Abstract
Oral squamous cell carcinoma (OSCC) is one of the most common cancers worldwide. Aberrations in miRNA regulation are known to play important roles in OSCC pathogenesis. miR-187 was shown to be up-regulated in head and neck malignancies in our previous screening. This study further investigated the oncogenic potential, clinical implications, and targets of miR-187 in OSCC. We observed that miR-187 increased oncogenicity, particularly migration, of OSCC cells. miR-187 expression increased the xenografic tumorigenicity and metastasis in mice. In addition, metastatic human OSCC had higher miR-187 expression than did non-metastatic tumors. Through vigorous screening, we confirmed BarH-like Homeobox 2 (BARX2) gene as an miR-187 target. BARX2 expression suppressed the migration, invasion, anchorage-independent colony formation, and orthotopic tumorigenesis of OSCC cells. The migratory phenotype and neck metastasis induced by miR-187 was rescued by BARX2 expression. BARX2 expression was down-regulated in the vast majority of OSCC, and this down-regulation was particularly conspicuous in tumors with advanced nodal metastasis. In addition, plasma miR-187 was significantly higher in OSCC patients than in normal individuals. This study highlights the roles of miR-187-BARX2 in driving the carcinogenesis of OSCC. The results suggest that miR-187 is a potential serological marker for OSCC and that targeting of miR-187 might prove effective in attenuating nodal metastasis.
Keywords: BARX2, carcinoma, invasion, metastasis, miR-187
INTRODUCTION
Head and neck squamous cell carcinoma (HNSCC), including oral SCC (OSCC), is the 6th most prevalent malignancy worldwide [1–3]. Previous studies have shown that microRNAs (miRNAs) can control the initiation or progression of HNSCC or OSCC, acting either as oncogenes or tumor suppressors [2–10]. MicroRNAs are a class of short, endogenous, non-coding RNAs that negatively regulate target gene expression [11, 12]. As of today, there are 1881 predicted miRNA species in human in miRBase database (version 21). One individual miRNA can regulate a diverse set of target genes, with approximately 60% of the human protein-coding genes regulated by miRNAs. Thus, miRNAs play crucial roles in the modulation of physiological processes and pathogenesis [13]. Disruption in miRNA expression could affects the malignancy and survival of OSCC by interfering with the regulation of processes such as apoptosis, invasion, drug resistance, stemness, hypoxia and signaling pathways, and others [4, 7, 14].
The miR-187 gene is located on chromosome 18q12.2. Its overexpression has been observed in thyroid cancer, possibly serving as a clinical diagnostic marker [15], in breast cancer,, where it has been demonstrated and has been shows to is correlated with a more aggressive, invasive cancer phenotype as well as poor patient outcome and lower survival [16]. miR-187 also represses the tumor-suppressor gene disabled homolog-2 (Dab2) in ovarian cancers [17]. Increased miR-187 expression was observed in patients with ovarian and gall bladder cancer [18, 19]. In addition, low miR-187 expression defines the sensitivity of ovarian cancers to taxol therapy [20]. However, studies have also revealed that miR-187 is suppressive to some malignancies, including prostate and pancreatic carcinomas as well as clear renal cell carcinoma [21–25]. Therefore, the functional role of miR-187 in different cancers may be paradoxical. In addition to its role in oncogenesis, miR-187 takes part in the regulation of inflammation, cell stemness, and insulin secretion [26–28]. miR-187 was shown in our preliminary screening study as the 3rd most conspicuously up-regulated miRNA in HNSCC [7]. However, the oncogenic role of miR-187 and its target gene in OSCC have been unknown.
BARX2 (aka, BarH-like Homeobox 2; homeobox protein BarH-like) is located on chromosome 11q24-25 [29]. Frequent loss of 11q23-25 loci has been reported in ovarian cancers and HNSCC [30, 31]. BARX2 is known to be involved in cytoskeletal organization, cell adhesion, growth factor signaling, and transcriptional regulation, and it acts as a transcription factor [32]. BARX2 is also involved in the development of craniofacial structures, salivary glands, hair follicles, and the squamous epithelium of the tongue and esophagus [33–36]. Moreover, BARX2 is reported to be down-regulated in ovarian cancer and hepatocellular carcinoma, indicating that it normally acts as a tumor suppressor [30, 37, 38]. In this study, we investigated the oncogenic role of miR-187 by targeting the BARX2 tumor suppressor in OSCC.
RESULTS
Increased miR-187 expression in OSCC tumors and patient plasma
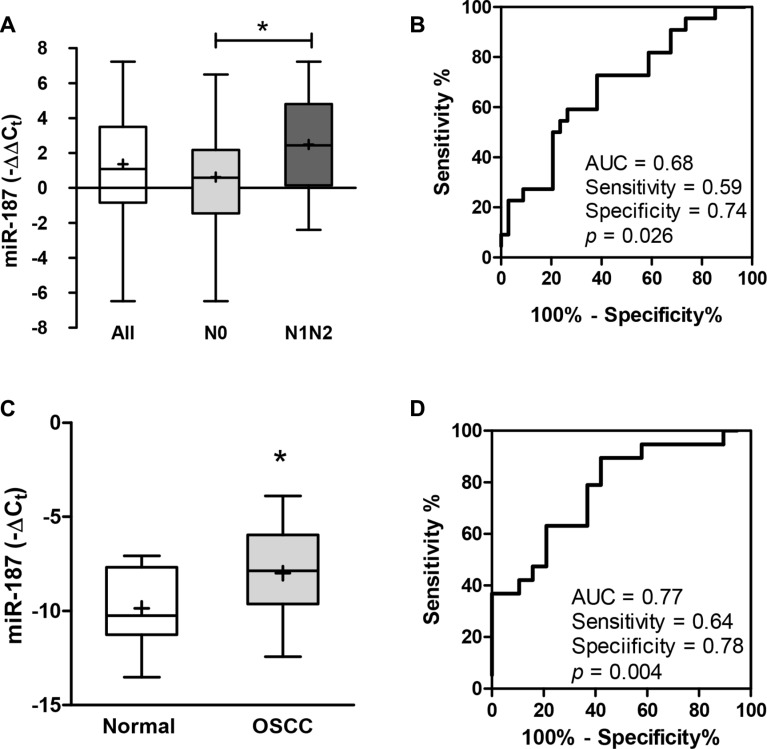
To explore the expression of miR-187, 56 OSCC tumors and their paired NCMTs were subjected to qRT-PCR analysis (Table S1). Up-regulation of miR-187 was found in 40 (71%) of OSCC tumor tissues. In addition, a significant increase in miR-187 expression was noted in tumors with nodal metastasis relative to tumors without node involvement (Figure 1A). ROC analyses indicated that miR-187 expression in OSCC had a predictive power of 0.68 for distinguishing metastatic from non-metastatic states (Figure 1B). miR-187 expression was not associated with other clinicopathological parameters. To investigate examine the feasibility of using diagnostic value of the plasma level of miR-187, we compared the plasma levels as a diagnostic marker, plasma samples were collected of miR-187 from in OSCC patients and healthy controls. A significant increase in −ΔCt in patients with OSCC relative to controls was noted (Figure 1C). ROC analysis indicated that the plasma miR-187 level had a predictive power of 0.77 for distinguishing malignant from non-malignant states (Figure 1D).
Figure 1. Up-regulation of miR-187 expression in OSCC.
(A, C) Box and whiskers plots illustrating the expression of miR-187 in tumor tissue pairs (A) and plasma (C) detected by qRT-PCR analysis. Un-paired t-test; *p < 0.05. (B, D) ROC analysis. (B) Comparison across tumors without metastasis and those with metastasis. (D) Comparison of plasma samples from control individuals and OSCC patients. AUC, area under curve.
miR-187 expression increased OSCC oncogenicity
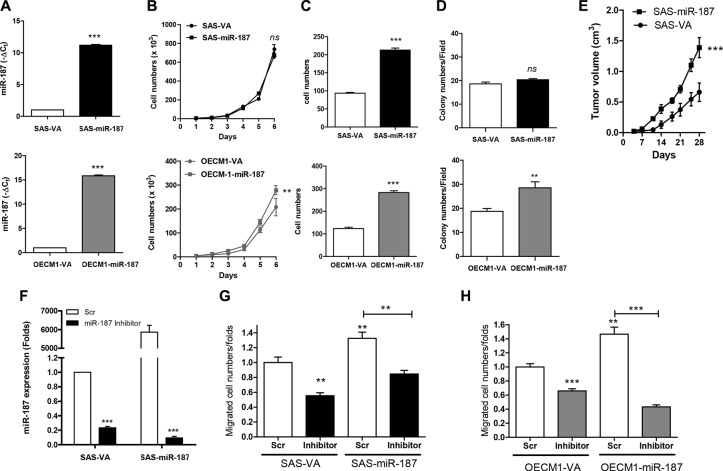
Cell subclones expressing miR-187 were established in SAS and OECM1 cells, which have high and low endogenous miR-187 expression, respectively (Figure 2A). These subclones were designated SAS-miR-187 and OECM1-miR-187. The miR-187 expression in SAS-miR-187 and OECM1-miR-187 subclone increased ~11.2 and ~15.9 folds relative to respective control subclones. Exogenous miR-187 expression did not significantly change the proliferation (Figure 2B, upper) or AIG (Figure 2D, upper) of SAS cells. However, exogenous miR-187 expression significantly increased the migration (Figure 2C, upper) and xenografic tumor growth (Figure 2E) in SAS-miR-187 subclones. The proliferation, migration, and AIG in OECM1-miR-187 subclone was higher than control subclone (Figure 2B–2D, lower). Upon treatment with an miR-187 inhibitor, endogenous and exogenous miR-187 expression was drastically suppressed (Figure 2F). The increased cell migration associated with endogenous and exogenous miR-187 expression was decreased by miR-187 inhibition in both SAS (Figure 2G) and OECM1 cells (Figure 2H). Overall, miR-187 expression increased the oncogenicity of OSCC cells, especially in the OECM1 cell line. The phenotype of increased migration was particularly consistent across different OSCC cells.
Figure 2. miR-187 modulates oncogenicity of OSCC cells.
(A) miR-187 expression. (B–D) Assays of proliferation, migration and AIG, respectively. (A–D), Upper, SAS cell subclone; Lower, OECM1 cell subclone. (E) Subcutaneous tumorigenesis assay for SAS cell subclone. (F, G) Treatment with miR-187 inhibitor in SAS cell subclone. (F) miR-187 expression; (G) Migration assay. (H) Migration assay of OECM1 cell subclone following the treatment with miR-187 inhibitor. Data are presented as the mean ± SE. ns, not significant; **p <0.01; ***p < 0.001; two-way ANOVA or Mann–Whitney test.
Identification of BARX2 as the target gene of miR-187 in OSCC
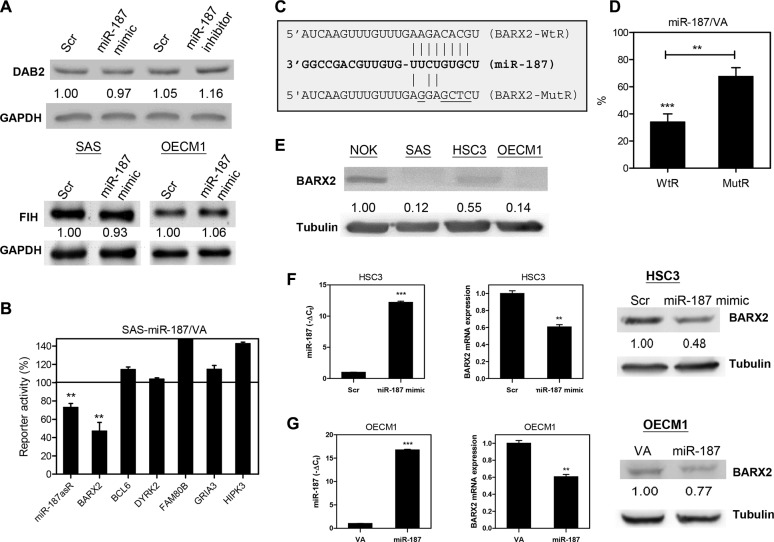
Dab2 was reported to be a target of miR-187 in ovarian cancers [17]. FIH, a tumor suppressor gene in HNSCC [7], was predicted to be a target of miR-187. However, our preliminary Western blot analyses have excluded these genes as miR-187 targets in OSCC cells (Figure 3A). To generate reporters for target screening, we cloned sequence fragments of the predicted miR-187 target genes BARX2, BCL6, DYRK2, FAM80B, GRIA3, and HIPK3. The assays were validated in side-by-side analysis showing the suppression of miR-187asR activity (Figure 3B). Of the potential targets, BARX2 emerged as a target of miR-187, as the reporter activity decreased significantly, to 46% of control reporter activity in SAS-miR-187 cell subclones (Figure 3B). Figure 3C illustrates the complementarity between the BARX2 3′UTR sequence and miR-187. We then generated BARX2 MutR, in which the targeted sites were mutated. Reporter assays of SAS-miR-187 cell subclone indicated that miR-187 repressed the reporter activity by directly targeting the wild type sequence and that the mutation partially reverted the repression (Figure 3D). OSCC cell lines had lower BARX2 expression than did NOK cells (Figure 3E). BARX2 expression was about 4-fold higher in HSC3 than in SAS and OECM1 cells. To confirm the targeting of miR-187 on BARX2, HSC3 cells were treated with miR-187 mimic for 48 hr. Exogenous miR-187 expression (Figure 3F, left) was associated with decreased BARX2 mRNA expression (Figure 3F, middle) and BARX2 protein expression (Figure 3F, right) in HSC3 cells. Similarly, the OECM1-miR-187 cell subclone also had lower BARX2 expression (Figure 3G).
Figure 3. miR-187 targets BARX2 in OSCC.
(A, E) Western blot analysis. (A) Upper, The treatment with miR-187 mimic does not affect the Dab2 expression in SAS cells. Lower, miR-187 mimic treatment does not affect the FIH expression in OSCC cells. (B, D) Reporter assay. (B) Reporters for various genes. The results indicated that the activity of BARX2 reporters was repressed by ~55% in SAS-miR-187 cell subclone relative to control. The miR-187asR activity is decreased for ~28%. (C) Schematic diagram to show the complimentarity between miR-187 and BARX2-WtR, and BARX2-MutR. (D) Suppression of BARX2- WtR activity in SAS-miR-187 cell subclones is partially reversed in BARX2-MutR. (E) BARX2 protein expression in oral keratinocytes. OSCC cell lines exhibited lower BARX2 expression than did NOK cells. (F, G) HSC3 and OECM1 cell subclones, respectively. (F) Up-regulation of miR-187 expression in HSC3 cells (left) following treatment with miR-187 mimic is associated with decreased BARX2 mRNA expression (middle) and protein expression (right). (G) Up-regulation of miR-187 expression in OECM1-miR-187 cell subclone (left) is associated with decreased BARX2 mRNA expression (middle) and protein expression (right). Numbers below pictures represent normalized values. Data are presented as the mean ± SE. ns, not significant; **p < 0.01; ***p < 0.001; Mann–Whitney test.
Knockdown of BARX2 expression increased OSCC oncogenicity
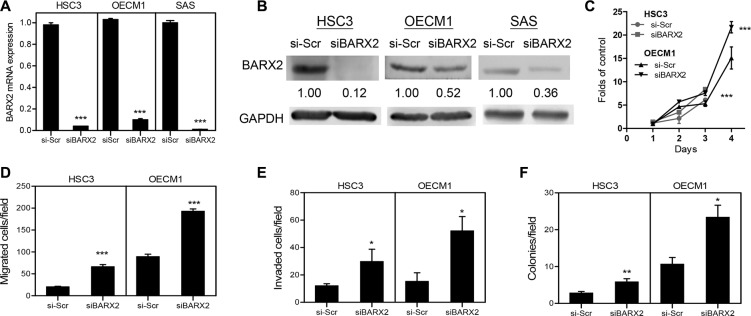
OSCC cells were treated with siBARX2 oligonucleotides for 48 h. qRT-PCR analysis revealed the significant down-regulation of the BARX2 mRNA expression clear down-regulation of BARX2 mRNA expression (Figure 4A) and protein expression (Figure 4B) in OSCC cells. BARX2 knockdown was greater in HSC3 cells than in OECM1 cells and was associated with a significant increase in proliferation (Figure 4C), migration (Figure 4D), invasion (Figure 4E) and AIG (Figure 4F) in both HSC3 and OECM1 cells.
Figure 4. siBARX2 treatment enhances oncogenicity.
(A, B) qRT-PCR analysis and Western blot analysis. Treatment of OSCC cells with siBARX2 oligonucleotide leads to decreased BARX2 mRNA expression (A) and protein expression (B). (C–F) proliferation, migration, invasion, and AIG analysis in HSC3 and OECM1 cells treatment with siBARX2 oligonucleotide. Numbers below pictures represent normalized values. Data are presented as the mean ± SE. *p < 0.05; **p < 0.01; ***p < 0.001; two-way ANOVA or Mann–Whitney test.
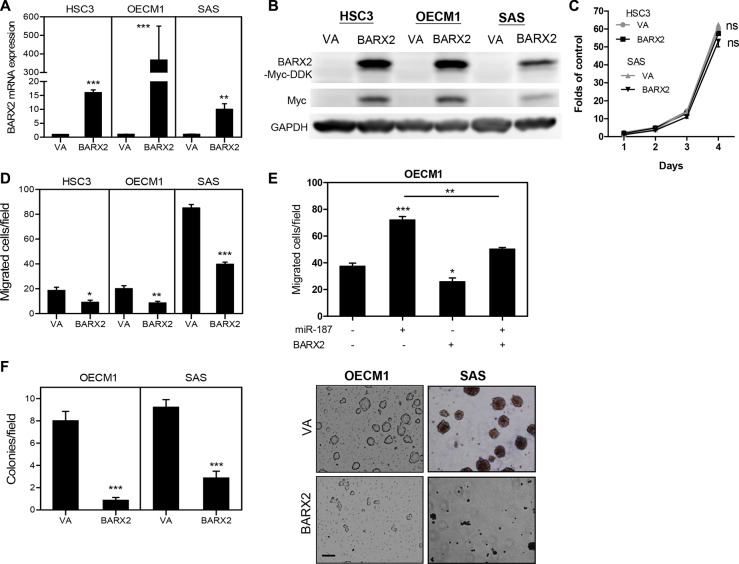
Exogenous BARX2 expression decreased OSCC oncogenicity
As BARX2 knockdown promoted oncogenicity, the effects of BARX2 in suppressing oncogenicity were further tested in OSCC cell subclones expressing the BARX2-myc-DDK fusion protein (Figure 5A, 5B). Exogenous BARX2 expression had no effect on proliferation (Figure 5C) but decreased the migration of OSCC cells (Figure 5D). OECM1 cells expressing BARX2 were treated with miR-187 mimic. Data showing that miR-187-induced cell migration was attenuated by BARX2 (Figure 5E) support the notion that miR-187 targeted BARX2 to modulate OSCC cell migration. Exogenous BARX2 expression also decreased the AIG of OSCC cells (Figure 5F).
Figure 5. OSCC cells expressing BARX2 exhibit decreased oncogenicity.
(A, B) qRT-PCR analysis and Western blot analysis. BARX2 mRNA expression (A) and exogenous BARX2-myc-DDK protein expression (in B) are increased in OSCC cell subclones with stable exogenous BARX2 expression. (C, D) proliferation and migration. (E) Migration assay in OECM1 cells. BARX2 expression reverted miR-187-induced cell migration. (F) AIG assays. Representative fields of colonies (right) and quantitation (left); bar, 50 μm. Data are presented as the mean ± SE. ns, not significant;*p < 0.05; **p < 0.01; ***p < 0.001; two-way ANOVA or Mann–Whitney test.
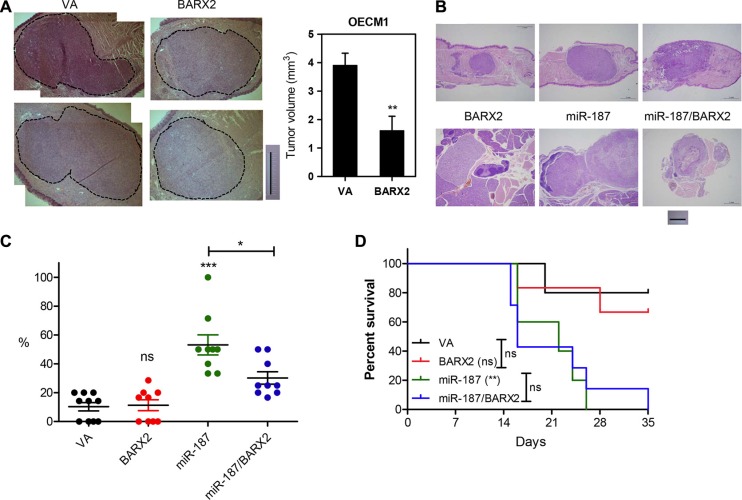
Exogenous BARX2 decreased orthotopic tumorigenesis of OECM1 cells
Orthotopic tongue tumorigenic assays were carried out on OECM1 cell subclones expressing BARX2 [10]. Exogenous BARX2 expression significantly reduced the growth of orthotopic xenografts (Figure 6A), supporting the hypothesis that BARX2 is a suppressor of OSCC oncogenicity, particularly in regard to cell mobility. Although there was significant difference in the primary tumor burden, gross evaluation of dissected neck tissue did not reveal potential metastasis in mice. Moreover, the survival rate of OECM1 cell subclones expressing BARX2 is not different from controls.
Figure 6. Analysis of tumor xenografts in athymic mice.
(A) Orthotopic tongue xenografts in OECM1 cell subclones. Illustrations of representative tongue sections (left). Dotted lines indicate tumor areas; bar, 1 mm. Quantitation of tumor volume (right). (B–D) Orthotopic tongue xenografts and neck metastasis in SAS cell subclones. (B) Illustrations of representative sections of primary tumors in tongue and neck nodal lesions. (C) Dot plot of metastatic rate of cell subclones. Mann–Whitney test. (D) Kaplan-Meier survival analysis of cell subclones. Data are presented as the mean ± SE. ns, not significant; *p < 0.05; **p < 0.01; ***p < 0.001.
Exogenous BARX2 rescued miR-187-enhanced neck metastasis of SAS cells
The tissue analysis revealed the primary xenografic tumor growth of SAS cell subclones in tongue and the metastatic involvement in neck lymph nodes (Figure 6B). Since the advanced tumor may confound the metastasis rate, mice carrying primary tumors > 20 mm3 were excluded for metastasis analysis [10]. In animals carrying relatively smaller tumors, miR-187 expression significantly increased the neck metastasis, and this increase can be rescued by concordant BARX2 expression (Figure 6C). However, exogenous BARX2 expression did not affect the metastatic rates of control cell subclone. The survival rate of mice carrying the xenografts of SAS-miR-187 cell subclones was drastically decreased comparing to controls (Figure 6D). Exogenous BARX2 expression did not seem to affect the survival in this animal model.
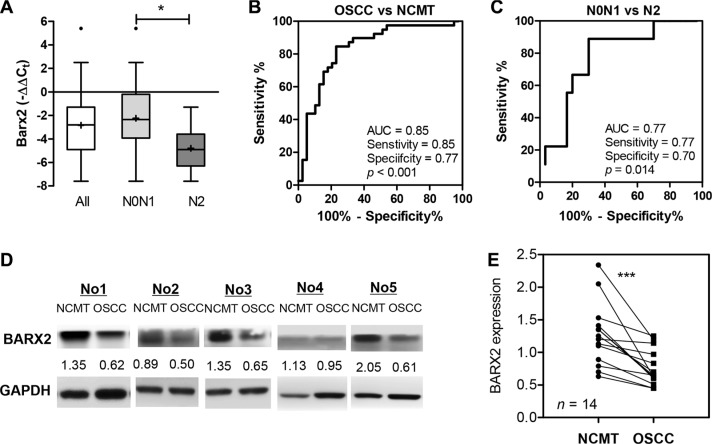
Decreased BARX2 expression is associated with the advanced nodal metastasis of OSCC
The expression of BARX2 in tumors and their paired NCMTs were analyzed using qRT-PCR analysis and Western blot analysis. A reduced expression of BARX2 mRNA was found in tumor tissues relative to the surrounding tissue. Although there was no significant difference in BARX2 mRNA expression on metastatic or recurrent tumors, a significant decrease in miR-187 expression was seen in tumors with advanced nodal metastasis (N2) relative to contrasting tumors (Figure 7A). ROC analysis indicated that BARX2 mRNA expression in OSCC had a predictive power of 0.85 for distinguishing OSCC from NCMT (Figure 7B), and a power of 0.77 for distinguishing the most severe metastatic status from other states (Figure 7C). Analysis of BARX2 protein expression in 14 available tissue pairs also revealed significant down-regulation of BARX2 protein expression in OSCC tumors (Figure 7D, 7E).
Figure 7. Down-regulation of BARX2 expression in OSCC.
(A) Box and whiskers plots illustrating the down-regulation of BARX2 mRNA expression in tumor tissue pairs as detected by qRT-PCR analysis. The down-regulation was particularly evident in tumors with advanced neck metastasis (N2). (B, C) ROC analysis. (B) Comparison between NCMT and tumors. (C) Comparison across tumors with N2 grade nodal metastasis and others. AUC, area under curve. (D, E) BARX2 protein expression in OSCC. (D) Western blot analysis of BARX2 protein expression in 5 representative NCMT and OSCC sample pairs. Numbers below pictures represent normalized values. (E) Quantitation of 14 sample pairs. Paired t-test; ***p < 0.01.
DISCUSSION
miR-187 has diverse expression across malignancies and plays differential oncogenic roles in different types of tumors [15, 16, 18–25]. Our previous screening study identified miR-187 up-regulation in HNSCC [7], and the present study further confirms the up-regulation of miR-187 in OSCC tissues and correlates miR-187 up-regulation with lymph node metastasis. Functional clues support the oncogenicity of miR-187 in OSCC, particularly with respect to enhancing tumor cell migration. Exogenous miR-187 expression levels differ between cell lines, which may underlie the differences in its potential to promote proliferation and AIG. miR-187 expression also increases the metastatic rate of xenografic tumors and this is associated with worse host survival. The findings in animal models seem to be consistent with the clinical implications.
Many oncogenic miRNAs are potential serological markers of HNSCC or OSCC [2, 10, 39], and our preliminary analysis further supports that plasma miR-187 is a potential marker of OSCC. Further study is required to confirm the efficacy of miR-187 as a non-invasive marker of oral premalignant disorders [40]. miR-21, Let-7 family members, miR-133a and other miRNAs are involved in the metastasis machinery of HNSCC by targeting regulators of cell stemness or mobility [3–6, 8, 9]. It would be important to specify the enrichment of these aberrant miRNAs with miR-187 in promoting tumor metastasis in future study.
Few studies have investigated the relationship of miR-187 targets to tumorigenesis [7, 17, 24]. The targets and mechanisms associated with miR-187 in OSCC pathogenesis are unknown. Studies have excluded Dab2, FIH suppressors, and other predicted genes as miR-187 targets [7, 17]. BARX2 is involved in regulating squamous epithelium and craniofacial development [33–36] and is a suppressor of several type of neoplasms [30, 37, 38]. This study provides novel evidence that BARX2 is a target of miR-187 in OSCC. Loss of heterozygosity in loci containing 11q24.2, which harbors the BARX2 gene, is frequent in HNSCC [41]. We observed that OSCC cell lines exhibited decreased BARX2 expression compared to that of NOK cells, possibly due to allele loss or targeting by endogenously-expressed miR-187 in cancer cells. Furthermore, knockdown and overexpression experiments indicate suppressor activity of BARX2 in multiple OSCC cell lines. Our findings suggest that in addition to chromosomal deletion, epigenetic regulation by oncogenic miRNAs such as miR-187 is also able to repress BARX2 in OSCC pathogenesis.
With modest up-regulation of miR-187 in OSCC, qRT-PCR analysis and Western blot unequivocally demonstrated a drastic down-regulation of BARX2 expression in OSCC tumors relative to paired normal mucosa. This decreased expression could be caused by the combined effects of BARX2 deletion and the targeting effects of miR-187, since miR-187 was not greatly up-regulated in cancer cells relative to normal cells. Intriguingly, BARX2 down-regulation was particularly apparent in tumors with advanced nodal metastasis. In agreement with this fact, BARX2 expression reduces the tumor metastasis in xenografic tumors exhibiting highest metastatic potential. This finding suggests that BARX2 expression could be a marker for OSCC metastasis and that restoration of BARX2 could be used as a therapeutic regimen to intercept the progression of OSCC. Acting as a transcription factor, BARX2 is reported to control the expression of adhesion molecules and cytoskeletal elements [32, 36, 37], which may underlie its repressive activity against cell mobility and invasion. Although BARX2 expression reduces the tumor burden of xenografts, it is unable to decrease the mortality of animals significantly, its targets and functional mechanisms in OSCC tumors remain to be identified. Alternatively, the rather low abundance of exogenous BARX2 expression in SAS cells or the relatively smaller sample being analyzed in this study could have impeded the analytical power.
This study suggests that miR-187 may contribute to OSCC progression through suppression of BARX2 expression and indicates the clinical importance of the miR-187–BARX2 cascade in tumor metastasis. A further understanding of the signals that up-regulate miR-187 expression and downstream effectors of BARX2 is needed to clarify the role of this regulatory axis in neoplastic processes. Such information will be valuable for the development of diagnostic tools and therapies.
MATERIALS AND METHODS
Tissue and blood samples
The surgical specimens consisted of the primary OSCC tumors together with paired non-cancerous matched tissues (NCMTs) (Table S1). Blood samples (5 mL) were collected from patients one week before surgery and from 19 sex- and age-matched control subjects with no oral disease or malignancy. These samples were collected after obtaining written informed consent, and this study was approved by The Institutional Review Board (IRB) of Mackay Memorial Hospital with IRB approval numbers 09MMHIS146 and 10MMHIS185.
Cell culture, reagents and phenotypic analysis
The HSC3, OECM1, and SAS OSCC cell lines, 293FT cells, phoenix package cells, and NOK cells established in our laboratory were cultured as previously described [7, 42]. An miR-187 mimic, miR-187 inhibitor, and scramble (Scr) control were purchased from Applied Biosystems (Foster City, CA, USA). The siBARX2 oligonucleotide and scramble (si-Scr) control oligonucleotide were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). TransFectin Lipid Reagent (BioRad Laboratories, Hercules, CA, USA) was used as the transfection reagent. Analyses of oncogenic phenotypes, including cell proliferation, transwell migration assay, cell invasion and anchorage-independent growth (AIG) were carried out according to previously published protocols [7, 10, 42].
Establishment of cell subclones with miR-187 or BARX2 expression
A lentivirus carrying the pre-miR-187 sequence and a red fluorescence (RFP) tag was purchased from Biosetta (San Diego, CA, USA). Red fluorescence in cells indicated infection. OSCC cell subclones with stable miR-187 expression and controls were established by puromycin selection. The coding sequence of BARX2 together with Myc-DDK tag were cloned into pBabe-puro retroviral vector to produce the pBabe-BARX2-myc-DDK construct. Cell subclones with BARX2 expression were established by viral infection and puromycin selection. In addition, cell subclones with exogenous miR-187 and BARX2 expression (designated miR-187/BARX2) were established after viral infection, puromycin selection and fluorescence sorting.
qRT-PCR analysis
Tri-reagent was used to isolate total RNA, which was reverse transcribed to produce the corresponding cDNAs. The TaqMan miRNA assay kit was used to quantify the expression of miR-187, BARX2, and other genes according to the manufacturer's instructions (Applied Biosystems). Let7a small nuclear RNA, U6B, or GAPDH were used as an internal controls
Western blot analysis
Cell lysates (60 μg) were subjected to Western blot analysis using various primary antibodies (Table S2) and secondary antibodies (Chemicon Int., Billerica, MA) according to previously described protocols [7, 42]. Normalization of the signals to internal GAPDH or tubulin was used to generate relative expression values.
Prediction of targets
Targetscan in silico (http://www.targetscan.org/) version 6.2 was used to predict the potential targets of miR-187 [7].
Reporter plasmid construction and assays
To test the targeting activity of miR-187, fragments of 3′ UTR sequences of BARX2, BCL6, DYRK2, FAM80B, GRIA3, and HIPK3 containing predicted miR-187 target sites were amplified by PCR (Table S3) and cloned into the pCMV-LacZ plasmid [7]. In addition, the antisense sequence of miR-187 was cloned to generate miR-187asR as a positive control reporter. The wild type 3′UTR sequence of BARX2 was also cloned into the pMIR-REPORT vector (Life Technologies, Grand Island, NY, USA) (Table S4) to generate the BARX2 WtR reporter [43]. A mutant reporter (MutR) construct was obtained from the BARX2 WtR reporter by replacing the original sequence AGACACG at the target site of BARX2 3′UTR with GGAGCUC to create a new Sac I restriction enzyme digestion site. LacZ activity, firefly luciferase activity, or normalization to transfection efficiency was used to represent reporter activity.
Tumorigenesis and metastasis assays
For xenografic tumorigenesis, 2.5 × 105 SAS cell subclones were injected subcutaneously into the flank of 6–8-week-old athymic mice for 4 weeks. For orthotopic induction of xenografts in tongue, 3 × 105 OECM1 cell subclones were injected into the central portion of tongue in mice for 2 weeks. After sacrifice of the animals, the resected tumors, tongues and neck tissues were photographed, and examined grossly and histopathologically. Tumor volumes were calculated using the formula: volume = 0.5 × a × b2, where a and b were the long and short diameters of the tumors, respectively [7].
For induction of neck nodal metastasis, 3 × 105 SAS cell subclones were injected into the central portion of the tongue of 6–8-week-old athymic mice. The mice were sacrificed at the 5th week after inoculation or at the time points when body weight loss for more than 30% [10]. Indian ink was injected into the tongue tissue to facilitate node identification. In addition, the head and neck region was photographed under an illuminating device (LT9500 Illumatool TLS; Lightools Research, Encinitas, CA) to detect positive nodes. The whole tongues were resected and subjected to histopathological evaluation. Neck tissues achieved by radical dissection were embedded for histopathological examination. The animal study was approved by the Institutional Animal Care and Use Committee of Mackay Memorial Hospital.
Statistical analysis
The t-test, Mann–Whitney test, and two-way ANOVA were used to compare differences between variants. The extent to which variants could be used to distinguish disease status was determined using receiver operating characteristic (ROC) analysis. The area under the curve (AUC) showed the discriminative ability. Kaplan-Meier analysis was used for the comparison of survival between groups. A difference was considered statistically significant when p < 0.05.
SUPPLEMENTARY MATERIALS FIGURES
ACKNOWLEDGMENTS AND FUNDING
We acknowledge the helps from Professor Wan-Chun Li and Dr. Li-Han Lin. This study was supported by grant MOST-99-2628-B-010-013-MY3, MOST-101-2314-B-010-050-MY3, MOST-102-2314-B-195-007-MY3 and MOST103-2314-B-010-024-MY3 from the Ministry of Science and Technology, and Aim for the Top University Plan from the Department of Education, Taiwan. MMH-E-104-12 from MacKay Memorial Hospital, Taipei.
Footnotes
CONFLICTS OF INTEREST
Authors declare no conflicts of interest.
REFERENCES
- 1.Warnakulasuriya S. Global epidemiology of oral and oropharyngeal cancer. Oral Oncol. 2009;45:309–16. doi: 10.1016/j.oraloncology.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 2.Yeh LY, Liu CJ, Wong YK, Chang C, Lin SC, Chang KW. miR-372 inhibits p62 in head and neck squamous cell carcinoma in vitro and in vivo. Oncotarget. 2015;6:6062–75. doi: 10.18632/oncotarget.3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tu HF, Lin SC, Chang KW. MicroRNA aberrances in head and neck cancer: pathogenetic and clinical significance. Curr Opin Otolaryngol Head Neck Surg. 2013;21:104–11. doi: 10.1097/MOO.0b013e32835e1d6e. [DOI] [PubMed] [Google Scholar]
- 4.Childs G, Fazzari M, Kung G, Kawachi N, Brandwein-Gensler M, McLemore M, Chen Q, Burk RD, Smith RV, Prystowsky MB, Belbin TJ, Schlecht NF. Low-level expression of microRNAs let-7d and miR-205 are prognostic markers of head and neck squamous cell carcinoma. Am J Pathol. 2009;174:736–45. doi: 10.2353/ajpath.2009.080731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kinoshita T, Nohata N, Fuse M, Hanazawa T, Kikkawa N, Fujimura L, Watanabe-Takano H, Yamada Y, Yoshino H, Enokida H, Nakagawa M, Okamoto Y, Seki N. Tumor suppressive microRNA-133a regulates novel targets: moesin contributes to cancer cell proliferation and invasion in head and neck squamous cell carcinoma. Biochem Biophys Res Commun. 2012;418:378–83. doi: 10.1016/j.bbrc.2012.01.030. [DOI] [PubMed] [Google Scholar]
- 6.Chien CS, Wang ML, Chu PY, Chang YL, Liu WH, Yu CC, Lan YT, Huang PI, Lee YY, Chen YW, Lo WL, Chiou SH. Lin28B/Let-7 Regulates Expression of Oct4 and Sox2 and Reprograms Oral Squamous Cell Carcinoma Cells to a Stem-like State. Cancer Res. 2015;75:2553–65. doi: 10.1158/0008-5472.CAN-14-2215. [DOI] [PubMed] [Google Scholar]
- 7.Liu CJ, Tsai MM, Hung PS, Kao SY, Liu TY, Wu KJ, Chiou SH, Lin SC, Chang KW. miR-31 ablates expression of the HIF regulatory factor FIH to activate the HIF pathway in head and neck carcinoma. Cancer Res. 2010;70:1635–44. doi: 10.1158/0008-5472.CAN-09-2291. [DOI] [PubMed] [Google Scholar]
- 8.Bourguignon LY, Earle C, Wong G, Spevak CC, Krueger K. Stem cell marker (Nanog) and Stat-3 signaling promote MicroRNA-21 expression and chemoresistance in hyaluronan/CD44-activated head and neck squamous cell carcinoma cells. Oncogene. 2012;31:149–60. doi: 10.1038/onc.2011.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang WH, Lan HY, Huang CH, Tai SK, Tzeng CH, Kao SY, Wu KJ, Hung MC, Yang MH. RAC1 activation mediates Twist1-induced cancer cell migration. Nat Cell Biol. 2012;14:366–74. doi: 10.1038/ncb2455. [DOI] [PubMed] [Google Scholar]
- 10.Liu CJ, Shen WG, Peng SY, Cheng HW, Kao SY, Lin SC, Chang KW. miR-134 induces oncogenicity and metastasis in head and neck carcinoma through targeting WWOX gene. Int J Cancer. 2014;134:811–21. doi: 10.1002/ijc.28358. [DOI] [PubMed] [Google Scholar]
- 11.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dalmay T, Edwards DR. MicroRNAs and the hallmarks of cancer. Oncogene. 2006;25:6170–5. doi: 10.1038/sj.onc.1209911. [DOI] [PubMed] [Google Scholar]
- 13.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 14.Gee HE, Camps C, Buffa FM, Patiar S, Winter SC, Betts G, Homer J, Corbridge R, Cox G, West CM, Ragoussis J, Harris AL. hsa-mir-210 is a marker of tumor hypoxia and a prognostic factor in head and neck cancer. Cancer. 2010;116:2148–58. doi: 10.1002/cncr.25009. [DOI] [PubMed] [Google Scholar]
- 15.Nikiforova MN, Tseng GC, Steward D, Diorio D, Nikiforov YE. MicroRNA expression profiling of thyroid tumors: biological significance and diagnostic utility. J Clin Endocrinol Metab. 2008;93:1600–8. doi: 10.1210/jc.2007-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mulrane L, Madden SF, Brennan DJ, Gremel G, McGee SF, McNally S, Martin F, Crown JP, Jirstrom K, Higgins DG, Gallagher WM, O'Connor DP. miR-187 is an independent prognostic factor in breast cancer and confers increased invasive potential in vitro. Clin Cancer Res. 2012;18:6702–13. doi: 10.1158/1078-0432.CCR-12-1420. [DOI] [PubMed] [Google Scholar]
- 17.Chao A, Lin CY, Lee YS, Tsai CL, Wei PC, Hsueh S, Wu TI, Tsai CN, Wang CJ, Chao AS, Wang TH, Lai CH. Regulation of ovarian cancer progression by microRNA-187 through targeting Disabled homolog-2. Oncogene. 2012;31:764–75. doi: 10.1038/onc.2011.269. [DOI] [PubMed] [Google Scholar]
- 18.Li G, Pu Y. MicroRNA signatures in total peripheral blood of gallbladder cancer patients. Tumour Biol. 2015;36:6985–90. doi: 10.1007/s13277-015-3412-4. [DOI] [PubMed] [Google Scholar]
- 19.Chao A, Lai CH, Chen HC, Lin CY, Tsai CL, Tang YH, Huang HJ, Lin CT, Chen MY, Huang KG, Chou HH, Chang TC, Chen SJ, et al. Serum microRNAs in clear cell carcinoma of the ovary. Taiwan J Obstet Gynecol. 2014;53:536–41. doi: 10.1016/j.tjog.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 20.Kim YW, Kim EY, Jeon D, Liu JL, Kim HS, Choi JW, Ahn WS. Differential microRNA expression signatures and cell type-specific association with Taxol resistance in ovarian cancer cells. Drug Des Devel Ther. 2014;8:293–314. doi: 10.2147/DDDT.S51969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fuse M, Kojima S, Enokida H, Chiyomaru T, Yoshino H, Nohata N, Kinoshita T, Sakamoto S, Naya Y, Nakagawa M, Ichikawa T, Seki N. Tumor suppressive microRNAs (miR-222 and miR-31) regulate molecular pathways based on microRNA expression signature in prostate cancer. J Hum Genet. 2012;57:691–9. doi: 10.1038/jhg.2012.95. [DOI] [PubMed] [Google Scholar]
- 22.Casanova-Salas I, Rubio-Briones J, Calatrava A, Mancarella C, Masia E, Casanova J, Fernandez-Serra A, Rubio L, Ramirez-Backhaus M, Arminan A, Dominguez-Escrig J, Martinez F, Garcia-Casado Z, et al. Identification of miR-187 and miR-182 as biomarkers of early diagnosis and prognosis in patients with prostate cancer treated with radical prostatectomy. J Urol. 2014;192:252–9. doi: 10.1016/j.juro.2014.01.107. [DOI] [PubMed] [Google Scholar]
- 23.Schultz NA, Andersen KK, Roslind A, Willenbrock H, Wojdemann M, Johansen JS. Prognostic microRNAs in cancer tissue from patients operated for pancreatic cancer—five microRNAs in a prognostic index. World J Surg. 2012;36:2699–707. doi: 10.1007/s00268-012-1705-y. [DOI] [PubMed] [Google Scholar]
- 24.Casanova-Salas I, Masia E, Arminan A, Calatrava A, Mancarella C, Rubio-Briones J, Scotlandi K, Vicent MJ, Lopez-Guerrero JA. MiR-187 targets the androgen-regulated gene ALDH1A3 in prostate cancer. PLoS One. 2015;10:e0125576. doi: 10.1371/journal.pone.0125576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao J, Lei T, Xu C, Li H, Ma W, Yang Y, Fan S, Liu Y. MicroRNA-187 down-regulated in clear cell renal cell carcinoma and associated with lower survival inhibits cell growth and migration though targeting B7-H3. Biochem Biophys Res Commun. 2013;438:439–44. doi: 10.1016/j.bbrc.2013.07.095. [DOI] [PubMed] [Google Scholar]
- 26.Rossato M, Curtale G, Tamassia N, Castellucci M, Mori L, Gasperini S, Mariotti B, De Luca M, Mirolo M, Cassatella MA, Locati M, Bazzoni F. IL-10-induced microRNA-187 negatively regulates TNF-alpha IL-6 and IL-12p40 production in TLR4-stimulated monocytes. Proc Natl Acad Sci USA. 2012;109:E3101–10. doi: 10.1073/pnas.1209100109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Razak SR, Ueno K, Takayama N, Nariai N, Nagasaki M, Saito R, Koso H, Lai CY, Murakami M, Tsuji K, Michiue T, Nakauchi H, Otsu M, et al. Profiling of microRNA in human and mouse ES and iPS cells reveals overlapping but distinct microRNA expression patterns. PLoS One. 2013;8:e73532. doi: 10.1371/journal.pone.0073532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Locke JM, da Silva Xavier G, Dawe HR, Rutter GA, Harries LW. Increased expression of miR-187 in human islets from individuals with type 2 diabetes is associated with reduced glucose-stimulated insulin secretion. Diabetologia. 2014;57:122–128. doi: 10.1007/s00125-013-3089-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hjalt TA, Murray JC. The human BARX2 gene: genomic structure chromosomal localization and single nucleotide polymorphisms. Genomics. 1999;62:456–459. doi: 10.1006/geno.1999.6037. [DOI] [PubMed] [Google Scholar]
- 30.Sellar GC, Watt KP, Li L, Nelkin BD, Rabiasz GJ, Porteous DJ, Smyth JF, Gabra H. The homeobox gene BARX2 can modulate cisplatin sensitivity in human epithelial ovarian cancer. Int J Oncol. 2002;21:929–33. [PubMed] [Google Scholar]
- 31.Tan D, Wiseman S, Zhou Y, Li Q, Ward P, Slocum HK, Alrawi S, Loree T, Hicks W, Rigual N, Anderson G, Stoler D. Definition of a region of loss of heterozygosity at chromosome 11q23 3-25 in head and neck squamous cell carcinoma using laser capture microdissection technique. Diagn Mol Pathol. 2004;13:33–40. doi: 10.1097/00019606-200403000-00006. [DOI] [PubMed] [Google Scholar]
- 32.Stevens TA, Iacovoni JS, Edelman DB, Meech R. Identification of novel binding elements and gene targets for the homeodomain protein BARX2. J Biol Chem. 2004;279:14520–30. doi: 10.1074/jbc.M310259200. [DOI] [PubMed] [Google Scholar]
- 33.Jones FS, Kioussi C, Copertino DW, Kallunki P, Holst BD, Edelman GM. Barx2 a new homeobox gene of the Bar class is expressed in neural and craniofacial structures during development. Proc Natl Acad Sci USA. 1997;94:2632–7. doi: 10.1073/pnas.94.6.2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sander G, Bawden CS, Hynd PI, Nesci A, Rogers G, Powell BC. Expression of the homeobox gene Barx2 in wool follicle development. J Invest Dermatol. 2000;115:753–6. doi: 10.1046/j.1523-1747.2000.00122.x. [DOI] [PubMed] [Google Scholar]
- 35.Naka T, Yokose S. Immunohistochemical localization of barx2 in the developing fetal mouse submandibular glands. Acta Histochem Cytochem. 2009;42:47–53. doi: 10.1267/ahc.08027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olson LE, Zhang J, Taylor H, Rose DW, Rosenfeld MG. Barx2 functions through distinct corepressor classes to regulate hair follicle remodeling. Proc Natl Acad Sci USA. 2005;102:3708–13. doi: 10.1073/pnas.0500519102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sellar GC, Li L, Watt KP, Nelkin BD, Rabiasz GJ, Stronach EA, Miller EP, Porteous DJ, Smyth JF, Gabra H. BARX2 induces cadherin 6 expression and is a functional suppressor of ovarian cancer progression. Cancer Res. 2001;61:6977–81. [PubMed] [Google Scholar]
- 38.Zhang Y, Zhang JX, Huang LL, He LJ, Liao YJ, Lai YR, Deng HX, Tian XP, Kung HF, Xie D, Zhu SL. Low expression of BARX2 in human primary hepatocellular carcinoma correlates with metastasis and predicts poor prognosis. Hepatol Res. 2015;45:228–37. doi: 10.1111/hepr.12340. [DOI] [PubMed] [Google Scholar]
- 39.Liu CJ, Kao SY, Tu HF, Tsai MM, Chang KW, Lin SC. Increase of microRNA miR-31 level in plasma could be a potential marker of oral cancer. Oral Dis. 2010;16:360–4. doi: 10.1111/j.1601-0825.2009.01646.x. [DOI] [PubMed] [Google Scholar]
- 40.Hung KF, Liu CJ, Chiu PC, Lin JS, Chang KW, Shih WY, Kao SY, Tu HF. MicroRNA-31 upregulation predicts increased risk of progression of oral potentially malignant disorder. Oral Oncol. 2016;53:42–7. doi: 10.1016/j.oraloncology.2015.11.017. [DOI] [PubMed] [Google Scholar]
- 41.Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–6. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 42.Hung PS, Tu HF, Kao SY, Yang CC, Liu CJ, Huang TY, Chang KW, Lin SC. miR-31 is upregulated in oral premalignant epithelium and contributes to the immortalization of normal oral keratinocytes. Carcinogenesis. 2014;35:1162–71. doi: 10.1093/carcin/bgu024. [DOI] [PubMed] [Google Scholar]
- 43.Tseng SH, Yang CC, Yu EH, Chang C, Lee YS, Liu CJ, Chang KW, Lin SC. K14-EGFP-miR-31 transgenic mice have high susceptibility to chemical-induced squamous cell tumorigenesis that is associating with Ku80 repression. Int J Cancer. 2015;136:1263–75. doi: 10.1002/ijc.29106. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.