Summary
Over the past two decades, researchers studying both microbial and host cell communities have gained an appreciation for the ability of bacteria to produce, regulate, and functionally utilize outer membrane vesicles (OMVs) as a means to survive and interact with their cellular and acellular environments. Common ground has emerged, as it appears that vesicle production is an environmentally-controlled and specific secretion process, however it has been challenging to discover the principles that govern fundamentals of vesicle-mediated transport. Namely, there does not appear to be a single mechanism modulating OMV export, nor universal “markers” for OMV cargo incorporation, nor particular host cell responses common to treatment with all OMVs. Given the diversity of species studied, their differences in envelope architecture and composition, the diversity of environmentally regulated bacterial processes, and the variety of interactions between bacteria and their abiotic and biotic environments, this is hardly surprising. Nevertheless, the ability of bacteria to control exported material in the context of a packaged insoluble particle, a vesicle, is emerging as a significant contribution to bacterial viability, biofilm communities, and bacterial-host interactions. In this review, we focus on detailing important, recent findings regarding the content and functional differences in bacterially secreted vesicles that are influenced by growth conditions.
Introduction
The process of OMV-mediated secretion
Recent reviews have focused on mechanistic models of OMV production (Kulp et al., 2010, Bonnington et al., 2013, Klein et al., 2014, Kulkarni et al., 2014a, Haurat et al., 2015, Schwechheimer et al., 2015a), and therefore this topic is not the focus here. Overall, the process appears simple: OMVs form from where the outer membrane (OM) buds outward and these portions of the bacterial envelope are released in the form of membrane vesicles enclosing soluble components. However, many studies now support the concept that the process of OMV generation is controlled and regulated. In a variety of cases where OMVs have been carefully characterized, OMVs have been found to exhibit a) selective protein and lipid content compared to envelope, b) variance in the amount and content of OMV released depending on growth conditions, and c) gene-dependent variation in production levels and content (Bernadac et al., 1998, Berlanda Scorza et al., 2008, Bager et al., 2013, Bai et al., 2014, Elhenawy et al., 2014, Cahill et al., 2015, Kulp et al., 2015, Mantri et al., 2015, Elhenawy et al., 2016, Roier et al., 2016). This is in contrast to the generation of vesicles which result from the encapsulation of soluble material by lysed bacterial membranes whose function in biofilms has been described recently (Turnbull et al., 2016).
Many mechanistic questions remain regarding the OMV-mediated secretion process. For instance, what type of energy is used to generate scission? Is a specific, conserved (set of) protein(s) needed for this process? Is cargo incorporation an active process, or does cargo selectivity result from particular sites of the envelope that are more prone to budding as a result of heterogeneity in the membrane and/or envelope composition? Other types of vesicles and extensions of the envelope are noted to exist from Gram-negative bacteria, e.g. inner-outer membrane vesicles (I-OMVs), nanopods, nanotubes, and nanowires (Schwechheimer et al., 2015a) but these are not discussed further here. How these structures mechanistically and functionally relate to OMVs also remains unknown.
Current challenges and goals
Progress to establish commonalities in the OMV field is hampered due to the variety of analytical methods used to study OMV generation and composition. For instance, there are inconsistencies between studies in the quantification methods used to measure OMV production in mutants or under specific growth conditions. The quantitative methods include counting particles by particle tracking, FACS, confocal microscopy, or transmission electron microscopy techniques (Van Der Pol et al., 2010, Devos et al., 2015); measurement by absorbance of phospholipids in the sample after chloroform extraction (Stewart, 1980, Schertzer et al., 2010, Wessel et al., 2013); quantification of lipid content using lipid probes such as FM4–64 (McBroom et al., 2006); and quantification of protein content by SDS-PAGE staining and densitometry of outer membrane protein, or assays of total protein by Bradford reagent or bicinchoninic-acid assay (BCA) (Sharpe et al., 2011, Chen et al., 2016). Each of these methods has strengths and weaknesses. OMV counting is limited by particle size and the ability to distinguish between protein complexes and vesicles. Growth conditions and genetic mutations can change lipid to protein ratios and composition in the bacterial membrane, skewing lipid- or protein-only based quantitative comparisons of OMV production. Furthermore, samples may not consist of pure OMVs so the use of total protein quantification in samples is reliable only if OMV purity has been established. Depending on the preparative method or culture conditions, cell-free supernatants may be contaminated with protein from lysed cells, flagellin, cytoplasm, or cytoplasmic membrane that co-pellet with OMVs upon ultracentrifugation. Density gradient purification of OMVs reduces contaminants but also impacts quantitative yield, therefore it is most useful for quantitation as a means to identify which protein bands should be used in SDS-PAGE densitometry measurements of a less pure sample. Finally, OMV production data also must be normalized to the number of bacteria in the culture, either by utilizing optical density (OD) or colony forming units (CFU). A combination of quantitative techniques is often useful to establish OMV production. For instance, OMV lipid quantification can be coupled to measurements of outer membrane protein (OMP) by SDS-PAGE and densitometry, and normalized to bacterial counts (Chutkan et al., 2013, Schwechheimer et al., 2015b, Roier et al., 2016).
The lack of a universal methodology extends to studies of OMV composition. There is a growing need to establish a “baseline” OMV protein and lipid composition for a particular bacterial species grown in a specific growth phase and media conditions (Table 1). This would allow invaluable comparisons of OMV composition, function, and production from the same species in different media conditions, or of OMVs produced by different species in the same growth phase. Also, in order to identify OMV cargo enrichment and subsequently study the mechanics and functional consequences of cargo selectivity, OMV content must be compared with purified outer membrane (OM) and periplasmic fractions using quantitative proteomic and lipidomic methods, which are typically cost-prohibitive. However, with the current rate of technical progress, these goals are becoming increasingly realistic.
Table 1.
Summary of recent OMV proteomic analyses1
| Species | Size (nm)2 |
Growth Phase3 |
Medium | Proteome4 | Functional Analysis5 |
Immunological Activity Analysis6 |
Reference |
|---|---|---|---|---|---|---|---|
|
Acinetobacter baumanii Clinical isolate A38 |
30 – 140 |
early stationary |
LB broth | 148 | ✓ | ✓ | (Li et al., 2015) |
|
A. baumanii Clinical isolate 5806 |
138 | ✓ | ✓ | ||||
|
A. baumanii ATCC 19606 |
40 – 70 | log | LB broth | ND | ✓ | (Jun et al., 2013) | |
|
A. baumanii ATCC 17978 |
ND | in host | Mouse pneumonia model |
112 | ✓ | (Jin et al., 2011) | |
|
A.baumanii Clinical isolate DU202 |
20 – 160 |
early stationary |
LB broth | 132 | ✓ | (Kwon So, 2009) | |
|
A. radioresistens Clinical isolate MMC5 |
10 – 150 |
early stationary |
LB broth | 71 | ✓ | (Fulsundar et al., 2015) | |
|
Aggregatibacter Actinomycetemcom itans serotype e strain, 173 |
ND | NS | Blood agar plates |
151 | (Kieselbach et al., 2015) | ||
|
Bacteroides fragilis NCTC 9343 |
30 – 80 | stationary | Supplement ed BHI, or supplement ed basal medium |
115 | ✓ | (Elhenawy et al., 2014) | |
|
B thetaiotaomicron VPI-5482 |
ND | stationary | 21 | ||||
|
Campylobacter jejuni ATCC 700819 |
50 | mid log | BHI | 134 | (Jang et al., 2013) | ||
|
Escherichia coli MG1655 |
80 – 100 | stationary | LB broth | 316 | ✓ | (Kulkarni et al., 2015) | |
| E. coli Nissle 1917 | 20 – 60 | early log | LB broth | 192 | (Aguilera et al., 2014) | ||
| E. coli DH5α | 20 – 40 | stationary | LB broth | 141 | (Lee et al., 2007) | ||
| Delftia sp. Cs1–4* | ND | biofilm | Defined mineral salts medium |
19 | (Shetty et al., 2011) | ||
|
Francisella novicida ATCC 15482 |
43 – 125 |
stationary | 0.1%l- cysteine HCl BHI broth |
416 | ✓ | (Pierson et al., 2011) | |
|
F. philomiragia ATCC 25015 |
stationary | Chamberlai n’s medium |
238 | ✓ | |||
|
F. novicida ATCC 15482 |
50 – 300, OM tubes: 0.3–1.5 µm |
log, early stationary |
BHI | 99, 286 | ✓ | ✓ | (McCaig et al., 2013) |
|
Haemophilus influenzae Clinical isolate 86-028NP |
20 – 200 |
stationary | BHI | 142 | ✓ | (Sharpe et al., 2011) | |
|
H. influenza (several mutants) |
≤ 100 | late exponential |
BHI broth | 163 | ✓ | (Roier et al., 2016) | |
|
H. parasuis Nagasaki |
50 – 200 |
early stationary |
Supplement ed BHI (liquid and plate grown) |
86 (liquid), 250 (plate grown) |
✓ | (McCaig et al., 2016) | |
| H. parasuis D74 | 93 (liquid), 251 (plate grown) |
||||||
|
Helicobacter pylori |
50 – 110 |
early stationary |
Blood agar plates of Brucella agar, Brucella broth |
306 | ✓ | (Olofsson et al., 2010) | |
|
Klebsiella pneumoniae ATCC 13883 |
20 – 200 |
early stationary |
LB broth | 159 | ✓ | ✓ | (Lee et al., 2012) |
|
Mannheimia haemolytica serotype S1 strain 89010807N |
10 – 20 | early stationary |
BHI | 226 | ✓ | (Ayalew et al., 2013) | |
|
Moraxella catarrhalis |
50 – 150 |
stationary | BHI | 57 | ✓ | (Schaar et al., 2011) | |
|
Mycobacterium tuberculosis H37Rv |
20 – 250 |
early stationary |
Minimal media |
287 | (Lee et al., 2015) | ||
| M. bovis BCG | 40 – 250 |
10 days | Defined minimal media? |
66 | ✓ | ✓ | (Prados-Rosales et al., 2011) |
| M. smegmatis | 64 | ✓ | ✓ | ||||
|
Myxococcus xanthus DK1622 WT |
30 – 100 | 18 hrs in plates Vegetative / Development tal cells |
CTT medium |
107/124 | (Kahnt et al., 2010) | ||
| M. xanthus | ND | late log | DCY-rich medium |
75 | (Whitworth et al., 2015) | ||
|
Neisseria gonorrhoeae§ |
ND | late log | GCBL Medium+ |
168 | (Zielke et al., 2014) | ||
|
N. meningitidis (MC58csb− and 2120csc− strains) |
ND | 10 h | gonococci agar |
172 | (Lappann et al., 2013) | ||
|
Porphyromonas gingivalis W50 |
60 – 120 |
log | Supplement ed Tryptic soy- enriched BHI broth |
151 | (Veith et al., 2014) |
||
|
P. gingivalis ATCC 53978 |
|||||||
|
P. gingivalis 33277 |
ND | log | Tryptic soy broth |
67 | (Mantri et al., 2015) |
||
| P. gingivalis W83 | 70 | ||||||
|
Prochlorococcus MIT9313 |
70 – 100 |
mid/late log | Constant light flux conditions, Pro99 media |
27 | (Biller et al., 2014) | ||
|
Prochlorococcus MED4 |
ND | 40 | ✓ | ||||
|
Pseudomonas aeruginosa |
~<200 nm | in biofilm | LB broth | 76 | (Toyofuku et al., 2012) | ||
| stationary | 194 | ||||||
|
P. aeruginosa PAO1 |
ND | biofilm (24, 48, and 96 hrs) |
Tryptic soy broth |
18, 78, 98 | ✓ | (Park et al., 2015) | |
|
P. aeruginosa PAO1 |
50 – 250 |
early log | Lysogeny broth |
338 | (Choi et al., 2011) | ||
| P. putida KT2440 | 25 – 75 | log | LB broth | 243 | ✓ | (Choi et al., 2014) | |
| P. putida KT2440 | Minimal medium + 10 mM succinate |
359 | |||||
| P. putida KT2440 | Minimal medium + 5 mM benzoate |
456 | |||||
| P. syringae Lz4W | 60 – 100 |
stationary | Antarctic bacterial medium |
429 | ✓ | (Kulkarni et al., 2014b) | |
|
P. syringae pv. Tomato T1 |
120 – 125 |
early stationary |
King’s broth medium |
139 | (Chowdhury et al., 2013) | ||
|
Shewanella vesiculosa |
25 – 250 |
late log | Tryptic soy agar |
46 | (Perez-Cruz et al., 2013) | ||
|
Shigella flexneri 2a str |
50 – 200 |
log | Tryptic soy broth |
148 | (Chen et al., 2014) | ||
|
Strenotrophomona s maltophilia ATCC13637 |
ND | early stationary |
LB broth | 274 | (Ferrer-Navarro et al., 2016) | ||
|
S. maltophilia M30 |
ND | early stationary |
133 | ||||
|
S. maltophilia 44/98 (LMG 26824) |
NS | early stationary |
LB broth | 234 | (Devos et al., 2015) | ||
|
Vibrio cholerae biotype strain C6706 |
ND | OD600 = ~0.9 | Hepes- buffered LB |
90 | (Altindis et al., 2014) |
From publications in 2010–2016.
Diameter range or average.
Growth phase when OMVs were isolated for analysis.
Total proteins identified in sample, studies where only most abundant were identified are not included in this table.
One or more of the following: OMVs were found to have some activity via supplementation of OMVs to another culture; OMVs conferred some sort of protection (e.g. antibiotic tolerance); OMVs were tested for cytotoxicity.
OMVs elicited immune response (e.g. cytokine secretion) when incubated with mammalian cells.
Strain is flagella production mutant
See reference for details
Many OMV studies on this bacterium are focused on detergent-derived vesicles and not included in this table
ND, not determined; NS, not shown; LB, Luria-Bertani; BHI, Brain-heart infusion; DCY, Double casitone yeast
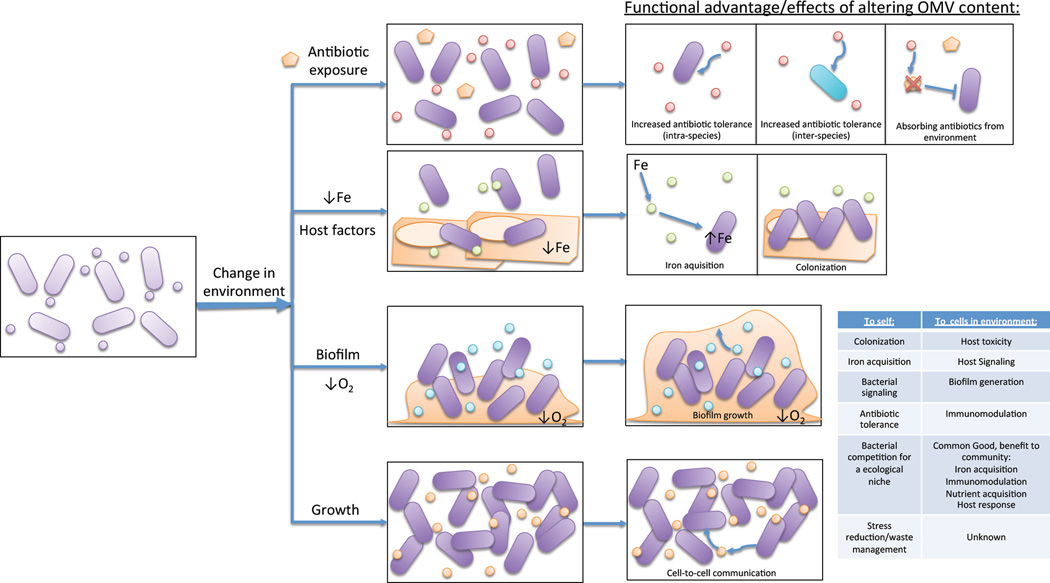
A major goal in the field is to elucidate how differing bacterial growth conditions impact OMV functional properties such as enabling viability, host cell adhesion, cytotoxicity, and inter-species communication. This review will exclusively focus on studies in which functional analyses of vesicles grown in specific conditions (e.g. a change in media or antibiotic exposure) revealed important roles for vesicles or vesicle secretion in that particular environment (Fig. 1, Table 2).
Figure 1. Environment shifts elicit changes in functional roles for OMVs.
Environment changes such as antibiotic exposure, release of host factors, low iron, transition to biofilm lifestyle, anaerobic conditions, and changes in growth phases can cause modulation of OMV production. In many cases, vesicles produced upon the shift in environmental conditions exhibit functional roles that can help the mother cell or other bystander cells adapt to the new environment, survive future stressors, and/or interact with host cells or co-inhabitant cells differently.
Table 2.
Environmental changes demonstrated to affect vesiculation (e.g. levels, activity or contents)
| Reference | |
|---|---|
| Media | (Bager et al., 2013, Choi et al., 2014) |
| Growth phase | (Tashiro et al., 2010) |
| Biofilm vs planktonic mode | (Grande et al., 2015, Park et al., 2015) |
| Temperature | (Macdonald et al., 2013, Fulsundar et al., 2014) |
| Host cell factor | (Ballok et al., 2014, Metruccio et al., 2016) |
| Antibiotic exposure | (Maredia et al., 2012, Macdonald et al., 2013, Devos et al., 2015, Devos et al., 2016) |
| Anoxic conditions | (Toyofuku et al., 2013) |
| Iron depletion | (Keenan et al., 2000) |
Environment controls OMV content and production
Achieving a simple definition of OMV-related activities, even for those derived from a single species, will be impossible without including a caveat for growth conditions. Growth phase, solid or liquid media, or the composition of the media, are well-known to affect virulence-associated bacterial traits such as adherence and toxin secretion, and impact membrane composition. Similarly, OMVs are complex entities composed of environmentally and temporally-controlled factors. Therefore, we can best focus on elucidating the functional importance of OMVs produced in a particular environment to bacteria growing in that setting. Here we highlight some recent examples demonstrating the environmental control of OMVs.
Iron availability
Iron is an essential element for bacterial survival, however, its bioavailability is strictly controlled within the host. Several reviews have highlighted the importance of iron acquisition during infection (Caza et al., 2013, Ferreira et al., 2016) and in light of recent evidence, OMVs might potentially serve as vehicles for iron uptake during host invasion.
Evidence for environmental influence over OMV production for M. tuberculosis comes from a study by Rosales et al. in which vesiculation was assayed in conditions of high and low iron concentration using minimal media (Prados-Rosales et al., 2014). While OMV size did not change significantly between both conditions, vesicle production was enhanced in iron-limiting conditions. Furthermore, OMVs from low iron conditions were enriched for the lipid siderophore mycobactin whereas its precursor, with lower iron binding affinity, was the main siderophore found in OMVs from high iron conditions. Therefore, iron deprivation stimulated the production of OMVs enriched in the siderophore mycobactin which was also able to deliver iron to iron deficient mutants of M. tuberculosis. The selective packaging of mycobactin in vesicles grown in iron limiting conditions and the ability of these vesicles to donate iron and support the growth of mutants deficient in iron uptake, suggest a crucial role for OMVs during stringent conditions.
In addition, many proteins associated with iron acquisition were found in biofilm OMVs of P. aeruginosa (Toyofuku et al., 2012). Given that iron limiting conditions are also known to inhibit biofilm formation (Singh et al., 2002, Cai et al., 2010), iron uptake mechanisms in the OMVs might also be important for the transition from a planktonic to a biofilm lifestyle, allowing the bacteria to thrive in this type of community.
In the case of H. pylori, it was observed that iron-limiting conditions also caused a marked increase in vesicle production and influenced the expression of the OMV-associated virulence factor VacA. Reduced levels of VacA were found in vesicles from iron limiting conditions and iron salt was able to restore this decrease (Keenan et al., 2000). Similarly, studies of OMV production by Haemophilus influenzae, E. coli, and Vibrio cholerae, found iron-limiting conditions corresponded to increased OMV production, and that a deletion in the iron-sensitive fur transcription factor also lead to increased OMV levels (Roier et al., 2016). In this case, it was found that the absence of the transcriptional activating activity of fur caused a decrease in iron-activated genes such as vacJ and yrbE which control phospholipid transport. Phospholipid and fatty acid analysis of the OMVs and OM from wildtype and mutants of these genes in H. influenzae suggested that vacJ-and yrbE-dependent changes in envelope lipid composition could be the cause of the observed iron-regulated OMV formation. Together, these findings point to a link between iron availability and modulation of both vesicle production and cargo.
Biofilm vs planktonic lifestyle
Many studies highlight differences in both planktonic and biofilm OMV proteomes of P. aeruginosa (Toyofuku et al., 2012, Park et al., 2015). In particular, a recent proteomic analysis revealed differences in the amount of the antibiotic secondary metabolite and pigment, pyocyanin, in OMVs over time (Park et al., 2015). Vesicles from late biofilm cultures (96 hrs) were found to contain higher levels of pyocyanin, in comparison to OMVs from early planktonic cultures (24 hrs). Given that in early planktonic cultures, supernatants had low levels of pyocyanin as did later biofilm supernatants, the authors argue OMVs are serving as a reservoir for pyocyanin in the context of biofilms. Consistent with this finding is the enrichment for phenazine biosynthetic proteins in early biofilms in comparison to planktonic cultures (Park et al., 2015). Although the quantification of pyocyanin in OMVs might point to a possible mechanism for the delivery of this toxin, it is yet to be determined whether pyocyanin within the vesicles can interact or inhibit growth of competing bacterial species.
In the case of H. pylori biofilms, OMVs seem to play more of a structural role. Extracellular DNA (eDNA) is a component of H. pylori biofilms and using PicoGreen staining, Grande et al. observed increased levels of double stranded DNA (dsDNA) associated with biofilm OMVs in comparison to planktonic OMVs (Grande et al., 2015). It is argued that the association of eDNA with vesicle membranes explain the protection observed for H. pylori biofilms treated with DNase I (Grande et al., 2011). Thus, as bacterial cells transition from a planktonic to a biofilm lifestyle, OMVs could not only have a role in toxin storage but also in protection of eDNA from degradation, maintaining the integrity of the extracellular matrix of the biofilm. It is important to mention that although many studies have detected DNA as well as RNA as a component of OMVs (Table 3), there is still a great deal of debate regarding how nucleic acids enter into OMVs, their association with OMVs, and their role in host-microbe interactions.
Table 3.
Recent studies where lipid analysis was performed on OMVs and/or DNA was detected in vesicle samples.
| Species | Lipid Analysis | DNA Presence | Reference |
|---|---|---|---|
|
Acinetobacter radioresistens MMC5 strain |
✓* | (Fulsundar et al., 2015) | |
| A. baylyi | ✓ | (Fulsundar et al., 2014) | |
| Ahrensia kielensis | ✓ | (Hagemann et al., 2013) | |
| Bacteroides fragilis | ✓ | (Elhenawy et al., 2014) | |
| B. thetaiotaomicron | ✓ | ||
| Delftia sp. Cs1–4 | ✓ | (Shetty et al., 2011, Shetty et al., 2014) | |
| Francisella novicida ATCC 15482 | ✓ | (Pierson et al., 2011) | |
| F. philomiragia ATCC 25015 | ✓ | ||
| Haemophilus influenzae | ✓ | (Sharpe et al., 2011) | |
|
Haemophilus influenzae (several mutants) |
✓✓ | (Roier et al., 2016) | |
| Helicobacter pylori | ✓✓ | (Olofsson et al., 2010) | |
| Kingella knigae | ✓✓ | (Maldonado et al., 2011) | |
| Moraxella catarrhalis | ✓✓ | (Schaar et al., 2011) | |
| Mycobacterium bovis BCG | ✓✓ | ✓✓ | (Prados-Rosales et al., 2011) |
| M. smegmatis | ✓✓ | ✓✓ | |
| Prochlorococcus sp. | ✓✓ | ✓✓ | (Biller et al., 2014) |
| Pseudoalteromonas marina | ✓✓ | (Hagemann et al., 2013) | |
| Pseudomonas aeruginosa | ✓✓ | (Tashiro et al., 2011) | |
| P. syringae Lz4W | ✓✓ | (Kulkarni et al., 2014b) | |
| P. syringae pv. Tomato T1 | ✓✓ | (Chowdhury et al., 2013) | |
| Porphyromonias gingivalis | ✓✓ | (Ho et al., 2015) | |
| Shewanella vesiculosa | 2713✓ | (Frias et al., 2010, Perez-Cruz et al., 2013) | |
| Shigella flexneri 2a str | (Chen et al., 2014) |
Only fatty acid analysis
Oxygen availability
Oxygen availability impacts bacterial physiology in biofilms as well as in liquid anaerobic conditions, and the ability of oxygen to modulate OMV production has been studied most extensively in P. aeruginosa. Most studies of OMV production in P. aeruginosa use aerobically grown cultures in which the Pseudomonas quinolone signal (PQS) which is identified to be involved in P. aeruginosa OMV production (Mashburn-Warren et al., 2008) is active. However, Toyofuku et al. focused on monitoring OMV production under denitrifying conditions (Toyofuku et al., 2013). P. aeruginosa undergo anaerobic respiration via denitrification, utilizing nitrates or oxidized forms of nitrogen (Arai, 2011, Arat et al., 2015). This is particularly relevant during biofilm growth, due to the generation of anaerobic microenvironments because of inherent oxygen gradients within the biofilm (Xu et al., 1998, Werner et al., 2004).
Firstly, the authors determined that P. aeruginosa is able to produce OMVs in the absence of oxygen, conditions lacking PQS. They further observed marked differences in OMV production under anoxic conditions in different media. While anoxic cultures of P. aeruginosa produced very few vesicles in brain heart infusion (BHI) medium, anoxic Luria Bertani (LB) broth-grown cultures produced up to six-fold more vesicles in comparison to the aerobic cultures (Toyofuku et al., 2013), as measured by FM4–64 dye incorporation. This reduction in vesicle production is consistent with a previous study that revealed reduced vesicle production under anaerobic conditions in BHI (Schertzer et al., 2010) despite the fact that the vesicle isolation and purification methods for these two studies were slightly different. When the same group examined the anaerobic OMV proteome, various pyocin-related proteins were found in the vesicles while pyocin structures were not observed. Pyocins are bacteriocins produced by P. aeruginosa which are toxic to other strains of this species. Additional experiments with mutants also uncovered a connection between OMVs and pyocin production under denitrifying conditions (Toyofuku et al., 2013). In the anaerobic conditions tested, nitric oxide (NO) was identified as an inducer of vesicle production given that a mutant for nitrite reductase (NIR) displayed a decrease in pyocin production as well as vesicle production. In accordance with these results, a microarray analysis by Chang et al. revealed oxidative stress induced pyocin genes in P. aeruginosa (Chang et al., 2005). Furthermore, treatment of P. aeruginosa with hydrogen peroxide has been shown to cause an increase in OMV production (Macdonald et al., 2013). These results are all consistent with the idea that oxygen availability modulates OMV production, however, it has yet to be determined whether the enrichment of pyocin synthesis proteins in OMVs provides P. aeruginosa vesicles with particular functions such as modulating stress resistance or pyocin production.
SOS response and antibiotic exposure
Bacteria encounter antibiotics in their native as well as human host environments, and antibiotics often result in activation of stress responses which can be tied to OMV production and function. A recent study by Maredia et al, revealed that the SOS response in P. aeruginosa increases OMV production (Maredia et al., 2012). A wildtype strain treated with ciprofloxacin to induce the SOS response, experienced a 100-fold increase in OMVs, as measured by Bradford assay, in comparison to untreated cells. Tests with a LexA (SOS repressor) noncleavable strain indicated that both the SOS response and the antibiotic itself contributed to the increase in OMV levels. To exclude the possibility of co-precipitated proteins in the OMV samples, Maredia et al. also performed a lipid analysis which revealed that ciprofloxacin treated wildtype OMVs were heavier than those of the lexAN mutant strain (Maredia et al., 2012). Moreover, a proteomic analysis identified SOS-regulated proteins in OMVs of antibiotic-treated cells. The authors suggest then that in the presence of antibiotics, OMV production is enhanced and SOS plays a role in such a mechanism.
Whether ciprofloxacin-induced OMVs improve bacterial viability or resistance in the presence of the antibiotics was not studied by Mareida et al, however another study demonstrated that OMVs can improve survival by absorbing antibiotics in the environment (Manning et al., 2011). A hypervesiculating mutant strain grew better than wildtype when treated with cyclic cationic antimicrobial peptides (AMPs) polymyxin B and colistin, but not when treated with antibiotics that target peptidoglycan synthesis and protein synthesis. Therefore, hypervesiculation was found to be protective against antibiotics that more specifically target the outer membrane. Furthermore, treatment of wildtype cultures with polymyxin B or colistin in the presence of purified OMVs resulted in increased survival in comparison to cultures without OMVs. Once again, OMVs were not protective against ampicillin, ceftriaxone, or tetracycline. In the same study, purified OMVs from an enterotoxigenic strain of E. coli (ETEC) or from K12 increased growth of ETEC in the presence of polymyxin B (Manning et al., 2011). This is consistent with a model in which antibiotic exposure induces the production of vesicles that interact with the stressor molecules and confer protection, increasing survival.
In a consequent set of studies, treatment of S. maltophilia with the β-lactam antibiotic imipenem was found to not only increase in OMV production, but also, analysis of the OMV proteome demonstrated that the vesicles contained β-lactamase (Devos et al., 2015, Devos et al., 2016). The presence of this molecule within the OMVs points to a protective role for vesicles when cells are faced with antibiotic stress. Whether the enzyme is selectively enriched in these OMVs or whether its increased abundance results from the bulk-flow incorporation of an increased amount of enzyme in the periplasm due to stress, is unknown.
The β-lactamase-containing OMVs observed by Devos et al. were further shown to be capable of improving bacterial survival during antibiotic exposure (Devos et al., 2016). Ampicillin-related vesicle functions have been studied for several other bacteria and are also consistent with their ability to confer protection to self or bystander bacteria. In the case of S. aureus, addition of β-lactamase-containing vesicles from this Gram-positive bacterium mediated survival of ampicillin-susceptible E. coli DH5α and S. aureus strains in the presence of ampicillin. The same effect was observed for S. enterica serovar enteridis, E. coli O157:H7 and S. epidermis ATCC 12228 on LB supplemented with ampicillin (Lee et al., 2013). Notably, vesicle-mediated resistance was antibiotic-specific. Only when OMV “donor” E. coli DH5α cells were transformed with a plasmid encoding β-lactamase, were the OMVs able to mediate the survival of an E. coli DH5α ampicillin-susceptible strain (Lee et al., 2013).
Recently, the Devos group also treated bacterial cultures with penicillin G to induce the packaging of β-lactamase into S. maltophilia vesicles (Devos et al., 2016). OMVs from these stimulated cultures where then added to cultures of P. aeruginosa, and B. cenocepacia, bacterial species known to cohabitate with S. maltophilia. For both species, OMVs caused a 100-fold increase in the minimum inhibitory concentrations for other β-lactam antibiotics (imipenem, amoxicillin, and ticarcilin) (Devos et al., 2016). These results demonstrate that OMVs of resistant bacteria exposed to antibiotics can package active antibiotic-degrading enzymes, and reduce the antibiotic susceptibility of bystander pathogens.
Host factors
During invasion and colonization, bacteria encounter a variety of host factors that can potentially affect OMV production as well as content and cytotoxicity. For instance, CFTR inhibitory factor (Cif) is commonly found in the P. aeruginosa OMV proteome (Bomberger et al., 2009). Cif is an epoxide hydrolase enzyme, capable of altering the recycling of CFTR, a mammalian ABC transporter relevant to the development of cystic fibrosis (Bomberger et al., 2011). It has been previously established that this virulence factor is regulated by direct binding and repression by CifR, an epoxide-responsive repressor (Ballok et al., 2012). Epoxides are toxic compounds secreted as by-products of eukaryotic metabolism with roles in cellular signaling. Notably, human-derived epoxides are produced by leukocytes in the lung, a niche for P. aeruginosa during infection (Hayakawa et al., 1986, Gómez et al., 2007). Given that Cif is known to be packaged into OMVs of P. aeruginosa, Ballock et al. set to examine whether epoxides mediate the packaging of Cif into vesicles. Treatment with a model epoxide, epibromohydrin (EBH) induced changes in OMV shape and density, however, more interestingly, it altered the packaging of proteins into the vesicles, supporting the idea of selective sorting of cargo into OMVs. Furthermore, additional experiments revealed that vesicles formed in the presence of EBH had reduced epithelial cell cytotoxicity. Therefore, during host invasion, exposure to human-derived epoxides could cause significant changes to bacterial vesicles that can influence host-pathogen interactions.
Besides the lungs, P. aeruginosa can colonize humans in other sites and is known to be the most common cause of corneal infection in contact-lens wearers (Stapleton et al., 1995). However, the mechanism of infection has not yet been fully elucidated (Tam et al., 2010, Evans et al., 2013). Given differences in models of bacterial infection in the presence and absence of contact lenses, several studies have indicated that bacteria can release additional virulence factors in the eye-lens environment (Tam et al., 2010). In experiments with P. aeruginosa exposed to human tear fluid, Metruccio et al. revealed that OMV production is induced in the presence of both solutions in comparison to buffer only and epithelial cell lysates controls (Metruccio et al., 2016). As abundant components of human tear fluid, lysozyme and lactoferrin were tested, and, lysozyme specifically, was found to trigger OMV production to levels comparable to tear fluid. OMVs from the lysozyme-induced conditions also had a similar protein-banding pattern as human tear-induced vesicles (Metruccio et al., 2016). In lactate dehydrogenase (LDH) release assays to measure host cell lysis, these lysozyme-induced OMVs induced cytotoxicity of corneal epithelial cells to a greater degree than vesicles from uninduced conditions. These cytotoxicity results were consistent with in an in vivo model of mouse corneas. In the same in vivo study, recruitment of Ly6G/C myeloid cells was also observed as a result of the vesicle treatment. Furthermore, intact lysozyme-induced vesicles promoted association of P. aeruginosa to mouse corneas in an ex vivo model (Metruccio et al., 2016). These results suggest that the ocular environment can modulate the release of OMVs by P. aeruginosa during a corneal infection and affect cell adhesion as well as cytotoxicity towards host cells.
Considering that Gram-negative bacteria can alter the structural composition of LPS present in their OM in response to various environmental stimuli, a system was developed to track the native dynamics of lipid A change in Salmonella enterica serovar Typhimurium following environmental shift to PhoP/Q- and PmrA/B-inducing conditions which mimic effects of host cell infection and colonization (Bonnington et al., 2016). Growth conditions altering pH and Mg concentrations influenced OMV production, size, and lipid A content. Interestingly, lipid A content of OMVs did not fit with a stochastic model of content selection, revealing the significant OM retention as well as OMV enrichment of lipid A species in the different host-like conditions. The concept that OMV composition and production reflects changes occurring in the OM during host-induced OM remodeling was also studied by an investigation of the effect of overexpression of PagL, a PhoP/Q controlled OM-localized lipid A deacylase in S. Typhimurium (Elhenawy et al., 2016). They found that deacylated lipid A was found exclusively in the OMVs, not the OM, of cells overexpressing PagL. This result suggests that PagL activity is extremely localized to small patches of the OM, or that PagL is active only after OMVs are formed. The former possibility is supported by the fact that PagL overexpression also corresponded to increased levels of OMVs, and these data lead the authors to suggest that the deacylated LPS alters the OM geometry to promote budding.
Growth phase and media composition
Several studies have examined changes in OMV production and cargo under various growth conditions in addition to altering iron concentrations which were discussed in an earlier section. Whereas data from OMVs produced by the P. putida DOT-T1E strain in particular, revealed few changes to the OMV proteome under varying conditions (Baumgarten et al., 2012a, Baumgarten et al., 2012b), studies of other strains and bacterial species have demonstrated media- and growth phase-dependent changes in OMV proteomes and production. For example, a study by Choi et al. on the proteome of P. putida KT2440 (See Table 1), assessed OMV production of cells grown in different media: LB broth, Minimal medium with 10 mM succinate, or minimal medium with 5 mM benzoate. OMV production in LB media was found to be more than three-fold increased than in the other two conditions. Interestingly, OMVs from cells grown in benzoate-containing media contained proteins involved in benzoate degradation. However, it was not determined here whether the vesicles from the different conditions conferred some sort of protection against stress-inducing conditions. On the other hand, they did observe that OMVs derived from LB-rich media decreased cell viability of A549 carcinomic human alveolar basal epithelial cells, but it was not determined whether vesicles from the other two conditions are also able to confer cytotoxicity (Choi et al., 2014). In a study of G. anatis, changes in medium composition also affected vesicle production. Addition of 1 mM EDTA to BHI medium or growth in RPMI 1640 HEPES medium caused a reduction in OMV production in comparison to growth in BHI (Bager et al., 2013). Again, it remains unknown whether the reduction in OMV production is a functional advantage to cells grown in those specific conditions.
The impact of growth conditions on OMV production and function should also be thought of in a temporal context, as bacteria alter their composition throughout their cell cycle, and OMVs are likely to be similarly affected. While OMVs are produced at all stages of bacterial growth, the maximum rate of vesicle production has been known to occur at the end of log phase for various species of bacteria (Chatterjee et al., 1967, Hoekstra et al., 1976, Gamazo et al., 1987). Interestingly, P. aeruginosa vesicles from different growth phases also exhibit altered physical properties. For instance, vesicles from stationary phase cultures contained higher levels of PQS and were found to be more negatively charged in comparison to OMVs from the exponential phase (Tashiro et al., 2010). Also, based on the idea that vesicles may potentially play a role in cell to cell communication (Kadurugamuwa et al., 1995), the same study employed FTIC labeled vesicles to test whether growth phase impacted association of vesicles with P. aeruginosa cells and observed that OMVs from stationary cells more readily associated with the bacterial cells. This could be explained by the fact that more negatively-charged vesicles have an increased potential to form salt-bridges to the Ca2+ or Mg2+ surface of bacteria (Tashiro et al., 2010). Therefore, as bacteria transition between growth phase, vesicles can acquire inherently different properties that may influence their interactions with neighboring cells and this might be crucial for cell-cell communication.
Concluding remarks
The process of OMV biogenesis has gained much recent attention given the potential of vesicles for vaccine development and their role in bacterial pathogenesis (Baker et al., 2014, Toyofuku et al., 2015, van der Pol et al., 2015). Consequently, many studies have focused on vesicle engineering to design OMVs suited for drug and antigen delivery while others have characterized vesicle protein, lipid, and nucleic acid composition (See Tables 1 and 3) in order to better understand bacterial physiology and improve on current antimicrobial therapeutic approaches. While there has been a substantial and increasing number of proteomic studies of bacterial OMVs, there is a concomitant need to characterize the function of vesicles produced by bacteria grown under various physiologically-relevant conditions. It is well established in biology that environmental changes affect bacterial gene expression, metabolism, and composition, allowing adaptation to new conditions. The studies highlighted in this review indicate that such environmental conditions similarly affect not only the process of vesiculation, both the amount of vesicles and their composition, but also the specific functional roles for the vesicles (Fig 1). Uncovering functions for vesicles produced under specific conditions will not only help us understand how bacteria thrive in different environments, but will also allow us to define specific conditions in which vesicles play a crucial role in bacterial survival, further defining their role in bacterial pathogenesis.
Acknowledgments
The authors acknowledge research support from NIH grant R01-GM099471 and are indebted to Carmen Schwechheimer for assembling data for Tables 1 and 3.
References
- Aguilera L, Toloza L, Giménez R, Odena A, Oliveira E, Aguilar J, et al. Proteomic analysis of outer membrane vesicles from the probiotic strain Escherichia coli Nissle 1917. Proteomics. 2014;14:222–229. doi: 10.1002/pmic.201300328. [DOI] [PubMed] [Google Scholar]
- Altindis E, Fu Y, Mekalanos JJ. Proteomic analysis of Vibrio cholerae outer membrane vesicles. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E1548–E1556. doi: 10.1073/pnas.1403683111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai H. Regulation and function of versatile aerobic and anaerobic respiratory metabolism in Pseudomonas aeruginosa. Frontiers in Microbiology. 2011;2 doi: 10.3389/fmicb.2011.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arat S, Bullerjahn GS, Laubenbacher R. A network biology approach to denitrification in Pseudomonas aeruginosa. PLos One. 2015;10 doi: 10.1371/journal.pone.0118235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayalew S, Confer AW, Shrestha B, Wilson AE, Montelongo M. Proteomic analysis and immunogenicity of Mannheimia haemolytica vesicles. Clin Vaccine Immunol. 2013;20:191–196. doi: 10.1128/CVI.00622-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bager RJ, Persson G, Nesta B, Soriani M, Serino L, Jeppsson M, et al. Outer membrane vesicles reflect environmental cues in Gallibacterium anatis. Veterinary microbiology. 2013;167:565–572. doi: 10.1016/j.vetmic.2013.09.005. [DOI] [PubMed] [Google Scholar]
- Bai J, Kim SI, Ryu S, Yoon H. Identification and characterization of outer membrane vesicle-associated proteins in Salmonella enterica serovar Typhimurium. Infection and immunity. 2014;82:4001–4010. doi: 10.1128/IAI.01416-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker JL, Chen L, Rosenthal JA, Putnam D, DeLisa MP. Microbial biosynthesis of designer outer membrane vesicles. Current opinion in biotechnology. 2014;29:76–84. doi: 10.1016/j.copbio.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballok AE, Bahl CD, Dolben EL, Lindsay AK, St. Laurent JD, Hogan DA, et al. Epoxide-Mediated CifR Repression of cif Gene Expression Utilizes Two Binding Sites in Pseudomonas aeruginosa. Journal of Bacteriology. 2012;194:5315–5324. doi: 10.1128/JB.00984-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballok AE, Filkins LM, Bomberger JM, Stanton BA, O’Toole GA. Epoxide-Mediated Differential Packaging of Cif and Other Virulence Factors into Outer Membrane Vesicles. Journal of Bacteriology. 2014;196:3633–3642. doi: 10.1128/JB.01760-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgarten T, Sperling S, Seifert J, von Bergen M, Steiniger F, Wick LY, Heipieper HJ. Membrane Vesicle Formation as a Multiple-Stress Response Mechanism Enhances Pseudomonas putida DOT-T1E Cell Surface Hydrophobicity and Biofilm Formation. Applied and Environmental Microbiology. 2012a;78:6217–6224. doi: 10.1128/AEM.01525-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgarten T, Vazquez J, Bastisch C, Veron W, Feuilloley MGJ, Nietzsche S, et al. Alkanols and chlorophenols cause different physiological adaptive responses on the level of cell surface properties and membrane vesicle formation in Pseudomonas putida DOT-T1E. Applied Microbiology and Biotechnology. 2012b;93:837–845. doi: 10.1007/s00253-011-3442-9. [DOI] [PubMed] [Google Scholar]
- Berlanda Scorza F, Doro F, Rodríguez-Ortega MJ, Stella M, Liberatori S, Taddei AR, et al. Proteomics characterization of outer membrane vesicles from the extraintestinal pathogenic Escherichia coli DeltatolR IHE3034 mutant. Molecular & cellular proteomics : MCP. 2008;7:473–485. doi: 10.1074/mcp.M700295-MCP200. [DOI] [PubMed] [Google Scholar]
- Bernadac A, Gavioli M Fau -Lazzaroni JC, Lazzaroni Jc Fau -Raina S, Raina S Fau -Lloubes R, Lloubes R. Escherichia coli tol-pal mutants form outer membrane vesicles. Journal of bacteriology. 1998;180:4872–4878. doi: 10.1128/jb.180.18.4872-4878.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biller SJ, Schubotz F, Roggensack SE, Thompson AW, Summons RE, Chisholm SW. Bacterial vesicles in marine ecosystems. Science. 2014;343:183–186. doi: 10.1126/science.1243457. [DOI] [PubMed] [Google Scholar]
- Bomberger JM, Maceachran DP, Coutermarsh BA, Ye S, O’Toole GA, Stanton BA. Long-distance delivery of bacterial virulence factors by Pseudomonas aeruginosa outer membrane vesicles. PLoS pathogens. 2009;5:e1000382–e1000382. doi: 10.1371/journal.ppat.1000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomberger JM, Ye S, Maceachran DP, Koeppen K, Barnaby RL, O’Toole GA, Stanton BA. A Pseudomonas aeruginosa toxin that hijacks the host ubiquitin proteolytic system. PLoS pathogens. 2011;7:e1001325. doi: 10.1371/journal.ppat.1001325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnington KE, Kuehn MJ. Protein selection and export via outer membrane vesicles. Biochim Biophys Acta. 2013;1873 doi: 10.1016/j.bbamcr.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnington KE, Kuehn MJ. Outer membrane vesicle production facilitates LPS remodeling and outer membrane maintenance in Salmonella during environmental transitions. mBio. 2016 doi: 10.1128/mBio.01532-16. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill BK, Seeley KW, Gutel D, Ellis TN. Klebsiella pneumoniae O antigen loss alters the outer membrane protein composition and the selective packaging of proteins into secreted outer membrane vesicles. Microbiol Res. 2015;180:1–10. doi: 10.1016/j.micres.2015.06.012. [DOI] [PubMed] [Google Scholar]
- Cai Y, Wang R, An M-M, Liang B-B. Iron-Depletion prevents biofilm formation in Pseudomonas Aeruginosa through twitching mobility and quorum sensing. Brazilian Journal of Microbiology. 2010;41:37–41. doi: 10.1590/S1517-83822010000100008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caza M, Kronstad JW. Shared and distinct mechanisms of iron acquisition by bacterial and fungal pathogens of humans. Front Cell Infect Microbiol. 2013;3:80. doi: 10.3389/fcimb.2013.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W, Small DA, Toghrol F, Bentley WE. Microarray analysis of Pseudomonas aeruginosa reveals induction of pyocin genes in response to hydrogen peroxide. BMC Genomics. 2005;6:115. doi: 10.1186/1471-2164-6-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee SN, Das J. Electron microscopic observations on the excretion of cell-wall material by Vibrio cholerae. J Gen Microbiol. 1967;49:1–11. doi: 10.1099/00221287-49-1-1. [DOI] [PubMed] [Google Scholar]
- Chen L, Valentine JL, Huang C-J, Endicott CE, Moeller TD, Rasmussen JA, et al. Outer membrane vesicles displaying engineered glycotopes elicit protective antibodies. Proceedings of the National Academy of Sciences. 2016;113:E3609–E3618. doi: 10.1073/pnas.1518311113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Liu L, Fu H, Wei C, Jin Q. Comparative proteomic analysis of outer membrane vesicles from Shigella flexneri under different culture conditions. Biochem Biophys Res Commun. 2014;453:696–702. doi: 10.1016/j.bbrc.2014.09.142. [DOI] [PubMed] [Google Scholar]
- Choi C-W, Park EC, Yun SH, Lee S-Y, Lee YG, Hong Y, et al. Proteomic characterization of the outer membrane vesicle of Pseudomonas putida KT2440. Journal of proteome research. 2014;13:4298–4309. doi: 10.1021/pr500411d. [DOI] [PubMed] [Google Scholar]
- Choi DS, Kim DK, Choi SJ, Lee J, Choi JP, Rho S, et al. Proteomic analysis of outer membrane vesicles derived from Pseudomonas aeruginosa. Proteomics. 2011;11:3424–3429. doi: 10.1002/pmic.201000212. [DOI] [PubMed] [Google Scholar]
- Chowdhury C, Jagannadham MV. Virulence factors are released in association with outer membrane vesicles of Pseudomonas syringae pv. tomato T1 during normal growth. Biochim Biophys Acta. 2013;1834:231–239. doi: 10.1016/j.bbapap.2012.09.015. [DOI] [PubMed] [Google Scholar]
- Chutkan H, Macdonald I, Manning A, Kuehn MJ. Quantitative and qualitative preparations of bacterial outer membrane vesicles. Methods Mol Biol. 2013;966:259–272. doi: 10.1007/978-1-62703-245-2_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos S, Stremersch S, Raemdonck K, Braeckmans K, Devreese B. Intra- and Interspecies Effects of Outer Membrane Vesicles from Stenotrophomonas maltophilia on beta-Lactam Resistance. Antimicrobial Agents and Chemotherapy. 2016;60:2516–2518. doi: 10.1128/AAC.02171-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos S, Van Oudenhove L, Stremersch S, Van Putte W, De Rycke R, Van Driessche G, et al. The effect of imipenem and diffusible signaling factors on the secretion of outer membrane vesicles and associated Ax21 proteins in Stenotrophomonas maltophilia. Frontiers in microbiology. 2015;6:298–298. doi: 10.3389/fmicb.2015.00298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhenawy W, Bording-Jorgensen M, Valguarnera E, Haurat MF, Wine E, Feldman MF. LPS Remodeling Triggers Formation of Outer Membrane Vesicles in Salmonella. MBio. 2016;7 doi: 10.1128/mBio.00940-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhenawy W, Debelyy MO, Feldman MF. Preferential packing of acidic glycosidases and proteases into bacteroides outer membrane vesicles. MBio. 2014;5 doi: 10.1128/mBio.00909-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans DJ, Fleiszig SMJ. Why Does the Healthy Cornea Resist Pseudomonas aeruginosa Infection? American journal of ophthalmology. 2013;155:961–970. doi: 10.1016/j.ajo.2013.03.001. e962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira D, Seca AMLCGAD, Silva AMS. Targeting human pathogenic bacteria by siderophores: A proteomics review. J Proteomics. 2016;S1874–3919:30128–30122. doi: 10.1016/j.jprot.2016.04.006. [DOI] [PubMed] [Google Scholar]
- Ferrer-Navarro M, Torrent G, Mongiardini E, Conchillo-Solé O, Gibert I, Daura X. Proteomic analysis of outer membrane proteins and vesicles of a clinical isolate and a collection strain of Stenotrophomonas maltophilia. Journal of proteomics. 2016;142:122–129. doi: 10.1016/j.jprot.2016.05.001. [DOI] [PubMed] [Google Scholar]
- Frias A, Manresa A, de Oliveira E, Lopez-Iglesias C, Mercade E. Membrane vesicles: a common feature in the extracellular matter of cold-adapted antarctic bacteria. Microb Ecol. 2010;59:476–486. doi: 10.1007/s00248-009-9622-9. [DOI] [PubMed] [Google Scholar]
- Fulsundar S, Harms K, Flaten GE, Johnsen PJ, Chopade BA, Nielsen KM. Gene transfer potential of outer membrane vesicles of Acinetobacter baylyi and effects of stress on vesiculation. Applied and environmental microbiology. 2014;80:3469–3483. doi: 10.1128/AEM.04248-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulsundar S, Kulkarni HM, Jagannadham MV, Nair R, Keerthi S, Sant P, et al. Molecular characterization of outer membrane vesicles released from Acinetobacter radioresistens and their potential roles in pathogenesis. Microbial pathogenesis. 2015;83–84:12–22. doi: 10.1016/j.micpath.2015.04.005. [DOI] [PubMed] [Google Scholar]
- Gamazo C, Moriyón I. Release of outer membrane fragments by exponentially growing Brucella melitensis cells. Infection and Immunity. 1987;55:609–615. doi: 10.1128/iai.55.3.609-615.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez MI, Prince A. Opportunistic infections in lung disease: Pseudomonas infections in cystic fibrosis. Curr Opin Pharmacol. 2007;7:244–251. doi: 10.1016/j.coph.2006.12.005. [DOI] [PubMed] [Google Scholar]
- Grande R, Di Giulio M, Bessa LJ, Di Campli E, Baffoni M, Guarnieri S, Cellini L. Extracellular DNA in Helicobacter pylori biofilm: a backstairs rumour. J Appl Microbiol. 2011;110:490–498. doi: 10.1111/j.1365-2672.2010.04911.x. [DOI] [PubMed] [Google Scholar]
- Grande R, Di Marcantonio MC, Robuffo I, Pompilio A, Celia C, Di Marzio L, et al. Helicobacter pylori ATCC 43629/NCTC 11639 Outer Membrane Vesicles (OMVs) from Biofilm and Planktonic Phase Associated with Extracellular DNA (eDNA) Front Microbiol. 2015;6:1369. doi: 10.3389/fmicb.2015.01369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemann SJ. DNA-bearing membrane vesicles produced by Ahrensia kielensis and Pseudoalteromonas marina. Microbiol. 2013 doi: 10.1002/jobm.201300376. [DOI] [PubMed] [Google Scholar]
- Haurat MF, Elhenawy W, Feldman MF. Prokaryotic membrane vesicles: new insights on biogenesis and biological roles. Biological Chemistry. 2015;396:95–109. doi: 10.1515/hsz-2014-0183. [DOI] [PubMed] [Google Scholar]
- Hayakawa M, Sugiyama S, Takamura T, Yokoo K, Iwata M, Suzuki K, et al. Neutrophils biosynthesize leukotoxin, 9, 10-epoxy-12-octadecenoate. Biochem Biophys Res Commun. 1986;137:424–430. doi: 10.1016/0006-291x(86)91227-1. [DOI] [PubMed] [Google Scholar]
- Ho M-H, Chen C-H, Goodwin JS, Wang B-Y, Xie H. Functional Advantages of Porphyromonas gingivalis Vesicles. PLoS ONE. 2015;10:e0123448. doi: 10.1371/journal.pone.0123448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra D, van der Laan JW, de Leij L, Witholt B. Release of outer membrane fragments from normally growing Escherichia coli. Biochimica et biophysica acta. 1976;455:889–899. doi: 10.1016/0005-2736(76)90058-4. [DOI] [PubMed] [Google Scholar]
- Jang KS, Sweredoski MJ, Graham RL, Hess S, Clemons WM., Jr Comprehensive proteomic profiling of outer membrane vesicles from Campylobacter jejuni. J Proteomics. 2013;98:90–98. doi: 10.1016/j.jprot.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin JS, Moon DC, Choi C-W, Kim S-I. A Pathogenic Potential of Acinetobacter baumannii-Derived Membrane Vesicles. Journal of Analytical Science & Technology. 2011 [Google Scholar]
- Jun SH, Lee JH, Kim BR, Kim SI, Park TI, Lee JC, Lee YC. Acinetobacter baumannii outer membrane vesicles elicit a potent innate immune response via membrane proteins. PloS one. 2013;8:e71751–e71751. doi: 10.1371/journal.pone.0071751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadurugamuwa JL, Beveridge TJ. Virulence factors are released from Pseudomonas aeruginosa in association with membrane vesicles during normal growth and exposure to gentamicin: a novel mechanism of enzyme secretion. Journal of bacteriology. 1995;177:3998–4008. doi: 10.1128/jb.177.14.3998-4008.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahnt J, Aguiluz K, Koch J, Treuner-Lange A, Konovalova A, Huntley S, et al. Profiling the outer membrane proteome during growth and development of the social bacterium Myxococcus xanthus by selective biotinylation and analyses of outer membrane vesicles. Journal of proteome research. 2010;9:5197–5208. doi: 10.1021/pr1004983. [DOI] [PubMed] [Google Scholar]
- Keenan JI, Allardyce RA. Iron influences the expression of Helicobacter pylori outer membrane vesicle-associated virulence factors. Eur J Gastroenterol Hepatol. 2000;12:1267–1273. doi: 10.1097/00042737-200012120-00002. [DOI] [PubMed] [Google Scholar]
- Kieselbach T, Zijnge V, Granström E, Oscarsson J. Proteomics of Aggregatibacter actinomycetemcomitans Outer Membrane Vesicles. PLoS ONE. 2015;10:e0138591. doi: 10.1371/journal.pone.0138591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein G, Kobylak N, Lindner B, Stupak A, Raina S. Assembly of lipopolysaccharide in Escherichia coli requires the essential LapB heat shock protein. The Journal of biological chemistry. 2014;289:14829–14853. doi: 10.1074/jbc.M113.539494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni HM, Jagannadham MV. Biogenesis and multifaceted roles of outer membrane vesicles from Gram-negative bacteria. Microbiology. 2014a;1++0:2109–2121. doi: 10.1099/mic.0.079400-0. [DOI] [PubMed] [Google Scholar]
- Kulkarni HM, Nagaraj R, Jagannadham MV. Protective role of E. coli outer membrane vesicles against antibiotics. Microbiological research. 2015;181:1–7. doi: 10.1016/j.micres.2015.07.008. [DOI] [PubMed] [Google Scholar]
- Kulkarni HM, Swamy Ch , Jagannadham MV. Molecular Characterization and Functional Analysis of Outer Membrane Vesicles from the Antarctic Bacterium Pseudomonas syringae Suggest a Possible Response to Environmental Conditions. J Proteome Res. 2014b;13:1345–1358. doi: 10.1021/pr4009223. [DOI] [PubMed] [Google Scholar]
- Kulp A, Kuehn MJ. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annual review of microbiology. 2010;64:163–184. doi: 10.1146/annurev.micro.091208.073413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulp AJ, Sun B, Ai T, Manning AJ, Orench-Rivera N, Schmid AK, Kuehn MJ. Genome-Wide Assessment of Outer Membrane Vesicle Production in Escherichia coli. PLoS One. 2015;10:e0139200. doi: 10.1371/journal.pone.0139200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon SoGYSLJCKSI. Proteome analysis of outer membrane vesicles from a clinical Acinetobacter baumannii isolate. FEMS Microbiol Lett. 2009;297:150–156. doi: 10.1111/j.1574-6968.2009.01669.x. [DOI] [PubMed] [Google Scholar]
- Lappann M, Otto A, Becher D, Vogel U. Comparative proteome analysis of spontaneous outer membrane vesicles and purified outer membranes of Neisseria meningitidis. Journal of bacteriology. 2013;195:4425–4435. doi: 10.1128/JB.00625-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E-Y, Bang JY, Park GW, Choi D-S, Kang JS, Kim H-J, et al. Global proteomic profiling of native outer membrane vesicles derived from Escherichia coli. Proteomics. 2007;7:3143–3153. doi: 10.1002/pmic.200700196. [DOI] [PubMed] [Google Scholar]
- Lee J, Kim S-H, Choi D-S, Lee JS, Kim D-K, Go G, et al. Proteomic analysis of extracellular vesicles derived from Mycobacterium tuberculosis. Proteomics. 2015;15:3331–3337. doi: 10.1002/pmic.201500037. [DOI] [PubMed] [Google Scholar]
- Lee J, Lee E-Y, Kim S-H, Kim D-K, Park K-S, Kim KP, et al. Staphylococcus aureus extracellular vesicles carry biologically active β-lactamase. Antimicrobial agents and chemotherapy. 2013;57:2589–2595. doi: 10.1128/AAC.00522-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JC, Lee EJ, Lee JH, Jun SH, Choi CW, Kim SI, et al. Klebsiella pneumoniae secretes outer membrane vesicles that induce the innate immune response. FEMS Microbiol Lett. 2012;331:17–24. doi: 10.1111/j.1574-6968.2012.02549.x. [DOI] [PubMed] [Google Scholar]
- Li Z-T, Zhang R-L, Bi X-G, Xu L, Fan M, Xie D, et al. Outer membrane vesicles isolated from two clinical Acinetobacter baumannii strains exhibit different toxicity and proteome characteristics. Microbial pathogenesis. 2015;81:46–52. doi: 10.1016/j.micpath.2015.03.009. [DOI] [PubMed] [Google Scholar]
- Macdonald IA, Kuehn MJ. Stress-induced outer membrane vesicle production by Pseudomonas aeruginosa. Journal of bacteriology. 2013;195:2971–2981. doi: 10.1128/JB.02267-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado R, Wei R, Kachlany SC, Kazi M, Balashova NV. Cytotoxic effects of Kingella kingae outer membrane vesicles on human cells. Microb Pathog. 2011;51:22–30. doi: 10.1016/j.micpath.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning AJ, Kuehn MJ. Contribution of bacterial outer membrane vesicles to innate bacterial defense. BMC Microbiology. 2011;258 doi: 10.1186/1471-2180-11-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantri CK, Chen C-H, Dong X, Goodwin JS, Pratap S, Paromov V, Xie H. Fimbriae-mediated outer membrane vesicle production and invasion of Porphyromonas gingivalis. MicrobiologyOpen. 2015;4:53–65. doi: 10.1002/mbo3.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maredia R, Devineni N, Lentz P, Dallo SF, Yu J, Guentzel N, et al. Vesiculation from Pseudomonas aeruginosa under SOS. TheScientificWorldJournal. 2012;2012:402919. doi: 10.1100/2012/402919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashburn-Warren L, Howe J, Garidel P, Richter W, Steiniger F, Roessle M, et al. Interaction of quorum signals with outer membrane lipids: insights into prokaryotic membrane vesicle formation. Molecular Microbiology. 2008;69:491–502. doi: 10.1111/j.1365-2958.2008.06302.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBroom AJ, Johnson AP, Vemulapalli S, Kuehn MJ. Outer membrane vesicle production by Escherichia coli is independent of membrane instability. Journal of bacteriology. 2006;188:5385–5392. doi: 10.1128/JB.00498-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaig WD, Koller A, Thanassi DG. Production of outer membrane vesicles and outer membrane tubes by Francisella novicida. J Bacteriol. 2013;195:1120–1132. doi: 10.1128/JB.02007-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaig WD, Loving CL, Hughes HR, Brockmeier SL. Characterization and Vaccine Potential of Outer Membrane Vesicles Produced by Haemophilus parasuis. PLoS ONE. 2016;11:e0149132. doi: 10.1371/journal.pone.0149132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metruccio MME, Evans DJ, Gabriel MM, Kadurugamuwa JL, Fleiszig SMJ. Pseudomonas aeruginosa outer membrane vesicles triggered by human mucosal fluid and lysozyme can prime host tissue surfaces for bacterial adhesion. Frontiers in Microbiology. 2016;7 doi: 10.3389/fmicb.2016.00871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olofsson A, Vallstrom A, Petzold K, Tegtmeyer N, Schleucher J, Carlsson S, et al. Biochemical and functional characterization of Helicobacter pylori vesicles. Mol Microbiol. 2010;77:1539–1555. doi: 10.1111/j.1365-2958.2010.07307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park AJ, Murphy K, Surette MD, Bandoro C, Krieger JR, Taylor P, Khursigara CM. Tracking the Dynamic Relationship between Cellular Systems and Extracellular Subproteomes in Pseudomonas aeruginosa Biofilms. J Proteome Res. 2015;14:4524–4537. doi: 10.1021/acs.jproteome.5b00262. [DOI] [PubMed] [Google Scholar]
- Perez-Cruz C, Carrion O, Delgado L, Martinez G, Lopez-Iglesias C, Mercade E. New type of outer membrane vesicle produced by the Gram-negative bacterium Shewanella vesiculosa M7T: implications for DNA content. Appl Environ Microbiol. 2013;79:1874–1881. doi: 10.1128/AEM.03657-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson T, Matrakas D, Taylor YU, Manyam G, Morozov VN, Zhou W, van Hoek ML. Proteomic characterization and functional analysis of outer membrane vesicles of Francisella novicida suggests possible role in virulence and use as a vaccine. Journal of proteome research. 2011;10:954–967. doi: 10.1021/pr1009756. [DOI] [PubMed] [Google Scholar]
- Prados-Rosales R, Baena A, Martinez LR, Luque-Garcia J, Kalscheuer R, Veeraraghavan U, et al. Mycobacteria release active membrane vesicles that modulate immune responses in a TLR2-dependent manner in mice. The Journal of clinical investigation. 2011;121:1471–1483. doi: 10.1172/JCI44261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prados-Rosales R, Weinrick BC, Piqué DG, Jacobs WR, Casadevall A, Rodriguez GM. Role for Mycobacterium tuberculosis membrane vesicles in iron acquisition. Journal of bacteriology. 2014;196:1250–1256. doi: 10.1128/JB.01090-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roier S, Zingl FG, Cakar F, Durakovic S, Kohl P, Eichmann TO, et al. A novel mechanism for the biogenesis of outer membrane vesicles in Gram-negative bacteria. Nat Commun. 2016;7:10515. doi: 10.1038/ncomms10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaar V, Nordstrom T, Morgelin M, Riesbeck K. Moraxella catarrhalis outer membrane vesicles carry beta-lactamase and promote survival of Streptococcus pneumoniae and Haemophilus influenzae by inactivating amoxicillin. Antimicrob Agents Chemother. 2011;55:3845–3853. doi: 10.1128/AAC.01772-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schertzer JW, Brown SA, Whiteley M. Oxygen Levels Rapidly Modulate Pseudomonas aeruginosa Social Behaviors via Substrate Limitation of PqsH. Molecular microbiology. 2010;77:1527–1538. doi: 10.1111/j.1365-2958.2010.07303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer C, Kuehn MJ. Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functions. Nat Rev Micro. 2015a;13:605–619. doi: 10.1038/nrmicro3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer C, Rodriguez DL, Kuehn MJ. NlpI-mediated modulation of outer membrane vesicle production through peptidoglycan dynamics in Escherichia coli. Microbiologyopen. 2015b;4:375–389. doi: 10.1002/mbo3.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe SW, Kuehn MJ, Mason KM. Elicitation of epithelial cell-derived immune effectors by outer membrane vesicles of nontypeable Haemophilus influenzae. Infection and immunity. 2011;79:4361–4369. doi: 10.1128/IAI.05332-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shetty A, Chen S, Tocheva EI, Jensen GJ, Hickey WJ. Nanopods: a new bacterial structure and mechanism for deployment of outer membrane vesicles. PloS one. 2011;6:e20725. doi: 10.1371/journal.pone.0020725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shetty A, Hickey WJ. Effects of outer membrane vesicle formation, surface-layer production and nanopod development on the metabolism of phenanthrene by Delftia acidovorans Cs1–4. PloS one. 2014;9:e92143. doi: 10.1371/journal.pone.0092143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh PK, Parsek MR, Greenberg EP, Welsh MJ. A component of innate immunity prevents bacterial biofilm development. Nature. 2002;417:552–555. doi: 10.1038/417552a. [DOI] [PubMed] [Google Scholar]
- Stapleton F, Dart J. Pseudomonas keratitis associated with biofilm formation on a disposable soft contact lens. The British Journal of Ophthalmology. 1995;79:864–865. doi: 10.1136/bjo.79.9.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart JC. Colorimetric determination of phospholipids with ammonium ferrothiocyanate. Analytical Biochemistry. 1980;104:10–14. doi: 10.1016/0003-2697(80)90269-9. [DOI] [PubMed] [Google Scholar]
- Tam C, Mun JJ, Evans DJ, Fleiszig SMJ. The Impact of Inoculation Parameters on the Pathogenesis of Contact Lens-Related Infectious Keratitis. Investigative Ophthalmology & Visual Science. 2010;51:3100–3106. doi: 10.1167/iovs.09-4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro Y, Ichikawa S, Shimizu M, Toyofuku M, Takaya N, Nakajima-Kambe T, et al. Variation of Physiochemical Properties and Cell Association Activity of Membrane Vesicles with Growth Phase in Pseudomonas aeruginosa. Applied and Environmental Microbiology. 2010;76:3732–3739. doi: 10.1128/AEM.02794-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro Y, Inagaki A, Shimizu M, Ichikawa S, Takaya N, Nakajima-Kambe T, et al. Characterization of Phospholipids in Membrane Vesicles Derived from Pseudomonas aeruginosa. Bioscience, Biotechnology, and Biochemistry. 2011;75:605–607. doi: 10.1271/bbb.100754. [DOI] [PubMed] [Google Scholar]
- Toyofuku M, Roschitzki B, Riedel K, Eberl L. Identification of proteins associated with the Pseudomonas aeruginosa biofilm extracellular matrix. Journal of proteome research. 2012;11:4906–4915. doi: 10.1021/pr300395j. [DOI] [PubMed] [Google Scholar]
- Toyofuku M, Tashiro Y, Hasegawa Y, Kurosawa M, Nomura N. Bacterial membrane vesicles, an overlooked environmental colloid: Biology, environmental perspectives and applications. Advances in Colloid and Interface Science. 2015;226 Part A:65–77. doi: 10.1016/j.cis.2015.08.013. [DOI] [PubMed] [Google Scholar]
- Toyofuku M, Zhou S, Sawada I, Takaya N, Uchiyama H, Nomura N. Membrane vesicle formation is associated with pyocin production under denitrifying conditions in Pseudomonas aeruginosa PAO1. Environ Microbiol. 2013;16:2927–2938. doi: 10.1111/1462-2920.12260. [DOI] [PubMed] [Google Scholar]
- Turnbull L, Toyofuku M, Hynen AL, Kurosawa M, Pessi G, Petty NK, et al. Explosive cell lysis as a mechanism for the biogenesis of bacterial membrane vesicles and biofilms. Nature Communications. 2016;14 doi: 10.1038/ncomms11220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Pol E, Hoekstra AG, Sturk A, Otto C, Van Leeuwen TG, Nieuwland R. Optical and non-optical methods for detection and characterization of microparticles and exosomes. Journal of Thrombosis and Haemostasis. 2010;8:2596–2607. doi: 10.1111/j.1538-7836.2010.04074.x. [DOI] [PubMed] [Google Scholar]
- van der Pol L, Stork M, van der Ley P. Outer membrane vesicles as platform vaccine technology. Biotechnology Journal. 2015;10:1689–1706. doi: 10.1002/biot.201400395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veith PD, Chen Y-Y, Gorasia DG, Chen D, Glew MD, O’Brien-Simpson NM, et al. Porphyromonas gingivalis outer membrane vesicles exclusively contain outer membrane and periplasmic proteins and carry a cargo enriched with virulence factors. Journal of proteome research. 2014;13:2420–2432. doi: 10.1021/pr401227e. [DOI] [PubMed] [Google Scholar]
- Werner E, Roe F, Bugnicourt A, Franklin MJAHSM, et al. Stratified growth in Pseudomonas aeruginosa biofilms. Applied and environmental microbiology. 2004;70:6188–6196. doi: 10.1128/AEM.70.10.6188-6196.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessel AK, Liew J, Kwon T, Marcotte EM, Whiteley M. Role of Pseudomonas aeruginosa peptidoglycan-associated outer membrane proteins in vesicle formation. Journal of bacteriology. 2013;195:213–219. doi: 10.1128/JB.01253-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitworth DE, Slade SE, Mironas A. Composition of distinct sub-proteomes in Myxococcus xanthus: metabolic cost and amino acid availability. Amino acids. 2015;47:2521–2531. doi: 10.1007/s00726-015-2042-x. [DOI] [PubMed] [Google Scholar]
- Xu KD, Stewart PS, Xia F, Huang CT, McFeters GA. Spatial physiological heterogeneity in Pseudomonas aeruginosa biofilm is determined by oxygen availability. Appl Environ Microbiol. 1998;64:4035–4039. doi: 10.1128/aem.64.10.4035-4039.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielke RA, Wierzbicki IH, Weber JV, Gafken PR, Sikora AE. Quantitative proteomics of the Neisseria gonorrhoeae cell envelope and membrane vesicles for the discovery of potential therapeutic targets. Mol Cell Proteomics. 2014 doi: 10.1074/mcp.M113.029538. [DOI] [PMC free article] [PubMed] [Google Scholar]