Abstract
Decellularization is a promising new method to prepare natural matrices for tissue regeneration. Successful decellularization has been reported using various tissues including skin, tendon, and cartilage, though studies using hard tissue such as bone are lacking. In this study, we aimed to define the optimal experimental parameters to decellularize natural bone matrix using 0.5% sodium dodecyl sulfate and 0.1% NH4OH. Then, the effects of decellularized bone matrix on rat mesenchymal stem cell proliferation, osteogenic gene expression, and osteogenic differentiations in a two-dimensional culture system were investigated. Decellularized bone was also evaluated with regard to cytotoxicity, biochemical, and mechanical characteristics in vitro. Evidence of complete decellularization was shown through hematoxylin and eosin staining and DNA measurements. Decellularized bone matrix displayed a cytocompatible property, conserved structure, mechanical strength, and mineral content comparable to natural bone. To study new bone formation, implantation of decellularized bone matrix particles seeded with rat mesenchymal stem cells was conducted using an orthotopic in vivo model. After 3 months post-implantation into a critical-sized defect in rat calvaria, new bone was formed around decellularized bone matrix particles and also merged with new bone between decellularized bone matrix particles. New bone formation was analyzed with micro computed tomography, mineral apposition rate, and histomorphometry. Decellularized bone matrix stimulated mesenchymal stem cell proliferation and osteogenic differentiation in vitro and in vivo, achieving effective bone regeneration and thereby serving as a promising biological bone graft.
Keywords: Decellularized bone matrix, mesenchymal stem cell, orthotopic, mineral apposition rate, histomorphometry
Introduction
Bone grafts and substitutes command a tremendous global market, standing at 3.02 billion US dollars in 2014 with projections reaching 3.48 billion dollars by the end of 2023.1 Among the commonly used scaffolding biomaterials, the autograft is ubiquitously referred to as the gold standard for tissue engineering and regenerative medicine applications, despite the drawbacks of donor site morbidity and limited availability. Allografts and xenografts can alternatively be used, yet they have the potential risks of recipient immune system recognition and rejection as well as disease transmission.2,3 For these reasons, there is growing interest regarding the ideal scaffolding for bone tissue engineering, which requires initial mechanical strength, structural and chemical compositions comparable to natural bone, nonimmunogenicity, produces effective bone induction from host tissues, and has potential remodeling capability. Ceramic biomaterials such as hydroxyapatite (HA), calcium phosphate, and bioactive glasses are widely used at present to replace damaged bone. However, there are two general issues with ceramic scaffolds in terms of mechanical strength and biodegradability, which are the two most important characteristics as scaffolds in bone tissue engineering. HA and calcium phosphates are bioinert and mechanically strong materials, whereas bioactive glasses are biodegradable and fragile. Biodegradation profile may increase the chance of failure in modeling new matrix formation and material degradation.4
Decellularization of hard tissue presents a promising scaffolding alternative. Originally inspired to prevent the immune response in organ transplantation, Gilbert et al.5 developed the process of decellularization using small intestine submucosa (SIS). Decellularization is the process of removing cells from a tissue or an organ, preserving the complex mixture of structural and functional proteins that constitute the extracellular matrix (ECM) framework. This process creates a natural scaffold material for cell growth, cell differentiation, and tissue regeneration while also eliminating the adverse immune response through repopulating the matrix with a patient’s own cells. A wide range of tissues including skin, bladder, cornea, blood vessel, heart valve, liver, nerve, tendon, and cartilage have been studied for their decellularization capability, with some in transition to the preclinical trial stage or already in clinical application.6–8 However, few reports exist regarding decellularization of hard tissue such as cortical bone. Decellularized bone matrix (DecBM) has not been well studied until recently.
Yoshihide Hashimoto et al. reported the first use of hydrostatic pressure to obtain decellularized porcine femur using hydrostatic pressure. After DecBM subcutaneous implantation in rats, cell infiltration with neovascularization was achieved after 6 months. The DecBM also promoted the osteogenic differentiation of mesenchymal stem cells (MSCs) in vitro.9 Marcos-Campos et al. compared MSC osteogenesis in decellularized bone of different densities, concluding that DecBM density is negatively correlated with pore size and porosity and positively correlated with the compressive elastic modulus. Cellular infiltration was observed after 5 weeks of MSC culture in the medium density of trabecular DecBM.10 These studies indicate that DecBM can be recellularized, via repopulating with seeded cells, serving as a promising bone scaffolding material with advantageous qualities to address the current biomaterial limitations and meet growing market demands. To achieve this, we propose use of DecBM for cortical bone regeneration through orthotopic site implantation. As few decellularization protocols currently exist and no reports have utilized DecBM for cortical bone regeneration by implantation into the orthotopic site, our study is both novel and has potential to meet an unmet and growing need. Although Yoshihide et al. employed decellularized porcine, cortical bone tissue regeneration was assessed ectopically under the skin.
Previously, we evaluated the mechanical strength of rat calvaria before and after decellularization using three-point bending. The results indicated that the chemical decellularization process had little effect on DecBM physical properties and that DecBM maintains ECM orientations specific to bone that could not be easily synthesized in vitro.11 DecBM is therefore an excellent candidate to serve as an ideal bone grafting material as it displays the required strength to withstand loads and generate adequate forces for movement while maintaining a structural composition of natural bone. In this study, we established a simple decellularization method for rat calvaria DecBM as the grafting material. We hypothesized that DecBM can induce osteogenic differentiation of rat mesenchymal stem cells (rMSCs) in vitro and in vivo new bone formation (NBF) to confirm DecBM to be an excellent grafting material alternative for bone regeneration. In vitro effects of DecBM were evaluated by culturing rMSCs with DecBM particles to achieve proliferation and osteogenic differentiation. DecBM analysis included examining the cytocompatibility, biochemical composition of ECM, histology, ultrastructure, and mechanical properties. After implantation in a critical-sized defect for 12 weeks, further investigation of NBF was assessed by micro computed tomography (microCT), mineral apposition rate (MAR), and histomorphometry.
Materials and methods
All animal experimental procedures were approved by the Institutional Animal Care and Use Committee (IACUC) at the University of North Carolina, Chapel Hill.
Decellularization of bone matrix
To prepare the DecBM, the rat calvaria were harvested using a trephine burr (8 mm in diameter) and washed with deionized water for 1 h, frequently exchanging the water. DecBM decellularization was done in a solution containing 0.5% sodium dodecyl sulfate (SDS; Sigma–Aldrich, St Louis, MO, USA) and 0.1% ammonium hydroxide (NH4OH; Sigma–Aldrich) using a mechanical shaker at room temperature. The decellularization solution was replaced every 36 h for 3 weeks. The DecBM samples were then repeatedly washed with deionized water until SDS was completely removed from the matrix. Completion of the decellularization was confirmed by the histological methods of hematoxylin and eosin (H&E) staining and DNA assays. The decellularized calvaria were broken into particles, with particles larger than 2 mm in their longest dimension selected for in vitro and in vivo experiment. The DecBM samples were lyophilized before sterilizing with cold ethylene oxide gas prior to implantation. To test for residual cells, decellularized calvaria (25 mg) were frozen in liquid nitrogen and then mechanically pulverized. DNA was extracted from the powdered calvaria using the QIAamp DNA kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. DNAs were run on a 1% agarose gel to identify the bands. Gel images were acquired by ImageQuant LAS 4000 (GE, Piscataway, NJ, USA) (Figure 1).
Figure 1.
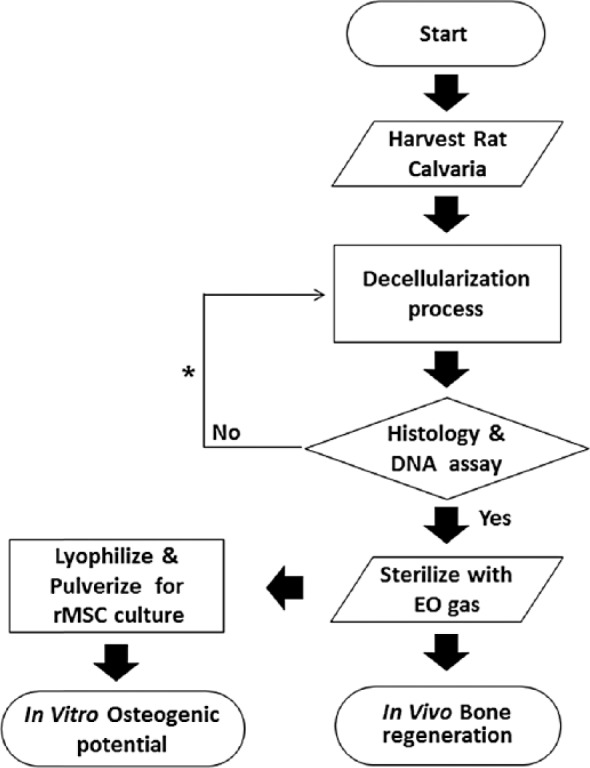
Schematic of study.
Scanning electron microscopy and energy-dispersive X-ray analysis
Both natural bone matrix (NBM) and DecBM were fixed in a cacodylate/saccharose buffer solution (0.05 M/0.6 M; pH 7.4) for 2 h at room temperature. After that, the samples were critical point-dried by dehydrating in an ethanol-graded series. Then sample cross section was sputter-coated with gold in a vacuum and imaged using a Hitachi S-4700 cold cathode field emission scanning electron microscopy (SEM; Hitachi High Technologies America, Inc., Schaumburg, Illinois USA). An energy-dispersive X-ray (EDX) analysis was performed to examine at least three random regions on sample’s cross section, which were analyzed by INCA operator software (Oxford Instruments Analytical, High Wycombe, UK).
Isolation and characterization for rMSCs
Sprague-Dawley rat (3 weeks old, male) femurs were aseptically removed, followed by removal of both femur ends, to then flush out bone marrow using growth media, Dulbecco’s modified Eagle medium (DMEM) basal media supplemented with 10% (v/v) fetal bovine serum (FBS), freshly prepared ascorbic acid (50 µg/mL), 1% penicillin and streptomycin, and 250 µL GlutaMax (Invitrogen, Carlsbad, CA, USA). The flushed cells were collected in 100-cm2 culture dishes with 10 mL growth media and cultured at 37°C in a humidified atmosphere of 5% CO2. Cells were allowed to attach for 4 days, with non-adherent cells removed by exchanging the growth media. The culture medium was changed every 3 days.
For colony-forming unit (CFU) assays, rMSCs were diluted in growth media and plated at approximately 10 cells/cm2 in 35-mm tissue culture dishes. After incubation for 14 days, the cells were washed with phosphate-buffered saline (PBS) and stained with 0.5% Crystal Violet in methanol for 5 min. Immunostaining was performed for the rMSCs on the monolayer culture for CD44, CD90, CD34, and CD45. Monolayer cultures (100,000 cells/35 mm dish) were fixed with 4% paraformaldehyde (PFA), rinsed with deionized water, treated with 0.3% H2O2 for 30 min, blocked with 10% FBS in PBS for 30 min, and washed vigorously with 0.4% Tween-20 in PBS solution three times. After blocking for 30 min, the cells were incubated overnight at 4°C with the anti-(CD44, CD90, CD34, and CD45) (BD Science, San Jose, CA, USA) antibodies (1:500 ratio), for 1 h with the secondary antibodies conjugated to fluorescein isothiocyanate (FITC) and Texas Red (Millipore, Billerica, MA USA), and 4′,6-diamidino-2-phenylindole (DAPI) for nuclei staining.
To evaluate osteogenic capacity, rMSCs were treated with osteogenic media for 3 weeks with the medium exchanged every 3 days. Osteogenic media are composed of culture media supplemented with 0.1 M dexamethasone (Sigma–Aldrich), 10 mM Beta-glycerolphosphate (Sigma–Aldrich), and 0.2 mM ascorbic acid (Sigma–Aldrich). To induce chondrogenic differentiation, rMSCs were transferred into 15 mL polypropylene tubes and centrifuged at 1000 r/min for 5 min to form a pellet and then treated with chondrogenic media (STEMCELL Technologies, Vancouver, BC, Canada) for 3 weeks with fresh media supplied every 3 days. Cell pellets were imbedded in O.C.T (Sakura, Japan) and frozen at −80°C. The frozen block was sectioned into 5 µm slices using a Cryotome (Leica Biosystems CM3050 S, Richmond, IL, USA) and stained using Safranin O counterstained with fast green. To induce adipogenic differentiation, rMSCs were treated with adipogenic medium (STEMCELL Technologies, Vancouver, BC, Canada) for 3 weeks with the media exchanged every 3 days for 21 days. Cells were fixed with 10% formalin and stained with Oil Red O staining to detect fat droplets. At passage 10, rMSCs were characterized prior to use for in vitro and in vivo studies.
Three-point bending test
DecBM and fresh NBM serving as a control were excised from calvaria and stored in PBS until the three-point bending test was performed. Specimens were cut along the sagittal direction to yield one strip per specimen with a width of 2 mm and a length of 10 mm in a rectangular shape in preparation for testing. Ten tissue strips in each group were tested using three-point bending by Instron (model 4204; Canton, MA, USA) at a 2-mm/min crosshead speed. The failure point of each specimen was then determined by analysis of the stress–strain curves.
Total protein analysis
Both fresh and decellularized calvaria (25 mg) were lyophilized and then mechanically pulverized. Total protein was extracted using 4% SDS and measured by using Pierce BCA Protein Assay Kit (Thermo Fisher Scientific Inc., Rockford, IL, USA).
Live and dead and proliferation assays
The Live/Dead Assay Kit (Molecular Probes, Leiden, The Netherlands) was used to examine the viability of rMSCs cultured on a 35-mm dish with the addition of DecBM particles, as instructed by the company protocol. Cell viability was assessed under a fluorescence microscope, as calcein is detected as green fluorescence in live cells and ethidium homodimer-1 (EthD-1) is detected as red fluorescence in dead cells.
Plating of rMSCs with and without DecBM particles was done in 12-well plates at a density of 50,000 cells per well. The proliferation of the rMSCs in growth media was conducted using the MTS (3-(4, 5-dimethylthiazol-2-yl)-5-(3 carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium) Cell Proliferation Assay Kit (Promega Co., Madison, WI, USA) via the company’s instructions. MTS was reacted with cells at 37°C for 1 h. After transferring the solution into a 96-well plate, the absorbance was measured on days 1, 3, 5, and 7 at 490 nm using a plate reader (BioRad, Hercules, CA, USA).11
Real-time polymerase chain reaction
After culturing rMSCs in growth media with and without DecBM particles for 4 or 7 days, RNA was extracted using Trizol reagent (Invitrogen). Then, complementary DNA (cDNA) was synthesized using iScript Kit (BioRad) by following the company’s protocol. Real-time polymerase chain reaction (PCR) was performed using the 7200 Fast Real-Time PCR System (Applied Biosystems, Bedford, MA, USA) to determine messenger RNA (mRNA) expression of each osteogenic-specific gene (Table 1). All probes used to detect target genes were labeled with SYGR (Life Technologies, Carlsbad, CA, USA). Cycling conditions consisted of initial denaturation at 94°C for 5 min, followed by 40 cycles consisting of 10 s of denaturation at 94°C, 15 s of annealing at 60°C, and 15 s of elongation at 72°C. The ΔΔCt method was used to calculate the relative levels of gene expression. Real-time PCR was repeated three times and each sample was triplicated in each experiment.
Table 1.
Primers for real-time PCR for osteogenic gene expression.
| Target | Primer sequence (5′−3′) | Size (bp) |
|---|---|---|
| ALP | F: AACCAGACACAAGCATTCC R: GCCTTTGAGGTTTTTGGTCA |
200 |
| BSP | F: CCGGCCACGTACTTTCTT R: TGCACTGGAAACCGTTTCAGA |
66 |
| OC | F: CTGACCTCACAGATGCCAA R: GGTCTGATAGTCTGTCACAA |
185 |
| GAPDH | F: TGAGGTGACCGCATCTTCTTG R: TGGTAACCAGGCGTCCGATA |
101 |
PCR: polymerase chain reaction; ALP: alkaline phosphatase; BSP: bone sialoprotein; OC: osteocalcin; DAPDH: glyceraldehyde 3-phosphate dehydrogenase.
Mineralization
To analyze mineralization, rMSCs were seeded at a density of 100,000 cells per well with or without DecBM particles in 12-well culture dishes with fresh osteogenic media supplied every 3 days for 28 days. The cells were harvested on days 10, 17, and 28 for fixation in 95% ethanol for 30 min at room temperature. The fixed cells were washed with PBS and stained with 1% Alizarin Red S solution (pH 4.2) for 10 min at room temperature. Quantitative analysis was performed by elution with 10% (w/v) cetylpyridinium chloride for 10 min at room temperature and the optical density (OD) was measured at 570 nm.
In vivo bone formation
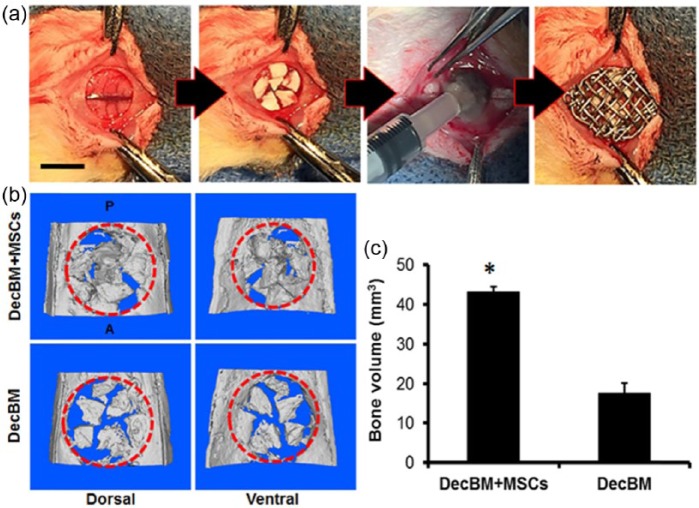
Differentiated rMSCs were grown in osteogenic media for 14 days and seeded with DecBM particles (n = 3). Two test groups (DecBM + rMSCs and DecBM only) with five Sprague-Dawley rats (Charles River, Wilmington, MA, USA; about 250–300 g, 7 weeks) in each group were used to study in vivo bone formation. The detailed surgical procedure and the fluorochrome injections for studying the MAR have been well described in our previous study.11 Only modification we made in this study was utilization of titanium mesh to prevent dislocating DecBM particles from the defect site (Figure 6(a)).
Figure 6.
(a) Images of surgical implantation of DecBM into the critical-sized defect of rat calvaria. (b) MicroCT images of critical-sized calvarial defects after 12 weeks of implantation with DecBM and DecBM with rMSCs, with the red circle indicating the defect site (8 mm in diameter). (c) Bone volume of the defect site after 12 weeks of implantation was quantified (n = 3, *p < 0.05).
Rats were sacrificed at 12 weeks post-surgery and followed by calvaria removal, trimming, and preservation of the implanted sites before fixation in 10% formalin for 7 days at 4°C. The specimens were then stored in 70% isopropyl alcohol at 4°C for further analysis on the NBF. The detailed methods for the microCT, slide preparation, image acquisition and calculation for MAR, and histomorphometric analysis including Stevenel’s Blue/Van Gieson staining were well described in our previous study.11
Statistics
All results were analyzed by analysis of variance (ANOVA) and represented as mean ± standard deviation to define whether differences between each group were significant or not. When the p-value was <0.05, the differences were considered significant.
Results
Decellularization of rat calvaria
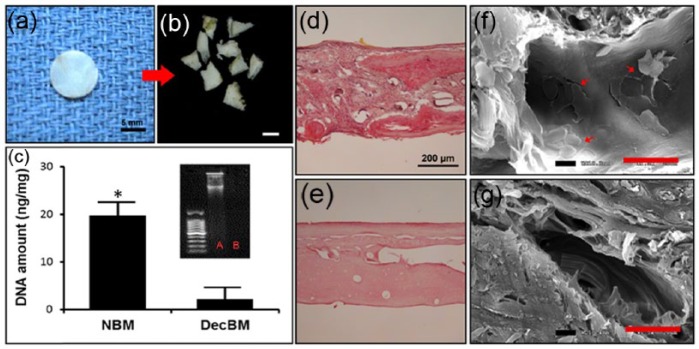
Rat calvaria were successfully decellularized following treatment with SDS and NH4OH after 3 weeks. The decellularized calvaria conserved their original structural configuration and gross examination indicated the calvaria coloration turned to pale white (Figure 2(a)). Decellularized calvaria were then broken down into pieces and particles larger than 2500 µm in the shortest dimension were selected for (Figure 2(b)). H&E staining revealed no residual cells present in DecBM (Figure 2(e)), whereas the presence of numerous cells in NBM was revealed (Figure 2(d)). The DNA content was quantified from of the dry weight of both NBM and DecBM using NanoDrop 2000 (Thermo Scientific, Wilmington, DE, USA). The DNA content in NBM was 19.71 ± 2.9 ng/mg, while the DNA content in the DecBM group was 2.05 ± 2.38 ng/mg, indicating a significant reduction (*p < 0.05) in nuclear material of the calvaria which was further confirmed by gel electrophoresis data (Figure 2(C)). SEM further revealed that similar matrix morphology was exhibited in both NBM (Figure (f)) and DecBM (Figure (g)). In NBM, cells were adhered to and infiltrated into the matrix, while cells were absent in DecBM and thus indicative of effective calvarial bone decellularization.
Figure 2.
Gross image of decellularized rat calvaria (a) before and (b) after breaking into particles, with particles larger than 2 mm in their longest dimension selected for; scale: 2.5 mm. (c) DNA quantification in nanograms/microliter of natural bone matrix (NBM) and decellularized bone matrix (DecBM), with gel electrophoresis to detect residual DNA before (lane A) and after decellularization (lane B); *p < 0.05. Hematoxylin and eosin staining of calvaria (d) before and (e) after decellularization. SEM images for visualizing (f) the presence of cells in calvaria before and after decellularization, with a maintained ECM structure and (g) no cell presence evident after decellularization; scale bar: 20 µm.
Isolation and characterization of bone marrow rMSCs
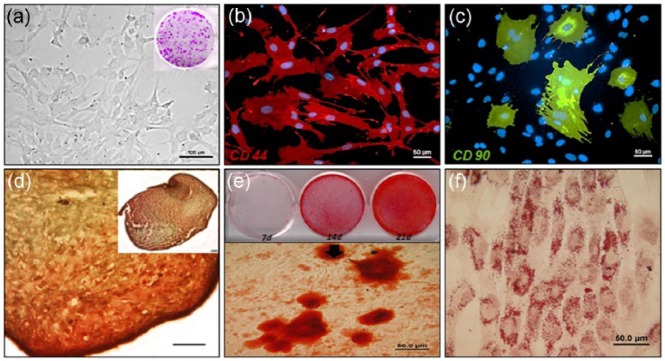
rMSC characterization was performed before use of the primary cultured cells from bone marrow, with the rMSC phenotype determined by differential interference contrast (DIC) imaging (Figure 3(a)). Single MSCs were assessed for their colony-forming capability. In the immunocytochemical analysis, rMSCs were positive for CD44 and CD90 antibodies (Figure 3(b) and (c)) and negative for CD45 and CD34 (data not shown) antibodies. Safranin O staining was positive for the cartilage-like matrix produced by the rMSC aggregate after 21 days of differentiation under chondrogenic induction media. Lacunae were also visible in the matrix (Figure 3(d)). Mineralization was evident in rMSCs differentiated for 21 days in osteogenic media and no mineralization was observed in the control of rMSCs in growth media. Alizarin Red S staining clearly stained mineral nodules by binding to calcium (Figure 3(e)). The lipid vacuoles were clearly visualized by Oil Red O staining in rMSC after 21 days of differentiation under adipogenic induction medium but not in the control of rMSCs in growth media (Figure 3(f)).
Figure 3.
Characterization of rat MSCs (rMSCs) isolated from bone marrow. (a) Microscopic image of rMSCs plated at approximately 10 cells/cm2 in 35-mm tissue culture dishes and analyzed with colony-forming unit (CFU) assays. rMSCs were stained with (b) CD44 antibody conjugated with Texas Red fluorescent dye and (c) CD90 antibody conjugated with FITC. (d) Image indicates that rMSCs were able to differentiate into chondrogenic lineage after 3D aggregation culture for 21 days; scale bar: 200 µm; (e) osteogenic linage after 21 days and (f) adipogenic linage after 21 days.
In vitro mechanical, biochemical, and viability assessment for DecBM
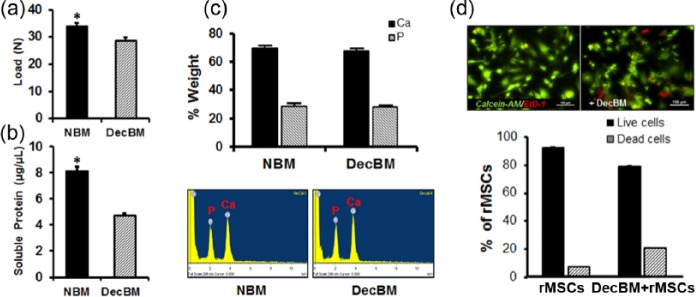
The mean values of the three-point bending test were 33.86 ± 1.28 and 28.61 ± 1.25 N for NBM and DecBM of calvaria, respectively (Figure 4(a)). The decellularization process decreased the mechanical strength by 15.51% (n = 7, *p < 0.05) in DecBM compared to NBM. In addition, the amounts of soluble proteins were significantly reduced by 41.75% from 8.12 ± 0.35 to 4.73 ± 0.15 µg/µL after the decellularization process (Figure 4(b)). A quantitative assessment of bone matrix was carried out by EDX analysis of the NBM and DecBM. The EDX spectra are shown in Figure 4(c) along with the atomic percentages of elements in minerals of NBM and DecBM. Almost identical percentages of Ca and P were found for the DecBM (63.44% and 34.04%) in comparison with NBM (64.95% and 34.74%). The EDX results showed that the decellularization process preserved mineralized tissue with a Ca:P ratio of 1.87 for NBM with a Ca:P ratio of 1.86 for DecBM. Live/dead staining showed 91.82% ± 2.12% live (green) cells and 8.75% ± 2.63% dead (red) cells in the MSC control culture, while the culture with DecBM particles showed 79.63% ± 1.89% live cells and 21.3% ± 1.54% dead cells (Figure 4(d)).
Figure 4.
(a) Three-point bending tests were performed on NBM and DecBM of calvaria (n = 7, *p < 0.05). (b) Total soluble protein was quantified before and after decellularized calvaria (n = 5, *p < 0.05). (c) For EDX analysis, SEM images of NBM and DecBM were acquired, and chemical element composition was analyzed on three randomly selected areas. (d) The Live/Dead Assay was performed to measure material toxicity after the decellularization process, with viable cells staining as green by Calcein-AM and dead cells staining as red by EtD-1.
Effects of DecBM on proliferation, gene expression, and mineralization
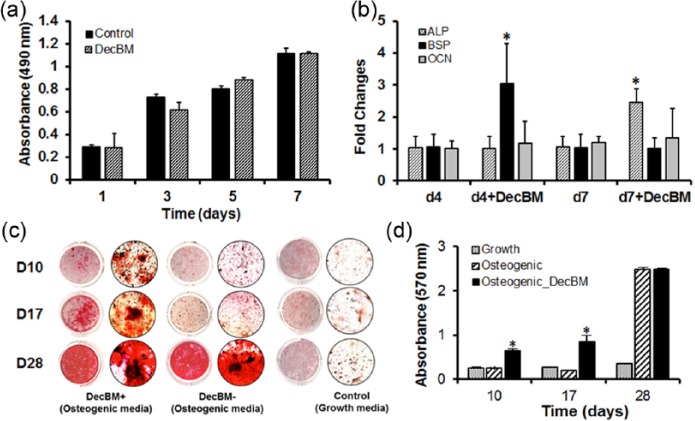
MTS activity of the rMSCs was measured on days 1, 3, 5, and 7. The rMSCs both with and without DecBM particles represented almost the same growth rate up to day 7 without significant difference in the proliferation rate (p > 0.05), except that the OD measurement in the rMSCs with DecBM particles’ group (0.61 ± 0.07) on day 3 was lower than that of the rMSC-only group (0.73 ± 0.03, *p < 0.05) on day 3 (Figure 5(a)). The short-term growth results indicated that DecBM particles had little adverse effects on the rMSC proliferation.
Figure 5.
In vitro assessment of osteogenic differentiation of rMSCs by DecBM. (a) MTS proliferation assays were performed on days 1, 3, 5, and 7 in growth media containing 10% FBS in 12-well plates with n = 5 measurements from three independent samples per group. The osteogenic gene expression of rMSCs was evaluated by culturing with and without DecBM, then using real-time PCR analysis to detect ALP, BSP, and OCN genes. (b) Real-time PCR data were normalized with GAPDH expression (n = 3 per group, *p < 0.05). (c) Mineral formation was detected by Alizarin Red S staining after culturing rMSCs with or without DecBM under osteogenic media, with growth media serving as the control. (c) Microscopic images confirmed the ability of mineralization by rMSCs at 10, 17, and 28 days of osteogenic differentiation. (d) Alizarin Red S–stained particles were quantified by the CPC extraction method, with the absorbance of the extracted solution measured at 570 nm (n = 5, *p < 0.05).
Real-time PCR analysis was performed to determine whether osteogenic gene expression is increased in rMSCs due to addition of DecBM particles. The osteogenic gene expression of rMSCs with DecBM particles in growth media showed a significant increase in BSP gene expression (3.03 ± 1.26-fold) on day 4 and ALP gene expression (2.45 ± 0.41-fold) on day 7 in comparison with the gene expression of rMSC-only (1.05 ± 0.41-fold on day 4 and 1.05 ± 0.42-fold on day 7). The OCN genes showed small increase from 1.99 ± 0.92-fold to 1.34 ± 0.43-fold in the gene expression level for 7 days (Figure 5(b)).
The mineralization of rMSCs with DecBM differentiated in a monolayer was tested by Alizarin Red S staining and CPC extraction on the calcium nodules after 10, 17, and 28 days of osteogenic differentiation. Mineral nodules in the control plate without DecBM appeared after 7 days of osteogenic differentiation and increased over time up to 28 days (Figure 5(c)). Differentiation of rMSCs with DecBM particles resulted in significantly higher mineralization (0.85 ± 0.15) than the rMSCs without DecBM (0.21 ± 0.15), as indicated by Alizarin Red S staining on day 17 (*p < 0.05). Mineralization was also induced using rMSCs with DecBM particles under growth media, although the differentiation time was twice longer than under osteogenic media (data not shown). However, the level of mineralization reached 2.49 ± 0.03 in the rMSCs with DecBM particles’ group and 2.48 ± 0.04 in the rMSC-only group by day 28 under osteogenic differentiation conditions (Figure 5(d)).
Post-implantation evaluation by microCT, MAR, and histomorphometry
After 12 weeks post-implantation, the entire calvaria were explanted for microCT, MAR, and histomorphometric analysis. MicroCT analysis indicated reconstructed calvaria in three dimensions (3D; Figure 6(a)). NBF was calculated to be 43.24 ± 1.21 mm3 for the rMSCs with DecBM group and 17.59 ± 2.58 mm3 for the DecBM-only group (Figure 6(c)). Although figures of empty defect and autograft group were not shown, their volumes were 10.06 ± 1.93 and 49.72 ± 2.48 mm3, respectively. As indicated by the higher volume, there was greater formation of new bone observed in the center of defect in the rMSCs with DecBM group. The DecBM-only group showed most DecBM grafts merged to the surrounding host bone, not to the center of defect (Figure 6(b)). Overall, the rMSCs with DecBM particles had greater bone regeneration in the defect site versus DecBM-only.
Evaluation of the bone formation rate using MAR was originally tried to be assessed by in the defect region of each sample from the rMSCs with DecBM group and the DecBM-only group. However, the difficulty of visualization of fluorochrome inside pores and inconsistent distribution in the defect site was unable to calculate MAR with accuracy.
After fluorochrome imaging, the nondecalcified resin sections were stained with Stevenel’s Blue and Van Gieson to identify NBF (red color) in the defect site. Histological evaluation demonstrated that the defect within the DecBM with rMSCs’ group was filled with new bone that bridged with the host bone through the center of the defect by 12 weeks (Figure 7(b)). The newly formed bone around the DecBM particles in the rMSCs with DecBM group was well integrated at the interface between the host bone and DecBM particles in the defect. In the DecBM-only group, NBF was moderate, evident primarily at the periphery of the defect and without much integration with DecBM particles (Figure 7(d)). In addition, there was an absence of NBF in the defect area with intervening fibrous tissue and almost full regeneration in the defect area with autografts (data not shown). In Figure 7(e), quantitative measurements of NBF demonstrated 68.73% ± 7.31% bone regeneration for the DecBM with rMSCs group, 42.36% ± 5.45% for the DecBM-only group, 21.52% ± 4.21% for the empty defect group, and 89.13% ± 9.03% for the autograft group (figure of empty defect and autograft groups not shown).
Figure 7.
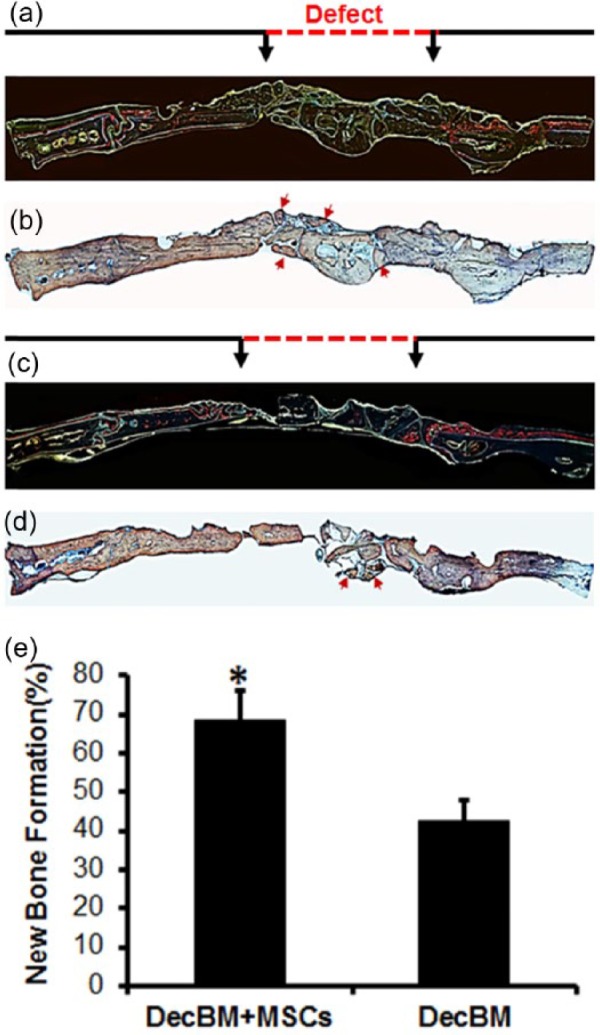
Fluorescent and histological section of the defect area after 12 weeks of implantation. Fluorescence images labeled with (a and c) calcein and Alizarin Red S dye were acquired before staining with (b and d) Steven Blue and Van Gieson. (e) The area of new bone formation (NBF) was quantified in percentage using Image J software, *p < 0.05.
Discussion
DecBM is a naturally derived biomaterial formed by removing cellular components from the bone matrix. This biomaterial is expected to be an ideal scaffolding candidate for hard tissue regeneration due to its excellent mechanical and structural profile, comparable to natural bone, and lack of immunogenicity in host tissue. In addition, natural ECM is known to preserve bioactive molecules such as growth factors and cytokines even after decellularization process in various tissues.12–15 In this study, we developed allogenic natural bone substitutes with an optimal decellularization method and tested for the osteogenic potential in vitro and in vivo.
Effective decellularization depends on the type of tissue, selection of decellularizing solution, and decellularization cycle, which is focused on preservation of the ECM structure with complete removal of cellular components. For optimization of complete tissue decellularization, it is necessary to establish the protocol for each specific target tissue based on their structural, biochemical, and physiological characteristics. Tissue should be permeable to allow for decellularizing solution to penetrate into the tissue in order to remove cellular organelles and nucleic acids. The exact mechanism of the decellularization of bone is not known yet because of the technical limitations of processing dense tissue and its sophisticated microstructure. In previous studies, bone permeability was predicted by injecting different dyes of varying particle sizes.16 After measuring the fluid pathways in cortical bone in vitro, it was determined that the primary pathways are the haversian and Volkmann’s canals and secondary pathways are small channels such as the canalicular and lacunar spaces.17
Discarding the cells from the bone matrix is critical, as it prevents humoral immune reactions against membrane proteins of the cells. Removing DNA from the bone matrix is also critical because this DNA may stimulate immune reactions by activating cytokine production and B-cell immunoglobulin secretion after allogenic or xenogeneic implantation.18 Therefore, recellularizing DecBM with a recipient’s own cells can eliminate the potential adverse immune reaction. Moreover, both rodent and human MSCs have been reported to have immune-tolerance characteristics (immunomodulatory effects), which can facilitate the use of MSCs as allogenic donor cells with potential clinical implications.19 In addition, several studies reported that ECM is conserved among species and tolerated well even by xenogeneic recipients.20–23 This enables a wide variety of ECM scaffolds to become commercially available for tissue engineering applications.
Regarding recellularization, almost no cells were observed inside the critical size defect (8 mm in diameter) of decellularized calvaria bone in our previous study.11 In contrast, many nuclei were observed inside the DecBM particles (2500–3500 µm) after 12 weeks of implantation. We predicted that many accesses for cell infiltration may have been created during the process of particle breakdown. The results from Hashimoto et al. provide additional support for growth factor and ECM component preservation, which remain in active form after the decellularization process and can promote the infiltration and osteogenic differentiation of MSCs.9
Our results depicted in Figure 4 indicate that the mechanical strength was decreased by 15.51% after the decellularization process. This difference was possibly caused by a loss of soluble protein by SDS. Although SDS was more effective than nonionic agents to remove soluble proteins from the matrix in such a compact tissue such as bone, it may have removed structural proteins in the matrix. However, the EDX results show that there was little difference in Ca:P ratio after the decellularization process with a Ca:P ratio of 1.8, similar to the 1.7 ratio seen in natural bone. The EDX analysis demonstrated the decellularization did not have any significant effect on the inorganic composition in bone matrix and that DecBM can provide mechanical strength comparable to natural bone.
MSC cultures with DecBM particles were evaluated by live/dead assaying to observe any residual toxic effects after the decellularization process. As an increased percentage of cells were found to be dead after 3 days, with 21.3% ± 1.54% dead cells evident in the rMSC culture with DecBM compared to 8.75% ± 2.63% dead cells in the control rMSC culture, there is likely a minor cytotoxic effect occurring on rMSCs by DecBM. Alternatively, this increased cytotoxicity may be caused by incomplete washing to remove the decellularizing solution. The physical disturbance from direct contact of the DecBM particles on the rMSC culture could also increase the percentage of dead cells. Although the Live/Dead assay showed that DecBM displays minor toxic effects on rMSCs, it had no effect on rMSC proliferation, which provides further support for the safety of DecBM usage as a bone grafting biomaterial.
To further understand and investigate the cellular effects of DecBM, osteogenic differentiation was examined, including osteogenic gene expression and mineralization. DecBM was found to induce BSP, ALP, and OCN gene expression. Expression levels were investigated under growth media to observe the sole effect of DecBM on these osteogenic genes, thereby eliminating other osteogenic differentiation stimulators contained in osteogenic media. Evidence for the stimulatory effect of DecBM on mineralization was evident in histological analysis data regarding Alizarin Red S staining of mineral nodules. The rMSCs with DecBM group showed more intense and larger nodule formation than the group without DecBM up to 21 days, then equalized by day 28. We predict the stimulatory effect of DecBM on rMSCs may be due to either calcium or phosphate, based on the evidence of positive Alizarin Red S staining.
At 12 weeks post-implantation, calvaria were resected and evaluated by microCT, MAR, and histomorphometry. In many cases, the microCT obtained from the dorsal and ventral scans indicated various degrees of bone formation. The possible reason for this is because of uneven distribution of rMSCs with DecBM on the defect site. Therefore, more vigorous seeding techniques will be necessary for future studies. The two possible ways of preparing DecBM scaffolds for effective cell seeding are the creation of pores throughout the DecBM or processing DecBM into small particles and reconstituting using a crosslinking method. In both cases, the mechanical property associated with structural integrity is key to resolve for effective bone regeneration.
Many critical factors are still unknown for the optimal bone regeneration using DecBM, such as (1) whether cells should be osteogenically differentiated or undifferentiated during cell seeding process, (2) are exogenous growth factors (bone morphogenetic protein (BMP) and vascular endothelial growth factor (VEGF)) necessary? (3) what are the ideal cell numbers? and (4) can dynamic seeding using bioreactor significantly improve bone regeneration? Future studies using DecBM as bone graft can be significantly improved by considering the above factors.
Conclusion
In this study, the optimal decellularization process was defined to prepare biological bone graft material, DecBM for bone regeneration. DecBM can stimulate rMSC’s osteogenic differentiation in vitro, and seeding it with rMSCs yielded a synergic effect to enhance bone regeneration in rat calvarial critical-sized defects. Further studies are needed to adapt optimal architectures of DecBM for finding alternative applications as a scaffold in bone regeneration.
Footnotes
Declaration of conflicting interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported, in part, by NIH/NIDCR K08DE018695 and R01DE022816.
References
- 1. Bone Grafts And Substitutes Market Analysis By Material (Natural - Autografts, Allografts; Synthetic - Ceramic, Composite, Polymer, Bone Morphogenetic Proteins (BMP)), By Application (Craniomaxillofacial, Dental, Foot & Ankle, Joint Reconstruction, Long Bone, Spinal Fusion) Forecasts To 2024. October 2016, Report ID: GVR-1-68038-154-2.
- 2. Li L, Zhou G, Wang Y, et al. Controlled dual delivery of BMP-2 and dexamethasone by nanoparticle-embedded electrospun nanofibers for the efficient repair of critical-sized rat calvarial defect. Biomaterials 2015; 37: 218–229. [DOI] [PubMed] [Google Scholar]
- 3. Delloye C, Cornu O, Barbier O. Bone allografts: what they can offer and what they cannot. J Bone Joint Surg Br 2007; 89: 574–579. [DOI] [PubMed] [Google Scholar]
- 4. Thavornyutikarn B, Chantarapanich N, Sitthiseripratip K, et al. Bone tissue engineering scaffolding: computer-aided scaffolding techniques. Prog Biomater 2014; 3: 61–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gilbert TW, Sellaro TL, Badylak SF. Decellularization of tissues and organs. Biomaterials 2006; 27: 3675–3683. [DOI] [PubMed] [Google Scholar]
- 6. Badylak SF. Xenogeneic extracellular matrix as a scaffold for tissue reconstruction. Transpl Immunol 2004; 12: 367–377. [DOI] [PubMed] [Google Scholar]
- 7. Dellgren G, Eriksson M, Brodin LA, et al. The extended Biocor stentless aortic bioprosthesis. Early clinical experience. Scand Cardiovasc J 1999; 33: 259–264. [DOI] [PubMed] [Google Scholar]
- 8. Harper C. Permacol: clinical experience with a new biomaterial. Hosp Med 2001; 62: 90–95. [DOI] [PubMed] [Google Scholar]
- 9. Hashimoto Y, Funamoto S, Kimura T, et al. The effect of decellularized bone/bone marrow produced by high-hydrostatic pressurization on the osteogenic differentiation of mesenchymal stem cells. Biomaterials 2011; 32: 7060–7067. [DOI] [PubMed] [Google Scholar]
- 10. Marcos-Campos I, Marolt D, Petridis P, et al. Bone scaffold architecture modulates the development of mineralized bone matrix by human embryonic stem cells. Biomaterials 2012; 33: 8329–8342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lee DJ, Padilla R, Zhang H, et al. Biological assessment of a calcium silicate incorporated hydroxyapatite-gelatin nanocomposite: a comparison to decellularized bone matrix. Biomed Res Int 2014; 2014: 837524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rahman S, Patel Y, Murray J, et al. Novel hepatocyte growth factor (HGF) binding domains on fibronectin and vitronectin coordinate a distinct and amplified Met-integrin induced signalling pathway in endothelial cells. BMC Cell Biol 2005; 6: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Comoglio PM, Boccaccio C, Trusolino L. Interactions between growth factor receptors and adhesion molecules: breaking the rules. Curr Opin Cell Biol 2003; 15: 565–571. [DOI] [PubMed] [Google Scholar]
- 14. Crapo PM, Medberry CJ, Reing JE, et al. Biologic scaffolds composed of central nervous system extracellular matrix. Biomaterials 2012; 33: 3539–3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ng CP, Mohamed Sharif AR, Heath DE. Enhanced ex vivo expansion of adult mesenchymal stem cells by fetal mesenchymal stem cell ECM. Biomaterials 2014; 35: 4046–4057. [DOI] [PubMed] [Google Scholar]
- 16. Beno T, Yoon YJ, Cowin S, et al. Estimation of bone permeability using accurate microstructural measurements. J Biomech 2006; 39: 2378–2387. [DOI] [PubMed] [Google Scholar]
- 17. Wang L, Ciani C, Stephen BD, et al. Delineating bone’s interstitial fluid pathway in vivo. Bone 2004; 34: 499–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pisetsky DS. DNA and the immune system. Ann Intern Med 1997; 126: 169–171. [DOI] [PubMed] [Google Scholar]
- 19. Nauta AJ, Fibbe WE. Immunomodulatory properties of mesenchymal stromal cells. Blood 2007; 110: 3499–3506. [DOI] [PubMed] [Google Scholar]
- 20. Bernard MP, Chu ML, Myers JC, et al. Nucleotide sequences of complementary deoxyribonucleic acids for the pro alpha 1 chain of human type I procollagen. Statistical evaluation of structures that are conserved during evolution. Biochemistry 1983; 22: 5213–5223. [DOI] [PubMed] [Google Scholar]
- 21. Bernard MP, Myers JC, Chu ML, et al. Structure of a cDNA for the pro alpha 2 chain of human type I procollagen. Comparison with chick cDNA for pro alpha 2(I) identifies structurally conserved features of the protein and the gene. Biochemistry 1983; 22: 1139–1145. [DOI] [PubMed] [Google Scholar]
- 22. Constantinou CD, Jimenez SA. Structure of cDNAs encoding the triple-helical domain of murine alpha 2 (VI) collagen chain and comparison to human and chick homologues. Use of polymerase chain reaction and partially degenerate oligonucleotide for generation of novel cDNA clones. Matrix 1991; 11: 1–9. [DOI] [PubMed] [Google Scholar]
- 23. Exposito JY, D’Alessio M, Solursh M, et al. Sea urchin collagen evolutionarily homologous to vertebrate pro-alpha 2(I) collagen. J Biol Chem 1992; 267: 15559–15562. [PubMed] [Google Scholar]