (p < 0.05) were successfully identified (Fig. S17–S24†). Each of these eight unique free N-terminal Cys-containing peptides has a false positive rate less than 1%, suggesting the effectiveness of our fishing strategy. As we mentioned above, the fragment ions of fished N-terminal peptides should start with the b1 ion of 363.034. In this work, eight N-terminal Cys peptides were fished and identified, whose peptide sequences, isoelectric points (pIs), and MWs are listed in Table 1.
Table 1. Eight unique free N-terminal Cys-containing peptides identified in AF in this work.
| Protein | Peptide | Length | pI | MW |
| K0100_HUMAN | CFLHLP | 6 | 6.73 | 728.37 |
| MUC19_HUMAN | CGSQCTCQ | 8 | 5.50 | 828.26 |
| FRITZ_HUMAN | CHQMSFCL | 8 | 6.72 | 967.37 |
| ZSWM5_HUMAN | CILLLEGGP | 9 | 4.00 | 913.49 |
| PCSK5_HUMAN | CKTCNGSATLCTSCPK | 16 | 8.53 | 1615.68 |
| CO4A1_HUMAN | CNGTKCERGPLGPPG | 15 | 8.22 | 1438.70 |
| LAMA5_HUMAN | CQHNTCGGTCDRC | 13 | 6.71 | 1396.47 |
| FBN3_HUMAN | CVVPIC | 6 | 5.51 | 632.30 |
After fishing and identifying eight N-terminal Cys peptides in AF, we also identified the candidate peptidases that cleave their corresponding proteins to yield the peptides. For example, by searching the peptidase database MEROPS,47 we identified a number of candidate peptidases that would likely cleave the Ser1178-Cys1179 or the Lys1121-Gly1122 site of PCSK5_HUAMN (Table S2†). According to Table S2,† matrix metallopeptidase-2 (MMP-2) is the only peptidase that was predicted to be able to cleave both the above two sites of PCSK5_HUAMN to yield the peptide CKTCNGSATLCTSCPK.48 Therefore, we concluded that the peptide CKTCNGSATLCTSCPK fished by 1 was the cleaved product of PCSK5_HUAMN by MMP-2 (Table S2†). For further function analysis, we predicted the peptide structures using bioinformatic analysis. The sequence-based prediction of the secondary and supersecondary structures of proteins (or peptides) is of great interest to biologists and has found wide applications in numerous areas relating to protein structure and function. Our results indicated that three of the “fished” eight peptides, including peptide CKTCNGSATLCTSCPK, have C-coil structures (Fig. S25†). Coincidentally, using Simple Modular Architecture Research Tool (SMART) and the protein sequence to predict the protein domains of these eight peptide-associating proteins,49 we found that the above mentioned three peptides were also located in the functional domain regions of their corresponding proteins (Fig. S26†). For example, peptide CQHNTCGGTCDRC was identified in the region of the Laminin EGF-like domain (EGF_Lam) of LAMA5_HUMAN, which contains abundant cysteines to form multiple disulfide bonds.
Identification of a bioactive peptide in the eight “fished” peptides
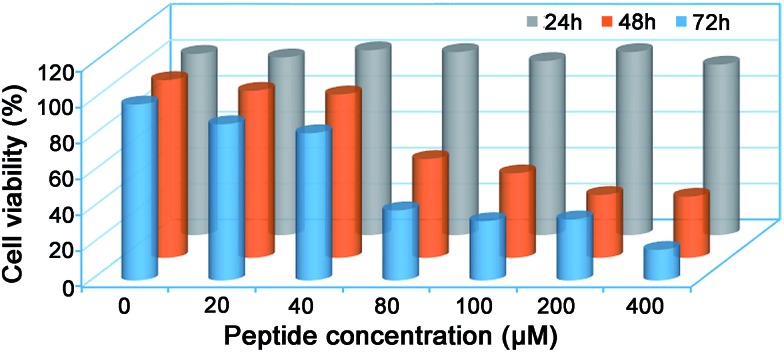
Since many of the biologically active peptides contain one or multiple Cys, which are believed to contribute to the activity of the peptides,22–24 we proposed that some of our “fished” N-terminal Cys peptides might process biofunctions. Generally, bioactive peptides in AF are defined as those that selectively inhibit the growth of endometrial cancer cells but not normal endometrial cells; we thus investigated the cytotoxicity of the eight “fished” peptides on uterine endometrial cancer cells and normal cells. Before incubating the peptides with cells, we chemically synthesized and modified the peptides by sequentially tagging a transactivator of transcription (TAT, YGRKKRRQRRR) and a nuclear localization signal (NLS, RKRRK) to the N-terminal of each peptide and capped its C-terminal with an amide bond. For example, CKTCNGSATLCTSCPK was modified to be TAT-NLS-CKTCNGSATLCTSCPK-NH2. TAT is a cell-penetrating peptide from the human immunodeficiency virus, and it can help to deliver proteins, DNA, RNA, or nanoparticles to the cytoplasm in a short time with extremely high efficiency.50,51 After this modification, each of the eight peptides was assured of being efficiently delivered into the cells. The cell counting kit-8 (CCK-8, Sigma) assay indicated that only the modified peptide TAT-NLS-CKTCNGSATLCTSCPK-NH2 in the eight peptides inhibited the growth of uterine endometrial cancer HEC-1-B cells, but not the normal endometrial cells such as AA-CELL-24 (Fig. S27 and S28†). In detail, the peptide had half-inhibitory concentrations (IC50) on HEC-1-B cells of 108.6 ± 5.3 μM and 66.0 ± 0.9 μM for 48 h and 72 h, respectively (Fig. 5). To further investigate the inhibitory mechanism of peptide TAT-NLS-CKTCNGSATLCTSCPK-NH2 on endometrial cancer HEC-1-B cells, after the cells were incubated with 80 μM TAT-NLS-CKTCNGSATLCTSCPK-NH2 for 24 h and stained with FITC Annexin V and propidium iodide, we applied the fixed cells for flow cytometry analysis. The results indicated that, compared with control cells, peptide-treated cells showed increases of the early-apoptotic stage (Q3) from 4.5% to 28.0% and late-apoptotic stage (Q2) from 2.13% to 12.2% (Fig. S29†). But in the necrosis stage (Q1), the peptide-treated cells (0.055%) did not show an obvious change compared with the control cells (0.031%) (Fig. S29†). All these data above suggest that the peptide TAT-NLS-CKTCNGSATLCTSCPK-NH2 induces the death of endometrial cancer HEC-1-B cells via a typical apoptotic pathway.
Fig. 5. Cell viability of endometrial cancer HEC-1-B cells treated with peptide TAT-NLS-CKTCNGSATLCTSCPK-NH2 for 24 h, 48 h, and 72 h, respectively. Each value represents the average value from three independent experiments.
Conclusions
In summary, employing InESI-MS and our home-built micro-reactor setup coupled to InESI, we successfully uncovered the mechanism underlying the CBT-Cys click condensation reaction and obtained its second order reaction rate constants at different pH values. Using 15N-Cys as one reactant, we determined that the nitrogen atom in the second thiazole ring of the product Aminoluciferin originated from Cys, with which the mechanism underlying this click reaction was proposed. Using our home-built micro-reactor setup coupled to the MS inlet, the crucial Intermediate 2 in the mechanistic scheme of this reaction was successfully intercepted and structurally characterized for the first time. These InESI-MS and tandem MS evidences corroborated the proposed mechanism. Moreover, using the ion chronograms of the reactant Cys (m/z 122) to monitor the reaction in real time, we obtained the curves of 1/[Cys] vs. reaction time at different pH values. Linear regression analyses of the plots gave the second-order reaction rate constants. For example, the second-order reaction rate constant of this click reaction at pH 7.4 was calculated to be 59.7 M–1 s–1 with our method, which is consistent in order of magnitude with that recently reported, calculated by the conventional HPLC method (26.8 M–1 s–1). After mechanistic study of the CBT-Cys click reaction, we rationally designed a small molecular CBT derivative 1 and developed a facile method of using this solid phase CBT to fish N-terminal Cys peptides in real clinic samples. Using a synthesized N-terminal Cys peptide 2 as the standard and mass spectrometry analysis in a single automated operation, we found our method could be applied to fish and quantitatively detect N-terminal Cys peptides within a wide amount range (100 amol to 10 nmol) with very high sensitivity. Using this method, we identified eight unique N-terminal Cys peptides in AF with a false positive rate less than 1%. Proteomic analysis indicated that three of the eight peptides have C-coil structures and coincidentally are located in the functional domain regions of their corresponding proteins. By tagging a TAT and NLS to each peptide, we studied the cytotoxicity of the eight modified peptides on uterine endometrial cancer cells and normal cells. The CCK-8 assay indicated that only one modified peptide (i.e., TAT-NLS-CKTCNGSATLCTSCPK-NH2) inhibited the growth of uterine endometrial cancer HEC-1-B cells, but not the normal endometrial cells such as AA-CELL-24. Flow cytometry analysis indicated that the peptide induces the death of endometrial cancer HEC-1-B cells via an apoptotic pathway. By selectively isolating N-terminal Cys-containing peptides from a real biological sample, our method significantly reduces the complexity of the peptide mixture in the sample. For example, a theoretical tryptic digest of the entire yeast proteome (6113 proteins) yields 344 855 peptides, among which only 30 619 peptides contain a Cys residue while N-terminal Cys-containing peptides are additionally quite fewer. Thus, if our method is applied for the isolation of the N-terminal Cys peptides in yeast, the complexity of its peptide mixture will be significantly reduced, while its protein quantification and identification can still be achieved. With its mechanism satisfactorily elucidated and kinetic parameters obtained with our InESI-MS method, as well as using this CBT-Cys click reaction to identify bioactive N-terminal Cys peptides in real clinical samples, we anticipate that this CBT-Cys click condensation reaction could be applied more widely for the facile isolation, site-specific identification, and quantification of N-terminal Cys-containing peptides in complex biological samples.
Acknowledgments
This work was supported by Collaborative Innovation Center of Suzhou Nano Science and Technology, the Major program of Development Foundation of Hefei Center for Physical Science and Technology, Hefei Science Center CAS (2015HSC-UP012), Recruitment Program of Global Experts, and the National Natural Science Foundation of China (Grants U1532144, 31400727, 21375121 and 21475121).
Footnotes
References
- Lowe A. B., Hoyle C. E., Bowman C. N. J. Mater. Chem. 2010;20:4745–4750. [Google Scholar]
- Truong V. X., Ablett M. P., Richardson S. M., Hoyland J. A., Dove A. P. J. Am. Chem. Soc. 2015;137:1618–1622. doi: 10.1021/ja511681s. [DOI] [PubMed] [Google Scholar]
- Hawker C. J., Wooley K. L. Science. 2005;309:1200–1205. doi: 10.1126/science.1109778. [DOI] [PubMed] [Google Scholar]
- Rajendran A., Endo M., Sugiyama H. Angew. Chem., Int. Ed. 2012;51:874–890. doi: 10.1002/anie.201102113. [DOI] [PubMed] [Google Scholar]
- Ye D. J., Shuhendler A. J., Pandit P., Brewer K. D., Tee S. S., Cui L. N., Tikhomirov G., Rutt B., Rao J. H. Chem. Sci. 2014;5:3845–3852. doi: 10.1039/C4SC01392A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speers A. E., Adam G. C., Cravatt B. F. J. Am. Chem. Soc. 2003;125:4686–4687. doi: 10.1021/ja034490h. [DOI] [PubMed] [Google Scholar]
- Kolb H. C., Finn M. G., Sharpless K. B. Angew. Chem., Int. Ed. 2001;40:2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Yuan Y., Li D., Zhang J., Chen X. M., Zhang C., Ding Z. L., Wang L., Zhang X. Q., Yuan J. H., Li Y. M., Kang Y. B., Liang G. L. Chem. Sci. 2015;6:6425–6431. doi: 10.1039/c5sc01315a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G., Ren H., Rao J. Nat. Chem. 2010;2:54–60. doi: 10.1038/nchem.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Q., Li Q., Yuan Q., Li L., Hai Z., Liu S., Liang G. Anal. Chem. 2015;87:3460–3466. doi: 10.1021/ac504836a. [DOI] [PubMed] [Google Scholar]
- Baskin J. M., Prescher J. A., Laughlin S. T., Agard N. J., Chang P. V., Miller I. A., Lo A., Codelli J. A., Bertozzi C. R. Proc. Natl. Acad. Sci. U. S. A. 2007;104:16793–16797. doi: 10.1073/pnas.0707090104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y., Sun H., Ge S., Wang M., Zhao H., Wang L., An L., Zhang J., Zhang H., Hu B., Wang J., Liang G. ACS Nano. 2015;9:761–768. doi: 10.1021/nn5062657. [DOI] [PubMed] [Google Scholar]
- Dragulescu-Andrasi A., Kothapalli S.-R., Tikhomirov G. A., Rao J., Gambhir S. S. J. Am. Chem. Soc. 2013;135:11015–11022. doi: 10.1021/ja4010078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G., Ronald J., Chen Y., Ye D., Pandit P., Ma M. L., Rutt B., Rao J. Angew. Chem., Int. Ed. 2011;50:6283–6286. doi: 10.1002/anie.201007018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai X. Z., Ho C. J. H., Aw J., Attia A. B. E., Mu J., Wang Y., Wang X. Y., Wang Y., Liu X. G., Chen H. B., Gao M. Y., Chen X. Y., Yeow E. K. L., Liu G., Olivo M., Xing B. G. Nat. Commun. 2016;7:10432. doi: 10.1038/ncomms10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Miao Q., Zou P., Liu L., Wang X., An L., Zhang X., Qian X., Luo S., Liang G. Theranostics. 2015;5:1058–1067. doi: 10.7150/thno.11758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y., Ge S. C., Sun H. B., Dong X. J., Zhao H. X., An L. N., Zhang J., Wang J. F., Hu B., Liang G. L. ACS Nano. 2015;9:5117–5124. doi: 10.1021/acsnano.5b00287. [DOI] [PubMed] [Google Scholar]
- Yuan Y., Wang L., Du W., Ding Z., Zhang J., Han T., An L., Zhang H., Liang G. Angew. Chem., Int. Ed. 2015;54:9700–9704. doi: 10.1002/anie.201504329. [DOI] [PubMed] [Google Scholar]
- Liu S., Tang A., Xie M., Zhao Y., Jiang J., Liang G. Angew. Chem., Int. Ed. 2015;54:3639–3642. doi: 10.1002/anie.201409952. [DOI] [PubMed] [Google Scholar]
- Yuan Y., Liang G. Org. Biomol. Chem. 2014;12:865–871. doi: 10.1039/c3ob41241e. [DOI] [PubMed] [Google Scholar]
- Sato Y., Inaba K. FEBS J. 2012;279:2262–2271. doi: 10.1111/j.1742-4658.2012.08593.x. [DOI] [PubMed] [Google Scholar]
- Antelmann H., Helmann J. D. Antioxid. Redox Signaling. 2011;14:1049–1063. doi: 10.1089/ars.2010.3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandes N., Schmitt S., Jakob U. Antioxid. Redox Signaling. 2009;11:997–1014. doi: 10.1089/ars.2008.2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jortzik E., Wang L., Becker K. Antioxid. Redox Signaling. 2012;17:657–673. doi: 10.1089/ars.2011.4266. [DOI] [PubMed] [Google Scholar]
- Ren D., Julka S., Inerowicz H. D., Regnier F. E. Anal. Chem. 2004;76:4522–4530. doi: 10.1021/ac0354645. [DOI] [PubMed] [Google Scholar]
- Paulech J., Solis N., Edwards A. V., Puckeridge M., White M. Y., Cordwell S. J. Anal. Chem. 2013;85:3774–3780. doi: 10.1021/ac400166e. [DOI] [PubMed] [Google Scholar]
- Yao Y. T., Huang J. F., Cheng K., Pan Y. B., Qin H. Q., Ye M. L., Zou H. F. Anal. Chem. 2015;87:11353–11360. doi: 10.1021/acs.analchem.5b02711. [DOI] [PubMed] [Google Scholar]
- Santos L. S. Eur. J. Org. Chem. 2008;2008:235–253. [Google Scholar]
- Vikse K. L., Ahmadi Z., Manning C. C., Harrington D. A., McIndoe J. S. Angew. Chem., Int. Ed. 2011;50:8304–8306. doi: 10.1002/anie.201102630. [DOI] [PubMed] [Google Scholar]
- Brown T. A., Chen H., Zare R. N. Angew. Chem., Int. Ed. 2015;54:11183–11185. doi: 10.1002/anie.201506316. [DOI] [PubMed] [Google Scholar]
- Brown T. A., Chen H., Zare R. N. J. Am. Chem. Soc. 2015;137:7274–7277. doi: 10.1021/jacs.5b03862. [DOI] [PubMed] [Google Scholar]
- Yan X., Sokol E., Li X., Li G., Xu S., Cooks R. G. Angew. Chem., Int. Ed. 2014;53:5931–5935. doi: 10.1002/anie.201310493. [DOI] [PubMed] [Google Scholar]
- Iacobucci C., Reale S., De Angelis F. Angew. Chem., Int. Ed. 2016;55:2980–2993. doi: 10.1002/anie.201507088. [DOI] [PubMed] [Google Scholar]
- Hinderling C., Adlhart C., Chen P. Angew. Chem., Int. Ed. 1998;37:2685–2689. doi: 10.1002/(SICI)1521-3773(19981016)37:19<2685::AID-ANIE2685>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Iacobucci C., Reale S., Gal J. F., De Angelis F. Angew. Chem., Int. Ed. 2015;54:3065–3068. doi: 10.1002/anie.201410301. [DOI] [PubMed] [Google Scholar]
- van den Broek I., Blokland M., Nessen M. A., Sterk S. Mass Spectrom. Rev. 2015;34:571–594. doi: 10.1002/mas.21419. [DOI] [PubMed] [Google Scholar]
- Mayne J., Ning Z., Zhang X., Starr A. E., Chen R., Deeke S., Chiang C.-K., Xu B., Wen M., Cheng K. Anal. Chem. 2016;88:95–121. doi: 10.1021/acs.analchem.5b04230. [DOI] [PubMed] [Google Scholar]
- Huang G. M., Li G. T., Ducan J., Ouyang Z., Cooks R. G. Angew. Chem., Int. Ed. 2011;50:2503–2506. doi: 10.1002/anie.201007819. [DOI] [PubMed] [Google Scholar]
- Huang G. M., Li G. T., Cooks R. G. Angew. Chem., Int. Ed. 2011;50:9907–9910. doi: 10.1002/anie.201103687. [DOI] [PubMed] [Google Scholar]
- Li G. Y., Huang G. M. J. Mass Spectrom. 2014;49:639–645. doi: 10.1002/jms.3385. [DOI] [PubMed] [Google Scholar]
- Ren H., Xiao F., Zhan K., Kim Y. P., Xie H., Xia Z., Rao J. Angew. Chem., Int. Ed. 2009;48:9658–9662. doi: 10.1002/anie.200903627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon J., Shen B., Xiong L., Miao Z., Lee K. H., Rao J., Chin F. T. Bioconjugate Chem. 2012;23:1902–1908. doi: 10.1021/bc300273m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabane H., Lamazzi C., Thiery V., Pierre A., Leonce S., Pfeiffer B., Renard P., Guillaumet G., Besson T. J. Enzyme Inhib. Med. Chem. 2003;18:167–174. doi: 10.1080/1475636032000069837. [DOI] [PubMed] [Google Scholar]
- Suckau D., Resemann A., Schuerenberg M., Hufnagel P., Franzen J., Holle A. Anal. Bioanal. Chem. 2003;376:952–965. doi: 10.1007/s00216-003-2057-0. [DOI] [PubMed] [Google Scholar]
- Vesce F., Pavan B., Lunghi L., Giovannini G., Scapoli C., Piffanelli A., Biondi C. Obstet. Gynecol. 2004;103:108–113. doi: 10.1097/01.AOG.0000101282.38902.13. [DOI] [PubMed] [Google Scholar]
- Orczyk-Pawiłowicz M., Floriański J., Zalewski J., Kątnik-Prastowska I. Glycoconjugate J. 2005;22:433–442. doi: 10.1007/s10719-005-4437-5. [DOI] [PubMed] [Google Scholar]
- Rawlings N. D., Barrett A. J., Bateman A. Nucleic Acids Res. 2012;40:343–350. doi: 10.1093/nar/gkr987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.-G., Lone A. M., Nolte W. M., Saghatelian A. Proc. Natl. Acad. Sci. U. S. A. 2012;109:8523–8527. doi: 10.1073/pnas.1203195109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Doerks T., Bork P. Nucleic Acids Res. 2015;43:D257–D260. doi: 10.1093/nar/gku949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagahara H., Vocero-Akbani A. M., Snyder E. L., Ho A., Latham D. G., Lissy N. A., Becker-Hapak M., Ezhevsky S. A., Dowdy S. F. Nat. Med. 1998;4:1449–1452. doi: 10.1038/4042. [DOI] [PubMed] [Google Scholar]
- Gump J. M., Dowdy S. F. Trends Mol. Med. 2007;13:443–448. doi: 10.1016/j.molmed.2007.08.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.